Abstract
Ubiquitin-regulated protein degradation is a critical regulatory mechanism that controls a wide range of biological processes in plants. Here, we report that OsDIS1 (for Oryza sativa drought-induced SINA protein 1), a C3HC4 RING finger E3 ligase, is involved in drought-stress signal transduction in rice (O. sativa). The expression of OsDIS1 was up-regulated by drought treatment. In vitro ubiquitination assays showed that OsDIS1 possessed E3 ubiquitin ligase activity and that the conserved region of the RING finger was required for the activity. Transient expression assays in Nicotiana benthamiana leaves and rice protoplasts indicated that OsDIS1 was localized predominantly in the nucleus. Overexpression of OsDIS1 reduced drought tolerance in transgenic rice plants, while RNA interference silencing of OsDIS1 enhanced drought tolerance. Microarray analysis revealed that a large number of drought-responsive genes were induced or suppressed in the OsDIS1 overexpression plants under normal and drought conditions. Yeast two-hybrid screening showed that OsDIS1 interacted with OsNek6 (for O. sativa NIMA-related kinase 6), a tubulin complex-related serine/threonine protein kinase. Coexpression assays in N. benthamiana leaves indicated that OsNek6 was degraded by OsDIS1 via the 26S proteasome-dependent pathway and that this degradation was abolished by the OsDIS1(H71Y) mutation, which is essential for its E3 ligase activity. Together, these results demonstrate that OsDIS1 plays a negative role in drought stress tolerance through transcriptional regulation of diverse stress-related genes and possibly through posttranslational regulation of OsNek6 in rice.
Unlike animals, plants are sessile and may be subjected to diverse environmental stresses throughout their life cycle. Among these environmental stresses, drought stress is the primary cause for reductions in crop yield (Boyer, 1982; Cushman and Bohnert, 2000; Luo, 2010). Recent research has revealed that drought stress can cause a series of physiological and biochemical responses, such as stomatal closure, suppression of cell division and elongation, and inhibition of photosynthesis (Shinozaki and Yamaguchi-Shinozaki, 2007). To survive drought stress, plants have evolved complicated mechanisms to trigger a suite of physiological, cellular, and molecular responses (Fujita et al., 2006; Shinozaki and Yamaguchi-Shinozaki, 2007). A vast array of drought-induced and -suppressed genes are involved in these responses, which can act at transcriptional, posttranscriptional, epigenetic, and posttranslational levels in plants (Hirayama and Shinozaki, 2010).
The ubiquitin/26S proteasome system (UPS) is one of the most prominent mechanisms that plants use to control growth and development and to respond to biotic and abiotic stresses (Smalle and Vierstra, 2004; Santner and Estelle, 2010). Genome-wide studies have predicted that up to 6% of the Arabidopsis (Arabidopsis thaliana) proteome is involved in the UPS, which contains thousands of interacting proteins (Vierstra, 2009). The UPS consists of three key enzymes, ubiquitin-activating enzymes (E1), ubiquitin-conjugating enzymes (E2), and ubiquitin ligases (E3), which coordinately attach the 76-amino acid small protein ubiquitin to candidate substrates (Bachmair et al., 2001; Dreher and Callis, 2007; Vierstra, 2009). Depending on the number (single or multiple) of ubiquitins attached to the substrates and on the manner in which multiple ubiquitins are attached to the substrates, the ubiquitin-mediated protein modifications can be classified into monoubiquitination, multiubiquitination, and polyubiquitination (Hochstrasser, 2006; Mukhopadhyay and Riezman, 2007; Vierstra, 2009). Among them, only polyubiquitination via Lys-48 is involved in proteasomal degradation (Smalle and Vierstra, 2004; Vierstra, 2009). Other ubiquitination forms can modify their substrates to further affect substrate activity, stability, protein-protein interaction, or sublocalization (Mukhopadhyay and Riezman, 2007). These diversified regulatory mechanisms function in nearly all aspects of plant life, including the cell cycle, embryogenesis, photomorphogenesis, hormone signaling, and biotic and abiotic stress responses (Vierstra, 2003; Smalle and Vierstra, 2004; Zeng et al., 2006; Dreher and Callis, 2007; Santner and Estelle, 2010).
The function of E3 ubiquitin ligases in response to drought stress has been well documented in Arabidopsis. For example, the XERICO (Greek for “drought tolerant”) gene, encoding a RING-H2 zinc finger E3 ubiquitin ligase, positively regulates the drought response at the adult stage by increasing abscisic acid (ABA) biosynthesis (Ko et al., 2006). We previously reported that overexpression of the RING E3 ligase gene SDIR1 enhances drought tolerance by positively regulating the ABA signaling pathway (Zhang et al., 2007). AtAIRP1, a C3H2C3-type RING E3 ubiquitin ligase, also positively regulates drought response in an ABA-dependent manner (Ryu et al., 2010). Except for these three ABA-related E3 ligases, six other E3 ligases are also involved in drought responses via the UPS in both positive and negative manners. The U-box E3 ubiquitin ligase CaPUB1 negatively regulates the water-stress signaling pathway by ubiquitinating RPN6 in Arabidopsis (Cho et al., 2006). The two U-box E3 ubiquitin ligases, PUB22 and PUB23, negatively regulate the drought signaling in a coordinated manner by ubiquitinating cytosolic RPN12a (Cho et al., 2008). DRIP1 and DRIP2 negatively regulate drought-responsive gene expression by targeting DREB2A to the 26S proteasome (Qin et al., 2008). In contrast, Rma1H1, a RING-type membrane-anchor E3 ubiquitin ligase, positively regulates drought stress by inhibiting aquaporin trafficking to the plasma membrane by proteasomal degradation (Lee et al., 2009).
Heterogeneous expression of the Arabidopsis SDIR1 gene in rice (Oryza sativa) leads to enhanced drought tolerance (Zhang et al., 2008), suggesting that E3 ubiquitin ligases may play a role in drought response in rice. The rice genome contains at least 1,332 members of E3 ubiquitin ligases as reported in the PlantsUPS database (Du et al., 2009), and many of them are induced or repressed under drought stress according to the data deposited in the public microarray databases. Two recent studies show that heterogeneous expression of OsBIRF1, a benzothiadiazole-induced RING finger E3 ligase gene in Nicotiana benthamiana, has pleiotropic effects on plant growth and increases drought tolerance (Liu et al., 2008), whereas knockout of OsDSG1, a RING finger E3 ligase gene in rice, leads to delayed germination and enhanced drought tolerance (Park et al., 2010b). However, the biological functions of most rice E3 ubiquitin ligase genes in drought response are still unknown.
To investigate the functions of E3 ubiquitin ligase genes in drought stress in rice, we performed in silico gene expression analysis and identified an E3 ligase gene called OsDIS1 (for O. sativa drought-induced SINA protein 1), which is induced by drought stress as reported in the public rice microarray databases. We found that the OsDIS1 overexpression plants showed reduced drought tolerance and, conversely, that OsDIS1 RNA interference (RNAi) plants showed enhanced drought tolerance. To investigate the possible mechanisms involved in the OsDIS1-mediated drought-stress pathway, both microarray and yeast two-hybrid assays were performed. Our results indicate that OsDIS1 plays a negative role in drought stress through transcriptional and posttranslational regulation of diverse stress-related genes in rice.
RESULTS
Identification and Characterization of the OsDIS1 Gene
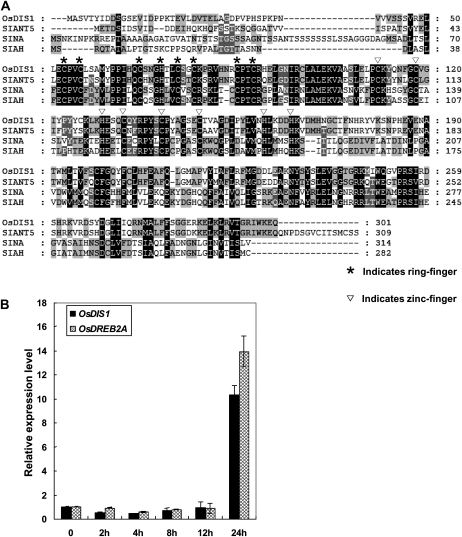
To identify E3 ubiquitin ligase genes involved in the drought-stress response in rice, we analyzed their expression patterns using microarray data deposited in the public databases (https://www.genevestigator.com/gv/index.jsp and http://signal.salk.edu/cgi-bin/RiceGE; Zimmermann et al., 2004, 2008). Among the analyzed 378 RING finger-type and 76 U-box-type E3 ubiquitin ligase genes in rice, we identified a drought-induced C3HC4 RING finger gene (LOC_Os03g24040). The deduced protein of this gene contains conserved RING finger and zinc finger motifs and has high sequence identity (82%) with SINAT5 in Arabidopsis (Fig. 1A; Xie et al., 2002), which was identified as a homolog of Drosophila SINA (Carthew and Rubin, 1990). Thus, we named the gene OsDIS1 in this study. To confirm the microarray expression results for OsDIS1, real-time PCR analysis was conducted with the OsDIS1 gene-specific primers using RNA isolated from 7-d-old seedlings before and after drought treatment. In the reactions, OsDREB2A was used as a positive control for the drought treatment (Matsukura et al., 2010). Similar to OsDREB2A, OsDIS1 was highly induced 24 h after the drought treatment (Fig. 1B), suggesting that OsDIS1 is involved in the drought response in rice.
Figure 1.
Identification and expression analysis of the OsDIS1 gene in rice. A, Protein sequence alignment between OsDIS1, SINAT5 (Arabidopsis), SINA (Drosophila), and SIAH (human). B, Real-time PCR analysis of the OsDIS1 expression level under drought treatment. Ubq was used as an internal control, and OsDREB2A was used as a positive control for drought stress.
A genome-wide search identified five paralogous SINA proteins of OsDIS1 in the rice genome. The amino acid identity between OsDIS1 and its paralogs is 91% with Os07g46560, 83% with Os02g19140, 83% with Os05g14860, 83% with Os01g13370, and 71% with Os02g03620. Further alignment between rice and Arabidopsis SINA proteins revealed that except for the N terminus, all the SINA members in rice and Arabidopsis had highly conserved sequences, and all contained the conserved RING finger and zinc finger motifs (Supplemental Fig. S1A). Phylogenetic analysis showed that each of the following protein pairs belongs to the same clade: SINAT1 and SINAT2, SINAT4 and SINAT5, and OsDIS1 and Os07g46560 (Supplemental Fig. S1B); this supports the inference of a possible duplication of these genes in both genomes (Wang et al., 2008). Based on pairwise distance analysis, OsDIS1 has a minimal distance of 0.0725 with Os0746560 in rice and 0.2624 with SINAT5 in Arabidopsis, suggesting that Os07g46560 is the closest paralog of OsDIS1 in rice and SINAT5 is the closest ortholog of OsDIS1 in Arabidopsis (Supplemental Fig. S1C).
OsDIS1 Is a Functional E3 Ubiquitin Ligase and Requires the Intact RING Finger Domain for Its E3 Ligase Activity
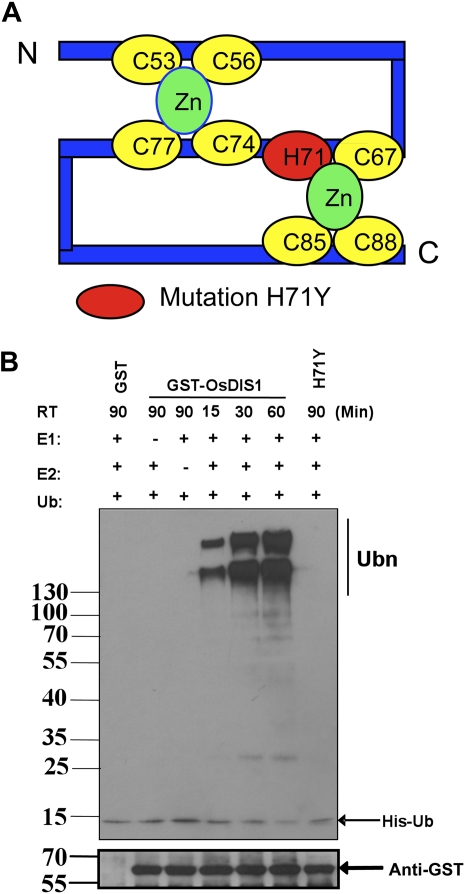
Previous research had shown that many RING motif-containing proteins function as ubiquitin E3 ligases (Stone et al., 2005). The N terminus of OsDIS1 contains a C3HC4-type RING finger motif (53–88 amino acids) with conserved Cys and His residues (Fig. 2A). To investigate whether OsDIS1 is a functional E3 ligase enzyme, we produced OsDIS1 in Escherichia coli as a fusion with the glutathione S-transferase (GST) tag and purified the tagged protein with GST affinity beads. In the presence of ubiquitin, ATP, wheat (Triticum aestivum) E1, and human E2 (UBCH5b), GST-OsDIS1 generated a high molecular self-ubiquitinated form in a time-dependent manner compared with the controls (Fig. 2B). In contrast, the purified GST and the combinations lacking E1 or E2 had no ubiquitinated signal even when incubated for a longer time, indicating that OsDIS1 has E3 ubiquitin ligase activity.
Figure 2.
E3 ubiquitin ligase activity assay of OsDIS1 and the RING finger mutant proteins. A, Scheme of OsDIS1 C3HC4 RING finger composition and the mutated amino acid in the RING finger. B, E3 ubiquitin ligase activity of OsDIS1. GST-OsDIS1 and its mutant form GST-OsDIS1(H71Y) fusion proteins were assayed for E3 activity in the presence of wheat E1 (GI: 136632), human E2 (UBCH5b), and 6xHis tag ubiquitin (Ub). The numbers at left denote the molecular masses of marker proteins in kilodaltons. RT represents the reaction time in the ubiquitination assay. GST was used as a negative control. Samples were resolved on a 10% SDS-PAGE gel. Nickel-horseradish peroxidase was used to detect His tag ubiquitin (top panel). The bottom panel indicates the presence of GST-OsDIS1/GST-OsDIS1(H71Y) proteins in the assay using anti-GST antibody. [See online article for color version of this figure.]
The intact RING finger domain is necessary for the E3 ligase activity of RING-type E3 ligase proteins (Xie et al., 2002; Stone et al., 2006; Zhang et al., 2007). To test whether the intact RING finger domain of OsDIS1 is needed for its E3 ligase activity, we produced a mutant construct in which His-71 was changed to Tyr-71 (H71Y) to disrupt the RING domain (Fig. 2A). Indeed, the OsDIS1(H71Y) mutant protein lost its E3 ligase activity even if it was incubated for a longer time (Fig. 2B), suggesting that an intact RING motif is necessary for the E3 ligase activity of OsDIS1.
OsDIS1 Is Localized Predominantly in the Nucleus
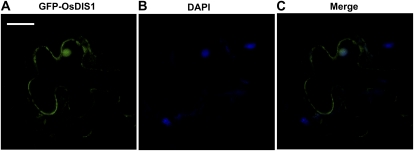
To investigate the subcellular localization of OsDIS1, we introduced the full-length cDNA of OsDIS1 into both pGDG and pGDR vectors, which contain a cauliflower mosaic virus promoter-driven GFP and the DsRed gene, respectively (Goodin et al., 2002). Subcellular localization assays were conducted in both N. benthamiana leaves and rice protoplasts. Localization of the fusion protein was then determined by visualization with a confocal microscope, and the position of the nucleus of the N. benthamiana epidermal cell was confirmed with 4′,6-diamidino-2-phenylindole (DAPI) staining (Fig. 3). Transient expression assays in N. benthamiana leaves indicated that the GFP-OsDIS1 fusion protein was localized predominantly in the nucleus, as well as some in the cytoplasm and plasma membrane (Fig. 3), which is consistent with the distribution of SINAT5 in Arabidopsis (Xie et al., 2002). The localization of OsDIS1 in N. benthamiana leaves was confirmed by its subcellular localization in rice protoplasts (Supplemental Fig. S2).
Figure 3.
Subcellular localization of the OsDIS1 protein in N. benthamiana leaves. The Agrobacterium strain carrying the GFP-OsDIS1 plasmid was infiltrated into N. benthamiana leaves, and the transformed leaves were analyzed by confocal microscopy 48 h after agroinfiltration. A, Fluorescence image of GFP-OsDIS1. B, Image of a nucleus stained with DAPI. C, Merged image. Bar = 50 μm.
OsDIS1 Negatively Regulates the Drought Response in Rice
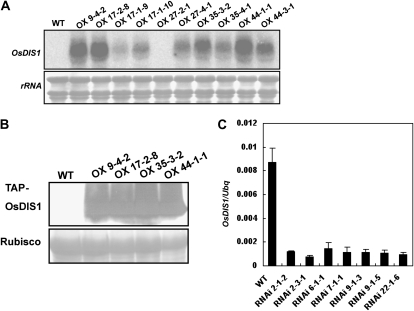
To explore the biological function of OsDIS1, we created OsDIS1 overexpression and RNAi constructs, which were transformed into the japonica cv Nipponbare by Agrobacterium tumefaciens-mediated transformation (Qu et al., 2006). The expression of OsDIS1 in the overexpression homozygous lines was first analyzed by RNA gel blot using the whole coding sequence of OsDIS1 as the probe (Fig. 4A). Four independent homozygous transgenic lines (OX 9-4-2, OX 17-2-8, OX 35-3-2, and OX 44-1-1) with obvious high OsDIS1 expression were chosen for further western blot analysis. In all four lines, a strong accumulation of the TAP-OsDIS1 fusion protein was detected by peroxidase anti-peroxidase antibody (Fig. 4B). The expression of OsDIS1 in the OsDIS1 RNAi homozygous lines was first analyzed by reverse transcription (RT)-PCR (data not shown) and then confirmed by real-time PCR (Fig. 4C). Four lines (RNAi 2-1-2, RNAi 7-1-1, RNAi 9-1-3, and RNAi 22-1-6) with significantly decreased expression of OsDIS1 were chosen for further analysis. Real-time PCR analysis revealed that the expression of OsDIS1’s five paralogs was not affected in both overexpression and RNAi lines using five pairs of gene-specific primers (data not shown).
Figure 4.
Expression analysis of OsDIS1 T3 homozygous overexpression plants and RNAi plants. A, The expression of OsDIS1 in the overexpression plants as determined by RNA gel blot. Total RNA (10 μg per sample) was loaded onto a 1.2% agarose gel, and the nylon membrane was hybridized with a 32P-labeled OsDIS1 probe (full-length coding sequence). The 28s rRNA was used as a loading control. OX denotes OsDIS1 overexpression transgenic lines. B, The expression of OsDIS1 in the overexpression plants as determined by western blot. The total protein was prepared from 10-d-old rice seedlings and subjected to SDS-PAGE, followed by immunoblot analysis using peroxidase anti-peroxidase antibody. Rubisco protein stained with Ponceau S was used as a loading control. C, The expression of OsDIS1 in the RNAi plants as determined by real-time PCR. The relative expression level of OsDIS1 was compared in the RNAi transgenic plants and the wild type (WT) using Ubq as an internal control.
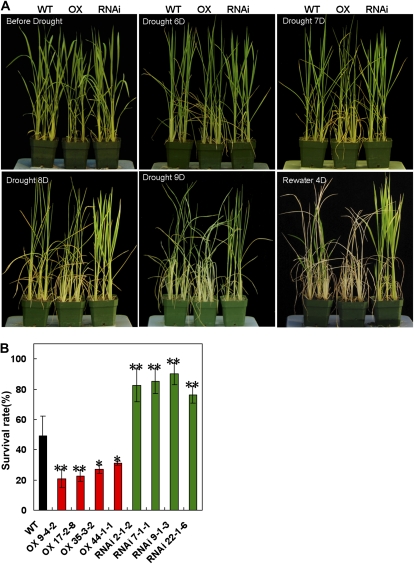
The drought response of the OsDIS1 overexpression and RNAi lines was evaluated with 4-week-old transgenic and wild-type Nipponbare plants. Under our greenhouse conditions and without drought stress, the growth of both types of OsDIS1 transgenic plants was similar to that of the wild-type plants (Fig. 5A, Before Drought). Then, these plants were removed from the trays and kept on a bench to induce drought stress. After 6 d without watering, the leaves of the OsDIS1 overexpression plants began to roll but the leaves of the OsDIS1 RNAi plants and wild-type plants had no obvious changes (Fig. 5A, Drought 6D). After 7 d without watering, the leaves of all overexpression plants were curled, while those of the wild-type plants were only beginning to show the drought-stress phenotypes (Fig. 5A, Drought 7D). On day 9, all of the overexpression plants and most of the wild-type plants had wilted, while the leaves of the RNAi plants had just begun to curl (Fig. 5A, Drought 9D). At the end of 9 d, all plants were moved back to trays with water. After 4 d of recovery in water, about 75% to 90% of the RNAi plants recovered from the stress and started to grow (Fig. 5, A, Rewater 4D, and B), but only about 50% of the wild-type plants and 20% to 30% of the overexpression plants survived (Fig. 5, A, Rewater 4D [P < 0.05], and B). These results demonstrate that OsDIS1 has a negative role in drought tolerance in rice.
Figure 5.
Phenotypes of the OsDIS1 overexpression and RNAi transgenic plants in response to drought treatment. A, Drought-stress treatment of 4-week-old plants. The OsDIS1 transgenic plants as well as wild-type (WT) control plants were grown in soil in a tray with water for 4 weeks and then removed from the tray for drought treatment. The plants were watered 4 d after the treatment. B, Survival rate of the OsDIS1 transgenic plants 4 d after watering. Each data point is the average of at least three replications. ** t test, with P < 0.01; * t test, with P < 0.05.
Global Gene Expression Analysis of the OsDIS1 Overexpression Plants under Normal and Drought-Stress Conditions
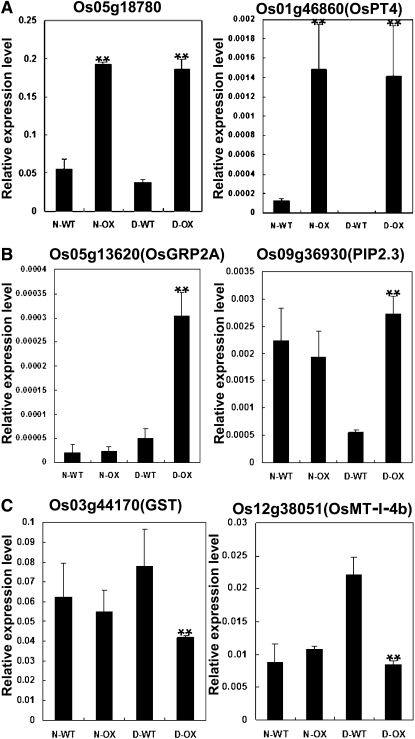
To identify genes involved in the OsDIS1-mediated drought-responsive pathway, we performed microarray analysis of the OsDIS1 overexpression and wild-type plants under both normal and drought-stress conditions using the Agilent rice Genechip. Seven-day-old plants of the OsDIS1 overexpression line 9-4-2 as well as the wild-type plants were used in the drought treatment. The expression of the drought-related gene OsLEA3 was highly induced, suggesting that the drought treatment was effective (Supplemental Fig. S3A). The microarray analysis showed that 435 genes had at least a 2-fold change in the OsDIS1 overexpression plants compared with the wild-type plants under normal conditions (Supplemental Fig. S3B). Among them, 135 genes were up-regulated and 300 genes were down-regulated (Supplemental Tables S1 and S2). For example, Os05g18780, which encodes an RNase P-related protein, and Os01g46860, which encodes a high-affinity inorganic phosphate transporter 11 protein that is similar to OsPT4 (Paszkowski et al., 2002; Ai et al., 2009), were specifically induced in the overexpression plants. Meanwhile, 260 genes were specifically up-regulated and 255 genes were specifically down-regulated 2-fold in the overexpression plants compared with the wild-type plants under drought conditions (Supplemental Fig. S3C; Supplemental Tables S3 and S4). Many of these genes with differential expression under drought stress encode stress-related proteins and regulatory factors (Shinozaki and Yamaguchi-Shinozaki, 2007), such as protease inhibitor/seed storage/LTP family protein precursors, peroxidase precursors, universal stress proteins, amino acid metabolism enzymes and carrier proteins, transcription factors (WRKY, MYB, and AP2), and protein kinase.
We then performed real-time PCR to confirm the differential expression of some of these genes under both normal and drought conditions. The analysis showed that Os01g46860 and Os05g18780 were specifically induced in the OsDIS1 overexpression plants under normal and drought conditions (P < 0.01; Fig. 6A). OsGRP2A and PIP2.3, whose Arabidopsis homologs act as negative regulators of drought response (Aharon et al., 2003; Jang et al., 2007; Kim et al., 2008), were induced in the OsDIS1 overexpression plants only under drought conditions (P < 0.01; Fig. 6B). Conversely, GST and OsMT-I-4b, whose homologs act as positive regulators of the drought response in Nicotiana tabacum and rice, respectively (Zhou et al., 2006; Yang et al., 2009; George et al., 2010; Ji et al., 2010), were repressed in the OsDIS1 overexpression plants under drought conditions (P < 0.01; Fig. 6C). Those results indicated that OsDIS1 negatively regulates the drought response by modulating the expression of diverse stress-related genes in rice.
Figure 6.
Real-time PCR confirmation of differentially expressed genes in the OsDIS1 overexpression plants under normal and drought conditions. A, Specifically induced genes in the OsDIS1 overexpression plants under normal conditions. B, Induced genes in the OsDIS1 overexpression plants under drought conditions. C, Suppressed genes in the OsDIS1 overexpression plants under drought conditions. N denotes the normal condition, D denotes the drought condition, WT denotes wild-type plants, and OX denotes OsDIS1 overexpression transgenic lines. ** t test, with P < 0.01; * t test, with P < 0.05.
OsDIS1 Interaction with OsNek6, a Ser/Thr Protein Kinase
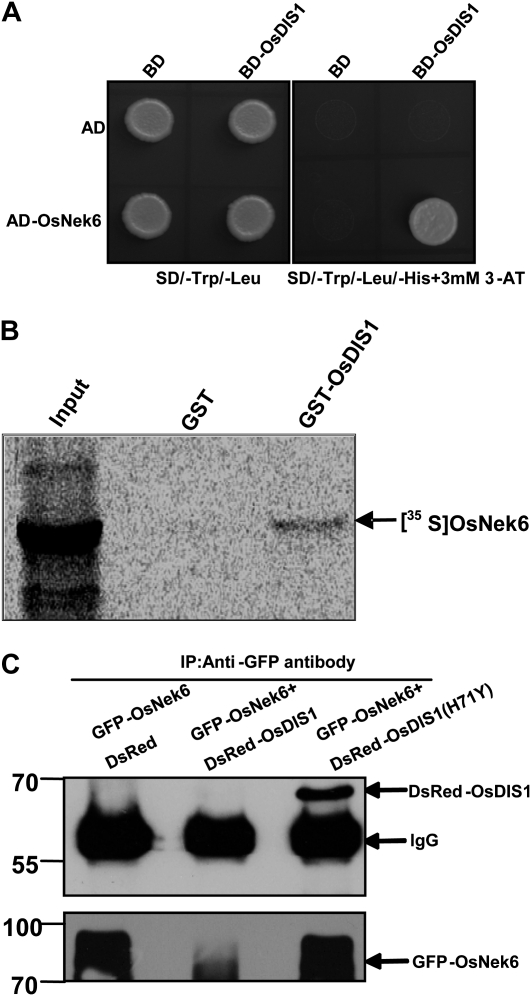
To identify proteins that interact with OsDIS1, we screened a rice cDNA library using OsDIS1 as the bait in the yeast two-hybrid assay. After two rounds of cDNA library screening, a candidate gene was identified. Cotransformation of this gene with OsDIS1 confirmed their interaction in yeast (Fig. 7A). Sequence analysis showed that the gene encodes a protein highly similar to the Ser/Thr protein kinase Nek4-like protein from human. We called this gene OsNek6 because it was first named OsNek6 by Vigneault et al. (2007) and that name was later adopted by Fujii et al. (2009).
Figure 7.
OsDIS1 interaction with OsNek6 in vitro and in vivo. A, Yeast two-hybrid assay. OsDIS1 and OsNek6 were cloned into pGBKT7 and pGADT7 vectors, respectively. The derived constructs were transformed into the yeast strain HF7c. The transformed yeast cells were plated onto the SD/−His/−Trp/−Leu medium including 3 mm 3-amino-1,2,4,-triazole (3-AT). AD, pGADT7 vector that contains an activation domain; BD, pGBKT7 vector that contains a binding domain. B, In vitro GST pull-down assay for OsDIS1 and OsNek6. The [35S]OsNek6 protein was in vitro transcribed and translated using the TNT Coupled Wheat Germ Extract Systems. GST-OsDIS1 protein was expressed in E. coli BL21 and purified. [35S]OsNek6 protein (3 μL) was incubated with 1 μg of GST-OsDIS1 protein at room temperature for 1 h. The GST protein was used as a negative control, and 1 μL of [35S]Met-labeled OsNek6 protein was used as input. C, In vivo coimmunoprecipitation assay for OsDIS1 and OsNek6. The protein of the N. benthamiana tissues coinfiltrated with plasmids of the DsRed/DsRed-OsDIS1/DsRed-OsDIS1(H71Y) and GFP-OsNek6 combinations was extracted and immunoprecipitated with anti-GFP antibody. Immunoblot analysis was performed using anti-DsRed antibody and anti-GFP antibody. The numbers at left denote the molecular masses of marker proteins in kilodaltons.
To verify the interaction between OsDIS1 and OsNek6, we performed an in vitro GST pull-down assay using the purified GST-OsDIS1 fusion proteins and [35S]Met-labeled OsNek6 proteins. Autoradiography exposure revealed that GST-OsDIS1 pulled down OsNek6 but not the control GST in vitro (Fig. 7B).
To further confirm the GST pull-down result, we performed an in vivo coimmunoprecipitation experiment using the agroinfiltration method. The Agrobacterium strains containing the GFP-OsNek6 and DsRed-OsDIS1 plasmids were coinfiltrated into N. benthamiana leaves with the GFP-OsNek6 and DsRed combination as a control. Three days after agroinfiltration, the protein extracts were immunoprecipitated with anti-GFP antibody followed by immunoblotting using anti-DsRed and anti-GFP antibodies. As shown in Figure 7C (bottom panel), a strong GFP-OsNek6 band was detected in the DsRed and GFP-OsNek6 combination, and only a weak GFP-OsNek6 band was detected in the DsRed-OsDIS1 and GFP-OsNek6 combination, by anti-GFP antibody. At the same time, almost no DsRed-OsDIS1 protein was visualized in the DsRed-OsDIS1 and GFP-OsNek6 combination (top panel). Interestingly, when we replaced OsDIS1 with OsDIS1(H71Y) in the coprecipitation assay, GFP-OsNek6 had a similar expression level with the DsRed and GFP-OsNek6 combination; however, DsRed-OsDIS1(H71Y) was effectively coprecipitated, as detected by anti-DsRed antibody (Fig. 7C). Together, these results suggest that OsDIS1 interacts with OsNek6 in vivo.
OsNek6 Sublocalization and Colocalization with OsDIS1
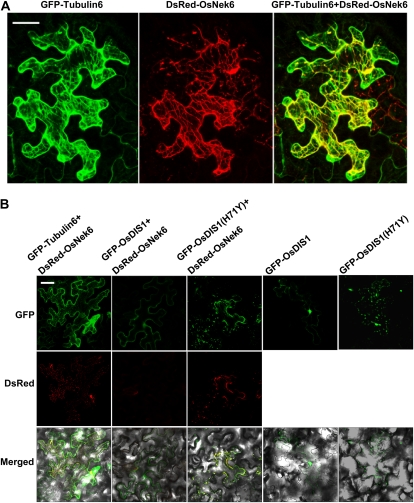
AtNek6, an OsNek6 homolog in Arabidopsis, is involved in cellular morphogenesis and is sublocalized in the tubulin complex (Motose et al., 2008). To determine OsNek6 sublocalization, we coinfiltrated the Agrobacterium strains carrying the DsRed-OsNek6 plasmid and the GFP-Tubulin6 plasmid (a microtubule marker; Nakamura et al., 2004) into N. benthamiana leaves. Confocal image analysis showed that the DsRed-OsNek6 fusion protein was detected in the plasma membrane as well as in some areas in the cytoplasm where the branching points of the filamentous structures were present, which had a good colocalization with GFP-Tubulin6 (Fig. 8A); this is similar to AtNek6 sublocalization in Arabidopsis (Motose et al., 2008). These results indicate that OsNek6 is sublocalized in cytoplasm related to the tubulin complex.
Figure 8.
OsNek6 sublocalization and cosublocalization with OsDIS1. A, OsNek6 sublocalization with GFP-Tubulin6. The Agrobacterium strains carrying the DsRed-OsNek6 and GFP-Tubulin6 plasmids were coinfiltrated into N. benthamiana, and the transformed leaves were analyzed by confocal microscopy 48 h later. Bar = 50 μm. B, OsNek6 colocalization with OsDIS1. The Agrobacterium strains carrying the GFP-OsDIS1 and DsRed-OsNek6 plasmids were coinfiltrated into N. benthamiana leaves. The GFP-Tubulin6 and GFP-OsDIS1(H71Y) plasmids were used as controls. The infiltrated leaves were analyzed by confocal microscopy 48 h after agroinfiltration. Bar = 50 μm.
To investigate whether OsNek6 overlaps with OsDIS1 in sublocalization, we coinfiltrated the Agrobacterium strains carrying the GFP-OsDIS1 and DsRed-OsNek6 plasmids using the GFP-Tubulin6 and GFP-OsDIS1(H71Y) plasmids as controls. Only a weak DsRed-OsNek6 signal was observed when GFP-OsDIS1 and DsRed-OsNek6 were coexpressed in the same cells (Fig. 8B). In contrast, an intermediate DsRed-OsNek6 fluorescence signal was detected in both the GFP-Tubulin6 and DsRed-OsNek6 and the GFP-OsDIS1(H71Y), and DsRed-OsNek6 combinations (Fig. 8B), indicating that OsNek6 was not able to coexist with OsDIS1, which is consistent with the coimmunoprecipitation experiment (Fig. 7C). Furthermore, the GFP-OsDIS1(H71Y) mutant protein had a different sublocalization pattern with GFP-OsDIS1 but had good colocalization with DsRed-OsNek6. Based on these results, we speculate that OsNek6 might be degraded by OsDIS1 via the ubiquitination pathway and that this degradation could be abolished by the mutation in OsDIS1(H71Y), which causes the loss of its E3 ubiquitin activity and alterations in the protein sublocalization (Fig. 2B).
OsDIS1 Promotes OsNek6 Degradation in Vivo
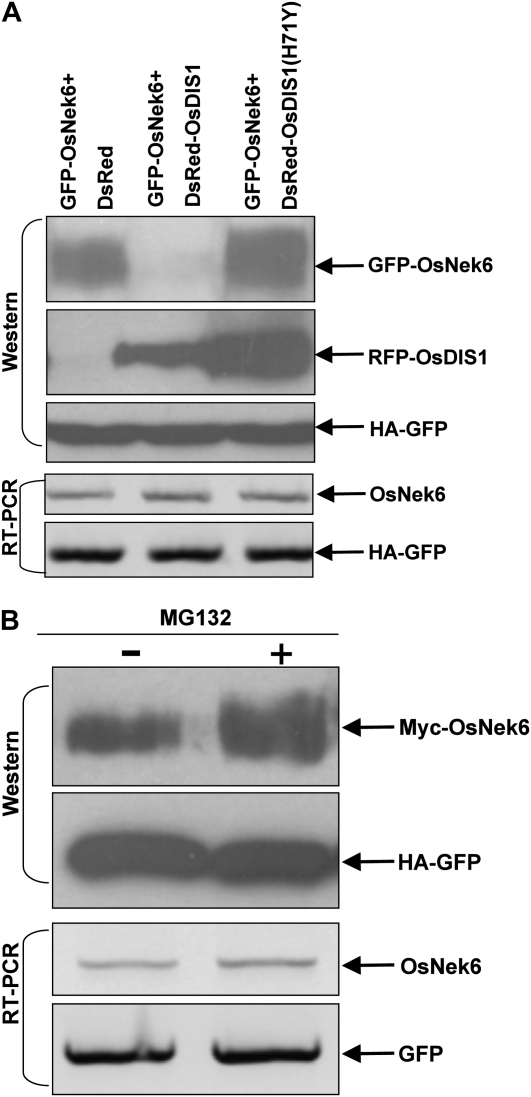
To test the hypothesis that OsNek6 is degraded by OsDIS1 in vivo, we coinfiltrated the Agrobacterium strains containing the plasmids into N. benthamiana leaves with similar coprecipitation combinations used in the above in vivo assays (Fig. 7C), except that hemagglutinin (HA)-GFP plasmid was added as an internal control (Liu et al., 2010). Western-blot analysis revealed that the GFP-OsNek6 protein had a relatively high-level expression in the GFP-OsNek6 and DsRed combination (Fig. 9A). However, the main GFP-OsNek6 band was not detected in the GFP-OsNek6 and DsRed-OsDIS1 combination (Fig. 9A). Furthermore, in the GFP-OsNek6 and DsRed-OsDIS1(H71Y) combination, GFP-OsNek6 protein expression was similar to that of the GFP-OsNek6 and DsRed combination (Fig. 9A). Meanwhile, the expression of DsRed-OsDIS1 and DsRed-OsDIS1(H71Y) was detected by the anti-DsRed antibody, and the internal control HA-GFP protein showed a similar expression in all three combinations (Fig. 9A). RT-PCR analysis showed that the OsNek6 and HA-GFP genes in the infiltrated tissues had similar expression levels in these three combinations (Fig. 9A). Together, these results indicated that OsDIS1 promotes OsNek6 degradation in vivo while OsDIS1(H71Y) loses degradation ability due to the lack of E3 ubiquitin ligase activity (Fig. 2B).
Figure 9.
OsDIS1 promotion of OsNek6 degradation in vivo. A, OsNek6 degradation assay. The Agrobacterium strains carrying the DsRed-OsDIS1 and GFP-OsNek6 plasmids were coinfiltrated into N. benthamiana, with the DsRed and DsRed-OsDIS1(H71Y) plasmids as controls. The HA-GFP plasmid was used as an internal control for protein synthesis. Three days after infiltration, samples were collected for protein and RNA extraction. B, Inhibition of OsNek6 degradation by MG132. A 50 μm solution of MG132 was infiltrated into N. benthamiana 24 h before sampling with DMSO as a control. The extracted protein and RNA were used for western-blot and RT-PCR analyses, respectively.
Because E3 ubiquitin ligases degrade their substrate proteins mainly via the 26S proteasome-dependent pathway (Xie et al., 2002; Liu et al., 2010), we investigated whether OsNek6 degradation is affected by MG132, a proteasome inhibitor. Western-blot analysis showed that after 24 h, OsNek6 accumulation was greater with the MG132 treatment than with the mock dimethyl sulfoxide (DMSO) treatment. At the same time, the internal control HA-GFP had an equal protein expression, as well as at the transcript level of OsNek6 and HA-GFP, revealed by RT-PCR (Fig. 9B). Combined with the above in vivo degradation data (Fig. 9A), we conclude that OsNek6 is degraded by OsDIS1 in vivo via the 26S proteasome-dependent pathway.
DISCUSSION
The SINAT E3 Ligase OsDIS1 Is a Novel Negative Regulator of Drought Response in Rice
In plants, the first identified SINA member was Arabidopsis SINAT5, which interacts with the transcription factor NAC1 and promotes its degradation to attenuate auxin signals (Xie et al., 2002). SINAT5 is also involved in flowering-time regulation by ubiquitinating LHY in Arabidopsis (Park et al., 2010a). The Arabidopsis SINAT2 interacts with RAP2.2 and functions in the carotenogenesis of leaves (Welsch et al., 2007). The ectopically expressed SINAT5 dominant negative form SINAT5 (C49S) in Medicago truncatula affects plant growth and nodulation (Den Herder et al., 2008). Although there are six SINAT-like genes in rice, 10 in Populus trichocarpa, at least six in Zea mays, and at least one in Physcomitrella patens (Wang et al., 2008), none of their functions has been elucidated. In this study, we identified and characterized the OsDIS1 gene in rice, which is the ortholog of the SINAT5 gene in Arabidopsis (Xie et al., 2002). We found that OsDIS1 is induced by drought stress and that the protein has E3 ligase activity. Silencing of the gene in transgenic rice leads to enhanced drought tolerance whereas overexpression of the gene leads to increased drought sensitivity, indicating a negative role in drought response. To the best of our knowledge, this is the first SINA gene to be functionally characterized in rice and the first SINA gene determined to be involved in the drought response in plants.
As climate changes and global warming are inevitable in the future, drought will be one of the most severe limitations for rice production in many regions of the world. As shown in Figure 5A, the OsDIS1 RNAi plants can recover well after a 9-d drought stress. The recovered plants grew and produced seeds normally. These results demonstrated the potential application of this gene in engineering drought-tolerant plants for rice production. It will be worthwhile to check if there is a correlation between a low expression level of the OsDIS1 gene in rice germplasm and enhanced drought tolerance. If a positive result is obtained, the expression level of OsDIS1 can be used as a functional marker for breeding line selection. Alternatively, RNAi transgenic plants with reduced OsDIS1 expression can be generated for rice production if transgenic rice cultivars are allowed to grow in the field.
OsDIS1 Is Involved in the Rice Drought Response via Regulating Drought-Responsive Genes
To better understand the molecular basis of OsDIS1-mediated drought tolerance in rice, we first compared the global expression profiles of OsDIS1 overexpression and wild-type plants under normal conditions (Supplemental Fig. S3B; Supplemental Tables S1 and S2). The microarray analysis showed that OsDIS1 can affect not only the expression of a large number of abiotic stress-related genes but also genes involved in pollen development, disease resistance, and cell wall development; this indicates that OsDIS1 may be involved in various cellular processes in rice. For example, Os01g46860, which encodes a high-affinity inorganic phosphate transporter 11 protein (Paszkowski et al., 2002; Ai et al., 2009), was specifically induced in the OsDIS1 overexpression plants (Fig. 6A), suggesting that OsDIS1 may participate in nutritional equilibrium regulation in rice. We also performed microarray analysis of the OsDIS1 overexpression and wild-type plants under drought conditions (Supplemental Fig. S3C; Supplemental Tables S3 and S4). As was previously reported for OsbZIP23 and DSM1 transgenic rice (Xiang et al., 2008; Ning et al., 2010), many genes in the OsDIS1 overexpression plants showed differential expression under drought treatment. These genes encode stress-related proteins and regulatory factors (Shinozaki and Yamaguchi-Shinozaki, 2007), such as protease inhibitor/seed storage/LTP family protein precursors, peroxidase precursors, universal stress proteins, amino acid metabolism enzymes and carrier proteins, transcription factors (WRKY, MYB, and AP2), and protein kinase. Among them, the induced gene OsGRP2A (Fig. 6B) is the homolog of GRP7 in Arabidopsis, which regulates stomatal opening in response to drought stress (Kim et al., 2008). Whether OsGRP2A also has a similar function in rice is under investigation. In addition, the suppressed gene OsMT-I-4b (Fig. 6C), a homolog of OsMT1a (a type 1 metallothionein), acts as a positive regulator in drought tolerance in rice by directly participating in the reactive oxygen species-scavenging pathway (Yang et al., 2009). Because the zinc finger protein DST positively regulates drought tolerance by affecting stomatal opening and closing in rice via a direct modulation of genes related to hydrogen peroxide homeostasis (Huang et al., 2009), we speculate that OsDIS1 may be a negative regulator of drought-stress signaling by the suppression of the reactive oxygen species-scavenging pathway and stomatal opening in rice. Except for interacting with OsNek6, it is possible that the E3 ligase OsDIS1 acts as a negative regulator of the drought response by polyubiquitination or monoubiquitination of drought-related transcription factors. Degradation by polyubiquitination or structural modifications by monoubiquitination of these transcription factors could lead to a rapid expression change of the downstream genes in the drought signaling pathway in rice.
OsDIS1 Interacts with OsNek6 and Promotes Its Degradation in Vivo
In plants, the Nek genes are highly expressed in meristems and are involved in early organ development and cell cycle regulation (Pnueli et al., 2001; Cloutier et al., 2005). A genome-wide analysis showed that there are seven Nek members in Arabidopsis, six members in rice, and nine members in P. trichocarpa (Vigneault et al., 2007). The phylogenetic analysis demonstrated that plant Nek family members are highly conserved and have a similar exon-intron structure, indicating that they might be derived from large-scale duplication events (Vigneault et al., 2007). Until now, the functions of only a few Neks have been identified in plants. For example, the rice OsNek3 gene is involved in cytoplasmic male sterility, which is preferentially expressed in mature pollen, downstream of DCW11, and interacts with OsPLIM2b (Fujii et al., 2009). Another gene, AtNek6 in Arabidopsis, is associated with microtubules and functions in epidermal cell morphogenesis via its kinase activity and interaction with ARK1 and ARK2 (Motose et al., 2008; Sakai et al., 2008). Moreover, AtNek6 also interacts with ARIA and is involved in the response to ABA and high osmolarity during seed germination (Lee et al., 2010). These results indicate that Neks have multiple functions in plant growth and responses to stress. We identified OsNek6 in the yeast two-hybrid screens using OsDIS1 as the bait (Fig. 7A). Although the GST pull-down assay demonstrated that OsDIS1 physically interacts with OsNek6 in vitro (Fig. 7B), the in vivo coimmunoprecipitation assay showed that OsDIS1 was not able to coexist with OsNek6 while the mutant OsDIS1(H71Y) did (Fig. 7C); this result indicated that OsDIS1, as a ubiquitin E3 ligase, might promote OsNek6 degradation. This speculation was confirmed by the cosublocalization assay for OsDIS1 and OsNek6 in N. benthamiana (Fig. 8B). An in vivo degradation assay also showed that OsDIS1 can promote OsNek6 degradation while the OsDIS1(H71Y) mutant cannot (Fig. 9A). In addition, our sublocalization assays indicated that OsDIS1 is localized predominantly in the nucleus, as well as some in the cytoplasm and plasma membrane (Fig. 3; Supplemental Fig. S2), and OsNek6 is mainly localized in the tubulin complex of cytoplasm (Fig. 8A). Interestingly, the interaction between these two proteins seems to occur in the tubulin complex (Fig. 8B). The mutant OsDIS1(H71Y) is also exported to the same location (Fig. 8B, right panel). It is possible that OsDIS1 is imported from the nucleus to the cytoplasm to interact with OsNek6, because it has two nuclear export signals in amino acid positions 98 to 100 and 107 to 109. Alternatively, OsNek6 interacts with OsDIS1 in the nucleus and brings it out of the nucleus to the tubulin complex. The reason why GFP-OsDIS1(H71Y) has a different sublocalization with GFP-OsDIS1 and the real interaction relationship between OsDIS1 and OsNek6 need further investigation. Since OsDIS1 likely causes OsNek6 degradation, they may have opposite functions in drought stress regulation. Therefore, it is possible that overexpression of OsNek6 may lead to enhanced drought tolerance and that RNAi silencing may lead to enhanced drought sensitivity in transgenic rice. Further functional analysis of the OsNek6 gene using reverse genetic approaches will provide evidence for these speculations.
MATERIALS AND METHODS
Plant Materials and Stress Treatment
Rice (Oryza sativa japonica ‘Nipponbare’) and Nicotiana benthamiana were used in this study. Rice seeds were surface disinfected in 75% ethanol for 1 min and in 2% sodium hypochlorite for 35 min. The germinated seeds were transferred to soil as described previously by Vega-Sanchez et al. (2008). Rice seedlings were grown under a 14-h-light/10-h-dark photoperiod in the greenhouse at approximately 30°C in the day and approximately 25°C at night. N. benthamiana plants were grown under a 16-h-light/8-h-dark photoperiod at 22°C with 70% relative humidity in a growth chamber. About 1- to 1.5-month-old N. benthamiana plants were used for agroinfiltration experiments. For gene expression analysis, 7-d-old rice seedlings cultured on half-strength Murashige and Skoog (MS) medium plus 3% Suc were transferred onto filter papers to induce drought stress and sampled at 0, 2, 4, 8, 12, and 24 h after treatment.
RNA Isolation, RT-PCR, Real-Time PCR, and RNA Gel-Blot Analyses
Total RNA was extracted from rice and N. benthamiana leaves using Trizol (Invitrogen) according to the manufacturer’s instructions. To examine the OsNek6 transcriptional level in the infiltrated N. benthamiana leaves by RT-PCR, the RNA samples (approximately 2 μg) were treated with DNase I (Invitrogen) and subjected to the RT system (Promega). PCR amplification was preformed using the OsNek6-specific forward (RT Fw, 5′-ATGTGCCCAGAAATACTAG-3′) and reverse (RT Rev, 5′-CTACGTAAGTTTTGGTGACCC-3′) primers. The expression of HA-GFP was used as an internal control. To check the OsDIS1 expression level in Nipponbare in response to drought stress and OsDIS1 RNAi transgenic plants and to confirm the microarray data by real-time PCR, about 2 μg of rice total RNA was treated by DNase I (Invitrogen) and used for first-strand cDNA synthesis by the RT system (Promega). The reaction products were diluted 5-fold and used as the template for real-time PCR. Real-time PCR was performed with the Bio-Rad iQ5 Gradient Real Time PCR system (Bio-Rad) with the following reaction conditions: 95°C for 4 min, then 40 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 20 s. The rice Ubiquitin (Ubq) gene was used as the reference gene to normalize target gene expression. Primers used in real-time PCR are listed in Supplemental Table S5. To examine the OsDIS1 expression by RNA gel blot, 10 μg of total RNA from each sample was separated on 1.2% (w/v) agarose formaldehyde gels, transferred to Hybond-N nylon membranes (Amersham Pharmacia Biotech), and then hybridized with a 32P-labeled full-length OsDIS1 fragment. The 28s rRNA was used as a loading control.
E3 Ubiquitin Ligase Activity Assay
The full-length cDNA of OsDIS1 was cloned into the pGEX-6P-1 vector, fused to the N-terminal GST, and expressed in Escherichia coli strain BL21. The GST-OsDIS1 fusion proteins were prepared according to the instruction manual (Amersham Biosciences). For E3 ubiquitin ligase activity assay of the fusion proteins, the reaction used crude extract containing recombinant wheat (Triticum aestivum) E1 (GI: 136632), human E2 (UBCH5b; approximately 40 ng), purified E3 GST-OsDIS1 (approximately 1 μg), and purified Arabidopsis (Arabidopsis thaliana) ubiquitin (a ubiquitin monomer of UBQ14; At4g02890; approximately 2 μg) fused with the His tag. The reaction was incubated at 30°C for 15, 30, 60, and 90 min. The OsDIS1 mutant that contains a mutation in the RING finger domain (H71Y) was prepared using the Quickchange site-directed mutagenesis system (Stratagene). The sequences of the primer pair used for the preparation of the His-71→Tyr-71 mutant were as follows: M1F (5′-CATCAGTGCTCTAATGGTTATACATTGTGCTCTGGATGC-3′) and M1R (5′-GCATCCAGAGCACAATGTATAACCATTAGAGCACTGATG-3′). The in vitro E3 ligase assays were performed as described previously (Xie et al., 2002; Zhang et al., 2007).
GFP and DsRed Fusion Vectors and Subcellular Localization
The coding sequences of both OsDIS1 and OsNek6 genes were directionally cloned into the pGDG and pGDR vectors, which contain a cauliflower mosaic virus promoter-driven GFP and DsRed gene, respectively (Goodin et al., 2002). For subcellular localization in N. benthamiana leaves, an Agrobacterium tumefaciens strain carrying the GFP-OsDIS1, GFP-OsDIS1(H71Y), GFP-Tubulin6, or DsRed-OsNek6 plasmid was infiltrated into N. benthamiana leaves individually or in pairs and analyzed by confocal microscopy 48 h after agroinfiltration, as described previously (Goodin et al., 2002). For detection of the nucleus, samples were stained with 50 μg mL−1 DAPI for 5 min. For subcellular localization in rice protoplasts, 10 μg of GFP/DsRed or GFP/DsRed-OsDIS1 plasmids was transformed into protoplasts isolated from rice seedlings and analyzed by confocal microscopy after 16 h of incubation following transfection, as described previously (Chen et al., 2006).
Generation of the OsDIS1 Transgenic Rice
For the overexpression construct, the full-length cDNA of OsDIS1 was inserted into the Ubix.nc1300.ntap.gck vector under the control of the Zea mays ubiquitin promoter, in which the TAP tag is located after the promoter and before the OsDIS1 gene (Chern et al., 2005). The RNAi construct was built in the pANDA vector to silence OsDIS1 using the unique N-terminal region (Miki and Shimamoto, 2004). All the constructs were transformed into Nipponbare using the Agrobacterium-mediated method (Qu et al., 2006). Transgenic rice lines and their progeny were grown in the greenhouse. The seeds of T3 homozygous overexpression and RNAi lines were selected on half-strength MS medium plus 3% Suc containing 50 mg L−1 hygromycin before they were subjected to molecular and phenotypic analyses.
Drought-Stress Treatments for the OsDIS1 Transgenic Lines
The seeds of the T3 generation homozygous OsDIS1 overexpression and RNAi transgenic lines as well as wild-type seeds were geminated on half-strength MS medium plus 3% Suc. Seven-day-old seedlings were planted in small pots (five plants per pot, and five pots for each line) that contained the same amount of soil, and the pots, which had holes in the bottom, were placed in large trays that held 5 to 6 cm of water in the greenhouse (30°C during the day and 25°C at night with natural light). The water in the trays maintained the moisture in the pots. When the plants were 4 weeks old, they were moved out of the trays and placed on a bench in the greenhouse for drought treatment; they were not watered. Drought phenotypes of the treated plants were observed on days 6, 7, and 8 after water was withheld. Nine days after the treatment, the plants were moved back to the large trays, which had 5 to 6 cm of water, for recovery. The survival rate of the transgenic lines and wild-type plants was recorded 4 d after rehydration. The drought stress experiment was performed at least three times.
Microarray Analysis
The OsDIS1 overexpression 9-4-2 plants and the wild-type plants were cultured on half-strength MS medium plus 3% Suc for 7 d. About half of the plants were sampled as the untreated control for RNA isolation, and the rest were transferred with half-strength MS medium onto filter papers to induce drought stress. When the leaves of the OsDIS1 overexpression plants began to show drought-stress phenotypes, we collected leaves for RNA isolation. OsLEA3 was used as a positive control for the drought treatment. Total RNA was extracted using Trizol (Invitrogen) according to the manufacturer’s instructions. Two independent replicates were used for each sample. cDNA synthesis, purification, labeling, and chip hybridization were performed by ShanghaiBio using the Agilent Genechip. The chip hybridization results were scanned by the Agilent Microarray Scanner System and were normalized and analyzed with Agilent Feature Extraction software. Genes with more than 2-fold change in the overexpression plants compared with the wild-type plants were selected. The expression pattern of some differentially expressed genes was further confirmed by real-time PCR.
Yeast Two-Hybrid Screening
The ProQuest Two-Hybrid System (Invitrogen) was used to screen for OsDIS1-interacting protein(s), as described previously (Vega-Sanchez et al., 2008). The coding sequence of OsDIS1 was cloned into the bait vector pDBLeu. The yeast strain MaV203 was used for the transformation, and the cDNA library of 3-week-old rice seedlings was used for the screening. After two rounds of cDNA library screening, one of the positive clones was identified as OsNek6. To confirm the interaction between OsDIS1 and OsNek6, the coding sequences of OsDIS1 and OsNek6 were cloned to pGBKT7 and pGADT7 vectors, respectively. The yeast strain HF7c (including two reporter genes: lacZ and his) was used. The yeast cells were plated onto synthetic dextrose (SD)/−Trp/−Leu medium to confirm the cotransformation. At the same time, the yeast cells were plated onto SD/−His/−Trp/−Leu containing 3 mm 3-amino-1,2,4,-triazole medium to screen for candidate interactors.
In Vitro Pull-Down Assay
The full-length cDNA of OsNek6 was cloned into the pTNT vector, and then in vitro transcription and translation were performed using the TNT Coupled Wheat Germ Extract Systems (Promega) to generate [35S]Met-labeled OsNek6 proteins. The coding sequence of OsDIS1 was cloned into the pGEX-6P-1 vector, and the fusion protein was prepared according to the instruction manual (Amersham Biosciences). In vitro GST pull-down assay was conducted as described by Lai et al. (2009) with the following modifications. [35S]Met-labeled OsNek6 protein (3 μL) was inoculated with 1 μg of GST-OsDIS1 protein in GST-binding buffer at room temperature for 1 h. The GST protein was used as a control in the assay. After incubation and washing, the mixed proteins as well as 1 μL of [35S]Met-labeled OsNek6 protein as the input were subjected to SDS-PAGE; then the blot was exposed and scanned.
In Vivo Coimmunoprecipitation Experiments
The protein from the N. benthamiana leaves coinfiltrated with the DsRed/DsRed-OsDIS1/DsRed-OsDIS1(H71Y) plasmids and GFP-OsNek6 plasmids was extracted with native extraction buffer (Suc buffer) as described by Liu et al. (2010). A 15-μL volume of anti-GFP antibody (Roche) was added to 1 mL of cell lysates with 50 μm MG132 to prevent protein degradation. Then, binding was carried out at 4°C for 4 h with gentle shaking and was followed by the addition of 20 μL of protein G agarose beads (Millipore). After 3 h of incubation at 4°C, the precipitated samples were washed, separated by SDS-PAGE, and subjected to immunoblot analysis using anti-DsRed antibody (BGI) and anti-GFP antibody (Roche).
In Vivo Degradation Assay and the MG132 Treatment
For in vivo degradation assay, we followed the protocol described by Liu et al. (2010). We coinfiltrated the Agrobacterium strains carrying the DsRed-OsDIS1 and GFP-OsNek6 plasmids into N. benthamiana leaves. The DsRed and DsRed-OsDIS1(H71Y) plasmids were used as the controls, and the HA-GFP plasmid was added as an internal control. Three days after infiltration, samples were collected for protein and RNA extraction. For protein level analysis, the extracts were analyzed using anti-DsRed antibody (BGI), anti-GFP antibody (Roche), and anti-HA antibody (Santa Cruz). For RNA level expression analysis, RT-PCR was performed.
For the MG132 treatment, 24 h before sampling, MG132 with a final concentration of 50 μm was infiltrated into N. benthamiana leaves, and DMSO was used as a control. The harvested tissues were used for western-blot and RT-PCR analyses.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number AK058336.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. SINA proteins in rice and Arabidopsis.
Supplemental Figure S2. Subcellular localization of the OsDIS1 protein in rice protoplasts.
Supplemental Figure S3. Drought treatment for microarray and linear model analysis.
Supplemental Table S1. Up-regulated genes in the OsDIS1 overexpression transgenic plants under normal conditions.
Supplemental Table S2. Down-regulated genes in the OsDIS1 overexpression transgenic plants under normal conditions.
Supplemental Table S3. Up-regulated genes in the OsDIS1 overexpression transgenic plants under normal and drought conditions.
Supplemental Table S4. Down-regulated genes in the OsDIS1 overexpression transgenic plants under normal and drought conditions.
Supplemental Table S5. Primers used for real-time PCR.
Acknowledgments
We thank Dr. Ko Shimamoto (Nara Institute of Science and Technology) and Dr. Pamela Ronald (University of California, Davis) for kindly providing the pANDA and Ubix.nc1300.ntap.gck vectors for rice transformation, respectively, as well as Dr. Michael M. Goodin (University of Kentucky) for kindly providing the pGDG and pGDR plasmids for subcellular localization.
References
- Aharon R, Shahak Y, Wininger S, Bendov R, Kapulnik Y, Galili G. (2003) Overexpression of a plasma membrane aquaporin in transgenic tobacco improves plant vigor under favorable growth conditions but not under drought or salt stress. Plant Cell 15: 439–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai P, Sun S, Zhao J, Fan X, Xin W, Guo Q, Yu L, Shen Q, Wu P, Miller AJ, et al. (2009) Two rice phosphate transporters, OsPht1;2 and OsPht1;6, have different functions and kinetic properties in uptake and translocation. Plant J 57: 798–809 [DOI] [PubMed] [Google Scholar]
- Bachmair A, Novatchkova M, Potuschak T, Eisenhaber F. (2001) Ubiquitylation in plants: a post-genomic look at a post-translational modification. Trends Plant Sci 6: 463–470 [DOI] [PubMed] [Google Scholar]
- Boyer JS. (1982) Plant productivity and environment. Science 218: 443–448 [DOI] [PubMed] [Google Scholar]
- Carthew RW, Rubin GM. (1990) Seven in absentia, a gene required for specification of R7 cell fate in the Drosophila eye. Cell 63: 561–577 [DOI] [PubMed] [Google Scholar]
- Chen S, Tao L, Zeng L, Vega-Sanchez ME, Umemura K, Wang GL. (2006) A highly efficient transient protoplast system for analyzing defence gene expression and protein-protein interactions in rice. Mol Plant Pathol 7: 417–427 [DOI] [PubMed] [Google Scholar]
- Chern M, Canlas PE, Fitzgerald HA, Ronald PC. (2005) Rice NRR, a negative regulator of disease resistance, interacts with Arabidopsis NPR1 and rice NH1. Plant J 43: 623–635 [DOI] [PubMed] [Google Scholar]
- Cho SK, Chung HS, Ryu MY, Park MJ, Lee MM, Bahk YY, Kim J, Pai HS, Kim WT. (2006) Heterologous expression and molecular and cellular characterization of CaPUB1 encoding a hot pepper U-box E3 ubiquitin ligase homolog. Plant Physiol 142: 1664–1682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SK, Ryu MY, Song C, Kwak JM, Kim WT. (2008) Arabidopsis PUB22 and PUB23 are homologous U-box E3 ubiquitin ligases that play combinatory roles in response to drought stress. Plant Cell 20: 1899–1914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloutier M, Vigneault F, Lachance D, Seguin A. (2005) Characterization of a poplar NIMA-related kinase PNek1 and its potential role in meristematic activity. FEBS Lett 579: 4659–4665 [DOI] [PubMed] [Google Scholar]
- Cushman JC, Bohnert HJ. (2000) Genomic approaches to plant stress tolerance. Curr Opin Plant Biol 3: 117–124 [DOI] [PubMed] [Google Scholar]
- Den Herder G, De Keyser A, De Rycke R, Rombauts S, Van de Velde W, Clemente MR, Verplancke C, Mergaert P, Kondorosi E, Holsters M, et al. (2008) Seven in absentia proteins affect plant growth and nodulation in Medicago truncatula. Plant Physiol 148: 369–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Zhou X, Li L, Su Z. (2009) PlantsUPS: a database of plants’ ubiquitin proteasome system. BMC Genomics 10: 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreher K, Callis J. (2007) Ubiquitin, hormones and biotic stress in plants. Ann Bot (Lond) 99: 787–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii S, Yamada M, Toriyama K. (2009) Cytoplasmic male sterility-related protein kinase, OsNek3, is regulated downstream of mitochondrial protein phosphatase 2C, DCW11. Plant Cell Physiol 50: 828–837 [DOI] [PubMed] [Google Scholar]
- Fujita M, Fujita Y, Noutoshi Y, Takahashi F, Narusaka Y, Yamaguchi-Shinozaki K, Shinozaki K. (2006) Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Curr Opin Plant Biol 9: 436–442 [DOI] [PubMed] [Google Scholar]
- George S, Venkataraman G, Parida A. (2010) A chloroplast-localized and auxin-induced glutathione S-transferase from phreatophyte Prosopis juliflora confer drought tolerance on tobacco. J Plant Physiol 167: 311–318 [DOI] [PubMed] [Google Scholar]
- Goodin MM, Dietzgen RG, Schichnes D, Ruzin S, Jackson AO. (2002) pGD vectors: versatile tools for the expression of green and red fluorescent protein fusions in agroinfiltrated plant leaves. Plant J 31: 375–383 [DOI] [PubMed] [Google Scholar]
- Hirayama T, Shinozaki K. (2010) Research on plant abiotic stress responses in the post-genome era: past, present and future. Plant J 61: 1041–1052 [DOI] [PubMed] [Google Scholar]
- Hochstrasser M. (2006) Lingering mysteries of ubiquitin-chain assembly. Cell 124: 27–34 [DOI] [PubMed] [Google Scholar]
- Huang XY, Chao DY, Gao JP, Zhu MZ, Shi M, Lin HX. (2009) A previously unknown zinc finger protein, DST, regulates drought and salt tolerance in rice via stomatal aperture control. Genes Dev 23: 1805–1817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang JY, Lee SH, Rhee JY, Chung GC, Ahn SJ, Kang H. (2007) Transgenic Arabidopsis and tobacco plants overexpressing an aquaporin respond differently to various abiotic stresses. Plant Mol Biol 64: 621–632 [DOI] [PubMed] [Google Scholar]
- Ji W, Zhu Y, Li Y, Yang L, Zhao X, Cai H, Bai X. (2010) Over-expression of a glutathione S-transferase gene, GsGST, from wild soybean (Glycine soja) enhances drought and salt tolerance in transgenic tobacco. Biotechnol Lett 32: 1173–1179 [DOI] [PubMed] [Google Scholar]
- Kim JS, Jung HJ, Lee HJ, Kim KA, Goh CH, Woo Y, Oh SH, Han YS, Kang H. (2008) Glycine-rich RNA-binding protein 7 affects abiotic stress responses by regulating stomata opening and closing in Arabidopsis thaliana. Plant J 55: 455–466 [DOI] [PubMed] [Google Scholar]
- Ko JH, Yang SH, Han KH. (2006) Upregulation of an Arabidopsis RING-H2 gene, XERICO, confers drought tolerance through increased abscisic acid biosynthesis. Plant J 47: 343–355 [DOI] [PubMed] [Google Scholar]
- Lai J, Chen H, Teng K, Zhao Q, Zhang Z, Li Y, Liang L, Xia R, Wu Y, Guo H, et al. (2009) RKP, a RING finger E3 ligase induced by BSCTV C4 protein, affects geminivirus infection by regulation of the plant cell cycle. Plant J 57: 905–917 [DOI] [PubMed] [Google Scholar]
- Lee HK, Cho SK, Son O, Xu Z, Hwang I, Kim WT. (2009) Drought stress-induced Rma1H1, a RING membrane-anchor E3 ubiquitin ligase homolog, regulates aquaporin levels via ubiquitination in transgenic Arabidopsis plants. Plant Cell 21: 622–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SJ, Cho DI, Kang JY, Kim MD, Kim SY. (2010) AtNEK6 interacts with ARIA and is involved in ABA response during seed germination. Mol Cells 29: 559–566 [DOI] [PubMed] [Google Scholar]
- Liu H, Zhang H, Yang Y, Li G, Yang Y, Wang X, Basnayake BM, Li D, Song F. (2008) Functional analysis reveals pleiotropic effects of rice RING-H2 finger protein gene OsBIRF1 on regulation of growth and defense responses against abiotic and biotic stresses. Plant Mol Biol 68: 17–30 [DOI] [PubMed] [Google Scholar]
- Liu L, Zhang Y, Tang S, Zhao Q, Zhang Z, Zhang H, Dong L, Guo H, Xie Q. (2010) An efficient system to detect protein ubiquitination by agroinfiltration in Nicotiana benthamiana. Plant J 61: 893–903 [DOI] [PubMed] [Google Scholar]
- Luo LJ. (2010) Breeding for water-saving and drought-resistance rice (WDR) in China. J Exp Bot 61: 3509–3517 [DOI] [PubMed] [Google Scholar]
- Matsukura S, Mizoi J, Yoshida T, Todaka D, Ito Y, Maruyama K, Shinozaki K, Yamaguchi-Shinozaki K. (2010) Comprehensive analysis of rice DREB2-type genes that encode transcription factors involved in the expression of abiotic stress-responsive genes. Mol Genet Genomics 283: 185–196 [DOI] [PubMed] [Google Scholar]
- Miki D, Shimamoto K. (2004) Simple RNAi vectors for stable and transient suppression of gene function in rice. Plant Cell Physiol 45: 490–495 [DOI] [PubMed] [Google Scholar]
- Motose H, Tominaga R, Wada T, Sugiyama M, Watanabe Y. (2008) A NIMA-related protein kinase suppresses ectopic outgrowth of epidermal cells through its kinase activity and the association with microtubules. Plant J 54: 829–844 [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay D, Riezman H. (2007) Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science 315: 201–205 [DOI] [PubMed] [Google Scholar]
- Nakamura M, Naoi K, Shoji T, Hashimoto T. (2004) Low concentrations of propyzamide and oryzalin alter microtubule dynamics in Arabidopsis epidermal cells. Plant Cell Physiol 45: 1330–1334 [DOI] [PubMed] [Google Scholar]
- Ning J, Li X, Hicks LM, Xiong L. (2010) A Raf-like MAPKKK gene DSM1 mediates drought resistance through reactive oxygen species scavenging in rice. Plant Physiol 152: 876–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park BS, Eo HJ, Jang IC, Kang HG, Song JT, Seo HS. (2010a) Ubiquitination of LHY by SINAT5 regulates flowering time and is inhibited by DET1. Biochem Biophys Res Commun 398: 242–246 [DOI] [PubMed] [Google Scholar]
- Park GG, Park JJ, Yoon J, Yu SN, An G. (2010b) A RING finger E3 ligase gene, Oryza sativa Delayed Seed Germination 1 (OsDSG1), controls seed germination and stress responses in rice. Plant Mol Biol 74: 467–478 [DOI] [PubMed] [Google Scholar]
- Paszkowski U, Kroken S, Roux C, Briggs SP. (2002) Rice phosphate transporters include an evolutionarily divergent gene specifically activated in arbuscular mycorrhizal symbiosis. Proc Natl Acad Sci USA 99: 13324–13329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pnueli L, Gutfinger T, Hareven D, Ben-Naim O, Ron N, Adir N, Lifschitz E. (2001) Tomato SP-interacting proteins define a conserved signaling system that regulates shoot architecture and flowering. Plant Cell 13: 2687–2702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin F, Sakuma Y, Tran LS, Maruyama K, Kidokoro S, Fujita Y, Fujita M, Umezawa T, Sawano Y, Miyazono K, et al. (2008) Arabidopsis DREB2A-interacting proteins function as RING E3 ligases and negatively regulate plant drought stress-responsive gene expression. Plant Cell 20: 1693–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu S, Liu G, Zhou B, Bellizzi M, Zeng L, Dai L, Han B, Wang GL. (2006) The broad-spectrum blast resistance gene Pi9 encodes a nucleotide-binding site-leucine-rich repeat protein and is a member of a multigene family in rice. Genetics 172: 1901–1914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu MY, Cho SK, Kim WT. (2010) The Arabidopsis C3H2C3-type RING E3 ubiquitin ligase AtAIRP1 is a positive regulator of an abscisic acid-dependent response to drought stress. Plant Physiol 154: 1983–1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai T, Honing H, Nishioka M, Uehara Y, Takahashi M, Fujisawa N, Saji K, Seki M, Shinozaki K, Jones MA, et al. (2008) Armadillo repeat-containing kinesins and a NIMA-related kinase are required for epidermal-cell morphogenesis in Arabidopsis. Plant J 53: 157–171 [DOI] [PubMed] [Google Scholar]
- Santner A, Estelle M. (2010) The ubiquitin-proteasome system regulates plant hormone signaling. Plant J 61: 1029–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K. (2007) Gene networks involved in drought stress response and tolerance. J Exp Bot 58: 221–227 [DOI] [PubMed] [Google Scholar]
- Smalle J, Vierstra RD. (2004) The ubiquitin 26S proteasome proteolytic pathway. Annu Rev Plant Biol 55: 555–590 [DOI] [PubMed] [Google Scholar]
- Stone SL, Hauksdottir H, Troy A, Herschleb J, Kraft E, Callis J. (2005) Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis. Plant Physiol 137: 13–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone SL, Williams LA, Farmer LM, Vierstra RD, Callis J. (2006) KEEP ON GOING, a RING E3 ligase essential for Arabidopsis growth and development, is involved in abscisic acid signaling. Plant Cell 18: 3415–3428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega-Sanchez ME, Zeng L, Chen S, Leung H, Wang GL. (2008) SPIN1, a K homology domain protein negatively regulated and ubiquitinated by the E3 ubiquitin ligase SPL11, is involved in flowering time control in rice. Plant Cell 20: 1456–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra RD. (2003) The ubiquitin/26S proteasome pathway, the complex last chapter in the life of many plant proteins. Trends Plant Sci 8: 135–142 [DOI] [PubMed] [Google Scholar]
- Vierstra RD. (2009) The ubiquitin-26S proteasome system at the nexus of plant biology. Nat Rev Mol Cell Biol 10: 385–397 [DOI] [PubMed] [Google Scholar]
- Vigneault F, Lachance D, Cloutier M, Pelletier G, Levasseur C, Seguin A. (2007) Members of the plant NIMA-related kinases are involved in organ development and vascularization in poplar, Arabidopsis and rice. Plant J 51: 575–588 [DOI] [PubMed] [Google Scholar]
- Wang M, Jin Y, Fu J, Zhu Y, Zheng J, Hu J, Wang G. (2008) Genome-wide analysis of SINA family in plants and their phylogenetic relationships. DNA Seq 19: 206–216 [DOI] [PubMed] [Google Scholar]
- Welsch R, Maass D, Voegel T, Dellapenna D, Beyer P. (2007) Transcription factor RAP2.2 and its interacting partner SINAT2: stable elements in the carotenogenesis of Arabidopsis leaves. Plant Physiol 145: 1073–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Tang N, Du H, Ye H, Xiong L. (2008) Characterization of OsbZIP23 as a key player of the basic leucine zipper transcription factor family for conferring abscisic acid sensitivity and salinity and drought tolerance in rice. Plant Physiol 148: 1938–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q, Guo HS, Dallman G, Fang S, Weissman AM, Chua NH. (2002) SINAT5 promotes ubiquitin-related degradation of NAC1 to attenuate auxin signals. Nature 419: 167–170 [DOI] [PubMed] [Google Scholar]
- Yang Z, Wu Y, Li Y, Ling HQ, Chu C. (2009) OsMT1a, a type 1 metallothionein, plays the pivotal role in zinc homeostasis and drought tolerance in rice. Plant Mol Biol 70: 219–229 [DOI] [PubMed] [Google Scholar]
- Zeng LR, Vega-Sanchez ME, Zhu T, Wang GL. (2006) Ubiquitination-mediated protein degradation and modification: an emerging theme in plant-microbe interactions. Cell Res 16: 413–426 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Li Y, Gao T, Zhu H, Wang DJ, Zhang HW, Ning YS, Liu LJ, Wu YR, Chu CC, et al. (2008) Arabidopsis SDIR1 enhances drought tolerance in crop plants. Biosci Biotechnol Biochem 72: 2251–2254 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Yang C, Li Y, Zheng N, Chen H, Zhao Q, Gao T, Guo H, Xie Q. (2007) SDIR1 is a RING finger E3 ligase that positively regulates stress-responsive abscisic acid signaling in Arabidopsis. Plant Cell 19: 1912–1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G, Xu Y, Li J, Yang L, Liu JY. (2006) Molecular analyses of the metallothionein gene family in rice (Oryza sativa L.). J Biochem Mol Biol 39: 595–606 [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. (2004) GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Laule O, Schmitz J, Hruz T, Bleuler S, Gruissem W. (2008) Genevestigator transcriptome meta-analysis and biomarker search using rice and barley gene expression databases. Mol Plant 1: 851–857 [DOI] [PubMed] [Google Scholar]