Abstract
The strong regulation of plant carbon allocation and growth by trehalose metabolism is important for our understanding of the mechanisms that determine growth and yield, with obvious applications in crop improvement. To gain further insight on the growth arrest by trehalose feeding, we first established that starch-deficient seedlings of the plastidic phosphoglucomutase1 mutant were similarly affected as the wild type on trehalose. Starch accumulation in the source cotyledons, therefore, did not cause starvation and consequent growth arrest in the growing zones. We then screened the FOX collection of Arabidopsis (Arabidopsis thaliana) expressing full-length cDNAs for seedling resistance to 100 mm trehalose. Three independent transgenic lines were identified with dominant segregation of the trehalose resistance trait that overexpress the bZIP11 (for basic region/leucine zipper motif) transcription factor. The resistance of these lines to trehalose could not be explained simply through enhanced trehalase activity or through inhibition of bZIP11 translation. Instead, trehalose-6-phosphate (T6P) accumulation was much increased in bZIP11-overexpressing lines, suggesting that these lines may be insensitive to the effects of T6P. T6P is known to inhibit the central stress-integrating kinase SnRK1 (KIN10) activity. We confirmed that this holds true in extracts from seedlings grown on trehalose, then showed that two independent transgenic lines overexpressing KIN10 were insensitive to trehalose. Moreover, the expression of marker genes known to be jointly controlled by SnRK1 activity and bZIP11 was consistent with low SnRK1 or bZIP11 activity in seedlings on trehalose. These results reveal an astonishing case of primary metabolite control over growth by way of the SnRK1 signaling pathway involving T6P, SnRK1, and bZIP11.
Trehalose (α,α-1,1-linked d-glucopyranosyl d-glucopyranoside) inhibits growth and is toxic when fed to seedlings of the dodder vine Cuscuta reflexa (Veluthambi et al., 1982a, 1982b) or Arabidopsis (Arabidopsis thaliana; Wingler et al., 2000; Fritzius et al., 2001; Schluepmann et al., 2004; Ramon et al., 2007; Schluepmann and Paul, 2009). This effect is somewhat surprising, as trehalose is a common sugar and synthesized in many organisms at high concentrations, functioning as a carbon source and stress protection compound. In plants, the trehalose pathway is ubiquitous and indispensable, but typically trehalose does not accumulate to high levels. A major function of the pathway in sugar signaling and the regulation of growth and development has been elucidated in plants in recent years (Eastmond et al., 2002; Schluepmann et al., 2003; Satoh-Nagasawa et al., 2006; Zhang et al., 2009; Paul et al., 2010). The impact of trehalose feeding on growth is most likely related to this signaling function (Schluepmann et al., 2004). However, the precise mode of action of growth inhibition by trehalose is not known.
A striking feature of Arabidopsis seedlings grown on trehalose is starch accumulation in the source tissues, while no starch accumulates in the tip of the roots in the columella, representing a reversal of carbon allocation. Starch accumulation in the cotyledons can be explained through up-regulation of ADPglucose pyrophosphorylase (AGPase) transcriptionally (Wingler et al., 2000) and through AGPase redox activation (Kolbe et al., 2005) and also by inhibition of starch degradation (Ramon et al., 2007). However, interestingly, this effect is found only in source tissues such as cotyledons and not in sink tissues such as the columella of root tips, which no longer accumulate starch and appear to be starving due to a lack of sufficient carbohydrate (Wingler et al., 2000). Feeding metabolizable sugar in combination with trehalose rescues growth, and it would appear that the primary effect of trehalose on growth is related to the utilization of sugar. This is also a feature of trehalose toxicity in cut dodder shoots, where trehalose and Suc uptake and accumulation were studied (Veluthambi et al., 1982a, 1982b). Radiolabeled trehalose accumulated evenly throughout the shoot. Trehalose accumulation is associated with a decrease in the radiolabeled Suc accumulation and starch content of shoot tips as well as growth arrest in the growing zone of the shoot tips (Veluthambi et al., 1982a).
Growth arrest of Arabidopsis seedlings on medium with 100 mm trehalose was previously attributed to trehalose-6-phosphate (T6P) accumulation under these particular conditions (Schluepmann et al., 2004). T6P has emerged as a powerful signal molecule in plants, a target of which has been identified as SnRK1 of the SNF1/AMPK group of protein kinases (Zhang et al., 2009; Paul et al., 2010; Martinez-Barajas et al., 2011). SNF1-related protein kinases perform a fundamental role in the physiological response of cells to energy limitation and starvation of carbon source through the regulation of pathways and processes involved in metabolism, growth, and development (Hardie, 2007; Polge and Thomas, 2007; Halford and Hey, 2009). SnRK1 integrates stress, sugar, and specific developmental signals (Baena-González et al., 2007; Baena-González and Sheen, 2008; Baena-González, 2010). T6P inhibits SnRK1 from all plant tissues so far tested except for mature leaves. This can be explained through the requirement of an intermediary factor present in growing tissues but not in mature leaves and is consistent with the view that T6P promotes anabolic processes associated with growing tissues (Zhang et al., 2009). While T6P promotes growth on Suc (Zhang et al., 2009; Paul et al., 2010), T6P inhibits growth on trehalose (Schluepmann et al., 2004). SnRK1 activity was proposed to be inactivated by sugars (Baena-González et al., 2007), consistent with the observation that Suc feeding causes a rise in T6P concentrations (Schluepmann et al., 2003; Lunn et al., 2006), which subsequently inhibit SnRK1 activity, promoting growth. The link between SnRK1 and T6P is significant, as it establishes T6P as a signaling metabolite integrating carbon metabolism with the activity of enzymes and gene expression reprogramming controlled by this central protein kinase (Zhang et al., 2009; Paul et al., 2010). It is quite possible that growth arrest on trehalose is mediated through SnRK1.
Expression of Asparagine Synthase1 (ASN1) has previously been used as a reporter of SnRK1 activity; the expression of ASN1 increases when the catalytic subunit of SnRK1, KIN10, is overexpressed (Baena-González et al., 2007). The promoter of ASN1 was shown to contain a G-box sequence known to be bound by bZIP (for basic region/Leu zipper motif) called the G-box binding factor transcription factor. Coexpression of several bZIP transcription factors with KIN10 was shown to potentiate the expression of ASN1 (Baena-González et al., 2007). bZIP transcription factors regulate a range of processes in growth and development in relation to the environment, including stress responses (Jakoby et al., 2002). A model was proposed by which several stress signals including hypoxia and darkness converge through SnRK1 signaling and are coordinated in part by the bZIP transcription factors 54, 18, 63, 1, 38, and 11 (Baena-González et al., 2007). It is not known whether these bZIP proteins are phosphorylation targets of SnRK1 and/or if bZIP gene expression is controlled by SnRK1 directly. bZIP54, -18, -63, -1, and -38 gene expression is induced while bZIP11 is repressed when KIN10 is increased (Supplemental Fig. S1A; Baena-González et al., 2007) or when seedlings are starving; the opposite is true when seedlings have increased carbon access through sugar or CO2 feeding (Supplemental Fig. S1B). The potentiation of KIN10-induced ASN1 expression when KIN10 and bZIP11 are coexpressed suggests that the SnRK1 and bZIP11 controls of ASN1 expression interact.
The changes in gene expression after KIN10 overexpression are comparable to those obtained during carbon starvation regimes and opposite those under high supply of Glc, Suc, or CO2 (Supplemental Fig. S1B). The S1 class of bZIP transcription factors consists of bZIP1, -2, -11, -44, and -53 and appears to be expressed specifically in sinks such as young leaves, anthers, and seeds (Rook et al., 1998; Weltmeier et al., 2009); their expression is also affected by abiotic stresses in a tissue-specific manner (Kilian et al., 2007). S1-class bZIPs are generally regulated at the posttranscriptional level by Suc-induced repression of translation (SIRT) at a conserved upstream open reading frame (uORF) located in the 5′ region of the mRNA (Wiese et al., 2004; Hanson et al., 2008; Weltmeier et al., 2009). bZIP11 has been proposed to alter nitrogen metabolism by controlling the expression of ASN1 and Proline Dehydrogenase (PDH; Hanson et al., 2008), and changes in bZIP1 alter the transcriptional response to the carbon-nitrogen ratio (Kang et al., 2010; Obertello et al., 2010). While T6P and SnRK1 as well as bZIP11 and SnRK1 have been linked, a connection between all three components in the regulation of growth has not previously been established.
The strong regulation of carbon allocation and growth by the trehalose pathway is important for our understanding of mechanisms that determine plant growth and yield, with obvious application in crop improvement. To gain further insight on the growth arrest by trehalose feeding, we first established that starch-deficient seedlings of the plastidic phosphoglucomutase1 (pgm1) mutant are similarly affected as the wild type on trehalose. Starch accumulation in the cotyledons, the source tissue, therefore, does not cause starvation and consequent growth arrest in the growing zones. We then screened the FOX collection of Arabidopsis expressing full-length cDNAs behind the cauliflower mosaic virus 35S promoter (Ichikawa et al., 2006) for seedling resistance to 100 mm trehalose. Three independent transgenic lines from different pools of the collection were identified with dominant segregation of the trehalose resistance trait that overexpressed the bZIP11 transcription factor. The resistance of these lines to trehalose could not be explained through enhanced trehalase activity or through inhibition of bZIP11 translation. Instead, T6P accumulation was much increased in bZIP11-overexpressing lines, suggesting that these lines may be insensitive to the effects of T6P. T6P is known to inhibit SnRK1 activity, and we confirmed that this holds true in extracts from seedlings grown on trehalose, then we showed that two independent transgenic lines overexpressing KIN10 were insensitive to trehalose. Moreover, expression of a set of marker genes known to be jointly controlled by SnRK1 activity and bZIP11 was found to be consistent with low SnRK1 or bZIP11 activity in seedlings on trehalose. These results were consistent with the existence of a growth-regulating pathway involving T6P, SnRK1 activity, and bZIP11, regulating growth in the growing zones of Arabidopsis seedlings.
RESULTS
Primary and Secondary Screening of the FOX Collection on Trehalose
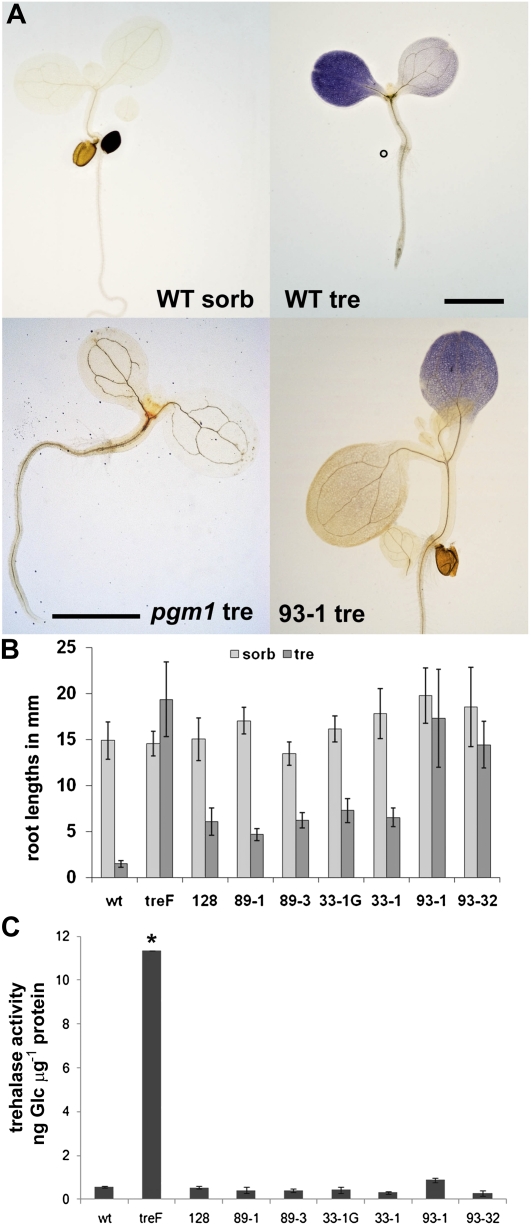
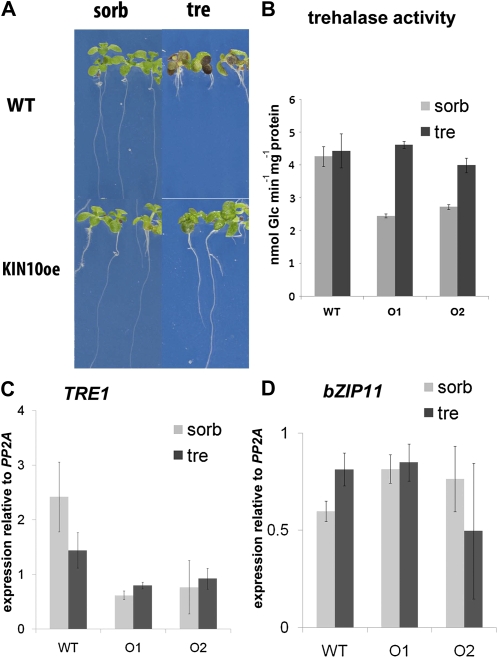
Trehalose at 100 mm in half-strength Murashige and Skoog (MS) medium inhibited the growth of Arabidopsis seedlings of all accessions thus far tested. Seedlings germinated on this medium developed short roots less than 3 mm long after 14 d, and the leaf primordia did not extend into leaves (Fig. 1, A, WT tre, and B wt) compared with normal development on 100 mm sorbitol osmoticum control (Fig. 1, A, WT sorb, and B, wt). Starch accumulated in large amounts in one or both cotyledons in seedlings grown on trehalose (Fig. 1A, WT tre); such massive accumulation of carbon as starch in the source tissues of seedlings could possibly cause starvation in the growing zones of seedlings. The starchless pgm1 mutant (Caspar et al., 1985), however, was inhibited on trehalose in a similar manner to the wild type (Fig. 1A, pgm1 tre). Thus, starch accumulation in the source tissues of seedlings, the cotyledons, does not cause growth arrest on trehalose.
Figure 1.
Carbon allocation, growth, and trehalase activity of seedlings on 100 mm trehalose. Seedlings were grown under long-day conditions for 14 d on medium with 100 mm sorbitol osmoticum control (sorb) or trehalose (tre). A, Starch staining. Seedlings were harvested at midday, stained with Lugol, and mounted in chloral hydrate. WT, Wild-type Col-0 seedlings; pgm1, seedlings lacking Plastidic Phosphoglucomutase1 (Caspar et al., 1985); 93-1, seedlings from FOX line 93-1. Bars = 3 mm. B, Root lengths. Average root lengths from more than 20 seedlings of the different genotypes with sd are shown. treF, Seedlings overexpressing E. coli trehalase treF; 128, 89-1, 89-3, 33-1G, 33-1, 93-1, and 93-32, lines from the FOX collection of FOX pools 128, 89, 33, and 93. C, Trehalase activity in extracts of seedlings grown on trehalose for 14 d. The data are averages with sd of three independent extracts. * P < 0.050 by ANOVA.
To further understand the mechanism of growth arrest on trehalose using a non-hypothesis-driven approach, we used a genetic screen: seedlings from the Arabidopsis FOX collection (Ichikawa et al., 2006) were screened for growth on 100 mm trehalose. The entire collection was partitioned in 141 pools each containing 20 to 30 T1 seed from 100 independent transgenic lines; primary screening was carried out twice with 1,000 seeds per plate for all 141 pools. The germination frequency varied between 100% and as low as 30%, indicating that not all transgenic lines were assayed for their resistance to trehalose and thus that the screen did not test all the cDNAs of the FOX collection. T1 seedlings during the primary screening were chosen that displayed significantly longer roots than the wild type and leaf primordia (Fig. 1A, 93-1 tre). Secondary screening of T2 retained only lines where seedling roots after 14 d of growth on trehalose were at least 3-fold longer than the wild type and of the same length as the wild type on osmoticum control (Fig. 1B). In the case of lines 93-1 and 93-32 from pool 93, seedling root lengths were nearly as long on trehalose as they were on sorbitol (Fig. 1B). Seedlings from line 70 were assayed at a later stage because trehalose-resistant seedlings obtained from pool 70 were difficult to grow, and after rescreening only one plant grew to set seed. The T2 seedlings from lines 93-3, 33-1, and 70 were 100% resistant to trehalose, while those from 93-1 were not and thus were still segregating.
An obvious cause of resistance to trehalose would be increased trehalase activity. Expression of the Escherichia coli cytosolic trehalase in Arabidopsis seedlings resulted in seedlings with high trehalase activity that thrive on trehalose (Fig. 1, B, treF, and C, treF). Therefore, activity of the Arabidopsis trehalase (TRE1) was assayed in lines identified during the secondary screening. We tested several membrane and cell wall preparations from wild-type seedlings, but trehalase activities in these fractions were below detection levels. Instead, activity was readily detected in the soluble fraction and was not significantly different in FOX lines compared with the wild type, except for a small increase from pool 93 (ANOVA, P = 0.052). These activities were 10- to 20-fold lower compared with seedlings expressing E. coli trehalase treF (Fig. 1C).
Overexpression of the Full-Length cDNA of bZIP11 Is Linked with Resistance to Trehalose
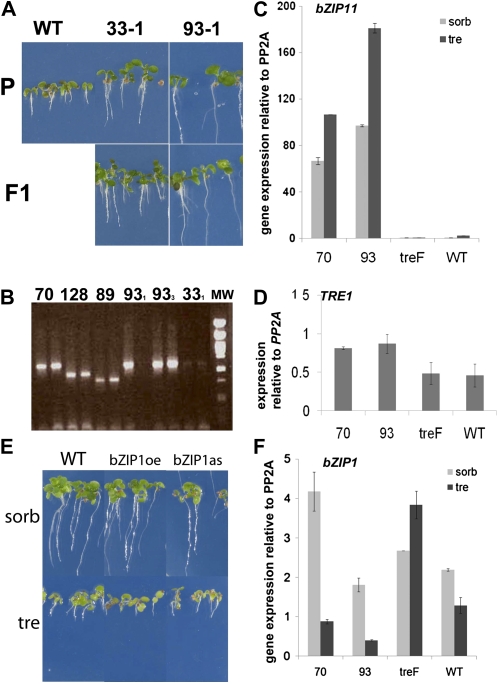
Seedlings from the FOX lines were backcrossed into the wild type, and resistance to trehalose of F1 generation seedlings was evaluated for each line. F1 seedlings of lines 33-1, 93-1, and 70 were resistant to trehalose, indicating that the trehalose resistance in these lines segregates as a dominant trait (Fig. 2A, 33-1 and 93-1). The cDNA present in the T-DNAs in the heterozygous F1, therefore, caused trehalose resistance. The cDNA was then amplified using primers on the flanking sequences of the FOX vector for two different plants from each line (Fig. 2B). Fragments obtained from plants of the lines 33-1, 33-3, 70, 93-1, and 93-3 were of the same length. Subsequent cloning and sequencing revealed that these fragments all contained the full-length cDNA of bZIP11. The lines were from three different pools, pools 33, 70, and 93. Therefore, the lines represented three independent transgenic events selected through screening that linked the presence of the bZIP11 cDNA with trehalose resistance.
Figure 2.
Characterization of the independent FOX lines expressing bZIP11 cDNA from pools 33, 93, and 70. A, Trehalose resistance is a dominant trait, as shown for lines 33-1 and 93-1. P, Seedlings from the parental lines: WT, the wild type; 33-1, FOX line 33-1; 93-1, FOX line 93-1. F1 represents the first generation from FOX line crosses with the wild type. B, PCR amplification of the FOX cDNA using DNA template from plants of lines 70, 128, 89, 93-1, and 33-1. MW, Molecular weight marker λ Pst1. C, Expression of bZIP11 in 14-d-old seedlings from FOX lines 70 and 93, the line expressing the E. coli trehalase treF (treF), and wild-type Col-0. D, Expression of the TRE1 trehalase in the genotypes from C. E, Seedlings with altered expression of bZIP1 do not grow on medium with 100 m trehalose (tre) compared with sorbitol (sorb). bZIP1oe and bZIP1as, bZIP1-overexpressing and antisense lines, as described by Kang et al. (2010). F, Expression of bZIP1 in seedlings of FOX lines 70 and 93, the line expressing treF, and the wild type grown on medium with sorbitol or trehalose. Expression was determined by Q-PCR and is given relative to PP2A (At1g13320). Error bars represent sd of three replicates.
Expression of bZIP11 cDNA was tremendously increased in the FOX lines containing the bZIP11 cDNA compared with the wild type (Fig. 2C). In seedlings on trehalose, AtTRE1 expression was less than 2-fold increased compared with the wild type (Fig. 2D; ANOVA, P = 0.014) and very much less increased than bZIP11 expression. Overexpression or antisense expression of bZIP1 in lines previously characterized (Kang et al., 2010) did not yield resistance to trehalose, suggesting that the function of bZIP11 could not be replaced by the other S1-class bZIP, bZIP1 (Fig. 2E). bZIP1 expression was high in treF (E. coli trehalase) expressors on trehalose compared with the wild type or bZIP11 expressors (Fig. 2F). Even though trehalase activity determinations suggested that the mechanism of trehalose resistance in bZIP11 overexpressors differed from that in treF expressors, we still wondered if the in vitro assays of trehalase (Fig. 1C) were conclusive.
AtTRE1 Is Not Required for bZIP11-Mediated Trehalose Resistance
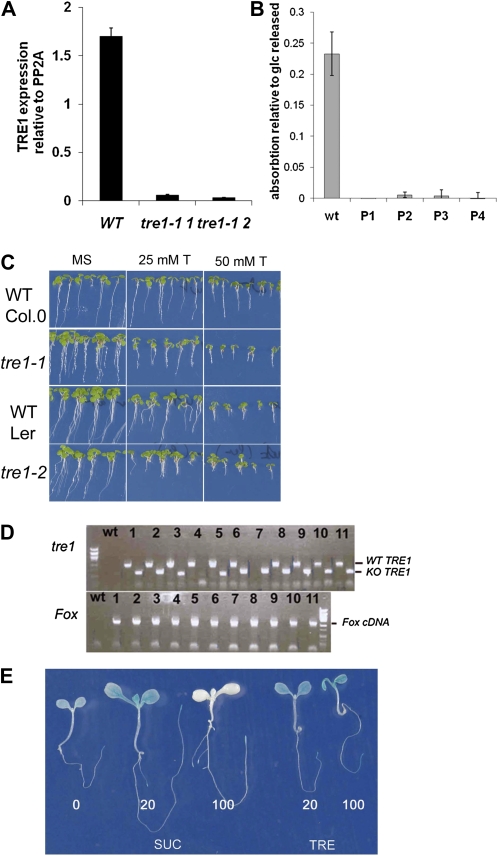
In further confirmation that TRE1 did not underlie resistance to trehalose in FOX lines expressing bZIP11, a tre1 knockout was characterized from the SALK collection (Salk 147073c; Alonso et al., 2003). PCR with primers on the left border of the T-DNA and in the start of the gene confirmed that the T-DNA was located 5′ to the ATG of TRE1. Sequencing of the insertion site revealed an additional 25-bp insertion in the 5′ region and confirmed the T-DNA insertion site. Gene expression quantification with quantitative (Q)-PCR further verified that the insertion causes a dramatic decrease in TRE1 mRNA (Fig. 3A). Trehalase activity in seedlings and flowers of the tre1-1 line was below detection (Fig. 3B). tre1-1 was more susceptible to 25 and 50 mm trehalose than the wild type from the Columbia (Col-0) accession (Fig. 3C), showing that TRE1 does contribute to the relative tolerance of wild-type seedlings to low levels of trehalose. The tre1-1 seedlings were similarly sensitive to trehalose as the previously characterized tre1-2 in the Landsberg erecta (Ler) accession (Fig. 3C; Vandesteene, 2009). The F1 seedlings from the cross of tre1-1 with the selected FOX lines were resistant to 100 mm trehalose (shown for the cross with FOX 93 in Supplemental Fig. S2). When analyzing the genotype of F2 seedlings with roots at least 3-fold longer than the wild type, F2 seedlings could be found that were homozygous for the tre1-1 T-DNA insertion (Fig. 3D). The trehalose resistance phenotype of seedlings homozygous for tre1-1 and overexpressing bZIP11 is shown in Supplemental Figure S2. Together, the data showed that AtTRE1 may contribute to but is not required for resistance to 100 mm trehalose and cannot explain the rescue of seedlings on trehalose by bZIP11.
Figure 3.
The roles of trehalase (TRE1) and of uORF2 in the FOX lines growing on trehalose. Error bars represent sd of three replicates. A, Expression of TRE1 in wild-type Col-0 (WT) and seedlings from two different plants of the tre1-1 line (Salk 147073c). B, Trehalase activity in flowers from the wild type and several plants from the tre1-1 line (P1–P4). C, tre1-1 and tre1-2 seedling growth compared with their respective wild types. Growth was on MS medium without (MS) or with 25 and 50 mm trehalose (25 mm T and 50 mm T). D, Genotype analysis of long root seedlings in the F2 generation of the cross 93-1 with the wild type. DNA from the wild type and 11 different seedlings (1–10) was used as the template. PCR was carried out to amplify the wild-type sequence of TRE1 (WT TRE1) or the T-DNA insertion at the TRE1 locus (KO TRE1) on the top agarose gel (tre1). PCR was also carried out to amplify the FOX cDNA on the bottom gel (Fox cDNA). E, Unlike on Suc, translational repression of bZIP11 does not occur on trehalose. Seedlings expressing the 5′ mRNA uORFs of the bZIP11 mRNA fused to the GUS gene were grown for 7 d on MS medium, transferred for 48 h to medium with Suc (SUC) or trehalose (TRE) at 0, 20, or 100 mm (0, 20, and 100), and then stained for GUS activity.
Translational Repression at uORF2 Does Not Occur on Trehalose, Unlike on Suc
bZIP11 cDNA is known to contain several uORFs in the 5′ region (Wiese et al., 2004; Weltmeier et al., 2009). uORF2 is conserved in the plant kingdom and found in all the S1-class bZIP mRNAs. uORF2 mediates SIRT of the bZIP11 protein. The mechanism may also inhibit bZIP11 protein synthesis in seedlings grown on trehalose. To test this possibility, seedlings of the UBQ10:5′UTR-GUS/GFP-I line (Wiese et al., 2004) that express the 5′ UTR-containing uORF2 in front of GUS/GFP were grown for 7 d and then transferred to medium supplemented with either Suc or trehalose for 48 h before GUS staining. GUS staining confirmed SIRT on Suc medium supplemented with 100 mm Suc throughout all tissues of the seedling (Fig. 3E, 100 SUC). On trehalose medium, however, repression of translation was not observed (Fig. 3E, 100 TRE). Therefore, we concluded that seedlings from the FOX lines translate bZIP11 unrestricted by SIRT when grown on trehalose.
bZIP11-Overexpressing Seedlings Accumulate T6P
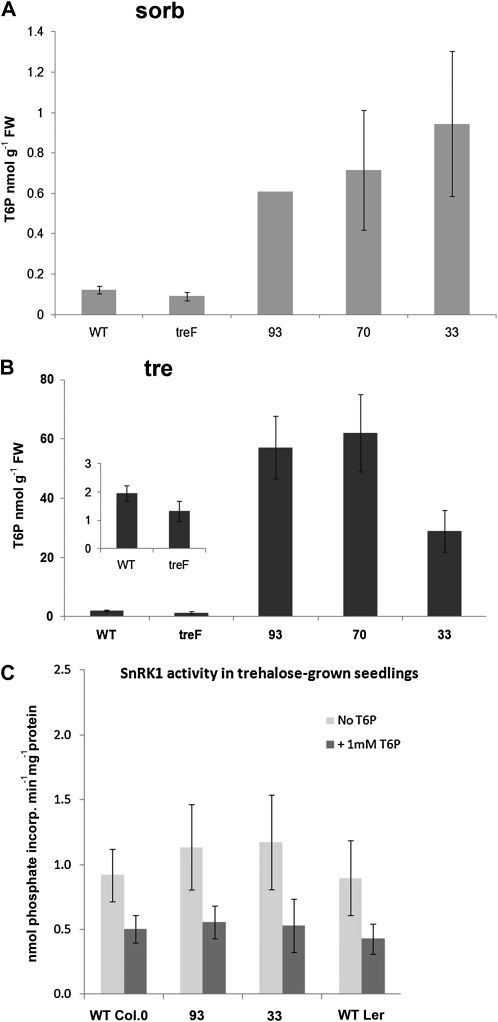
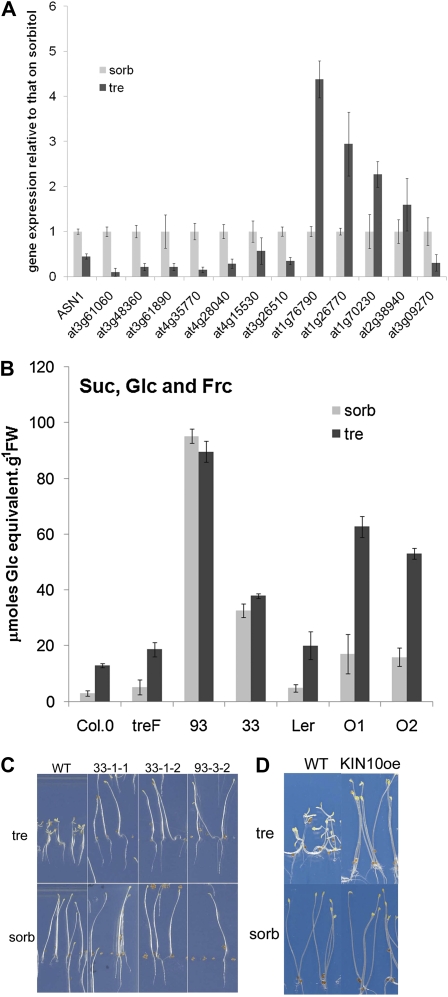
Wild-type seedlings growing on 100 mm trehalose accumulate T6P (Schluepmann et al., 2004), which has been linked to growth arrest under these particular conditions. We wondered whether the FOX line seedlings were insensitive to T6P or had just a low accumulation of T6P. T6P in seedlings grown for 14 d on osmoticum control was 0.6 to 1 nmol g−1 fresh weight in bZIP11 overexpressors, 6- to 10-fold higher than in the wild type (Fig. 4A). On trehalose medium, T6P accumulated in the wild type to 1.8 nmol g−1 fresh weight, as expected. T6P accumulation in bZIP11 overexpressors, however, was much higher, reaching over 60 nmol g−1 fresh weight (Fig. 4B). Therefore, the data showed that bZIP11 overexpression causes T6P accumulation.
Figure 4.
T6P accumulation and in vitro T6P inhibition of SnRK1 in seedlings grown on trehalose. WT, Wild-type Col-0; TreF, seedlings expressing E. coli trehalase treF; 93, 70, and 33, seedlings from FOX lines 93, 70, and 33, respectively; FW, fresh weight. A, Seedlings grown on osmoticum control for 14 d (100 mm sorbitol [sorb]). B, Seedlings grown on 100 mm trehalose for 14 d. C, SnRK1 activity assayed using the AMARA peptide in 14-d-old seedlings grown on 100 mm trehalose from wild-type accession Col-0, FOX lines 93 and 33, and wild-type accession Ler. Error bars represent se of three biological replicates.
In contrast, T6P levels in seedlings expressing E. coli trehalase treF were similar to the wild type under control growth conditions. On trehalose, T6P levels in seedlings expressing treF accumulated to 1.5 nmol g−1 fresh weight, a somewhat lesser extent than in the wild type, suggesting that when trehalase is expressed it is unlikely that all of the trehalose is cleaved. The different accumulation of T6P in bZIP11 compared with treF-expressing seedlings further supported the results, which showed that trehalose resistance by bZIP11 is independent of trehalase. Instead, the results point to the possibility that bZIP11 overexpression renders seedlings less susceptible to T6P accumulation.
T6P is known to inhibit seedling SnRK1 activity when seedlings are grown under normal conditions (Zhang et al., 2009). To test whether SnRK1 is inhibited by T6P in seedlings grown on trehalose, extracts from wild-type Arabidopsis seedlings grown in 100 mm trehalose were prepared, from which small-Mr compounds were removed using a desalting procedure. SnRK1 activity was then assayed in the absence or presence of 1 mm T6P. The results confirmed that SnRK1 from trehalose-grown seedlings was significantly inhibited by T6P (Fig. 4C). In addition, SnRK1 activity assayed from extracts of bZIP11-overexpressing seedlings (93 and 33) was similar to that of the wild type (Fig. 4C). Overexpression of bZIP11 thus did not cause large changes in SnRK1 activity. Therefore, we hypothesized that the T6P accumulation and inhibition of SnRK1 in seedlings expressing constitutively high levels of bZIP11 may be overcome, because bZIP11 likely controls a subset of genes responsive to SnRK1 activity. To test this possibility, we first needed to establish whether SnRK1 activity limits the growth of seedlings on trehalose.
KIN10-Overexpressing Seedlings Grew on 100 mm Trehalose
To test whether reduced SnRK1 activity underlies the growth inhibition on trehalose, two lines, O1 and O2, overexpressing KIN10 with high SnRK1 activity, previously characterized in detail, were used (Baena-González et al., 2007). Seedlings of both lines were resistant to 100 mm trehalose (Fig. 5A; only line O2 shown). SnRK1 activity, therefore, likely is limiting when wild-type seedlings are grown on trehalose. This result further indicated that T6P inhibition of SnRK1 likely underlies the growth arrest on trehalose and genetically confirmed previous results obtained by combining a biochemical assay with gene expression profiling (Zhang et al., 2009).
Figure 5.
KIN10-overexpressing seedlings grow on trehalose without increased trehalase or bZIP11 expression. Seedlings were grown on trehalose (tre) compared with osmoticum control (sorbitol [sorb]) for 14 d. A, Phenotype of the Ler wild type (WT) and KIN10 overexpression line O1. B, Trehalase activity in extracts from the Ler wild type and the lines overexpressing KIN10 (O1 and O2). C, TRE1 expression determined relative to PP2A by Q-PCR in the genotypes from B. D, bZIP11 expression. The levels are averages of three biological replicates, and error bars represent sd.
As with the bZIP11 overexpressors, KIN10 overexpression did not cause a significant induction of AtTRE1 activity (Fig. 5B) or of trehalase expression in seedling extracts (Fig. 5C). In addition, KIN10 overexpression did not affect the expression of bZIP11 (Fig. 5D); neither did bZIP11 overexpression affect SnRK1 activity (Fig. 4C) or KIN10 expression (Supplemental Fig. S3). Therefore, the data suggested that the SnRK1 and bZIP11 interaction described previously (Baena-González et al., 2007) in seedlings on trehalose is likely posttranscriptional, with SnRK1 activity changes altering bZIP11 subcellular localization or activity.
Regulation of Targets Common to bZIP11 and SnRK1
ASN1 is one of the known targets of SnRK1, and its expression was much reduced in the wild type grown on trehalose compared with sorbitol, thus confirming that SnRK1 activity is likely inhibited by T6P on trehalose (Fig. 6A, ASN1).
Figure 6.
Do KIN10 and bZIP11 act in the same pathway? A, Expression of targets common to KIN10 and bZIP11. Seedlings of the wild type were grown for 14 d on 100 mm either sorbitol (sorb) or trehalose (tre) and collected at midday. Expression was determined by Q-PCR relative to PP2A, then normalized to the level of expression on sorbitol. B, Soluble sugars Suc, Glc, and Fru in the seedlings with the genotype wild type (Col-0), treF expressors (treF), bZIP11 expressors from line 93 (93) and 33 (33), wild-type Ler (Ler), and KIN10-overexpressing lines O1 and O2. In A and B, levels are averages of three biological replicates, and error bars represent sd. FW, Fresh weight. C, Phenotypes of bZIP11-expressing seedlings on trehalose in continuous darkness. After 78 h at 4°C, seed were exposed to light and 22°C for 6 h, then grown for 14 d in continuous darkness on medium with 100 mm either sorbitol or trehalose. WT, Wild type Col-0; 33-1-1 and 33-1-2, seeds from two plants of FOX line 33-1; 93-3-2, seeds from FOX line 93. D, Phenotype of KIN10-expressing seedlings on trehalose in continuous darkness. Seeds were treated as in C. WT, Wild type Ler; KIN10oe, seeds from the O2 line (Baena-González et al., 2007).
It is known that bZIP11 potentiates the induction of DIN6 (ASN1) gene expression when SnRK1 activity is increased by KIN10 overexpression in protoplasts (Baena-González et al., 2007). We compared genes known to be controlled by a 6-h transient expression of KIN10 in protoplasts (511 up, 521 down; Baena-González et al., 2007) with those known to be controlled in whole seedlings by the nuclear transfer of bZIP11 after 2 h (167 up, 96 down; Hanson et al., 2008). Table I lists the genes jointly controlled by KIN10 and bZIP11. In spite of the different conditions used to identify the regulation of gene expression, a large proportion of the bZIP11-induced genes, 32 out of 167, were also induced by KIN10. These include four genes of the seven confirmed to be regulated by bZIP11 under its endogenous promoter: ASN1 (At3g47340), PRODH (At3g30775), BT2 (At3g48360), and PGPD14 (At5g22920). Nine genes from the 96 genes repressed by bZIP11 were also repressed by KIN10; only three genes from the total of 261 genes compared appeared differentially regulated by bZIP11 and KIN10. Results obtained from this comparison were thus consistent with the hypothesis that bZIP11 may be a target of SnRK1 regulation and may mediate a part of the output of SnRK1 signaling; they further showed that bZIP11 additionally regulates the transcription of a set of genes not included in regulation by SnRK1.
Table I. Genes jointly controlled by SnRK1 and bZIP11.
Two hundred sixty-one differentially expressed genes at 2 h after nuclear transfer of bZIP11 (Hanson et al., 2008) were compared with the 1,021 genes altered by 6 h of transient KIN10 expression in protoplasts (Baena-González et al., 2007). The commonly regulated genes are listed with The Arabidopsis Information Resource annotations (May 2011).
| Identifier | Annotation |
| Induced | |
| At1g02660 | α/β-Hydrolase superfamily protein; putative triglyceride lipase activity |
| At1g10070 | BCAT-2 chloroplast branched-chain amino acid aminotransferase |
| At1g18460 | α/β-Hydrolase superfamily putative lipase family |
| At1g32170 | XTR4 xyloglucan endotransglycosylase-related protein |
| At1g62510 | Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein |
| At1g64660 | MGL cytosolic Met γ-lyase |
| At2g25200 | Plant protein of unknown function (DUF868) |
| At2g30600 | BTB/POZ domain-containing protein; involved in cell adhesion |
| At2g32150 | Haloacid dehalogenase-like hydrolase (HAD) superfamily protein; nitrate responsive |
| At2g33380 | CALEOSIN3 calcium binding, induced by NaCl, abscisic acid, and desiccation |
| At2g36220 | Expressed protein |
| At2g38400 | AGT3 Ala:glyoxylate aminotransferase 2 homolog |
| At2g39570 | ACT domain-containing protein; functions in amino acid binding |
| At2g47770 | TSPO (outer membrane Trp-rich sensory protein)-related |
| At3g13450 | DIN4 branched-chain α-keto acid dehydrogenase E1 β |
| At3g26510 | Octicosapeptide/Phox/Bem1p family protein |
| At3g30775a | ERD5, PRO1, PRODH, Pro dehydrogenase |
| At3g47340a | ASN1 Gln-dependent Asn synthetase |
| At3g48360a | BT2 (AtBT-2) component of the TAC1-mediated telomerase activation pathway |
| At3g57520 | RS2, SIP2 raffinose-specific α-galactosidase |
| At3g61060 | PP2-A13, phloem protein 2-A13 |
| At3g61890 | HB-12, homeobox-Leu zipper protein HB-12 |
| At4g15530 | PPDK pyruvate,orthophosphate dikinase |
| At4g28040 | Nodulin MtN21/EamA-like transporter family protein (drug/metabolite transporter) |
| At4g35770 | SEN1 senescence-associated protein |
| At5g04040 | SDP1, triacylglycerol lipase that is involved in storage lipid breakdown |
| At5g18670 | BMY3, BAM9 glycosyl hydrolase family 14 (β-amylase) |
| At5g22920a | CHY-type/CTCHY-type/RING-type zinc finger protein |
| At5g49360 | BXL1, bifunctional β-d-xylosidase/α-l-arabinofuranosidase |
| At5g53590 | SAUR-like auxin-responsive protein family |
| At5g66170 | STR18, encodes a thiosulfate sulfurtransferase/rhodanase |
| At5g66650 | Protein of unknown function DUF607 |
| Repressed | |
| At1g26770 | EXP10 (expansins[α-expansin gene family]): expansin 10 |
| At1g64060 | RbohF(respiratory burst oxidase family, cytochrome b558-H+channel) |
| At1g69530 | EXP1 (α-expansin gene family) |
| At1g70230 | TBL27, trichome birefringence-like, plant-specific DUF231 |
| At1g76790 | IGMT5, indole glucosinolate O-methyl transferase 5 |
| At2g16660 | Major facilitator superfamily protein, endomembrane system |
| At2g38170 | CAX1, high-affinity vacuolar calcium antiporter |
| At2g38940 | PHT1;4, member of the Pht1 family of phosphate transporters |
| At3g09270 | GSTU8 glutathione transferase belonging to the τ-class of GSTs |
| Oppositely regulated | |
| At2g14170 | ALDH6B2, methylmalonate-semialdehyde dehydrogenase |
| At3g57040 | ARR9 response regulator A type; cytokinin signaling |
| At5g17760 | P-loop-containing nucleoside triphosphate hydrolases superfamily protein |
Also in the list of seven genes that are induced by bZIP11 under the control of its own promoter.
To test whether KIN10- and bZIP11-mediated rescue on trehalose may involve genes that are commonly controlled by both, we assayed the expression of a randomly chosen array of jointly induced (eight) or repressed (five) genes in wild-type seedlings grown on trehalose compared with sorbitol (Fig. 6A). All but one of the eight genes jointly induced by bZIP11 and KIN10 were significantly repressed on trehalose. Furthermore, three of the five jointly repressed genes were significantly induced on trehalose compared with sorbitol. We conclude that a large proportion of genes jointly regulated by KIN10 and bZIP11 respond on trehalose as if KIN10 or bZIP11 was less active.
To compare the downstream events in KIN10 and bZIP11 overexpression, the levels of soluble sugars (Glc, Fru, and Suc) were compared with those found in the wild type and in treF expressors (Fig. 6B). Similar results were found for hexoses and Suc, and data are presented as total soluble sugar. Soluble sugars accumulated in wild-type Col-0 or Ler seedlings grown on trehalose and in treF seedlings that were included for comparison. The data were consistent with carbon accumulation in source tissues, while sugars were not used for sink growth. Strikingly, bZIP11 overexpression lines 93 or 33 had 10 to 20 times higher soluble sugar than the wild type (Fig. 6B, 93 and 33) irrespective of the medium. Sugars were also elevated (3-fold compared with the wild type) in KIN10 overexpressors, and this increase was even more on trehalose (Fig. 6B, O1 and O2).
Arrest of hypocotyl elongation when wild-type seedlings were grown in continuous darkness was another effect of feeding trehalose (Fig. 6, C and D, WT tre). We thus concluded that trehalose feeding not only stopped the use of carbon fixed by photosynthesis but also of carbon from seedling reserves needed for the hypocotyl growth in the etiolation response. Overexpression of bZIP11 (Fig. 6C) or KIN10 (Fig. 6D) was able to suppress hypocotyl growth inhibition by trehalose in the dark. This observation suggested that KIN10 and bZIP11 rescue T6P inhibition of SnRK1 regardless of the source of carbon fueling the growth of sinks. Therefore, convergence of the pathways involving KIN10 and bZIP11 in the control of carbon utilization for growth is likely.
DISCUSSION
Trehalose, a widespread disaccharide synthesized in all nonvertebrate organisms, functions widely as a carbon source and stress protection compound. When fed exogenously to plants, however, it can have a surprisingly strong inhibitory effect on growth and carbon allocation (Veluthambi et al., 1982a, 1982b; Wingler et al., 2000). The trehalose pathway in plants has evolved a role that is distinct from other organisms in the regulation of metabolism in relation to growth and development (Schluepmann and Paul, 2009; Zhang et al., 2009; Paul et al., 2010). The signal transduction mechanisms involved in trehalose signaling, therefore, are of great interest as potential targets for crop improvement. Here, we provide evidence that growth arrest by T6P is an astonishing case of primary metabolite control over growth by way of the SnRK1 signaling pathway also involving bZIP11. Our work highlights the importance of bZIP11 and SnRK1 specifically in the growth response to trehalose and more generally in the regulation of growth by carbon availability to growing sinks.
Growth Arrest Is Not Due to Excessive Starch Accumulation But to Impaired Utilization of Sugar
The phenotype of Arabidopsis seedlings on trehalose is interesting because it involves a large change in carbon allocation within the seedling in addition to growth arrest. Starch accumulates in cotyledons, while growing sinks appear carbon limited. Starchless pgm1 mutants are similarly impaired as the wild type on trehalose, which rules out the possibility that sinks are carbon limited because starch accumulation sequesters available carbon. Feeding metabolizable sugar in combination with trehalose alleviates growth arrest; therefore, growth arrest is likely due to insufficient carbon in growing sinks (Schluepmann et al., 2004). Feeding trehalose could affect Suc transport, with high levels of trehalose displacing Suc at the Suc transporters and so reducing phloem loading and unloading processes. Growth arrest by trehalose, however, already occurs at trehalose concentrations of 25 mm when trehalose is fed in combination with the trehalase inhibitor validamycin A (Wingler et al., 2000). At these concentrations, trehalose is unlikely to have an effect on Suc transport just by displacing Suc. In contrast, T6P has previously been shown to cause the growth inhibition on trehalose and to have an important signaling function (Schluepmann et al., 2004; Kolbe et al., 2005; Zhang et al., 2009). One of these signaling functions is in plastid redox processes (Kolbe et al., 2005; Geigenberger, 2011). Starch accumulation in trehalose-fed leaves of Arabidopsis is caused by T6P accumulation stimulating AGPase redox activation in plastids (Kolbe et al., 2005). This is consistent with the massive accumulation of starch seen in cotyledons of seedlings. The pgm1 mutants show that the effect of T6P on AGPase redox activation in cotyledons is not involved in the mechanism leading to growth arrest.
The Transcription Factor bZIP11 Overcomes Growth Inhibition on Trehalose
The FOX collection was utilized to screen for the rescue of growth on trehalose. Strikingly, we isolated three independent transgenic lines overexpressing bZIP11, a transcription factor shown to be important in the regulation of growth (Hanson et al., 2008). Translation of bZIP11 is repressed by Suc (Wiese et al., 2004); however, no such effect was produced by trehalose. A simple way of explaining rescue on trehalose is through the breakdown of trehalose by trehalase (Schluepmann et al., 2004). However, trehalase activity was not elevated in seedlings overexpressing bZIP11, and neither did backcrossing bZIP11 into a trehalase knockout compromise the rescue of seedlings by bZIP11, showing that bZIP11 rescues growth on trehalose without increasing the breakdown of the exogenously supplied trehalose. This implies that bZIP11 is part of a growth regulatory process that can be invoked to overcome the growth inhibition normally imposed by trehalose. The results obtained differ from those described by Ma et al. (2011), where bZIP11 protein was fused to a nucleus-targeting domain and where targeting of the fusion protein to the nucleus induced the expression and activity of TRE1. TRE1 was not detected as a target of native bZIP11 (Hanson et al., 2008), however, and this is consistent with the results obtained with the FOX lines here. bZIP11 is expressed along the vasculature (Rook et al., 1998) and is proposed to regulate nitrogen metabolism by controlling the expression of ASN1 and PDH (Hanson et al., 2008). Sugar supply induces the expression of bZIP11; therefore, it was proposed that bZIP11 is important to relate nitrogen and carbon metabolism. Plants overexpressing KIN10 with active SnRK1 and primed to respond to starvation conditions consistently have reduced expression of bZIP11 (Baena-González et al., 2007).
bZIP11 Overexpression Results in Unprecedented Levels of T6P Recorded in Arabidopsis
T6P accumulation in plants grown on trehalose was previously shown to be causally related to growth inhibition (Schluepmann et al., 2004). This was shown through specific reduction in T6P content through the expression of a T6P hydrolase that rescued growth. Therefore, it was surprising that bZIP11 overexpressors on trehalose exhibited huge accumulation of T6P to the highest levels so far reported for Arabidopsis (Fig. 4B). Sugars were also elevated in bZIP11 (Fig. 5E). A strong relationship between Suc (Lunn et al., 2006) and Suc and hexoses (Martinez-Barajas et al., 2011) and T6P has previously been found. It is thus possible that elevated T6P can be accounted for as a response to elevated sugar found in bZIP11 overexpressors compared with the wild type. Given the recent evidence that SnRK1 is a target of T6P (Zhang et al., 2009; Paul et al., 2010), that SnRK1 would be inhibited by T6P accumulation on trehalose (Fig. 4C), and that bZIP11 overexpression increases the impact of endogenous SnRK1 activity (Baena-González et al., 2007), we hypothesized that high SnRK1 activity could also be a mechanism of rescue of growth on trehalose.
KIN10 Overexpression Rescues Growth on Trehalose
SnRK1 is essential for carbon utilization in growth. KIN10 overexpression increases seedling growth compared with the wild type when carbon availability is limiting, whereas seedlings with low SnRK1 activity through KIN10 antisense thrive when carbon availability is high (Baena-González et al., 2007). We found that, as with bZIP11, overexpression of SnRK1 also rescues seedling growth on trehalose (Fig. 5A). Like seedlings expressing bZIP11, KIN10 expressors also accumulate free sugars compared with either the wild type or treF when they are grown in osmoticum control conditions (Fig. 5E). This would imply that SnRK1 too is part of a mechanism that regulates growth in the presence of trehalose and that T6P, through the inhibition of SnRK1, prevents growth on trehalose. This conclusion is supported by previous work that shows the inhibition of SnRK1 by T6P in Arabidopsis seedlings, by direct in vitro assay, and indirectly by profiling known targets of SnRK1 in seedlings with altered T6P steady state (Zhang et al., 2009).
Interaction of bZIP11 and SnRK1
A subset of S-class bZIP transcription factors are proposed to mediate some of the transcriptional reprogramming in response to altered SnRK1 activity (Baena-González et al., 2007). The mechanism linking kinase activity signaling and transcriptional control by S-class bZIP proteins is not known, but it possibly involves phosphorylation of the bZIP protein, which would explain the potentiation of the SnRK1 response when KIN10 and bZIP11 are overexpressed simultaneously in protoplasts (Baena-González et al., 2007). Recombinant bZIP EEL/DPBF4 and ABI5 were substrates of SnRK1 after immunoprecipitation of the KIN10GFP protein fusion (Bitrián et al., 2011). We confirm KIN10 and bZIP11 interaction when seedlings are on trehalose: (1) overexpression of bZIP11 has the effect of overcoming the effects of low SnRK1 activity on trehalose; (2) the majority of genes tested that are jointly controlled by KIN10 and bZIP11 are expressed in a manner consistent with low activity of either KIN10 or bZIP11 in wild-type seedlings on trehalose (Table I; Fig. 6A); (3) under different conditions, KIN10 and bZIP11 permit growth of sinks in seedlings on trehalose; and (4) T6P accumulates in bZIP11-overexpressing seedlings grown on sorbitol. Comparison of data from Baena-González et al. (2007) and Hanson et al. (2008) shows that 44 genes may be jointly regulated by bZIP11 and KIN10 (Table I). Constitutive and high expression of bZIP11, therefore, may counteract T6P inhibition of SnRK1 activity on trehalose and impact at least a subset of the targets that are commonly regulated by bZIP11 and KIN10. These targets of bZIP11 are likely important for the control of carbon utilization for growth.
S1-class bZIP transcription factors are thought to act redundantly but have a differential expression responsiveness and pattern (Weltmeier et al., 2006, 2009; Hanson et al., 2008). Seedlings overexpressing or with antisense to bZIP1 (Kang et al., 2010) were sensitive to trehalose, however (Fig. 2E), suggesting that bZIP11 has a different function from bZIP1. Control of bZIP11 gene expression is opposite that of the other S-class bZIP capable of potentiating the effect of KIN10 expression: bZIP11 is repressed under carbon starvation conditions or by high SnRK1 activity (Baena-González et al., 2007; Supplemental Fig. S1). bZIP11 expression, furthermore, is induced by increased carbon availability, and this is consistent with the results presented here, where bZIP11 is required for carbon utilization for growth.
The arrest of hypocotyl growth by trehalose in seedlings in the dark was not reported previously. Seedlings on the combination of trehalose and validamycin in the dark were reported not to accumulate starch; therefore, it was concluded that carbon accumulating as starch in light-grown seedlings on trehalose was of photosynthetic origin (Wingler et al., 2000). Inhibition of hypocotyl growth in dark-grown seedlings on trehalose, and suppression thereof by overexpression of either KIN10 or bZIP11, indicate that the pathway of growth inhibition by trehalose in the dark and in the light likely involves the same mechanism. In the dark, gluconeogenesis from lipids stored in both the endosperm and the cotyledons fuels hypocotyl growth (Penfield et al., 2004). In light or dark, therefore, KIN10/bZIP11 activities are limiting the growth of sink tissues in seedlings as T6P accumulates on trehalose. Inhibition of the growth of sink tissues when SnRK1 was reduced by antisense inhibition has previously been reported in barley (Hordeum vulgare) pollen grains and in potato (Solanum tuberosum; Purcell et al., 1998; Zhang et al., 2001). Importantly, the work with segregating pollen grains by Zhang et al. (2001) shows that the effect is in the cells where SnRK1 is low and is not due to low SnRK1 in surrounding tissue.
A Possible Model of Interactions That Control Carbon Availability in Growing Sinks
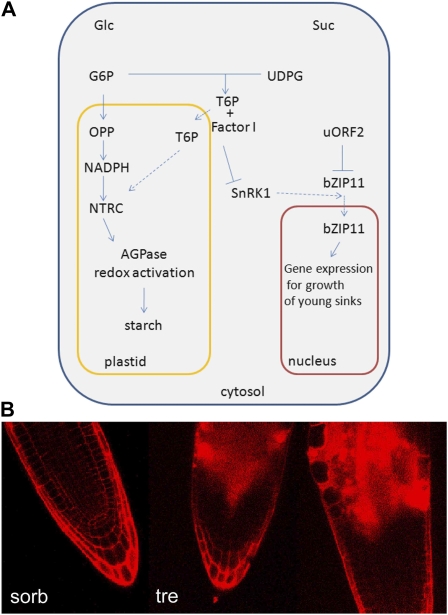
T6P inhibition of SnRK1 could be part of a regulatory loop that relates SnRK1 activity with the amount of Suc (Fig. 7A). In this regulatory loop, Suc-induced T6P increase inhibits SnRK1 when Suc is plentiful. As Suc decreases, T6P decreases and active SnRK1 signals nutrient stress such that more carbon is then made available to the heterotrophic growing cells. This regulatory loop could explain why both too little and too much of (either SnRK1 or) T6P are growth inhibitory (Schluepmann et al., 2003, 2004; Debast et al., 2011). It also would explain the reported correlation between levels of Suc and T6P (Lunn et al., 2006; Martinez-Barajas et al., 2011).
Figure 7.
A, Model of interactions affecting growth and starch accumulation on trehalose when T6P accumulates. Glc and Suc feeding cause AGPase redox activation and thus starch synthesis by different pathways that are likely also relevant for the growth responses to these sugars (Tiessen et al., 2003; Michalska et al., 2009; Geigenberger, 2011). When feeding Glc, T6P does not accumulate (T.L. Delatte, P. Sedijani, and H. Schluepmann, unpublished data), and Glc-6-P (G6P) in plastids is shunted through the oxidative part of the pentose phosphate pathway (OPP), generating NADPH for NADPH-thioredoxin reductase C (NTRC)-dependent reduction of AGPase and thus activation. In contrast, feeding Suc or trehalose leads to T6P increase, which acts upon AGPase redox by an unknown mechanism (Schluepmann et al., 2004; Kolbe et al., 2005; Lunn et al., 2006; Michalska et al., 2009). Suc inhibits the translation of bZIP11 by way of uORF2 (Wiese et al., 2004), but trehalose does not. When feeding trehalose, T6P accumulates. T6P accumulation inhibits SnRK1; this inhibition of SnRK1 depends on an intermediary factor I, present in young tissues (Zhang et al. 2009). Possibly, SnRK1 phosphorylation activates bZIP11 transfer to the nucleus or complexing of the transcription factor in such a way that bZIP11 controls part of the SnRK1 output that is required for growth. Thus, when T6P accumulates and inhibits SnRK1 in young tissues, overexpression of bZIP11 may act as a surrogate for SnRK1. B, Antisense SnRK1 restricted to individual pollen of barley in particular (Zhang et al., 2001) but also work in developing potato tubers (Purcell et al., 1998) show that SnRK1 is required in growing heterotrophic cells for growth and starch accumulation. Therefore, it is possible that SnRK1 is needed to respond to nutrient stress so as to make carbon available in growing sinks. SnRK1 inhibition (by artificially increasing T6P when feeding trehalose or by antisense SnRK1) would thus uncouple growth from carbon starvation responses, leading to the swollen cells observed in the growing zones of roots of Arabidopsis seedlings on trehalose (tre) compared with sorbitol (sorb). Root tips were stained with propidium iodide in water immediately prior to visualization with the confocal microscope. [See online article for color version of this figure.]
Glc and Suc feeding cause AGPase redox activation and thus increased starch synthesis by two different pathways that are likely also relevant for the growth responses to these sugars (Tiessen et al., 2003; Michalska et al., 2009; Geigenberger, 2011). When feeding Glc, T6P does not accumulate (T.L. Delatte, P. Sedijani, and H. Schluepmann, unpublished data) and Glc-6-phosphate is shunted through the plastidic oxidative part of pentose phosphate pathway, generating NADPH for NADPH thioredoxin reductase C-dependent reduction of AGPase and thus activation (Michalska et al., 2009). In contrast, feeding Suc or trehalose leads to T6P increase (Schluepmann et al., 2004; Lunn et al., 2006). T6P increase may result from the increased amount of the substrate uridine diphosphate Glc when Suc is cleaved by Suc synthase. Alternatively, T6P increase in response to Suc feeding may result from a sensing/signaling system affecting either T6P synthesis or degradation. The Suc pathway to AGPase redox activation was shown to depend upon SnRK1 activity (Tiessen et al., 2003), but we have yet to discover at which step SnRK1 is required. SnRK1 may not be required when trehalose-grown seedlings convert the carbon fixed by chloroplasts into starch; alternatively, factor I may be absent in cotyledons. A number of class II T6P synthases (TPS), including TPS5, are likely targets of SnRK1, but until now these enzymes were not shown to synthesize T6P (Harthill et al., 2006). When feeding Suc or trehalose, T6P also causes NADPH thioredoxin reductase C-dependent AGPase redox activation in the chloroplasts (Kolbe et al., 2005; Michalska et al., 2009). Although the interaction with redox signaling may be an important aspect of T6P signaling, we have shown here that T6P accumulation on trehalose and the consequent accumulation of starch in cotyledons does not cause growth arrest.
When feeding trehalose, T6P accumulation inhibits SnRK1. Inhibition of SnRK1 depends on an intermediary factor (factor I) present in seedling extracts but not in leaf extracts (Zhang et al., 2009). Possibly, SnRK1 activates bZIP11 transfer to the nucleus or complexing of the transcription factor in such a way that bZIP11 controls a part of the SnRK1 output that is required for growth. Thus, when T6P accumulates and inhibits SnRK1, overexpression of bZIP11 may act as a surrogate for SnRK1 in the growing zones. Antisense SnRK1 restricted to individual pollen of barley in particular (Zhang et al., 2001) but also work in developing potato tubers (Purcell et al., 1998) suggest that SnRK1 is required in individual cells of growing sinks for growth and starch accumulation.
In short-term trehalose feeding experiments of potato slices, AGPase redox activation was also found, and it was dependent on the presence of SnRK1 (Kolbe et al., 2005). SnRK1, therefore, may coordinate substrate availability for starch synthesis and AGPase redox activation in sink tissues. In this respect, it is important to take into account that the subcellular localization of SnRK1’s catalytic subunit KIN10 was shown to differ in the different tissues of the seedlings (Bitrián et al., 2011): this may also change SnRK1 susceptibility to T6P. In growing zones of root and shoot, KIN10 was reported in the nucleus, while in the hypocotyls, it was not. It is thus possible that in growing sinks, SnRK1 is required to signal nutrient stress so as to activate processes that will make carbon available to growing cells. We conclude that SnRK1 inhibition (by artificially increasing T6P when feeding trehalose or by antisense SnRK1) may uncouple carbon starvation from growth responses, leading to the swollen cells observed in the growing zones of roots of Arabidopsis seedlings on trehalose compared with sorbitol (Fig. 7B). More research is needed to understand the precise role of bZIP11 in the carbon allocation responses and to understand where and how SnRK1 as well as bZIP11 overexpression lead to the accumulation of free sugars in seedlings.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Lines in the Arabidopsis (Arabidopsis thaliana) Col-0 accession are as follows: Tref line 42 (Schluepmann et al., 2003), tre1-1 is Salk 147073c (http://signal.salk.edu; Alonso et al., 2003), FOX lines (Ichikawa et al., 2006), the bZIP1 overexpression and antisense lines (Kang et al., 2010), and finally the line to test translational repression of bZIP11 that contains the 5′ untranslated leader of bZIP11 mRNA fused to GUS/GFP under the control of the UBIQUITIN10 promoter (UBQ10:5′UTR-GUS/GFP; Wiese et al., 2004). Lines in the Ler accession are as follows: tre1-2 (Vandesteene, 2009) and KIN10 O1 and O2 (Baena-González et al., 2007). Seeds were generally vapor-phase sterilized (Clough and Bent, 1998), plated on medium, and cold treated at 4°C for 72 h in darkness. The medium used for growth of seedlings was generally agar-solidified (0.8%, w/v) half-strength MS (Murashige and Skoog, 1962) with 100 mm filter-sterilized sorbitol or trehalose. The plates were then transferred for growth at 22°C in a long-day light cycle (16-h-light/8-h-dark) regime with 100 μmol m−2 s−1 light intensity and 80% humidity. When grown in the dark, seedlings were first exposed to 6 h of light at 22°C to promote germination before transfer to darkness at 22°C.
Seedling Screening for Trehalose Resistance
Seeds from the Arabidopsis FOX collection (Ichikawa et al., 2006; T1 generation) were collected using a bamboo skewer with a small pinhole at one end serving as a measure for 20 to 30 seeds; collection was at the Plant Science Center, RIKEN, in Yokohama, Japan. Twenty to 30 T1 seeds from 100 different transgenic lines were pooled in a single pool; 141 pools covered the entire collection. Seedlings from different pools are therefore necessarily from different transgenic events. During the primary screen, 1,000 seeds per pool of 100 independent transgenic lines were screened twice on plates containing half-strength MS salts and vitamins supplemented with 100 mm trehalose. The germination frequency varied between 100% and 30%. Seedlings were chosen over a period of 1 to 3 weeks of growth that displayed longer roots and more growth of leaf primordia than the wild type (Fig. 1A, 93-1 tre). For seven of the pools, more than one seedling of the pool (one to six seedlings) was identified to be resistant. The seedlings were transferred to soil and grown for T2 seed set; a total of 157 seedlings were grown, resulting in seeds of 121 lines for a secondary screen. Seedlings recovered from pool 70 mostly died upon transfer to soil; after repeated screening, only one plant survived to set seed and was dealt with later in a separate secondary screening. Secondary screening was carried out on the T2 generation seeds for each line by testing seedling growth after 14 d on 100 mm trehalose and 100 mm sorbitol to test the specificity of the resistance to trehalose and on medium with 12 μg L−1 hygromycin B to evaluate the expression of the marker gene associated with the presence of the T-DNA insertion. Lines from six different pools were retained after the secondary screening, with average root length unchanged on the sorbitol control but at least 3-fold longer than the wild type on trehalose medium. Lines obtained from different pools necessarily are of independent transgenic origins, while lines obtained from the same pool likely are not. FOX lines from the T2 generation were subsequently grown to generate T3 and T4 seed stock with 100% trehalose resistance and homozygous for the presence of bZIP11 cDNA.
Lugol and GUS Staining
Seedlings were grown for 14 d in a long-day light cycle. Seedlings were harvested at midday, then washed in 70% ethanol before starch staining with 43.4 mm KI/5.7 mm I2 (Lugol solution from Sigma; Wingler et al., 2000) and mounted in a mixture of chloral hydrate:glycerol:water (8:1:2, v/v). To test the inhibition of bZIP11 translation, seedlings were grown in a long-day light cycle for 7 d on MS medium, then transferred for 48 h to MS supplemented with either Suc or trehalose at 0, 20, or 100 mm concentration. GUS staining was in GUS buffer (0.1 m NaH2PO4, 0.1 m Na2HPO4, 0.5 mm ferricyanide, 0.5 mm ferrocyanide, 10 mm EDTA, 0.1% [v/v] Triton X-100, and 1 mg mL−1 5-bromo-4-chloro-3-indolyl-β-d-GlcA) according to Wiese et al. (2004). The experiment was repeated with seedlings grown for 14 d on 100 mm either Suc or trehalose with results similar to those obtained after only 48 h of incubation.
Propidium Iodide Staining of Roots, and Confocal Microscopy
Seedlings were grown for 5 d on MS supplemented with 100 mm either sorbitol or trehalose. Seedlings were mounted with the root only between the coverslips and in 10 μm propidium iodide, and mounting was immediately before visualization with the confocal microscope (numerical aperture 63, 1.4 Plan Apochromat water immersion objective, Leica SP2 inverted laser confocal microscope with argon 488-nm laser excitation, dichroic 488/543/633, and emission settings of 562–588 nm).
DNA Extractions, PCR, and Sequencing
To genotype the F2 generation seedlings obtained from the crosses of the wild type with FOX lines, seedlings were grown for 14 d on MS with 100 mm trehalose, then DNA was extracted from a single seedling with a long root; the DNA extraction was as described previously (Cheung et al., 1993). Primers GS4 and GS6 (Supplemental Table S1) were used for PRC amplification of the cDNA in the FOX lines as described by Ichikawa et al. (2006). Genotyping of tre1-1 was as recommended (http://signal.salk.edu) using primers LBb1, LP1tre1, RP1tre1, LP2tre1, and RP2tre1 (Supplemental Table S1).
Assays for Trehalase Activity
Trehalase activity was assayed as described previously with some modifications (Brodmann et al., 2002). Seedlings (T2) were grown on trehalose medium, and short-root wild-type seedlings or seedlings with long roots from the FOX lines were used. Seedlings were pooled as 70 mg fresh weight and ground in 50 μL of buffer (0.1 m MES/KOH [pH 6.3], 1 mm EDTA, 1 mm phenylmethylsulfonyl fluoride, 1 mm dithiothreitol, and 0.01% [v/v] Triton X-100) at 4°C. Subsequently, 100 μL of buffer was added, and the crude extract was mixed and then centrifuged for 5 min, 13,000 rpm at 4°C, to remove the insoluble fraction. Sugars in the soluble supernatant were then removed by repeated dilution and concentration of the proteins over a 10-kd-cutoff cellulose membrane (Amicon Ultra, 0.5-mL 10K Ultracel; Millipore); dilution was with three consecutive additions of 300 μL of 20 mm MES/KOH (pH 6.3). The final volume of extract was adjusted to 50 μL. To assay trehalase, extract (10 μL) was incubated in triplicate with 15 mm trehalose for 1 h at 37°C and then boiled for 10 min. Alternatively, control assays were boiled immediately. Glc released from trehalose was quantified (Enzytec D-Glc kit; Scil Diagnostics) as the difference between boiled controls and samples that were assayed for 1 h. Values are averages from three biological replicates with sd.
T6P Determinations
Seedlings were grown under long-day growth conditions for 14 d. Five replicate samples of 50 mg each were harvested at midday by snap freezing. Lactose-1-phosphate (5 nmol) was added as an internal standard. Materials were then ground frozen, extracted, and analyzed as described (Delatte et al., 2009). Briefly, seedling extracts obtained by the subsequent liquid-liquid and solid-phase extractions were reconstituted in water and analyzed by anion-exchange chromatography combined with electrospray ionization mass spectrometry. The method provided baseline resolution of T6P and allowed its specific detection at mass-to-charge ratio 421 with good linearity. T6P concentrations were inferred from a five-point calibration curve using the signal obtained for the internal standard to correct for potential recovery losses.
Assays for SnRK1 Activity
Seedlings were grown for 14 d in 100 mm sorbitol or trehalose and snap frozen at midday as 100-mg fresh weight replicates. Three biological replicates for each data point were each ground frozen and extracted, and T6P as well as sugars were removed by the desalting procedure described previously (Zhang et al., 2009). SnRK1 activity determinations used the AMARA peptide as substrate.
Sugar Determinations
Soluble sugars were extracted from samples of 14-d-old seedlings; samples were three biological replicates for each data point. The extraction procedure was as described previously (Schluepmann et al., 2003). Suc, Glc, and Fru were assayed enzymatically (Enzytec D-Glc/D-Fru/Suc kit; Scil Diagnostics).
RNA Extraction and Q-PCR
RNA was extracted from flash-frozen seedlings collected at midday (50 mg fresh weight). Samples were in triplicate biological replicates for each data point, and the extraction protocol was according to the instruction manual (Spectrum Plant Total RNA Kit; Sigma-Aldrich). DNase treatment, reverse transcription, and Q-PCR were as described previously (Hanson et al., 2008). Primers used for the Q-PCR are described in Supplemental Table S2. Primers from the genes jointly targeted by KIN10 and bZIP11 were taken from the CATMA site (http://www.catma.org/).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. SnRK1 and sugar-responsive expression of bZIP transcription factors bZIP1 and bZIP11 from the S class.
Supplemental Figure S2. Trehalose resistance after crossing tre1-1 with FOX line 93.
Supplemental Figure S3. KIN10 expression in FOX lines 70 and 93.
Supplemental Table S1. PCR primers for characterization and cloning.
Supplemental Table S2. Primers for Q-PCR.
Acknowledgments
We thank Dr. Jyan-Chyun Jang (Department of Horticulture and Crop Science and Plant Biotechnology Center, Ohio State University) for providing the bZIP1-overexpressing and antisense lines. We thank Prof. Patrick van Dijck and Dr. Lies Vandesteene (Katholieke Universiteit Leuven) for providing the tre1-2 line in the Ler background and Dr. Elena Baena-Gonzalez (Instituto Gulbenkian de Ciência) for providing the KIN10-overexpressing lines. We thank Fritz Kind and Ronald Leito (Department of Biology, Utrecht University) for support in photography. In addition, we thank two anonymous reviewers for pertinent comments.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen HM, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Baena-González E. (2010) Energy signaling in the regulation of gene expression during stress. Mol Plant 3: 300–313 [DOI] [PubMed] [Google Scholar]
- Baena-González E, Rolland F, Thevelein JM, Sheen J. (2007) A central integrator of transcription networks in plant stress and energy signalling. Nature 448: 938–942 [DOI] [PubMed] [Google Scholar]
- Baena-González E, Sheen J. (2008) Convergent energy and stress signaling. Trends Plant Sci 13: 474–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitrián M, Roodbarkelari F, Horváth M, Koncz C. (2011) BAC-recombineering for studying plant gene regulation: developmental control and cellular localization of SnRK1 kinase subunits. Plant J 65: 829–842 [DOI] [PubMed] [Google Scholar]
- Brodmann A, Schuller A, Ludwig-Müller J, Aeschbacher RA, Wiemken A, Boller T, Wingler A. (2002) Induction of trehalase in Arabidopsis plants infected with the trehalose-producing pathogen Plasmodiophora brassicae. Mol Plant Microbe Interact 15: 693–700 [DOI] [PubMed] [Google Scholar]
- Caspar T, Huber SC, Somerville C. (1985) Alterations in growth, photosynthesis, and respiration in a starchless mutant of Arabidopsis thaliana (L.) deficient in chloroplast phosphoglucomutase activity. Plant Physiol 79: 11–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung WY, Hubert N, Landry BS. (1993) A simple and rapid DNA microextraction method for plant, animal, and insect suitable for RAPD and other PCR analyses. PCR Methods Appl 3: 69–70 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Debast S, Nunes-Nesi A, Hajirezaei MR, Hofmann J, Sonnewald U, Fernie AR, Börnke F. (2011) Altering trehalose-6-phosphate content in transgenic potato tubers affects tuber growth and alters responsiveness to hormones during sprouting. Plant Physiol (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delatte TL, Selman MH, Schluepmann H, Somsen GW, Smeekens SC, de Jong GJ. (2009) Determination of trehalose-6-phosphate in Arabidopsis seedlings by successive extractions followed by anion exchange chromatography-mass spectrometry. Anal Biochem 389: 12–17 [DOI] [PubMed] [Google Scholar]
- Eastmond PJ, van Dijken AJ, Spielman M, Kerr A, Tissier AF, Dickinson HG, Jones JD, Smeekens SC, Graham IA. (2002) Trehalose-6-phosphate synthase 1, which catalyses the first step in trehalose synthesis, is essential for Arabidopsis embryo maturation. Plant J 29: 225–235 [DOI] [PubMed] [Google Scholar]
- Fritzius T, Aeschbacher R, Wiemken A, Wingler A. (2001) Induction of ApL3 expression by trehalose complements the starch-deficient Arabidopsis mutant adg2-1 lacking ApL1, the large subunit of ADP-glucose pyrophosphorylase. Plant Physiol 126: 883–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geigenberger P. (2011) Regulation of starch biosynthesis in response to a fluctuating environment. Plant Physiol 155: 1566–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halford NG, Hey SJ. (2009) Snf1-related protein kinases (SnRKs) act within an intricate network that links metabolic and stress signalling in plants. Biochem J 419: 247–259 [DOI] [PubMed] [Google Scholar]
- Hanson J, Hanssen M, Wiese A, Hendriks MM, Smeekens S. (2008) The sucrose regulated transcription factor bZIP11 affects amino acid metabolism by regulating the expression of ASPARAGINE SYNTHETASE1 and PROLINE DEHYDROGENASE2. Plant J 53: 935–949 [DOI] [PubMed] [Google Scholar]
- Hardie DG. (2007) AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nat Rev Mol Cell Biol 8: 774–785 [DOI] [PubMed] [Google Scholar]
- Harthill JE, Meek SE, Morrice N, Peggie MW, Borch J, Wong BH, Mackintosh C. (2006) Phosphorylation and 14-3-3 binding of Arabidopsis trehalose-phosphate synthase 5 in response to 2-deoxyglucose. Plant J 47: 211–223 [DOI] [PubMed] [Google Scholar]
- Ichikawa T, Nakazawa M, Kawashima M, Iizumi H, Kuroda H, Kondou Y, Tsuhara Y, Suzuki K, Ishikawa A, Seki M, et al. (2006) The FOX hunting system: an alternative gain-of-function gene hunting technique. Plant J 48: 974–985 [DOI] [PubMed] [Google Scholar]
- Jakoby M, Weisshaar B, Dröge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, Parcy F. (2002) bZIP transcription factors in Arabidopsis. Trends Plant Sci 7: 106–111 [DOI] [PubMed] [Google Scholar]
- Kang SG, Price J, Lin PC, Hong JC, Jang JC. (2010) The Arabidopsis bZIP1 transcription factor is involved in sugar signaling, protein networking, and DNA binding. Mol Plant 3: 361–373 [DOI] [PubMed] [Google Scholar]
- Kilian J, Whitehead D, Horak J, Wanke D, Weinl S, Batistic O, D’Angelo C, Bornberg-Bauer E, Kudla J, Harter K. (2007) The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J 50: 347–363 [DOI] [PubMed] [Google Scholar]
- Kolbe A, Tiessen A, Schluepmann H, Paul M, Ulrich S, Geigenberger P. (2005) Trehalose 6-phosphate regulates starch synthesis via posttranslational redox activation of ADP-glucose pyrophosphorylase. Proc Natl Acad Sci USA 102: 11118–11123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunn JE, Feil R, Hendriks JH, Gibon Y, Morcuende R, Osuna D, Scheible WR, Carillo P, Hajirezaei MR, Stitt M. (2006) Sugar-induced increases in trehalose 6-phosphate are correlated with redox activation of ADPglucose pyrophosphorylase and higher rates of starch synthesis in Arabidopsis thaliana. Biochem J 397: 139–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Hanssen M, Lundgren K, Hernández L, Delatte T, Ehlert A, Liu CM, Schluepmann H, Dröge-Laser W, Moritz T, et al. (2011) The sucrose-regulated Arabidopsis transcription factor bZIP11 reprograms metabolism and regulates trehalose metabolism. New Phytol 191: 733–745 [DOI] [PubMed] [Google Scholar]
- Martinez-Barajas E, Delatte T, Schluepmann H, de Jong GJ, Somsen GW, Nunes C, Primavesi LF, Coello P, Mitchell RAC, Paul MJ. (2011) Wheat grain development is characterized by remarkable trehalose 6-phosphate accumulation pregrain filling: tissue distribution and relationship to SNF1-related protein kinase1 activity. Plant Physiol 156: 373–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalska J, Zauber H, Buchanan BB, Cejudo FJ, Geigenberger P. (2009) NTRC links built-in thioredoxin to light and sucrose in regulating starch synthesis in chloroplasts and amyloplasts. Proc Natl Acad Sci USA 106: 9908–9913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. (1962) A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol Plant 15: 473–479 [Google Scholar]
- Obertello M, Krouk G, Katari MS, Runko SJ, Coruzzi GM. (2010) Modeling the global effect of the basic-leucine zipper transcription factor 1 (bZIP1) on nitrogen and light regulation in Arabidopsis. BMC Syst Biol 4: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul MJ, Jhurreea D, Zhang Y, Primavesi LF, Delatte T, Schluepmann H, Wingler A. (2010) Upregulation of biosynthetic processes associated with growth by trehalose 6-phosphate. Plant Signal Behav 5: 386–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield S, Rylott EL, Gilday AD, Graham S, Larson TR, Graham IA. (2004) Reserve mobilization in the Arabidopsis endosperm fuels hypocotyl elongation in the dark, is independent of abscisic acid, and requires PHOSPHOENOLPYRUVATE CARBOXYKINASE1. Plant Cell 16: 2705–2718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polge C, Thomas M. (2007) SNF1/AMPK/SnRK1 kinases, global regulators at the heart of energy control? Trends Plant Sci 12: 20–28 [DOI] [PubMed] [Google Scholar]
- Purcell P, Smith AM, Halford NG. (1998) Antisense expression of a sucrose nonfermenting-1-related protein kinase sequence in potato results in decreased expression of sucrose synthase in tubers and loss of sucrose-inducibility of sucrose synthase transcripts in leaves. Plant J 14: 195–202 [Google Scholar]
- Ramon M, Rolland F, Thevelein JM, Van Dijck P, Leyman B. (2007) ABI4 mediates the effects of exogenous trehalose on Arabidopsis growth and starch breakdown. Plant Mol Biol 63: 195–206 [DOI] [PubMed] [Google Scholar]
- Rook F, Gerrits N, Kortstee A, van Kampen M, Borrias M, Weisbeek P, Smeekens S. (1998) Sucrose-specific signalling represses translation of the Arabidopsis ATB2 bZIP transcription factor gene. Plant J 15: 253–263 [DOI] [PubMed] [Google Scholar]
- Satoh-Nagasawa N, Nagasawa N, Malcomber S, Sakai H, Jackson D. (2006) A trehalose metabolic enzyme controls inflorescence architecture in maize. Nature 441: 227–230 [DOI] [PubMed] [Google Scholar]
- Schluepmann H, Paul MJ. (2009) Trehalose metabolites in Arabidopsis: elusive, active and central. The Arabidopsis Book 7: e0122, doi/10.1199/tab.0122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluepmann H, Pellny T, van Dijken A, Smeekens S, Paul M. (2003) Trehalose 6-phosphate is indispensable for carbohydrate utilization and growth in Arabidopsis thaliana. Proc Natl Acad Sci USA 100: 6849–6854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluepmann H, van Dijken A, Aghdasi M, Wobbes B, Paul M, Smeekens S. (2004) Trehalose mediated growth inhibition of Arabidopsis seedlings is due to trehalose-6-phosphate accumulation. Plant Physiol 135: 879–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiessen A, Prescha K, Branscheid A, Palacios N, McKibbin R, Halford NG, Geigenberger P. (2003) Evidence that SNF1-related kinase and hexokinase are involved in separate sugar-signalling pathways modulating post-translational redox activation of ADP-glucose pyrophosphorylase in potato tubers. Plant J 35: 490–500 [DOI] [PubMed] [Google Scholar]
- Vandesteene L. (2009) Functional analysis of the trehalose metabolism gene family in Arabidopsis thaliana. PhD thesis. Katholieke Universiteit Leuven, Leuven, Belgium [Google Scholar]
- Veluthambi K, Mahadevan S, Maheshwari R. (1982a) Trehalose toxicity in Cuscuta reflexa: sucrose content decreases in shoot tips upon trehalose feeding. Plant Physiol 69: 1247–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veluthambi K, Mahadevan S, Maheshwari R. (1982b) Trehalose toxicity in Cuscuta reflexa: cell wall synthesis is inhibited upon trehalose feeding. Plant Physiol 70: 686–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weltmeier F, Ehlert A, Mayer CS, Dietrich K, Wang X, Schütze K, Alonso R, Harter K, Vicente-Carbajosa J, Dröge-Laser W. (2006) Combinatorial control of Arabidopsis proline dehydrogenase transcription by specific heterodimerisation of bZIP transcription factors. EMBO J 25: 3133–3143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weltmeier F, Rahmani F, Ehlert A, Dietrich K, Schütze K, Wang X, Chaban C, Hanson J, Teige M, Harter K, et al. (2009) Expression patterns within the Arabidopsis C/S1 bZIP transcription factor network: availability of heterodimerization partners controls gene expression during stress response and development. Plant Mol Biol 69: 107–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese A, Elzinga N, Wobbes B, Smeekens S. (2004) A conserved upstream open reading frame mediates sucrose-induced repression of translation. Plant Cell 16: 1717–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingler A, Fritzius T, Wiemken A, Boller T, Aeschbacher RA. (2000) Trehalose induces the ADP-glucose pyrophosphorylase gene, ApL3, and starch synthesis in Arabidopsis. Plant Physiol 124: 105–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Primavesi LF, Jhurreea D, Andralojc PJ, Mitchell RA, Powers SJ, Schluepmann H, Delatte T, Wingler A, Paul MJ. (2009) Inhibition of SNF1-related protein kinase1 activity and regulation of metabolic pathways by trehalose-6-phosphate. Plant Physiol 149: 1860–1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Shewry PR, Jones H, Barcelo P, Lazzeri PA, Halford NG. (2001) Expression of antisense SnRK1 protein kinase sequence causes abnormal pollen development and male sterility in transgenic barley. Plant J 28: 431–441 [DOI] [PubMed] [Google Scholar]