Abstract
The formation of nitrogen-fixing nodules in legumes is tightly controlled by a long-distance signaling system in which nodulating roots signal to shoot tissues to suppress further nodulation. A screen for supernodulating Medicago truncatula mutants defective in this regulatory behavior yielded loss-of-function alleles of a gene designated ROOT DETERMINED NODULATION1 (RDN1). Grafting experiments demonstrated that RDN1 regulatory function occurs in the roots, not the shoots, and is essential for normal nodule number regulation. The RDN1 gene, Medtr5g089520, was identified by genetic mapping, transcript profiling, and phenotypic rescue by expression of the wild-type gene in rdn1 mutants. A mutation in a putative RDN1 ortholog was also identified in the supernodulating nod3 mutant of pea (Pisum sativum). RDN1 is predicted to encode a 357-amino acid protein of unknown function. The RDN1 promoter drives expression in the vascular cylinder, suggesting RDN1 may be involved in initiating, responding to, or transporting vascular signals. RDN1 is a member of a small, uncharacterized, highly conserved gene family unique to green plants, including algae, that we have named the RDN family.
Legume plants benefit from their symbiosis with rhizobial bacteria because the bacteria are able to fix molecular nitrogen and share it with the plant, allowing legumes to grow under nitrogen-limiting conditions. In exchange, the plant provides the rhizobia residing in root nodules with fixed carbon from photosynthesis. The interaction is complex and involves multiple layers of regulation by both partners. Genetic analysis of nodulation, initially begun because of the potential for agricultural improvement offered by understanding nitrogen-fixing symbioses, has revealed regulators relevant both to nodule formation and to nonleguminous plants (Kouchi et al., 2010).
The establishment of the symbiosis follows a similar pattern in most legumes. Legume roots secrete flavonoid signals into the rhizosphere. Rhizobia respond to flavonoids by producing a lipochitin oligosaccharide termed Nod factor. Perception of species-specific Nod factor by the compatible species of legume triggers Ca2+ spiking in root hair cells and induces changes in gene expression. Perception also results in a physical response; the plant root hair cell curls to sequester the bacteria. In indeterminate nodulators such as pea (Pisum sativum) and alfalfa (Medicago sativa) the inner cortical cells leave the G0 stage of the cell cycle and begin to divide. At the same time, the plant forms a structure called an infection thread, which allows the trapped, dividing bacteria to pass through the root hair and epidermal and outer cortical cells to be released in symbiosomes within the dividing inner cortical cells. The resulting structure, called a nodule, establishes the physical and biochemical environment to support nitrogen fixation (for review, see Oldroyd and Downie, 2008).
Because the maintenance of active nodules has an energy cost to the plant estimated at 12 to 17 g of carbon per gram of nitrogen obtained (Crawford et al., 2000), regulation of nodule number by the plant is presumed important to balance the need for fixed nitrogen with the cost of supporting the rhizobia. In addition to regulating nodule initiation based on available nitrogen status, the plant regulates spatial location of the nodules and the number of nodules that form in a given symbiotic interaction (for review, see Ferguson et al., 2010). In wild-type plants, early nodules suppress the development of later nodules (autoregulation of nodulation [AON]; Caetano-Anollés and Gresshoff, 1991). Grafting experiments demonstrated that AON involves whole plant signal transduction as well as local signaling events (Delves et al., 1986). Genetic analysis of AON has identified several mutants with an increased number of nodules, often accompanied by an inability to regulate nodule number based on nitrogen status and by abnormalities in root length and lateral root formation. The nodules formed on these mutants have normal morphology and are able to fix nitrogen.
Genes corresponding to these mutants can be divided into those with disruptions in genes that regulate nodule number from the shoot (shoot-controlled supernodulators) and those with a point of action in the root (root-controlled supernodulators). For some of these supernodulators, the corresponding gene has been cloned, while others are presently identified only by phenotype. Additional genes are likely to be involved in the pathway, evidenced by nodulation phenotypes that result from gene overexpression, but are not yet represented by mutations in the genes themselves.
The first AON gene cloned, HAR1 in Lotus japonicus (ortholog Sym29 in pea), encodes a Leu-rich repeat receptor-like kinase (LRR-RLK) with homology to the Arabidopsis (Arabidopsis thaliana) meristematic regulator CLV1. HAR1 functions in the shoots to regulate nodulation (Krusell et al., 2002; Nishimura et al., 2002a). Orthologs in soybean (Glycine max; NARK; Searle et al., 2003) and Medicago truncatula (SUNN; Schnabel et al., 2005) have also been identified. Plants with mutations in these genes display shortened roots, excessive nodules (5- to 10-fold more than wild-type plants), nodulation in the presence of nitrate levels that prevent nodulation in wild-type plants, and in some cases excessive lateral roots (Carroll et al., 1985; Sagan and Duc, 1996; Wopereis et al., 2000; Schnabel et al., 2005). Identified as an independent genetic lesion, the lss shoot-controlled supernodulator in M. truncatula has greatly reduced SUNN expression (Schnabel et al., 2010). Another gene encoding an LRR-RLK kinase involved in shoot regulation of nodulation, KLAVIER (KLV) in L. japonicus, has recently been identified (Miyazawa et al., 2010). The klv mutant, like har1 mutants, supernodulates and is able to nodulate in the presence of abundant nitrate. Additionally, the klv mutant has dwarf shoots and roots, altered vascular and floral development, and delayed flowering (Oka-Kira et al., 2005). Shoot-controlled supernodulators with similar nodulation phenotypes but for which the molecular identity is unknown include ntsn in bean (Phaseolus vulgaris; Park and Buttery, 1989) and sym28 in pea (Sagan and Duc, 1996).
A number of root-controlled AON loci have been identified by mutational analysis, but only one, the M. truncatula EIN2 ortholog SICKLE, has been cloned (Penmetsa et al., 2008). The supernodulation phenotype of sickle mutants, which have disrupted ethylene signaling, demonstrates the role of ethylene in controlling nodulation. The mutants rdh1, tml, and plenty of L. japonicus and nod3 of pea-like har1/sym29/nark/sunn, form short roots with excessive nodules and nodulate in the presence of nitrate, but the nodulation phenotype of a grafted plant depends on the genotype of the root, not the shoot (Postma et al., 1988; Ishikawa et al., 2008; Magori et al., 2009; Yoshida et al., 2010). None of these mutants appear to have a defect in ethylene signaling.
The astray mutant of L. japonicus has approximately twice the nodules of wild-type plants (Nishimura et al., 2002c), which is termed enhanced rather than super nodulation. Also in contrast, nodulation in this mutant is sensitive to nitrate in the same degree as wild type. ASTRAY encodes a basic Leu zipper protein with a RING-finger motif, but whether it acts in the shoot or root has not been reported (Nishimura et al., 2002b).
Overexpression of nodulation-induced CLE peptides (Okamoto, et al., 2009; Mortier et al., 2010) has been shown to reduce nodule number. In L. japonicus, overexpression of LjCLE-RS1 or LjCLE-RS2 systemically reduces nodule number in a HAR1-dependent manner (Okamoto et al., 2009), while in soybean overexpression of the CLE peptides RIC1, RIC2, or NIC1 systemically reduce nodulation in a NARK-dependent manner (Reid et al., 2011). Similar effects of MtCLE12 or MtCLE13 overexpression are seen in M. truncatula (Mortier et al., 2010). Additionally overexpression of MtCLE12 and MtCLE13 in roots impacts shoot growth, allowing speculation that the CLE peptides act as long-distance signaling molecules. However, long-distance transport of CLE peptides in any system has not been demonstrated.
Plant hormones have also been shown to be involved in nodule number regulation. The sunn-1 mutant has a defect in long-distance auxin transport that may affect nodule number (van Noorden et al., 2006); cytokinin receptor mutations can suppress the nodule number defect of har1 (Murray et al., 2007) and sunn-1 (E. Schnabel and J. Frugoli, unpublished data); and inducing abscisic acid insensitivity by expression of a dominant negative allele of Arabidopsis ABSCISIC ACID INSENSITIVE1 results in hypernodulation (Ding et al., 2008). Methyl jasmonate and brassinosteroid have also been implicated in nodule number regulation (Nakagawa and Kawaguchi, 2006; Terakado et al., 2006).
Here we report the cloning of a gene from M. truncatula and its ortholog in pea with an essential root-localized function in AON. The ROOT DETERMINED NODULATION1 (MtRDN1) gene and PsNOD3 are members of a previously uncharacterized gene family conserved across the plant kingdom from green algae to higher plants. RDN1 encodes a protein of unknown function that appears to be expressed at low levels in the vasculature of M. truncatula. Although RDN1 is involved in the legume AON signal transduction pathway, the high level of conservation of RDN family genes throughout the green plant lineage suggests a role for RDN family proteins in basic plant function.
RESULTS
Identification and Mapping of a Root-Controlled Supernodulation Locus in M. truncatula
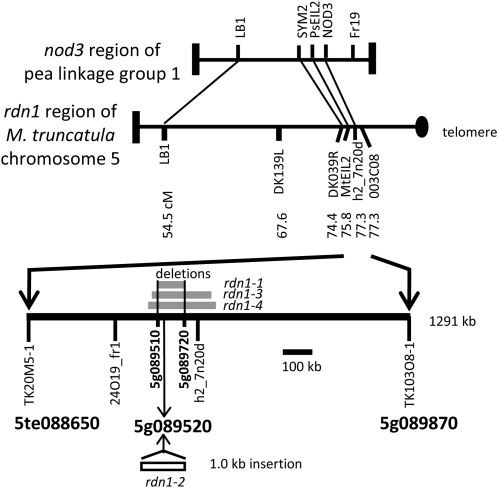
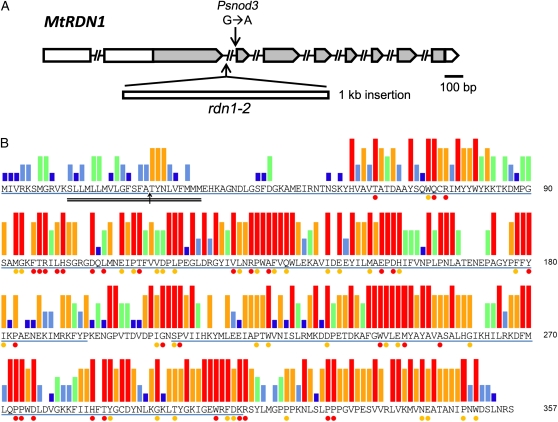
Supernodulating mutants of M. truncatula were identified by a visual screen of fast neutron bombardment M2 seedlings for nodulation phenotypes. Grafting experiments demonstrated that for four mutants (GY15-2E3, D39-13F-V1, D39-1H-T2, and D39-9X-V2) the supernodulation phenotype was conferred by the root tissue (Table I). None displayed the delayed petal senescence phenotype seen in plants with lesions in the SICKLE gene (Penmetsa and Cook, 1997), the only root-determined supernodulation locus reported thus far in M. truncatula, and therefore these new mutants were presumed to define at least one new supernodulation locus in M. truncatula and have been designated rdn1 mutants. Genetic mapping was performed for three of the mutants. In each case the supernodulation phenotype cosegregated with markers on the bottom of chromosome 5 to a region syntenic with the Psnod3 supernodulation locus region (Fig. 1; Temnykh et al., 1995; Gualtieri et al., 2002). A 1,291-kb region centered on marker h2_7n20d was delineated for mutant D39-1H-T2 (rdn1-3), an approximately 2,900-kb region for mutant D39-13F-V1 (rdn1-2), and much larger region for GY15-2E3 (rdn1-1).
Table I. Grafting experiments to determine site of action of RDN1.
Number of nodules formed on grafted seedlings 7 to 10 d after inoculation with S. medicae ABS7. Shown is average nodule number per seedling ± se (no. of seedlings).
| Mutant | Allele | Mutant Shoot/Wild-Type Root | Wild-Type Shoot/Mutant Root |
| GY15-2E3 | rdn1-1 | 11 ± 1.7 (7) | 34 ± 7.4 (8) |
| D39-13F-V1 | rdn1-2 | 13 ± 1.0 (15) | 49 ± 14.0 (3) |
| D39-1H-T2 | rdn1-3 | 9 ± 1.0 (11) | 63 ± 2.5 (7) |
| D39-9X-V2 | rdn1-4 | 12 ± 2.1 (8) | 56 ± 4.9 (5) |
Figure 1.
Positional cloning of rdn1 alleles. The rdn1 locus was mapped to the distal end of the long arm of chromosome 5 using publically available markers. The alignment of the rdn1 region to the syntenic region of the nod3 supernodulation locus of pea is shown. Fine mapping defined a 1,291-kb region for the rdn1 locus flanked by markers TK20M5-1 and TK103O8-1. Multigene deletions found in three of the rdn1 mutants (rdn1-1, rdn1-3, and rdn1-4) are indicated with gray boxes. The 103-kb region missing in common between the three alleles (indicated with black vertical lines) spans predicted genes Medtr5g089510 through Medtr5g089720 (IMGAG v3.5). Medtr5g089520 was found to be altered in rdn1-2 by an indel. cM, Centimorgan.
Transcript profiling of the rdn1-1 mutant versus A17 on the GeneChip Medicago Genome Array (Mitra et al., 2004) identified three genes in the 1,291-kb mapped region with significantly reduced signals in the mutant. PCR analysis revealed that rdn1-1, rdn1-3, and rdn1-4 have large nested deletions of 103, 209, and approximately 240 kb, respectively, spanning two of these genes (Fig. 1). In the genomic segment corresponding to the shortest deletion, 17 genes have been annotated by the International Medicago Genome Annotation Group (IMGAG v3.5). In the rdn1-2 mutant one of these 17 candidate genes (Medtr5g089520) is altered by a 1-kb indel. These data are consistent with the rdn1 mutants representing four alleles of a gene we designate RDN1.
rdn1 Mutants Have Increased Nodulation and Reduced Root Growth
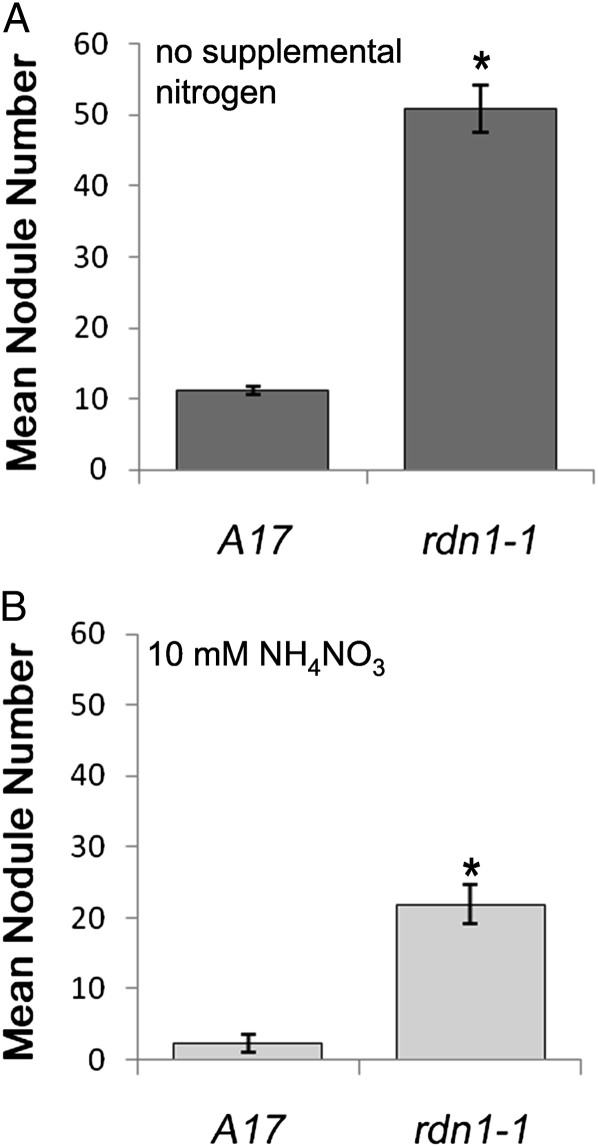
The extent of nodulation of the rdn1-1 mutant was compared to that of wild type using an aeroponic growth chamber. Seedlings were grown under nitrogen-limiting conditions, which favor the development of nodules, and assessed 10 d after inoculation with the compatible rhizobia Sinorhizobium medicae strain ABS7 (Bekki et al., 1987). An average of 5 times more nodules formed on rdn1-1 than on wild type (Fig. 2A).
Figure 2.
Supernodulation phenotype of rdn1-1. Seedlings of the two genotypes were grown together in an aeroponic chamber containing nutrient solution lacking nitrogen (A) or supplemented with10 mm NH4NO3 (B) and were assayed after 14 d of growth (10 d post inoculation with S. medicae). Nodules per plant (means ± se) are shown (A, 40 to 45 plants of each genotype from three combined experimental replicates; B, 17 to 20 plants per genotype from two combined experimental replicates). Student’s t tests were used to determine significance of differences from wild type (*P < 0.0001).
Because previously isolated supernodulation mutants, such as the sunn mutants, have been reported to have nitrate-tolerant nodulation, nodulation of rdn1-1 in the presence of supplemental nitrogen was evaluated (Fig. 2B). Aeroponic growth chambers supplemented with 10 mm NH4NO3 were run side by side with the nitrogen-limited growth chambers. Under these conditions, wild-type roots had very limited nodulation, with the majority of seedlings producing no nodules. In contrast, rdn1-1 produced abundant numbers of nodules, although the numbers formed were less than what was seen in plants grown without NH4NO3. We conclude that supplemental nitrogen has a moderate suppressive effect on nodulation in rdn1-1 similar to what has been seen previously in sunn mutants.
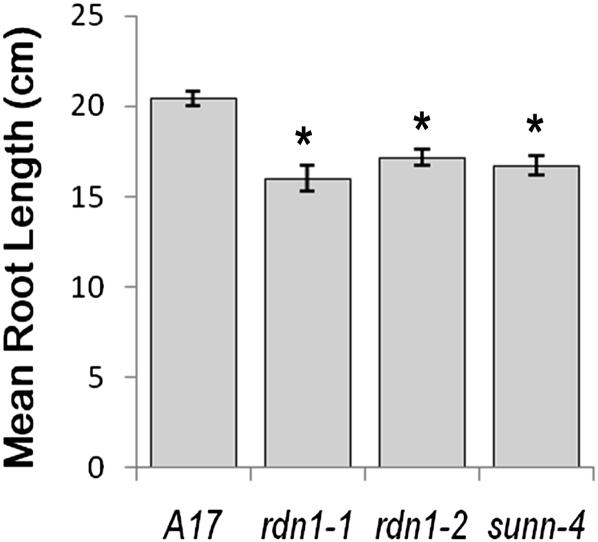
Root length in sunn mutants is shorter than in wild-type plants even in the absence of rhizobia (Schnabel et al., 2005, 2010) presumably due to shorter cells (van Noorden et al., 2006). Root growth in rdn1 mutants was evaluated to see if disruption of the RDN1 locus also impacts root length. The growth rate of rdn1-1 and rdn1-2 roots was less than that of wild-type roots and resembled that of sunn-4 roots (Fig. 3). No other obvious gross morphological differences were noted between rdn1 mutants and wild-type plants. Phenotypes similar to those of rdn1-1 were observed for the other rdn1 alleles (data not shown).
Figure 3.
Root growth in rdn1 mutants compared to wild type (A17) and the sunn-4 supernodulation mutant. Root lengths of seedlings were measured after 14 d of growth in an aeroponic chamber containing nutrient solution supplemented with 5 mm NH4NO3 as a nitrogen source. Means ± se are shown. n = 8 plants per genotype. Student’s t tests were used to determine the significance of differences from wild type (*P < 0.001).
Rescue of the rdn1 Phenotype
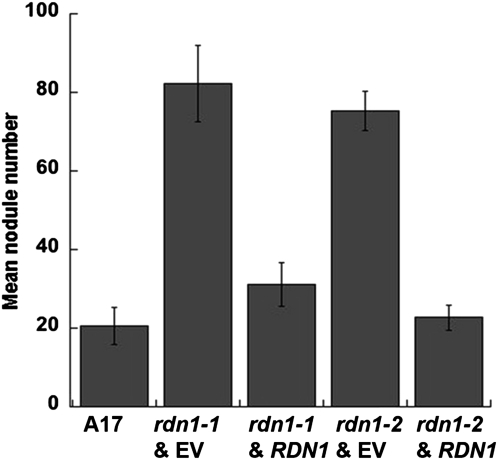
Based on mapping, transcript profiling, and PCR analysis we identified a candidate gene, Medtr5g089520, for RDN1. To verify that Medtr5g089520 was RDN1, the full-length coding sequence driven by the cauliflower mosaic virus 35S promoter (35Spro:RDN1) was introduced into rdn1-1 and rdn1-2 mutant roots by Agrobacterium rhizogenes-mediated transformation using a T-DNA vector also carrying a UBQ10pro:DsRed1 reporter (see “Materials and Methods”). The same vector carrying only the reporter was used as a control. Plants transformed by this method can produce both transgenic and nontransgenic roots. All nontransgenic roots, i.e. those lacking DsRed1 reporter fluorescence, were removed prior to inoculation with S. medicae for nodulation assessment. The rdn1-1 and rdn1-2 mutants transformed with control T-DNA produced, respectively, 82 ± 10 and 75 ± 5 nodules per plant whereas transformed with the RDN1 candidate gene T-DNA they produced only 31 ± 6 and 23 ± 3 nodules per plant, respectively (Fig. 4; Supplemental Fig. S1). This level of nodulation in the presence of the Medtr5g089520 transgene was similar to the nodulation of wild-type roots (21 ± 5). Because restored expression of Medtr5g089520 was sufficient to restore nodule number regulation in rdn1 mutants, we conclude that it includes the segment corresponding to the rdn1 mutant locus.
Figure 4.
Rescue of the rdn1 phenotype with Medtr5g089520 (RDN1) cDNA. Mean nodule number per plant ± se on seedlings with transformed hairy roots. Nontransformed roots (DSRed negative) were trimmed off prior to inoculation. A17 with empty vector (EV), n = 18; rdn1-1 with EV, n = 25; rdn1-1 with 35Spro:RDN1, n = 10; rdn1-2 with EV, n = 25; rdn1-2 with 35Spro:RDN1, n = 38.
Analysis of the RDN1 Gene Sequence
The sequence of RDN1 cDNA amplified from root tissue shows an open reading frame (ORF) of 1,071 bases. The cDNA included several hundred bases of sequence preceding this ORF, suggesting a long 5′ untranslated region. Further PCR analysis localized the transcription start site to a region between positions −872 and −689 relative to the predicted translation start site and identified a 157-bp intron at positions −419 to −263. The predicted 5′ untranslated region includes several potential start codons, one with an ORF of 177 bases and the rest with ORFs of 51 bases or less. Alignment of the cDNA sequence with the genomic sequence shows a 7.4-kb gene consisting of nine exons and eight introns predicted to encode a 357-amino acid protein (Fig. 5).
Figure 5.
RDN1 gene structure and predicted RDN1 protein sequence. A, A schematic representation of the exon and intron structure of the MtRDN1 gene. Gray boxes indicate coding sequence, white boxes indicate untranslated sequences, and intervening lines indicate introns. The rdn1-2 allele has a 9-bp deletion replaced with a 1,017-bp sequence in the second intron. The Psnod3 mutant has a G-to-A transition altering a 3′ intron splice site in the pea ortholog of RDN1. B, The deduced amino acid sequence of MtRDN1. The conflicting SignalP 3.0 predicted signal peptide cleavage site (arrow) and TMHMM 2.0 predicted transmembrane domain (double underline) are indicated. The two predicted globular domains of MtRDN1 are underlined in blue. The strength of sequence conservation in RDN family members is shown for 43 aligned sequences from 12 species of land plants as colored bars above each amino acid (percent identity: red, 100%; orange, 80%–99%; green, 60%–79%; light blue, 40%–59%; dark blue, 20%–39%). Sequence conservation between MtRDN1 and predicted algal sequences is shown below each amino acid as colored dots for identity between MtRDN1 and sequences from both clusters of algal RDN family members (100% identity with 13 aligned sequences from four species of algae, red dots) and between MtRDN1 and the algal RDN family members in the more closely related cluster (five aligned sequences from three species of algae, orange dots). For description of and relationships between the sequences used to analyze conservation refer to Figure 7 and Supplemental Table S1.
The RDN1 coding sequence has an ATG sequence at the seventh codon position in our annotation that is the predicted start site in the IMGAG annotation of Medtr5g089520. We predict the more 5′ start because of the high level of conservation of the six amino acids predicted by these codons among RDN1-like sequences we have identified in other organisms.
Three of the rdn1 alleles are null alleles with multigene deletions, and the remaining allele, rdn1-2, harbors an indel within intron 2. Sequence analysis of the indel region revealed that a 9-bp segment in the middle of the 1,148-bp intron 2 had been replaced with a 1,071-bp sequence of unknown origin (Fig. 5A). Because it was not clear that this alteration of intron 2 would impact gene function, we first determined if rdn1-2 produced RDN1 mRNA. Root cDNA from rdn1-2 was analyzed by PCR and RDN1 transcript was detected (Supplemental Fig. S2). To determine if RDN1 mRNA levels were altered in the mutant, cDNAs from rdn1-2 and wild-type roots were analyzed using reverse transcription quantitative PCR (RT-qPCR) by amplifying a fragment from within exon 2 to measure overall transcript levels. The level of RDN1 cDNA was approximately 75% lower in rdn1-2 than in wild type (Fig. 6). We speculated that the insertion within intron 2 could have an impact on the splicing dynamics of RDN1 mRNA such that a significant portion of the RDN1 mRNA in rdn1-2 mutants would be incompletely spliced. Therefore, to assess splicing of RDN1 mRNA, the levels of RDN1 cDNA from which intron 2 had been spliced were measured by RT-qPCR using primers spanning from exon 2 to exon 3. Splicing of RDN1 mRNA was severely reduced in rdn1-2 compared to wild type, with over 500-fold less spliced transcript detected in the mutant than in wild type.
Figure 6.
Reduced RDN1 expression in rdn1-2. A, A schematic representation of the MtRDN1 gene showing locations of primers used in RT-qPCR analysis. Indicated are coding regions (black boxes), untranslated regions (white boxes), introns (black lines), and primers a through d (arrowheads). Due to the large size of the second intron (2.9 kb in wild type and 3.9 kb in rdn1-2), the PCR product from primers c and d is only amplified from cDNA if the intron has been spliced out. B, The abundance of RDN1 mRNA in rdn1-2 compared to wild type as estimated by RT-qPCR using the primers indicated in A. Overall abundance (left section) and splicing of the second intron (right section) are shown. The level of RDN1 transcript was normalized to the expression level of the reference gene Secret Agent for each cDNA sample using the Pffafl method (Pfaffl, 2001). The normalized levels of PCR products from A17 were defined as 1. The values represent the average of three independent biological replicates (for rdn1-2: means ± se).
The similarity between rdn1 mutants and the pea nod3 mutant, as well as the location of the mutated loci in syntenic regions, led us to speculate that RDN1 and NOD3 were orthologs. We identified a homolog of MtRDN1 in the Psnod3 mutant and its parental cultivar Rondo by PCR. The sequence of the predicted protein is 90% identical to the MtRDN1 sequence. The Psnod3 allele has a point mutation at the 3′ end of the first coding region intron that results in the production of an mRNA with an altered splice site (Fig. 5A; Supplemental Fig. S3). The predicted protein from Psnod3 includes the first 126 amino acids of the wild-type protein followed by two novel amino acids and a premature stop codon.
RDN1 Is Predicted to Encode a Protein of the Endosomal System with Unknown Function
Analysis of the predicted 357-amino acid RDN1 protein using TargetP 1.0 and SignalP 3.0 (Bendtsen et al., 2004; Emanuelsson et al., 2007) suggests that RDN1 enters the secretory pathway and has a 24-amino acid cleaved signal peptide. In contrast, the topology prediction software TMHMM 2.0 (Krogh et al., 2001) and the transmembrane topology and signal peptide predictor Phobius (Käll et al., 2004) instead find the N-terminal sequence to be a transmembrane domain, not a cleaved signal peptide. No potential glycosylphosphatidylinositol lipid anchoring sites were detected by the big-PI Plant Predictor (Eisenhaber et al., 2003). No endoplasmic reticulum retention signal was detected as determined by ScanProsite analysis (http://www.expasy.ch/tools/scanprosite/). The predicted protein contains no conserved characterized domains and does not resemble any previously characterized protein. GlobPlot 2.3 (http://globplot.embl.de; Linding et al., 2003), which identifies regions of globularity and disorder within protein sequences, predicts two globular domains in RDN1 (Fig. 5B). The C-terminal region of RDN1 contains a Pro-rich segment (PPPX5PPPXXP).
RDN1 Is a Member of a Small Uncharacterized Plant Gene Family
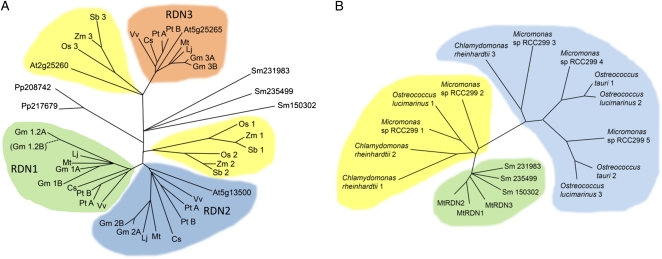
BLAST searches revealed that RDN1 is a member of a small gene family conserved throughout the land plants and green algae. No members of the gene family were detected outside of the Viridiplantae. Three RDN family genes were found in the M. truncatula genome and EST databases including MtRDN1 and two other closely related genes designated MtRDN2 (Medtr8g039290) and MtRDN3, encoding predicted proteins of 361 and 360 amino acids, respectively, which are 63% and 74% identical to MtRDN1.
Predicted RDN-related sequences were analyzed from the sequenced genomes of 11 other land plant species including the dicots Arabidopsis, L. japonicus, soybean, cucumber (Cucumus sativus), poplar (Populus trichocarpa), and grape (Vitus vinifera); the monocots rice (Oryza sativa), maize (Zea mays), and sorghum (Sorghum bicolor); the moss Physcomitrella patens; and the lycophyte Selaginella moellendorfii. A total of 43 predicted RDN family genes were identified (Supplemental Table S1). The deduced protein sequences were aligned (Supplemental Fig. S4) and subjected to phylogenetic analysis (Fig. 7A). Predicted RDN family proteins ranged in size from 344 to 380 amino acids, with most of the variability in size and much of the sequence variation occurring in the amino terminal signal peptide region. The predicted proteins from dicots clustered into three groups, designated the RDN1, RDN2, and RDN3 groups. As was seen for M. truncatula, in L. japonicus, cucumber, and grape a single member of each group was found. In poplar and soybean, which have undergone major genome duplication events, pairs of genes were found for each group; the soybean genome has an additional apparent duplication of the RDN1 group genes with two pairs of RDN1 group sequences (genes GmRDN1A, GmRDN1B, and GmRDN1.2A, and the pseudogene GmRDN1.2B). Arabidopsis was the only analyzed dicot species for which an RDN1 group gene was not found.
Figure 7.
Phylogeny of the RDN protein family. Phylogenetic relationships between predicted RDN proteins in land plants (A) and in algae (B) derived using the neighbor-joining method. Branches supported by at least 50% of the bootstrap replicates (n = 1,000) are shown. Genes were identified by BLAST analysis of the sequenced genomes of the higher plants M. truncatula (Mt), soybean (Gm), L. japonicus (Lj), cucumber (Cs), poplar (Pt), grape (Vv), Arabidopsis (At), rice (Os), sorghum (Sb), and maize (Zm); the moss P. patens (Pp); the lycophyte S. moellendorfii (Sm); and the green algae C. reinhardtii, Micromonas sp. RCC299, O. tauri, and O. lucimarinus. The gene names and sequence sources are given in Supplemental Table S1. [See online article for color version of this figure.]
Three RDN family genes were found in each of the monocots rice, maize, and sorghum. The inferred protein sequences clustered into two groups distinct from the dicot RDN groups. The lycophyte S. moellendorfi and moss P. patens have RDN family members that cluster outside of the dicot and monocot groups.
The degree of sequence conservation between the predicted RDN family proteins of land plants is striking. Among the 43 sequences analyzed, amino acid identity ranged from 55% to 98%. Within the RDN1 group, the proteins were 74% to 95% identical. Similar levels of conservation were found within the RDN2 group. Sequence conservation among the RDN3 group members was somewhat higher (83%–98%). Of the 357 amino acids of the MtRDN1 sequence, 104 were identical in the other 42 RDN sequences (Fig. 5B). An additional 87 amino acids were identical in at least 80% of the RDN family sequences. This corresponds to approximately 60% of predicted mature RDN family protein residues matching in over 80% of RDN family proteins.
Thirteen predicted RDN1-related genes were identified in the sequenced genomes of the green algae Chlamydomonas reinhardtii, Micromonas sp. RCC299, Ostreococcus tauri, and Ostreococcus lucimarinus (Supplemental Table S1). The predicted algal RDN family proteins ranged in size from 409 to 697 amino acids, which was larger than for any of the identified land plant sequences. Longer predicted N-terminal regions in the algal sequences accounted for much of the difference. An alignment of predicted RDN family proteins from algae, M. truncatula, and S. moellendorffii beginning at amino acid 60 of MtRDN1 was used for analysis and generation of a phylogenetic tree (Fig. 7B; Supplemental Fig. S5). Among the algal sequences divergence was greater than for the land plant sequences. For example, over this aligned region the three predicted proteins from C. reinhardtii were only 26% to 50% identical to each other compared to over 73% identity among the MtRDN proteins. The predicted algal proteins were separated into two main clusters composed of sequences approximately 50% and approximately 35% identical to MtRDN1. Amino acids highly conserved between the algal sequences and MtRDN1 are shown in Figure 5B.
RDN1 Expression
We determined where RDN1 was expressed in vivo, to gain preliminary clues about function. RDN1 expression was detected in both roots and shoots of seedlings by PCR from cDNA (Supplemental Fig. S2). RDN1 is represented on the Affymetrix Gene Chip Medicago Genome Array by probe set Mtr.42387.1.S1_at. Examination of microarray data using the M. truncatula Gene Expression Atlas revealed widespread low-level expression of RDN1 with roots appearing to have higher levels of RDN1 mRNA than shoots (Benedito et al., 2008). RDN1 expression in roots did not vary during the course of nodulation in this dataset.
The other two MtRDN genes are also represented on the microarray. Each exhibited expression throughout the plant at higher levels than for RDN1. Compared to RDN1, signal intensities were generally 2- to 5-fold higher for RDN2 (Mtr.40743.1.S1_at) and over 10-fold higher for RDN3 (Mtr.45545.1.S1_at and Mtr.13077.1.S1_at). Expression of these two genes also appeared to be higher in the roots than in the shoots.
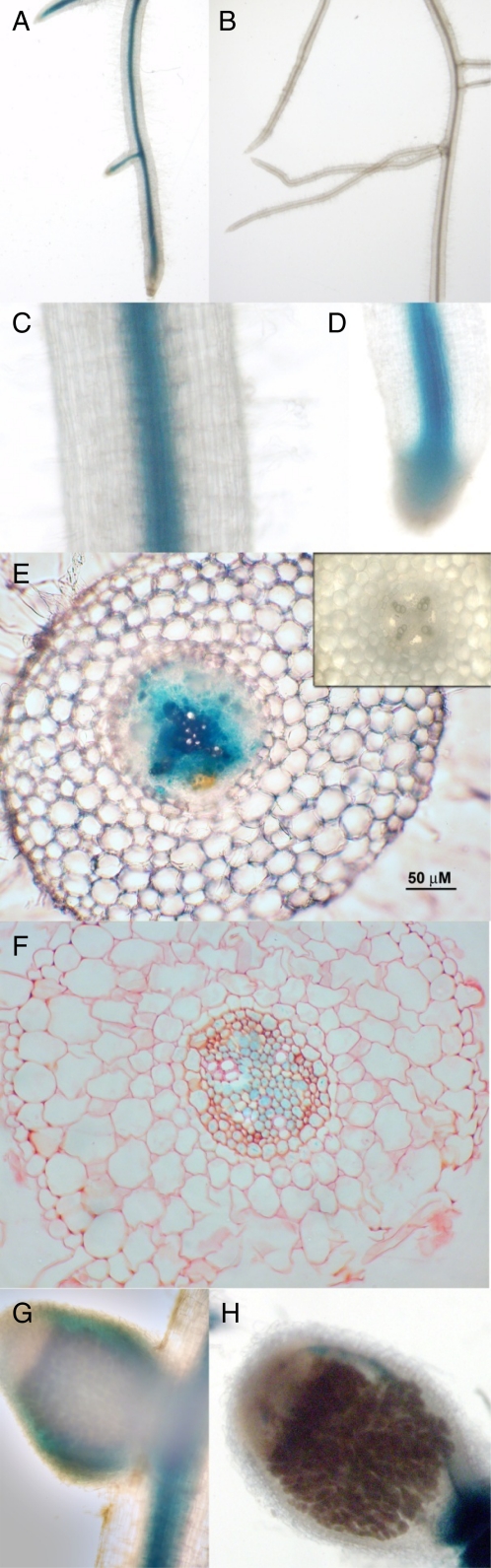
Localization of RDN1 expression within M. truncatula roots was evaluated by use of a reporter construct introduced by A. rhizogenes-mediated transformation. The upstream region of the MtRDN1 gene was used to drive expression of the GUS gene using 2.1 kb of promoter sequence (RDN1-2.1pro:GUS). GUS activity was detected in the vascular cylinder throughout the root (Fig. 8, A, C–F). The staining was specific to the presence of the RDN1 promoter construct as evidenced by a lack of staining in roots transformed with a construct lacking the GUS gene (Fig. 8B). The GUS staining appeared most intense in the area of the xylem, although other regions of the vascular cylinder also often showed staining. No activity was detected in mature nodules besides the expression in the nodule vasculature, which is located on the periphery of the nodule (Fig. 8, G and H).
Figure 8.
Localization of RDN1 promoter activity using a promoter:GUS construct. Roots transformed with T-DNA carrying RDN1-2.1pro:GUS (A and C–H) or lacking the GUS gene (B and E, inset) were incubated with GUS detection reagent. A, GUS activity was detected in the central cylinder throughout RDN1-2.1pro:GUS transformed roots. B, Without the GUS gene, roots show no staining. C and D, Close up of root tissue (C) and root tip (D). E, Transverse root cross section with staining apparent in the vasculature. Negative control shows no background staining (inset). F, Transverse root cross section (3 μm) of fixed and counterstained tissue. GUS staining is present in most regions of the vascular cylinder, including the endodermal layer. G and H, Intact nodule (G) and nodule cross section (H) showing GUS activity only in the vasculature around the nodule following extended incubation (36 h) with GUS detection reagent. Scale bars where shown are 50 μm.
DISCUSSION
rdn1 Mutants
Four mutant alleles of a previously uncharacterized M. truncatula nodulation regulation locus, designated RDN1, were identified. The RDN1 gene was cloned and confirmed by mapping, transcript profiling, and rescue of mutant phenotype by the candidate gene in rdn1 mutant roots. Three of the alleles have deletions of greater than 100 kb encompassing the RDN1 gene and represent null alleles; the other allele has an indel within an intron that dramatically reduces production of mature RDN1 (Figs. 1 and 6). The effect of insertions in introns has been observed in other systems. In Arabidopsis mutants with T-DNA insertions located within introns, transcript levels were shown to be impacted in nearly all cases (Wang, 2008).
RDN1 lies on chromosome 5 in a region syntenic with the region of the pea nod3 root-controlled supernodulation locus. We found a corresponding pea gene with over 90% sequence identity to MtRDN1 that is mutated in the nod3 mutant, indicating that this apparently orthologous gene is the NOD3 gene (Figs. 1 and 5A).
AON shows similarities in distinct groups of legumes although these show differences in nodule development and structure. For example, the orthologous LRR-RLKs SUNN, SYM29, HAR1, and NARK control AON in the shoots of M. truncatula and pea, which form indeterminate nodules, and in L. japonicus and soybean, which form determinate nodules. It might be expected that other proteins involved in AON, such as RDN1, would be similar between legumes that form indeterminate and determinate nodules. We have identified a putative RDN1 ortholog in the L. japonicus genome database located on chromosome 2 in a region with synteny to the RDN1 region of M. truncatula (Cannon et al., 2006). LjRDN1 lies within the 1.2-cM region of chromosome 2 defined for the L. japonicus PLENTY root-controlled supernodulation locus (Yoshida et al., 2010), suggesting that the plenty mutant phenotype could be caused by a lesion in LjRDN1.
Disruption of the RDN1 locus causes an approximately 5-fold increase in root nodulation of seedlings. The rdn1 mutants behave similarly to sunn mutants in nodulation and root growth behaviors: Nodulation under nitrogen-limiting conditions is abundant while the presence of available nitrogen has a moderately suppressive effect on nodule formation; the roots are shorter than wild-type roots (Figs. 2 and 3). In contrast to SUNN that exerts its effect in the shoots, the role of RDN1 in regulating nodule number is in the roots (Table I).
AON requires transmission of a signal from nodulating roots to the shoot and then the relay of information to the whole root system. Such long-distance communication presumably involves the movement of signaling molecules through the vascular cylinder. The promoter of the SUNN gene is active in the vasculature throughout the plant (A. Karve and J. Frugoli, unpublished data), as has also been found for its orthologs LjHAR1 and GmNARK, reported as active predominantly in the phloem (Nontachaiyapoom et al., 2007). Reporter gene analysis in roots shows that like the SUNN promoter, the RDN1 promoter appears to be active in cells of the vasculature although its activity appears to be more widespread than the SUNN promoter (Fig. 8). RDN1 message is also detected in shoots and although our promoter activity assay was limited to root tissues, we expect RDN1 promoter activity in the shoots to be located in the vasculature as well.
Evidence that RDN1 may act in the production or transmission of the AON signal and not in responding to the shoot-derived inhibitor comes from experiments using a pea supernodulating line with the recessive RisfixC mutation, which by mapping and phenotype represents an allele of nod3 (Novak, 2010). Grafted plants possessing a large RisfixC main root system and smaller wild-type adventitious roots produced high numbers of nodules on both the mutant roots and wild-type roots. This observation suggested to the authors that the root system, composed mainly of mutant roots, was unable to send a signal sufficient to trigger the AON response in the shoot, thereby allowing the wild-type roots to produce excessive numbers of nodules.
RDN1 Protein and Related Proteins
The RDN1 gene is predicted to encode a protein of 357 amino acids composed of a secretory signal sequence, two uncharacterized globular domains, and a Pro-rich segment (Fig. 5B). Its cellular function is unknown. We obtained two conflicting structural predictions: one that RDN1 is released as a soluble protein into the endoplasmic reticulum lumen following cleavage of its signal sequence and the other that RDN1 is an integral membrane protein with a single transmembrane domain. However, the predicted localization of RDN proteins to membranes is supported by the identification of the Arabidopsis RDN3 family protein At5g25265 in proteomics analyses in plasma membrane (Marmagne et al., 2007; Mitra et al., 2009) and vacuolar fractions (Carter, et al., 2004; Jaquinod et al., 2007). Determining the subcellular location of RDN1 experimentally would provide an important clue as to the function of the protein, such as whether RDN1 is secreted, located in the plasma membrane, or targeted elsewhere in the cell.
RDN1-related sequences were found in the genomes of all plants examined, including those of green algae, and represent a gene family we have designated the RDN family. All of the putative RDN family proteins identified are predicted by TargetP to have signal sequences, although, as was found for MtRDN1, there are conflicting predictions for whether the signal sequences are cleaved. Three RDN family genes were found in most land plant genomes. For example, M. truncatula has two genes similar to MtRDN1, named MtRDN2 and MtRDN3, which are both predicted to encode proteins approximately 70% identical and over 80% similar to MtRDN1. Predicted dicot RDN family proteins clustered into three groups represented by the three MtRDN proteins (Fig. 7). Predicted monocot RDN family proteins clustered separately into two groups. The structure of the RDN family phylogenetic tree suggests the existence of an RDN gene family prior to the divergence of monocots and dicots, followed by duplications forming the RDN1 and RDN2 groups of the dicots and forming one of the monocot clusters.
Arabidopsis does not have an RDN1 ortholog but does have an additional RDN family gene, At2g25260, divergent from other dicot RDN family genes. Analysis of database sequences from other plants of the order Brassicacales revealed no RDN1 orthologs in close relatives of Arabidopsis in the family Brassicaceae, but a gene similar to At2g25260 was found, while Carica papaya, a member of the family Caricaceae, possesses RDN1, RDN2, and RDN3 homologs like the other dicot lineages. This suggests that loss of RDN1 and appearance of the At2g25260 type gene occurred in the Brassicaceae lineage.
The predicted RDN family proteins from land plants are highly conserved. Over 60% of the amino acids of the predicted mature proteins are highly conserved among family members with over 80% of the family members identical at those positions. Many residues (10%) are also invariant in the algal RDN family sequences.
The conservation of RDN family genes across the Viridiplantae, including unicellular algae, suggests a basic cellular function for RDN family proteins. Of the nearly 7,000 protein families identified in the Chlamydomonas genome, 172 families appear to be unique to the green plants, with almost two-thirds of these proteins predicted to be chloroplast localized (Merchant et al., 2007). The RDN family is one of only 61 identified green plant-specific protein families whose members are not predicted to be localized to chloroplasts and one of only 10 whose members are predicted to have secretory signal sequences.
Expression of RDN Family Genes
A survey of the major organ systems of M. truncatula (Benedito et al., 2008; He et al., 2009) using the Medicago Gene Expression Array revealed that the three RDN genes are expressed in all organs examined, including leaves, stems, flowers, vegetative buds, roots, nodules, pods, and seeds (Supplemental Fig. S6). The abundance of the message appears to vary between the genes with RDN1 detected only at low levels, RDN2 and RDN3 at higher levels.
Similarly, using the Arabidopsis Electronic Fluorescent Pictograph browser for whole genome tiling array data (Winter et al., 2007; Laubinger et al., 2008), the Arabidopsis RDN2 and RDN3 family genes At5g13500 and At5g25265 were found to be expressed in most tissues examined (roots, leaves, shoot apex, cotyledons, hypocotyls, seeds, flowers, young siliques) with the RDN3 family gene being expressed at higher levels (Supplemental Fig. S3). In contrast, At2g25260, the Arabidopsis RDN family gene without an ortholog in the other dicots, was detected primarily in roots and shoot apex inflorescences at high and moderate levels, respectively, and only weakly in other tissues.
In microarray analyses of fluorescently sorted cells from the roots of a series of GFP-marked lines, the Arabidopsis RDN2 family gene was most strongly expressed in the root hair cell lineage with strong expression also throughout the elongation zone. Strong elongation zone expression was also observed in another study (Brady et al., 2007; Dinneny et al., 2008). Thus, it appears that RDN family proteins are expressed in most plant organs, with perhaps higher levels of expression in certain cell types such as vascular cells for MtRDN1 and root hair cells for the Arabidopsis RDN2 family gene.
Consistent with the demonstrated role of MtRDN1 in AON and its expression in vascular tissue, we propose that MtRDN1 is involved in initiating, responding to, or transporting vascular signals and that this vascular signaling function of RDN1-related proteins may be present in all dicots. Furthermore, the wide conservation of RDN family genes across the green plants, including unicellular algae, suggests other conserved molecular functions for the RDN family proteins in the plant cell.
MATERIALS AND METHODS
Plant Materials
Preparation of Medicago truncatula seeds and growth of plants in aeroponic chambers was performed as described in Schnabel et al. (2010). For all other assays, scarified and imbibed seeds were vernalized in the dark at 4°C for 2 d on Harrison Modified Farheus agar (Huo et al., 2006) covered with Whatman filter paper. Following overnight germination at room temperature, seedlings were transferred to plates half covered with filter paper with the radicals on the paper and the cotyledons on the uncovered medium, placed vertically in a growth chamber at 25°C with a 16-h photoperiod for 5 d, and then used for experiments. For all nodulations experiments the strain Sinorhizobium medicae strain ABS7 was used (Bekki et al., 1987).
The rdn1 mutants were identified from independent M2 seed pools collected from fast neutron bombarded M1 seeds of M. truncatula ‘Jemalong’. M3 plants were rescreened in an aeroponic chamber for nodulation phenotype and in the greenhouse for petal senescence. Lines used for detailed analyses were backcrossed to A17 wild type three times (rdn1-1) or once (rdn1-2).
A near-isogenic line of the pea (Pisum sativum ‘Rondo’) mutant nod3 backcrossed into pea cv Juneau (nod3I, PI 598367) and pea cv Rondo were obtained through the National Plant Germplasm System (http://www.ars-grin.gov/).
Mapping of rdn and Sequence Analysis
The F2 self-pollinated progeny from crosses of rdn1 mutants GY15-2E, D39-1H-T2, and D39-13F-V1 to M. truncatula ecotype A20 were used for genetic mapping of the rdn1 locus. DNA from F2 individuals with high nodule numbers was evaluated for the segregation of cleaved amplified polymorphic sequence and other markers as previously described (Schnabel et al., 2010; Supplemental Table S2). For each mutant, 80 to 195 supernodulating F2 plants were tested. For D39-1H-T2 and D39-13F-V1, supernodulating plants represented approximately 25% of the F2 progeny as expected for a single recessive locus. In the F2 progeny of the GY15-2E mapping cross, fewer than expected supernodulating plants were observed (approximately 10%); recombination around the rdn1 locus was suppressed at least 5-fold; and skewed marker segregation was observed, with cosegregation of markers from the long arms of chromosomes 5 and 8. These data suggest a genomic rearrangement within the GY15-2E mutant.
The Gene Chip Medicago Genome Array (Affymetrix) was used for oligonucleotide hybridization experiments comparing transcript profiles of buffer-inoculated wild-type A17 roots with buffer-inoculated rdn1-1 mutant roots. Methods were as described by Mitra et al. (2004).
The search for RDN family members used BLAST algorithms blastp and tblastn (Altschul et al., 1990). Sequences from selected organisms with sequenced genomes were used for analysis. No sequences with a probability relationship to MtRDN1 of e < 3 were found outside of the Viridiplantae.
Phylogenetic Analysis
RDN family predicted protein sequences were aligned using the ClustalW algorithm of MegAlign in Lasergene 7.1.0 (DNAStar). Phylogenetic trees were constructed with MEGA version 4 (Tamura et al., 2007) using the neighbor-joining method with 1,000 bootstrap replicates. Branches supported by at least 500 of the 1,000 bootstrap replicates are shown in Figure 7.
RNA Isolation and RT-qPCR
RNA was isolated from plant tissues using the Qiagen RNeasy mini kit (http://www.qiagen.com), treated with RQ1 RNase-Free DNase (http://www.promega.com) followed by phenol and chloroform extractions and ethanol precipitation, and quantified spectrophotometrically. cDNA was synthesized in 20-μL reactions from 0.5 to 2 μg RNA using random hexanucleotide primers and Superscript Reverse Transcriptase II (http://www.invitrogen.com) following the manufacturer’s recommendations. The absence of genomic DNA in cDNAs was verified by PCR with primers JF1330 and JF1331, specific for a noncoding region near Medtr5g089720. The expression of RDN1 and the specificity of RDN1 primers was evaluated by visualizing PCR products from genomic DNA and cDNA on 1% tris-borate-EDTA gels following PCR amplification (Supplemental Fig. S2).
Expression levels were quantified in an iQ5 thermocycler (http://www.bio-rad.com) using PerfeCTa SYBR green supermix (Quanta Biosciences). Expression levels and splicing of RDN1 transcripts were assessed using primers for amplifying within exon 2 (primers qPCR-a and qPCR-b) and from exon 2 to exon 3 (primers qPCR-c and qPCR-d). Levels of cDNA were normalized using the Secret Agent gene as a reference (Kuppusamy et al., 2004) and ratios were calculated using the Pfaffl method (Pfaffl, 2001). Three independent biological replicates were evaluated. For each cDNA three technical replicates were performed and the values averaged. The efficiency of each primer pair was assessed by use of a dilution series. In all runs, the primer pairs for exon 2, exon 2 to exon 3, and Secret Agent had measured efficiencies of 2.0, 2.0, and 1.8, respectively. Across the biological replicates, threshold cycles for all products from the cDNAs fell within the valid range of the standard curves with the exception of spliced products from rdn1-2 that had high Ct values.
Generation of Transgenic Hairy Roots and Histochemical Analysis
For expression of RDN1 in transgenic plants, a fragment of RDN1 cDNA was PCR amplified from M. truncatula cDNA using primers RDN1cDNA-A and RDN1cDNA-B and cloned downstream of the cauliflower mosaic virus 35S promoter in pC-DsRED2 using KpnI and XhoI. The vector pC-DsRed2 was constructed from pCAMBIA0390 by replacing a portion of the polylinker with the polylinker region of pCAMBIA3201 (EcoRI to PstI), adding the AscI/HindIII UBQ10pro:DsRed1 fragment of pRedRoot (Limpens et al., 2004), and adding additional restriction sites to the polylinker by ligating an EcoRI/MluI/XhoI adaptor into the EcoRI site. For promoter activity analysis, 3.3- and 2.1-kb fragments from upstream of the predicted RDN1 translation start site were amplified by PCR from M. truncatula genomic DNA using primers P2.1-F and P2.1-R and primers P3.3-F and P3.3-R, respectively, and cloned using NcoI and EcoRI into pC2381ES (Huo et al., 2006). The coding and promoter sequences in the binary vector were confirmed by sequencing.
Prepared seedlings were transformed as previously described (Limpens et al., 2004) using Agrobacterium rhizogenes strain ARqua1 (Quandt et al., 1993) containing the appropriate binary vector. Seedlings were maintained on plates until sufficient root tissue had grown. For nodulation experiments, nontransgenic roots (those lacking DsRed fluorescence) were trimmed off prior to transfer of plants to pots of perlite mixed with Harrison Modified Farheus medium without nitrate. After 5 d of nitrogen starvation, plants were flood inoculated with S. medicae (OD600 = 0.1) and nodules were counted 21 d later.
For promoter experiments, transformed tissue was washed twice in 0.1 m Na2HPO4/NaH2PO4, pH 7.2 for 15 min and GUS activity was localized based on a protocol by Jefferson and others (Jefferson et al., 1987). Samples were infiltrated with substrate under vacuum for 30 min and incubated at 37°C for 18 h, unless otherwise indicated. Where indicated, roots were fixed in 5% glutaraldehyde/0.1 m sodium phosphate buffer, pH 7.2 for 2 h under vacuum. Serial ethanol dehydration was then performed by increasing the concentration (10%, 30%, 50%, 70%, 90%, and 100%) at room temperature for 10 min each. Samples were embedded in Technovit 7100 resin (Heraeus Kulzer) using the manufacturer’s instruction. Sections were prepared using a RM2165 microtome (Leica Microsystems), dried onto glass slides at 42°C, and counterstained for 1 min in 1% aqueous saffronin-O solution. Slides were washed briefly with water, dried, and mounted in permount (Fisher Scientific). Tissue was photographed using a Zeiss Axiostar plus microscope and a Nikon E600 microscope with a Retiga EXi FAST monochrome CCD 12-bit camera.
Accession numbers are as follows: MtRDN1 mRNA (GU580937), PsNOD3 gene (GU580938), and PsNOD3 mRNA (GU580939).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Nodulation on rdn1 roots rescued with the Medtr5g089520 (RDN1) cDNA.
Supplemental Figure S2. PCR analysis of RDN1 expression.
Supplemental Figure S3. The structure of the PsNOD3 coding region in pea cultivar Rondo and the derived mutant nod3.
Supplemental Figure S4. Alignment of the predicted sequences of RDN family proteins from 12 land plant species.
Supplemental Figure S5. Alignment of the predicted sequences of RDN family proteins from the green algae with RDN protein sequences from land plants.
Supplemental Figure S6. Expression levels of RDN family genes of M. truncatula and Arabidopsis in various tissues.
Supplemental Table S1. RDN family genes identified in the genomes of 12 land plants and four algae.
Supplemental Table S2. Primers used in RDN1 mapping, T-DNA vector preparation, and RT-qPCR.
Note Added in Proof
Recently, mutations in a CLV2-like gene were found in pea sym28 shoot-controlled supernodulation mutants. Similarly, in Lotus japonicus, a mutant with a lesion in a CLV2-like gene had increased nodule number (Krusell L, Sato N, Fukuhara I, Koch BE, Grossmann C, Okamoto S, Oka-Kira E, Otsubo Y, Aubert G, Nakagawa T, et al [2011] The Clavata2 genes of pea and Lotus japonicus affect autoregulation of nodulation. Plant J 65: 861–871). This finding adds to the number of known genes involved in nodule number regulation
Acknowledgments
We would like to thank Leah Howell, Christine Gianniny, Kyle Ames, and Sherri-Hughes Murphree for preliminary help in the mapping of rdn1; Steve Ellis and Nancy Korn for assistance in use of the Clemson Animal and Veterinary Sciences Histology Core Facility; Harry Kurtz, Jr. for use of his microscope; and Clarice Coyne of the U.S. Department of Agriculture Western Regional Plant Introduction Station for providing seeds of pea lines. This manuscript is Technical Contribution number 5890 of the Clemson Experiment Station.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Bekki A, Trichant JC, Rigaud J. (1987) Nitrogen fixation (C2H2 reduction) by Medicago nodules and bacteroids under sodium chloride stress. Physiol Plant 71: 61–67 [Google Scholar]
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S. (2004) Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 340: 783–795 [DOI] [PubMed] [Google Scholar]
- Benedito VA, Torres-Jerez I, Murray JD, Andriankaja A, Allen S, Kakar K, Wandrey M, Verdier J, Zuber H, Ott T, et al. (2008) A gene expression atlas of the model legume Medicago truncatula. Plant J 55: 504–513 [DOI] [PubMed] [Google Scholar]
- Brady SM, Orlando DA, Lee JY, Wang JY, Koch J, Dinneny JR, Mace D, Ohler U, Benfey PN. (2007) A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318: 801–806 [DOI] [PubMed] [Google Scholar]
- Caetano-Anollés G, Gresshoff PM. (1991) Plant genetic control of nodulation. Annu Rev Microbiol 45: 345–382 [DOI] [PubMed] [Google Scholar]
- Cannon SB, Sterck L, Rombauts S, Sato S, Cheung F, Gouzy J, Wang X, Mudge J, Vasdewani J, Schiex T, et al. (2006) Legume genome evolution viewed through the Medicago truncatula and Lotus japonicus genomes. Proc Natl Acad Sci USA 103: 14959–14964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll BJ, McNeil DL, Gresshoff PM. (1985) A supernodulation and nitrate-tolerant symbiotic (nts) soybean mutant. Plant Physiol 78: 34–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter C, Pan S, Zouhar J, Avila EL, Girke T, Raikhel NV. (2004) The vegetative vacuole proteome of Arabidopsis thaliana reveals predicted and unexpected proteins. Plant Cell 16: 3285–3303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford N, Kahn M, Leustek T, Long S. (2000). Nitrogen and Sulfur. American Association of Plant Biologists, Rockville, MD [Google Scholar]
- Delves AC, Mathews A, Day DA, Carter AS, Carroll BJ, Gresshoff PM. (1986) Regulation of the soybean-Rhizobium nodule symbiosis by shoot and root factors. Plant Physiol 82: 588–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Kalo P, Yendrek C, Sun J, Liang Y, Marsh JF, Harris JM, Oldroyd GE. (2008) Abscisic acid coordinates nod factor and cytokinin signaling during the regulation of nodulation in Medicago truncatula. Plant Cell 20: 2681–2695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinneny JR, Long TA, Wang JY, Jung JW, Mace D, Pointer S, Barron C, Brady SM, Schiefelbein J, Benfey PN. (2008) Cell identity mediates the response of Arabidopsis roots to abiotic stress. Science 320: 942–945 [DOI] [PubMed] [Google Scholar]
- Eisenhaber B, Wildpaner M, Schultz CJ, Borner GH, Dupree P, Eisenhaber F. (2003) Glycosylphosphatidylinositol lipid anchoring of plant proteins: sensitive prediction from sequence- and genome-wide studies for Arabidopsis and rice. Plant Physiol 133: 1691–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Brunak S, von Heijne G, Nielsen H. (2007) Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc 2: 953–971 [DOI] [PubMed] [Google Scholar]
- Ferguson BJ, Indrasumunar A, Hayashi S, Lin MH, Lin YH, Reid DE, Gresshoff PM. (2010) Molecular analysis of legume nodule development and autoregulation. J Integr Plant Biol 52: 61–76 [DOI] [PubMed] [Google Scholar]
- Gualtieri G, Kulikova O, Limpens E, Kim DJ, Cook DR, Bisselin T, Geurts R. (2002) Microsynteny between pea and Medicago truncatula in the SYM2 region. Plant Mol Biol 50: 225–235 [DOI] [PubMed] [Google Scholar]
- He J, Benedito VA, Wang M, Murray JD, Zhao PX, Tang Y, Udvardi MK. (2009) The Medicago truncatula gene expression atlas web server. BMC Bioinformatics 10: 441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo X, Schnabel E, Hughes K, Frugoli J. (2006) RNAi phenotypes and the localization of a protein:GUS fusion imply a role for Medicago truncatula PIN genes in nodulation. J Plant Growth Regul 25: 156–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa K, Yokota K, Li Y, Wang Y, Liu C, Suzuki S, Aono T, Oyaizu H. (2008) Isolation of a novel root-determined hypernodulation mutant rdh1 of Lotus japonicus. Soil Sci Plant Nutr 54: 259–263 [Google Scholar]
- Jaquinod M, Villiers F, Kieffer-Jaquinod S, Hugouvieux V, Bruley C, Garin J, Bourguignon J. (2007) A proteomics dissection of Arabidopsis thaliana vacuoles isolated from cell culture. Mol Cell Proteomics 6: 394–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Käll L, Krogh A, Sonnhammer EL. (2004) A combined transmembrane topology and signal peptide prediction method. J Mol Biol 338: 1027–1036 [DOI] [PubMed] [Google Scholar]
- Kouchi H, Imaizumi-Anraku H, Hayashi M, Hakoyama T, Nakagawa T, Umehara Y, Suganuma N, Kawaguchi M. (2010) How many peas in a pod? Legume genes responsible for mutualistic symbioses underground. Plant Cell Physiol 51: 1381–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL. (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305: 567–580 [DOI] [PubMed] [Google Scholar]
- Krusell L, Madsen LH, Sato S, Aubert G, Genua A, Szczyglowski K, Duc G, Kaneko T, Tabata S, de Bruijn F, et al. (2002) Shoot control of root development and nodulation is mediated by a receptor-like kinase. Nature 420: 422–426 [DOI] [PubMed] [Google Scholar]
- Kuppusamy KT, Endre G, Prabhu R, Penmetsa RV, Veereshlingam H, Cook DR, Dickstein R, Vandenbosch KA. (2004) LIN, a Medicago truncatula gene required for nodule differentiation and persistence of rhizobial infections. Plant Physiol 136: 3682–3691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laubinger S, Zeller G, Henz S, Sachsenberg T, Widmer C, Naouar N, Vuylsteke M, Scholkopf B, Ratsch G, Weigel D. (2008) At-TAX: a whole genome tiling array resource for developmental expression analysis and transcript identification in Arabidopsis thaliana. Genome Biol 9: R112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limpens E, Ramos J, Franken C, Raz V, Compaan B, Franssen H, Bisseling T, Geurts R. (2004) RNA interference in Agrobacterium rhizogenes-transformed roots of Arabidopsis and Medicago truncatula. J Exp Bot 55: 983–992 [DOI] [PubMed] [Google Scholar]
- Linding R, Russell RB, Neduva V, Gibson TJ. (2003) GlobPlot: exploring protein sequences for globularity and disorder. Nucleic Acids Res 31: 3701–3708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magori S, Oka-Kira E, Shibata S, Umehara Y, Kouchi H, Hase Y, Tanaka A, Sato S, Tabata S, Kawaguchi M. (2009) Too much love, a root regulator associated with the long-distance control of nodulation in Lotus japonicus. Mol Plant Microbe Interact 22: 259–268 [DOI] [PubMed] [Google Scholar]
- Marmagne A, Ferro M, Meinnel T, Bruley C, Kuhn L, Garin J, Barbier-Brygoo H, Ephritikhine G. (2007) A high content in lipid-modified peripheral proteins and integral receptor kinases features in the arabidopsis plasma membrane proteome. Mol Cell Proteomics 6: 1980–1996 [DOI] [PubMed] [Google Scholar]
- Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Maréchal-Drouard L, et al. (2007) The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318: 245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra RM, Gleason CA, Edwards A, Hadfield J, Downie JA, Oldroyd GE, Long SR. (2004) A Ca2+/calmodulin-dependent protein kinase required for symbiotic nodule development: gene identification by transcript-based cloning. Proc Natl Acad Sci USA 101: 4701–4705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra SK, Walters BT, Clouse SD, Goshe MB. (2009) An efficient organic solvent based extraction method for the proteomic analysis of Arabidopsis plasma membranes. J Proteome Res 8: 2752–2767 [DOI] [PubMed] [Google Scholar]
- Miyazawa H, Oka-Kira E, Sato N, Takahashi H, Wu GJ, Sato S, Hayashi M, Betsuyaku S, Nakazono M, Tabata S, et al. (2010) The receptor-like kinase KLAVIER mediates systemic regulation of nodulation and non-symbiotic shoot development in Lotus japonicus. Development 137: 4317–4325 [DOI] [PubMed] [Google Scholar]
- Mortier V, Den Herder G, Whitford R, Van de Velde W, Rombauts S, D’Haeseleer K, Holsters M, Goormachtig S. (2010) CLE peptides control Medicago truncatula nodulation locally and systemically. Plant Physiol 153: 222–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray JD, Karas BJ, Sato S, Tabata S, Amyot L, Szczyglowski K. (2007) A cytokinin perception mutant colonized by Rhizobium in the absence of nodule organogenesis. Science 315: 101–104 [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Kawaguchi M. (2006) Shoot-applied MeJA suppresses root nodulation in Lotus japonicus. Plant Cell Physiol 47: 176–180 [DOI] [PubMed] [Google Scholar]
- Nishimura R, Hayashi M, Wu GJ, Kouchi H, Imaizumi-Anraku H, Murakami Y, Kawasaki S, Akao S, Ohmori M, Nagasawa M, et al. (2002a) HAR1 mediates systemic regulation of symbiotic organ development. Nature 420: 426–429 [DOI] [PubMed] [Google Scholar]
- Nishimura R, Ohmori M, Fujita H, Kawaguchi M. (2002b) A Lotus basic leucine zipper protein with a RING-finger motif negatively regulates the developmental program of nodulation. Proc Natl Acad Sci USA 99: 15206–15210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura R, Ohmori M, Kawaguchi M. (2002c) The novel symbiotic phenotype of enhanced-nodulating mutant of Lotus japonicus: astray mutant is an early nodulating mutant with wider nodulation zone. Plant Cell Physiol 43: 853–859 [DOI] [PubMed] [Google Scholar]
- Nontachaiyapoom S, Scott PT, Men AE, Kinkema M, Schenk PM, Gresshoff PM. (2007) Promoters of orthologous Glycine max and Lotus japonicus nodulation autoregulation genes interchangeably drive phloem-specific expression in transgenic plants. Mol Plant Microbe Interact 20: 769–780 [DOI] [PubMed] [Google Scholar]
- Novak K. (2010) Early action of pea symbiotic gene NOD3 is confirmed by adventitious root phenotype. Plant Sci 179: 472–478 [DOI] [PubMed] [Google Scholar]
- Oka-Kira E, Tateno K, Miura K, Haga T, Hayashi M, Harada K, Sato S, Tabata S, Shikazono N, Tanaka A, et al. (2005) klavier (klv), a novel hypernodulation mutant of Lotus japonicus affected in vascular tissue organization and floral induction. Plant J 44: 505–515 [DOI] [PubMed] [Google Scholar]
- Okamoto S, Ohnishi E, Sato S, Takahashi H, Nakazono M, Tabata S, Kawaguchi M. (2009) Nod factor/nitrate-induced CLE genes that drive HAR1-mediated systemic regulation of nodulation. Plant Cell Physiol 50: 67–77 [DOI] [PubMed] [Google Scholar]
- Oldroyd GE, Downie JA. (2008) Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu Rev Plant Biol 59: 519–546 [DOI] [PubMed] [Google Scholar]
- Park S, Buttery B. (1989) Inheritance of nlterate-tolerant supernodulation in EMS-induced mutants of common bean (Phaseolus vulgaris L.). J Hered 80: 486–488 [Google Scholar]
- Penmetsa RV, Cook DR. (1997) A legume ethylene-insensitive mutant hyperinfected by its rhizobial symbiont. Science 275: 527–530 [DOI] [PubMed] [Google Scholar]
- Penmetsa RV, Uribe P, Anderson J, Lichtenzveig J, Gish JC, Nam YW, Engstrom E, Xu K, Sckisel G, Pereira M, et al. (2008) The Medicago truncatula ortholog of Arabidopsis EIN2, sickle, is a negative regulator of symbiotic and pathogenic microbial associations. Plant J 55: 580–595 [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postma J, Jacobsen E, Feenstra W. (1988) Three pea mutants with an altered nodulation studied by genetic analysis and grafting. J Plant Physiol 132: 424–430 [Google Scholar]
- Quandt HJ, Puehler A, Broer I. (1993) Transgenic root nodules of Vicia hirsuta: a fast and efficient system for the study of gene expression in indeterminate-type nodules. Mol Plant Microbe Interact 6: 699–706 [Google Scholar]
- Reid DE, Ferguson BJ, Gresshoff PM. (2011) Inoculation- and nitrate-induced CLE peptides of soybean control NARK-dependent nodule formation. Mol Plant Microbe Interact 24: 606–618 [DOI] [PubMed] [Google Scholar]
- Sagan M, Duc G. (1996) Sym28 and Sym29, two new genes involved in regulation of nodulation in pea (Pisum sativum L.). Symbiosis 20: 229–245 [Google Scholar]
- Schnabel E, Journet EP, de Carvalho-Niebel F, Duc G, Frugoli J. (2005) The Medicago truncatula SUNN gene encodes a CLV1-like leucine-rich repeat receptor kinase that regulates nodule number and root length. Plant Mol Biol 58: 809–822 [DOI] [PubMed] [Google Scholar]
- Schnabel E, Mukherjee A, Smith L, Kassaw T, Long S, Frugoli J. (2010) The lss supernodulation mutant of Medicago truncatula reduces expression of the SUNN gene. Plant Physiol 154: 1390–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle IR, Men AE, Laniya TS, Buzas DM, Iturbe-Ormaetxe I, Carroll BJ, Gresshoff PM. (2003) Long-distance signaling in nodulation directed by a CLAVATA1-like receptor kinase. Science 299: 109–112 [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Temnykh S, Kneen B, Weeden N, LaRue T. (1995) Localization of nod-3, a gene conditioning hypernodulation, and identification of a novel translocation in Pisum sativum L. cv. Rhondo. J Hered 86: 303–305 [Google Scholar]
- Terakado J, Yoneyama T, Fujihara S. (2006) Shoot-applied polyamines suppress nodule formation in soybean (Glycine max). J Plant Physiol 163: 497–505 [DOI] [PubMed] [Google Scholar]
- van Noorden GE, Ross JJ, Reid JB, Rolfe BG, Mathesius U. (2006) Defective long-distance auxin transport regulation in the Medicago truncatula super numeric nodules mutant. Plant Physiol 140: 1494–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YH. (2008) How effective is T-DNA insertional mutagenesis in Arabidopsis? J Biochem Tech 1: 11–20 [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wopereis J, Pajuelo E, Dazzo FB, Jiang Q, Gresshoff PM, De Bruijn FJ, Stougaard J, Szczyglowski K. (2000) Short root mutant of Lotus japonicus with a dramatically altered symbiotic phenotype. Plant J 23: 97–114 [DOI] [PubMed] [Google Scholar]
- Yoshida C, Funayama-Noguchi S, Kawaguchi M. (2010) plenty, a novel hypernodulation mutant in Lotus japonicus. Plant Cell Physiol 51: 1425–1435 [DOI] [PubMed] [Google Scholar]