Abstract
Several angiosperm plant genomes, including Arabidopsis (Arabidopsis thaliana), rice (Oryza sativa), poplar (Populus trichocarpa), and grapevine (Vitis vinifera), have been sequenced, but the lack of reference genomes in gymnosperm phyla reduces our understanding of plant evolution and restricts the potential impacts of genomics research. A gene catalog was developed for the conifer tree Picea glauca (white spruce) through large-scale expressed sequence tag sequencing and full-length cDNA sequencing to facilitate genome characterizations, comparative genomics, and gene mapping. The resource incorporates new and publicly available sequences into 27,720 cDNA clusters, 23,589 of which are represented by full-length insert cDNAs. Expressed sequence tags, mate-pair cDNA clone analysis, and custom sequencing were integrated through an iterative process to improve the accuracy of clustering outcomes. The entire catalog spans 30 Mb of unique transcribed sequence. We estimated that the P. glauca nuclear genome contains up to 32,520 transcribed genes owing to incomplete, partially sequenced, and unsampled transcripts and that its transcriptome could span up to 47 Mb. These estimates are in the same range as the Arabidopsis and rice transcriptomes. Next-generation methods confirmed and enhanced the catalog by providing deeper coverage for rare transcripts, by extending many incomplete clusters, and by augmenting the overall transcriptome coverage to 38 Mb of unique sequence. Genomic sample sequencing at 8.5% of the 19.8-Gb P. glauca genome identified 1,495 clusters representing highly repeated sequences among the cDNA clusters. With a conifer transcriptome in full view, functional and protein domain annotations clearly highlighted the divergences between conifers and angiosperms, likely reflecting their respective evolutionary paths.
Angiosperms are the most diverse and widely studied among the five major phyla of seed plants, the Spermatophyta. They are also the only group of plants with sequenced genomes, which includes the model plant Arabidopsis (Arabidopsis thaliana; Arabidopsis Genome Initiative, 2000) and cultivated plants like rice (Oryza sativa; Goff et al., 2002; Yu et al., 2002), grapevine (Vitis vinifera; Jaillon et al., 2007), and poplar (Populus trichocarpa; Tuskan et al., 2006), among others. The lack of reference genomes in other phyla representing the gymnosperms reduces the potential for insights into plant evolution and for impacts of genomics research.
Conifers are the largest and most ubiquitous group of gymnosperms. They are woody perennials that shape many northern hemisphere ecosystems and support large industries through the provision of wood, fiber, and energy. Sequencing conifer genomes is relevant owing to their taxonomic position, their ecological significance, and their economic importance, but it has been delayed by the very large size of their genomes (i.e. 15–30 Gb; Ohri and Khoshoo, 1986; Murray et al., 2010; Morgante and De Paoli, 2011).
Full knowledge of genes, namely protein-coding sequences, is the most fundamental outcome of genome sequencing projects. Identifying the gene complement of any genome opens up numerous opportunities to enhance our understanding of biological functions and their evolution. Genome sequencing and large-scale cDNA analysis are the major approaches that have been deployed to predict genes and obtain transcribed sequences, respectively. Because conifer genomes are extremely large, investigations of conifers including pines (Pinus spp.), spruces (Picea spp.), Douglas fir (Pseudotsuga menziesii) and Japanese cedar (Cryptomeria japonica) have primarily focused on transcript sequencing and analysis. Clustering of ESTs obtained from capillary (Sanger) sequencing has been used to infer putative unigenes or transcript sets (Allona et al., 1998; Kirst et al., 2003; Pavy et al., 2005; Cairney et al., 2006; Lorenz et al., 2006; Liang et al., 2007; Futamura et al., 2008; Ralph et al., 2008; Li et al., 2009). Recent high-throughput 454 pyrosequencing on GS-FLX instruments has followed a similar strategy in Pinus contorta (Parchman et al., 2010). While EST clustering is a cost-effective way to obtain large data sets of transcribed sequences, it is notorious for producing more sequences than there are transcribed genes (Kawai et al., 2001; Vettore et al., 2003), and the outcomes may be highly variable (MacKay and Dean, 2011). Futamura et al. (2008) used a mate-paired analysis to improve clustering and identify a set of full-length cDNA inserts. Regardless of the experimental approaches and the goals of genome projects, full-length (FL)-cDNA sequencing remains a gold standard for assisting genome-wide characterization and annotation of eukaryotic genes. FL-cDNA analysis was only reported in one conifer, Picea sitchensis, where 6,464 cDNA sequences were finished with the use of custom primers (Ralph et al., 2008).
In this report, we aimed to produce a resource that accurately and more completely describes the transcribed gene catalog of a major conifer tree. To reach this goal, we clustered ESTs from numerous cDNA libraries and used mate-pair analysis of cDNA clones together with full-length sequencing to obtain 23,589 unique full-length insert cDNAs (FLICs) from Picea glauca (white spruce). Next-generation (Next-Gen) sequencing technologies were used to extend our sampling of the transcriptome and obtain deeper coverage of the sequence clusters. Genomic sample sequencing of 1.69 Gb also proved useful to help characterize the set of cDNA clusters. This P. glauca FLIC resource has already contributed to accelerating investigations of gene expression and genetic diversity in P. glauca (Beaulieu et al., 2011; Pelgas et al., 2011) and in Picea marianna (black spruce; Prunier et al., 2011) and will likely enhance the outcomes of conifer genome sequencing initiatives.
RESULTS
A clustering and cDNA sequence analysis process was developed with the aim of correctly grouping multiple transcripts of the same gene. An iterative approach was used to deal with several rounds of production of Sanger EST sequences (Fig. 1A). Analyses featured the full exploitation of cDNA clone information. Each cDNA clone is unique, originates from a single transcript, exists as a frozen stock, and can be used to generate multiple sequence reads. This clone-level analysis allows proper use of directional 5′ and 3′ sequencing, and multiple reads from a single clone can be assembled together to produce a higher quality sequence than those of individual reads. The final outcome of this process being a gene catalog of P. glauca, we refer to it as GCAT-Pgl.
Figure 1.
The GCAT process applied to P. glauca. A, Overview of the EST, clone, and FL-cDNA analyses steps. The clone analysis and the FL-cDNA sequencing enable iterative clustering, ultimately helping to optimize gene models, annotations, and downstream applications. B, Size distribution and sequence completion of the representative cDNA clones for the 27,720 cDNA clusters. For incomplete clones, a minimum length was estimated based on available sequence.
The P. glauca Gene Catalog Comprises 23,589 Unique FLICs
The gene catalog was developed by using EST data from 42 P. glauca cDNA libraries (Supplemental Table S1). A total of 146,616 P. glauca high-quality ESTs were produced (from libraries GQ028–GQ041) and analyzed together with 125,556 previously described ESTs from the Arborea (www.arborea.ulaval.ca; Pavy et al., 2005) and Treenomix (www.treenomix.ca; Ralph et al., 2008) research programs (Supplemental Table S1). In total, these 272,172 ESTs represent 201,405 distinct cDNA clones (Table I).
Table I. Sequence completion of cDNA clones and cluster representative clones.
| Type | No. | Fraction |
| % | ||
| cDNA clones (all) | 201,405 | |
| Complete (FLIC) | 131,148 | 65.1 |
| 5′ and 3′ Reads | 4,270 | 2.1 |
| 5′ Reads only | 35,150 | 17.5 |
| 3′ Reads only | 30,837 | 15.3 |
| Genes (clusters) | 27,720 | |
| Complete (FLIC) | 23,589 | 85.1 |
| 5′ and 3′ Reads | 577 | 2.1 |
| 5′ Reads only | 861 | 3.1 |
| 3′ Reads only | 2,693 | 9.7 |
A critical step in the GCAT process was the production of clone sequences by orienting 5′ and 3′ ESTs and assembling all ESTs from the same cDNA into a higher quality sequence representing that transcript. Clone sequences were further analyzed for sequence composition, base quality, and cloning context in order to eliminate chimeric constructs and only use oriented high-quality sequences during the clustering step. Clone sequences were also used to assess insert size distribution, gene discovery, and sequence coverage in order to direct the strategy of cDNA sequencing.
In order to represent clusters by their most informative sequence, we identified the most 5′ cDNA in each cluster and sought to obtain the FLIC sequence of such clones through directed and internal sequencing steps. These steps were repeated to produce FLIC sequences for most of the cluster representative clones, merging and splitting some clusters at each iteration.
The resulting gene catalog is a grouping of cDNA clone sequences into 27,720 unique cDNA clusters, each estimated to represent a distinct gene (Table I). A reference clone was selected for each cluster for annotation and for assessing transcript completion. In total, 23,589 of the clusters (85%) are represented by a FLIC. The RNA transcripts encompass a wide range of lengths, with several hundred above 2,000 nucleotides (Fig. 1B). The entire catalog currently spans 30.15 Mb of sequence.
The majority of clusters (61%) contain at least two distinct cDNA clones, and the average is 7.3 clones; however, the median is only two clones, 39% of the clusters contain a single clone, and 18% contain 10 or more clones (Table II). The low sequence redundancy indicated by these cluster composition data may be attributed to the wide diversity of the cDNA libraries as well as the efficacy of cDNA normalization (Supplemental Tables S1 and S2).
Table II. Number of clones per cluster and validations by Next-Gen sequencing.
| Sanger Sequencing: Cluster Depth |
Clusters Matched by 454 Sequences (GS-FLX)a |
Clusters Matched by Illumina Sequenesa |
Total Next-Gen Matchesa |
||||||
| Clones per Cluster | No. of Clusters(% of Total) | 0 | 1–5 | >5 | 0 | 1–5 | >5 | 0 | ≥1 |
| 1 | 10,941 (39%) | 2,522 | 2,389 | 6,030 | 2,057 | 802 | 8,082 | 1,584 | 9,357 |
| 2–9 | 11,275 (41%) | 479 | 832 | 9,964 | 405 | 220 | 10,650 | 254 | 11,021 |
| 10–49 | 4,979 (18%) | 40 | 42 | 4,897 | 41 | 22 | 4,916 | 28 | 4,951 |
| 50–99 | 416 (1.5%) | 2 | 1 | 413 | 1 | 2 | 413 | 1 | 415 |
| ≥100 | 109 (0.4%) | 0 | 0 | 109 | 0 | 1 | 108 | 0 | 109 |
| Total (per no. of reads) | 3,043 | 3,264 | 21,413 | 2,504 | 1,047 | 24,169 | 1,867 | 25,853 | |
| Representation reads | 11% | 12% | 77% | 9% | 4% | 87% | 7% | 93% | |
Number of quality sequences matching a given cDNA cluster (classes of 0 reads, 1–5 reads, or >5 reads) for GS-FLX, Illumina, or GS-FLX and Illumina data combined (0 or ≥1 reads). Library details are in Supplemental Table S8.
Sequence Similarity with Plant Genomes Enables Extensive Functional Annotation
The P. glauca cDNA clusters were annotated based on the detection of open reading frames (ORFs) and based on sequence similarity searches (see “Materials and Methods”). Sequence similarity to conifer sequences was found for 22,255 clusters (80%) at the protein level and 23,900 clusters (86%) at the nucleotide level, based on the large transcriptome data sets of P. sitchensis and Pinus taeda (Supplemental Table S3). Overall protein similarity with the angiosperm genomes was significantly lower: 16,453 (59%) for Arabidopsis, 16,996 (61%) for poplar, and 16,093 (58%) for rice. A total of 49 clusters were identified as contaminating chloroplast-encoded sequences based on the P. sitchensis chloroplast genome (Cronn et al., 2008).
Considering protein-level similarity to the above plants, 23,262 (84%) of the GCAT-Pgl clusters had at least one hit and 54% had a putative function assigned (Table III). Significant similarity hits were much more frequent for clusters with long representative sequences, as nearly all of the sequences above 1,000 bp (96% of 13,311) matched a known sequence, and 73% of them were assigned a putative function (Table III). These data show that functional annotation in conifers can be significantly improved by obtaining more complete sequence information for transcribed sequences. The internal sequencing described in this report was effective at fully capturing many long cDNA clones, at generating a high proportion of FLICs, and ultimately at improving annotation. The longest 100 cDNA clones were all estimated to be above 2,800 nucleotides, and 75% of these were completely sequenced (Supplemental Table S4). The presence of a gap in 25% of these long clones suggested that the FLICs are still somewhat biased toward shorter transcripts. It was also observed that the most sampled sequences had high levels of functional annotation. The 100 clusters with the deepest coverage (all more than 102 clones, all with several FLICs) gave strong matches with plant sequences of known function, predominantly encoding basic housekeeping functions such as protein synthesis and turnover or photosynthesis (Supplemental Table S4), with only 10% that could not be assigned a putative function.
Table III. Sequence similarity and functional annotations according to cluster length based on matches to Arabidopsis, poplar, or rice annotated genome sequences.
| Type | Length Classes |
||||
| cDNA clusters (bp) | All clusters | <501 | 501–1,000 | 1,001–1,999 | >2,000 |
| No. of clusters | 27,720 | 2,429 | 1,1903 | 1,1903 | 1,408 |
| Total sequence (unique; Mb) | 30.15 | 0.92 | 9.27 | 16.7 | 3.26 |
| Similar to a known plant sequence | 84% | 52% | 78% | 95% | 98% |
| Putative function assigned | 54% | 16% | 41% | 71% | 83% |
| Transcribed locus, unknown or predicted proteina | 30% | 36% | 37% | 23% | 15% |
| Unassigned (no significant hit) | 16% | 48% | 22% | 5% | 2% |
The only significant hits had no assigned function (unknown proteins, predicted proteins, transcribed loci [undefined], unknown mRNA, and transcribed loci weakly or moderately similar to a known or predicted protein).
The Distribution of Protein Domains Points to Divergent Evolution with Angiosperms
Similarity to known protein domains was searched using Pfam-A models (Finn et al., 2010) in the 26,217 sequences predicted to contain an ORF (Table IV). The protein families and clans were nearly identical in number and largely overlapping between spruce, Arabidopsis, and rice (Table V) and poplar and grapevine (Supplemental Table S6); however, both the number and the proportion of spruce sequences (59%) matching Pfam domains were lower than observed in Arabidopsis (78%) and rice (69%). The identification of fewer members per family on average may result from incomplete cDNA sequences, smaller gene families, rare transcripts not being sampled, or genomic DNA contamination in the EST data set.
Table IV. RNA transcript sequence completion, CDS status, and transcriptome coverage.
| Classa | CDS Coverage | Genes | Mb | FLIC cDNAs | FLIC Average Length | Estimated Missing Mbb |
| % | ||||||
| 1 | Complete confirmed (protein) | 5,837 | 7.97 | 96 | 1,365 | 0 |
| 2 | Complete predicted (protein) | 3,941 | 5.68 | 90 | 1,419 | 0 |
| 3 | Complete predicted (ORF) | 1,830 | 1.49 | 94 | 812 | 0 |
| 4 | Incomplete (protein) | 7,949 | 8.67 | 78 | 1,130 | 8.07 |
| 5 | Incomplete (ORF) | 6,654 | 5.63 | 77 | 839 | 3.53 |
| 6 | No CDS or ORF detected | 1,509 | 0.71 | 91 | 462 | 0 |
Classes 1 to 3 are similar to a reference protein (Arabidopsis, rice, poplar, grapevine; SwissProt BLASTX E-value = 1e-10): class 1, complete confirmed if the CDS is similar over the entire protein; class 2, complete predicted if the CDS is similar over part of the protein but the transcript extends long enough on each side to cover the protein length; class 3, incomplete if the ORF does not extend enough beyond the similarity region to cover the protein length. Classes 4 to 6 show no similarity to Arabidopsis proteins: class 4, complete predicted if the ORF starts with a Met upstream stop codon in a transcript; class 5, incomplete if the ORF starts at the beginning of a transcript sequence but the first Met is well downstream or missing; class 6, no ORF detected.
Missing transcriptome sequence was estimated based on sequence alignments with the closest reference homolog and statistics of class 1 clusters used as a reference. Class 2 and 3 clusters with a predicted complete CDS were estimated not to lack any sequence in 5′ or 3′ UTRs. For incomplete clones of class 4, reference protein matches were used to estimate 6.29 Mb of coding sequence, 1.2 Mb of 5′ UTR, and 0.58 Mb of 3′ UTR. The estimate for class 5 was based on the average transcript statistics in class 1.
Table V. Pfam-A assignations in P. glauca, Arabidopsis, and rice.
| Parameter | P. glauca | Arabidopsisa | Ricea |
| Genes or cDNA clusters | 27,720b | 27,379 | 28,292 |
| Transcriptome sequence (Mb) | 41 | 40 | 49 |
| Pfam families | 3,498c | 3,594 | 3,440 |
| Pfam clans | 2,309 | 2,417 | 2,312 |
| Genes with Pfam-A, no. | 15,508c | 21,233 | 19,629 |
| Genes with Pfam-A, proportion | 59%b | 78% | 69% |
| Transcript size | 1,365d | 1,326 | 1,507 |
| CDS | 867 | 1,021 | 1,096 |
| 5′ UTR size | 202d | 101 | 121 |
| 3′ UTR size | 322d | 204 | 290 |
The number of genes in Arabidopsis is based on TAIR (version 9) and that in rice is based on RefSeq (version 43).
The analysis only considered the 26,217 sequences containing one or more ORFs.
The assignation to Pfam-A families (version 24.0) was done in two steps: (1) direct assignation: 13,971 gene clusters with direct assignation to 3,162 Pfam-A families; (2) indirect assignation of an additional 1,037 clusters by association with a TAIR homolog added another 186 Pfam-A families (see “Materials and Methods”).
The estimation was conducted using class 1, 2, and 3 cDNA clusters (Table IV), which are biased toward short transcripts.
The occurrence of Pfam domains revealed classes of proteins that are likely to be overrepresented, underrepresented (Fig. 2), or conserved in spruce (Supplemental Table S5). The FAD/NAD(P)-binding Rossmann fold superfamily (CL0063) appeared highly conserved, as it comprised 1,924 spruce sequences representing 114 domains, compared with 1,920 sequences and 115 domains in Arabidopsis and 1,770 sequences and 108 domains in rice. In contrast, 28 protein domains were statistically overrepresented in spruce. Many of them could be linked to metabolic processes or to stress response (Fig. 2A). Most of the overrepresented stress response proteins were related to osmotic stress, such as dehydrins, abscisic acid/water deficit stress-induced protein, late embryogenesis, and the AWPM-19-like family (involved in cold tolerance). The metabolic process proteins were very diverse. They included a few secondary metabolism enzymes (e.g. O-methyltransferases) and a few carbohydrate metabolism enzymes (e.g. glycosyl hydrolases family 16, found in xyloglucan endotransglycosylase).
Figure 2.
Occurrence and classification of protein family domains in P. glauca relative to angiosperms. The total number of P. glauca cDNA clusters containing each of the Pfam-A domains was compared with Arabidopsis, rice, and poplar genes (for entire list, see Supplemental Table S6) by using normalized angiosperm data to account for the number of overall genes in each species. The number of overrepresented (A) and underrepresented (B) protein domains in P. glauca was determined by χ2 testing (P < 0.05, with Bonferroni correction) for Pfam domains found six times or more in at least one of the species compared and with a 50% difference between the species. The Pfam domains that were statistically different in at least two out of three comparisons were classified into major biological groups based upon TAIR annotations of Arabidopsis homologs (Gene Ontology process) and Interpro and Pfam descriptions. DUF, Domains of unknown function.
Protein domains from type I transposable elements (retroelements) were overrepresented, whereas type II transposable elements were underrepresented, which is consistent with recent literature (Morgante and De Paoli, 2011). Specific domains of unknown function also fell into both categories, while many other domains of unknown function were variable or lineage specific among angiosperms.
The underrepresented protein domains (41 in total) included large protein families involved in cellular processes (e.g. F-box and associated FBA1 and FBA3 domains involved in ubiquitination), signaling (e.g. protein kinases), and transcription (Fig. 2B). Underrepresented domains were also part of different classes of cell wall-related proteins (e.g. extensin-like region, glycosyl hydrolases family 28, and plant invertase/pectin methylesterase inhibitor), a few biotic interaction proteins (lectin binding), and the S-locus glycoprotein family (36–198 loci in angiosperms), which appears to be nearly absent from Picea.
The total number of P. glauca sequences containing one of the 39 Pfam domains found in plant transcription factors (TFs) and the representation of most of these domains were approximately half those of Arabidopsis, rice, poplar, and grapevine (Supplemental Table S7). The B3 DNA-binding domain found in the ABI3/VP1 transcription factor was the most underrepresented in spruce, with 15% to 20% of the number of angiosperm sequences. The only overrepresented TF domains in spruce were the histone-like TFs (CBF/NF-Y) and archaeal histones (PF00808) found in plant Hap proteins. In contrast, the relative representation of each TF domain among the total TF domains was largely conserved between spruce and angiosperm plants (Fig. 3).
Figure 3.
Relative frequencies of major Pfam domains found in TF. Frequencies were determined for Pfam-A domains with hits in three or more genes in P. glauca and calculated relative to the total number of genes containing TF Pfam domains within each of the species (P. glauca (Pgl), poplar (Ptr), Arabidopsis (Ath), rice (Osa), and grapevine (Vvi). Stars indicate frequencies that are significantly different from Arabidopsis and rice.
The Size of the Spruce Transcriptome May Be Estimated from Full-Length Inserts and mRNAs
We used the clone completion (Table I) and gene annotation information to assess the mRNA status of P. glauca cDNA clusters (Table IV). Full-length mRNA status was achieved when a representative transcript sequence contained a complete protein coding sequence (CDS) as well as some 5′ untranslated region (UTR) and 3′ UTR sequences. We assessed the CDS status of GCAT gene sequences as follows: first, sequences with BLASTX similarity to a reference protein in Arabidopsis, rice, poplar, grapevine, or SwissProt were classified, with regard to length and similarity of the match, into three classes: complete confirmed (class 1), complete predicted (class 2), and incomplete (class 4). Genes without similarity to a reference protein were classified according to whether an ORF could be predicted or not and the features of the predicted ORF (classes 3, 5, and 6).
Based on these criteria, 11,608 (49.2%) of the FLICs were estimated to contain a complete CDS (in classes 1–3). The average mRNA size was deduced from classes 1 and 2 to be 1,396 nucleotides, with an average CDS of 871 bp (290 predicted amino acids), compared with CDS of 1,021 bp in Arabidopsis and 1,096 in rice (Table V), consistent with the sampling bias toward shorter FLICs and indicating that spruce has longer 3′ and 5′ UTRs.
The GC content of ORFs in class 1 sequences was determined as 46.6%, which is very similar to Arabidopsis (45%; Alexandrov et al., 2006) and significantly lower than maize (Zea mays; 58%; Alexandrov et al., 2009). The 47% GC content of the third codon position was also similar to Arabidopsis, whereas maize (and other monocots) has much higher and more variable GC contents in the third position (Alexandrov et al., 2009).
The P. glauca gene catalog contains 27,720 cDNA clusters, representing 30.15 Mb of unique sequence. The majority of clusters (23,589) are represented by a FLIC, but 12,736 (54%) of them lacked a complete CDS and 1,509 of them completely lacked any ORF. We used the sequence completion status (Table I) and CDS status (Table IV) to estimate the full size of the P. glauca transcriptome sampled to date (Table IV). The transcriptome represented in the P. glauca gene catalog was predicted to span 40.75 Mb (i.e. 0.20% of the P. glauca genome reported to be 19.8 Gb by Ohri and Khoshoo [1986]). We thus estimate that 11.60 Mb as missing (Table IV).
Next-Gen Sequencing Enhances and Extends the Gene Catalog
The Sanger-based gene catalog was validated and augmented by cDNA analysis with 454 sequencers (GS-FLX; Roche) and by RNA sequencing (RNA-Seq) with GA-II (Illumina) sequencers (see “Materials and Methods”). For libraries and samples sequenced, see Supplemental Table S8. A total of 7.42 m GS-FLX and 59.5 m GA (RNA-Seq) high-quality sequences were obtained and mapped to the cDNA clusters.
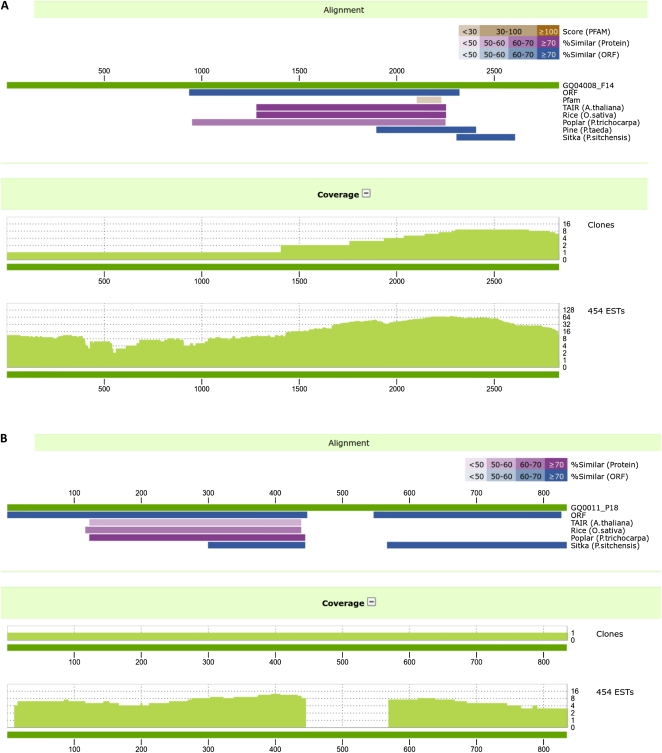
The Next-Gen sequences provided independent validations of the Sanger-derived sequences (Table II). They were obtained in part from separate libraries and tissue samples (Supplemental Table S8). Low coverage of cDNA sequences is a common feature of Sanger-based EST catalogs due to factors such as limited sampling of rare transcripts and partial sequencing. A total of 93% (25,853) of the P. glauca clusters were matched by at least one Next-Gen sequence, with 98% (16,496) for multiple-clone clusters and 86% (9,357) for the singletons (data not shown). Figure 4 illustrates how GS-FLX sequences directly matching cDNA clones were useful to confirm sequence structure in areas of low coverage, such as 5′ ends of transcripts (Fig. 4A) or to help delineate unspliced introns (Fig. 4B). The Next-Gen sequences also extended many incomplete clones and clusters. A total of 2.9 Mb was added through extensions at the 5′ and 3′ ends and by filling gaps in clusters with nonoverlapping 5′ and 3′ reads (Table VI).
Figure 4.
Coverage and validation of EST clusters with GS-FLX (454) ESTs. A, Illustration of the validation of cDNA clusters with low clone coverage in the 5′ proximal region (GQ04008_F14; CC3HC4-type RING finger protein sequence). B, Confirmation of an unspliced intron in a unique cDNA clone that was suggested by sequence similarity analyses (GQ0011_p18; RING 1A sequence, containing a putative unspliced intron).
Table VI. Contribution of Next-Gen sequencing to extending the P. glauca gene catalog.
| Parameter | No. | Average Length | Total |
| bp | Mb | ||
| Extension of GCAT clusters | |||
| 3′ Clone extensions | 9,012 | 176 | 1.58 |
| 5′ Clone extensions | 7,844 | 151 | 1.19 |
| Gaps filled | 69 | 184 | 0.012 |
| Unattached sequences | 20,945 | 243 | 5.0 |
The discovery of genes not represented in the Sanger-based gene catalog was assessed in the Next-Gen sequencing data using several approaches. First, we determined the proportion of Next-Gen sequences that mapped to existing clusters based on direct sequence overlap. Depending on the library, 87% to 92% of the GS-FLX reads mapped to the GCAT-Pgl clusters, except for library GQ043 to GQ045, which gave 37% representation, indicating likely contamination; we were able to identify the contaminating sequences as predominantly derived from genomic P. glauca DNA (Supplemental Table S3). The GA-II reads were represented in 80% of the GCAT-Pgl clusters. The lower frequency of matches observed with GA reads is consistent with the lack of 3′ bias in RNA-Seq compared with available P. glauca ESTs and the shorter sequence lengths. Consistent with this expectation, the GA-II match rate increased to 87% after the Sanger-based cDNA clusters were extended with the directly matching GS-FLX reads.
Second, the orphan GS-FLX sequences (not mapped to a cDNA cluster) were assembled and produced 3,129 contigs longer than 400 bases and containing two or more reads. Approximately 35% (1,146) of these orphan contigs produced a strong match to a Pinaceae putative unique transcript from PlantGDB (Table VII). The number of similar sequences (more than 85% identity) ranged from 88 to 567 depending on the species. Third, the contigs and high-quality singletons obtained from orphan GS-FLX data that were confirmed by RNA-Seq were also checked for similarity to plant proteins (TBLASTX). A total of 20,549 contigs ranging from 64 to 586 bp and spanning a total of 5.0 Mb were thus identified as putative transcriptome sequence (data not shown); however, the majority of the best matches were against proteins also matched by Sanger-based cDNA clusters, indicating that the set of orphan sequences likely represents missing sequence for many of the same genes as well as a smaller fraction of unsampled genes.
Table VII. Similarity of orphan Next-Gen sequences with conifer transcripts in public databases.
A total of 3,129 P. glauca contigs (>400 bp, >2 reads) that were obtained by cDNA assembly from the orphan GS-FLX reads (that could not be mapped to any GCAT-Pgl cluster; see “Materials and Methods”) were compared with other Pinaceae transcriptomes, producing 1,146 unique hits.
| Parameter |
Picea |
Pinus |
|||||
| P. abies | P. engelmannii × P. glauca | P. glauca | P. sitchensis | P. contorta | P. pinaster | P. taeda | |
| PUTs in PlantGDBa | 5,184 | 13,880 | 29,178 | 12,901 | 13,570 | 15,648 | 72,829 |
| no. of hits | |||||||
| Identity | |||||||
| >95% | 68 | 188 | 67 | 490 | 15 | 22 | 67 |
| >90% | 82 | 206 | 88 | 541 | 126 | 110 | 368 |
| >85% | 88 | 216 | 92 | 567 | 194 | 140 | 547 |
| >70% (total) | 91 | 223 | 96 | 593 | 593 | 214 | 578 |
| Average | |||||||
| Identity (%) | 96 | 98 | 96 | 97 | 90 | 91 | 91 |
| Length (bp) | 307 | 446 | 424 | 470 | 539 | 376 | 478 |
| Score | 504 | 768 | 706 | 799 | 685 | 503 | 625 |
Putative unique transcripts (PUTs) from the Plant Genome Database project of the versions described in “Materials and Methods” (http://www.plantgdb.org/prj/ESTCluster/progress.php).
Genomic Sample Sequencing Identifies Repeated Sequences in the cDNA Data Set
Sample sequencing of the P. glauca genome based on 4.91 million GS-FLX reads totaling 1.69 Gb and representing genome coverage of 8.5% was used to further characterize the gene catalog and specifically to detect putative repetitive sequences. Retroelements and noncharacterized repetitive elements were estimated to make up 75% of the 18-Gb Picea abies genome (Morgante and De Paoli, 2011). Over 20,000 complete copies of the gymny retroelement were estimated to be in the 22-Gb P. taeda genome (Morse et al., 2009). Therefore, identifying repetitive sequences is useful to assess genomic contamination. A total of 1,490 cDNA cluster sequences produced strong alignments with numerous genomics sequences and were represented by only one or two ESTs that lacked similarity to plant proteins (Table VIII). They included 1,195 clusters recognized by 40 or more genomic sequence reads (Table VIII) and 223 clusters with more than 1,000 significant alignments to genomic sequence data (data not shown). The majority of these sequences contained one or several short ORFs (68% in class 5) or had no ORF (15% in class 6; Table IV). Only 7% had an ORF containing a Pfam domain, and most of the matches were weakly similar to retroelement proteins such as reverse transcriptases, RNase H, and integrases. Despite these observations, a majority of the sequences were similar to putatively transcribed sequences from at least one other conifer tree, such as P. taeda or P. sitchensis; therefore, some of them may represent bona fide transcripts from retroelements or from unknown high-copy genes.
Table VIII. Identification of putative repetitive genomic sequences in the cDNA data set.
| Type of Sequence | No. of cDNA Clusters | Sample Sequencinga | Poisson Probabilityb | Genomic Copy No. Estimatec | Occurrence in the cDNA Data Set | Similarity to Arabidopsis Proteins |
| Highly repetitive | 1,195 | 40+ | 4.84 e-68 | 400+ | One or two ESTs | None |
| Moderately repetitive | 295 | 30+ | 9.38 e-48 | 300–399 | Only in GQ043–GQ045d | None |
Number of sequences that matched the cluster sequence (98% or more).
The Poisson probabilities are for the number of matches observed (30 or 40) if the sequence is a single-copy gene. The probability of a single match is 0.333, assuming random sampling and gene distribution and given a 0.085× genome coverage, the average length of the clusters (1,348), and the average length of the genomic sequences (344 bp) but without accounting for introns.
Based on sample sequencing data assuming a random sampling and 0.085× coverage of the P. glauca genome.
These libraries were determined to contain a large fraction of contaminating genomic DNA.
DISCUSSION
Development of a FL-cDNA Transcriptome Resource for Conifer Genomics
The development of a P. glauca gene catalog creates a resource that will enhance our understanding of the conifer transcriptome and facilitate other genomic investigations, including genome sequencing and comparative genomics. Large-scale FL-cDNA sequencing represents a proven approach to most effectively support these objectives. Previous cDNA analyses in conifers used large-scale clustering of end reads (5′, 3′, or both) to infer putative unigenes or transcript sets, primarily from P. taeda (Allona et al., 1998; Kirst et al., 2003; Cairney et al., 2006; Lorenz et al., 2006; Liang et al., 2007), Pinus radiata (Li et al., 2009), Picea species (Ralph et al., 2008), P. glauca (Pavy et al., 2005), and Cryptomeria japonica (Futamura et al., 2008). The only exception to this approach was the full-length analysis of 6,464 P. sitchensis cDNA clones (Ralph et al., 2008). Futamura et al. (2008) used a mate-paired analysis to improve clustering and identify a set of full-length cDNA inserts. Mate-pair analysis is routinely used in FL-cDNA sequencing; for example, it contributed in the recent characterization of the maize genome (Soderlund et al., 2009). Here, the P. glauca clone analyses integrated mate pairs (nonoverlapping 5′ and 3′ reads) from the same cDNA insert and internal sequencing in an iterative process to improve the accuracy of clustering outcomes. The clone analysis helped to verify error models in our data set (data not shown), and full-length analysis helped to verify sequence orientation and to improve sequence annotations. Each cluster was represented by a single cDNA insert rather than a consensus sequence, with the aim of minimizing potential informatics artifacts and of reducing ambiguous clustering or underclustering.
The advantages associated with FL-cDNA have been abundantly illustrated for aiding genome assembly and annotation (Haas et al., 2002) and for characterizing transcriptomes (Alexandrov et al., 2006). Numerous EST assembly methods and integrated platforms have been developed and optimized (Lee et al., 2007; Forment et al., 2008). However, by their nature, EST assemblies typically overestimate the number of genes in transcriptome samples. The ESTs from the same locus may fail to assemble owing to several factors, including alternate splicing, multiple polyadenylation sites, sequence polymorphism, and sequencing errors. Levels of redundancy after EST assembly have been estimated to range from 20% to 22% even under optimal conditions (Kawai et al., 2001; Vettore et al., 2003). The outcomes of EST clustering also vary depending on the objectives and the analytical approach, especially in uncharacterized genomes. For example, conifers with large EST data sets (P. taeda, P. glauca, and P. sitchensis) produced Unigene builds ranging from 18,709 to 19,825 (National Center for Biotechnology Information; Unigene assemblies accessed in December 2010) but gave assemblies of putative unique transcripts of 31,054 to 72,829 (PlantGDB; Dong et al., 2004). Sequence data from multiple species also inflate the number of unique entities resulting from sequence assemblies, as observed with the 75,758 tentative consensus sequences for spruce (Dana-Farber Cancer Institute Spruce Gene Index, release 4.0; Quackenbush et al., 2001) and the 69,968 pine tentative consensus sequences (DFCI Pine Gene Index, release 8.0).
Until quality drafts of conifer genomes become available, FL-cDNAs provide us with models that most accurately represent the gene-coding portion of the genome. As such, they are essential to facilitate downstream applications of genomics. For example, tracking sequence polymorphisms such as single-nucleotide polymorphisms has become central to understanding the relationship between genotype and phenotype targeted by association genetics or other approaches (Namroud et al., 2008; Pavy et al., 2008; Eckert et al., 2009; Beaulieu et al., 2011; Neale and Kremer, 2011). Without adequate reference sequences, allelic and paralogous variants may be confounded and lead to erroneous genomic inferences and representations (Ritland et al., 2011), which is a particularly acute problem in undomesticated and highly heterozygous organisms such as conifer trees. Applications such as RNA-Seq and short-read clustering will also be facilitated by the availability of accurate cDNA models to be used in reference assembly methods.
Next-Gen sequences from two types of technologies (GS-FLX and GA-II) were used to confirm and annotate Sanger clustering results, to obtain deeper coverage, and to extend the transcriptome sampling. Recent studies involving uncharacterized genomes such as P. contorta (Parchman et al., 2010) and European oaks (Quercus petraea and Quercus robur; Ueno et al., 2010) have shown that GS-FLX is efficient for transcriptome sequencing, but clustering methods are prone to generate exceedingly large numbers of unique sequences upon assembly, particularly when multiple individuals are pooled for sequencing. Even when integrating Sanger and GS-FLX reads in a hybrid analysis, the assembly of oak sequences yielded a minimum of 116,000 unique sequence elements (Ueno et al., 2010). Assembly of GS-FLX data alone generated short contigs averaging 353 bp in Eucalyptus (Novaes et al., 2008; de novo assembly) and 500 bp in pine (Parchman et al., 2010; combined de novo and reference assembly). Our results indicate that sequences under 1,000 bp are suboptimal for annotation of conifer genes. Kumar and Blaxter (2010) tested several transcriptome assembly tools and approaches and reported that the best results were obtained in a stepwise use of two different assembly programs. Similarly, for P. glauca, we mapped Next-Gen sequences to existing cDNA clusters, and only the unmapped sequences were used for new assemblies. With this approach, the assembly of orphan spruce sequences produced over 20,000 unique but relatively short sequences that did not directly overlap with the cDNA clusters.
Genomic sample sequencing of P. glauca (8.5% of the genome) enabled the annotation of 1% of the clones as putative repetitive sequences. These sequences accounted for 5.4% of the clusters, owing to their uniqueness in the data set. These sequences represent potential genomic DNA contaminants, although some of them could represent bona fide transcripts. A recent report of GS-FLX P. contorta transcriptome sequencing identified approximately 6% of the sequences as putative retrotransposons (Parchman et al., 2010). The authors indicated that they were transcribed sequences found at a much higher frequency than in previous reports. Our observations lead us to suggest that they could also represent genomic DNA contaminants, which could be more difficult to account for when using Next-Gen methods, especially when poly(A) tail homopolymers are avoided by using random hexamers for reverse transcription.
A Conifer Transcriptome in Full View
Estimating the number of transcribed genes in a conifer genome is relevant for many downstream investigations and has been an object of interest and debate (for review, see MacKay and Dean, 2011). The GCAT-Pgl catalog is based on the sampling of over 200,000 cDNA clones from 42 diverse cDNA libraries. These sequences were grouped into 27,720 cDNA clusters spanning 30.15 Mb. The removal of 1,545 putative contaminating sequences (genomic and chloroplastic) gives 26,185 clusters spanning 29.35 Mb of the nuclear transcriptome. We estimated that 11.60 Mb was missing from the representative sequences containing a CDS or predicted ORF, owing to incomplete clones or partial sequencing. The transcriptome sampled by Sanger sequencing was thus predicted to be up to 40.75 Mb (Table IV), which is 2% more and 17% less than the gene space in Arabidopsis and rice, respectively (Table V). Taken together, these predictions suggest that several genes and transcripts were most likely not represented in the P. glauca catalog based on Sanger sequences.
The data reported here support different approaches to assess the overall size of the P. glauca transcriptome and thus estimate the number of missing sequences. First, the number of transcribed genes may be estimated based on the number of matching sequences in two independent sets of transcripts. This approach was used for human (Ewing and Green, 2000) and maize (Alexandrov et al., 2009) and may be applied to P. glauca, because the ESTs represented two independent sets of cDNA libraries (GQ001–GQ0041 and WS001–WS0034; Supplemental Table S1). The number of transcribed genes was estimated as 30,214 excluding putative genomic contaminants and 32,720 when they were included (see “Materials and Methods”). These estimates indicate that 83% to 85% of the expressed P. glauca genes have been sampled by Sanger sequencing. The 4,029 to 4,800 transcribed genes not represented could span 5.50 to 6.55 Mb (based on 1,365 bp of class 1 sequence), so the total transcriptome could be as large as 47.30 Mb, or 0.24% of the Picea genome.
Second, 7.78 Mb of new sequence data were discovered through analyses of Next-Gen sequence data that matched with P. glauca cDNA clusters or with known plant protein sequences. When combined, Sanger and Next-Gen sequencing data produced a total of up to 37.93 Mb of unique transcribed sequence. The data are consistent with our estimates of the overall transcriptome size. Future assemblies of transcript sequences integrating these different types of sequence data will help to improve overall transcriptome coverage and bring an even higher resolution to the number of transcribed genes.
Insights into Conifer Genome Structure and Evolution
Our highest estimate of 32,720 transcribed genes in the P. glauca genome is larger than Arabidopsis (Arabidopsis Genome Initiative, 2000) and is nearly identical to maize at 32,450 genes (Schnable et al., 2009). However, it remains at the lower end of the spectrum observed among angiosperms. From an evolutionary perspective, this finding is not surprising, and given the high degree of genome and sequence conservation among the Pinaceae (Ritland et al., 2011), it is likely to apply to other members of the family. Early studies hypothesized that the Pinaceae had many more genes (Kinlaw and Neale, 1997), ranging as high as 225,000 genes (Rabinowicz et al., 2005); however, evidence has not yet been obtained for genome-wide gene amplifications or polyploidization events that could result in such large numbers of transcribed genes (Morgante and De Paoli, 2011). In fact, it appears that the gene content may have been overestimated, for reasons that have been uncovered in recent reports. On the one hand, conifer genomes contain a substantial fraction of single-copy sequences (over 15%; i.e. 3 Gb), including more or less degenerate retroelements that may appear as unique sequences in the genome (Morgante and De Paoli, 2011). On the other hand, the fraction of the genomic sequences that share significant similarity with genes appears exceedingly large (Magbanua et al., 2011). It was reported to be roughly 3% in P. abies (Morgante and De Paoli, 2011), which is close to 600 Mb (our estimate), a size that is larger than most plant genomes sequenced to date. Such a finding appears to be at odds with genome evolution theory as presented by authors such as Lynch (2007) but may be explained in part by the rather large number of pseudogenes; for example, García-Gil (2008) found that pseudogenes were much more numerous than intact coding genes in the case of the phytochrome gene family in Scots pine (Pinus sylvestris). Large numbers of pseudogenes could have contributed to inflating indirect estimations of gene content (Rabinowicz et al., 2005) and to producing complex genomic hybridization patterns (Kinlaw and Neale, 1997).
Angiosperm genome sequences have been used to study the evolution of genome structure and of gene families. For example, the evolution of most TFs was shown to involve two major bursts of gain/expansion, coinciding with the water-to-land transition and the radiation of flowering plants (angiosperms; Lang et al., 2010). Given that their radiation largely predates that of flowering plants and their apparent slow rate of morphological evolution (Gernandt et al., 2011) and their conserved genome macrostructure (Ritland et al., 2011), it stands to reason that gymnosperms may have fewer TFs in their genomes than angiosperms. Conifers were previously shown to have fewer members overall and to lack members in specific subfamilies for TF families such as Knox I and HD-Zip-III and to have more numerous recent gene duplications in closely related subfamilies (Guillet-Claude et al., 2004; Côté et al., 2010) and in R2R3-MYBs (Bedon et al., 2010), among others. Consistent with the model presented by Lang and colleagues (2010), the occurrence and distribution of TF protein domains in the cDNA resource described here strongly suggest that lower overall representation applies to the majority of TF families (Fig. 3). Our data also suggest that the distribution of sequences among TF families is largely conserved between angiosperms and gymnosperms (conifers). These hypotheses may be fully tested through phylogenetic analyses and verified once a quality draft genome is obtained.
CONCLUSION
The cDNA resource described here will enable a host of downstream applications ranging from functional and evolutionary investigations to population and association genetic studies. Approximately 85% of the cDNA clusters are represented by fully sequenced inserts, and we estimated that the gene catalog represents up to 85% of the transcribed genes in the P. glauca genome. This coverage and the FL-cDNA approach have allowed us to begin describing the extent of conservation and divergence in transcribed genes between a representative gymnosperm (conifer) and angiosperms more systematically than was previously possible. More detailed investigations of protein families may help to uncover metabolic and regulatory mechanisms relevant to the unique biology of conifers and gymnosperms. The extent of the sequence data available for P. glauca will also be instrumental to leverage the throughput of Next-Gen sequencers for whole genome, exome analysis or RNA-Seq.
MATERIALS AND METHODS
Sanger and Next-Gen Sequencing of cDNA, RNA, and Genomic DNA
Nucleic Acid Isolation Methods and cDNA Libraries
For cDNA libraries, tissue sampling was as described (Bedon et al., 2007; for tissue descriptions, see Supplemental Table S1). RNA extractions were performed according to Chang et al. (1993). Total RNA was ethanol precipitated and shipped at room temperature to Evrogen for library synthesis. For libraries GQ028 to GQ041, cDNA (catalog no. CS-040) and normalized cDNA (catalog no. CS011-2B) libraries were constructed and directionally cloned into pAL17.3 by Evrogen using the SMART procedure (Zhu et al., 2001). Normalization of amplified cDNA was performed using the duplex-specific nuclease normalization method (Zhulidov et al., 2004). The normalization efficiency was monitored by quantitative PCR based on four reference genes (Supplemental Table S2). For 454 sequencing, four uncloned normalized cDNA libraries for Next-Gen sequencing (catalog no. CS010) were constructed by Evrogen (Supplemental Table S8). For RNA-Seq, total RNA was extracted from secondary xylem and secondary phloem of Picea glauca as described (Bedon et al., 2007). The samples were created by pooling the RNA from 20 3-year-old open-pollinated trees (Pavy et al., 2008a). For genomic sequencing, DNA was isolated using the NucleoSpin 96 Plant II extraction system (Macherey-Nagel) following the manufacturer’s instructions from the same P. glauca genotype as library F7701; quality analysis was as described (Pavy et al., 2008a).
Sequencing Methods for cDNA, RNA, and Genomic DNA
The Sanger cDNA sequences reported here (i.e. not reported previously by Pavy et al. [2005] or Ralph et al. [2008]) were generated at the McGill University and Génome Québec Innovation Centre. Briefly, the cDNA libraries were plated on Luria-Bertani agar medium containing ampicillin and Xgal. White colonies were picked onto 384-well plates containing 2× Luria-Bertani medium and 50% glycerol using a QPix2XT (Genetix) and grown overnight. Plasmid DNA was isolated using an alkaline lysis procedure and PALL filtration plates. Sanger sequencing was produced on 3730xl DNA Analyzers with the Big Dye Terminator sequencing kit (Applied Biosystems). The full-length sequencing of clones was performed as described above using a combination of 5′ (vector), 3′ [vector or poly(A)], and internal (clone-specific) oligonucleotides. The 454 reads were obtained on GS-FLX sequencers (Roche) from different service providers using standard or titanium protocols (Supplemental Table S8). Normalized amplified cDNAs obtained from Evrogen or genomic DNA were fragmented and used as starting material for shotgun library synthesis, library amplification onto beads by emulsion PCR, and sequencing following manufacturer-recommended procedures. For RNA-Seq, total RNA was quantified by Bioanalyzer (Agilent) and then submitted to the standard protocol for RNA-Seq library construction and sequencing on the Genome Analyzer (GAIIx; Illumina).
Analysis of Sanger cDNA Sequencing Data
Production of High-Quality ESTs and cDNA Clone Sequences
Base calling and quality scoring were performed by Phred (Ewing and Green, 1998). Vector identification was performed by cross-match and followed by vector-specific detection of cloning features, adapter sequences, and poly(A) tails, which were clipped to retain six adenines. The resulting insert sequences were submitted to a quality filter requiring 100 consecutive high-complexity bases above a Phred score of 20 and then to an identification step for bacterial and fungal contamination removal.
Clone sequences were produced by orienting ESTs and assembling all ESTs from the same clone using CAP3 (Huang and Madan, 1999), followed by an analysis of the cloning context (gathered from the EST production step) to eliminate chimeras and determine the sequence completion status of the clone (5′ end, 3′ end, 5′–3′ with a gap, and full-length insert).
Iterative Clustering of cDNA Clone Sequences
Clones were grouped into gene clusters using an iterative clustering scheme. First, pairwise sequence comparisons using Nuclear (Gydle) were used to group clones having 97% or more sequence identity over 100 bases of their clusterable regions and no large region of dissimilarity (defined by 150 bases with identity between 85% and 94%). Second, we proceeded with several iterations of clone addition and clone sequence modification as a result of adding sequence reads to existing clones. These modifications resulted in the merging and splitting of clusters based on human and automated curation steps.
Cluster Representative Sequences and Full-Length Sequencing
The selection of the most 5′ cDNA was based on pairwise sequence alignments using Nuclear (Gydle) between clone sequences. Many clusters had a selected clone that was already complete, owing to the length of Sanger reads (800 bases) and the large number of clones with 5′ and 3′ reads (enough to span over 1.5 kb). The clones with incomplete sequence were reracked and submitted to end sequencing for verification and extension, then to internal sequencing steps with the use of custom internal primers if needed. Each round of sequence completion was followed by clone reassemblies and a cluster analysis, merging or splitting clusters, and selecting an alternative clone whenever necessary. In total, 11,421 clones were thus selected and 18,350 additional sequences were produced, resulting in a FLIC for 9,812 of these clones, bringing the number of clusters to 27,720 and the number of clusters with a FLIC to 23,589.
Functional Annotations
Gene sequences (i.e. cluster representative clone sequences) were characterized as follows. Protein-level similarity searches were performed with BLASTX (E-value ≤ 1e-10) against proteins of Arabidopsis (Arabidopsis thaliana; The Arabidopsis Information Resource [TAIR] version 9), rice (Oryza sativa; RefSeq version 43), poplar (Populus trichocarpa; version 43), and grapevine (Vitis vinifera; RefSeq version 43) and with TBLASTX (E-value ≤ 1e-10) against representative transcript sequences of Pinus taeda (UniGene build 11) and Picea sitchensis (UniGene build 15). Nucleotide-level searches were also performed using Nuclear (Gydle) against EST sequences of P. taeda (identity ≥ 85%; 186,000 ESTs) and P. sitchensis (identity ≥ 95%; 293,000 ESTs), the different identity criteria reflecting the average transcript similarity with P. glauca.
For ORF identification in sequences not found to contain a complete CDS and possibly containing frame shifts, gene sequences were translated in all six frames and the resulting ORFs were screened as follows. The first ORF on the plus strand was clipped to the first Met if it appeared in the first 300 bases of the sequence. The ORFs on the plus strand were retained if the longest ORF was larger than n amino acids (n = 40), in which case all other ORFs of length n/2 or longer were also kept. On the minus strand, the longest ORF was retained only if it was longer than all ORFs on the plus strand.
Clusters were assigned protein family domains in two steps. First, ORFs from gene sequences were assigned directly against Pfam-A (version 24.0) using HMMER (version 3.0; domain-specific thresholds). Second, indirect Pfam assignations were made for those genes that gave no direct Pfam match but had a TAIR homolog that both qualified the gene sequence as an incomplete CDS and itself contained a Pfam domain.
Estimation of the Number of Transcribed Genes in the P. glauca Genome
The number of transcribed genes was estimated based on the method developed by Ewing and Green (2000) to estimate the number of human genes. This method was also applied to estimate the number of transcripts in the maize (Zea mays) genome (Alexandrov et al., 2009), such that: m = (n1/N)(n2/N)n = n1n2/N, where m is the total number of genes, n1 is the number of genes in set 1, n2 is the number of genes in set 2, and N is the number of genes shared in both sets. The sequence data sets (set 1, GQ001–GQ041; set 2, WS001–WS034) represent two independent samplings of the P. glauca transcriptome derived from separate biological materials and cDNA libraries (Supplemental Table S1). The estimate of m (total number of transcribed genes) was based on 27,720 clusters, or 26,185 after removal of putative genomic and chloroplastic contaminating sequences (see “Results”). The two sets represent 20,849 (n1) and 17,215 (n2) clusters, respectively, with 11,879 clusters in common (N).
Analysis of Next-Gen cDNA and RNA Sequence Data
Sequence Processing and Mapping onto cDNA Clusters
Next-Gen reads from 454 and RNA-Seq were base called with the manufacturer’s software to produce FASTQ files. Sequence filtering was performed using Gydle software, by detection of cloning features, adapter sequences, and poly(A) tails (clipped to retain six adenines) followed by composition filtering. Sequences containing 50 or more high-complexity bases of high quality (90% scores over Q20) were kept. After identification and removal of bacterial, chloroplast, and ribosomal sequences, the resulting ESTs were aligned to cDNA cluster sequences using Nuclear (Gydle), requiring a minimum of 95% identity over 50 bases for assignment to their best hit.
Clustering and Analysis of Orphan GS-FLX Sequences
The 454 sequences that could not be mapped to a GCAT cluster (371,372 reads; 89 Mb in total) were assembled using the genomic mode of Newbler (version 2.3; Roche); 183,369 reads were assembled into 11,032 contigs and 179,437 singletons. First, the contigs were used as queries to search for transcribed conifer homologs using BLAST (E value < 1e-10) in PlantGDB (i.e. assemblies 157a for P. taeda and Picea engelmannii × P. glauca; 175a for P. glauca, P. sitchensis, Picea abies, and Pinus contorta; and 177a for Pinus pinaster). Second, both the contigs and singletons were used for sequence similarity matches against Arabidopsis, rice, poplar, and grapevine using TBLASTX, as described above.
Analysis of Genomic Sample Sequencing Data
Reads were trimmed using the default settings and then assembled using the genomic mode assembly of Newbler. The assembler eliminated 1.19 million reads based on quality criteria, placed 2.44 million reads into 264,420 contigs, leaving 2.47 million singletons that were longer than 100 bases of quality sequence of high complexity. These contigs and singletons were mapped to the GCAT clusters using Nuclear (Gydle) as described for the cDNA sequences.
The Sanger-produced ESTs are in dbEST (accession nos. CK434215–CK445168, CO472624–CO490610, DV970116–DV998428, EX305736–EX445576, GE471531–GE483395, GO360018–GO372194, GO653068–GO653136, GR222328–GR223664, GR435961–GR435991, GR953522–GR954913, GT051873–GT052669, and GT738463–GT740210); the FLICs are in GenBank (accession nos. BT10331–BT120001); and the Next-Gen sequences are in the National Center for Biotechnology Information short-read archive (accession no. SRA0293921). The cluster compositions (ESTS and clones) are shown in the supplemental tables. The sequence is compiled in a single fasta file and a quality file, and cluster compositions are available for download at www.Aborea.ulaval.ca.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. Descriptions, ESTs, clones, and clusters of cDNA libraries.
Supplemental Table S2. Assessment of normalization efficiency in cDNA libraries.
Supplemental Table S3. Functional annotations of P. glauca cDNA clusters based on sequence similarity with major angiosperm genes and proteins and conifer cDNAs.
Supplemental Table S4. Clusters with the longest sequence (100) and deepest coverage (100).
Supplemental Table S5. Protein domains in Pgl cDNA clusters based on Pfam-A models.
Supplemental Table S6. Distribution of Pfam domains in P. glauca and major angiosperm plants.
Supplemental Table S7. Distribution of TF Pfam domains in P. glauca and major angiosperm plants.
Supplemental Table S8. Next-Gen libraries and sequences.
Acknowledgments
We are grateful for the provision of trace sequence files from the Treenomix project by J. Bohlmann and collaborators (University of British Columbia). We thank V. Jacobi, I. Giguère, S. Caron, F. Gagnon, and S. Blais (Laval University) and C.G. Allen (University of Alberta) for RNA and DNA isolations, and we thank A. Villeneuve and D. Sinnett for Sanger sequencing work (McGill University and Genome Québec Innovation Center). Biological materials were provided by N. Isabel (Canadian Forest Service) and J. Beaulieu (Canadian Wood Fibre Center).
References
- Alexandrov NN, Brover VV, Freidin S, Troukhan ME, Tatarinova TV, Zhang H, Swaller TJ, Lu YP, Bouck J, Flavell RB, et al. (2009) Insights into corn genes derived from large-scale cDNA sequencing. Plant Mol Biol 69: 179–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov NN, Troukhan ME, Brover VV, Tatarinova T, Flavell RB, Feldmann KA. (2006) Features of Arabidopsis genes and genome discovered using full-length cDNAs. Plant Mol Biol 60: 69–85 [DOI] [PubMed] [Google Scholar]
- Allona I, Quinn M, Shoop E, Swope K, St Cyr S, Carlis J, Riedl J, Retzel E, Campbell MM, Sederoff R, et al. (1998) Analysis of xylem formation in pine by cDNA sequencing. Proc Natl Acad Sci USA 95: 9693–9698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Beaulieu J, Doerksen T, Boyle B, Clément S, Deslauriers M, Beauseigle S, Blais S, Poulin P-L, Lenz P, Caron S, et al. (2011) Association genetics of wood physical traits in the conifer white spruce and relationships with gene expression. Genetics 188: 197–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedon F, Bomal C, Caron S, Levasseur C, Boyle B, Mansfield SD, Schmidt A, Gershenzon J, Grima-Pettenati J, Séguin A, et al. (2010) Subgroup 4 R2R3-MYBs in conifer trees: gene family expansion and contribution to the isoprenoid- and flavonoid-oriented responses. J Exp Bot 61: 3847–3864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedon F, Grima-Pettenati J, Mackay J. (2007) Conifer R2R3-MYB transcription factors: sequence analyses and gene expression in wood-forming tissues of white spruce (Picea glauca). BMC Plant Biol 7: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairney J, Zheng L, Cowels A, Hsiao J, Zismann V, Liu J, Ouyang S, Thibaud-Nissen F, Hamilton J, Childs K, et al. (2006) Expressed sequence tags from loblolly pine embryos reveal similarities with angiosperm embryogenesis. Plant Mol Biol 62: 485–501 [DOI] [PubMed] [Google Scholar]
- Chang S, Puryear JD, Cairney J. (1993) A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep 11: 113–116 [Google Scholar]
- Côté CL, Boileau F, Roy V, Ouellet M, Levasseur C, Morency MJ, Cooke JE, Séguin A, MacKay J. (2010) Gene family structure, expression and functional analysis of HD-Zip III genes in angiosperm and gymnosperm forest trees. BMC Plant Biol 10: 273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronn R, Liston A, Parks M, Gernandt DS, Shen R, Mockler T. (2008) Multiplex sequencing of plant chloroplast genomes using Solexa sequencing-by-synthesis technology. Nucleic Acids Res 36: e122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Q, Schlueter SD, Brendel V. (2004) PlantGDB, plant genome database and analysis tools. Nucleic Acids Res (Database issue) 32: D354–D359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert AJ, Pande B, Ersoz ES, Wright NH, Rashbrook VK, Nicolet CM, Neale DB. (2009) High-throughput genotyping and mapping of single nucleotide polymorphisms in loblolly pine (Pinus taeda L). Tree Genet Genomes 5: 225–234 [Google Scholar]
- Ewing B, Green P. (1998) Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res 8: 186–194 [PubMed] [Google Scholar]
- Ewing B, Green P. (2000) Analysis of expressed sequence tags indicates 35,000 human genes. Nat Genet 25: 232–234 [DOI] [PubMed] [Google Scholar]
- Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunesekaran P, Ceric G, Forslund K, et al. (2010) The Pfam protein families database. Nucleic Acids Res (Database issue) 38: D211–D222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forment J, Gilabert F, Robles A, Conejero V, Nuez F, Blanca JM. (2008) EST2uni: an open, parallel tool for automated EST analysis and database creation, with a data mining Web interface and microarray expression data integration. BMC Bioinformatics 9: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futamura N, Totoki Y, Toyoda A, Igasaki T, Nanjo T, Seki M, Sakaki Y, Mari A, Shinozaki K, Shinohara K. (2008) Characterization of expressed sequence tags from a full-length enriched cDNA library of Cryptomeria japonica male strobili. BMC Genomics 9: 383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Gil MR. (2008) Evolutionary aspects of functional and pseudogene members of the phytochrome gene family in Scots pine. J Mol Evol 67: 222–232 [DOI] [PubMed] [Google Scholar]
- Gernandt DS, Willyard A, Syring JV, Liston A. (2011) The conifers (Pinophyta). Plomion C, Bousquet J, Kole C, , Genetics, Genomics and Breeding of Conifer Trees. Edenbridge Science Publishers and CRC Press, New York: (in press) [Google Scholar]
- Goff SA, Ricke D, Lan T-H, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H, et al. (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296: 92–100 [DOI] [PubMed] [Google Scholar]
- Guillet-Claude C, Isabel N, Pelgas B, Bousquet J. (2004) The evolutionary implications of knox-I gene duplications in conifers: correlated evidence from phylogeny, gene mapping, and analysis of functional divergence. Mol Biol Evol 21: 2232–2245 [DOI] [PubMed] [Google Scholar]
- Haas BJ, Volfovsky N, Town CD, Troukhan M, Alexandrov N, Feldmann KA, Flavell RB, White O, Salzberg SL. (2002) Full-length messenger RNA sequences greatly improve genome annotation. Genome Biol 3: research00291–research002912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Madan A. (1999) CAP3: a DNA sequence assembly program. Genome Res 9: 868–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillon O, Aury JM, Noel B, Policriti A, Clepet C, Casagrande A, Choisne N, Aubourg S, Vitulo N, Jubin C, et al. (2007) The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449: 463–467 [DOI] [PubMed] [Google Scholar]
- Kawai J, Shinagawa A, Shibata K, Yoshino M, Itoh M, Ishii Y, Arakawa T, Hara A, Fukunishi Y, Konno H, et al. (2001) Functional annotation of a full-length mouse cDNA collection. Nature 409: 685–690 [DOI] [PubMed] [Google Scholar]
- Kinlaw CS, Neale DB. (1997) Complex gene families in pine genomes. Trends Plant Sci 2: 356–359 [Google Scholar]
- Kirst M, Johnson AF, Baucom C, Ulrich E, Hubbard K, Staggs R, Paule C, Retzel E, Whetten R, Sederoff R. (2003) Apparent homology of expressed genes from wood-forming tissues of loblolly pine (Pinus taeda L.) with Arabidopsis thaliana. Proc Natl Acad Sci USA 100: 7383–7388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Blaxter ML. (2010) Comparing de novo assemblers for 454 transcriptome data. BMC Genomics 11: 571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang D, Weiche B, Timmerhaus G, Richardt S, Riaño-Pachón DM, Corrêa LGG, Reski R, Mueller-Roeber B, Rensing SA. (2010) Genome-wide phylogenetic comparative analysis of plant transcriptional regulation: a timeline of loss, gain, expansion, and correlation with complexity. Genome Biol Evol 2: 488–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B, Hong T, Byun SJ, Woo T, Choi YJ. (2007) ESTpass: a Web-based server for processing and annotating expressed sequence tag (EST) sequences. Nucleic Acids Res (Web Server issue) 35: W159–W162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Wu HX, Dillon SK, Southerton SG. (2009) Generation and analysis of expressed sequence tags from six developing xylem libraries in Pinus radiata D. Don. BMC Genomics 10: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C, Wang G, Liu L, Ji G, Fang L, Liu Y, Carter K, Webb JS, Dean JFD. (2007) ConiferEST: an integrated bioinformatics system for data reprocessing and mining of conifer expressed sequence tags (ESTs). BMC Genomics 8: 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz WW, Sun F, Liang C, Kolychev DS, Wang H, Zhao X, Cordonnier-Pratt M-M, Pratt LH, Dean JFD. (2006) Water stress-responsive genes in loblolly pine (Pinus taeda) roots identified by analyses of expressed sequence tag libraries. Tree Physiol 26: 1–16 [DOI] [PubMed] [Google Scholar]
- Lynch M. (2007) The Origins of Genome Architecture. Sinauer Associates, Sunderland, MA [Google Scholar]
- MacKay JJ, Dean JFD. (2011) Transcriptomics. Plomion C, Bousquet J, Kole C, , Genetics, Genomics and Breeding of Conifer Trees. Edenbridge Science Publishers and CRC Press, New York: (in press) [Google Scholar]
- Magbanua ZV, Ozkan S, Bartlett BD, Chouvarine P, Saski CA, Liston AI, Cronn RC, Nelson CD, Peterson DG. (2011) Adventures in the enormous: a 1.8 million clone BAC library for the 21.7 Gb genome of loblolly pine. PLoS ONE 6: e16214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgante M, De Paoli E. (2011) Toward the conifer genome sequence. Plomion C, Bousquet J, Kole C, , Genetics, Genomics and Breeding of Conifer Trees. Edenbridge Science Publishers and CRC Press, New York: (in press) [Google Scholar]
- Morse AM, Peterson DG, Islam-Faridi MN, Smith KE, Magbanua Z, Garcia SA, Kubisiak TL, Amerson HV, Carlson JE, Nelson CD, et al. (2009) Evolution of genome size and complexity in Pinus. PLoS ONE 4: e4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray BG, Leitch IJ, Bennett MD. (2010) Gymnosperm DNA C-values database, release 4.0, December 2010. http://www.kew.org/cvalues/
- Namroud M-C, Beaulieu J, Juge N, Laroche J, Bousquet J. (2008) Scanning the genome for gene single nucleotide polymorphisms involved in adaptive population differentiation in white spruce. Mol Ecol 17: 3599–3613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale DB, Kremer A. (2011) Forest tree genomics: growing resources and applications. Nat Rev Genet 12: 111–122 [DOI] [PubMed] [Google Scholar]
- Novaes E, Drost DR, Farmerie WG, Pappas GJ, Jr, Grattapaglia D, Sederoff RR, Kirst M. (2008) High-throughput gene and SNP discovery in Eucalyptus grandis, an uncharacterized genome. BMC Genomics 9: 312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohri D, Khoshoo TN. (1986) Genome size in gymnosperms. Plant Syst Evol 153: 119–132 [Google Scholar]
- Parchman TL, Geist KS, Grahnen JA, Benkman CW, Buerkle CA. (2010) Transcriptome sequencing in an ecologically important tree species: assembly, annotation, and marker discovery. BMC Genomics 11: 180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavy N, Boyle B, Nelson C, Paule C, Giguère I, Caron S, Parsons LS, Dallaire N, Bedon F, Bérubé H, et al. (2008a) Identification of conserved core xylem gene sets: conifer cDNA microarray development, transcript profiling and computational analyses. New Phytol 180: 766–786 [DOI] [PubMed] [Google Scholar]
- Pavy N, Paule C, Parsons L, Crow JA, Morency M-J, Cooke J, Johnson JE, Noumen E, Guillet-Claude C, Butterfield Y, et al. (2005) Generation, annotation, analysis and database integration of 16,500 white spruce EST clusters. BMC Genomics 6: 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavy N, Pelgas B, Beauseigle S, Blais S, Gagnon F, Gosselin I, Lamothe M, Isabel N, Bousquet J. (2008) Enhancing genetic mapping of complex genomes through the design of highly-multiplexed SNP arrays: application to the large and unsequenced genomes of white spruce and black spruce. BMC Genomics 9: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelgas B, Bousquet J, Meirmans PG, Ritland K, Isabel N. (2011) QTL mapping in white spruce: gene maps and genomic regions underlying adaptive traits across pedigrees, years and environments. BMC Genomics 12: 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prunier J, Laroche J, Beaulieu J, Bousquet J. (2011) Scanning the genome for gene SNPs related to climate adaptation and estimating selection at the molecular level in boreal black spruce. Mol Ecol 20: 1702–1716 [DOI] [PubMed] [Google Scholar]
- Quackenbush J, Cho J, Lee D, Liang F, Holt I, Karamycheva S, Parvizi B, Pertea G, Sultana R, White J. (2001) The TIGR Gene Indices: analysis of gene transcript sequences in highly sampled eukaryotic species. Nucleic Acids Res 29: 159–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowicz PD, Citek R, Budiman MA, Nunberg A, Bedell JA, Lakey N, O’Shaughnessy AL, Nascimento LU, McCombie WR, Martienssen RA. (2005) Differential methylation of genes and repeats in land plants. Genome Res 15: 1431–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph SG, Chun HJ, Kolosova N, Cooper D, Oddy C, Ritland CE, Kirkpatrick R, Moore R, Barber S, Holt RA, et al. (2008) A conifer genomics resource of 200,000 spruce (Picea spp.) ESTs and 6,464 high-quality, sequence-finished full-length cDNAs for Sitka spruce (Picea sitchensis). BMC Genomics 9: 484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritland K, Krutovsky K, Tsumura Y, Pelgas B, Isabel N, Bousquet J. (2011) Genetic mapping in conifers. Plomion C, Bousquet J, Kole C, , Genetics, Genomics and Breeding of Conifers Trees. Edenbridge Science Publishers and CRC Press, New York: (in press) [Google Scholar]
- Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, Liang C, Zhang J, Fulton L, Graves TA, et al. (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326: 1112–1115 [DOI] [PubMed] [Google Scholar]
- Soderlund C, Descour A, Kudrna D, Bomhoff M, Boyd L, Currie J, Angelova A, Collura K, Wissotski M, Ashley E, et al. (2009) Sequencing, mapping, and analysis of 27,455 maize full-length cDNAs. PLoS Genet 5: e1000740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov A, et al. (2006) The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313: 1596–1604 [DOI] [PubMed] [Google Scholar]
- Ueno S, Le Provost G, Léger V, Klopp C, Noirot C, Frigerio J-M, Salin F, Salse J, Abrouk M, Murat F, et al. (2010) Bioinformatic analysis of ESTs collected by Sanger and pyrosequencing methods for a keystone forest tree species: oak. BMC Genomics 11: 650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vettore AL, da Silva FR, Kemper EL, Souza GM, da Silva AM, Ferro MIT, Henrique-Silva F, Giglioti ÉA, Lemos MVF, Coutinho LL, et al. (2003) Analysis and functional annotation of an expressed sequence tag collection for tropical crop sugarcane. Genome Res 13: 2725–2735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Hu S, Wang J, Wong GK, Li S, Liu B, Deng Y, Dai L, Zhou Y, Zhang X, et al. (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296: 79–92 [DOI] [PubMed] [Google Scholar]
- Zhu YY, Machleder EM, Chenchik A, Li R, Siebert PD. (2001) Reverse transcriptase template switching: a SMART approach for full-length cDNA library construction. Biotechniques 30: 892–897 [DOI] [PubMed] [Google Scholar]
- Zhulidov PA, Bogdanova EA, Shcheglov AS, Vagner LL, Khaspekov GL, Kozhemyako VB, Matz MV, Meleshkevitch E, Moroz LL, Lukyanov SA, et al. (2004) Simple cDNA normalization using kamchatka crab duplex-specific nuclease. Nucleic Acids Res 32: e37. [DOI] [PMC free article] [PubMed] [Google Scholar]