Abstract
The herbicide glyphosate inhibits the shikimate pathway of the synthesis of amino acids such as phenylalanine, tyrosine, and tryptophan. However, much uncertainty remains concerning precisely how glyphosate kills plants or affects cellular redox homeostasis and related processes in glyphosate-sensitive and glyphosate-resistant crop plants. To address this issue, we performed an integrated study of photosynthesis, leaf proteomes, amino acid profiles, and redox profiles in the glyphosate-sensitive soybean (Glycine max) genotype PAN809 and glyphosate-resistant Roundup Ready Soybean (RRS). RRS leaves accumulated much more glyphosate than the sensitive line but showed relatively few changes in amino acid metabolism. Photosynthesis was unaffected by glyphosate in RRS leaves, but decreased abundance of photosynthesis/photorespiratory pathway proteins was observed together with oxidation of major redox pools. While treatment of a sensitive genotype with glyphosate rapidly inhibited photosynthesis and triggered the appearance of a nitrogen-rich amino acid profile, there was no evidence of oxidation of the redox pools. There was, however, an increase in starvation-associated and defense proteins. We conclude that glyphosate-dependent inhibition of soybean leaf metabolism leads to the induction of defense proteins without sustained oxidation. Conversely, the accumulation of high levels of glyphosate in RRS enhances cellular oxidation, possibly through mechanisms involving stimulation of the photorespiratory pathway.
The concept that the pathways of primary nitrogen assimilation in leaves are closely associated with respiration, photosynthesis, and photorespiration through interactions with carbon metabolism and requirements for energy (ATP) and reducing power (NADH and NADPH) has long been accepted, but our understanding of many of the mechanistic details of coordinate regulation is far from complete (Foyer et al., 2003). In particular, uncertainties remain concerning how the photosynthetic cell balances energy-producing and energy-requiring processes in order to maintain cellular redox status, particularly in stressful conditions (Foyer et al., 2009). For example, there are conflicting reports in the literature concerning the effects of nitrogen supply on cellular redox status and antioxidant enzymes, with some studies reporting that antioxidant enzyme activities increase as the nitrogen supply is increased (Medici et al., 2004) while other studies show either no effect (Domínguez-Valdivia et al., 2007) or that plants experiencing nitrogen deficiency have a higher abundance of antioxidant enzymes (Ramalho et al., 1998). Several lines of evidence suggest that a close relationship exists between cellular redox state and amino acid metabolism. First, the enrichment of the loss of complex I in the tobacco (Nicotiana tabacum) cytoplasmic male sterile II mutants leads to a nitrogen-rich phenotype that is characterized by a general increase in amino acids and organic acids (Dutilleul et al., 2005). Second, an increase in the capacity for chloroplast GSH synthesis led to a general increase in amino acids such as Val, Leu, Ile, Lys, and Tyr (Noctor et al., 1998). Third, a large number of enzymes involved in amino acid metabolism have been identified as potential thioredoxin and glutaredoxin targets (Lee et al., 2010). However, relatively little is known about the mechanistic relationships between amino acid metabolism and cellular redox homeostasis. Exposure to the herbicide glyphosate (N-[phosphonomethyl]glycine), which prevents the production of aromatic amino acids, was found to enhance the accumulation of antioxidant enzymes such as ascorbate peroxidase, glutathione S-transferase [GST], and a copper/zinc superoxide dismutase (Cu-Zn SOD), suggesting that glyphosate treatment might result in oxidative stress (Ahsan et al., 2008).
GSH (γ-l-glutamyl-l-cysteinyl-glycine) is at the heart of the complex antioxidant network of plants that acts to control reactive oxygen species accumulation and to facilitate appropriate cellular redox signaling and defense (Noctor et al., 2011). GSH is also important in herbicide detoxification, particularly with regard to the conjugation of xenobiotics via the glutathione S-transferase system (Dixon and Edwards, 2010). In legumes such as soybean (Glycine max), a homolog of GSH, called homoglutathione (γ-Glu-Cys-β-Ala; hGSH), functions alongside of or, in some species, completely replaces GSH (Klapheck, 1988). The relative proportions of GSH and hGSH that are produced vary considerably according to species and tissue type, but in soybean leaves, hGSH represents over 99.9% of the total hGSH plus GSH pool (Klapheck, 1988). The pathway of GSH and hGSH synthesis involves two ATP-dependent steps. In the first reaction, common to the synthesis of both molecules, γ-glutamyl-Cys is formed from Glu and Cys by γ-glutamylcysteinyl synthetase. In the second reaction, either Gly or β-Ala is added to the C-terminal site of γ-glutamyl-Cys by GSH synthetase or hGSH synthetase, respectively (Frendo et al., 1999, 2001). Although GSH and hGSH are synthesized by distinct enzymes encoded by different genes (Frendo et al., 2001), the homologs are considered to fulfill similar functions in regulating cellular redox state (Noctor et al., 2011). The disulfide forms of both hGSH and another glutathione homolog, γ-Glu-Cys-Ser, are reducible by glutathione reductase (Klapheck, 1988). There is a strong positive correlation between nitrogenase activity and nodule ascorbate and (h)GSH contents in legume nodules (Dalton et al., 1993; Matamoros et al., 2003; Groten et al., 2005, 2006). Depletion of the (h)GSH pool in Medicago truncatula inhibits root nodule formation, indicating that both thiols play a crucial role in this process (Frendo et al., 2005). Resistance to the herbicide fomesafen was conferred on tobacco by the expression of a hGSH synthetase together with a hGSH-preferring GST from the fomesafen-resistant species, soybean (Skipsey et al., 2005).
Aromatic amino acids are synthesized by the shikimate pathway, which generates chorismate from which Trp, Tyr, and Phe are produced via branched pathways. As the starting point for the synthesis of phenylpropanoids, the production of Phe in particular is an important portal for downstream biosyntheses. A key step in the synthesis of chorismate is catalyzed by the chloroplastic enzyme 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS), which converts shikimate-3-phosphate and phosphoenolpyruvate to 5-enolpyruvyl-shikimate-3-phosphate. EPSPS is specifically inhibited by glyphosate, which is currently considered to be the most important herbicide in the world (Powles, 2008). This herbicide has become widely used in many parts of the world after the commercialization of glyphosate-resistant crops, which have constitutive overexpression of a highly efficient glyphosate-resistant form of EPSPS. This has led to the evolution of resistance in weedy species that endangers the continued success of transgenic glyphosate-resistant crops (Powles, 2008). A recently described mechanism of glyphosate resistance involves a large amplification of the EPSPS gene on multiple chromosomes (Gaines et al., 2010).
Despite the widespread use of glyphosate in agriculture, major questions remain concerning the precise mechanisms through which glyphosate kills plants and, even more importantly, how this herbicide affects cell metabolism and physiology in glyphosate-resistant plants. This study was undertaken in order to characterize the interactions between amino acid metabolism through the shikimate pathway and cellular redox state, as determined by changes in the physiologically relevant redox pools of ascorbate/dehydroascorbate, hGSH/oxidized glutathione homolog (hGSSG), and NAD(P)H/NAD(P). We compared the effects of glyphosate in the glyphosate-sensitive soybean genotype PAN809 (PAN) and glyphosate-resistant Roundup Ready Soybean (RRS), which express the Agrobacterium tumefaciens strain CP4 5-enolpyruvylshikimate-3-phosphate synthase that is resistant to inhibition by glyphosate. We undertook this work on soybean because it is a major crop species: genetically modified lines currently occupy more than 75% of the acreage planted globally. The data presented here provide new information concerning the influence of amino acid metabolism on the abundance of photosynthetic proteins and cellular redox status. We show that glyphosate induces defense proteins in the absence of oxidative stress in the sensitive line. However, the RRS plants accumulate much higher levels of glyphosate than the sensitive line, and this is associated with enhanced cellular oxidation and specific enhancement of proteins associated with photorespiration.
RESULTS
Amino Acid Profiling of Glyphosate Effects
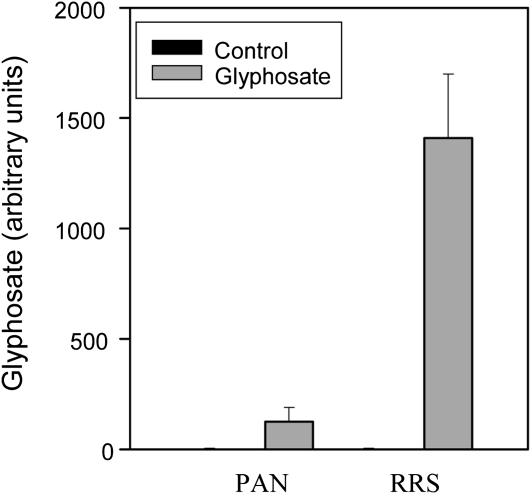
Analysis of primary amines using HPLC-fluorescence analysis with the widely used reagent o-phthalaldialdehyde revealed the presence of a peak that was only detected in glyphosate-treated plants. Although further analyses will be required to confirm its chemical identity, this peak was taken to be glyphosate based on coelution with a glyphosate standard. The peak was readily detectable in the sensitive PAN leaves 5 d after spraying but was over 10-fold higher in the resistant RRS leaves than in the PAN leaves at the same harvest point (Fig. 1).
Figure 1.
A comparison of the levels of glyphosate accumulation in the leaves of glyphosate-sensitive (PAN) and glyphosate-resistant (RRS) soybean genotypes, 5 d after spraying.
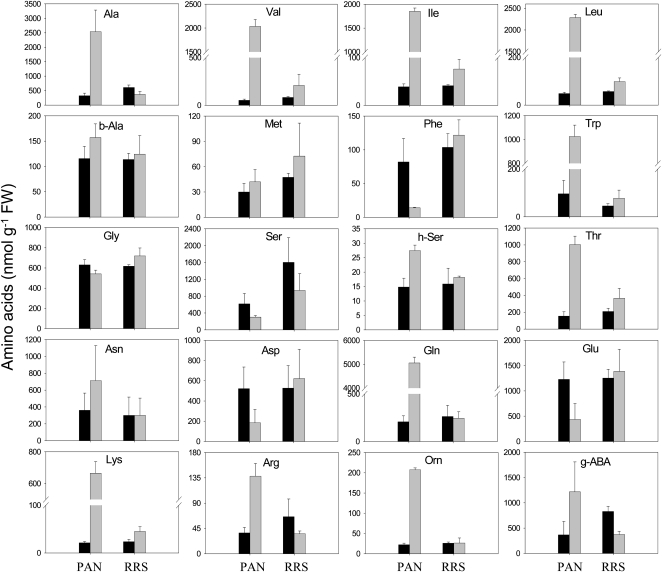
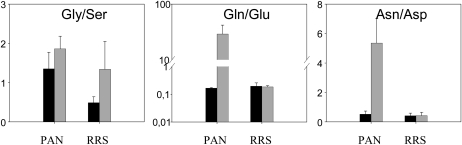
The glyphosate treatment led to a large (3-fold) increase in the total free amino acid pools of the PAN leaves (data not shown). Moreover, glyphosate had marked differential effects on the amino acid composition of the PAN leaves: the abundance of Phe, Glu, and Asp was decreased, while Gln, the primary product of ammonia (re)assimilation, was increased more than 20-fold as a result of glyphosate treatment (Fig. 2). The Gln-to-Glu ratio increased almost 100-fold in the glyphosate-treated PAN leaves (Fig. 3). This effect was not accompanied by marked changes in other amino acids involved in photorespiration, and the Gly-to-Ser ratio was not significantly changed (Fig. 3). However, the increase in Gln was accompanied by strongly increased pools of another major amino acid, Ala, and also by minor amino acids synthesized through shikimate-independent pathways. These included Thr, Lys, and the three branched-chain amino acids Val, Leu, and Ile. Of nonprotein amino acids detected, Orn was also increased by glyphosate (Fig. 2). This was accompanied by a 3-fold increase in Arg. Strikingly, Trp also showed the same pattern (i.e. this amino acid behaved antagonistically to Phe; Fig. 2).
Figure 2.
The effects of glyphosate on the abundance of leaf amino acids in glyphosate-sensitive (PAN) and glyphosate-resistant (RRS) soybean genotypes, 5 d after spraying. Black bars, controls; gray bars, glyphosate treated. FW, Fresh weight; g-ABA, γ-aminobutyric acid.
Figure 3.
The effect of glyphosate on the Gly-to-Ser, Gln-to-Glu, and Asn-to-Asp ratios in the leaves of glyphosate-sensitive (PAN) and glyphosate-resistant (RRS) soybean genotypes, 5 d after spraying. Black bars, control plants; gray bars, glyphosate-treated plants.
Despite the accumulation of glyphosate to much higher levels in RRS leaves compared with PAN (Fig. 1), glyphosate had only a small effect on the RRS leaf amino acid profile, and there was no significant change in total free amino acids (data not shown). The glyphosate treatment produced increases in certain amino acids that were qualitatively similar to those observed in PAN (Fig. 2). For example, similar trends were observed for Val, Ile, Leu, Trp, Thr, and Lys in glyphosate-treated PAN and RRS leaves. However, unlike in PAN leaves, these effects in RRS were not accompanied by a decrease in Phe or Glu or by an increase in Gln and Ala (Fig. 2).
Proteomic Analyses
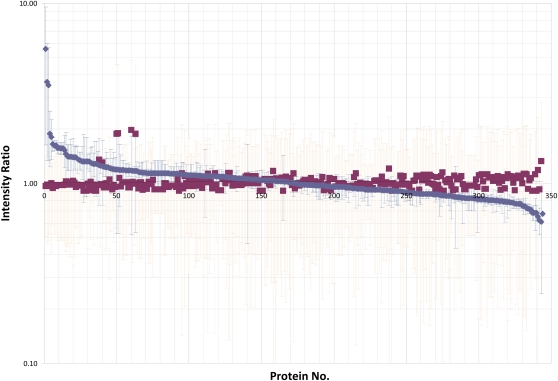
To assess the effect of glyphosate on PAN leaves, we performed a comparative analysis of the leaf proteome using tandem mass tag (TMT) 6-plex isobaric labeling (Fig. 4). By searching the soybean database (version 0.1c), 359 quantifiable gene products (see “Materials and Methods”) were identified with a false discovery rate of 1%. It is remarkable that the glyphosate-dependent changes in the PAN proteome are surprisingly subtle and that only a small number of proteins appear to be changed significantly (Table I). Eight proteins were up-regulated more than 1.4-fold in response to the treatment with glyphosate (Table I; Supplemental Table S1). Of these, the most marked effect was on the starvation-associated protein, SAM22. Proteins that were increased in response to glyphosate treatment include stress-response proteins such as the gene product of Gm0101x00067, a protein that is involved in disease resistance, and Gm0253x00027, which is a predicted member of the heat shock protein70 family. Other proteins that were changed in abundance following the glyphosate treatment include Gm0062x00004, encoding Met synthase, and Gm0097x00143, a gene with homology to O-methyltransferases (Table I).
Figure 4.
A comparison of the ratios of proteins in the leaves of glyphosate-sensitive (PAN) plants, 5 d after spraying with either water (blue squares and blue error bars) or glyphosate (red squares and red error bars). Red squares represent averages of three different reporter ion ratios stemming from the derivatization of water-control soybean leaves with TMT reagents 126, 127, and 128. Blue squares represent reporter ion ratios for glyphosate-treated soybean leaves derivatized with TMT reagents 129, 130, and 131. Red error bars represent se.
Table I. Proteins that are regulated in response to glyphosate treatment in glyphosate-sensitive soybean plants.
Proteins from control PAN leaves were labeled with TMT reagents 126, 127, and 128. Glyphosate-treated PAN plants were labeled with TMT reagents 129, 130, and 131. The average control ratio is the average of the ratios 127:126, 128:127, and 126:128; se control is the se for these three ratios. The glyphosate-treated (Gly)/control ratio is the average of the ratios 129:126, 130:126, and 131:126, with the se for these three ratios.
| Protein Identifier | Protein Description | Ratio Gly/Control | se Gly/Control | Average Control | se Control | Mascot Protein Score | Matched Spectra |
| Gm0068x00308 | Stress-induced protein SAM22; AltName: starvation-associated message; AltName: allergen = Gly m 4 | 5.61 | 2.28 | 1.06 | 0.27 | 1,137 | 90 |
| Gm0101x00067 | Disease resistance-responsive protein related/dirigent protein-related | 1.90 | 0.37 | 1.06 | 0.25 | 94 | 14 |
| Gm0062x00004 | Vitamin-B12-independent Met synthase, 5-methyltetrahydropteroyltri-Glu-homo-Cys | 1.83 | 0.45 | 1.09 | 0.34 | 218 | 15 |
| Gm0253x00027 | Predicted: similar to HSC70-1 (heat shock cognate 70-kD protein1); ATP-binding isoform 1 (Vitis vinifera) | 1.67 | 0.15 | 1.04 | 0.2 | 77 | 21 |
| Gm0072x00084 | Nucleoside diphosphate kinase 1; AltName: nucleoside diphosphate kinase I; short = NDP kinase I; short = NDPK I; short = NDK I | 1.47 | 0.46 | 1.12 | 0.35 | 94 | 10 |
| Gm0009x00167 | Chloroplast Cu/Zn SOD (Gossypium arboreum) | 1.42 | 0.35 | 1.11 | 0.36 | 272 | 17 |
| Gm0097x00143 | O-Methyltransferase, putative (Ricinus communis) | 1.42 | 0.11 | 1.1 | 0.36 | 38 | 8 |
| Gm0038x00256.1 | Peroxisomal 3-ketoacyl-CoA thiolase precursor | 1.41 | 0.13 | 1.04 | 0.22 | 75 | 7 |
| Gm0041x00326 | CP12 (Pisum sativum) | 0.63 | 0.21 | 1.42 | 1.48 | 49 | 17 |
| Gm0026x00493 | 14-kD Pro-rich protein DC2.15 precursor, putative | 0.64 | 0.06 | 1.02 | 0.20 | 959 | 54 |
| Gm0467x00004 | PSI P700 apoprotein A2 (Glycine max) | 0.67 | 0.01 | 1.28 | 1.14 | 86 | 20 |
| Gm0025x00404 | Germin-like protein (Pisum sativum) | 0.70 | 0.04 | 1.00 | 0.02 | 1,234 | 51 |
| Gm0025x00719 | Extracellular Ca2+-sensing receptor (Glycine max) | 0.71 | 0.08 | 1.21 | 0.82 | 53 | 8 |
| Gm0188x00005 | Dehydroascorbate reductase (Glycine max) | 0.74 | 0.02 | 1.13 | 0.72 | 38 | 60 |
| Gm0152x00052 | FLA13 (fasciclin-like arabinogalactan protein 13 precursor; Arabidopsis thaliana) | 0.75 | 0.03 | 1.03 | 0.27 | 445 | 28 |
| Gm0262x00055 | PSII 47-kD protein (Glycine max) | 0.75 | 0.02 | 1.22 | 0.97 | 2,399 | 154 |
To assess the effect of glyphosate on RRS leaves, we performed a semiquantitative analysis based on normalized peptide counting and a gel-based liquid chromatography-tandem mass spectrometry (LC-MS/MS)-based analytical strategy. In this analysis, 384 proteins were identified in the lysates from leaves of plants treated with water alone, whereas 109 proteins were identified in leaves sprayed with glyphosate. Of these, 90 proteins were identical in both samples (Supplemental Table S2). The quantitative ratio of 25 of these proteins indicates a regulation of protein abundance in response to glyphosate treatment in RRS leaves (Table II). The relative amounts of several proteins involved in photosynthesis, including PSII core complex proteins and proteins that are part of light-harvesting complex II, were strongly increased. In contrast, several proteins involved in amino acid metabolism, including a Ser hydroxymethyltransferase, an Ala aminotransferase, and a Ser glyoxylate aminotransferase, were decreased in response to glyphosate treatment (Table II).
Table II. Proteins that are regulated in response to glyphosate treatment in glyphosate-resistant soybean leaves.
Glyphosate-treated and control RRS leaf extracts were analyzed on separate lanes using one-dimensional SDS-PAGE. Of the proteins that were present in both samples (90 proteins; Supplemental Table S2), those that are either up-regulated more than 4-fold or down-regulated more than 2-fold in response to glyphosate treatment are shown here. Ratio Gly/Control is the ratio of the normalized peptide count for a particular protein, as detailed in “Materials and Methods.”
| Protein Identifier | Protein Description | Ratio Gly/Control | log(e) | Total No. of Peptide Identification Events |
| Gm0005x00294 | Chlorophyll a/b-binding protein, putative (Ricinus communis) | 65.2 | −109.0 | 27 |
| Gm0026x00493 | Homologous to protease inhibitor/seed storage/lipid transfer protein (LTP) family protein (Arabidopsis thaliana) | 10.4 | −5.6 | 15 |
| Gm0013x00790 | Homologous to ENH1 (ENHANCER OF SOS3-1); metal ion binding (Arabidopsis thaliana) | 9.7 | −16.1 | 2 |
| Gm0070x00460 | Chlorophyll a/b-binding protein CP24 precursor (Vigna radiata) | 7.2 | −30.3 | 9 |
| Gm0009x00162 | Homologous to HTB4; DNA binding (Arabidopsis thaliana) | 7.2 | −11.4 | 3 |
| Gm0217x00074.1 | Carbonic anhydrase (Vigna radiata) | 6.9 | −120.0 | 27 |
| Gm0035x00448 | Ribulose-5-phosphate-3-epimerase (Pisum sativum) | 6.4 | −17.4 | 4 |
| Gm0007x00350 | PSII protein D1 (Glycine max) | 6.4 | −17.3 | 8 |
| Gm0121x00254 | Light-harvesting complex II protein Lhcb8 (Populus trichocarpa) | 6.3 | −59.4 | 13 |
| Gm0028x00225 | 60S ribosomal protein L11, putative (Ricinus communis) | 4.8 | −4.8 | 1 |
| Gm0050x00070 | Chlorophyll a/b binding protein CP29 (Vigna radiata) | 4.0 | −68.3 | 15 |
| Gm0279x00004 | PSII type I chlorophyll a/b-binding protein (Glycine max) | 4.0 | −54.0 | 20 |
| Gm0026x00074 | Os01g0869800 [Oryza sativa (japonica cultivar group)] | 4.0 | −51.6 | 9 |
| Gm0074x00038 | Gly dehydrogenase [decarboxylating], mitochondrial; AltName: Gly decarboxylase; AltName: Gly cleavage system P-protein | 0.1 | −190.0 | 38 |
| Gm0088x00166 | Chain A, crystal structure of soybean lipoxygenase B | 0.1 | −169.0 | 33 |
| Gm0029x00038 | Ser hydroxymethyltransferase, mitochondrial; Short = SHMT; Short = Ser methylase; AltName: Gly hydroxymethyltransferase | 0.2 | −236.0 | 51 |
| Gm0002x00324 | Translation elongation factor 1A-2 (Gossypium hirsutum) | 0.2 | −101.0 | 24 |
| Gm0206x00022 | Ala aminotransferase 3 (Glycine max) | 0.3 | −194.0 | 33 |
| Gm0025x00286 | Ferric leghemoglobin reductase 2 (Glycine max) | 0.3 | −238.0 | 32 |
| Gm0683x00003 | Cytochrome f (Glycine max) | 0.4 | −108.0 | 41 |
| Gm0004x00436 | Ser glyoxylate aminotransferase 2 (Glycine max) | 0.4 | −90.1 | 13 |
| Gm0017x00151.1 | 1-Deoxy-d-xylulose 5-phosphate reductoisomerase (Pueraria montana var lobata) | 0.4 | −80.9 | 13 |
| Gm0235x00046 | Rubisco large subunit-binding protein subunit α, chloroplastic; AltName: 60-kD chaperonin subunit α; AltName: CPN-60α | 0.4 | −199.0 | 24 |
| Gm0262x00024 | PSI P700 apoprotein A1 (Glycine max) | 0.4 | −105.0 | 45 |
| Gm0261x00028 | Monodehydroascorbate reductase; Short = MDAR; AltName: ascorbate free radical reductase; Short = AFR reductase | 0.5 | −69.0 | 9 |
Effect of Glyphosate on Photosynthesis
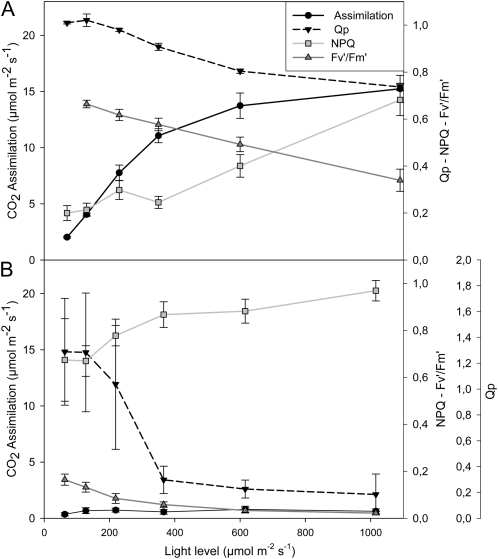
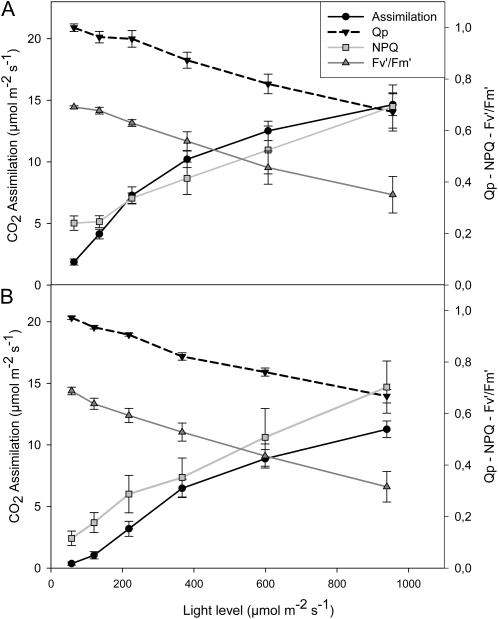
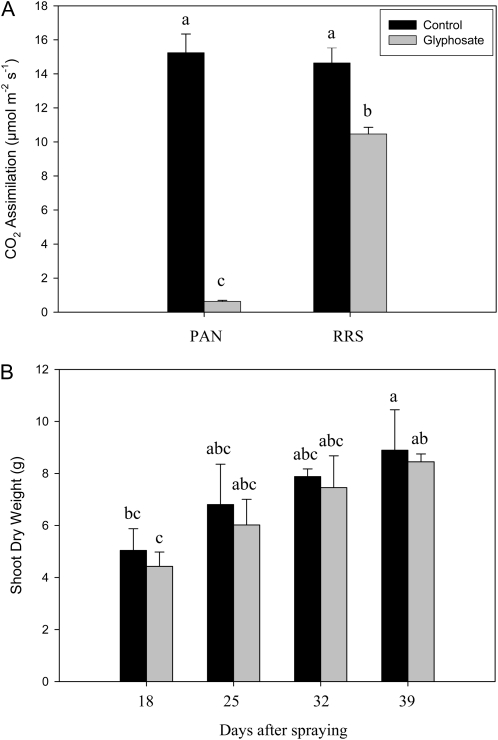
The effects of glyphosate on various parameters associated with photosynthesis were measured 24 h after spraying with water (Fig. 5A) or glyphosate (Fig. 5B). Leaf photosynthesis was rapidly inhibited in PAN leaves following spraying with glyphosate (Fig. 5). Light-response curves for photosynthesis (photosynthetic CO2 assimilation and the chlorophyll a fluorescence quenching parameters photochemical quenching [QP], nonphotochemical quenching [NPQ], and quantum yield of photosynthesis [Fv′/Fm′]) were determined 24 h after spraying with water (Fig. 5A) or glyphosate (Fig. 5B). This analysis revealed that even at high irradiance, photosynthetic CO2 assimilation rates in the glyphosate-treated PAN leaves remained close to the compensation point (Fig. 5). Similarly, glyphosate spraying of PAN leaves strongly impacted the irradiance curves for QP and NPQ of chlorophyll a fluorescence as well as the Fv′/Fm′ (Fig. 5). The application of glyphosate had much smaller effects on the light-response curves for photosynthesis in the RRS plants (Fig. 6). The light-response curves for photosynthetic CO2 assimilation and maximal photosynthesis rates were decreased by about 30%, when they were measured 24 h after spraying with glyphosate (Fig. 6B) compared with controls (Fig. 6A). Moreover, the chlorophyll content of the glyphosate-treated RRS leaves was lower (1.84 ± 0.17 mg−1 fresh weight) than the controls (2.95 ± 0.27 mg−1 fresh weight). However, the protein contents of the glyphosate-treated RRS leaves were not significantly different from the controls, and the kinetics of changes in QP, NPQ, and Fv′/Fm′ with increasing irradiance were not markedly changed in RRS leaves sprayed with glyphosate (Fig. 6B). Five days after spraying with glyphosate, photosynthetic CO2 assimilation rates were very low in PAN leaves compared with those sprayed with water alone (Fig. 7). While a significant decrease in photosynthesis was also observed in RRS leaves 5 d after the glyphosate treatment (Fig. 7A), the chlorophyll content of the glyphosate-treated leaves was not significantly different (2.17 ± 0.10 mg−1 fresh weight) than the controls (2.28 ± 0.23 mg−1 fresh weight). Photosynthetic CO2 fixation rates returned to values similar to controls within 7 d. To determine whether glyphosate-dependent decreases in photosynthetic CO2 fixation activity had an effect on biomass production in the RRS genotype, we measured the total shoot dry weights in the weeks following the application of glyphosate (Fig. 7B). While the glyphosate-treated RRS shoots showed a trend to lower biomass accumulation, no significant differences between treated and control shoots were observed within 3 weeks of the application of glyphosate (Fig. 7B). In contrast, the PAN shoots never recovered and had collapsed and were brown within 2 weeks of the application of glyphosate (data not shown).
Figure 5.
A comparison of photosynthetic CO2 assimilation rates and chlorophyll a fluorescence quenching analysis parameters (QP, NPQ, and Fv′/Fm′) in the leaves of glyphosate-sensitive soybean plants prior to glyphosate treatment (A) and 1 d after spraying with glyphosate (B).
Figure 6.
A comparison of photosynthetic CO2 assimilation rates and chlorophyll a fluorescence quenching analysis parameters (QP, NPQ, and Fv′/Fm′) in the leaves of glyphosate-resistant soybean plants prior glyphosate treatment (A) and 1 d after glyphosate spraying (B).
Figure 7.
A comparison of photosynthetic CO2 assimilation rates in the leaves of glyphosate-sensitive (PAN) and glyphosate-resistant (RRS) genotypes, 5 d after glyphosate spraying (A), and biomass accumulation (shoot dry weight) in the glyphosate-resistant soybean line in the days following treatment with glyphosate (B). Letters above the bars indicate statistical significance (P < 0.05).
Effect of Glyphosate on Cellular Redox State
In the absence of glyphosate, the ascorbate (ascorbate plus dehydroascorbate) and glutathione (hGSH plus hGSSG) pools were highly (over 90%) reduced in PAN and RRS leaves (Table III). The glyphosate treatment had little effect on the ascorbate pool of the PAN leaves, but there was a large increase in total hGSH 5 d after spraying, without any large effect on the hGSH-to-hGSSG ratio. The NAD(H) and NADP(H) pools were decreased as a result of the glyphosate treatment (Table III).
Table III. Effect of glyphosate on redox metabolites in leaves of glyphosate-sensitive and glyphosate-resistant soybean, 5 d after spraying.
| Metabolite | Glyphosate-Sensitive Line |
Glyphosate-Resistant Line |
||
| Control | Glyphosate | Control | Glyphosate | |
| Ascorbate (μmol mg−1 chlorophyll) | 8.1 ± 0.3 | 8.7 ± 0.5 | 10.4 ± 0.9 | 7.8 ± 1.3 |
| Dehydroascorbate (μmol mg−1 chlorophyll) | 0.8 ± 0.2 | 0.7 ± 0.1 | 3.4 ± 0.8 | 3.5 ± 1.7 |
| Ascorbate/dehydroascorbate | 10.3 | 12.1 | 3.1 | 2.2 |
| hGSH (nmol mg−1 chlorophyll) | 178.1 ± 16.8 | 456.2 ± 17.8 | 243.0 ± 23.7 | 212.6 ± 24.9 |
| hGSSG (nmol mg−1 chlorophyll) | 36.1 ± 6.7 | 78.6 ± 11.9 | 43.3 ± 13.3 | 114.8 ± 0.4 |
| hGSH/hGSSG | 9.9 | 11.6 | 11.2 | 3.7 |
| NADH (nmol mg−1 chlorophyll) | 1.4 ± 0.8 | 2.0 ± 0.4 | 2.2 ± 0.2 | 1.7 ± 0.4 |
| NAD (nmol mg−1 chlorophyll) | 34.4 ± 5.0 | 14.4 ± 4.0 | 24.3 ± 1.8 | 33.8 ± 0.1 |
| NAD/NADH | 24.0 | 7.1 | 11.0 | 20.0 |
| NADPH (nmol mg−1 chlorophyll) | 2.1 ± 0.7 | 2.7 ± 1.2 | 8.5 ± 0.9 | 9.8 ± 0.3 |
| NADP (nmol mg−1 chlorophyll) | 12.9 ± 0.5 | 5.8 ± 1.3 | 11.6 ± 1.3 | 10.3 ± 3.2 |
| NADP/NADPH | 6.1 | 2.1 | 1.4 | 1.1 |
The treatment with glyphosate caused slight decreases in the ascorbate pools of the RRS leaves. However, the total glutathione pool of the RRS leaves was oxidized as a result of the glyphosate treatment, and there was a marked decrease in the hGSH-tohGSSG ratio (Table III). This was accompanied by increases in the NAD(H) pool, but there was little change in the NADP(H) pool (Table III)
DISCUSSION
The widespread use of glyphosate in agriculture in combination with transgenic glyphosate-resistant crops worldwide has led to the evolution of glyphosate-resistant weeds (Powles, 2008). Despite the extensive use of this herbicide, major uncertainties remain concerning the precise mechanisms by which glyphosate kills plants and how it alters physiology and metabolism in glyphosate-resistant plants. Although we cannot fully answer this question, we have succeeded in specifying glyphosate-dependent cellular events. The application of glyphosate had marked but different effects on metabolism and leaf proteomes in sensitive and resistant soybean genotypes. In both lines, glyphosate has pronounced effects on cellular redox metabolism and the leaf proteome, but as we now discuss, these are clearly different.
Glyphosate Rapidly Inhibits Photosynthesis and Drives the Accumulation of Specific Amino Acids in Sensitive Plants
In leaves of the sensitive line, the treatments with glyphosate produced marked effects on amino acid composition. Decreases in Phe are consistent with the inhibition of the shikimate pathway. The associated changes in Glu and Asp likely reflect a shift toward the accumulation of nitrogen-rich amino acids synthesized from these early products of nitrogen assimilation. This interpretation receives support from the marked accumulation of Gln and Arg. Taken together, these patterns suggest that glyphosate triggers a nitrogen-rich amino acid profile, in which compounds containing two or more nitrogen atoms are enhanced at the expense of their precursors. This is reflected by the increase in both Gln-to-Glu and Asn-to-Asp ratios as well as the enhanced Arg and Orn pools. Other amino acids that accumulated were Ala and minor amino acids synthesized through shikimate-independent pathways, including Thr, Lys, Val, Leu, and Ile. Thus, deregulation of Phe synthesis causes the accumulation of amino acids synthesized through parallel pathways. Our data provide little evidence that nitrogen assimilation is strongly feedback inhibited in response to starvation for shikimate pathway products.
Glyphosate produced a rapid, marked, and sustained inhibition of photosynthetic CO2 assimilation in the glyphosate-sensitive line. This loss of CO2 assimilation capacity was accompanied by marked changes in the responses of the chlorophyll a fluorescence-quenching parameters to increasing irradiance. Values for QP and Fv′/Fm′ were much lower with respect to increasing irradiance, while NPQ reached high values even at low irradiance. These effects indicate a limited capacity for electron transport and increased release of excess excitation energy as heat, probably triggered by more limited use of ATP because of decreased metabolism. Together, the data show that glyphosate inhibition of the shikimate pathway rapidly causes the shutdown of photosynthesis. These findings are consistent with previous observations concerning the inhibitory effect of glyphosate on photosynthetic CO2 assimilation and chlorophyll fluorescence emission (Ireland et al., 1986). Glyphosate impairs photosynthesis through a perturbation of metabolism that affects both the Calvin cycle and chlorophyll biosynthesis (Kitchen et al., 1981; Ireland et al., 1986). In contrast, photosynthetic electron transport and light harvesting are not directly affected by the herbicide (Richard et al., 1979; Ireland et al., 1986). The observed decreases in photosynthesis-associated proteins (photosystem apoproteins, C12) that were observed 5 d after glyphosate treatment are consistent with the hypothesis that glyphosate causes a sustained loss of photosynthetic capacity and that this impairment involves the depletion of photosynthetic proteins. This was accompanied by large increases in the abundance of the starvation-associated SAM22 protein and in proteins associated with stress responses. The SAM22 protein belongs to the pathogenesis-related protein family and is responsible for severe allergic reactions in soybean (Ballmer-Weber and Vieths, 2008). It is widely induced under stress conditions, particularly starvation.
The abundance of chloroplast Cu/Zn-SOD was also increased as a result of EPSPS inhibition, in agreement with an earlier study on glyphosate-sensitive rice (Oryza sativa; Ahsan et al., 2008). These authors also reported that the glyphosate application caused an accumulation of reactive oxygen species in the rice leaves (Ahsan et al., 2008). In this study, the ratios of the reduced to oxidized forms of the major redox pools were similar in the water-treated and the glyphosate-treated leaves of the glyphosate-sensitive line 5 d after spraying with glyphosate. Taken together, these results suggest that glyphosate causes a transient but not sustained oxidation of the leaf cells that is sufficient to trigger an enhanced abundance of antioxidants (Cu/Zn-SOD, hGSH) that restores the cellular redox balance. A transient glyphosate-dependent oxidation in the treated glyphosate-sensitive soybean leaves would explain not only the enhanced abundance of chloroplast Cu/Zn-SOD but also the heat shock protein HSP70, because of the extensive cross talk between heat and oxidative stress signaling in plants (Davletova et al., 2005).
The abundance of tetrahydrofolate-independent Met synthase, which catalyzes the final step in Met biosynthesis, was also greatly increased as a result of the inhibition of EPSPS activity by glyphosate in the sensitive genotype. This enzyme is responsible for the production of Met from homo-Cys and forms part of the S-adenosyl-Met cycle. S-adenosyl-Met is a universal methyl donor (one-carbon metabolism) in a large range of methylation reactions. However, one-carbon metabolism also plays a critical role in DNA methylation and DNA synthesis. Thus, the enhanced abundance of Met synthase may suggest cross talk between amino acid metabolism and genetic and epigenetic processes.
Glyphosate Down-Regulates Defense Components and Proteins Associated with Photorespiration Independent of EPSPS Inhibition
Our parallel analysis of the resistant line allowed us to highlight the effects of glyphosate that are independent of EPSPS inhibition. In comparison with the marked effects of glyphosate on the amino acid composition of the leaves of the sensitive line, the glyphosate-resistant leaves showed relatively few changes in amino acid metabolism. However, some minor effects are worth noting. In RRS, Thr and Val were slightly increased by glyphosate, whereas the pools of the nonprotein amino acid, γ-aminobutyric acid, were somewhat decreased. Thus, amino acid metabolism is influenced by the presence of glyphosate even in the leaves of the glyphosate-resistant line.
Unlike the marked effects observed in the sensitive genotype, herbicide treatment had relatively little effect on photosynthesis in the glyphosate-resistant line. However, a small (30%) and transient inhibition of photosynthetic CO2 assimilation was observed in the resistant line 24 h after the application of glyphosate, and photosynthesis rates were still lower than in controls 5 d after the treatment. This inhibition may be due, at least in part, to the production of potentially harmful metabolites, such as aminomethylphosphonic acid, in glyphosate-resistant soybean (Reddy et al., 2004). Aminomethylphosphonic acid is considered to be less toxic than glyphosate, but its mode of action is unknown. Photosynthesis rates were only restored to values similar to the water-treated controls several weeks after spraying. This may explain why proteins involved in light harvesting and photosystem function were greatly enhanced in the leaves of the resistant genotype. Moreover, a number of proteins associated with the photorespiratory pathway, particularly Gly decarboxylase, Ser hydroxymethyltransferase, and Ser glyoxylate aminotransferase, were greatly decreased in the glyphosate-treated leaves of the resistant line. The differential expression of proteins associated with PSII and those associated with photorespiration is remarkable given that photosynthetic and photorespiratory components are generally expressed in a coordinated manner (Foyer et al., 2009). The initial small and transient glyphosate-induced inhibition of photosynthetic CO2 assimilation might have been sufficient to trigger the observed changes in the abundance of proteins associated with photosynthesis and photorespiration in the leaves of the resistant genotype, in which all the necessary amino acids remain readily available for protein synthesis. The enhanced abundance of photosynthetic proteins together with the decreased levels of photorespiratory proteins observed 5 d after glyphosate treatment may explain the recovery of photosynthesis and also the ability of the shoots to accumulate biomass at levels similar to controls within weeks of the application of glyphosate.
The application of glyphosate led to a marked increase in the hGSH pool in glyphosate-sensitive leaves but not in the leaves of the resistant genotype. This observation might suggest that hGSH metabolism is modulated in response to glyphosate-dependent alterations in the shikimate pathway. Studies on the effects of glyphosate on leaf gene expression patterns in soybean and other species suggest that glyphosate has little effect on gene expression. Of the few transcripts that are altered in abundance as a result of glyphosate treatment, most are involved with secondary metabolism or defense, and there is no change in transcripts associated with processes such as leaf senescence (Yu et al., 2007; Das et al., 2010). One of the 16 glyphosate-induced genes in Arabidopsis (Arabidopsis thaliana) and Brassica napus is a GST (GSTU24) that has been implicated in herbicide detoxification (Das et al., 2010). The observed increases in the total leaf hGSH pool suggest that up-regulation of this synthesis pathway is required to allow hGSH-dependent glyphosate detoxification pathways. The finding that hGSH accumulation is absent from the leaves of the resistant line, which accumulate high levels of glyphosate, suggests that the effect on cellular hGSH homeostasis is not due to the presence of glyphosate per se but is rather a consequence of its effects on amino acid metabolism. Additionally, the increase in hGSH in the sensitive line may have contributed to the accumulation of some of the amino acids, consistent with the effects observed in Populus with enhanced glutathione (Noctor et al., 1998).
While the glyphosate-dependent stimulation of hGSH metabolism was absent from the leaves of the resistant line, the ascorbate/dehydroascorbate and hGSH-to-hGSSG ratios were markedly decreased following the application of the herbicide. The sustained oxidation of both the ascorbate and hGSH pools after the herbicide treatment may be related to the very high levels of glyphosate detected in the leaves of the resistant plants compared with those of the sensitive line. It is possible, therefore, that the insensitivity of EPSPS to glyphosate in the resistant genotype decreases the ability of the plant to trigger glyphosate detoxification pathways. Thus, the herbicide accumulates to a level that triggers oxidative stress. Glyphosate is highly toxic to animal cells, which do not have the shikimic acid pathway, because it impairs detoxification systems, particularly by the inhibition of enzymes (Hietanen et al., 1983). Thus, a similar inhibition of cellular defense processes may occur in the herbicide-treated RRS plants, which become exposed to very high levels of glyphosate. The decreased abundance of monodehydroascorbate reductase revealed by the proteomics analysis of the leaves of the resistant line treated with glyphosate is consistent with the observed sustained oxidation of both the ascorbate pool and also with the negative effects of glyphosate on the cellular detoxification systems. The effects on leaves of the glyphosate-resistant line are probably linked to the extensive accumulation of glyphosate observed in this genotype, which was far in excess of that observed in the glyphosate-sensitive line.
MATERIALS AND METHODS
Plant Material
Seeds of the soybean (Glycine max) genotype PAN809 (van Heerden et al., 2003, 2008; Strauss et al., 2006) and commercial transgenic RRS plants obtained from Nidera (cv 4613; RRS) were sown in pots in Levington’s compost (Levington F2 plus Scotts Professional) and grown in controlled-environment chambers until the fourth leaf stage, at day/night temperatures of 24°C/19°C and an irradiance of 400 μmol m−2 s−1, with a 12-h-day/12-h-night cycle and twice-daily watering.
Glyphosate Treatments
For the glyphosate treatments, 20 mL of the concentrate Clinic Ace (41.5% glyphosate plus 8.1% Tallow alkylamine ethoxylate; Nufarm) was mixed with water to a total volume of 900 mL. Each plant at the fourth leaf stage was sprayed either with this glyphosate solution (4 mL per plant) or with water alone (controls). The glyphosate concentration in Clinic Ace solution is similar to Roundup (Monsanto), which contains 48% glyphosate.
Photosynthetic Gas-Exchange and Fluorescence Measurements
For the data shown in Figures 5 and 6, attached leaves from four plants were analyzed simultaneously in multichamber systems linked to infrared gas analyzers. Measurements of CO2 and water exchange under steady-state conditions were performed as described by Novitskaya et al. (2002). All experiments were conducted at 22°C and 50% relative humidity. Irradiance was provided by overhead flood lamps and adjusted by neutral density filters. Gas composition was controlled by a gas mixer supplying CO2 to the stated partial pressures, 21% oxygen and balance N2. Light-response curves for photosynthesis were performed at 350 μmol mol−1 CO2. At the start of the experiments, respiratory CO2 release was monitored for 20 min in the dark, and then irradiance was increased stepwise from 60 to 1,100 μmol m−2 s−1. Data were taken when photosynthetic rates reached a stable value after each increase in incident light level.
For the data shown in Figure 7, gas-exchange parameters were measured in a random sample of three fully expanded leaves on separate plants for each treatment using a CIRAS-1 Portable Photosynthesis System (Hitchin). Measurements were made with a photosynthetic photon flux density of 600 μmol m−2 s−1 and a CO2 level of 350 μmol mol−1 at 24.0°C ± 0.7°C.
Metabolite Extraction and Assays
Leaf discs were cut under water under the growth conditions, immediately frozen in liquid N2, and then maintained at −80°C until analysis. Unless otherwise stated, metabolites were extracted in 1 m HClO4; samples were mixed with acid while frozen. Upon thawing, aliquots were taken for the assay of chlorophyll as pheophytin, as described by Queval and Noctor (2007). Insoluble material was removed from the acid extracts by centrifugation, and the supernatant fractions were brought to pH 5 to 6 with K2CO3. Ascorbate and hGSH were measured in the same acid extracts. Total and reduced ascorbate were measured by the ascorbate oxidase-dependent decrease in A265 as described by Foyer et al. (1983). Total and oxidized hGSH were measured by the NADPH-driven glutathione-dependent reduction of 5,5′-dithiobis(2-nitrobenzoic acid) in the presence of NADPH and glutathione reductase as described by Noctor and Foyer (1998). This method can be used for the determination of hGSH, making the appropriate correction in assay sensitivity according to Klapheck (1988).
To analyze pyridine nucleotides, frozen material was ground in liquid nitrogen with a pestle and mortar, extracted with acid as described above (for NAD+ and NADP+) or in 0.2 m NaOH (for NADH and NADPH), and assayed as described by Queval and Noctor (2007). NADH and NADPH were neutralized with 0.2 n HCl to a final pH of 7 to 8. Pyridine nucleotides were assayed using the phenazine methosulfate-catalyzed reduction of dichlorophenolindophenol in the presence of ethanol and alcohol dehydrogenase (for NAD+ and NADH) or Glc-6-P and Glc-6-P dehydrogenase (for NADP+ and NADPH) as described by Queval and Noctor (2007).
For the analysis of amino acids by HPLC, extracts were prepared as described in detail by Noctor et al. (2007). Frozen material was ground in liquid nitrogen with a pestle and mortar and extracted into 80% methanol/20% water containing 100 μm γ-aminobutyrate as an internal standard. Following centrifugation, multiple aliquots were spin dried under vacuum and stored at −80°C. Aliquots were redissolved in water, centrifuged, and filtered into autosampler vials prior to automated precolumn derivatization with o-phthaldialdehyde. Amino acids labeled with o-phthaldialdehyde were separated by reversed-phase HPLC and identified by coelution with authentic standards. Amounts of amino acids were calculated on linear calibration curves generated for each standard. Values were corrected for the response of the internal standard (γ-aminobutyrate) and quantified on a tissue fresh weight basis.
Protein Estimation
Proteins that had precipitated in the pellet after acid extraction were solubilized using 120 mm sodium phosphate buffer with 1 mm EDTA (pH 7.6), and the total protein content was measured using a standard Bradford assay (Bradford, 1976).
Proteomic Analysis
Protein Sample Preparation
Leaf extracts were prepared from 8-week-old control and glyphosate-treated plants. Three technical replicates were prepared from each extraction. Proteins were precipitated after grinding leaf material (200 mg) in liquid nitrogen. Ground leaf material was incubated overnight at −20°C in 1 mL of the precipitation buffer containing TCA (10%, w/v) and β-mercaptoethanol (0.07%, v/v) in acetone. Precipitated protein was pelleted by centrifuging for 30 min at 4°C at 16,000g and was washed six times with ice-cold 90% acetone (v/v) and 0.07% β-mercaptoethanol (v/v) in water. Air-dried protein pellets were solubilized in 1 mL of extraction solution containing urea (8 m), 2% CHAPS (w/v), and dithiothreitol (20 mm) by sonication in an ultrasonic water bath for 1 h, with vortexing at 15-min intervals. Samples were then incubated at 4°C overnight with interval shaking in a Thermomixer (Eppendorf). Cell debris was removed by centrifugation for 30 min at 16,000g. Solubilized proteins were quantified using the Bradford assay (Bradford, 1976) and bovine serum albumin (Sigma) as a standard. A total of 420 μg of protein was removed from each sample and precipitated using the 2D Clean-Up Kit (GE Healthcare); the resulting pellet was dissolved in 100 μL of 1.5 m urea in 50 mm triethylammonium bicarbonate (TEAB; Fluka), pH 8.
Protein Reduction, Alkylation, and Digestion
To reduce the protein samples, 5 μL of 200 mm Tris(2-carboxyethyl)phosphine in 50 mm TEAB was added to each sample, and the mixture was incubated at 55°C for 1 h, then 5 μL of 375 mm iodoacetamide (AppliChem) in 50 mm TEAB (pH 8) was added to alkylate the proteins. The mixture was kept at 30°C in the dark for 30 min, then 290 μL of 50 mm TEAB was added. Next, 10 μL of trypsin solution (Promega) containing 10 μg of the enzyme was added to each sample, and the mixtures were incubated overnight at 37°C for complete protein digestion.
Isobaric Mass Tagging of Peptides
Labeling was carried out with a TMT Isobaric Mass Tagging Kit (ThermoFisher). The reagents were equilibrated to room temperature. First, 41 μL of acetonitrile was added to each of the six labeling reagents (TMT 126–131; 0.8 mg each). Then, TMT labeling reagent (40 μL) was added to each sample followed by 1 h of incubation at room temperature. After incubation, 23 μL of 5% hydroxylamine (Fluka) in 200 mm TEAB (v/v) was added to each sample and incubated for 15 min. Labeled samples were mixed prior to off-gel fractionation of the peptides.
Off-Gel Fractionation
For fractionation of the TMT-labeled peptides, an Agilent 3100 OFFGEL Fractionator with 24-well loading tray and immobilized pH gradient strip (pH 3–10 linear; GE Healthcare) was used. The volume of the mixed labeled peptide was made up to 3.6 mL by the addition of 8.4 m urea, 2.4 m thiourea, 0.08 m dithiothreitol, 6% (v/v) glycerol, and 1.0% (v/v) Pharmalyte, pH 3 to 10 (OFFGEL Stock Solution; GE Healthcare). The immobilized pH gradient strip was rehydrated for 15 min by adding 40 μL of the OFFGEL Stock Solution containing 0.002% (w/v) bromphenol blue. Aliquots (150 μL) of TMT-labeled peptides were added to each well of the fractionator’s loading tray. Fractionation was carried out at a constant current of 50.0 μA for about 100 h. Fractioned samples were collected separately in 1.5-mL microcentrifuge tubes and acidified by adding 10 μL of 5% trifluoroacetic acid (TFA) in water (v/v). Sample volumes were reduced in a SpeedVac to about 50 μL. LC-MS/MS analysis was then carried out using an LTQ XL orbitrap mass spectrometer (Thermo Scientific) coupled to an Ultimate 3000 nano-HPLC system. The reversed-phase LC system consisted of a desalting column (300 μm × 5 mm, PepMap C18 3 μm, 100 Å) and an analytical column (75 μm × 250 mm, PepMap C18 3 μm, 100 Å) with split solvent delivery (split ratio of 1:300). A Thermo Scientific nanospray II source was fitted with a 30-μm silica emitter tip (PicoTip; New Objective) and maintained at 1,100 V ion spray voltage. Peptide samples (5 μL) were loaded onto the trap column in 0.1% TFA at 20 μL min−1 for 3 min and eluted at 300 nL min−1 using a gradient from 0.05% formic acid in water (A) to 0.05% water in 80% acetonitrile (B). Using Excalibur 2.0.1, intact peptides were detected between mass-to-charge ratio 400 and 2,000 in the orbitrap at a resolution of 30,000 with external calibration. Maximum ion accumulation time allowed on the LTQ orbitrap was 1 s for all scan modes. Collision-induced dissociation (CID) spectra of the top three peptide ions were acquired at a normalized collision energy of 30, followed by higher energy C-trap dissociation (HCD) at a resolution of 7,500 with orbitrap acquisition. Dynamic exclusion was set with a repeat count of 2, a repeat time of 30 s, and an exclusion time of 3 min. The chromatography feature was enabled with a correlation area ratio of 1.0. Activation Q was set to 0.25 with a 30-ms activation time.
Protein Identification and Quantification
Using Proteome Explorer version 1.1 (Thermo Scientific), the orbitrap raw data were processed and peak lists were generated from the CID spectra (for protein identification) and from the HCD spectra (for quantitation). For identification, peak lists were submitted to an in-house installation of Mascot 2.2 searching the soybean database (version 0.1c) with the following parameters: MudPIT scoring, precursor mass tolerance of 10 ppm, fragment mass tolerance of 0.8 D, TMT 6-plex on peptide N termini and Lys and carbamidomethylation on Cys as static modifications, oxidation of Met, deamidation of Asn and Gly, and cyclization of peptide N-terminal Gly to pyro-Glu as dynamic modifications. The false discovery rate (based on searching a reversed decoy database) was set to 1%.
Quantification was carried out by ProteomeDiscoverer using six channels with a 20-ppm integration window around the accurate monoisotopic masses of the 6-plex TMT reporter ions. Isotope purity correction factors for each reporter ion (rows) and the mass differences (−2, −1, 0, 1, 2) were set according to the information given in Table IV. Raw quantitation values were used for the subsequent statistical analysis, and only unique peptides were used for the quantitation. Proteins in each channel were normalized to the protein median. The se of control samples was calculated from the se of the three ratios 126:127, 127:128, and 128:126. The se of glyphosate-treated samples was calculated from the se of the three ratios 129:130, 130:131, and 131:129.
Table IV. Isotopic abundance correction matrix for the TMT reagents used.
| Row Headers | −2 | −1 | 0 | 1 | 2 |
| 126 | 0.00 | 0.00 | 92.32 | 7.68 | 0.00 |
| 127 | 0.00 | 0.00 | 93.42 | 6.58 | 0.00 |
| 128 | 0.00 | 0.00 | 94.62 | 5.38 | 0.00 |
| 129 | 0.00 | 1.40 | 94.06 | 4.54 | 0.00 |
| 130 | 0.00 | 0.00 | 96.56 | 3.44 | 0.00 |
| 131 | 0.00 | 1.67 | 94.54 | 3.79 | 0.00 |
Sample Preparation for Gel-Based LC-MS/MS Analysis
For each sample, 200 mg of leaves was powdered in liquid nitrogen. Ground leaf material was incubated for 16 h at –20°C in precipitation solution (1 mL) containing TCA (10%, w/v) and β-mercaptoethanol (0.07%, v/v) in acetone. Precipitated protein was pelleted by centrifuging for 20 min at 4°C at 16,000g (5415R; Eppendorf) and washed six times with ice-cold washing solution containing acetone (90%, v/v) and β-mercaptoethanol (0.07%, v/v) in Milli-Q water. Proteins were solubilized in 1 mL of urea (8 m), CHAPS (2%, w/v), and dithiothreitol (9.3 mg mL−1) mix by sonication in an ultrasonic water bath for 1 h, followed by overnight incubation at room temperature for optimal protein solubilization. The samples were centrifuged at 16,000g, and the supernatant containing protein was transferred to a fresh microfuge tube. In order to concentrate the protein, 1 mL of 80% acetone was added to each sample to precipitate the proteins. After air drying, the pellet was redissolved in 50 μL of 1× NuPage LDS sample buffer (Invitrogen) by vortex and sonication in a cold-water bath for 15 min, then 25 μL of each sample was loaded on a Bis-Tris gel.
SDS-PAGE Separation of Proteins
Protein samples in loading buffer were heated for 5 min at 85°C, centrifuged, and then the soluble proteins were separated on a 1-mm, 4% to 12% Bis-Tris SDS-PAGE gel (NuPage; Invitrogen). The gel was stained using colloidal Coomassie blue (EZ Blue Gel Staining Reagent; Sigma). Each of the six lanes was cut into eight slices, and the resulting 48 samples were reduced, alkylated, in-gel digested with trypsin, and subjected to protein identification with an LTQ XL orbitrap mass spectrometer (Thermo Scientific).
Automated in-Gel Protein Reduction, Alkylation, and Trypsin Digestion
Each gel slice was mashed to small pieces using a pointed forceps. Minced gels containing protein samples were first transferred to microtiter plates and then subjected to digestion by a ProGest Protein Digester Robot (Genomic Solutions). Gel pieces were first soaked in 50 μL of 25 mm ammonium bicarbonate and then dehydrated using an equal volume of acetonitrile at 37°C. Dithiothreitol (40 μL of 10 mm) was added to each sample. After incubation at room temperature for 20 min, 30 μL of iodoacetamide was added and the samples were incubated at room temperature for 45 min. The samples were then washed with 50 μL of 25 mm ammonium bicarbonate and dehydrated with 50 μL of acetonitrile for 10 min. Trypsin (8 μg sample−1) was added to each sample, and the tubes were incubated for 14 h. Finally, the digestion was terminated with 10 μL of 5% TFA. Ten microliters of each sample was transferred to HPLC vials for further LC-MS/MS analysis.
LC-MS/MS Analysis of Gel-Derived Samples
LC-MS/MS analysis was carried out on an LTQ XL orbitrap mass spectrometer (Thermo Scientific) coupled to an Ultimate 3000 nano-HPLC system using essentially the same parameters as outlined for the analysis of TMT-derivatized samples derived from wild-type plants, with the difference that no HCD fragmentation was performed and that the top five ions from each survey scan were fragmented using CID.
Protein Identification and Quantitation
Using Proteome Explorer version 1.1 (Thermo Scientific), the orbitrap raw data were processed and peak lists were generated (Mascot generic format). Peak list files were submitted to an in-house installation of the global proteome machine (version 2009.04.4.1; www.thegpm.org), and the soybean database (version 01.c) was searched using X!Tandem and the following parameters: precursor mass error, 15 ppm; product ion mass error, 0.6 D; carbamidomethylation as a static modification; Met oxidation as a variable modification; two search refinement stages taking into account peptide phosphorylation, deamidation, methylation, deoxidation, and carbamidomethylation of amino acids other than Cys. For each protein, a normalized peptide count was calculated by dividing the total number of peptide spectra that contributed to the identification of that protein by the total number of identified peptide spectra in that sample. Relative protein concentrations for proteins detected in glyphosate-treated transgenic plants and in control transgenic plants were calculated by division of the normalized peptide counts for each protein (Supplemental Table S2).
Statistical Analysis
The gas-exchange data were analyzed by ANOVA. Data for all other physiological parameters were analyzed by Student’s t test or lsd test.
Sequence data from this article can be found in the GenBank/EMBL data libraries under the accession numbers in Supplemental Table S3.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. Proteomic analysis of glyphosate treatment of glyphosate-sensitive soybean leaves using 6-plex TMT.
Supplemental Table S2. Proteomic analysis of glyphosate treatment of glyphosate-resistant soybean leaves using normalized spectral counting.
Supplemental Table S3. Accession numbers.
References
- Ahsan N, Lee DG, Lee KW, Alam I, Lee SH, Bahk JD, Lee BH. (2008) Glyphosate-induced oxidative stress in rice leaves revealed by proteomic approach. Plant Physiol Biochem 46: 1062–1070 [DOI] [PubMed] [Google Scholar]
- Ballmer-Weber BK, Vieths S. (2008) Soy allergy in perspective. Curr Opin Allergy Clin Immunol 8: 270–275 [DOI] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Dalton DA, Langeberg L, Treneman NC. (1993) Correlations between the ascorbate-glutathione pathway and effectiveness in legume root nodules. Physiol Plant 87: 365–370 [Google Scholar]
- Das M, Reichman JR, Haberer G, Welzl G, Aceituno FF, Mader MT, Watrud LS, Pfleeger TG, Gutiérrez RA, Schäffner AR, et al. (2010) A composite transcriptional signature differentiates responses towards closely related herbicides in Arabidopsis thaliana and Brassica napus. Plant Mol Biol 72: 545–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davletova S, Rizhsky L, Liang H, Shengqiang Z, Oliver DJ, Coutu J, Shulaev V, Schlauch K, Mittler R. (2005) Cytosolic ascorbate peroxidase 1 is a central component of the reactive oxygen gene network of Arabidopsis. Plant Cell 17: 268–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon DP, Edwards R. (2010) Glutathione S-transferases. The Arabidopsis Book 8: e0131, doi/10.1199/tab.0131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domínguez-Valdivia MD, Aparicio-Tejo PM, Lamsfus C, Cruz C, Martins-Loução MA, Moran JF. (2007) Nitrogen nutrition and antioxidant metabolism in ammonium-tolerant and -sensitive plants. Physiol Plant 132: 359–369 [DOI] [PubMed] [Google Scholar]
- Dutilleul C, Lelarge C, Prioul JL, De Paepe R, Foyer CH, Noctor G. (2005) Mitochondria-driven changes in leaf NAD status exert a crucial influence on the control of nitrate assimilation and the integration of carbon and nitrogen metabolism. Plant Physiol 134: 64–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer C, Rowell J, Walker D. (1983) Measurement of the ascorbate content of spinach leaf protoplasts and chloroplasts during illumination. Planta 157: 239–244 [DOI] [PubMed] [Google Scholar]
- Foyer CH, Bloom A, Queval G, Noctor G. (2009) Photorespiratory metabolism: genes, mutants, energetics, and redox signaling. Annu Rev Plant Biol 60: 455–484 [DOI] [PubMed] [Google Scholar]
- Foyer CH, Parry M, Noctor G. (2003) Markers and signals associated with nitrogen assimilation in higher plants. J Exp Bot 54: 585–593 [DOI] [PubMed] [Google Scholar]
- Frendo P, Gallesi D, Turnbull R, Van de Sype G, Hérouart D, Puppo A. (1999) Localisation of glutathione and homoglutathione in Medicago truncatula is correlated to a differential expression of genes involved in their synthesis. Plant J 17: 215–219 [Google Scholar]
- Frendo P, Harrison J, Norman C, Hernandez-Jimenez MJ, Van de Sype G, Gilabert A, Puppo A. (2005) Glutathione and homoglutathione play a critical role in the nodulation process of Medicago truncatula. Mol Plant Microbe Interact 18: 254–259 [DOI] [PubMed] [Google Scholar]
- Frendo P, Jimenez MJ, Mathieu C, Duret L, Gallesi D, Van de Sype G, Herouart D, Puppo A. (2001) A Medicago truncatula homoglutathione synthetase is derived from glutathione synthetase by gene duplication. Plant Physiol 126: 1706–1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaines TA, Zhang W, Wang D, Bukuna B, Chisholma ST, Shanerd DL, Nissena SJ, Patzoldte WL, Tranele PJ, Culpepper AS, et al. (2010) Gene amplification confers glyphosate resistance in Amaranthus palmeri. Proc Natl Acad Sci USA 107: 1029–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groten K, Dutilleul C, van Heerden PDR, Vanacker N, Bernard S, Finkemeier I, Dietz KJ, Foyer CH. (2006) Redox regulation of peroxiredoxin and proteinases by ascorbate and thiols during pea root nodule senescence. FEBS Lett 580: 1269–1276 [DOI] [PubMed] [Google Scholar]
- Groten K, Vanacker H, Dutilleul C, Bastian F, Bernard S, Carzaniga R, Foyer CH. (2005) The roles of redox processes in pea nodule development and senescence. Plant Cell Environ 28: 1293–1304 [Google Scholar]
- Hietanen E, Linnainmaa K, Vainio H. (1983) Effects of phenoxyherbicides and glyphosate on the hepatic and intestinal biotransformation activities in the rat. Acta Pharm Toxicol 53: 103–112 [DOI] [PubMed] [Google Scholar]
- Ireland CR, Percival MP, Baker NR. (1986) Modification of the induction of photosynthesis in wheat by glyphosate, an inhibitor of amino acid metabolism. J Exp Bot 37: 299–308 [Google Scholar]
- Kitchen LM, Witt WW, Rieck CE. (1981) Inhibition of δ-aminolevulinic acid synthesis by glyphosate. Weed Sci 29: 571–577 [Google Scholar]
- Klapheck S. (1988) Homoglutathione: isolation, quantification and occurrence in legumes. Physiol Plant 74: 727–732 [Google Scholar]
- Lee H-M, Dietz KJ, Hofestadt R. (2010) Prediction of thioredoxin and glutaredoxin target proteins by identifying reversibly oxidised cysteinyl residues. J Integrat Bioinform 7: 130–141 [DOI] [PubMed] [Google Scholar]
- Matamoros MA, Dalton DA, Ramos J, Clemente MR, Rubio MC, Becana M. (2003) Biochemistry and molecular biology of antioxidants in the rhizobia-legume symbiosis. Plant Physiol 133: 499–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medici LO, Azevedo RA, Smith RJ, Lea PJ. (2004) The influence of nitrogen supply on antioxidant enzymes in plant roots. Funct Plant Biol 31: 1–9 [DOI] [PubMed] [Google Scholar]
- Noctor G, Arisi A-CM, Jouanin L, Foyer CH. (1998) Manipulation of glutathione and amino acid biosynthesis in the chloroplasts. Plant Physiol 118: 471–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor G, Bergot GL, Mauve C, Thominet D, Lelarge-Trouverie C, Prioul JL. (2007) A comparative study of amino acid measurement in leaf extracts by gas chromatography-time of flight-mass spectrometry and high performance liquid chromatography with fluorescence detection. Metabolomics 3: 161–174 [Google Scholar]
- Noctor G, Foyer CH. (1998) Ascorbate and glutathione: keeping active oxygen under control. Annu Rev Plant Physiol Plant Mol Biol 49: 249–279 [DOI] [PubMed] [Google Scholar]
- Noctor G, Queval G, Mhamdi A, Chaouch S, Foyer CH. (2011) Glutathione. The Arabidopsis Book 9: e0142, doi/10.1199/tab.0142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novitskaya L, Trevanion SJ, Driscoll S, Foyer CH, Noctor G. (2002) How does photorespiration modulate leaf amino acid contents? A dual approach through modelling and metabolite analysis. Plant Cell Environ 25: 821–835 [Google Scholar]
- Powles SB. (2008) Evolved glyphosate-resistant weeds around the world: lessons to be learnt. Pest Manag Sci 64: 360–365 [DOI] [PubMed] [Google Scholar]
- Queval G, Noctor G. (2007) A plate reader method for the measurement of NAD, NADP, glutathione, and ascorbate in tissue extracts: application to redox profiling during Arabidopsis rosette development. Anal Biochem 363: 58–69 [DOI] [PubMed] [Google Scholar]
- Ramalho JC, Camos PS, Teixeira MT, Nunes MA. (1998) Nitrogen-dependent changes in antioxidant system and in fatty acid composition of chloroplast membranes from Coffea arabica L. plants submitted to high irradiance. Plant Sci 135: 115–124 [Google Scholar]
- Reddy KN, Rimando AM, Duke SO. (2004) Aminomethylphosphonic acid, a metabolite of glyphosate, causes injury in glyphosate-treated, glyphosate-resistant soybean. J Agric Food Chem 52: 5139–5143 [DOI] [PubMed] [Google Scholar]
- Richard EP, Goss JR, Arntzen CJ. (1979) Glyphosate does not inhibit photosynthetic electron transport and phosphorylation in pea (Pisum sativum) chloroplasts. Weed Res 27: 684–688 [Google Scholar]
- Skipsey M, Cummins I, Andrews CJ, Jepson I, Edwards R. (2005) Manipulation of plant tolerance to herbicides through co-ordinated metabolic engineering of a detoxifying glutathione transferase and thiol cosubstrate. Plant Biotechnol J 3: 409–420 [DOI] [PubMed] [Google Scholar]
- Strauss AJ, Kruger GHJ, Strasser RJ, Van Heerden PDR. (2006) Ranking of dark chilling tolerance in soybean genotypes probed by the chlorophyll a fluorescence transient O-J-I-P. Environ Exp Bot 56: 147–157 [Google Scholar]
- van Heerden PDR, Kiddle G, Pellny TK, Mokwala PW, Jordaan A, Strauss AJ, de Beer M, Schlueter U, Kunert KJ, Foyer CH. (2008) Regulation of respiration and the oxygen diffusion barrier in soybean protect symbiotic nitrogen fixation from chilling-induced inhibition and shoots from premature senescence. Plant Physiol 148: 316–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heerden PDR, Tsimilli-Michael M, Kruger GHJ, Strasser RJ. (2003) Dark chilling effects on soybean genotypes during vegetative development: parallel studies of CO2 assimilation, chlorophyll a fluorescence kinetics O-J-I-P and nitrogen fixation. Physiol Plant 117: 476–491 [DOI] [PubMed] [Google Scholar]
- Yu W, Zhang R, Li R, Guo S. (2007) Isolation and characterization of glyphosate-regulated genes in soybean seedlings. Plant Sci 172: 497–504 [Google Scholar]