Abstract
Two different, essential Omp85 (Outer membrane protein, 85 kD)-related proteins exist in the outer envelope membrane of Arabidopsis (Arabidopsis thaliana) chloroplasts: Toc75 (Translocon at the outer envelope membrane of chloroplasts, 75 kD), encoded by atTOC75-III; and OEP80 (Outer Envelope Protein, 80 kD), encoded by AtOEP80/atTOC75-V. The atToc75-III protein is closely related to the originally identified pea (Pisum sativum) Toc75 protein, and it forms a preprotein translocation channel during chloroplast import; the AtOEP80 protein is considerably more divergent from pea Toc75, and its role is unknown. As knockout mutations for atTOC75-III and AtOEP80 are embryo lethal, we employed a dexamethasone-inducible RNA interference strategy (using the pOpOff2 vector) to conduct in vivo studies on the roles of these two proteins in older, postembryonic plants. We conducted comparative studies on plants silenced for atToc75-III (atToc75-III↓) or AtOEP80 (AtOEP80↓), as well as additional studies on a stable, atToc75-III missense allele (toc75-III-3/modifier of altered response to gravity1), and our results indicated that both proteins are important for chloroplast biogenesis at postembryonic stages of development. Moreover, both are important for photosynthetic and nonphotosynthetic development, albeit to different degrees: atToc75-III↓ phenotypes were considerably more severe than those of AtOEP80↓. Qualitative similarity between the atToc75-III↓ and AtOEP80↓ phenotypes may be linked to deficiencies in atToc75-III and other TOC proteins in AtOEP80↓ plants. Detailed analysis of atToc75-III↓ plants, by electron microscopy, immunoblotting, quantitative proteomics, and protein import assays, indicated that these plants are defective in relation to the biogenesis of both photosynthetic and nonphotosynthetic plastids and preproteins, confirming the earlier hypothesis that atToc75-III functions promiscuously in different substrate-specific import pathways.
Most proteins in chloroplasts are nucleus encoded, and are imported into the organelle following translation on free cytosolic ribosomes. They are synthesized as preproteins, each one with an N-terminal targeting sequence called a transit peptide that is cleaved off once targeting is completed (Soll and Schleiff, 2004; Kessler and Schnell, 2006; Smith, 2006; Inaba and Schnell, 2008; Jarvis, 2008). Chloroplasts are surrounded by a double membrane system called the envelope, which is impervious to the passive movement of macromolecules. Thus, protein import is mediated by the coordinate action of molecular machines in the outer and inner membranes, respectively called TOC and TIC (Translocon at the Outer/Inner envelope membrane of Chloroplasts). Mechanistically, there are many similarities between chloroplast protein import and transport into the other endosymbiotic organelle, the mitochondrion (Bolender et al., 2008). However, at the molecular level, the import machinery components of the two organelles are largely unrelated (Soll and Schleiff, 2004; Kessler and Schnell, 2006; Smith, 2006; Inaba and Schnell, 2008; Jarvis, 2008).
Biochemical studies using isolated pea (Pisum sativum) chloroplasts identified several components of the TOC/TIC apparatus. The core TOC components are Toc159, Toc34, and Toc75; the numbers indicate protein Mrs (Schnell et al., 1997). Toc159 and Toc34 are GTPases that control preprotein recognition, whereas Toc75 forms the outer envelope channel (Schnell et al., 1994; Tranel et al., 1995). Like the protein-conducting channel of the mitochondrial outer membrane (Tom40; Hill et al., 1998), Toc75 has a β-barrel structure. Studies on pea Toc75 reconstituted into artificial membranes revealed a pore size of approximately 14 to 26 Å (Hinnah et al., 2002), implying that preproteins must be mostly unfolded to pass through the channel; data also indicated that Toc75 can specifically recognize transit peptides without the aid of other TOC proteins, suggesting that its role is not confined to conductance. Toc75 homologs are widely distributed in different plants, including monocots and dicots, and can be found in different plastid types and throughout plant development (Tranel et al., 1995; Summer and Cline, 1999; Dávila-Aponte et al., 2003; Inoue and Potter, 2004).
Topological studies initially suggested that the 16 transmembrane β-strands of psToc75 are distributed along its length (Sveshnikova et al., 2000). However, it is now apparent that the protein has a two-domain structure with all of the β-strands at the C-terminal end (Ertel et al., 2005; Gentle et al., 2005; Bredemeier et al., 2007). In fact, Toc75 is a founder member of the ubiquitous Omp85 (Outer membrane protein, 85 kD) superfamily of proteins that possess this two-domain structure. Superfamily members include Omp85, which mediates outer membrane protein (OMP) biogenesis in bacteria, Sam50/Tob55, which mediates OMP insertion in mitochondria, and TpsB of bacterial two-partner secretion systems (Gentle et al., 2005). In addition to the C-terminal β-barrel channel domain, Omp85 superfamily proteins typically possess multiple N-terminal POTRA (polypeptide transport associated) repeats (Sánchez-Pulido et al., 2003; Gentle et al., 2005). The POTRA domain performs some auxiliary function, proposals for which include chaperone activity, channel gating, and client or partner recruitment (Ertel et al., 2005; Bos et al., 2007; Habib et al., 2007).
The Arabidopsis (Arabidopsis thaliana) genome contains multiple homologs for many TOC subunits; for example, there are two Toc34 homologous genes (atTOC33 and atTOC34) and four Toc159 homologs (atTOC159, atTOC132, atTOC120, and atTOC90; Jarvis, 2008). Studies on the roles of the different Toc isoforms revealed functional specialization: While atToc159 and atToc33 preferentially mediate the import of proteins involved in photosynthesis, atToc132, atToc120, and atToc34 mediate the import of lower abundance, housekeeping proteins (Bauer et al., 2000; Kubis et al., 2003, 2004; Ivanova et al., 2004). The existence of such substrate-specific import pathways may alleviate potential competition between different types of preprotein. Interestingly, there are also multiple Toc75 genes in Arabidopsis: two close Toc75 relatives, termed atTOC75-III and atTOC75-IV based on chromosomal location (Jackson-Constan and Keegstra, 2001; Baldwin et al., 2005), and one more distantly related gene, termed AtOEP80 (Outer Envelope Protein, 80 kD; or atTOC75-V), which is more similar to a cyanobacterial Toc75 homolog (Eckart et al., 2002; Inoue and Potter, 2004). Earlier work showed that the atTOC75-IV gene is expressed at very low levels, and that knockout mutant plants do not exhibit obvious developmental defects, suggesting that the atToc75-IV protein plays a relative minor, highly specialized role (Baldwin et al., 2005). By contrast, the atToc75-III and AtOEP80 proteins are essential for viability, since the corresponding knockout mutants abort during embryogenesis (Baldwin et al., 2005; Patel et al., 2008). While it is of importance to know that these proteins are essential, the nonviability of the knockout mutants beyond the embryo stage precluded their use for in vivo studies on the roles of the proteins during postembryonic growth. To circumvent this problem, we have employed inducible RNA interference (RNAi) technology to knockdown expression of the genes following completion of embryogenesis. Our results reveal important roles for both proteins at later stages of plant development.
RESULTS
Generation of the atTOC75-III and AtOEP80 RNAi Lines
Short, gene-specific regions (437 bp each) from the atTOC75-III and AtOEP80 coding sequences were carefully selected as targets for RNAi; these were shown to share no significant homology with other Arabidopsis sequences by BLAST analysis (Altschul et al., 1990). The selected sequences were inserted into the dexamethasone-inducible, hairpin RNAi vector, pOpOff2(hyg) (Wielopolska et al., 2005), and then the constructs (termed atToc75-III↓ and AtOEP80↓, respectively) were used to transform wild-type Arabidopsis plants. A total of 24 independent transformants were identified for each construct. From these transformants, several lines carrying a single insertion locus were identified by plating T2 families on selective medium (as indicated by a 3:1 segregation ratio). Analysis of these single-locus lines on dexamethasone-containing medium revealed that each construct was associated with a characteristic and distinct phenotype, as described below; these dexamethasone-induced phenotypes segregated with a 3:1 ratio, relative to the wild-type phenotype, indicating that they were caused by the relevant RNAi construct (Supplemental Table S1; Supplemental Fig. S1). Homozygous lines were identified in the T3 generation and propagated. The atToc75-III↓ number 6 and AtOEP80↓ number 7 lines were chosen for detailed analysis, as they each displayed a typical, heritable phenotype and exhibited comparable levels of down-regulation of the target gene. Control pOpOff2 transgenic lines did not display any of the characteristic phenotypes associated with atToc75-III↓ and AtOEP80↓ (Supplemental Appendix S1; Supplemental Figs. S2 and S3).
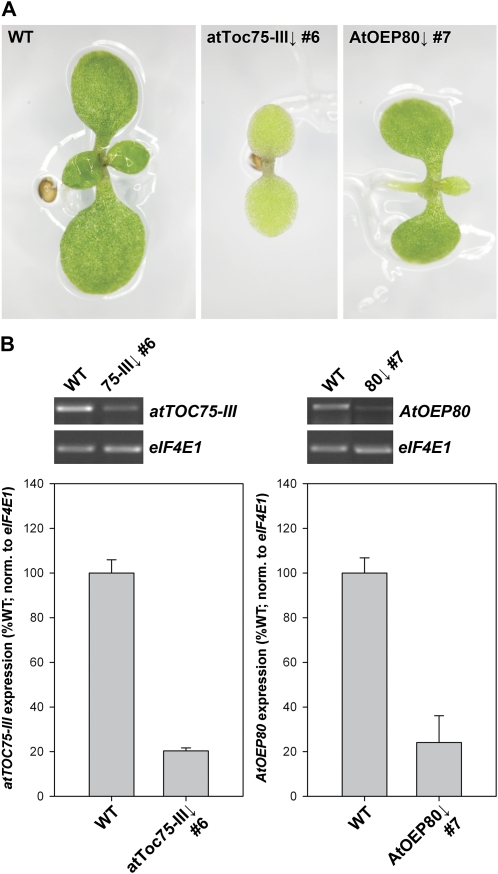
When grown on medium containing 50 μm dexamethasone for 10 d, atToc75-III↓ number 6 plants displayed strong chlorosis of the cotyledons and retarded growth (the first true leaves had not emerged at this stage) by comparison with wild-type plants grown under identical conditions (Fig. 1A). In contrast, while AtOEP80↓ number 7 plants were also smaller and paler than wild-type plants, the chlorosis of the cotyledons was much less severe and the plants were considerably more developed than the atToc75-III↓ plants. Interestingly, chlorosis in the AtOEP80↓ plants appeared to be more severe in the first true leaves than in the cotyledons (Fig. 1A). Semiquantitative reverse transcription (RT)-PCR revealed that the atToc75-III↓ number 6 and AtOEP80↓ number 7 seedlings exhibit very similar degrees of target gene down-regulation; expression was reduced to approximately 20% of the wild-type level in each case (Fig. 1B). Therefore, the much stronger phenotype of atToc75-III↓ plants did not appear to be related to RNAi construct efficacy, and was instead interpreted to be reflective of functional differences between atToc75-III and AtOEP80. Nonetheless, alternative explanations related to differing effects of the two constructs on target protein abundance, or to developmental differences in mRNA silencing not detected in our analysis, cannot be eliminated entirely and should be borne in mind.
Figure 1.
Appearance and molecular analysis of typical atToc75-III↓ and AtOEP80↓ RNAi lines. A, The appearance of 10-d-old seedlings of the indicated genotypes following growth on medium containing 50 μm dexamethasone. The transgenic lines shown are representative, and were selected following careful analyses of at least five independent transformants in each case. B, The expression level of the atTOC75-III or AtOEP80 gene in a respective, typical RNAi line, and in wild type, was analyzed by semiquantitative RT-PCR. Plants were grown for 10 d on medium containing 50 μm dexamethasone prior to analysis. Amplifications were nonsaturating and employed 21 PCR cycles for atTOC75-III and eIF4E1, and 23 cycles for AtOEP80. Bands were quantified using ImageQuant software. The charts show means (±sd) derived from three independent amplifications per genotype. Data for atTOC75-III and AtOEP80 were normalized to equivalent data for eIF4E1.
Assessment of Photosynthetic and Nonphotosynthetic Development in the atToc75-III↓ and AtOEP80↓ RNAi Lines
To shed further light on the in vivo functions of the atTOC75-III and AtOEP80 genes, we studied parameters of both photosynthetic and nonphotosynthetic development in the RNAi lines, in parallel. This comparative study aimed to elucidate the relative importance of each gene during different modes of development, as it has been found that different isoforms of some TOC components are specialized for either photosynthetic or nonphotosynthetic growth (Bauer et al., 2000; Jarvis, 2008). In general, we observed that atToc75-III↓ plants display defects in both photosynthetic and nonphotosynthetic development; the same was also true of the AtOEP80↓ plants, although their defects were much less severe such that the plants had an intermediate phenotype, between atToc75-III↓ and wild type (see below).
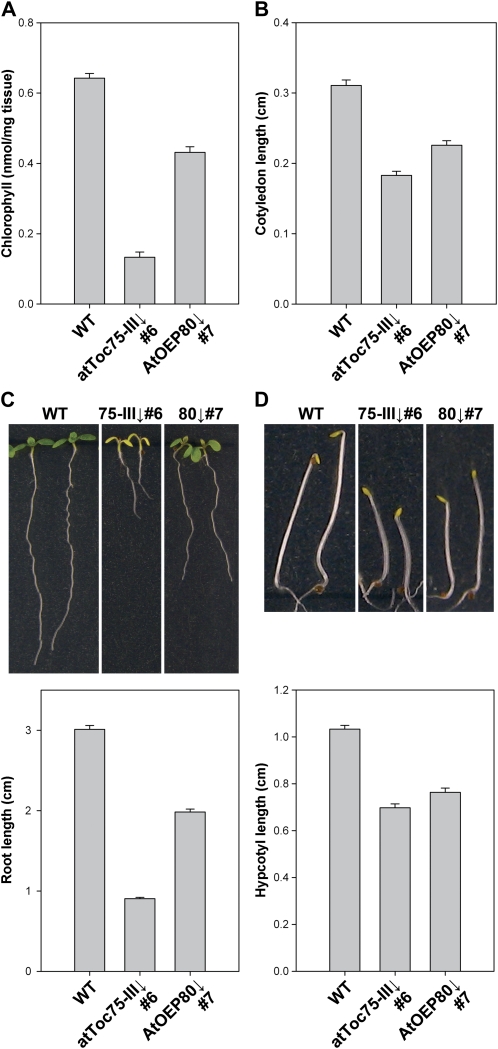
Chlorophyll content measurements done on 10-d-old seedlings grown on dexamethasone-containing medium revealed that atToc75-III↓ number 6 plants contain only approximately 21% of the wild-type pigment level (Fig. 2A), which is quantitatively similar to the chlorophyll deficiency seen in the atToc33 knockout mutant, plastid protein import1 (ppi1; Jarvis et al., 1998), or that in the atTic40 knockout mutant, tic40 (Bédard et al., 2007). In contrast, chlorophyll levels were much higher in AtOEP80↓ number 7 plants at approximately 67% of the normal level (Fig. 2A). Cotyledon length was assessed as an additional assay of photosynthetic growth (Fig. 2B), and similar results were obtained. Root length measurements essentially paralleled the chlorophyll and cotyledon data, with a strong defect in atToc75-III↓ plants (approximately 30% of wild type) and a considerably weaker defect in AtOEP80↓ plants (approximately 66% of wild type; Fig. 2C). These data imply that both proteins are important for photosynthetic and nonphotosynthetic development. A potential problem associated with using root length in light-grown plants as a measure of nonphotosynthetic development is that it may be influenced, indirectly, by defects in the photosynthetic tissues (for example, through a reduced yield of exported photosynthate). To address this issue, we measured hypocotyl lengths in dark-grown plants as an alternative assay of nonphotosynthetic growth (Fig. 2D). Again, significant growth defects were observed in both RNAi lines, although these defects were of reduced magnitude (approximately 68% of wild type in atToc75-III↓; approximately 74% of wild type in AtOEP80↓). This confirmed that the two proteins are directly important for nonphotosynthetic development.
Figure 2.
Parameters of photosynthetic and nonphotosynthetic growth in atToc75-III↓ and AtOEP80↓ plants. A, Plants of the indicated genotypes were grown on MS-dexamethasone medium for 10 d. Chlorophyll was determined photometrically. Values shown are means (±se) derived from three independent samples, each one containing 15 plants. Units are nmol chlorophyll a + b per mg fresh weight. B, Plants were grown on MS-dexamethasone agar plates for 10 d. Cotyledon length (from the shoot apex to the tip of the cotyledon) was quantified by analyzing digital images of the plants using SigmaScan Pro software (SPSS Inc.). Values shown are means (±se) derived from 31 to 36 independent measurements per genotype. C, Plants were grown side by side on vertically oriented MS-dexamethasone agar plates for 10 d. Primary root length was quantified by analyzing digital images of 10 independent plates, using SigmaScan Pro, each one carrying approximately 12 plants per genotype. Values shown are the means (±se) from 49 to 64 independent measurements per genotype. D, Plants were grown on MS-dexamethasone medium for 5 d in the dark. They were then laid down horizontally on agar medium for photography, and hypocotyl lengths were quantified by analyzing the digital images as described in the legend to section B. Values shown are means (±se) derived from 49 to 83 independent measurements per genotype.
Ultrastructural Analyses of Chloroplasts and Nongreen Plastids in the RNAi Lines
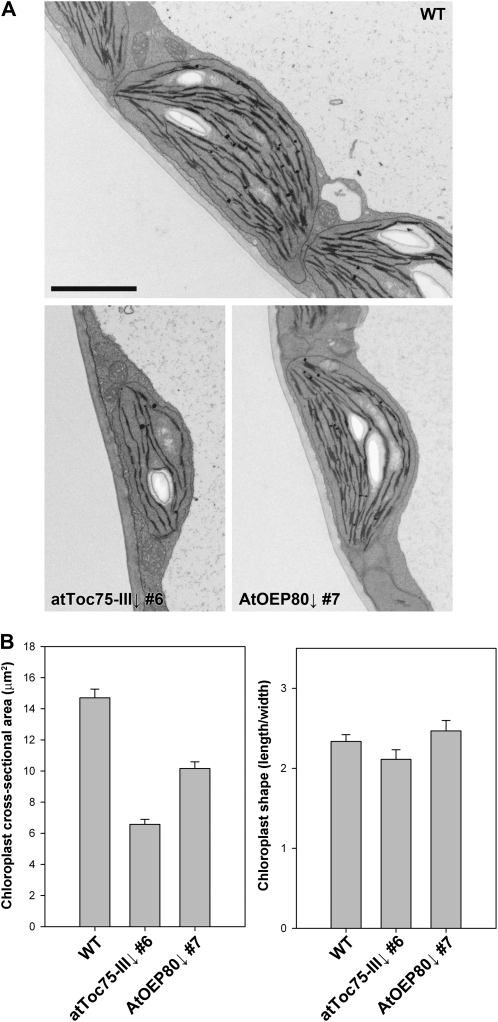
As a further comparison of the effects of the atToc75-III↓ and AtOEP80↓ transgenes on plastid development, we analyzed chloroplast ultrastructure in seedlings grown in the presence of dexamethasone (Fig. 3A). As expected, the chloroplasts of atToc75-III↓ plants were much smaller than those in wild type, and they had severely underdeveloped thylakoid networks. By contrast, chloroplasts in AtOEP80↓ plants were only slightly smaller than wild-type organelles, and they had quite well-developed thylakoid membranes (Fig. 3A). Quantitative analyses of the micrographs supported these conclusions (Fig. 3B, left side), and additionally indicated that chloroplast shape was not altered in the RNAi lines (Fig. 3B, right side). In this regard, atToc75-III↓ is more similar to ppi1 than to tic40, since the latter mutation causes a distinct change in chloroplast shape (Chou et al., 2003; Kovacheva et al., 2005); it is possible that this unique feature of the tic40 phenotype is a reflection of its additional role in inner membrane protein insertion (Chiu and Li, 2008). Overall, these data indicate that deficiencies in either atToc75-III or AtOEP80 affect the size and complexity of chloroplasts, but to different degrees.
Figure 3.
Chloroplast morphology in the atToc75-III↓ and AtOEP80↓ RNAi lines. A, Representative electron micrographs of wild-type, atToc75-III↓, and AtOEP80↓ chloroplasts. Cotyledons from 10-d-old plants of the indicated genotypes grown on MS-dexamethasone medium were analyzed. Approximately 10 whole-chloroplast micrographs from each of at least three independent plants per genotype were analyzed prior to selecting the images shown. Scale bar = 2.0 μm. B, Quantification of chloroplast electron micrographs. The length and width of each chloroplast in each of the micrographs described in the legend to A was measured. These values were used to estimate chloroplast cross-sectional area, shown in the left chart, and chloroplast shape (the length/width ratio), shown in the right chart. Values shown are means (±se) derived from measurements of 24 to 27 different chloroplasts per genotype.
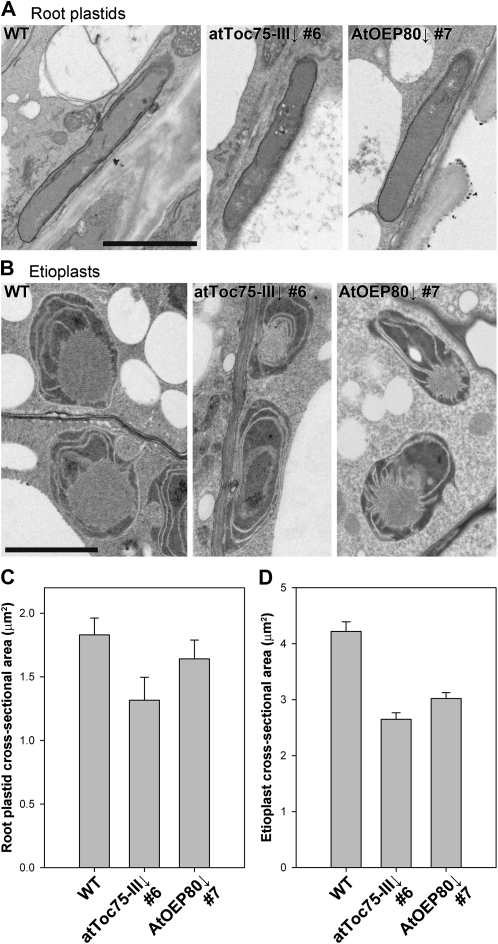
Results in Figure 2 suggested that atToc75-III and AtOEP80 may both be important for the development of nonphotosynthetic plastids as well as chloroplasts. To directly assess this possibility, we analyzed two different types of nongreen plastid in the atToc75-III↓ number 6 and AtOEP80↓ number 7 RNAi lines (following dexamethasone treatment) by electron microscopy: the leucoplasts of roots, and the etioplasts of dark-grown seedlings (Fig. 4). The results showed that both types of organelle are significantly underdeveloped in atToc75-III↓ plants, relative to those in wild type. While etioplasts were also significantly smaller in AtOEP80↓ plants, albeit to a lesser extent than those in atToc75-III↓, defects in AtOEP80↓ root plastids were more difficult to detect (this was most likely due to limitations of our method in relation to mild changes in plastids of this type, which have a high level of structural variability). To a considerable extent, these data parallel those from the developmental studies (Fig. 2), as can be seen by comparing the quantitative data in Figure 4, C and D with those in Figure 2, C and D, respectively. Overall, these data are consistent with the notion that both atToc75-III and AtOEP80 are important for the biogenesis of nonphotosynthetic plastids.
Figure 4.
Morphology of nonphotosynthetic plastids in atToc75-III↓ and AtOEP80↓ plants. A and B, Representative electron micrographs of root plastids (A) and etioplasts (B) in wild-type, atToc75-III↓ number 6, and AtOEP80↓ number 7 plants. Images were derived from plants grown on MS-dexamethasone medium, either in the light for 10 d (A) or in the dark for 5 d (B). Approximately five whole-organelle micrographs from each of at least three independent plants per genotype were analyzed carefully, in each case, and used to select the representative images shown. Scale bars = 2 μm. C and D, Quantification of root plastid (C) and etioplast (D) electron micrographs. Values shown are means (±se) derived from measurements of 14 to 37 different plastids per genotype, in each case.
Assessment of Chloroplast Protein Levels in the RNAi Lines
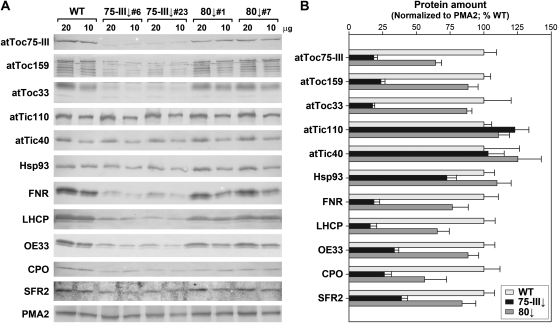
Next, we used immunoblotting to analyze the levels of several important proteins in atToc75-III↓ and AtOEP80↓ chloroplasts. Total protein extracts from plants grown in the presence of dexamethasone for 8 d were analyzed (Fig. 5). The atToc75-III↓ plants were found to be strongly deficient in the atToc75-III protein (approximately 18% of the wild-type level), which is as expected since it is the direct target of the RNAi construct. Levels of the major preprotein receptors, atToc159 and atToc33, were also significantly reduced; this is not surprising, as atToc75-III, atToc159, and atToc33 are all core components of the TOC complex, and it is possible that the depletion of just one of these destabilizes the complex with significant knockon effects in relation to other components. Moreover, Toc75 is known to be important for TOC receptor protein biogenesis (Wallas et al., 2003). Interestingly, all of these TOC proteins were also depleted in the AtOEP80↓ samples, with the strongest effect being observed for atToc75-III (approximately 64% of the wild-type level). This observation is consistent with the notion that AtOEP80 is important for the biogenesis of β-barrel proteins (e.g. atToc75-III), as was proposed previously (Schleiff and Soll, 2005; Hsu et al., 2008). Unfortunately, we were unable to assess the levels of AtOEP80 protein in the RNAi lines due to the absence of a suitable antibody; in our hands, the antibody raised against the pea ortholog (Eckart et al., 2002) did not detect a specific band in Arabidopsis extracts.
Figure 5.
Immunoblot analysis of chloroplast protein levels in atToc75-III↓ and AtOEP80↓ plants. A, Plants of the indicated genotypes were grown on MS-dexamethasone medium for 8 d; two different RNAi lines each (atToc75-III↓: numbers 6 and 23; AtOEP80↓: numbers 1 and 7) were employed to ensure acquisition of representative results. Total protein extracts were prepared, and two different loadings (20 and 10 μg) per genotype were analyzed by immunoblotting using antibodies against a variety of chloroplast proteins: these included components of the TOC (atToc75-III, atToc159, atToc33) and TIC (atTic110, atTic40, Hsp93) complexes, as well as representative photosynthetic and nonphotosynthetic proteins and a signal-anchored outer envelope membrane protein. Note that atToc159 is prone to degradation (Agne et al., 2010); the upper band shown is the intact protein. An antibody against the plasma membrane H+-ATPase, PMA2, was used to provide a loading control. B, All protein bands shown in A were quantified using Aida software. Data for PMA2 were used to normalize the results for the plastid proteins of interest. Values shown are means (±se) and are representative of repeated experiments; results for the two RNAi lines were similar and so were combined to yield a single mean value in each case.
Somewhat surprisingly, the chloroplast inner envelope proteins, atTic110 and atTic40, as well as the associated chaperone, Hsp93 (Heat shock protein, 93 kD), were not strongly affected by atToc75-III or AtOEP80 deficiency (Fig. 5). This may be related to the fact that these proteins are components of the TIC machinery of the inner membrane, and so are not direct partners of atToc75-III. The possibility that the TIC complex is a flexible, dynamic assembly may also be relevant (Kouranov et al., 1998), allowing its components to accumulate regardless of perturbations in translocon assembly. An alternative explanation is that levels of these important TIC proteins are maintained in an attempt to compensate for import inefficiencies caused by disruption of the TOC complex.
The abundant photosynthetic proteins, ferredoxin-NADP reductase (FNR), light-harvesting chlorophyll protein (LHCP), and oxygen-evolving complex 33-kD subunit (OE33), were all strongly depleted (to approximately 16%–34% of wild-type levels) in atToc75-III↓ plants, and to a lesser but nonetheless significant extent (approximately 65%–88% of wild-type levels) in AtOEP80↓ plants (Fig. 5). These results are consistent with the chlorotic phenotypes (Figs. 1A and 2A) and the chloroplast ultrastructural defects (Fig. 3) described earlier. In the case of atToc75-III↓, these deficiencies were almost certainly caused by disrupted import of the corresponding preproteins, due to a deficiency in their cognate translocation channel. In the case of AtOEP80↓, the cause is uncertain, although it may be linked to the aforementioned deficiency in atToc75-III and other TOC proteins. Abundance of the nonphotosynthetic chloroplast protein, coproporphyrinogen oxidase (CPO), which is involved in tetrapyrrole biosynthesis, was also significantly depleted in both atToc75-III↓ and AtOEP80↓ plants (Fig. 5); this is consistent with the results indicating that the two Omp85-related proteins are important for the development of nonphotosynthetic organs and plastids (Figs. 2–4). Finally, we observed a pronounced reduction in the abundance of the signal-anchored outer envelope membrane protein, SENSITIVE TO FREEZING2 (SFR2; Fourrier et al., 2008), in atToc75-III↓ plants. This implies that atToc75-III is important for the biogenesis of such proteins, which do not follow the canonical TOC-TIC import route (Hofmann and Theg, 2005), and is consistent with the earlier observations of Tu et al. (2004). A quantitatively smaller reduction in SFR2 abundance seen in AtOEP80↓ samples may be a consequence of the reduced atToc75-III levels in such plants.
Changes in the atToc75-III↓ chloroplast proteome were also assessed using difference gel electrophoresis (DIGE), employing the same procedures as used during analyses of the ppi1 chloroplast proteome (Kubis et al., 2003). As we were only interested in obtaining a snapshot of the proteome, we focused on just a few clearly resolved spots with strongly reduced levels in atToc75-III↓ organelles. In previous similar DIGE studies (Kubis et al., 2003, 2004), we normalized the wild-type and mutant samples on the basis of protein amounts. As the atToc75-III↓ chloroplasts are considerably smaller than those in the wild type (Fig. 3), we additionally employed an alternative normalization method based on organelle numbers. Quantitative data derived using each normalization method were obtained, and in both cases we detected depleted proteins belonging to photosynthetic and nonphotosynthetic categories (Supplemental Fig. S4). Thus, the results are consistent with the immunoblot data (Fig. 5), and support the notion that atToc75-III is important for the biogenesis of a broad cross section of preproteins.
Analysis of the Phenotype of an atToc75-III Missense Allele
The modifier of altered response to gravity1 (mar1) mutant was recently identified in a screen for modifiers of the altered response to gravity1 mutation (Stanga et al., 2009). The mar1 mutant was found to carry a point mutation in the atTOC75-III gene, which causes a single amino acid change (G658R) within the seventh predicted transmembrane β-strand of the channel domain (Ertel et al., 2005). In agreement with our results on atToc75-III↓ (Fig. 2A), the mar1 mutant was observed to exhibit a significant chlorophyll deficiency, although pigment concentrations (at approximately 70% of the wild-type level; Stanga et al., 2009) were considerably higher than those in the atToc75-III↓ number 6 line. To further corroborate the results obtained using atToc75-III↓ RNAi lines, we conducted a series of experiments on mar1, which shall henceforth be referred to as toc75-III-3 in accordance with the previously established nomenclature for atToc75-III mutants (Baldwin et al., 2005). An advantage of the toc75-III-3 mutant was that it enabled us to assess the consequences of atToc75-III defects during later stages of development, which was not possible using the RNAi lines, as pOpOff2-mediated silencing is impractical with mature plants on soil. Because toc75-III-3 was identified in the Wassilewskija ecotype (while the RNAi lines and nearly all other TOC/TIC mutants are of the Columbia-0 background), we first of all introgressed the mutation into Columbia-0 by seven consecutive backcrosses (achieving a theoretical ecotype purity of >99%). This Columbia-0-introgressed toc75-III-3 line was used in all subsequent studies.
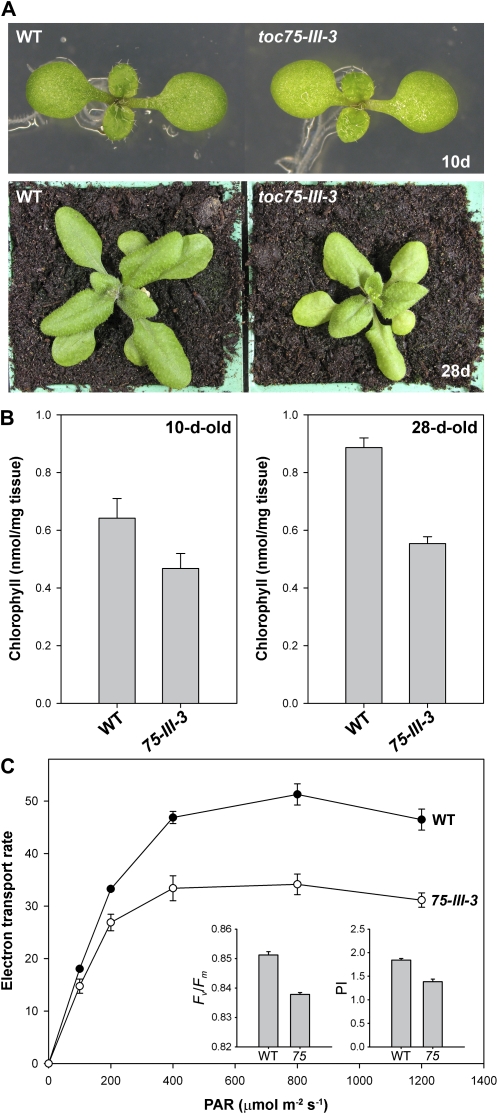
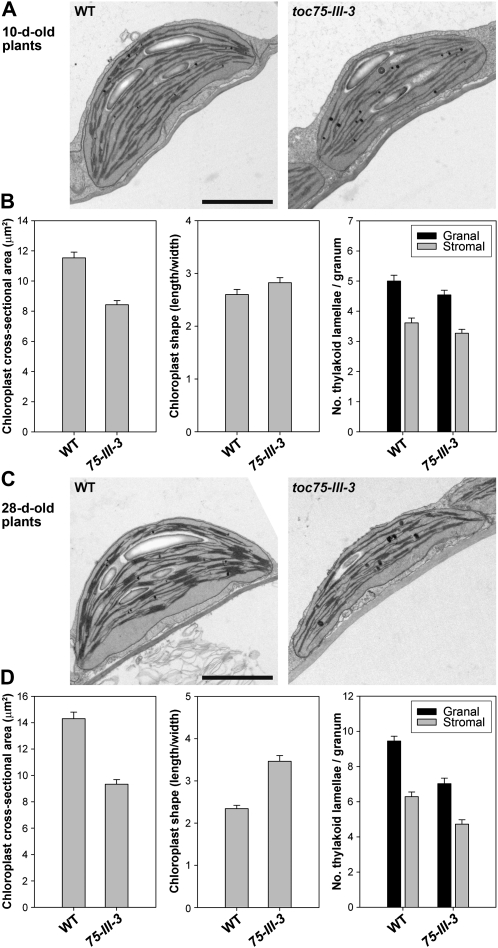
As shown in Figure 6A, toc75-III-3 plants were slightly paler than wild type throughout development, although the chlorosis was somewhat more apparent during later stages of development. Indeed, chlorophyll content measurements revealed a considerably greater pigment deficiency in older plants (73% of wild-type levels at 10 d; 62% of wild-type levels at 28 d; Fig. 6B). Moreover, chloroplast ultrastructural defects (i.e. cross-sectional area, and the numbers of granal and stromal lamellae per granum) were similarly more apparent in mature plants (Fig. 7, C and D) than in young seedlings (Fig. 7, A and B). Interestingly, mutant chloroplasts were somewhat elongated during later development (Fig. 7, C and D); the reason for this is unclear, but it may be related to the reductions in thylakoid development in the mutant.
Figure 6.
Analysis of photosynthetic development in the toc75-III-3 (mar1) mutant. A, Plants of the indicated genotypes were grown side by side, in vitro, for 10 d, and then photographed. Additional plants were transferred to soil on day 14 and allowed to grow for a further 2 weeks prior to photography on day 28. Representative plants are shown in each case. B, Chlorophyll concentrations in 10- and 28-d-old plants similar to those described in A were determined photometrically. Values shown are means (±se) derived from three independent samples, each one containing eight plants. Units are nmol chlorophyll a + b per mg fresh weight. C, Analysis of photosynthesis in the toc75-III-3 mutant. Light response curves of photosynthetic electron transport rates in wild-type and mutant plants were determined by measuring chlorophyll fluorescence. Values were recorded at different irradiances of photosynthetically active radiation (PAR), ranging from 0 to 1,200 μmol photons m−2 s−1. Units for the data shown are μmol electrons m−2 s−1, assuming 84% absorption of the incident light and equal distribution of the energy between the two photosystems (Meyer et al., 1997). The insets show PSII photochemical efficiency (Fv/Fm) and photosynthetic performance index readings for similar plants. Developmentally similar leaves (seventh or eighth true leaf) of 28-d-old plants grown under identical conditions were employed for all measurements. Values shown are means (±se) derived from five independent measurements.
Figure 7.
Chloroplast morphology at different developmental stages in the toc75-III-3 (mar1) mutant. A and C, Representative electron micrographs of wild-type and toc75-III-3 mutant chloroplasts. Cotyledons from 10-d-old plants grown in vitro (A), and true leaves from 28-d-old plants grown on soil (C), were analyzed. At least 10 whole-chloroplast micrographs from each of at least three independent plants per genotype were studied, in each case, prior to the selection of the images shown. Scale bars = 2.0 μm. B and D, Quantification of chloroplast electron micrographs derived from 10-d-old (B) and 28-d-old (D) plants. The length and width of each chloroplast in each of the micrographs described in the legend to A and C was measured. These values were used to estimate chloroplast cross-sectional area (shown in the charts at left), and chloroplast shape (the length/width ratio; shown in the central charts). Data shown in the charts at right indicate the extent of thylakoid membrane development in the chloroplasts at each developmental stage. Values shown are means (±se) derived from measurements of 30 to 40 different chloroplasts per genotype per age, or from the analysis of 26 to 36 different grana per genotype per age.
Overall, these data indicate that the atToc75-III protein is important throughout development, and suggest that it is at least as important in mature plants as in young seedlings. Photosynthetic performance measurements done on mature toc75-III-3 plants confirmed the important role of atToc75-III during later developmental stages (Fig. 6C). Finally, to corroborate the earlier conclusion that atToc75-III is important for both photosynthetic and nonphotosynthetic development (derived from the results in Fig. 2), we analyzed root length and hypocotyl elongation in toc75-III-3; both experiments revealed small but significant growth defects in the mutant (Supplemental Fig. S5), confirming the RNAi results and suggesting that atToc75-III is important for the biogenesis of nonphotosynthetic plastids and preproteins.
Analysis of Chloroplast Preprotein Import Rates in atToc75-III↓ Plants
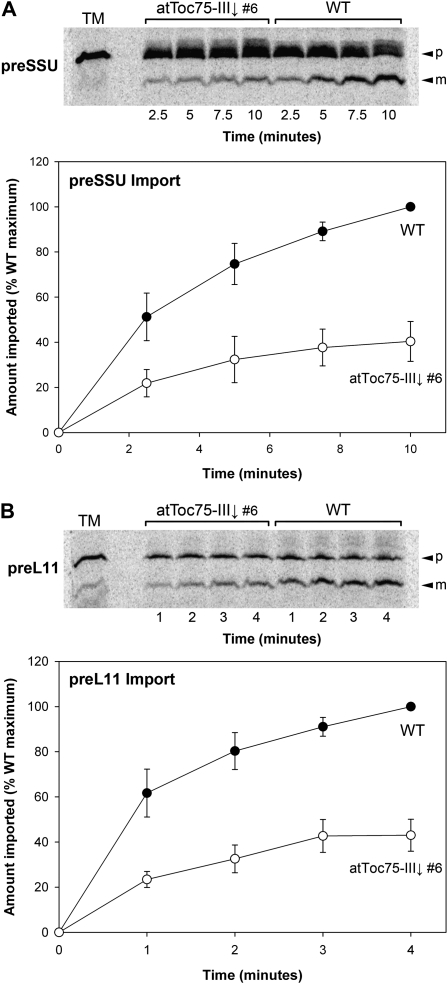
Experiments described thus far suggested that atToc75-III is important for the biogenesis of both photosynthetic and nonphotosynthetic preproteins. To obtain direct evidence in support of this hypothesis, we analyzed the import efficiencies of two different preproteins using chloroplasts isolated from the atToc75-III↓ number 6 RNAi line (which has a more severe visible phenotype than toc75-III-3 and so would be expected to give more clear import results). For these studies, we used the precursor of the small subunit of Rubisco (preSSU; a model photosynthetic preprotein, and an important component of photosynthetic carbon reactions) and the precursor of a 50S ribosomal subunit protein (preL11; a model nonphotosynthetic preprotein, and a component of the chloroplast’s endogenous genetic system). We conducted time-course experiments, and then quantified the imported, mature protein at each time point; the amount of imported protein in the knockdown chloroplasts was then expressed as a percentage of the amount imported into wild-type organelles at the last time point. As shown in Figure 8, import of the two tested preproteins was reduced to a roughly equivalent extent: The maximal amount of imported protein was down to approximately 40% of the wild-type level in atToc75-III↓ chloroplasts, for both SSU and L11. These results show that atToc75-III↓ chloroplasts do indeed have a reduced capacity to import both photosynthetic and nonphotosynthetic preproteins, strongly supporting the hypothesis that atToc75-III plays a general, non-substrate-specific role in plastid preprotein import. Similar experiments were not conducted using AtOEP80↓ chloroplasts, as the aforementioned TOC component deficiencies (Fig. 5) would be expected to have a strong, indirect influence on the results. Moreover, current evidence suggests that AtOEP80 does not play a direct role in chloroplast protein import (Eckart et al., 2002).
Figure 8.
Comparison of chloroplast import efficiencies in wild-type and atToc75-III↓ plants. Chloroplasts isolated from plants of the indicated genotypes (grown on MS-dexamethasone medium for 10 d) were used in protein import assays with [35S]-Met-labeled preproteins. One photosynthetic preprotein (preSSU [A]) and one nonphotosynthetic preprotein (preL11 [B]) was employed. Import was allowed to proceed for the indicated time periods, and then the samples were analyzed by SDS-PAGE and phosphorimaging. Results shown are representative of three independent experiments. TM indicates an aliquot of the translation mixture equivalent to 10% of the amount added to each import assay; p and m indicate the precursor and mature forms of the radiolabeled protein, respectively. Quantifications of all three experiments are shown in the graphs below the gel images. Mature protein bands at each time point were quantified, and then the data were expressed as percentages of the value for the final, wild-type time point in each experiment. The data shown are means (±se).
DISCUSSION
Previous studies revealed that the atToc75-III and AtOEP80/atToc75-V proteins are essential for viability during embryogenesis (Baldwin et al., 2005; Hust and Gutensohn, 2006; Patel et al., 2008). Interestingly, that earlier work revealed pronounced phenotype severity differences between the relevant toc75-III and oep80 knockout mutants. Whereas toc75-III knockouts were unable to pass the proembryo stage, such that the embryo proper never forms more than two cells, oep80 null mutants could progress much further and did not arrest until the globular stage (at which point the embryo proper is estimated to contain approximately 32–64 cells; Goldberg et al., 1994). These observations led to the suggestion that the two proteins perform distinct functions that become vital at different stages of embryo development (Hsu et al., 2008). The notion that the two proteins perform different roles is further supported by the data presented in this report (Figs. 1–3). While the phenotypes of the atToc75-III↓ and AtOEP80↓ RNAi lines indicated that both proteins are important for plastid biogenesis, there were large quantitative severity differences between the two phenotypes.
That atToc75-III acts as a preprotein import channel in the outer envelope membrane is a well-established fact (Kessler and Schnell, 2006; Jarvis, 2008). However, the nature of the function performed by AtOEP80 has remained elusive. One interesting proposal is that OEP80 is functionally analogous to most other members of the Omp85 superfamily (including Sam50/Tob55 in the mitochondrial outer membrane), in that it mediates the biogenesis of β-barrel proteins (Schleiff and Soll, 2005; Hsu et al., 2008). In this scenario, atToc75-III would be predicted to be one of the substrates of the putative AtOEP80-mediated assembly pathway; thus, one might expect a reduction in AtOEP80 activity to have broadly similar consequences to a deficiency in atToc75-III, although perhaps with developmentally delayed onset (as seen during embryogenesis, as discussed above) or a diminution in the magnitude of the effects. The fact the AtOEP80↓ phenotype closely parallels that of atToc75-III↓ plants, albeit with considerably reduced severity (Figs. 1–4), is therefore supportive of the aforementioned hypothesis. The fact that atToc75-III protein abundance is strongly reduced in AtOEP80↓ plants is also consistent with this model (Fig. 5). Nonetheless, an unequivocal description of AtOEP80 function must await further analysis.
Detailed analyses of the atToc75-III↓ RNAi lines, as well as of the hypomorphic mutant, toc75-III-3 (mar1), provided conclusive evidence that the atToc75-III channel is involved in the import of a broad range of functionally different preproteins, and that it is not specialized for either photosynthetic or nonphotosynthetic precursors (e.g. Fig. 8), in contrast with the TOC receptor proteins (Kessler and Schnell, 2006; Jarvis, 2008). These results support the notion that atToc75-III acts as a general import channel to which various receptor isoforms associate, differentially, to impart distinct client specificities. In this regard, our results provide genetic and functional support for the biochemically derived observations of Ivanova et al. (2004), who demonstrated that two distinct TOC complex subtypes (characterized by the receptor combinations, atToc159/atToc33 and atToc132-120/atToc34) share a common 75-kD immunoreactive component; the latter was almost certainly atToc75-III due to the fact that there are no other full-length, canonical Toc75 proteins in Arabidopsis (Inoue and Potter, 2004; Baldwin et al., 2005). The position of the amino acid substitution in the toc75-III-3 mutant, within one of the protein’s putative transmembrane β-strands (Ertel et al., 2005; Stanga et al., 2009), suggests that the lesion may affect structural and/or functional properties of the channel.
Our immunoblot data indicated that atToc75-III is required for the accumulation of the TOC receptors, atToc159 and atToc33, and that AtOEP80 is required for the accumulation of atToc75-III and (perhaps indirectly via its effect on atToc75-III) the TOC receptors (Fig. 5). These observations are consistent with the biochemical results of Wallas et al. (2003), who used proteoliposomes to show that Toc75 is needed for the membrane insertion of Toc159. The results are also in agreement with the notion of a TOC core complex, comprising Toc75, Toc159, and Toc34, as proposed by Schleiff et al. (2003) and Kikuchi et al. (2006); this implies that the three components associate cooperatively in a tightly bound structure, and so it is not surprising that a deficiency in the central, channel component has serious implications for complex integrity and the accumulation of the other members. In contrast, the fact that the levels of Tic110, Tic40, and Hsp93 are not significantly affected by Toc75 or OEP80 deficiency implies that TIC accumulation and/or assembly is not strictly dependent on TOC complex or OEP80 abundance.
Whereas most chloroplast proteins possess an N-terminal transit peptide for engagement of the TOC-TIC machinery (Kessler and Schnell, 2006; Jarvis, 2008), outer envelope membrane proteins generally do not, and are instead targeted by virtue of intrinsic targeting information in their hydrophobic transmembrane domains (Hofmann and Theg, 2005). Nonetheless, biochemical evidence indicating a role for Toc75 in the insertion of such outer membrane proteins was recently reported (Tu et al., 2004). Data presented in Figure 5 are consistent with this notion (and provide in vivo support for it), as they reveal a significant defect in the accumulation of the outer membrane protein, SFR2, in the atToc75-III↓ RNAi lines. Thus, it would appear that atToc75-III is a multifunctional protein, acting in multiple different TOC complexes in relation to the import of different preprotein categories, as well as in a separate pathway for the insertion of outer membrane proteins.
MATERIALS AND METHODS
Generation of RNAi Constructs and Transgenic Lines
Gene-specific sequences from atTOC75-III (735–1,171 bp of the CDS) and AtOEP80 (668–1,104 bp of the CDS) were amplified using primers that introduce partial attB recombination sites: atTOC75-III, 5′-AAA AAG CAG GCT GAG TAC ATG GCA ATC TGC-3′ and 5′-AGA AAG CTG GGT GCA CCA CAG GAA CTT GAG-3′; AtOEP80, 5′-AAA AAG CAG GCT TTG CTG TTG ATA CAC GAG-3′ and 5′-AGA AAG CTG GGT CAT GGC TAG CAC AGT GTC-3′. Secondary amplification using the following primers (both genes) completed the terminal attB sites: 5′-GGG GAC AAG TTT GTA CAA AAA GCA GGC T-3′ and 5′-GGG GAC CAC TTT GTA CAA GAA AGC TGG GT-3′. Amplified target sequences were inserted into the pOpOff2 (hyg) vector (Wielopolska et al., 2005) by Gateway recombination cloning (Invitrogen), via the pDONR201 donor vector. Constructs were stably introduced into wild-type Columbia-0 plants using Agrobacterium strain GV3101 (pMP90) and the floral-dip method (Clough and Bent, 1998). Transformants were identified by screening on Murashige and Skoog (MS) agar medium containing 15 μg/mL hygromycin B (plus 200 μg/mL cefotaxime sodium to eliminate agrobacterial contamination). Resulting T2 families were further screened on selective medium to identify lines with a single insertion locus (Supplemental Table S1). The T3 progenies of selected lines were analyzed on hygromycin-containing medium to identify homozygous lines, which were propagated and used for all subsequent experiments.
Plant Growth Conditions and Physiological Studies
With the exception of the original toc75-III-3 (mar1) mutant (Stanga et al., 2009), all Arabidopsis (Arabidopsis thaliana) plants were of the Columbia-0 ecotype. For in vitro growth, seeds were surface sterilized, sown on MS agar medium in petri plates, cold treated at 4°C, and thereafter kept in a growth chamber, as described previously (Aronsson and Jarvis, 2002). When required, the MS medium contained 50 μm dexamethasone (Sigma-Aldrich); dexamethasone was dissolved in ethanol to make a 20 mm stock solution. Different concentrations (10, 25, 50, and 100 μm dexamethasone) were evaluated in preliminary tests, using both atToc75-III↓ number 6 and AtOEP80↓ number 7. The lowest concentrations were less effective at inducing silencing, whereas 100 μm dexamethasone was toxic for the plants (growth of untransformed seedlings was inhibited; data not shown). The intermediate concentration of 50 μm induced efficient silencing and its toxicity was low. Control pOpOff2 transgenics appeared fully green on dexamethasone-containing medium and did not exhibit any of the various phenotypes associated with atToc75-III↓ or AtOEP80↓ (Supplemental Appendix S1; Supplemental Figs. S2 and S3). All plants were grown under a long-day cycle (16 h light, 8 h dark).
Root length measurements were conducted as described previously (Constan et al., 2004), using plants grown on vertically oriented MS agar plates containing dexamethasone under standard conditions for 9 d. Hypocotyl length measurements were done using plants grown on horizontal MS-dexamethasone plates kept in the dark for 5 d. Chlorophyll was extracted in N, N’-dimethylformamide and quantified photometrically as described previously (Constan et al., 2004). Photosynthetic electron transport rate, photochemical efficiency of PSII (Fv/Fm), and photosynthetic performance index were all determined by measuring chlorophyll fluorescence using a continuous excitation fluorimeter (Handy PEA; Hansatech Instruments), according to the manufacturer’s instructions (Meyer et al., 1997; Patel et al., 2008).
Basic Molecular Analyses
For RT-PCR, total RNA was extracted using an RNeasy plant mini kit (Qiagen), according to the manufacturer’s instructions. Downstream DNAse I treatment and RT steps were conducted as described previously (Kovacheva et al., 2005). Amplifications by PCR were performed using the following primers: atTOC75-III, 5′-CGT ATC TGG ATG GTG TTT ACA ATC-3′ and 5′-GGA ATT CTT AAT ACC TCT CTC CAA ATC GGA AGA AC-3′; AtOEP80, 5′-ATG CAT TGT CAC AAC GAT GA-3′ and 5′-TCT ACA TCC CTC TTC CCT TGA A-3′; eIF4E1 (eukaryotic translation initiation factor 4E1), 5′-AAA CAA TGG CGG TAG AAG ACA CTC-3′ and 5′-AAG ATT TGA GAG GTT TCA AGC GGT GTA AG-3′. Amplicons were resolved by agarose gel electrophoresis and visualized by staining with SYBR Safe (Invitrogen). Bands were quantified using ImageQuant (GE Healthcare) or Aida (Raytest) software.
Extraction of total cellular protein and immunoblotting were carried using standard procedures (Aronsson et al., 2003). Primary antibodies were all raised in rabbits, and have been described previously (Aronsson et al., 2003; Kubis et al., 2003; Bédard et al., 2007).
Transmission Electron Microscopy
For the analysis of the RNAi lines, plants were grown in vitro, either in the dark for 5 d or in the light for 10 d, on MS-dexamethasone medium and under standard conditions. For the analysis of the toc75-III-3 mutant, plants were grown in vitro for 10 d or on soil for 28 d, in the light and under standard conditions. Whole dark-grown plants were harvested for fixation under a green safe light. The cotyledons and the roots of the light-grown seedlings were also harvested, as were developmentally similar leaves (seventh or eighth true leaf) from the mature plants. The samples were analyzed by transmission electron microscopy as described previously (Aronsson et al., 2007) with some minor modifications: En-bloc staining with uranyl acetate was omitted; low viscosity resin (Agar Scientific) was used in place of Spurr’s resin. Midlamina cross sections of cotyledons or leaves were analyzed; for roots, a region approximately 5 mm back from the tip was sectioned. The length and width of each chloroplast or root plastid was measured using Adobe Photoshop software, using the measure tool; these values were then used to estimate organelle cross-sectional area using the following formula: π × 0.25 × length × width. Etioplast cross-sectional area was determined by counting pixel numbers using the lasso tool within Photoshop. In both cases, reference to an internal standard was used to convert the values into suitable units of length or area.
Chloroplast Isolation and Protein Import Assays
Arabidopsis cDNAs encoding L11 and RbcS1A (both in the pBluescript vector) were transcribed and translated using a coupled system containing rabbit reticulocyte lysate (TNT T7 Quick for PCR DNA; Promega), [35S]-Met, and T7 polymerase (Kovacheva et al., 2005). Chloroplast isolation and protein import assays were conducted using standard procedures, as described previously (Aronsson and Jarvis, 2002; Kubis et al., 2008).
The Arabidopsis Genome Initiative locus numbers for the major genes discussed in this article are as follows: atTOC75-III (At3g46740) and AtOEP80 (At5g19620). Sequence data can be found in the GenBank/EMBL data libraries under accession numbers NM_114541 and NM_121967, respectively.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Multiple independent transgenic lines for each RNAi construct share a characteristic visible phenotype.
Supplemental Figure S2. Basic characterization of the pOpOff2 control lines.
Supplemental Figure S3. Detailed characterization of photosynthetic and nonphotosynthetic growth in the pOpOff2 control lines.
Supplemental Figure S4. DIGE analysis of the atToc75-III↓↓ number 6 chloroplast proteome.
Supplemental Figure S5. Parameters of nonphotosynthetic growth in the toc75-III-3 (mar1) mutant.
Supplemental Table S1. Segregation of the T-DNA-associated hygromycin-resistance and chlorotic phenotypes in the T2 generation of selected RNAi lines.
Supplemental Appendix S1. Analysis of control pOpOff2 transgenic lines.
Acknowledgments
We thank Ramesh Patel for technical assistance and for introgressing toc75-III-3 into the Columbia-0 ecotype, Tony Wardle for further technical support, and Renata Feret and Michael Deery for assistance with the DIGE analysis. We are grateful to Natalie Allcock and Stefan Hyman for electron microscopy carried out within the Electron Microscopy Laboratory, University of Leicester. We also thank Chris Helliwell (CSIRO Plant Industry, Canberra, Australia) for providing the pOpOff2 vectors, Ian Moore (Oxford University, UK) for providing the pOpOff2(hyg)::LUC control transgenic line, Patrick Masson (University of Wisconsin, Madison, WI) for providing the toc75-III-3/mar1 mutant seeds, and Iris Meier (Ohio State University, Columbus, OH) for providing the pOpOff2(hyg)::RG(Δ) control transgenic line. We thank Marc Boutry (Université Catholique de Louvain, Belgium; PMA2), Bernhard Grimm (Humboldt University Berlin, Germany; CPO), Neil Hoffman (LHCP, OE33), Kentaro Inoue (University of California, Davis, CA; atToc75-III), Kenneth Keegstra (Michigan State University, East Lansing, MI; Hsp93, Tic110), Felix Kessler (Neuchâtel University, Switzerland; atToc159), Henrik Scheller (Lawrence Berkeley National Laboratory, Berkeley, CA; FNR), Jürgen Soll (Ludwig Maximilian University, Munich; Tic40, psToc75-V/OEP80), and Glenn Thorlby (SFR2) for providing antibodies.
References
- Agne B, Andrès C, Montandon C, Christ B, Ertan A, Jung F, Infanger S, Bischof S, Baginsky S, Kessler F. (2010) The acidic A-domain of Arabidopsis TOC159 occurs as a hyperphosphorylated protein. Plant Physiol 153: 1016–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Aronsson H, Boij P, Patel R, Wardle A, Töpel M, Jarvis P. (2007) Toc64/OEP64 is not essential for the efficient import of proteins into chloroplasts in Arabidopsis thaliana. Plant J 52: 53–68 [DOI] [PubMed] [Google Scholar]
- Aronsson H, Combe J, Jarvis P. (2003) Unusual nucleotide-binding properties of the chloroplast protein import receptor, atToc33. FEBS Lett 544: 79–85 [DOI] [PubMed] [Google Scholar]
- Aronsson H, Jarvis P. (2002) A simple method for isolating import-competent Arabidopsis chloroplasts. FEBS Lett 529: 215–220 [DOI] [PubMed] [Google Scholar]
- Baldwin A, Wardle A, Patel R, Dudley P, Park SK, Twell D, Inoue K, Jarvis P. (2005) A molecular-genetic study of the Arabidopsis Toc75 gene family. Plant Physiol 138: 715–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer J, Chen K, Hiltbunner A, Wehrli E, Eugster M, Schnell D, Kessler F. (2000) The major protein import receptor of plastids is essential for chloroplast biogenesis. Nature 403: 203–207 [DOI] [PubMed] [Google Scholar]
- Bédard J, Kubis S, Bimanadham S, Jarvis P. (2007) Functional similarity between the chloroplast translocon component, Tic40, and the human co-chaperone, Hsp70-interacting protein (Hip). J Biol Chem 282: 21404–21414 [DOI] [PubMed] [Google Scholar]
- Bolender N, Sickmann A, Wagner R, Meisinger C, Pfanner N. (2008) Multiple pathways for sorting mitochondrial precursor proteins. EMBO Rep 9: 42–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos MP, Robert V, Tommassen J. (2007) Functioning of outer membrane protein assembly factor Omp85 requires a single POTRA domain. EMBO Rep 8: 1149–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredemeier R, Schlegel T, Ertel F, Vojta A, Borissenko L, Bohnsack MT, Groll M, von Haeseler A, Schleiff E. (2007) Functional and phylogenetic properties of the pore-forming beta-barrel transporters of the Omp85 family. J Biol Chem 282: 1882–1890 [DOI] [PubMed] [Google Scholar]
- Chiu CC, Li HM. (2008) Tic40 is important for reinsertion of proteins from the chloroplast stroma into the inner membrane. Plant J 56: 793–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou ML, Fitzpatrick LM, Tu SL, Budziszewski G, Potter-Lewis S, Akita M, Levin JZ, Keegstra K, Li HM. (2003) Tic40, a membrane-anchored co-chaperone homolog in the chloroplast protein translocon. EMBO J 22: 2970–2980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Constan D, Patel R, Keegstra K, Jarvis P. (2004) An outer envelope membrane component of the plastid protein import apparatus plays an essential role in Arabidopsis. Plant J 38: 93–106 [DOI] [PubMed] [Google Scholar]
- Dávila-Aponte JA, Inoue K, Keegstra K. (2003) Two chloroplastic protein translocation components, Tic110 and Toc75, are conserved in different plastid types from multiple plant species. Plant Mol Biol 51: 175–181 [DOI] [PubMed] [Google Scholar]
- Eckart K, Eichacker L, Sohrt K, Schleiff E, Heins L, Soll J. (2002) A Toc75-like protein import channel is abundant in chloroplasts. EMBO Rep 3: 557–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ertel F, Mirus O, Bredemeier R, Moslavac S, Becker T, Schleiff E. (2005) The evolutionarily related beta-barrel polypeptide transporters from Pisum sativum and Nostoc PCC7120 contain two distinct functional domains. J Biol Chem 280: 28281–28289 [DOI] [PubMed] [Google Scholar]
- Fourrier N, Bédard J, Lopez-Juez E, Barbrook A, Bowyer J, Jarvis P, Warren G, Thorlby G. (2008) A role for SENSITIVE TO FREEZING2 in protecting chloroplasts against freeze-induced damage in Arabidopsis. Plant J 55: 734–745 [DOI] [PubMed] [Google Scholar]
- Gentle IE, Burri L, Lithgow T. (2005) Molecular architecture and function of the Omp85 family of proteins. Mol Microbiol 58: 1216–1225 [DOI] [PubMed] [Google Scholar]
- Goldberg RB, de Paiva G, Yadegari R. (1994) Plant embryogenesis: zygote to seed. Science 266: 605–614 [DOI] [PubMed] [Google Scholar]
- Habib SJ, Waizenegger T, Niewienda A, Paschen SA, Neupert W, Rapaport D. (2007) The N-terminal domain of Tob55 has a receptor-like function in the biogenesis of mitochondrial beta-barrel proteins. J Cell Biol 176: 77–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill K, Model K, Ryan MT, Dietmeier K, Martin F, Wagner R, Pfanner N. (1998) Tom40 forms the hydrophilic channel of the mitochondrial import pore for preproteins [see comment]. Nature 395: 516–521 [DOI] [PubMed] [Google Scholar]
- Hinnah SC, Wagner R, Sveshnikova N, Harrer R, Soll J. (2002) The chloroplast protein import channel Toc75: pore properties and interaction with transit peptides. Biophys J 83: 899–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann NR, Theg SM. (2005) Chloroplast outer membrane protein targeting and insertion. Trends Plant Sci 10: 450–457 [DOI] [PubMed] [Google Scholar]
- Hsu SC, Patel R, Bédard J, Jarvis P, Inoue K. (2008) Two distinct Omp85 paralogs in the chloroplast outer envelope membrane are essential for embryogenesis in Arabidopsis thaliana. Plant Signal Behav 3: 1134–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hust B, Gutensohn M. (2006) Deletion of core components of the plastid protein import machinery causes differential arrest of embryo development in Arabidopsis thaliana. Plant Biol (Stuttg) 8: 18–30 [DOI] [PubMed] [Google Scholar]
- Inaba T, Schnell DJ. (2008) Protein trafficking to plastids: one theme, many variations. Biochem J 413: 15–28 [DOI] [PubMed] [Google Scholar]
- Inoue K, Potter D. (2004) The chloroplastic protein translocation channel Toc75 and its paralog OEP80 represent two distinct protein families and are targeted to the chloroplastic outer envelope by different mechanisms. Plant J 39: 354–365 [DOI] [PubMed] [Google Scholar]
- Ivanova Y, Smith MD, Chen K, Schnell DJ. (2004) Members of the Toc159 import receptor family represent distinct pathways for protein targeting to plastids. Mol Biol Cell 15: 3379–3392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson-Constan D, Keegstra K. (2001) Arabidopsis genes encoding components of the chloroplastic protein import apparatus. Plant Physiol 125: 1567–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis P. (2008) Targeting of nucleus-encoded proteins to chloroplasts in plants. New Phytol 179: 257–285 [DOI] [PubMed] [Google Scholar]
- Jarvis P, Chen LJ, Li H, Peto CA, Fankhauser C, Chory J. (1998) An Arabidopsis mutant defective in the plastid general protein import apparatus. Science 282: 100–103 [DOI] [PubMed] [Google Scholar]
- Kessler F, Schnell DJ. (2006) The function and diversity of plastid protein import pathways: a multilane GTPase highway into plastids. Traffic 7: 248–257 [DOI] [PubMed] [Google Scholar]
- Kikuchi S, Hirohashi T, Nakai M. (2006) Characterization of the preprotein translocon at the outer envelope membrane of chloroplasts by blue native PAGE. Plant Cell Physiol 47: 363–371 [DOI] [PubMed] [Google Scholar]
- Kouranov A, Chen X, Fuks B, Schnell DJ. (1998) Tic20 and Tic22 are new components of the protein import apparatus at the chloroplast inner envelope membrane. J Cell Biol 143: 991–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacheva S, Bédard J, Patel R, Dudley P, Twell D, Ríos G, Koncz C, Jarvis P. (2005) In vivo studies on the roles of Tic110, Tic40 and Hsp93 during chloroplast protein import. Plant J 41: 412–428 [DOI] [PubMed] [Google Scholar]
- Kubis S, Baldwin A, Patel R, Razzaq A, Dupree P, Lilley K, Kurth J, Leister D, Jarvis P. (2003) The Arabidopsis ppi1 mutant is specifically defective in the expression, chloroplast import, and accumulation of photosynthetic proteins. Plant Cell 15: 1859–1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubis S, Patel R, Combe J, Bédard J, Kovacheva S, Lilley K, Biehl A, Leister D, Ríos G, Koncz C, et al. (2004) Functional specialization amongst the Arabidopsis Toc159 family of chloroplast protein import receptors. Plant Cell 16: 2059–2077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubis SE, Lilley KS, Jarvis P. (2008) Isolation and preparation of chloroplasts from Arabidopsis thaliana plants. Methods Mol Biol 425: 171–186 [DOI] [PubMed] [Google Scholar]
- Meyer U, Köllner B, Willenbrink J, Krause GHM. (1997) Physiological changes on agricultural crops induced by different ambient ozone exposure regimes. I. Effects on photosynthesis and assimilate allocation in spring wheat. New Phytol 136: 645–652 [DOI] [PubMed] [Google Scholar]
- Patel R, Hsu SC, Bédard J, Inoue K, Jarvis P. (2008) The Omp85-related chloroplast outer envelope protein OEP80 is essential for viability in Arabidopsis. Plant Physiol 148: 235–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Pulido L, Devos D, Genevrois S, Vicente M, Valencia A. (2003) POTRA: a conserved domain in the FtsQ family and a class of beta-barrel outer membrane proteins. Trends Biochem Sci 28: 523–526 [DOI] [PubMed] [Google Scholar]
- Schleiff E, Soll J. (2005) Membrane protein insertion: mixing eukaryotic and prokaryotic concepts. EMBO Rep 6: 1023–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleiff E, Soll J, Küchler M, Kühlbrandt W, Harrer R. (2003) Characterization of the translocon of the outer envelope of chloroplasts. J Cell Biol 160: 541–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell DJ, Blobel G, Keegstra K, Kessler F, Ko K, Soll J. (1997) A consensus nomenclature for the protein-import components of the chloroplast envelope. Trends Cell Biol 7: 303–304 [DOI] [PubMed] [Google Scholar]
- Schnell DJ, Kessler F, Blobel G. (1994) Isolation of components of the chloroplast protein import machinery. Science 266: 1007–1012 [DOI] [PubMed] [Google Scholar]
- Smith MD. (2006) Protein import into chloroplasts: an ever-evolving story. Can J Bot 84: 531–542 [Google Scholar]
- Soll J, Schleiff E. (2004) Protein import into chloroplasts. Nat Rev Mol Cell Biol 5: 198–208 [DOI] [PubMed] [Google Scholar]
- Stanga JP, Boonsirichai K, Sedbrook JC, Otegui MS, Masson PH. (2009) A role for the TOC complex in Arabidopsis root gravitropism. Plant Physiol 149: 1896–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summer EJ, Cline K. (1999) Red bell pepper chromoplasts exhibit in vitro import competency and membrane targeting of passenger proteins from the thylakoidal sec and DeltapH pathways but not the chloroplast signal recognition particle pathway. Plant Physiol 119: 575–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sveshnikova N, Grimm R, Soll J, Schleiff E. (2000) Topology studies of the chloroplast protein import channel Toc75. Biol Chem 381: 687–693 [DOI] [PubMed] [Google Scholar]
- Tranel PJ, Froehlich J, Goyal A, Keegstra K. (1995) A component of the chloroplastic protein import apparatus is targeted to the outer envelope membrane via a novel pathway. EMBO J 14: 2436–2446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu SL, Chen LJ, Smith MD, Su YS, Schnell DJ, Li HM. (2004) Import pathways of chloroplast interior proteins and the outer-membrane protein OEP14 converge at Toc75. Plant Cell 16: 2078–2088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallas TR, Smith MD, Sanchez-Nieto S, Schnell DJ. (2003) The roles of toc34 and toc75 in targeting the toc159 preprotein receptor to chloroplasts. J Biol Chem 278: 44289–44297 [DOI] [PubMed] [Google Scholar]
- Wielopolska A, Townley H, Moore I, Waterhouse P, Helliwell C. (2005) A high-throughput inducible RNAi vector for plants. Plant Biotechnol J 3: 583–590 [DOI] [PubMed] [Google Scholar]