Abstract
Plastid proteins that are encoded by the nuclear genome and synthesized in the cytosol undergo posttranslational targeting to plastids. Ankyrin repeat protein 2A (AKR2A) and AKR2B were recently shown to be involved in the targeting of proteins to the plastid outer envelope. However, it remains unknown whether other factors are involved in this process. In this study, we investigated a factor involved in AKR2A-mediated protein targeting to chloroplasts in Arabidopsis (Arabidopsis thaliana). Hsp17.8, a member of the class I (CI) cytosolic small heat shock proteins (sHsps), was identified in interactions with AKR2A. The interaction between Hsp17.8 and AKR2A was further confirmed by coimmunoprecipitation experiments. The carboxyl-terminal ankyrin repeat domain of AKR2A was responsible for AKR2A binding to Hsp17.8. Other CI cytosolic sHsps also interact with AKR2A to varying degrees. Additionally, Hsp17.8 binds to chloroplasts in vitro and enhances AKR2A binding to chloroplasts. HSP17.8 was expressed under normal growth conditions, and its expression increased after heat shock. Hsp17.8 exists as a dimer under normal physiological conditions, and it is converted to high oligomeric complexes, ranging from 240 kD to greater than 480 kD, after heat shock. High levels of Hsp17.8 together with AKR2A resulted in increased plastid targeting of Outer Envelope Protein7 (OEP7), a plastid outer envelope protein expressed as a green fluorescent protein fusion protein. In contrast, artificial microRNA suppression of HSP17.8 and closely related CI cytosolic sHSPs in protoplasts resulted in a reduction of OEP7:green fluorescent protein targeting to plastids. Based on these data, we propose that Hsp17.8 functions as an AKR2A cofactor in targeting membrane proteins to plastid outer membranes under normal physiological conditions.
In living organisms, high temperatures can damage various cellular processes. In particular, heat stress conditions can result in the denaturing of proteins that form highly cytotoxic nonspecific aggregates (Sharma et al., 2009). Thus, all organisms have evolved mechanisms to protect the cell under such stresses. One well-known response to heat stress is the production of a large number of proteins (Liberek et al., 2008). Among these is a group of proteins ranging between 15 and 45 kD. These proteins are characterized by an α-crystallin domain of approximately 90 amino acids flanked by a short C-terminal extension and an N-terminal arm of variable length (Sun et al., 2002; Sun and MacRae, 2005; Basha et al., 2006). Called small heat shock proteins (sHsps), these proteins possess chaperone activity, preventing heat stress-induced denatured proteins from forming nonspecific aggregates (Kirschner et al., 2000; Eyles and Gierasch, 2010). In addition to heat stress, sHsps are also induced by various abiotic and oxidative stresses (Sato and Yokoya, 2008).
These sHsps are found ubiquitously in all kingdoms of life, yet their number within an organism varies from two in Escherichia coli to 19 in Arabidopsis (Arabidopsis thaliana; Scharf et al., 2001). In general, the number of sHsps appears to be higher in plants. sHsps are classified into multiple subgroups based on sequence homology and subcellular localization (Scharf et al., 2001). For example, the 19 Arabidopsis sHsps are divided into 12 subgroups (Scharf et al., 2001; Sun et al., 2002; Siddique et al., 2008). Among them, seven classes, class I (CI) to CVII, contain sHsps localized in the cytoplasm and/or nucleus. In addition, sHsps are found in organelles, including plastids, mitochondria, peroxisomes, and the endoplasmic reticulum. With the exception of the mitochondrial Hsp22 in Drosophila melanogaster, these organellar sHsps are unique to plants.
In their native state, the majority of sHsps exist as large oligomers ranging from 12 to greater than 32 subunits (Lee et al., 1995, 1997; Helm et al., 1997; Stengel et al., 2010). As with larger Hsps, such as Hsp70 and Hsp40, sHsps display chaperone activity but are independent of ATP. Thus, the sHsp working mechanism differs from that of ATP-dependent Hsps (Lee et al., 1997; Lee and Vierling, 2000; van Montfort et al., 2001; Nakamoto and Vígh, 2007). sHsps do not directly facilitate the folding of heat-induced unfolded/denatured proteins. Instead, the sHsps capture denatured proteins, forming stable complexes that prevent irreversible aggregation. Subsequently, under favorable conditions, proteins captured by sHsps are released and refolded by ATP-dependent chaperone systems (Lee et al., 1997; Lee and Vierling, 2000). Although much information has been published, the exact mechanism of sHsps is not fully understood. In particular, the in vivo substrates of individual sHsps have not been identified. In most cases, sHsp chaperone activity is demonstrated using three artificial substrates: luciferase, malate dehydrogenase, and citrate synthase (Lee et al., 1997). The identification of in vivo substrates is crucial to understanding the physiological roles of sHsps (Basha et al., 2004). In addition to acting as molecular chaperones for unfolded proteins, sHsps have been reported to interact with lipids and to function in membrane quality control (Coucheney et al., 2005; Chowdary et al., 2007; Nakamoto and Vígh, 2007; Balogi et al., 2008). These findings expand the physiological roles of sHsps.
In plant cells, a large number of proteins are targeted posttranslationally to various chloroplast locations. Multiple pathways are involved depending on the final chloroplast location (Bruce, 2000; Dhanoa et al., 2010). Chloroplast interior proteins use transit peptides located at the N terminus as signal sequences. These transit peptides are recognized by Toc/Tic receptors on the chloroplast envelope (Agne and Kessler, 2009). The plastid proteins transit through the cytoplasm as unfolded proteins. Since the unfolded proteins are highly prone to the formation of nonspecific aggregates, the cytosolic levels of plastid precursors are tightly controlled by Hsc70-4, a member of the Hsp70 family (Lee et al., 2009). Outer envelope membrane proteins containing an N-terminal transmembrane domain (TMD) also need a chloroplast targeting signal. The TMD and its C-terminal-flanking positive region (CPR) function as the targeting signals of these proteins (Lee et al., 2001, 2004b). In this targeting pathway, AKR2A and AKR2B recognize the chloroplast outer membrane protein-targeting signals and deliver them to the chloroplast outer membranes (Bae et al., 2008).
To investigate the chloroplast outer membrane protein-targeting mechanism, we screened for AKR2A-interacting proteins using a protein pull-down approach. This resulted in the identification of Hsp17.8, a member of CI sHsps. Here, we demonstrate that Hsp17.8 interacts with both AKR2A and chloroplasts and enhances the chloroplast binding of AKR2A. Furthermore, higher levels of Hsp17.8 together with AKR2A enhance the targeting efficiency of membrane proteins to chloroplasts, whereas the suppression of HSP17.8 and closely related CI cytosolic sHSP genes using artificial microRNA (amiRNA) decreases the targeting efficiency of chloroplast membrane proteins.
RESULTS
Hsp17.8 Interacts with AKR2A
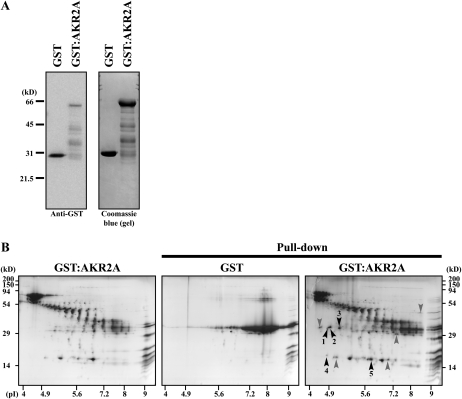
To gain insight into the molecular mechanism of protein targeting to chloroplast outer membranes, we identified proteins that interact with AKR2A. We generated a glutathione S-transferase (GST) fusion protein construct (GST:AKR2A) and expressed it in E. coli. GST:AKR2A and GST alone were purified from E. coli extracts. Purified GST:AKR2A was rather unstable and produced many degradation products (Fig. 1A). Purified GST:AKR2A was incubated with total soluble protein extracts of leaf tissues. Subsequently, proteins bound to GST:AKR2A were precipitated and analyzed using two-dimensional SDS-PAGE. As a control, GST alone was also included in the protein pull-down experiments. Figure 1B shows the two-dimensional images of proteins present in GST:AKR2A and GST control precipitates. Both samples yielded large numbers of proteins. The GST:AKR2A-specific proteins were identified and subjected to matrix-assisted laser-desorption ionization time of flight (MALDI-TOF) analysis for identification. Among these, one protein was identified as Hsp17.8, a protein belonging to the CI sHsps (Scharf et al., 2001; Sun et al., 2002; Basha et al., 2010). Other proteins identified in pull-down experiments are listed in Supplemental Table S1.
Figure 1.
Identification of Hsp17.8 by GST:AKR2A-mediated protein pull down in vitro. A, Purification of GST:AKR2A and GST alone. GST:AKR2A and GST alone were purified from E. coli extracts using glutathione agarose, and purified proteins were separated by SDS-PAGE and stained with Coomassie blue or detected with anti-GST antibody. Note that purified GST:AKR2A was rather unstable and produced many degradation products. B, GST:AKR2A immobilized onto glutathione agarose beads was incubated with total soluble protein extracts from leaf tissues, and proteins were precipitated with the beads. As a control, GST alone was included. Precipitates and GST:AKR2 (left panel) purified from E. coli extracts were analyzed using two-dimensional gel electrophoresis and stained with Coomassie blue. GST:AKR2A-specific spots were excised and subjected to MALDI-TOF analysis for identification. Approximately 50 μg of bead-bound GST (alone or as a fusion protein: GST:AKR2A) was used for pull-down assay. The protein spots indicated by arrowheads were subjected to MALDI-TOF analysis. Those that were identified are numbered and listed in Supplemental Table S1.
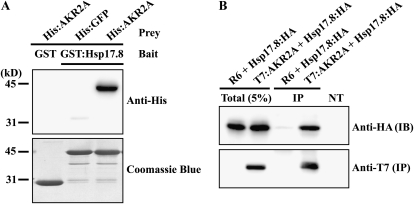
To confirm the interaction between Hsp17.8 and AKR2A, we generated a GST:Hsp17.8 fusion protein and expressed it in E. coli. Purified GST:Hsp17.8 was incubated with His-tagged AKR2A. GST alone and His:GFP were used as negative controls in the protein pull-down experiments. Proteins bound to GST:Hsp17.8 or the GST control were precipitated and subjected to western-blot analysis using anti-His antibody. GST:Hsp17.8 specifically precipitated His:AKR2A (Fig. 2A), confirming that Hsp17.8 interacts with AKR2A.
Figure 2.
Hsp17.8 interacts with AKR2A in vitro and in vivo. A, Interaction of Hsp17.8 with AKR2A in vitro. His:AKR2A and GST:Hsp17.8 were expressed in E. coli. Purified proteins (1 μg of His:AKR2A and 3 μg of GST:Hsp17.8) from E. coli extracts were incubated in the indicated combinations, and proteins were precipitated with glutathione agarose beads. The precipitates were separated by SDS-PAGE and analyzed by western blotting using anti-His antibody. In addition, the membrane was stained with Coomassie blue. As controls, His:GFP alone or GST alone was included in the analysis. B, Interaction of Hsp17.8 with AKR2A in vivo. HSP17.8:HA was transformed into protoplasts together with T7:AKR2A or the empty expression vector R6. Protein extracts were subjected to immunoprecipitation with anti-T7 antibody. The precipitates were analyzed by western blotting using anti-T7 and anti-HA antibodies. IB, Immunoblot; IP, immunoprecipitates; NT, protein extracts from nontransformed protoplasts; Total (5%), 5% of total protein extracts used for western-blot analysis.
To confirm that Hsp17.8 interacts with AKR2A in vivo, we performed coimmunoprecipitation experiments using protein extracts from protoplasts expressing the two proteins (Kirschner et al., 2000; Jin et al., 2001; Kim et al., 2001). At the same time, to rule out the possibility that the large GST domain at the N terminus of GST:Hsp17.8 contributes to the interaction, we tagged Hsp17.8 with a small-epitope hemagglutinin (HA) consisting of only nine amino acid residues at the C terminus (Hsp17.8:HA). HSP17.8:HA, together with T7-tagged AKR2A (T7:AKR2A) or the empty expression vector R6, was cotransformed into protoplasts. Protoplast protein extracts were subjected to immunoprecipitation using anti-T7 antibody. The immunoprecipitates were analyzed by western blotting using anti-T7 and anti-HA antibodies. Hsp17.8:HA was detected only in the pellet fraction from extracts containing both Hsp17.8:HA and T7:AKR2A (Fig. 2B), confirming that Hsp17.8 interacts with AKR2A in vivo. In addition, protein extracts from nontransformed protoplasts did not show any bands, thus confirming the specificity of antibody.
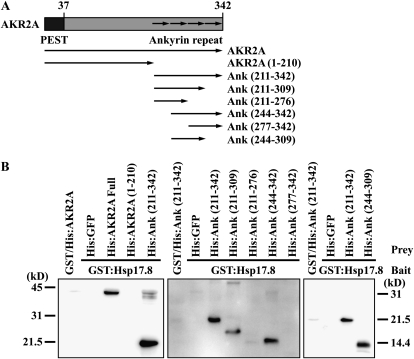
To further characterize the interaction between AKR2A and Hsp17.8, we defined the domain of AKR2A responsible for the interaction with Hsp17.8. In a previous study, we demonstrated that the N-terminal domain of AKR2A is involved in the binding of the targeting signal, TMD, plus the CPR flanking to the TMD in chloroplast outer envelope membrane proteins (Bae et al., 2008). Various AKR2A deletion mutants were generated (Fig. 3A) and expressed in E. coli as His-tagged proteins. Purified proteins were used for protein pull-down experiments with GST:Hsp17.8. The AKR2A C-terminal ankyrin repeat domain was sufficient for the Hsp17.8 interaction (Fig. 3B, left panel). To specifically define the minimal domain involved in the interaction, various ankryin repeat domain deletions were generated (Fig. 3A) and expressed as His-tagged fusion proteins. Again, deletion mutants were purified and used for protein pull-down experiments with GST:Hsp17.8 in vitro. Among these mutants, those missing the first and last ankyrin repeats displayed GST:Hsp17.8 binding (Fig. 3B, middle panel). These results indicate that the first and last ankyrin repeats are dispensable, while the central two ankyrin repeats are sufficient for the interaction between AKR2A and Hsp17.8. To confirm this idea, a new construct, His:Ank(244–309), that contained the two central ankyrin repeats was expressed in E. coli and used in protein pull-down experiments. Indeed, His:Ank(244–309) bound to GST:Hsp17.8 (Fig. 3B, right panel), confirming that the central two ankyrin repeats are sufficient for the interaction.
Figure 3.
The ankyrin repeat domain is involved in the Hsp17.8 interaction. A, Schemes of various constructs. The N-terminal PEST and C-terminal ankyrin repeats are indicated. The AKR2A domains used for the His-tagging constructs are indicated, with the amino acid positions in parentheses. B, In vitro pull-down experiments of AKR2A deletion mutants with Hsp17.8. GST:Hsp17.8 and His-tagged AKR2A constructs were expressed in E. coli and purified using affinity columns. Glutathione agarose beads with immobilized GST:Hsp17.8 (3 μg) were incubated with various His:AKR2 proteins (1 μg) at 4°C for 3 h. Proteins were precipitated with glutathione agarose beads. As a control, GST alone was included. The precipitates were separated by SDS-PAGE and analyzed by western blotting using anti-His antibody.
AKR2A Binds to Other CI Cytosolic sHsps to Varying Degrees
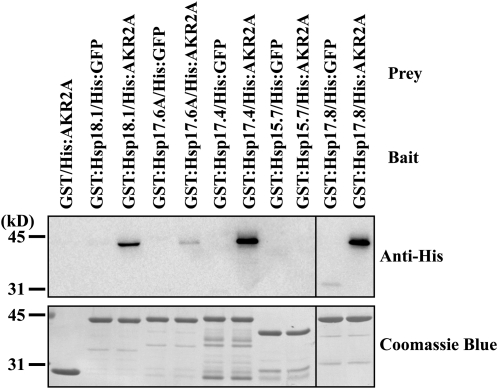
The Arabidopsis genome encodes 19 sHsps (Supplemental Fig. S1; Scharf et al., 2001; Sun et al., 2002; Siddique et al., 2008; Basha et al., 2010). Six of these sHsps, including Hsp17.8, belong to the CI cytosolic sHsps subgroup. To investigate whether AKR2A has any binding specificity among sHsps, protein pull-down experiments were performed with three additional CI cytosolic sHsps (Hsp17.4, Hsp17.6A, and Hsp18.1) together with peroxisomal Hsp15.7 as a negative control (Siddique et al., 2008). These sHsps were expressed as GST fusion proteins in E. coli. GST-fused sHsps immobilized to glutathione agarose beads were incubated with His:AKR2A. Proteins bound to the glutathione agarose beads were precipitated, and the precipitates were analyzed by western blotting using anti-His antibody. GST alone and His:GFP were included as negative controls. Among the sHsps tested, all three CI sHsps (Hsp17.4, Hsp18.1, and Hsp17.6A) displayed an interaction with AKR2A (Fig. 4). However, the amount of proteins detected in the pellets varied significantly depending on the individual CI sHsp: the AKR2A binding to Hsp17.4 was comparable to that of Hsp17.8, whereas Hsp18.1 and Hsp17.6A showed moderate and weak AKR2A binding, respectively. In contrast, peroxisomal Hsp15.7, used as a negative control, did not show any detectable level of AKR2A interaction. In the negative control samples, GST-tagged sHsps or GST alone did not show any binding to His:GFP or His:AKR2A, respectively, confirming the specificity of interactions between AKR2A and cytosolic sHsps. These results strongly suggest that AKR2A displays differential binding to members of CI cytosolic sHsps. In addition, these results are consistent with an AKR2A function in the cytosol (Bae et al., 2008).
Figure 4.
AKR2A displays differential binding to members of CI cytosolic sHsps. Various sHsps, four CI cytosolic sHsps and one peroxisomal sHsp, were expressed in E. coli as GST fusion proteins. GST-tagged sHsps (3 μg) or GST alone (3 μg) immobilized onto glutathione agarose beads were incubated with His:AKR2A or His:GFP (1 μg), and proteins were precipitated with the beads. The precipitates were analyzed by western blotting using anti-His antibody. Subsequently, the membrane was stained with Coomassie blue.
Hsp17.8 Increases the Amount of AKR2A Proteins That Bind to Chloroplasts
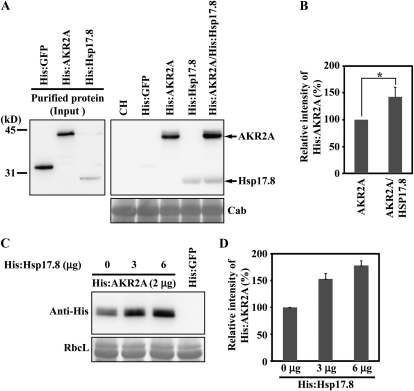
To gain insight into the Hsp17.8 physiological role, we first examined whether Hsp17.8 affects AKR2A binding to chloroplasts (Bae et al., 2008). We performed in vitro chloroplast binding experiments using His:AKR2A in the presence of His:Hsp17.8. Purified His:AKR2A and His:Hsp17.8 (Fig. 5A) were incubated with intact chloroplasts (Li and Chen, 1996; Tu and Li, 2000). The protein-chloroplast mixtures were subjected to low-speed centrifugation, so that only chloroplasts and proteins bound to chloroplasts were precipitated. Proteins in the pellet fraction were analyzed by western blotting using anti-His antibody. His:GFP was included as a negative control. In the presence of His:Hsp17.8, the amount of His:AKR2A bound to chloroplasts was increased approximately 142% compared with incubation without His:Hsp17.8 (Fig. 5B). This result indicates that Hsp17.8 enhances AKR2A binding to chloroplasts. His:GFP, used as a negative control, was not detected in the pellets. The level of endogenous chlorophyll a/b-binding proteins, used as a loading control, was equal among the samples, confirming that equal chloroplast amounts were used in the binding experiments.
Figure 5.
Hsp17.8 increases the amount of AKR2A proteins bound to chloroplasts. A and B, The effect of Hsp17.8 on the AKR2A binding to chloroplasts. A, His-tagged AKR2A or Hsp17.8 recombinant proteins were expressed in E. coli and purified using a nickel-nitrilotriacetic acid agarose affinity column (left panel). Purified recombinant proteins (2 μg) were incubated with intact chloroplasts purified from protoplasts. His:GFP (2 μg) was included as a control. After incubation on ice, chloroplasts were pelleted and proteins from the pellet fraction were analyzed by western blotting using anti-His antibody. As a control, 10% of total proteins (10%) was included in the western blotting. Subsequently, the blot was stained with Coomassie blue and chlorophyll a/b-binding protein (Cab) was used as a loading control. CH, Extracts from chloroplasts. B, To quantify the binding, the intensity of His:AKR2A and His:HSP17.8 was quantified using software equipped on the LAS3000 and is presented as relative values to the controls (His:AKR2A alone or His:HSP17.8 alone). AKR2A, His:AKR2A; HSP17.8, His:HSP17.8. The asterisk denotes a statistically significant difference compared with His:AKR2A alone (P < 0.01; n = 3). Error bars represent se (n = 3). C and D, The effect of Hsp17.8 concentrations on AKR2A binding to chloroplasts. C, Varying amounts of His:Hsp17.8 were incubated with a fixed amount of His:AKR2A (2 μg) and purified chloroplasts. Chloroplast-bound proteins were pelleted by low-speed centrifugation. His:GFP was used as a negative control for His:AKR2A. Pelleted proteins were analyzed by western blotting using anti-His antibody. Total proteins (10%) were included in the western-blot analysis. RbcL, Large subunit of the Rubisco complex, used as a loading control. D, To quantify binding, His:AKR2A intensity was measured and is presented as relative values to the control, His:Hsp17.8 (0 μg).
Figure 5A contains additional interesting data: the coprecipitation of His:Hsp17.8 with chloroplasts. When His:Hsp17.8 alone was incubated with intact chloroplasts in vitro, His:Hsp17.8 coprecipitated with chloroplasts (Fig. 5A, lane His:Hsp17.8). These data strongly suggest that His:Hsp17.8 interacts directly with chloroplasts. Furthermore, similar to His:AKR2A, the amount of His:Hsp17.8 that copurified with chloroplasts increased 138% when incubated with His:AKR2A. This result raised the possibility that Hsp17.8 binds to both AKR2A and chloroplasts. This interaction between Hsp17.8 and AKR2A may facilitate the binding of both of these proteins to chloroplasts.
To further confirm that Hsp17.8 enhances His:AKR2A binding to chloroplasts, varying amounts of His:Hsp17.8 were incubated with a fixed quantity of His:AKR2A. The protein-chloroplast incubation mixtures were subjected to low-speed centrifugation to precipitate only chloroplasts and proteins bound to chloroplasts. The pelleted fractions were analyzed by western blotting using anti-His antibody. The amount of His:AKR2A that coprecipitated with chloroplasts increased gradually with increasing concentrations of His:Hsp17.8 (Fig. 5, C and D). The large subunit of the Rubisco complex, used as a loading control, confirmed equal loading.
Hsp17.8 Exists as a Dimer under Normal Physiological Conditions and in Association with Chloroplasts
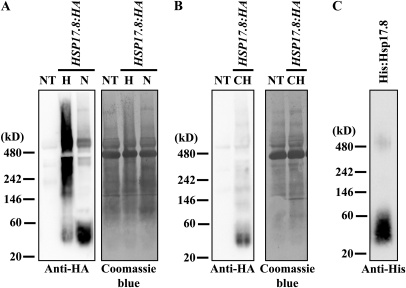
sHsps are large oligomers that contain 12 to more than 32 subunits (Lee et al., 1995, 1997; Helm et al., 1997; Stengel et al., 2010). In heat shock, sHsps form large complexes with unfolded proteins (Haslbeck et al., 2005). sHsps recognize the hydrophobic segments of unfolded proteins with no specificity (Nakamoto and Vígh, 2007). In contrast, Hsp17.8 specifically recognizes the C-terminal ankyrin repeat domain of AKR2A (Fig. 3B). The ankyrin repeat, a 33-residue sequence domain, is a very common protein-protein interaction motif (Mosavi et al., 2004). To gain insights into the interaction between AKR2A and Hsp17.8, we examined the oligomeric state of Hsp17.8 under normal physiological conditions using blue-native (BN)-PAGE (Kikuchi et al., 2006). Protein extracts from protoplasts transformed with HSP17.8:HA were separated by BN-PAGE and subjected to western-blot analysis using anti-HA antibody. Hsp17.8 was detected as a strong band at a molecular mass of 40 kD and as bands of lower intensity at high-molecular-mass positions (Fig. 6A). The 40-kD band implies that Hsp17.8 exists as a dimer under normal physiological conditions. Protein extracts from untransformed protoplasts did not show any signals, confirming the specificity of the antibody. In addition, upon heat shock, the Hsp17.8:HA dimer was converted to high-molecular-mass complexes ranging in size from 240 kD to greater than 480 kD, indicating that Hsp17.8 proteins can be converted to large oligomeric forms, as observed with other sHsps (Lee et al., 1995, 1997; Helm et al., 1997; Stengel et al., 2010).
Figure 6.
Hsp17.8 exists as dimers at normal physiological conditions and in association with chloroplasts. A and B, BN-polyacrylamide gel analysis of Hsp17.8:HA. A, Protein extracts from protoplasts transformed with HSP17.8:HA were separated by BN-PAGE using a 4% to 16% gradient gel and analyzed by western blotting using anti-HA antibody. In addition, protein extracts from protoplasts subjected to heat shock at 42°C for 30 min were included in the analysis. The membrane was stained with Coomassie blue. NT, Extracts from nontransformed protoplasts; N, normal condition; H, heat shock condition. B, Intact chloroplasts were purified from protoplasts transformed with HSP17.8:HA, and proteins that copurified with chloroplasts were separated by BN-PAGE and analyzed by western blotting using anti-HA antibody. The membrane was stained with Coomassie blue. NT, Protein extracts from nontransformed protoplasts; CH, proteins from purified chloroplasts. C, Dimer formation of His:Hsp17.8 in E. coli extracts. His:Hsp17.8 purified from E. coli extracts was separated by BN-PAGE and analyzed by western blotting using anti-His antibody.
Next, we examined the oligomeric state of Hsp17.8 when it is associated with chloroplasts. Intact chloroplasts were purified from protoplasts transformed with HSP17.8:HA, and proteins that copurified with chloroplasts were separated by BN-PAGE and subjected to western-blot analysis with anti-HA antibody. Again, Hsp17.8:HA was detected at 40 kD (Fig. 6B), indicating that Hsp17.8 binds to chloroplasts as a dimer. To further examine the biochemical properties of Hsp17.8, His:Hsp17.8 purified from E. coli extracts was analyzed by BN-PAGE and detected by anti-His antibody. His:Hsp17.8 was detected principally at 40 kD and secondarily at 480 kD, indicating that purified His:Hsp17.8 exists as a dimer under normal conditions.
HSP17.8 Is Expressed under Normal Growth Conditions without Heat Shock
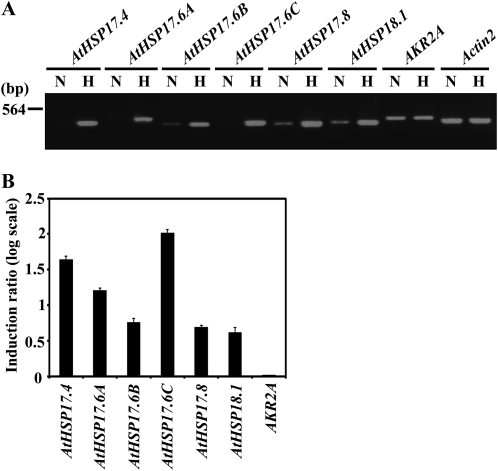
AKR2A is thought to mediate the targeting of membrane proteins to chloroplast outer envelopes (Bae et al., 2008). If Hsp17.8 plays a role in targeting membrane proteins to chloroplasts but not in the heat shock response, HSP17.8 should be expressed under normal growth conditions. Accordingly, we tested whether HSP17.8 is expressed under normal physiological conditions. Total RNAs from Arabidopsis seedlings treated with or without heat shock were used in semiquantitative reverse transcription (RT)-PCR. For comparison, five CI cytosolic sHSP genes were included in the analysis. In a previous study (Siddique et al., 2008), expression of HSP17.6C was examined by RT-PCR, which showed that HSP17.6C transcripts are not present under normal growth conditions but increased to high levels upon heat shock at 40°C. Consistent with the previous study, HSP17.6C was not detected in total RNA obtained from seedlings without heat shock under the semiquantitative RT-PCR condition (Fig. 7). Similarly, two other isoforms, HSP17.4 and HSP17.6A, were expressed at very low level without heat shock, thus being undetectable under the semiquantitative RT-PCR condition we used. By contrast, transcripts of three sHSP genes, HSP17.8, HSP18.1, and HSP17.6B, were detected in total RNA obtained from seedlings without heat shock under the same semiquantitative RT-PCR condition: HSP17.8 and HSP18.1 were expressed at moderate levels and HSP17.6B was expressed at a low level, indicating that certain isoforms, including HSP17.8, were expressed under normal growth conditions. Similarly, without heat shock, certain sHSP isoforms are expressed ubiquitously in all tissues, whereas others are expressed in a tissue-specific manner (Siddique et al., 2008). Hsp18.1 also interacted with AKR2A (Fig. 4), raising the possibility that Hsp18.1 may also work together with AKR2A in targeting membrane proteins to chloroplast outer membranes under physiological conditions. In addition, all six CI cytosolic sHSPs, including HSP17.8, were highly induced upon heat shock, and the degree of induction of HSP17.8 was comparable to that of HSP17.6C, indicating that Hsp17.8 may also function as a chaperone under heat stress conditions.
Figure 7.
Semiquantitative RT-PCR analysis of sHSPs and AKR2A transcript levels with or without heat shock. A, Total RNAs from 10-d-old Arabidopsis whole seedlings that had been treated with or without heat shock at 40°C for 1 h were analyzed by semiquantitative RT-PCR using gene-specific primers. Actin2 was included as an internal control. The PCR was performed at 94°C for 30 s, 57°C for 30 s, and 72°C for 30 s for 25 cycles. The PCR products were separated on agarose gels. N, Normal condition; H, heat shock condition. B, To quantify the induction of gene expression by heat shock, PCR band intensity was quantified by software on the LAS3000, and the ratio of gene expression under the heat shock condition to that under the normal condition was expressed at the log values. Error bars represent se (n = 3).
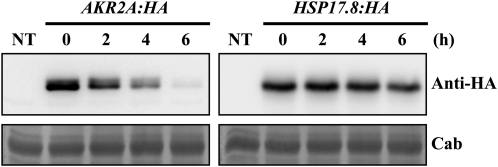
Since the results shown in Figures 4 and 5 strongly suggested that Hsp17.8 may work together with AKR2A, we compared the transcript level of AKR2A with that of HSP17.8 by semiquantitative RT-PCR analysis. The expression of AKR2A was not affected by heat shock treatment. Under normal growth conditions, the transcript level of AKR2 was higher than that of HSP17.8, suggesting that the AKR2A protein level would be higher than the Hsp17.8 protein level if the transcripts of the two genes are translated into proteins with the same efficiency. However, it is possible that the difference in the transcript levels between the two genes can be compensated by other mechanisms, such as a difference in the stability of the two proteins. Indeed, a PEST sequence is located in the N-terminal region of AKR2A (Bae et al., 2008). The PEST sequence, which is rich in Pro (P), Glu (E), Ser (S), and Thr (T), serves to target proteins for degradation and is associated with proteins that have a short intracellular half-life (Rogers et al., 1986). This observation raised the possibility that AKR2A may be a short-lived protein. Accordingly, we examined the half-lives of the two proteins. Protoplasts transformed with AKR2A:HA or HSP17.8:HA were treated with cycloheximide, an inhibitor of translation, 24 h after transformation. Subsequently, protein extracts were prepared at various time points after cycloheximide treatment and analyzed by western blotting using anti-HA antibody. The half-lives of AKR2A:HA and Hsp17.8:HA were approximately 3 h and over 6 h, respectively (Fig. 8), indicating that Hsp17.8 is much more stable than AKR2A. These results suggest that the low transcript level of HSP17.8 can be compensated by the longer half-life of the Hsp17.8 protein.
Figure 8.
Analysis of the half-life of Hsp17.8 and AKR2A in protoplasts. Protoplasts transformed with HSP17.8:HA or AKR2A:HA were treated with cycloheximide 24 h after transformation and then harvested at the time points indicated. Protein extracts from the transformed protoplasts were analyzed by western blotting using anti-HA antibody. To obtain the half-life of these proteins, the intensity of the protein bands was quantified using software on the LAS3000. NT, Extracts from nontransformed protoplasts; Cab, chlorophyll a/b-binding protein.
Coexpression of Hsp17.8:HA and AKR2A Increases the Targeting Efficiency of Outer Envelope Protein7:GFP to Chloroplasts
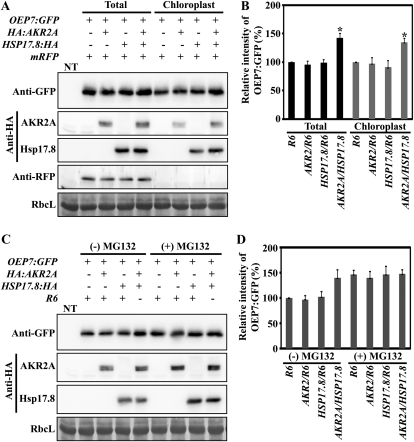
We examined the role of Hsp17.8 in AKR2A-mediated protein targeting to chloroplasts. In previous studies, it was shown that Outer Envelope Protein7 (OEP7) expressed as a GFP fusion protein (OEP7:GFP) is targeted to chloroplast outer membranes and that AKR2A mediates its targeting (Lee et al., 2001; Bae et al., 2008). Since sHsps are known to possess chaperone activity (Kirschner et al., 2000; Eyles and Gierasch, 2010), we first examined whether Hsp17.8 has any effect on the level of OEP7:GFP. In protoplasts, OEP7:GFP was cotransformed with HA:AKR2A, HSP17.8:HA, or both (HA:AKR2A plus HSP17.8:HA). The total plasmid amounts introduced into protoplasts were adjusted using the empty expression vector R6. Coexpression with AKR2A or Hsp17.8:HA individually did not affect the OEP7:GFP levels. However, in protoplasts transformed with three plasmids, OEP7:GFP, HA:AKR2A, and HSP17.8:HA, the amount of OEP7:GFP was increased to 143% compared with transformation with OEP7:GFP alone (Fig. 9, A and B). These results suggest that a portion of OEP7:GFP is subject to proteolytic degradation in protoplasts and that coexpression of both proteins, HA:AKR2A and Hsp17.8:HA, prevents OEP7:GFP degradation (Fig. 9, C and D).
Figure 9.
Hsp17.8 increases the targeting efficiency of OEP7:GFP to chloroplasts. A and B, The effect of Hsp17.8 on OEP7:GFP targeting to chloroplasts. A, Protoplasts were cotransformed with the indicated constructs, and proteins from the transformed protoplasts (Total) or from chloroplasts purified from transformed protoplasts (Chloroplast) were analyzed by western blotting using anti-GFP, anti-HA, and anti-RFP antibodies. The construct mRFP was included as a measure of transformation efficiency and contamination of cytosolic proteins in purified chloroplasts.. Note that anti-HA antibody detects both HA:AKR2A and Hsp17.8:HA. NT, Nontransformed protoplasts; RbcL, large subunit of the Rubisco complex, used as a loading control. B, To quantify the targeting efficiency, the intensity of OEP7:GFP was measured by software on the LAS3000 and is presented as relative values to control R6. Asterisks denote statistically significant differences compared with R6 or HSP17.8/R6 (P < 0.01; n = 3). Error bars represent se (n = 3). C and D, The effect of MG132 on the OEP7:GFP level. C, Transformed protoplasts (as described in A) were treated with (+) or without (−) MG132. Protein extracts were analyzed by western blotting using anti-GFP antibody. D, To quantify the OEP7:GFP level, the intensity of OEP7:GFP was measured and is presented as relative values to the control, R6/(−)MG132. Error bars represent se (n = 3).
To gain insights into the mechanism responsible for the higher OEP7:GFP levels when OEP7:GFP is coexpressed with both HA:AKR2A and Hsp17.8:HA, we examined the chloroplast-targeting efficiency of OEP7:GFP following the coexpression of both Hsp17.8 and AKR2A. Chloroplasts were purified from transformed protoplasts, and the amount of copurified OEP7:GFP was measured by western-blot analysis using anti-GFP antibody. When coexpressed with both HA:AKR2A and Hsp17.8:HA, the targeting efficiency of OEP7:GFP was increased to 132% compared with OEP7:GFP alone (Fig. 9, A and B). However, coexpression with HA:AKR2A or Hsp17.8:HA separately did not affect the OEP7:GFP levels targeted to chloroplasts, indicating that both Hsp17.8 and AKR2A are required to increase the amount of OEP7:GFP targeted to chloroplasts. These results imply that the OEP7:GFP levels in protoplasts are greater than the endogenous capacity of the chloroplast-targeting machinery and that excess amounts of OEP7:GFP are subject to degradation. However, higher levels of both AKR2A and Hsp17.8 may be able to handle the excess amount of OEP7:GFP in chloroplast targeting. This in turn results in the higher levels of OEP7:GFP targeting to chloroplasts. To test this possibility, we examined whether a portion of OEP7:GFP was subject to proteolytic degradation. Transformed protoplasts were treated with MG132, an inhibitor of the 26S proteasome (Lee and Goldberg, 1998), and OEP7:GFP protein levels were measured by western-blot analysis using anti-GFP antibody. Upon MG132 treatment, the OEP7:GFP levels in protoplasts transformed with OEP7:GFP alone or together with HA:AKR2A or HSP17.8:HA were elevated to those obtained from protoplasts transformed with all three constructs, OEP7:GFP, HA:AKR2A, and HSP17.8:HA (Fig. 9, C and D), confirming that a portion of OEP7:GFP was subjected to proteolytic degradation in protoplasts. Taken together, these results suggest that higher levels of both Hsp17.8 and AKR2A are necessary to prevent the degradation of OEP7:GFP and to increase the targeting efficiency of OEP7:GFP to chloroplasts.
Suppression of CI Cytosolic sHSP Transcript Levels by sHSP-CI amiRNA Reduces OEP7:GFP Chloroplast Targeting Efficiency
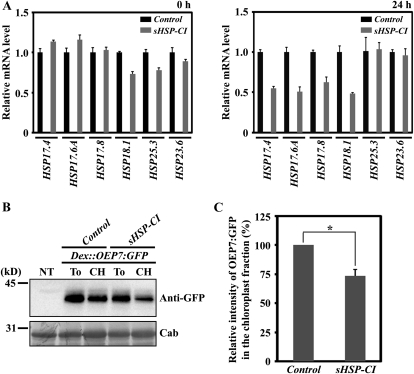
To further confirm the role of Hsp17.8 in protein targeting to chloroplast outer membranes, we investigated whether decreased Hsp17.8 levels have any detrimental effects on membrane protein-targeting efficiency to the chloroplast outer membranes. Since an hsp17.8 mutant was not available, we employed an amiRNA approach (Ossowski et al., 2008). This approach was recently used to successfully lower target protein levels in protoplasts (Kim and Somers, 2010). Because six CI cytosolic sHsps show high degrees of sequence similarity (Supplemental Fig. S2; Sun et al., 2001) and four of them interact to varying degrees with AKR2A, we suppressed the transcript levels of all six CI sHSP genes (HSP17.4, HSP17.6A, HSP17.6B, HSP17.6C, HSP17.8, and HSP18.1) simultaneously. This was accomplished using a 21-bp fragment (sHSP-CI amiRNA construct) that contained three to five base mismatches to all six CI cytosolic sHSP genes (Supplemental Fig. S3). Protoplasts were transformed with the sHSP-CI amiRNA construct or the control amiRNA vector, and sHSP transcript levels were measured in protoplast total RNA collected 0 and 24 h following transformation. Quantitative RT-PCR analysis was used to measure the levels of four CI cytosolic sHSP genes (HSP17.4, HSP17.6A, HSP17.8, and HSP18.1) and two organellar sHSP genes (plastid HSP25.3 and mitochondrial HSP23.6). At 0 h, the transcript levels of all examined sHSP genes ranged from 85% to 110% compared with the control transformed with control amiRNA (Fig. 10A, left panel). However, at 24 h post transformation, the transcript levels of the four CI cytosolic sHSP genes were reduced by 50% to 55% in comparison with the control amiRNA vector. The transcript levels of the two organellar sHSP genes (HSP25.3 and HSP23.6) were not affected (Fig. 10A, right panel). These results indicate that the sHSP-CI amiRNA construct specifically lowered CI cytosolic sHSP transcript levels.
Figure 10.
Suppression of CI cytosolic sHSP transcript levels decreases the chloroplast-targeting efficiency of OEP7:GFP in protoplasts. A, Real-time quantitative RT-PCR of various sHSP transcript levels in protoplasts. Protoplasts were transformed with the sHSP-CI amiRNA construct or control amiRNA vector. Total RNA extracted from the transformed protoplasts at 0 and 24 h after transformation was subjected to quantitative RT-PCR analysis using gene-specific primers. Quantitative RT-PCR was performed at 95°C for 15 s and 60°C for 1 min for 40 cycles. Actin8 was used as an internal control for the quantitative RT-PCR. Error bars represent se (n = 3). B and C, Effect of sHSP-CI amiRNA on the OEP7:GFP targeting to chloroplasts. B, Dex::OEP7:GFP was cotransformed into protoplasts together with sHSP-CI amiRNA or the control amiRNA vector. Transformed protoplasts were treated with dexamethasone 24 h after transformation and incubated for an additional 12 h. Subsequently, protoplasts were divided into two fractions: one fraction was used to purify chloroplasts by Percoll gradients, and the second fraction was used for total protein extracts. Proteins from the purified chloroplasts together with total protein extracts from the protoplasts were analyzed by western blotting using anti-GFP antibody. NT, Nontransformed protoplasts; To, total protein extracts; CH, protein extracts from purified chloroplasts; Cab, chlorophyll a/b-binding protein, used as a loading control. C, To quantify the targeting efficiency, the intensity of OEP7:GFP in the chloroplast fractions was measured and is presented as a relative value to that of control amiRNA vector. The asterisk denotes a statistically significant difference compared with control amiRNA vector (P < 0.01; n = 3). Error bars represent se (n = 3).
Next, we examined whether lower levels of CI cytosolic sHSP transcripts reduce OEP7:GFP chloroplast targeting efficiency. Dex::OEP7:GFP, a construct containing OEP7:GFP under the dexamethasone-inducible promoter (Aoyama and Chua, 1997), was cotransformed with sHSP-CI amiRNA or control amiRNA vector into protoplasts. The protoplasts were treated with dexamethasone 24 h after transformation and incubated for an additional 12 h. With these conditions, OEP7:GFP expression began 24 h after transformation, a time when the transcript levels of four CI cytosolic sHSP genes were reduced by 50% to 55% compared with the control. Protoplast protein extracts were analyzed by western blotting using anti-GFP antibody. In protoplasts transformed with the sHSP-CI amiRNA construct, the amount of OEP7:GFP copurified with chloroplasts was significantly reduced compared with the control amiRNA (Fig. 10B). When quantified, the targeting efficiency was reduced to 72% of the control level (Fig. 10C), confirming that CI cytosolic sHsps play a critical role in targeting OEP7:GFP to the chloroplasts.
Hsp17.8 Does Not Interact Directly with OEP7 in the Cytoplasm
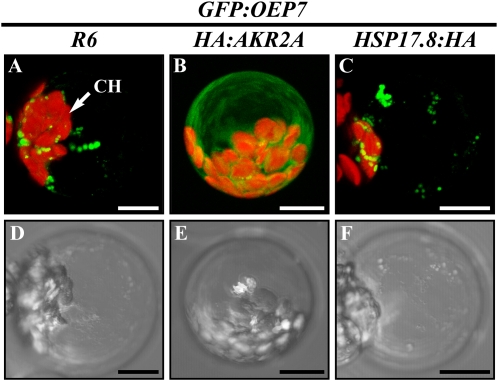
To examine further the role of Hsp17.8 in AKR2A-mediated protein targeting to chloroplast outer membranes, we tested whether Hsp17.8 also binds directly to proteins targeted to chloroplasts. In a previous study, the interaction between OEP7 and AKR2A in vivo was demonstrated using a fusion protein, GFP:OEP7 (Bae et al., 2008). When the GFP moiety was fused to the N terminus of OEP7, the fusion protein GFP:OEP7 was not targeted to chloroplasts but instead produced aggregates in the cytosol. However, when GFP:OEP7 was coexpressed with HA:AKR2A in protoplasts, HA:AKR2A prevented aggregate formation of GFP:OEP7 by binding to the hydrophobic TMD and the CPR in OEP7. In a similar approach, we tested whether Hsp17.8 interacts with OEP7. GFP:OEP7 was introduced into protoplasts together with HA:AKR2A, HSP17.8:HA, or R6 and the GFP pattern of GFP:OEP7 was examined. As reported previously (Bae et al., 2008), GFP:OEP7 alone produced a punctate staining pattern, indicating protein aggregates. In protoplasts cotransformed with GFP:OEP7 and HA:AKR2A, GFP:OEP7 produced a diffuse pattern (Fig. 11), indicating an interaction between AKR2A and OEP7. In contrast, in protoplasts cotransformed with GFP:OEP7 and HSP17.8:HA, GFP:OEP7 produced a punctate staining pattern, indicating protein aggregates. These results suggested that Hsp17.8 does not bind directly to OEP7.
Figure 11.
Hsp17.8 does not bind directly to OEP7 in the cytoplasm. GFP:OEP7 was introduced into protoplasts together with HA:AKR2A, HSP17.8:HA, or R6, and the GFP pattern of GFP:OEP7 was examined. Bottom panels show bright-field images. CH, Chloroplasts. Bars = 20 μm.
DISCUSSION
In this study, we demonstrated that Hsp17.8, one of the CI cytosolic sHsps in Arabidopsis, plays a role in membrane protein targeting to the chloroplast outer membrane. This conclusion is based on several lines of evidence. First, Hsp17.8 binds to both AKR2A and chloroplasts. Second, Hsp17.8 and AKR2A overexpression enhances the targeting efficiency of a membrane protein, OEP7:GFP, to chloroplasts, whereas decreased levels of Hsp17.8 and closely related CI cytosolic sHsps reduced the OEP7:GFP targeting efficiency to chloroplasts. In support of a role for Hsp17.8 in protein targeting to chloroplast outer membranes, HSP17.8 was expressed under normal growth conditions. In addition, under physiological conditions, Hsp17.8 existed primarily as a dimer in the cytosol and when it was associated with chloroplasts. Thus, under normal growth conditions, the Hsp17.8-assisted targeting of proteins to the chloroplast outer envelope membrane, as observed in this study, appears to be different from that observed previously for sHsps in the heat shock response (Lee et al., 1997; Lee and Vierling, 2000). When acting as chaperones in the heat shock response, sHsps bind to unfolded proteins in large complexes, thereby preventing them from forming nonspecific aggregates (Nakamoto and Vígh, 2007). sHsps may not display any specific binding with their substrates during these interactions. In fact, heat shock exposes hydrophobic domains in sHsps, and the proposed binding sites of sHsps are hydrophobic in character (Haslbeck et al., 2005), suggesting that hydrophobic interactions form the basis for interactions between sHsps and unfolded proteins. By contrast, Hsp17.8 recognizes the ankyrin repeat domain in AKR2A, implying a specific interaction between Hsp17.8 and AKR2A. This idea is supported by the fact that the ankyrin repeat is one of the most common protein-protein interaction motifs known (Mosavi et al., 2004). However, we cannot exclude the possibility that Hsp17.8 also functions as a chaperone under heat shock conditions, as has been observed with other sHsps (Lee et al., 1995, 1997; Helm et al., 1997; Stengel et al., 2010). Indeed, upon heat shock, the expression of HSP17.8 was strongly induced and Hsp17.8 was converted to high oligomeric forms with molecular masses ranging from 240 kD to greater than 480 kD.
In addition to Hsp17.8, various Hsps such as Hsp70 and Hsp90 have been identified to play a role in protein targeting to chloroplasts (Rial et al., 2000; Zhang and Glaser, 2002; Qbadou et al., 2006; Lee et al., 2009). However, the pathway that Hsp17.8 is involved in differs from those of other larger Hsps. The interaction of Hsp17.8 with AKR2A strongly suggests that Hsp17.8 is involved in targeting membrane proteins to chloroplast outer membranes. In contrast, other larger Hsps participate in the import of transit peptide-containing precursors into chloroplasts (Rial et al., 2000; Zhang and Glaser, 2002). For example, cytosolic Hsc70 interacts with the transit peptide of precursors, thereby keeping them in an unfolded, yet import-competent, state in the cytosol. Two stromal Hsps, cpHsc70 and Hsp93, are important for driving translocation into the stroma (Su and Li, 2010). In addition, Hsp90 is involved in delivering plastid precursors to the chloroplast import receptor Toc64 (Qbadou et al., 2006). Recently, Lee et al. (2009) showed that Hsc70-4 is indirectly involved in protein targeting to chloroplasts by controlling precursor levels in the cytosol. Thus, the role of Hsp17.8 is a novel function in protein targeting to chloroplasts.
A large number of membrane proteins are targeted to the outer membrane of chloroplasts after translation in the cytosol (Li et al., 1991; Hofmann and Theg, 2005). In this process, AKR2s mediate the targeting by binding to the targeting signals containing the hydrophobic TMD and the CPR. However, the mechanism by which AKR2s deliver proteins to the chloroplasts from the cytosol is not fully understood. Hsp17.8 binds both chloroplasts and AKR2A. AKR2A also has a chloroplast-binding ability (Bae et al., 2008). However, Hsp17.8 did not directly bind to chloroplast outer membrane proteins, at least when we used GFP:OEP7 as substrate. Thus, one possible scenario is that Hsp17.8 acts as a cofactor of AKR2s to facilitate the delivery of cargo proteins to chloroplasts by binding to both chloroplasts and AKR2s. Moreover, in this way, Hsp17.8 may play a role in solving the specificity issues of the targeting pathways involving AKR2s. In addition to chloroplast targeting, AKR2s are also implicated in the targeting of proteins to peroxisomes (Shen et al., 2010). Binding of Hsp17.8 to AKR2s may direct AKR2s to function specifically in the chloroplast-targeting pathway. Consistent with this hypothesis, increased levels of Hsp17.8 and AKR2A in protoplasts resulted in increased OEP7:GFP targeting to chloroplasts. In contrast, amiRNA-mediated suppression of HSP17.8 and other CI cytosolic sHSP genes in protoplasts resulted in reduced targeting efficiency. The identity of the Hsp17.8-binding partner located on the chloroplast outer membranes is unknown.
The Arabidopsis genome encodes six members of the CI cytosolic sHsps (Scharf et al., 2001; Sun et al., 2002; Basha et al., 2010). Thus, an important question for these CI cytosolic sHsps is whether they are functionally diverse. A few of them have been studied at molecular and biochemical levels. HSP17.6 is induced significantly by heat stress (Siddique et al., 2008). Furthermore, it interacts with the hydrophobic compound 1,1’-bi(4-anilino)naphthalene-5,5’-disulfonic acid at elevated temperatures, consistent with its role as a chaperone for unfolded protein. Indeed, the close homolog Hsp17.6C has been demonstrated to assist in the folding of denatured luciferase in vitro (Siddique et al., 2008). Other CI members (HSP17.4, HSP17.6A, and HSP18.1) are expressed in petals under normal growth conditions, implying a role in flower development (Dafny-Yelin et al., 2008). Two CI members, Hsp17.4 and Hsp17.6A, are also involved in acquired thermotolerance (Dafny-Yelin et al., 2008). Overexpression of Hsp17.6A enhances osmotolerance (Sun et al., 2001). Another member, HSP17.4, is induced by salt and drought stresses (Siddique et al., 2008), implying that this isoform may be involved in osmotic stress responses, as observed with Hsp17.6A. Thus, it appears that the functions of CI cytosolic sHsps overlap while also allowing for functional divergence. Consistent with this notion, AKR2A displayed differential binding affinities to some CI members: strong binding to Hsp17.8 and Hsp17.4, moderate binding to Hsp18.1, and weak binding to Hsp17.6. Thus, a subset of CI cytosolic sHsps may act as AKR2A cofactors and be involved in protein targeting to chloroplasts. However, despite their interactions with AKR2A in vitro, Hsp17.4 and Hsp18.1 were not identified in the original screening. One possibility is that they may not be expressed at high levels in leaf tissues under normal growth conditions. Indeed, HSP17.4 was nearly undetectable in leaf tissues under normal growth conditions. In addition, HSP18.1 was expressed at lower levels than HSP17.8 in rosette leaves (Arabidopsis eFP browser; http://bar.utoronto.ca/efp_arabidopsis/cgi-bin/efpWeb.cgi), and these levels may not have been high enough to detect its binding in protein pull-down assays.
sHsps are involved in diverse cellular defense responses against many different stresses, including high-temperature, oxidative, and osmotic stresses (Sato and Yokoya, 2008). Under nonstress conditions, sHsps are shown to play important roles in various stages of development, including flower, seed, and fruit development (Wehmeyer et al., 1996; Volkov et al., 2005; Dafny-Yelin et al., 2008). In this study, we provide evidence for a novel role of one CI cytosolic sHsp, Hsp17.8, as an AKR2A cofactor involved in targeting membrane proteins to chloroplasts.
MATERIALS AND METHODS
Growth of Plants
Arabidopsis (Arabidopsis thaliana) plants (ecotype Columbia) were grown on Murashige and Skoog plates supplemented with 1% Suc in a growth chamber at 20°C to 22°C under a 16-h/8-h light/dark cycle. Leaf or whole tissues were harvested from 10-d-old to 3-week-old plants and used immediately for protoplast isolation or total RNA extraction.
Construction of Plasmid DNAs
HSP15.7 (At5g37670), HSP17.4 (At3g46230), HSP17.6A (At1g59860), HSP17.8 (At1g07400), and HSP18.1 (At5g59720) were PCR isolated from an Arabidopsis cDNA library using gene-specific primer sets (Supplemental Table S1). PCR fragments were ligated into pGEX-5X-1 (GE Healthcare) for GST fusion constructs or pRSET-A (Invitrogen) for His-tagging constructs. HSP17.8:HA was PCR generated using the primers HSP17.8-HA-F and HSP17.8-HA-R (Supplemental Table S2).
Full-length AKR2A, various AKR2A deletion mutants, and GFP were generated through PCR approaches. The primer sets used were as follows: AKR2A-GST-F and AKR2A-GST/T7/HA-R for GST:AKR2A; AKR2A-His-F and AKR2A-His-R for His:AKR2A; AKR2A-His-F and AKR2A(210)-His-R for His:AKR2A(1–210); Ank(211)-His-F and AKR2A-His-R for His:Ank(211–342); Ank(211)-His-F and Ank(309)-His-R for His:Ank(211–309); Ank(211)-His-F and Ank(276)-His-R for His:Ank(211–276); Ank(244)-His-F and AKR2A-His-R for His:Ank(244–342); Ank(277)-His-F and AKR2A-His-R for His:Ank(277–342); Ank(244)-His-F and Ank(309)-His-R for His:Ank(244–309); and GFP-His-F and GFP-His-R for His:GFP. PCR products were ligated into pGEX-5X-1 (GE Healthcare) for GST fusion constructs or pRSET-A (Invitrogen) for His tagging. To generate N-terminal HA- or T7-tagged or C-terminal HA-tagged AKR2A, PCR was performed using AKR2A-T7/HA-F and AKR2A-GST/T7/HA-R for T7:AKR2A or HA:AKR2A and AKR2A-T7/HA-F and AKR2A-HA-R for AKR2A:HA. The PCR products were ligated into N-terminal HA or T7 tagging or C-terminal HA tagging vectors.
Construction of OEP7:GFP was described previously (Lee et al., 2001). To construct Dex::OEP7:GFP, OEP7:GFP was PCR amplified using OEP7-pTA7002-F and GFP-pTA7002-R primers and ligated into the pTA7002 vector.
The sHSP-CI amiRNA construct was designed using Web MicroRNA Designer 3 (http://wmd3.weigelworld.org). The sHSP-CI amiRNA was prepared by overlapping PCR as described previously (Schwab et al., 2006). The primer sets used were sHSP-CI-I miR-s, sHSP-CI-II miR-a, sHSP-CI-III miR*s, and sHSP-CI-IV miR*a, together with two vector primers (amiRNA-A and amiRNA-B) in the adjacent region that defines the amiRNA foldback. The resulting PCR fragment, including the full amiRNA foldback, was ligated into the vector pCsVMV-AmiR. The control amiRNA vector (pCsVMV-AmiR) was described previously (Kim and Somers, 2010). All PCR products were sequenced to confirm product accuracy.
Transient Expression of Proteins in Protoplasts
Arabidopsis protoplasts were isolated and transformed as described previously (Jin et al., 2001). For chemical treatments, transformed protoplasts were incubated with MG132 (30 μm; Calbiochem), dexamethasone (30 μm; Sigma), or cycloheximide (50 μm; Sigma).
RNA Extraction, Semiquantitative RT-PCR, and Gene Expression Analysis
To give heat shock, 10-d-old seedlings grown on Murashige and Skoog plates were preincubated at 40°C for 15 min and then at 22°C for 2 h. Subsequently, plants were subjected to a second heat treatment at 40°C for 1 h. Nonstress control plants were kept at 22°C. Total RNA was extracted using an RNAqueous kit (Ambion). TURBO DNase (Ambion) was used to remove any DNA contamination from the RNA samples, and extracted RNA was used for RT into cDNA using the High Capacity cDNA Reverse Transcription kit (AB).
Semiquantitative RT-PCR was performed using ExTaq polymerase (Takara) at 94°C for 30 s, 57°C for 30 s, and 72°C for 30 s for 25 cycles. The primer sets used were as follows: HSP17.4-RT-F and HSP17.4-RT-R for HSP17.4; HSP17.6A-RT-F and HSP17.6A-RT-R for HSP17.6A; HSP17.6B-RT-F and HSP17.6B-RT-R for HSP17.6B; HSP17.6C-RT-F and HSP17.6C-RT-R for HSP17.6C; HSP17.8-RT-F and HSP17.8-RT-R for HSP17.8; HSP18.1-RT-F and HSP18.1-RT-R for HSP18.1; AKR2A-RT-F and AKR2A-RT-R for AKR2A; and Actin2-RT-F and Actin2-RT-R for Actin2.
Quantitative real-time RT-PCR was performed with a SYBR Green kit (AB) to detect HSP17.4, HSP17.6A, HSP17.8, HSP18.2, HSP23.6, HSP25.3, and Actin8. Amplified samples were normalized with Actin8. The primers used were as follows: HSP17.4-F and HSP17.4-R for HSP17.4; HSP17.6A-F and HSP17.6A-R for HSP17.6A; HSP17.8-F and HSP17.8-R for HSP17.8; HSP18.1-F and HSP18.1-R for HSP18.1; HSP23.6-F and HSP23.6-R for HSP23.6; HSP25.3-F and HSP25.3-R for HSP25.3; and ACT8-F and ACT8-R for Actin8.
Coimmunoprecipitation
For coimmunoprecipitation, protoplasts transformed with expression constructs were lysed by sonication in immunoprecipitation buffer (20 mm Tris-HCl, pH 7.5, 150 mm NaCl, 3 mm MgCl2, 1 mm dithiothreitol (DTT), 1 mm EDTA, 0.5% Triton X-100, and 1× complete protease inhibitor cocktail [Roche]). The soluble proteins were prepared by centrifugation at 20,000g for 10 min. Samples were incubated with 2 μg of anti-T7 monoclonal antibody (Novagen) for 2 h at 4°C, followed by an additional incubation with 20 μL of protein A-Sepharose CL-4B beads (Amersham Biosciences) for 2 h at 4°C. Immunoprecipitates were washed three times with the immunoprecipitation buffer and subjected to immunoblot analysis with appropriate antibodies.
Recombinant Protein Purification and Protein Pull-Down Experiments
Escherichia coli BL21 (DE3) cells transformed with various constructs encoding GST-fused or His-fused recombinant proteins were cultured to an optical density at 600 nm of approximately 0.6. Induction of the protein was induced by adding 0.2 to 1 mm isopropyl-b-d-thiogalactoside at 37°C for 3 h. The GST or GST fusion proteins were bound to Immobilized Glutathione beads (Thermo Scientific) and washed several times with buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 5 mm EDTA, 1% Triton X-100, and 1× complete protease inhibitor cocktail [Roche]) for purification. The His-tagged proteins were bound to nickel-nitrilotriacetic acid agarose beads (Qiagen) and washed several times with washing buffer (50 mm NaH2PO4, pH 8.0, 300 mm NaCl, 10 mm imidazole, 1% Triton X-100, and 1× complete protease inhibitor cocktail [Roche]) for purification.
To pull down AKR2A-binding proteins, approximately 50 μg of GST (alone or as a fusion protein: GST:AKR2A) was bound to 100 μL of glutathione agarose beads and incubated with 10 mg of total soluble protein extracts for 3 h at 4°C. The beads were then washed several times with 10 mL of washing buffer (20 mm Tris-HCl, pH 7.5, 150 mm NaCl, 3 mm MgCl2, 5 mm EDTA, 1 mm DTT, 0.5% Triton X-100, 0.1% [v/v] Nonidet P-40, and 1× complete protease inhibitor cocktail [Roche]). The proteins bound to the beads were eluted and used for the two-dimensional analysis.
To study the interaction between the sHsps and AKR2A, in vitro protein pull-down experiments were performed. Briefly, GST alone or GST:sHsps (3 μg) immobilized onto glutathione beads was incubated with His-tagged recombinant proteins as prey in a binding buffer (20 mm Tris-HCl, pH 7.5, 150 mm NaCl, 3 mm MgCl2, 1 mm DTT, and 0.1% Triton X-100) at 4°C for 3 h. Beads were washed three times with the binding buffer. Subsequently, SDS-PAGE sample buffer was added to the beads, boiled, and analyzed by western blotting using an anti-His antibody.
Chloroplast Isolation and in Vitro Chloroplast-Binding Experiments
Intact chloroplasts were isolated from protoplasts using standard procedures on a Percoll gradient as described previously (Li and Chen, 1996; Tu and Li, 2000). Purified His-tagged recombinant proteins (2 μg) such as His:GFP, His:AKR2, and His:Hsp17.8 were incubated with intact chloroplasts (equivalent to 20 μg of chlorophyll) in 1 mL of binding buffer (50 mm HEPES-KOH, pH 7.6, 3 mm MgCl2, 330 mm sorbitol, 100 mm NaCl, 1 mm DTT, and 1× complete protease inhibitor cocktail [Roche]) on ice for 30 min. Subsequently, chloroplasts were pelleted by centrifugation (1,500g) for 5 min at 4°C. The pellet was washed with binding buffer, boiled with SDS-PAGE sample buffer, and subjected to western-blot analysis using an anti-His antibody.
BN-PAGE Analysis
The BN-PAGE analysis was performed as described previously (Kikuchi et al., 2006). Protoplasts were resuspended with the solubilization buffer (50 mm Bis-Tris-HCl, pH 7.0, 0.5 m aminocarproic acid, 10% [w/v] glycerol, 0.5% n-dodecyl b-d-maltoside, and 1× complete protease inhibitor cocktail [Roche]), incubated on ice for 10 min, and centrifuged at 20,000g. Insoluble materials were removed by ultracentrifugation at 100,000g for 10 min, and the supernatant was supplemented with Coomassie Brilliant Blue G-250 (SERVA). The samples were loaded on a 4% to 16% gradient gel (Native PAGE Novex 4-16% Bis-Tris Gel; Invitrogen). For BN-PAGE, the cathode tank buffer was 50 mm Tricine/15 mm Bis-Tris, pH 7.0, and 0.02% Coomassie Brilliant Blue G-250 and the anode tank buffer was 50 mm Bis-Tris, pH 7.0. Western-blot analysis was performed according to the manufacturer’s instructions using an anti-HA antibody.
Two-Dimensional Electrophoresis and MALDI-TOF Analyses
Isoelectric focusing and two-dimensional SDS-PAGE were performed as described previously (Lee et al., 2004a). Protein samples were applied to 23-cm immobilized pH gradient strips (nonlinear, pH gradient 4–10; Genomine). For isoelectric focusing, the voltage was linearly increased from 150 to 3,500 V during 3 h for sample entry followed by constant 3,500 V, with focusing complete after 96 kVh. Prior to the second dimension, strips were incubated for 10 min in equilibration buffer (50 mm Tris-Cl, pH 6.8, containing 6 m urea, 2% SDS, and 30% glycerol), first with 1% DTT and second with 2.5% iodoacetamide. This strips were inserted on 10% to 16% gradient SDS-PAGE gels (20 × 24 cm) using the Hoefer DALT 2-D system (Amersham Biosciences). The two-dimensional gels were silver stained as described previously (Oakley et al., 1980), but the fixing and sensitization step with glutaraldehyde was omitted.
The GST:AKR2A-specific protein spots were subjected to MALDI-TOF analysis for identification. Protein analysis was performed using an Ettan MALDI-TOF apparatus (Amersham Biosciences), and peptides were evaporated with an N2 laser at 337 nm using a delayed extraction approach. These peptides were accelerated with a 20-kV injection pulse for TOF analysis. Each spectrum was the cumulative average of 300 laser shots. The search software ProFound, developed by The Rockefeller University (http://prowl.rockefeller.edu/prowl-cgi/profound.exe), was used for protein identification by peptide mass fingerprinting, and spectra were calibrated with autodigested trypsin ion peak mass-to-charge ratio (842.510, 2,211.1046) as internal standards.
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: AKR2A (At4g35450), OEP7 (At3g52420), HSP15.7 (At5g37670), HSP17.4 (At3g46230), HSP17.6A (At1g59860), HSP17.6B (At2g29500), HSP17.6C (At1g53540), HSP17.8 (At1g07400), HSP18.1 (At5g59720), HSP23.6 (At4g25200), and HSP25.3 (At4g27670).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Phylogenetic tree of Arabidopsis sHsps.
Supplemental Figure S2. Amino acid sequence alignment of six CI cytosolic sHsps of Arabidopsis.
Supplemental Figure S3. Sequence alignment of sHSP-CI amiRNA.
Supplemental Table S1. List of proteins identified in pull down.
Supplemental Table S2. The sequences of primers used in this study.
References
- Agne B, Kessler F. (2009) Protein transport in organelles: the Toc complex way of preprotein import. FEBS J 276: 1156–1165 [DOI] [PubMed] [Google Scholar]
- Aoyama T, Chua NH. (1997) A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J 11: 605–612 [DOI] [PubMed] [Google Scholar]
- Bae W, Lee YJ, Kim DH, Lee J, Kim S, Sohn EJ, Hwang I. (2008) AKR2A-mediated import of chloroplast outer membrane proteins is essential for chloroplast biogenesis. Nat Cell Biol 10: 220–227 [DOI] [PubMed] [Google Scholar]
- Balogi Z, Cheregi O, Giese KC, Juhász K, Vierling E, Vass I, Vígh L, Horváth I. (2008) A mutant small heat shock protein with increased thylakoid association provides an elevated resistance against UV-B damage in Synechocystis 6803. J Biol Chem 283: 22983–22991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basha E, Friedrich KL, Vierling E. (2006) The N-terminal arm of small heat shock proteins is important for both chaperone activity and substrate specificity. J Biol Chem 281: 39943–39952 [DOI] [PubMed] [Google Scholar]
- Basha E, Jones C, Wysocki V, Vierling E. (2010) Mechanistic differences between two conserved classes of small heat shock proteins found in the plant cytosol. J Biol Chem 285: 11489–11497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basha E, Lee GJ, Breci LA, Hausrath AC, Buan NR, Giese KC, Vierling E. (2004) The identity of proteins associated with a small heat shock protein during heat stress in vivo indicates that these chaperones protect a wide range of cellular functions. J Biol Chem 279: 7566–7575 [DOI] [PubMed] [Google Scholar]
- Bruce BD. (2000) Chloroplast transit peptides: structure, function and evolution. Trends Cell Biol 10: 440–447 [DOI] [PubMed] [Google Scholar]
- Chowdary TK, Raman B, Ramakrishna T, Rao ChM. (2007) Interaction of mammalian Hsp22 with lipid membranes. Biochem J 401: 437–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coucheney F, Gal L, Beney L, Lherminier J, Gervais P, Guzzo J. (2005) A small HSP, Lo18, interacts with the cell membrane and modulates lipid physical state under heat shock conditions in a lactic acid bacterium. Biochim Biophys Acta 1720: 92–98 [DOI] [PubMed] [Google Scholar]
- Dafny-Yelin M, Tzfira T, Vainstein A, Adam Z. (2008) Non-redundant functions of sHSP-CIs in acquired thermotolerance and their role in early seed development in Arabidopsis. Plant Mol Biol 67: 363–373 [DOI] [PubMed] [Google Scholar]
- Dhanoa PK, Richardson LG, Smith MD, Gidda SK, Henderson MP, Andrews DW, Mullen RT. (2010) Distinct pathways mediate the sorting of tail-anchored proteins to the plastid outer envelope. PLoS ONE 5: e10098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyles SJ, Gierasch LM. (2010) Nature’s molecular sponges: small heat shock proteins grow into their chaperone roles. Proc Natl Acad Sci USA 107: 2727–2728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslbeck M, Miess A, Stromer T, Walter S, Buchner J. (2005) Disassembling protein aggregates in the yeast cytosol: the cooperation of Hsp26 with Ssa1 and Hsp104. J Biol Chem 280: 23861–23868 [DOI] [PubMed] [Google Scholar]
- Helm KW, Lee GJ, Vierling E. (1997) Expression and native structure of cytosolic class II small heat-shock proteins. Plant Physiol 114: 1477–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann NR, Theg SM. (2005) Chloroplast outer membrane protein targeting and insertion. Trends Plant Sci 10: 450–457 [DOI] [PubMed] [Google Scholar]
- Jin JB, Kim YA, Kim SJ, Lee SH, Kim DH, Cheong GW, Hwang I. (2001) A new dynamin-like protein, ADL6, is involved in trafficking from the trans-Golgi network to the central vacuole in Arabidopsis. Plant Cell 13: 1511–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi S, Hirohashi T, Nakai M. (2006) Characterization of the preprotein translocon at the outer envelope membrane of chloroplasts by blue native PAGE. Plant Cell Physiol 47: 363–371 [DOI] [PubMed] [Google Scholar]
- Kim DH, Eu YJ, Yoo CM, Kim YW, Pih KT, Jin JB, Kim SJ, Stenmark H, Hwang I. (2001) Trafficking of phosphatidylinositol 3-phosphate from the trans-Golgi network to the lumen of the central vacuole in plant cells. Plant Cell 13: 287–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Somers DE. (2010) Rapid assessment of gene function in the circadian clock using artificial microRNA in Arabidopsis mesophyll protoplasts. Plant Physiol 154: 611–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner M, Winkelhaus S, Thierfelder JM, Nover L. (2000) Transient expression and heat-stress-induced co-aggregation of endogenous and heterologous small heat-stress proteins in tobacco protoplasts. Plant J 24: 397–411 [DOI] [PubMed] [Google Scholar]
- Lee DH, Goldberg AL. (1998) Proteasome inhibitors: valuable new tools for cell biologists. Trends Cell Biol 8: 397–403 [DOI] [PubMed] [Google Scholar]
- Lee GJ, Pokala N, Vierling E. (1995) Structure and in vitro molecular chaperone activity of cytosolic small heat shock proteins from pea. J Biol Chem 270: 10432–10438 [DOI] [PubMed] [Google Scholar]
- Lee GJ, Roseman AM, Saibil HR, Vierling E. (1997) A small heat shock protein stably binds heat-denatured model substrates and can maintain a substrate in a folding-competent state. EMBO J 16: 659–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee GJ, Vierling E. (2000) A small heat shock protein cooperates with heat shock protein 70 systems to reactivate a heat-denatured protein. Plant Physiol 122: 189–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HW, Choe YH, Kim DK, Jung SY, Lee NG. (2004a) Proteomic analysis of a ferric uptake regulator mutant of Helicobacter pylori: regulation of Helicobacter pylori gene expression by ferric uptake regulator and iron. Proteomics 4: 2014–2027 [DOI] [PubMed] [Google Scholar]
- Lee S, Lee DW, Lee Y, Mayer U, Stierhof YD, Lee S, Jürgens G, Hwang I. (2009) Heat shock protein cognate 70-4 and an E3 ubiquitin ligase, CHIP, mediate plastid-destined precursor degradation through the ubiquitin-26S proteasome system in Arabidopsis. Plant Cell 21: 3984–4001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YJ, Kim DH, Kim YW, Hwang I. (2001) Identification of a signal that distinguishes between the chloroplast outer envelope membrane and the endomembrane system in vivo. Plant Cell 13: 2175–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YJ, Sohn EJ, Lee KH, Lee DW, Hwang I. (2004b) The transmembrane domain of AtToc64 and its C-terminal lysine-rich flanking region are targeting signals to the chloroplast outer envelope membrane [correction]. Mol Cells 17: 281–291 [PubMed] [Google Scholar]
- Li HM, Chen LJ. (1996) Protein targeting and integration signal for the chloroplastic outer envelope membrane. Plant Cell 8: 2117–2126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HM, Moore T, Keegstra K. (1991) Targeting of proteins to the outer envelope membrane uses a different pathway than transport into chloroplasts. Plant Cell 3: 709–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberek K, Lewandowska A, Zietkiewicz S. (2008) Chaperones in control of protein disaggregation. EMBO J 27: 328–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosavi LK, Cammett TJ, Desrosiers DC, Peng ZY. (2004) The ankyrin repeat as molecular architecture for protein recognition. Protein Sci 13: 1435–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamoto H, Vígh L. (2007) The small heat shock proteins and their clients. Cell Mol Life Sci 64: 294–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley BR, Kirsch DR, Morris NR. (1980) A simplified ultrasensitive silver stain for detecting proteins in polyacrylamide gels. Anal Biochem 105: 361–363 [DOI] [PubMed] [Google Scholar]
- Ossowski S, Schwab R, Weigel D. (2008) Gene silencing in plants using artificial microRNAs and other small RNAs. Plant J 53: 674–690 [DOI] [PubMed] [Google Scholar]
- Qbadou S, Becker T, Mirus O, Tews I, Soll J, Schleiff E. (2006) The molecular chaperone Hsp90 delivers precursor proteins to the chloroplast import receptor Toc64. EMBO J 25: 1836–1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rial DV, Arakaki AK, Ceccarelli EA. (2000) Interaction of the targeting sequence of chloroplast precursors with Hsp70 molecular chaperones. Eur J Biochem 267: 6239–6248 [DOI] [PubMed] [Google Scholar]
- Rogers S, Wells R, Rechsteiner M. (1986) Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science 234: 364–368 [DOI] [PubMed] [Google Scholar]
- Sato Y, Yokoya S. (2008) Enhanced tolerance to drought stress in transgenic rice plants overexpressing a small heat-shock protein, sHSP17.7. Plant Cell Rep 27: 329–334 [DOI] [PubMed] [Google Scholar]
- Scharf KD, Siddique M, Vierling E. (2001) The expanding family of Arabidopsis thaliana small heat stress proteins and a new family of proteins containing alpha-crystallin domains (Acd proteins). Cell Stress Chaperones 6: 225–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D. (2006) Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18: 1121–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma SK, Christen P, Goloubinoff P. (2009) Disaggregating chaperones: an unfolding story. Curr Protein Pept Sci 10: 432–446 [DOI] [PubMed] [Google Scholar]
- Shen G, Kuppu S, Venkataramani S, Wang J, Yan J, Qiu X, Zhang H. (2010) ANKYRIN REPEAT-CONTAINING PROTEIN 2A is an essential molecular chaperone for peroxisomal membrane-bound ASCORBATE PEROXIDASE3 in Arabidopsis. Plant Cell 22: 811–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddique M, Gernhard S, von Koskull-Döring P, Vierling E, Scharf KD. (2008) The plant sHSP superfamily: five new members in Arabidopsis thaliana with unexpected properties. Cell Stress Chaperones 13: 183–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stengel F, Baldwin AJ, Painter AJ, Jaya N, Basha E, Kay LE, Vierling E, Robinson CV, Benesch JL. (2010) Quaternary dynamics and plasticity underlie small heat shock protein chaperone function. Proc Natl Acad Sci USA 107: 2007–2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su PH, Li HM. (2010) Stromal Hsp70 is important for protein translocation into pea and Arabidopsis chloroplasts. Plant Cell 22: 1516–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Bernard C, van de Cotte B, Van Montagu M, Verbruggen N. (2001) At-HSP17.6A, encoding a small heat-shock protein in Arabidopsis, can enhance osmotolerance upon overexpression. Plant J 27: 407–415 [DOI] [PubMed] [Google Scholar]
- Sun W, Van Montagu M, Verbruggen N. (2002) Small heat shock proteins and stress tolerance in plants. Biochim Biophys Acta 1577: 1–9 [DOI] [PubMed] [Google Scholar]
- Sun Y, MacRae TH. (2005) Small heat shock proteins: molecular structure and chaperone function. Cell Mol Life Sci 62: 2460–2476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu SL, Li HM. (2000) Insertion of OEP14 into the outer envelope membrane is mediated by proteinaceous components of chloroplasts. Plant Cell 12: 1951–1960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Montfort RL, Basha E, Friedrich KL, Slingsby C, Vierling E. (2001) Crystal structure and assembly of a eukaryotic small heat shock protein. Nat Struct Biol 8: 1025–1030 [DOI] [PubMed] [Google Scholar]
- Volkov RA, Panchuk II, Schöffl F. (2005) Small heat shock proteins are differentially regulated during pollen development and following heat stress in tobacco. Plant Mol Biol 57: 487–502 [DOI] [PubMed] [Google Scholar]
- Wehmeyer N, Hernandez LD, Finkelstein RR, Vierling E. (1996) Synthesis of small heat-shock proteins is part of the developmental program of late seed maturation. Plant Physiol 112: 747–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XP, Glaser E. (2002) Interaction of plant mitochondrial and chloroplast signal peptides with the Hsp70 molecular chaperone. Trends Plant Sci 7: 14–21 [DOI] [PubMed] [Google Scholar]