Abstract
PHOSPHATE TRANSPORTER TRAFFIC FACILITATOR1 (PHF1) is known to regulate the plasma membrane localization of PHT1;1, a high-affinity inorganic phosphate (Pi) transporter in Arabidopsis (Arabidopsis thaliana). OsPHF1, a rice (Oryza sativa) gene homologous to AtPHF1, was isolated and found to regulate the localization of both low- and high-affinity Pi transporters to the plasma membrane. Three OsPHF1 allelic mutants carrying one-point mutations at the fifth WD-repeat motif and two at the transmembrane helix, respectively, showed arsenate resistance and severely reduced Pi accumulation. The data indicate that mutation of OsPHF1 results in the endoplasmic reticulum retention of the low-affinity Pi transporter OsPT2 and high-affinity Pi transporter OsPT8. Mutation of OsPHF1 also reduced Pi accumulation in plants exhibiting excessive shoot Pi accumulation due to the overexpression of OsPHR2. However, the transcript level of OsPHF1 itself is not controlled by OsPHR2. Overexpression of OsPHF1 increased Pi accumulation in both roots and shoots in a solution culture with Pi-supplied condition. These results indicate that the role of OsPHF1 is unique in the localization of both low- and high-affinity Pi transporters on the plasma membrane in rice and determines Pi uptake and translocation in rice. The similar function of PHF1 required to facilitate PHT1 transit through the endoplasmic reticulum between Arabidopsis and rice provides an example of expectations from what one would deduce from sequence comparisons to extend knowledge from Arabidopsis to crops.
Phosphorus is an essential macronutrient for plant growth and development. Plants acquire inorganic phosphate (Pi) directly from their environment by active absorption into the epidermal and cortical cells of the root via Pi transporters. After entry into the root cortical cells, Pi must eventually be loaded into the apoplastic space of the xylem, transported to the shoot, and then redistributed within the plant via Pi transporters (Schachtman et al., 1998).
As a constituent of nucleic acids, phospholipids, and cellular metabolites, living cells require millimolar amounts of Pi. However, most soil Pi is immobile and the Pi concentration available to roots is in micromolar quantities (Raghothama, 1999). To coordinate plant growth with the limited Pi availability, high-affinity Pi transporters have evolved to enable increased Pi acquisition from soils (Raghothama, 1999; Paszkowski et al., 2002; Rausch and Bucher, 2002; Ticconi and Abel, 2004). High-affinity plant Pi transporters were originally identified by sequence similarity with the high-affinity transporter of yeast (Saccharomyces cerevisiae), PHO84. Genes encoding some of these transporters are able to complement pho84 yeast mutants (Rausch and Bucher, 2002). These proteins belong to the PHOSPHATE TRANSPORTER1 (PHT1) family of Pi/H+ symporters (Rausch and Bucher, 2002). Nine PHT1 genes have been identified in Arabidopsis (Arabidopsis thaliana), and 13 PHT1 genes have been identified in rice (Oryza sativa; Goff et al., 2002; Rausch and Bucher, 2002).
Yeast cells starved for Pi activate feedback loops that regulate low- and high-affinity phosphate transport. In this manner, the interplay of positive and negative feedback loops leads to the bistability of phosphate transporter usage. Cells express predominantly either low- or high-affinity transporters, both of which have similar phosphate uptake capacities (Wykoff et al., 2007). Previous reports demonstrated that at least one member of the OsPHT1 family, OsPT2, is a low-affinity Pi transporter and may play a primary role during the Pi translocation process (Ai et al., 2009). OsPT2 is under the transcriptional control of OsPHR2, the functional ortholog of AtPHR1 in rice (Zhou et al., 2008). Recently, the function of OsPT8 as a high-affinity Pi transporter was reported (Jia et al., 2011). Overexpression of OsPT2 and OsPT8 causes excessive shoot Pi accumulation and results in a Pi toxicity phenotype, similar to the overexpression of OsPHR2 (Liu et al., 2010; Jia et al., 2011). These reports indicate that low- and high-affinity Pi transporters in plants and yeast may possess similar phosphate uptake capacities.
Similar to yeast PHO84, members of the PHT1 family are localized to the plasma membrane (Chiou et al., 2001; Shin et al., 2004). This indicates that the activity of Pi transporters is controlled through the secretory pathway trafficking to the plasma membrane. Several studies in yeast and animal cells have shown that the first crucial step in secretory trafficking is the exit from the endoplasmic reticulum (ER) to the Golgi apparatus. This step is mediated by COPII vesicles (Barlowe and Schekman, 1993; Barlowe, 2003). In Arabidopsis, the PHOSPHATE TRANSPORTER TRAFFIC FACILITATOR1 (PHF1) is reported to be an important factor for the localization of high-affinity Pi transporters to the plasma membrane (Gonzalez et al., 2005). Mutation of AtPHF1 specifically impairs Pi transport and results in the constitutive expression of many Pi starvation-induced genes, increased arsenate (As) resistance, and reduced Pi accumulation. Recently, PHF1 as a PHT1-specific facilitator required for PHT1 transiting through the ER to plasma membrane has been demonstrated. In the posttranslational regulation of PHT1, phosphorylation of PHT1 modulates ER exit and targeting to the plasma membrane that is conserved in PHT1 (Bayle et al., 2011). Additional evidence to the function of PHF1 in regulation of Pi uptake has been reported from its function involved in the excessive leaf Pi accumulation in nitrogen limitation adaptation (nla) mutant at low-NO3− supply (Kant et al., 2011). NLA gene is involved in adaptive responses to low-nitrogen conditions in Arabidopsis, where nla mutant plants display abrupt early senescence caused by excessive leaf Pi accumulation. Mutation of PHF1, as mutation of PHT1;1, can rescue the early senescence under low-nitrogen conditions of nla mutant.
Phylogenetic analysis revealed a single copy of PHF1 in Arabidopsis and two putative PHF1 homologs in rice (designated OsPHF1 and OsPHF1L). Although the function of PHF1 is crucial for the uptake and translocation of Pi, the functions of OsPHF1 and OsPHF1L in rice, the monocot model plant, have not yet been reported. This study reports the isolation and characterization of OsPHF1 and OsPHF1L in rice. Evidence supporting a role for OsPHF1 in regulating the localization of both low- and high-affinity Pi transporters to the plasma membrane is provided. Results also show that mutation of OsPHF1 reduces the excessive shoot Pi accumulation driven by the overexpression of OsPHR2. However, the transcript level of OsPHF1 is not controlled by OsPHR2. This work reveals that OsPHF1 is required for efficient low- and high-affinity Pi transporters targeting plasma membrane in rice, and determines the Pi uptake and translocation of Pi in rice plant.
RESULTS
Isolation and Characterization of the Mutants
As mutants defective in Pi uptake are tolerant to As, an ethylmethanesulfonate-generated mutant rice library (Japonica cv Nipponbare) was screened using a solution culture containing 30 μm As. Seedlings displaying tolerance to As were isolated after 7 d of growth in the As solution, after which tolerant seedlings were transferred to normal culture solution supplemented with 200 μm Pi for 2 weeks. The cellular Pi concentration in the leaves of these plants was then analyzed. Several plants exhibiting significantly lower Pi concentration were detected when compared to the wild-type plants (Fig. 1, A–D). To determine the genetic factor responsible for the defective Pi uptake, a cross was made between one mutant with extremely low-Pi concentrations in comparison to wild type and an Indica rice (Kasalath). Among the F2 offspring produced from this cross, the genetic segregation of the As tolerance indicated that the tolerance to As is controlled by a recessive single gene.
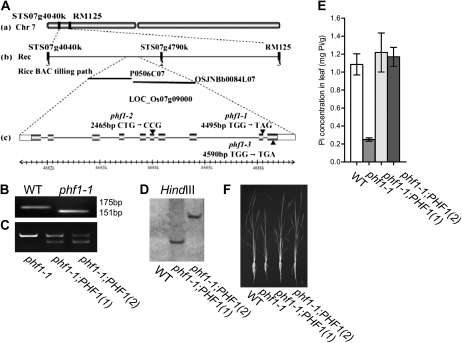
Figure 1.
Phenotypic characteristics and Pi concentration analysis of the Osphf1 mutants. A, Seedlings of the wild-type and three Osphf1 allelic mutants (phf1-1, phf1-2, and phf1-3) after growth in solution containing 30 μm As for 5 d. B, Growth performance of wild-type and Osphf1 allelic mutants grown in a solution culture with 200 μm Pi for 30 d. C and D, Cellular Pi concentration in the leaves (C) and roots (D) of wild-type and Osphf1 mutants. The data represents the mean of three replicates with sd indicated by error bars. E, RT-PCR analysis of the expression of Pi-responsive genes in wild-type and Osphf1-1 mutant roots. RNA was prepared from the roots of the 15-d-old seedlings grown in a solution culture with 200 μm Pi. The primers specific to the genes analyzed are listed in Supplemental Table S3. The transcript level of the Actin gene after 26 PCR cycles is used as a control.
Cloning and Characterization of the OsPHF1 Gene
To clone the mutated gene, seeds of the F2 population were grown in a solution culture containing 30 μm As for 7 d. Seedlings exhibiting As tolerance were selected for mapping the mutated gene. Using 100 F2 mutants, the mutated gene locus was initially mapped between two markers, STS07g4040K and RM125, on chromosome 7. Ultimately, the gene was mapped to a region of 750 kb flanked by STS07g4040K and STS07g4790K (Fig. 2A). Within this region, one gene homologous to AtPHF1 was annotated (LOC_Os07g09000). Sequencing analysis revealed one point mutation at 4,495 bp that resulted in a change from TGG to TAG. Consequently, a Trp residue was changed to a stop codon. Comparison of the genomic and cDNA sequences of the gene indicates that the gene contains 10 exons and nine introns. The identified point mutation occurs in the ninth exon (Fig. 2A). By sequencing the genomic DNA of other mutants exhibiting low-Pi concentrations, two allelic mutants were isolated. These mutants contain point mutations at the sixth exon resulting in a change from CTG to CCG (Leu to Pro) and at the 10th exon resulting in a change from TGG to TGA (Trp to stop codon), respectively (Fig. 2A). The three mutants were designated phf1-1, phf1-2, and phf1-3 (Fig. 2A). All three allelic mutants displayed a similar tolerance to As. Additionally, the mutant plants were smaller and had lower Pi concentrations in both the roots and shoots (Fig. 1, A–D). The constitutive expression of Pi starvation-induced genes (Hou et al., 2005; Aung et al., 2006; Bari et al., 2006) was also observed in the phf1-1 mutant (Fig. 1E). The phf1-1 point mutation creates a derived cleaved amplified polymorphic sequence (dCAPS) marker indicated by the digestion of the DNA fragment of the gene using XbaI (Supplemental Fig. S1). Using this dCAPS marker, the point mutation in the phf1-1 mutant was confirmed (Fig. 2B).
Figure 2.
Map-based cloning of OsPHF1 and complementation analysis. A, Map-based cloning of OsPHF1. OsPHF1 was mapped between STS07g4040K and STS07g4790K on chromosome 7 (a and b). The sequence surrounding the mutations (G-to-A transition, G-to-C transition, and G-to-A transition) in phf1-1, phf1-2, and phf1-3 is also shown (c). The exon structure of OsPHF1 is represented with boxes (light, untranslated; dark, coding region; c). B, PCR analysis using a pair of dCAPS primers to confirm the point mutation in Osphf1-1. The PCR products of OsPHF1 from the wild type and Osphf1-1 from the mutant were digested with XbaI. The primers are listed in Supplemental Table S1. C, The PCR products of OsPHF1 from Osphf1-1 and the two independent transgenic complementation lines using the primers flanking an intron of OsPHF1. One PCR product (597 bp) was obtained from Osphf1-1 and two PCR products (597 and 216 bp) were obtained from the complementation lines. The primers are listed in Supplemental Table S1. D, The two independent transgenic complementation lines indicated by Southern-blot analysis. E, Cellular Pi concentrations in leaves of 30-d-old wild type, Osphf1-1 mutant, and the transgenic complementation plants grown in a solution culture supplied with 200 μm Pi. F, Phenotype of 30-d-old plants of wild type, Osphf1-1 mutant, and two lines of complementation plants grown in a solution culture with 200 μm Pi.
For complementation of the phf1 mutant, the full-length coding sequence (CDS) of OsPHF1 (1,161 bp) was cloned into the binary vector pBI101.3 (Yue et al., 2010) and driven by the promoter of OsPHF1. The binary vectors were introduced into phf1-1 mutants using the Agrobacterium-mediated transformation method, as previously described (Chen et al., 2003). Two independent transgenic lines were confirmed by PCR analysis using primers flanking an intron of OsPHF1 (Fig. 2C) and Southern blot (Fig. 2D). The phenotype and cellular Pi concentration in leaves of the wild type, Osphf1-1 mutant, and the complementation lines (T2 plants) were investigated for 30-d-old plants grown in a solution culture supplied with 200 μm Pi (Fig. 2, E and F). The results showed that the Pi concentration in the transgenic plants was rescued to the level of the wild-type plants, which confirms that the impairment of Pi uptake ability in phf1-1 is caused by the functional loss of OsPHF1.
Overexpression of OsPHF1 Results in Excessive Pi Accumulation in Leaf and Root
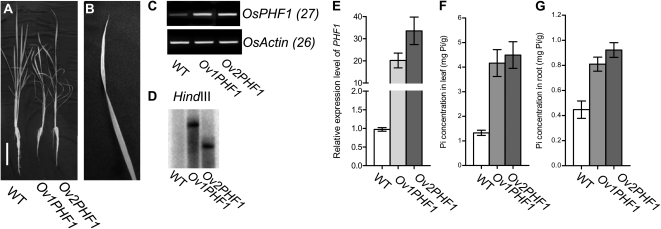
To further determine the function of OsPHF1 on Pi uptake and translocation, the transgenic lines with overexpression of OsPHF1 (OvPHF1) were developed for cellular Pi concentration analysis. Two independent transgenic lines with overexpression of OsPHF1 were determined by gene expression level and Southern-blot analysis (Fig. 3, C–E). The phenotype and Pi concentration analysis of OvPHF1 plants compared to the wild-type plants grown in a solution culture with Pi-supplied condition (200 μm Pi) were investigated. The results showed that the overexpression of OsPHF1 leads to a Pi excessive accumulation in both leaves and roots by about 3-fold of Pi concentration compared to wild type (Fig. 3, F and G). The leaf necrosis and plant growth inhibition of the OvPHF1 plants were observed (Fig. 3, A and B). In Arabidopsis, overexpression of PHF1 can slightly increase Pi concentration in leaves under low-NO3− condition compared to wild type (Kant et al., 2011). It is notable that we used solution culture with 200 μm Pi, which may lead to the higher Pi accumulation in OvPHF1 lines of rice than that of Arabidopsis lines. The different outcome between Arabidopsis and rice resulted from overexpression of PHF1 may also be caused by different regulation downstream of PHF1 that is required further investigation.
Figure 3.
Phenotype and cellular Pi concentration of the transgenic lines with overexpression of OsPHF1 (OvPHF1) grown under Pi-supplied condition (200 μm Pi) compared to the wild type (WT). A and B, Phenotype of the 30-d-old transgenic lines with overexpression of Ov1PHF1 grown under Pi-supplied condition compared to wild type. The arrowhead indicates the leaf necrosis of Ov1PHF1 plant and the enlarged image of leaf necrosis (B). Bar = 10 cm (A). C and E, RT-PCR (C) and qRT-PCR (E) analysis of the expression levels of two transgenic lines with overexpression of OsPHF1 (Ov1PHF1 and Ov2PHF1). D, Southern-blot analysis of the two independent transgenic lines with overexpression of OsPHF1. F and G, Cellular Pi concentrations in leaf and root of 30-d-old WT and the transgenic plants with overexpression of OsPHF1 grown in a solution culture supplied with 200 μm Pi.
Tissue Expression Pattern of OsPHF1
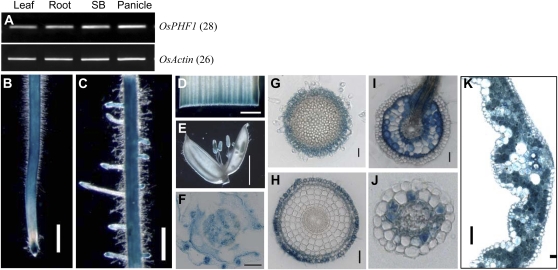
Reverse transcription (RT)-PCR analysis of OsPHF1 transcription indicates a constitutive expression pattern in plants (Fig. 4A). To detail the tissue-specific expression of OsPHF1, transgenic plants harboring a GUS reporter gene driven by the OsPHF1 promoter (−3,700 bp upstream of the start codon) were developed. The GUS staining results were consistent with RT-PCR analysis (Fig. 4, B–F). Examinations of tissue cross sections indicated that OsPHF1 is expressed in the root cap (Fig. 4G), along with expression in the epidermis, exodermis, and sclerenchyma in the elongation zone of the primary root (Fig. 4H). Within the root region where lateral roots emerge, OsPHF1 is expressed in the cortex (Fig. 4I). GUS staining was also detected in the cortex of lateral roots (Fig. 4J) and all the mesophyll cells of the leaf (Fig. 4K).
Figure 4.
Tissue-specific expression pattern of OsPHF1. A, RT-PCR analysis of OsPHF1 transcript levels in the leaf, root, stem base (SB), and panicle. The specific primers for OsPHF1: forward primer: CTC AGA ATA TTT CAT TGG CCG AGC; reverse primer: CCT AGA AAA GCG GCA ACA TTC AAT CTT C. B to E, GUS staining of the primary root (B), latearl roots (C), leaf (D), and flower (E) of the transgenic plants harboring the OsPHF1p:GUS fused gene. Bar = 0.5 mm (B and C). Bar = 5 mm (E). F to H, GUS-stained cross sections of stem base (F), root cap (G), root elongation zone (H), and root region where the lateral root emerged (I). Bar = 20 μm. Bar = 200 μm for F. J, Cross section of the GUS-stained lateral root. Bar = 20 μm. K, Cross section of GUS-stained leaf blade. Bar = 50 μm.
OsPHF1 Regulates the Plasma Membrane Localization of Low- and High-Affinity Pi Transporters
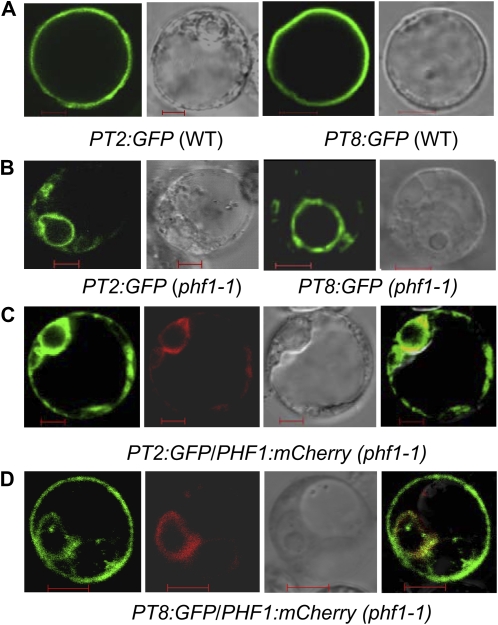
Previous reports demonstrated that the low-affinity Pi transporter OsPT2 and the high-affinity Pi transporter OsPT8 play important roles in the excessive accumulation of Pi in the shoots (Ai et al., 2009; Jia et al., 2011). To determine whether OsPHF1 regulates these Pi transporters, the subcellular localization of a constitutively expressed OsPHT1:GFP fusion protein was investigated in protoplast cells prepared from wild type and phf1-1 mutant. The results demonstrated that the mutation of OsPHF1 results in the retention of the investigated Pi transporters in the ER (Fig. 5, A and B). The ER location of PHF1 has been confirmed in Arabidopsis (Gonzalez et al., 2005; Bayle et al., 2011). Our data showed that OsPHF1 is also located in ER (Fig. 5C). To further confirm that the impairment of plasma membrane location of PT2 and PT8, the complementation of OsPHF1 to the plasma membrane location of PT2 and PT8 in Osphf1-1 mutant protoplasts was tested. The results indicate that the impairment of membrane location of PT2 and PT8 in phf1-1 mutant is rescued by the cotransformation of OsPHF1:mCherry (Fig. 5, C and D), which supports that OsPHF1 regulates the plasma membrane location of the high- and low-affinity Pi transporters in rice.
Figure 5.
OsPHF1 regulates the localization of low- and high-affinity Pi transporters (PT2 and PT8) to the protoplast membrane. A, Transient expression of fusion proteins PT2:GFP and PT8:GFP in wild-type protoplasts, showing the plasma membrane localization. Bar = 5 μm. B, Transient expression of PT2:GFP and PT8:GFP in Osphf1-1 protoplasts, showing their localization in the ER. Bar = 5 μm. C and D, Cotransient expression of OsPT2/PT8:GFP and OsPHF1:mCherry in Osphf1-1 protoplast. Bar = 5 μm. From left to right: PT2/8:GFP, OsPHF1:mCherry, bright field of the protoplasts, and emerged images.
Mutation of OsPHF1 Leads to a Reduction in OsPHR2-Driven Shoot Pi Accumulation; However, OsPHF1 Does Not Function Downstream of OsPHR2
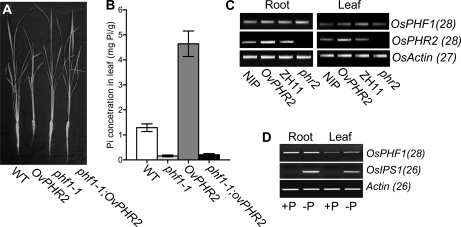
It has been suggested that AtPHF1 may be regulated by AtPHR1 (At3g52190; Gonzalez et al., 2005). To determine whether OsPHF1 is regulated by OsPHR2, a functional rice homolog of AtPHR1 (Zhou et al., 2008), plants overexpressing OsPHR2 in a phf1-1 background (designated as phf1-1;OvPHR2) were developed by crossing the phf1-1 mutant with transgenic plants overexpressing OsPHR2 (Zhou et al., 2008). The F3 lines of phf1-1;OvPHR2 were used for further analysis. Upon examination of plant growth in a solution culture supplemented with 200 μm Pi, the phf1-1;OvPHR2 plants were able to rescue the leaf necrosis and plant growth inhibition phenotype of the OvPHR2 plants (Fig. 6A). Additionally, the excessive shoot Pi accumulation exhibited by the OvPHR2 plants was reduced in the phf1-1;OvPHR2 plants to the level of the phf1-1 mutant (Fig. 6B).
Figure 6.
OsPHF1 is not regulated downstream of OsPHR2 at transcriptional level. A, Growth performance of 30-d-old plants of the wild type (WT), OvPHR2, Osphf1-1, and phf1-1;OvPHR2 plants grown in a solution culture supplied with 200 μm Pi. B, Cellular Pi concentration in the leaves of 30-d-old wild type, OvPHR2, Osphf1-1, and phf1-1;OvPHR2 grown in a solution culture supplied with 200 μm Pi. C, RT-PCR analysis of the expression levels of OsPHR2 and OsPHF1 in roots of the wild type (Nipponbare: NIP and Zhonghua 11: ZH11), OvPHR2 and phr2 mutant plants grown in a solution culture supplied with 200 mm Pi. The primers specific to the genes analyzed are listed in Supplemental Table S3. The transcript level of the Actin gene after 27 PCR cycles was used as a control. D, RT-PCR analysis of the expression levels of OsPHF1 and OsIPS1 in the root and leaf of the wild-type plants grown in a solution culture with Pi-supplied (+P) and Pi-deficient (−P) conditions. The primers specific to the genes analyzed are listed in Supplemental Table S3.
The transcript level of OsPHF1 is similar in roots under Pi-supplied and Pi-deficient conditions, while up-regulated in leaves under Pi-deficient condition (Fig. 6D). OvOsPHR2 can mimic the Pi-starvation signal (Zhou et al., 2008). To determine whether OsPHF1 is regulated downstream of OsPHR2 at transcription level, the expression levels of OsPHF1 in root and leaf of OvPHR2 and phr2 mutant (Supplemental Fig. S2) were investigated by RT-PCR analysis (Fig. 6C). The results indicate that the OsPHF1 transcript level is not regulated by OsPHR2, suggesting that the up-regulation of OsPHF1 in leaves is a response to low-Pi concentration but not the mimic Pi-starvation signal in OvPHR2 plants.
DISCUSSION
Mutations in OsPHF1 Result in the Impairment of Pi Uptake and Translocation
In Arabidopsis, mutations in PHF1 lead to increased As resistance and reduced Pi accumulation (Gonzalez et al., 2005). Recently a model for differential trafficking of PHT1 to the plasma membrane or vacuole as a function of phosphate concentration has been proposed where PHF1 acts on PHT1, upstream of vesicle coat protein COPII formation (Bayle et al., 2011). Hence, the investigation of PHF1 function in rice may provide information useful for the development of crops with enhanced Pi efficiency. In this study, three rice mutants carrying different point mutations in OsPHF1 were isolated. These allelic mutants exhibited a similar tolerance to As and a reduction of Pi concentration in both the roots and shoots (Fig. 1). Genetic complementation further confirmed that the mutation in OsPHF1 is responsible for the reduced Pi uptake and translocation in rice (Fig. 2). Thus, OsPHF1 is the functional homolog of AtPHF1.
Overexpression of OsPHF1 results in Pi excessive accumulation and the similar phenotype of leaf necrosis and plant growth inhibition in the transgenic plants under Pi-supplied condition (Fig. 3) as of overexpression of OsPHR2 and pho2 mutant. In OsPHR2-overexpressed plants and pho2 mutant plants, several Pi transporters are up-regulated (Wang et al., 2006; Zhou et al., 2008; Liu et al., 2010). The mutation of OsPHF1 under background of overexpression of OsPHR2 can reduce the accumulated Pi concentration to the level of the wild-type plants. These results provide additional evidence that OsPHF1 plays a role in Pi uptake and translocation in rice. It is noted that the increase of Pi accumulation driven by overexpression of PHF1 is only about 1.7-fold compared to wild type in Arabidopsis (Kant et al., 2011). This is probably because PHF1 is not generally limiting for Pi accumulation under the growth conditions used in the Arabidopsis experiments. In our case, the rice plants were grown in a solution culture with high-Pi level (200 μm Pi). Actually in a solution culture with low-Pi level (10 μm Pi), the increase in Pi concentration in the rice transgenic plants is much less than that in the high-level Pi solution culture.
The OsPHF1 protein has 47% amino acid sequence identity to Arabidopsis PHF1. Overall structural conservation, initially evaluated using the structure prediction metaserver GENESILICO (Kurowski and Bujnicki, 2003), predicted that PHF1 proteins contain seven WD40 repeats related to the C-terminal region of Tup1 (score 1e-81; Sprague et al., 2000). A transmembrane domain was also predicted in the C termini of PHF1 protein (Letunic et al., 2004). A premature stop codon in both Osphf1-1 and Osphf1-3 eliminates this transmembrane domain in the encoded proteins and likely results in a complete loss of function. A point mutation at 2,465 bp (the sixth exon) causing a change in the amino acid sequence (Leu-218 to Pro) in the fifth WD40 repeat domain in Osphf1-2 also eliminates the function of OsPHF1, implying that the WD40 repeat in PHF1 is crucial for its function (Supplemental Fig. S3).
Mutation of OsPHF1 Results in the ER Retention of Both Low- and High-Affinity Pi Transporters
Mutation of AtPHF1 was demonstrated to result in the ER retention of high-affinity Pi transporter (PHT1) as a PHT1-specific chaperone (Bayle et al., 2011). In rice, the low-affinity Pi transporter, OsPT2, has been reported to play an important role in the translocation of stored Pi within the plant (Ai et al., 2009). Moreover, OsPT2 makes a large contribution to the excessive accumulation of Pi in the shoots of OvPHR2 plants (Ai et al., 2009; Liu et al., 2010). The overexpression of OsPT2 and the high-affinity Pi transporter OsPT8 results in excessive Pi accumulation in the shoots (Liu et al., 2010; Jia et al., 2011). As-tolerant mutants carrying point mutations in OsPT8 (Supplemental Fig. S4) were also isolated in the mutant screens of this study, indicating that OsPT8 plays an important role in Pi uptake. Subcellular localization of fused proteins of OsPT2:GFP and OsPT8:GFP in protoplast cells indicated that both of the Pi transporters were retained in the ER of phf1-1 mutant cells (Fig. 5). Based on the similar Pi concentration observed in the phf1 mutants and phf1-1;OvPHR2 lines, along with the ER retention of the Pi transporters caused by the mutation of OsPHF1, we confirm the key role of OsPT2 and OsPT8 in Pi accumulation in roots and leaves is controlled by OsPHF1.
OsPHF1 May Play a Unique Role in Pi Uptake and Translocation in Rice
Phylogenetic analysis indicates a single copy of PHF1 in dicots and two copies of PHF1 (PHF1 and PHF1L) in monocots, including rice (Supplemental Fig. S5). This implies that the PHF1 in monocots experienced a duplication event after the divergence of monocots and dicots. The highly conserved WD40 repeat domain and transmembrane domain between OsPHF1 and OsPHF1L poses the possibility that OsPHF1L may have some function in Pi uptake ability. A T-DNA insertion within the OsPHF1L promoter (364 bp upstream of the start codon) results in the enhanced expression of OsPHF1L (Supplemental Fig. S5). It was reported that the multimerized transcriptional enhancers of the cauliflower mosaic virus 35S promoter, located next to the left border of T-DNA vector pGA2715, can significantly increase the expression of genes immediately adjacent to the inserted enhancer (Jeong et al., 2002). It is notable that the enhanced expression of OsPHF1L caused by the insertion of pGA2715 did not alter the Pi concentration in comparison to the wild type (Supplemental Fig. S5). In contrast, the overexpression of OsPHF1 results in the excessive accumulation of Pi in both the roots and leaves (Fig. 3). To confirm the function of OsPHF1L, we cotransferred the vectors with PT2:GFP fused gene and overexpressed OsPHF1L gene driven by 35S promoter into the protolasts of phf1-1 mutant, but no rescue of the plasma membrane localization of PT2 in phf1 was observed (Supplemental Fig. S5).
To examine whether function of OsPHF1L is not affected in the T-DNA insertional line, the sequence of full length of OsPHF1L in the mutant was compared with its parent (Zhonghua 11) and Nipponbare that was used for development of transgenic lines with overexpression of OsPHF1. The results showed that the gene structure of OsPHF1L in the mutant is completely the same as that in Zhonghua 11 and Nipponbare (Supplemental Fig. S6). From these results, OsPHF1 is hypothesized to play a unique role in Pi uptake and translocation. Recently, precise data demonstrate that PHF1 is required for efficient PHT1 targeting in Arabidopsis. Phosphoproteomic and mutagenesis analyses revealed modulation of PHT1;1 ER export by Ser-514 phosphorylation status (Bayle et al., 2011). Serial phosphorylation events of Ser residues 520 and 514 at the C end of PHT1;1 were found and the latter very likely prevents PHT1;1 exit from ER and its proper targeting to the plasma membrane. The conserved potential phosphorylation events of Ser-514 and ER exit motif at the C end between PHT1s between Arabidopsis and rice were found (Supplemental Fig. S7). These data strongly suggest that the role of OsPHF1 in exiting OsPHT1 from ER and its proper targeting to the plasma membrane is same as in Arabidopsis.
OsPHR2 Does Not Directly Control the Transcription of OsPHF1
AtPHF1, described as specifically responsive to Pi starvation, is expressed at a reduced level in the phr1 mutant (Gonzalez et al., 2005). It is proposed that PHF1 is under the direct transcriptional control of AtPHR1 (and/or other closely related homologs). This theory is based upon the fact that both the Arabidopsis and rice PHF1 genes share a conserved motif (GAATATCC) that conforms to the PHR1 core binding sequence (GNATATNC; Rubio et al., 2001) and is located within a 300-nucleotide region upstream of the start codon of the encoded protein (Gonzalez et al., 2005). OsPHR2 is a functional homolog of AtPHR1 in rice. The overexpression of OsPHR2 mimics Pi starvation, resulting in the constitutive expression of many Pi-starvation-responsive genes. Additionally, OsPT2 (a low-affinity Pi transporter), along with OsPT8 and OsPT10 (high-affinity transporters), is up-regulated in the roots of OvPHR2 plants. This leads to the excessive accumulation of shoot Pi (Zhou et al., 2008; Liu et al., 2010; Jia et al., 2011). Our data provide evidence that the expression of OsPHF1 was not changed in both OvPHR2 plants and the phr2 mutant (Fig. 6). Additionally, the transcript levels of OsPHF1 were not changed in the phf1-1 mutant, unlike other Pi-starvation-responsive genes (Liu et al., 2010). It has been reported that a PHR1 binding site is conserved in the promoter of rice and Arabidopsis (Gonzalez et al., 2005) and in parallelism with the reported functional redundancy between PHR1 members in Arabidopsis (Bustos et al., 2010). The functional redundancy of OsPHR2 in regulating PHF1 may explain for the case of phr2 mutant but it cannot be explained in the case of overexpression of OsPHR2. Therefore, the regulation upstream of OsPHF1 in rice may be different as in Arabidopsis.
In summary, this study identifies an AtPHF1 homolog in rice. Mutation of OsPHF1 results in the ER retention of both low- and high-affinity Pi transporters involved in Pi uptake and translocation. Although two copies of PHF1 (OsPHF1 and OsPHF1L) exist in rice, OsPHF1 is demonstrated to play an enough role in the localization of the Pi transporters to the plasma membrane. In addition, this study revealed differences in the transcriptional regulation of OsPHF1 and AtPHF1 by AtPHR1 homologs in rice and Arabidopsis, respectively. The similar function of PHF1 required to facilitate PHT1 transit through the ER between Arabidopsis and rice provides an example of expectations from what one would deduce from sequence comparisons to extend knowledge from Arabidopsis to crops. The finding that overexpression of OsPHF1 results in an increase of Pi accumulation in transgenic rice, which could not be observed in Arabidopsis highlights the importance of using more than one ecotype or species (genetic background) and growth condition to uncover the whole functional potential of any gene.
MATERIALS AND METHODS
Isolation of the phf1 and pt8 Mutants and Plant Growth Conditions
The phf1 and pt8 mutants were isolated by screening an ethylmethanesulfonate-generated rice (Oryza sativa) mutant library (Japonica cv Nipponbare) in a water culture containing 30 μm As. The tolerant seedlings exhibiting longer roots were transferred to a hydroponic culture (Yoshida et al., 1976) replacing FeCl3 with NaFe(III)-EDTA. The hydroponic culture supplemented with 200 μm Pi was adjusted to pH 5.5 using 1 m NaOH before use and the solution was replaced every 3 d. The hydroponically cultured mutants and wild-type Nipponbare were grown in a greenhouse with a 12 h day (30°C)/12 h night (22°C) photoperiod, approximately 200 μmol m−2 s−1 photon density, and approximately 60% humidity.
Map-Based Cloning of OsPHF1 and OsPT8
The phf1-1 and pt8-1 mutants were crossed with Kasalath, an Indica rice variety. F2 progeny plants exhibiting tolerance to As (30 μm) and reduced Pi concentration in the leaf were selected for genetic linkage analysis. Preliminary mapping was performed using simple sequence repeat markers distributed throughout the rice genome. For the fine mapping, sequence-tagged site primers were designed according to the DNA sequences of Indica and Japonica (http://www.ncbi.nlm.nih.gov). Primers used in the map-based cloning are listed in Supplemental Table S1.
Complementation Test
For complementation of the phf1 mutant, the full-length CDS of OsPHF1 (1,161 bp) was cloned by RT-PCR amplification. The CDS containing the entire OsPHF1 coding region was inserted into the modified binary vector pBI101.3 between XbaI and XhoI sites (Yue et al., 2010), and then the OsPHF1 promoter was amplified from Nipponbare genomic DNA and inserted before OsPHF1 coding region at the HindIII sites to generate the resulting binary vector. The binary vectors were introduced into phf1-1 mutants using the Agrobacterium-mediated transformation method, as previously described (Chen et al., 2003). Primers used in the cloning of CDS of OsPHF1 and the OsPHF1 promoter are listed in Supplemental Table S1. Southern-blotting analysis of complementation lines was performed by using neomycin phosphatransferase II-specific probe.
Identification of the T-DNA Insertional Mutants of phf1l and phr2
The phf1l insertion mutants (PFG_4A-02399.L) were obtained from the Crop Biotech Institute, Kyung Hee University, Republic of Korea. The homozygous phf1l mutant plants were identified using two PCR reactions as instructed on the RiceGE: Genome Express Database Web site (http://signal.salk.edu/cgi-bin/RiceGE).
The phr2 insertion mutants (RMD_04Z11NL88) were obtained from the Rice Mutant Database developed by the National Special Key Program on Rice Functional Genomics of China. The homozygous phr2 mutant plants were identified using two PCR reactions as instructed on the Web site (http://rmd.ncpgr.cn/). The T-DNA insertion sites were determined by sequencing analysis. The primers used for the identification of the T-DNA insertional phf1l and phr2 mutants are listed in Supplemental Table S2.
RNA Extraction, cDNA Preparation, RT-PCR, and Quantitative RT-PCR
Total RNAs were extracted using the RNAVzol reagent (Vigorous). Approximately 2 μg of total RNA was treated with DNase I and used to synthesize the first-strand cDNA using oligo(dT)18. The product of the first-strand cDNA served as the PCR reaction template. Semiquantitative RT-PCR was performed using a pair of gene-specific primers. Quantitative (q)RT-PCR was performed using a Roche SYBR green I kit on a LightCycler480 machine (Roche Diagnostics) according to the manufacturer’s instructions. Three replicates were performed for each gene. Rice Actin was included as an internal control in all analyses. The primers used for RT-PCR and qRT-PCR are listed in Supplemental Table S3.
Construction of Overexpression Constructs
For overexpression of OsPHF1 and OsPHF1L, the cDNA fragment of the WT(NIP) containing the OsPHF1 and OsPHF1L open reading frame were cloned into a binary vector, pCAMBIA 1300 (Yue et al., 2010) driven by the cauliflower mosaic virus 35S promoter, both by using the XbaI and XhoI sites. The transformation was conducted through Agrobacterium tumefaciens (strain EHA105)-mediated transformation as described by Chen et al. (2003). The primers used for PCR amplification are listed in Supplemental Table S4. Southern-blotting analysis of overexpression lines was performed by using hygromycin-resistant gene-specific probe.
Construction of PTs:GFP Fluorescent Fusion Constructs and OsPHF1:mCherry for Complementation in Protoplast
For the subcellular analysis of the OsPTs, the full-length CDS (excluding the stop codons) of OsPT2 (1,587 bp) and OsPT8 (1,626) was cloned by RT-PCR amplification. The CDS containing the OsPT2 gene was inserted into the modified pCAMBIA1300-35S-GFP vector using XbaI at both sides (Yue et al., 2010).The CDS of OsPT8 was cloned into the XbaI and XhoI sites of pBI221 (Miao and Jiang, 2007) in frame with and fused to GFP. The fusion sites were verified by sequencing, and the resulting constructs were purified and used for transient expression in rice protoplasts. For complementation of OsPTs trafficking to plasma membrane in phf1-1 protoplast, the ORF of OsPHF1 was cloned into the XhoI and EcoRI sites of pSAT4A-mCherry-N1 (http://www.arabidopsis.org/abrc/catalog/vector), and the resulting construct was cotransformed with PT2:GFP in phf1-1 protoplasts. Primers used in the cloning of the OsPTs and OsPHF1 are listed in Supplemental Table S4.
Construction of a GUS Fusion Construct
To develop the OsPHF1p::GUS vector, the OsPHF1 promoter was amplified from Nipponbare genomic DNA and inserted into a modified pBI101.3 (Yue et al., 2010) at the HindIII sites. The primers used for PCR amplification are listed in Supplemental Table S4. The construct was transformed into wild-type plants (Nipponbare) via A. tumefaciens EHA105 (Chen et al., 2003).
Gus Staining
Different portions of mature transgenic plants and seedlings were collected for the histochemical detection of GUS expression. The collected samples were incubated in a solution containing 50 mm sodium phosphate buffer (pH 7.0), 5 mm K3Fe(CN)6, 5 mm K4Fe(CN)6, 0.1% Triton X-100, and 1 mm X-Gluc at 37°C (Jefferson, 1989). Images were taken directly or under a stereomicroscope (SZX16, Olympus). Cross sections of various plant tissues were manually dissected under the stereomicroscope, and the procedures of staining, dehydrating, clearing, infiltrating, and embedding were performed according to Liu et al. (2005). The microtome sections were mounted on glass slides for imaging (Zeiss Axioskop).
Pi Concentration Determination
Pi measurement was conducted as previously described (Delhaize and Randall, 1995; Zhou et al., 2008). For Pi measurements, seedlings were grown in one-half Hoagland medium supplemented with 200 μm Pi for 4 weeks. The second and third fully extended leaves and the roots were collected for Pi determination using a continuous flow analyzer (SKALAR, SKALAR San plus system).
Transient Expression in Rice Protoplasts and Fluorescence Microscopic Imaging
Rice protoplast preparation and transfection followed previously described procedures (Miao and Jiang, 2007) with some modifications. The protoplast source for each experiment is indicated in Figure 5. Briefly, 0.2 mL of protoplast suspension (approximately 2 × 105 cells) was transfected with DNA of the various constructs (10 μg each). OsPHF1 was used as ER indicator and for Osphf1-1 complementation transient expression. Combination of PHF1:GFP with PTs:GFP was used in fusion vectors. After transfection, cells were cultured in protoplast medium (R2S + 0.4 m mannitol) overnight (approximately 12 h). Observations were made on a Nikon Eclipse 90i microscope (Nikon), and images were captured with a SPOT camera. Excitation and emission filters Ex460-500/DM505/BA510-560 and Ex516/10/DM575/BA590 (from Nikon) were used for GFP and RFP (Campbell et al., 2002). Protoplasts were observed under the 60× objective.
Statistical Analysis of Data
The data were analyzed using the Excel software (Microsoft) for average value, sd, and Student’s t test analysis.
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: OsPHF1, AK111777; OsPHF1L, AK111711; OsIPS1, AY568759; OsSQD2, AK060202; OsPHO2, AK067365; OsPHR2, AK100065; OsPT2, AF536962; and OsPT8, AF536968.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Primers for developing the dCAPS marker to detect the point mutation in the phf1-1 mutant.
Supplemental Figure S2. Identification of T-DNA insertional mutant phr2.
Supplemental Figure S3. Sequence alignment analysis of PHF1 protein homologs.
Supplemental Figure S4. Isolation of pt8 mutants and Pi concentration analysis in pt8 mutants.
Supplemental Figure S5. Functional analysis of OsPHF1L, the homolog of OsPHF1.
Supplemental Figure S6. Sequencing analysis of OsPHF1L gene in T-DNA insertional mutant of OsPHF1L.
Supplemental Figure S7. Sequence alignment of the C-terminal regions of 13 Arabidopsis and nine rice PHT1 transporters.
Supplemental Table S1. Primers used for the map-based cloning of OsPHF1 and OsPT8.
Supplemental Table S2. Primers used for the identification of the T-DNA insertional line of OvPHF1l and the phr2 mutant.
Supplemental Table S3. Primers used for RT-PCR and qRT-PCR analysis.
Supplemental Table S4. Primers used for the plasmid constructs.
Acknowledgments
The manuscript was edited by Dr. Karen Champ and Nicola Edwards.
References
- Ai P, Sun S, Zhao J, Fan X, Xin W, Guo Q, Yu L, Shen Q, Wu P, Miller AJ, et al. (2009) Two rice phosphate transporters, OsPht1;2 and OsPht1;6, have different functions and kinetic properties in uptake and translocation. Plant J 57: 798–809 [DOI] [PubMed] [Google Scholar]
- Aung K, Lin SI, Wu CC, Huang YT, Su CL, Chiou TJ. (2006) pho2, a phosphate overaccumulator, is caused by a nonsense mutation in a microRNA399 target gene. Plant Physiol 141: 1000–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bari R, Datt Pant B, Stitt M, Scheible WR. (2006) PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol 141: 988–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlowe C. (2003) Signals for COPII-dependent export from the ER: what’s the ticket out? Trends Cell Biol 13: 295–300 [DOI] [PubMed] [Google Scholar]
- Barlowe C, Schekman R. (1993) SEC12 encodes a guanine-nucleotide-exchange factor essential for transport vesicle budding from the ER. Nature 365: 347–349 [DOI] [PubMed] [Google Scholar]
- Bayle V, Arrighi JF, Creff A, Nespoulous C, Vialaret J, Rossignol M, Gonzalez E, Paz-Ares J, Nussaume L. (2011) Arabidopsis thaliana high-affinity phosphate transporters exhibit multiple levels of posttranslational regulation. Plant Cell 23: 1523–1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustos R, Castrillo G, Linhares F, Puga MI, Rubio V, Perez-Perez J, Solano R, Leyva A, Paz-Ares J. (2010) A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PLoS Genet 6: e1001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell RE, Tour O, Palmer AE, Steinbach PA, Baird GS, Zacharias DA, Tsien RY. (2002) A monomeric red fluorescent protein. Proc Natl Acad Sci USA 99: 7877–7882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Jin W, Wang M, Zhang F, Zhou J, Jia Q, Wu Y, Liu F, Wu P. (2003) Distribution and characterization of over 1000 T-DNA tags in rice genome. Plant J 36: 105–113 [DOI] [PubMed] [Google Scholar]
- Chiou TJ, Liu H, Harrison MJ. (2001) The spatial expression patterns of a phosphate transporter (MtPT1) from Medicago truncatula indicate a role in phosphate transport at the root/soil interface. Plant J 25: 281–293 [DOI] [PubMed] [Google Scholar]
- Delhaize E, Randall PJ. (1995) Characterization of a phosphate-accumulator mutant of Arabidopsis thaliana. Plant Physiol 107: 207–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff SA, Ricke D, Lan TH, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H, et al. (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296: 92–100 [DOI] [PubMed] [Google Scholar]
- Gonzalez E, Solano R, Rubio V, Leyva A, Paz-Ares J. (2005) PHOSPHATE TRANSPORTER TRAFFIC FACILITATOR1 is a plant-specific SEC12-related protein that enables the endoplasmic reticulum exit of a high-affinity phosphate transporter in Arabidopsis. Plant Cell 17: 3500–3512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou XL, Wu P, Jiao FC, Jia QJ, Chen HM, Yu J, Song XW, Yi KK. (2005) Regulation of the expression of OsIPS1 and OsIPS2 in rice via systemic and local Pi signalling and hormones. Plant Cell Environ 28: 353–364 [Google Scholar]
- Jefferson RA. (1989) The GUS reporter gene system. Nature 342: 837–838 [DOI] [PubMed] [Google Scholar]
- Jeong DH, An S, Kang HG, Moon S, Han JJ, Park S, Lee HS, An K, An G. (2002) T-DNA insertional mutagenesis for activation tagging in rice. Plant Physiol 130: 1636–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H, Ren H, Gu M, Zhao J, Sun S, Chen J, Wu P, Xu G. (2011) Phosphate transporter gene, OsPht1;8, is involved in phosphate homeostasis in rice. Plant Physiol 156: 1164–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kant S, Peng M, Rothstein SJ. (2011) Genetic regulation by NLA and microRNA827 for maintaining nitrate-dependent phosphate homeostasis in Arabidopsis. PLoS Genet 7: e1002021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurowski MA, Bujnicki JM. (2003) GeneSilico protein structure prediction meta-server. Nucleic Acids Res 31: 3305–3307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Copley RR, Schmidt S, Ciccarelli FD, Doerks T, Schultz J, Ponting CP, Bork P. (2004) SMART 4.0: towards genomic data integration. Nucleic Acids Res 32: D142–D144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Wang Z, Ren H, Shen C, Li Y, Ling HQ, Wu C, Lian X, Wu P. (2010) OsSPX1 suppresses function of OsPHR2 on expression of OsPT2 and phosphate homeostasis in shoots of rice. Plant J 65: 508–517 [DOI] [PubMed] [Google Scholar]
- Liu H, Wang S, Yu X, Yu J, He X, Zhang S, Shou H, Wu P. (2005) ARL1, a LOB-domain protein required for adventitious root formation in rice. Plant J 43: 47–56 [DOI] [PubMed] [Google Scholar]
- Miao Y, Jiang L. (2007) Transient expression of fluorescent fusion proteins in protoplasts of suspension cultured cells. Nat Protoc 2: 2348–2353 [DOI] [PubMed] [Google Scholar]
- Paszkowski U, Kroken S, Roux C, Briggs SP. (2002) Rice phosphate transporters include an evolutionarily divergent gene specifically activated in arbuscular mycorrhizal symbiosis. Proc Natl Acad Sci USA 99: 13324–13329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghothama KG. (1999) Phosphate acquisition. Annu Rev Plant Physiol Plant Mol Biol 50: 665–693 [DOI] [PubMed] [Google Scholar]
- Rausch C, Bucher M. (2002) Molecular mechanisms of phosphate transport in plants. Planta 216: 23–37 [DOI] [PubMed] [Google Scholar]
- Rubio V, Linhares F, Solano R, Martin AC, Iglesias J, Leyva A, Paz-Ares J. (2001) A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev 15: 2122–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schachtman DP, Reid RJ, Ayling SM. (1998) Phosphorus uptake by plants: from soil to cell. Plant Physiol 116: 447–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H, Shin HS, Dewbre GR, Harrison MJ. (2004) Phosphate transport in Arabidopsis: Pht1;1 and Pht1;4 play a major role in phosphate acquisition from both low- and high-phosphate environments. Plant J 39: 629–642 [DOI] [PubMed] [Google Scholar]
- Sprague ER, Redd MJ, Johnson AD, Wolberger C. (2000) Structure of the C-terminal domain of Tup1, a corepressor of transcription in yeast. EMBO J 19: 3016–3027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ticconi CA, Abel S. (2004) Short on phosphate: plant surveillance and countermeasures. Trends Plant Sci 9: 548–555 [DOI] [PubMed] [Google Scholar]
- Wang XM, Yi KK, Tao Y, Wang F, Wu ZC, Jiang D, Chen X, Zhu LH, Wu P. (2006) Cytokinin represses phosphate-starvation response through increasing of intracellular phosphate level. Plant Cell Environ 29: 1924–1935 [DOI] [PubMed] [Google Scholar]
- Wykoff DD, Rizvi AH, Raser JM, Margolin B, O'Shea EK. (2007) Positive feedback regulates switching of phosphate transporters in S. cerevisiae. Mol Cell 27: 1005–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S, Forno DA, Cock JH, Gomez KA. (1976) Laboratory Manual for Physiological Studies of Rice, Ed 3. International Rice Research Institute, Manila, The Philippines [Google Scholar]
- Yue R, Wang X, Chen J, Ma X, Zhang H, Mao C, Wu P. (2010) A rice stromal processing peptidase regulates chloroplast and root development. Plant Cell Physiol 51: 475–485 [DOI] [PubMed] [Google Scholar]
- Zhou J, Jiao F, Wu Z, Li Y, Wang X, He X, Zhong W, Wu P. (2008) OsPHR2 is involved in phosphate-starvation signaling and excessive phosphate accumulation in shoots of plants. Plant Physiol 146: 1673–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]