Abstract
To create a metabolic sink in the jasmonic acid (JA) pathway, we generated transgenic Nicotiana attenuata lines ectopically expressing Arabidopsis (Arabidopsis thaliana) jasmonic acid O-methyltransferase (35S-jmt) and additionally silenced in other lines the N. attenuata methyl jasmonate esterase (35S-jmt/ir-mje) to reduce the deesterification of methyl jasmonate (MeJA). Basal jasmonate levels did not differ between transgenic and wild-type plants; however, after wounding and elicitation with Manduca sexta oral secretions, the bursts of JA, jasmonoyl-isoleucine (JA-Ile), and their metabolites that are normally observed in the lamina, midvein, and petiole of elicited wild-type leaves were largely absent in both transformants but replaced by a burst of endogenous MeJA that accounted for almost half of the total elicited jasmonate pools. In these plants, MeJA became a metabolic sink that affected the jasmonate metabolic network and its spread to systemic leaves, with major effects on 12-oxo-phytodieonic acid, JA, and hydroxy-JA in petioles and on JA-Ile in laminas. Alterations in the size of jasmonate pools were most obvious in systemic tissues, especially petioles. Expression of threonine deaminase and trypsin proteinase inhibitor, two JA-inducible defense genes, was strongly decreased in both transgenic lines without influencing the expression of JA biosynthesis genes that were uncoupled from the wounding and elicitation with M. sexta oral secretions-elicited JA-Ile gradient in elicited leaves. Taken together, this study provides support for a central role of the vasculature in the propagation of jasmonates and new insights into the versatile spatiotemporal characteristics of the jasmonate metabolic network.
Plant cells, both those proximal to a wound site as well as those in more distal locations respond to tissue damage with large-scale changes in their transcription and metabolism, changes that help to promote survival of the entire plant (Reymond et al., 2000; Yan et al., 2007; Koo and Howe, 2009). Oxylipins, derived from fatty acid oxidation, play a central role in these responses: some function as direct defenses (Weber et al., 1999), others elicit the expression of defense-related genes (Browse, 2005; Gfeller et al., 2010b), or act as synomones in tritrophic interactions (Allmann and Baldwin, 2010). Jasmonic acid (JA) and its different derivatives collectively referred to as jasmonates, are the oxylipins whose biosynthesis and functions have been best characterized. They regulate responses to biotic and abiotic stresses (Baldwin et al., 1994; Devoto and Turner, 2005; Browse and Howe, 2008), but they also control different developmental processes throughout the life cycle of higher plants, including seed dormancy, flower morphogenesis, and fruit formation (Creelman and Mullet, 1995; Hause et al., 2000; Stintzi and Browse, 2000; Li et al., 2004).
Initial steps in jasmonate biosynthesis occur in plastids and are associated with the quick biochemical activation of specific lipases and oxidizing enzymes upon tissue damage. Trienoic fatty acids released from lipid membranes are first dioxygenated by 13-lipoxygenase enzymes to form the oxylipin 13(S)-hydroperoxy-(9Z,11E,15Z)-octadecatrienoic acid. The combined catalytic action of allene oxide synthase (AOS) and allene oxide cyclase (AOC) converts 13(S)-hydroperoxy-(9Z,11E,15Z)-octadecatrienoic acid to cis-12-oxo-phytodienoic-acid (OPDA). The next catalytic steps require the export of OPDA from plastids and its import into the peroxisome where it is reduced by OPDA reductase enzymes (OPR). Three subsequent cycles of β-oxidation finally yield JA, which is transported to the cytoplasm and serves as a precursor for the synthesis of a broad range of JA derivatives (Gfeller et al., 2010a). Our understanding of JA biosynthesis is largely biased toward the transcriptional regulation of JA biosynthesis genes (Howe and Schilmiller, 2002; Paschold et al., 2008) and several transcription factors modulating the accumulation of JA (Chung et al., 2008; Skibbe et al., 2008). However, fatty acid precursor availability is another essential determinant of the flux and metabolism of the JA pathway. Moreover the nature of the rate-limiting steps may also be context dependent. For instance, overexpression of AOS in Arabidopsis (Arabidopsis thaliana) and tobacco (Nicotiana tabacum) or AOC in tomato (Solanum lycopersicum) did not increase basal JA levels, but amplified the JA burst that is elicited in response to mechanical wounding (Laudert et al., 2000; Stenzel et al., 2003; Miersch et al., 2004). JA levels in leaves of many plant species increases within a few minutes after mechanical damage, which is too rapid to result entirely from the transcriptional regulation of JA biosynthesis genes. In Nicotiana attenuata, the model of this study, this early metabolic control is at least partly provided by the biochemical activation of a specific glycerolipase, NaGLA1, which supplies fatty acid substrate for herbivory-induced JA accumulation (Kallenbach et al., 2010).
Multiple competing enzymatic reactions controlling JA metabolism have been described. The resulting jasmonate signatures have been shown to differ between different tissues (Hause et al., 2000; Glauser et al., 2008), which suggest that different tissues may differentially metabolize JA. In tomato, much of the wound-induced JA synthesis has been proposed to take place in vascular tissues where JA biosynthesis enzymes seem to be preferentially located (Hause et al., 2003; Wasternack, 2007). Formation of JA’s amino acid conjugates such as jasmonoyl-Ile (JA-Ile) is mediated by JASMONATE RESISTANT (JAR) in Arabidopsis and its homologs in other plant species (Staswick et al., 2002; Staswick and Tiryaki, 2004; Kang et al., 2006). JA-Ile promotes the interaction of the F-box protein CORONATINE INSENSITIVE1 (COI1) with jasmonate ZIM-domain (JAZ) repressors and in turn, their targeting for proteolytic degradation in the proteasome (Chini et al., 2007; Thines et al., 2007). Less is known about the potential signaling functions of the different jasmonates produced upon wounding and whether the different reactions controlling their production also help regulating JA-Ile homeostasis.
Other modifications of JA include methylation to methyl jasmonate (MeJA), which increases JA’s volatility and capacity to cross membranes. Overexpression of O-methyltransferases targeting for instance auxin, GAs, or salicylic acid (SA), has been used to manipulate hormonal homeostasis and conversely uncover new signaling outcomes (Qin et al., 2005; Varbanova et al., 2007). In Arabidopsis, the expression of jasmonic acid O-methyltransferase (AtJMT) responsible for MeJA formation is developmentally regulated. Similar to JA biosynthesis and defense-related genes, AtJMT is also induced upon wounding or in response to JA application (Seo et al., 2001). Arabidopsis plants overexpressing AtJMT contain elevated levels of MeJA and normal levels of JA but display constitutively up-regulated expression of defense and JA biosynthesis-related genes (Seo et al., 2001).
The approach most commonly used to analyze jasmonate production in planta entails silencing or knockout mutations in JA biosynthesis genes. However, these loss-of-function approaches, while useful for the analysis of downstream consequences of interrupting specific steps in the JA pathway, per se cannot fully unravel the multiple control mechanisms that enable plants to constantly adjust their JA synthesis to its signaling output. Here, we redirect JA metabolism in N. attenuata toward the formation of MeJA, without disrupting the whole JA pathway, by ectopically expressing AtJMT (35S-jmt). We further reduced the deesterification of MeJA in 35S-jmt plants by additionally silencing in other lines (35S-jmt/ir-mje) the expression of N. attenuata methyl jasmonate esterase (NaMJE) and examined the consequences of these genetic manipulations on jasmonate kinetics, metabolic networks, pools, and downstream gene expression.
RESULTS
Generation of Transgenic Plants
In Arabidopsis, the homolog of Brassica campestris and Brassica rapa Nectarin1 genes has been shown to encode for a AtJMT (At1g19640; Seo et al., 2001). The AtJMT gene product methylates JA, but not OPDA, and its constitutive expression resulted in strongly increased basal endogenous MeJA levels (Seo et al., 2001). To reroute JA metabolism and evaluate the consequences for the regulation of the complete octadecanoid pathway, we generated stable transgenic N. attenuata lines expressing AtJMT in sense orientation under the control of the cauliflower mosaic virus 35S promoter. Two independently transformed homozygous 35S-jmt lines (35S-jmt-1 and 35S-jmt-2) with comparable alterations in floral style elongation (see below, Fig. 1B) were selected. The first line, 35S-jmt-1, harbored a single T-DNA insertion, the second one, 35S-jmt-2, harbored two detectable T-DNA insertions as revealed by Southern-blot analysis (Supplemental Fig. S1). To prevent MeJA deesterification, we further created transgenic lines ectopically expressing AtJMT in which the expression of NaMJE was additionally silenced. NaMJE had been previously cloned and functionally characterized in our group (Wu et al., 2008). As for 35S-jmt plants, two independently transformed homozygous 35S-jmt/ir-mje lines with similar alterations in flower morphology (35S-jmt/ir-mje-1 and 35S-jmt/ir-mje-2) were selected. The first line, 35S-jmt/ir-mje-1, harbored a single T-DNA insertion, the second line 35S-jmt/ir-mje-2 harbored three T-DNA insertions (Supplemental Fig. S1).
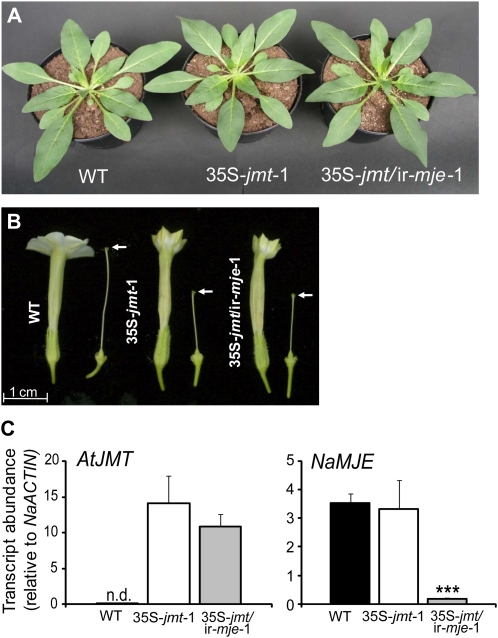
Figure 1.
N. attenuata plants ectopically expressing AtJMT (35S-jmt) or additionally silenced for the expression of NaMJE (35S-jmt/ir-mje) are indistinguishable from wild type during rosette stage growth but produce flowers with altered morphology. A, N. attenuata plants ectopically expressing AtJMT (35S-jmt-1) and additionally silenced for NaMJE (35S-jmt/ir-mje-1) do not differ from wild-type plants during rosette stage (approximately 30-d-old). B, Flowers of 35S-jmt and 35S-jmt/ir-mje plants have short styles (white arrows indicate stigma position) and closed corollas compared to wild-type plants. C, Relative transcript abundance (mean ± sd, n = 5) of AtJMT and NaMJE in leaf laminas of wild-type, 35S-jmt, and 35S-jmt/ir-mje-1 plants 1 h after mechanical wounding and application of M. sexta oral secretions to the wounds (W + OS). No AtJMT expression was detected (n.d.) in wild-type tissues after W + OS elicitation. NaMJE relative transcript levels in 35S-jmt/ir-mje-1 leaf laminas after W + OS elicitation were reduced to approximately 5% of wild-type levels but were unchanged in 35S-jmt-1.
All transgenic lines were indistinguishable from wild-type plants during rosette stage growth under controlled growth conditions (Fig. 1A). As is commonly reported for many mutants and transgenic lines altered in JA metabolism or signaling (Wasternack, 2007), flowers of 35S-jmt and 35S-jmt/ir-mje morphologically differed from those produced by wild-type plants. For all transgenic lines selected, style elongation was reduced to approximately half of that in wild-type flowers and the opening of the corolla limbs was impaired (Fig. 1B). Transcript measurements with gene-specific primers confirmed high relative expression levels of AtJMT in 35S-jmt-1 and 35S-jmt/ir-mje-1 but not in untransformed wild-type plants (Fig. 1C). After wounding and application of Manduca sexta oral secretions (W + OS), the expression of NaMJE was comparably high in 35S-jmt-1 and wild-type leaves and reduced by 95% in 35S-jmt/ir-mje-1 leaves.
35S-jmt and 35S-jmt/ir-mje Plants Have Altered JA Methylation and MeJA Demethylation Activities
We next verified that transgenic lines had deregulated JA methylation and MeJA demethylation activities. To assess AtJMT activity in vivo, we infiltrated 0.5 μg JA into wild-type, 35S-jmt-1, and 35S-jmt/ir-mje-1 leaf tissues and quantified increases in MeJA levels over a 45-min time period (Fig. 2A). Consistent with the low extent of damage inflicted by the infiltration technique, jasmonate (JA, MeJA, and JA-Ile) levels remained relatively low—barely exceeding basal levels—in the leaves of all three genotypes after infiltration of a control solution (mock infiltration, Fig. 2, A and B; Supplemental Fig. S2A). MeJA levels increased in JA-infiltrated leaf discs of 35S-jmt-1 and 35S-jmt/ir-mje-1—respectively, up to 60- and 170-fold the levels of mock-infiltrated leaves—but remained as low as after mock infiltration in wild-type leaves. The formation of other JA derivatives such as JA-Ile was antagonized compared to in wild-type leaves (Supplemental Fig. S2B). Consistent with different deesterification rates, MeJA accumulation was greater in 35S-jmt/ir-mje-1—attaining at 45 min 150 ng, 167-fold wild-type levels—than in 35S-jmt-1 plants, which attained at 45 min 30 ng, 32-fold wild-type levels.
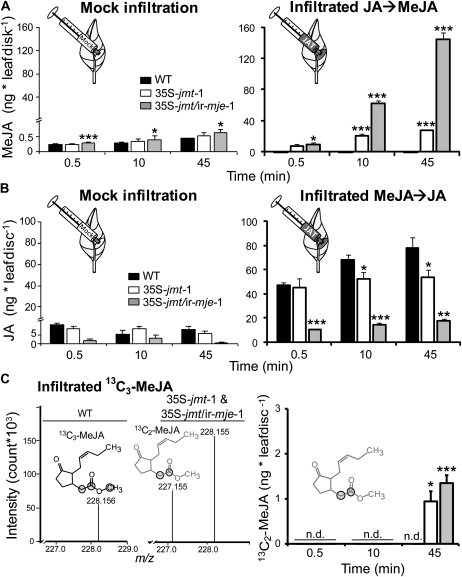
Figure 2.
N. attenuata 35S-jmt and 35S-jmt/ir-mje plants have altered JA-methylation and MeJA-demethylation activities. Leaf discs (0.4 cm2) were infiltrated with a control solution (Mock) and 0.5 μg of JA (A), unlabeled MeJA (B), or 0.25 μg synthetic MeJA labeled with 13C ([1, 2, 13-13C]MeJA; C). JA methylation (A), MeJA demethylation (B), and the remethylation of the subsequently released JA (C) were analyzed by quantifying 0.5, 10, and 45 min increases (mean ± sd, n = 5) in MeJA (JA → MeJA), JA (MeJA → JA), and [1, 2-13C]MeJA levels after infiltration in the elicited leaves. After JA infiltration, 35S-jmt-1 and 35S-jmt/ir-mje-1 leaves showed larger MeJA accumulations than wild type that came at the expense of other JA metabolites (Supplemental Fig. S2). MeJA deesterification was strongly impaired in 35S-jmt/ir-mje-1 and only slightly reduced in 35S-jmt-1 plants. Left section of C: high-resolution time-of-flight MS measurement of (calculated m/z = 228.158) [1, 2, 13-13C]MeJA-infiltrated leaf areas—zoom in of the spectral range m/z 227 to 229—revealed the production of a m/z signal at 227.155 characteristic for 13C2-MeJA ([1, 2-13C]MeJA, calculated m/z = 227.155) formed by remethylation of the deesterified 13C3-MeJA in 35S-jmt-1 and 35S-jmt/ir-mje-1, but not in wild-type leaf samples. 13C atoms are circled. Right section of C: [1, 2-13C]MeJA levels. Asterisks represent significant differences between wild-type and transgenic lines (unpaired t test; * P < 0.05, ** < 0.001, *** < 0.0001). n.d., Not detected.
The impairment of MeJA demethylation in 35S-jmt/ir-mje-1 was verified by measuring, as described above, the turnover of infiltrated MeJA (Fig. 2B). For simplicity, we do not describe here accumulation patterns of JA metabolites produced after MeJA deesterification (Supplemental Fig. S2B). While MeJA infiltration into wild-type and 35S-jmt-1 leaf laminas resulted in rapid (0.5 min after infiltration) increases of free JA levels, the release of JA was, at the same time, reduced by approximately 80% in MeJA-infiltrated leaf areas of 35S-jmt/ir-mje-1 (Fig. 2B): JA levels in 35S-jmt/ir-mje-1 attained 45 min after infiltration only 23% of the levels observed in wild type and translated into lower amounts of JA derivatives (Supplemental Fig. S2B). JA levels in MeJA-infiltrated 35S-jmt leaves were lower by up to 30% compared to wild type and the MeJA-to-JA conversion efficiencies did not differ among the three sampling times. These observations suggested that deesterified MeJA was quickly remethylated in 35S-jmt-1 leaves.
To evaluate the importance of this reaction in both 35S-jmt-1 and 35S-jmt/ir-mje-1, we performed the same experiment as above using 0.25 μg of synthetic MeJA labeled with 13C on the two first carbons and on the methylester group ([1, 2, 13-13C]MeJA). Similar differences were detected between 35S-jmt/ir-mje-1, 35S-jmt-1, and wild-type plants during [1, 2, 13-13C]MeJA infiltration (Fig. 2C), consistent with the above-mentioned results. High resolution mass measurement of [1, 2, 13-13C]MeJA-infiltrated leaf discs revealed the production of a mass-to-charge ratio (m/z) signal at 227.155 characteristic for [1, 2-13C]MeJA (absolute Δm/z calculated − m/z measured = 3.2 ppm) formed by remethylation of the deesterified [1, 2, 13-13C]MeJA in 35S-jmt-1 and 35S-jmt/ir-mje-1, but not in wild-type leaf samples (Fig. 2C). The exchange of the methylester 13C to 12C was monitored and [1, 2-13C]MeJA specifically quantified in infiltrated leaf areas. [1, 2-13C]MeJA increased 45 min after infiltration only in the transformants and represented up to 61% in 35S-jmt compared to only 28% in 35S-jmt/ir-mje-1 of the total labeled MeJA ([1, 2, 13-13C]MeJA and [1, 2-13C]MeJA; Fig. 2C).
AtJMT Ectopic Expression Depletes W + OS-Induced JA Levels
From the above results, we concluded that both transformants had increased JA methylation activities and that the flux of compounds through the methyl transferase out competed that through the intact MJE activity in 35S-jmt and the residual MJE activity in 35S-jmt/ir-mje-1. In N. attenuata, applying M. sexta oral secretions to puncture wounds in leaves (W + OS) is known to elicit a transient JA burst that wanes to basal levels in approximately 2 h. In comparison to JA and other jasmonates, the accumulation of MeJA remains marginal in resting tissues as well as after W + OS elicitation (Von Dahl and Baldwin, 2004). To confirm that AtJMT activity depleted W + OS-induced JA pools, we compared JA and MeJA basal and maximal levels (1 h after W + OS elicitation) in leaf laminas of wild type and the two 35S-jmt and 35S-jmt/ir-mje lines (Fig. 3). 35S-jmt and 35S-jmt/ir-mje plants failed to accumulate JA after W + OS elicitation: JA levels in these transformants were reduced to approximately 27% and 30%, respectively, of those in wild type. In contrast, although MeJA levels in wild type were about as low as before elicitation (0.03 nmol g−1 fresh weight [FW]−1), they reached 2.9 nmol g−1 FW−1 in 35S-jmt and 7.9 nmol g−1 FW−1 in 35S-jmt/ir-mje after W + OS elicitation. This shift of the JA flux toward methylation did not translate into increased MeJA emissions—less than 0.01% of endogenous maximum MeJA levels could be recovered from in the headspace of 35S-jmt and 35S-jmt/ir-mje leaves (Supplemental Fig. S3).
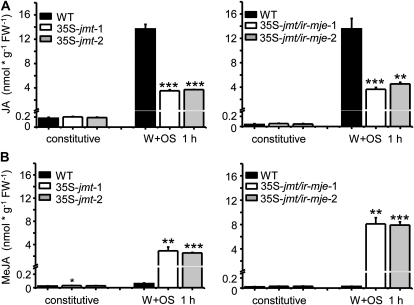
Figure 3.
AtJMT-catalyzed MeJA formation depletes W + OS-induced JA levels. Mean (±sd, n = 5) constitutive and induced JA (A) and MeJA (B) levels (nmol g−1 FW−1) after mechanical wounding and application of M. sexta oral secretions to the puncture wounds (OS elicitation) in laminas of wild-type, 35S-jmt (35S-jmt-1, 35S-jmt-2), and 35S-jmt/ir-mje (35S-jmt/ir-mje-1, 35S-jmt/ir-mje-2) plants. Prior to elicitation, JA and MeJA levels in leaf laminas of 35S-jmt and 35S-jmt/ir-mje lines did not differ from those in wild type. The JA burst observed in wild-type leaf laminas after W + OS elicitation was largely absent in 35S-jmt and 35S-jmt/ir-mje plants but was mirrored, due to AtJMT activity, by a large burst of MeJA. Asterisks represent significant differences between wild-type and transgenic lines (unpaired t test; * P < 0.05, ** < 0.001, *** < 0.0001).
Rerouting JA Metabolism in 35S-jmt and 35S-jmt/ir-mje Has Profound, But Differential, Effects on Jasmonate Spatiotemporal Accumulation Patterns
We examined large-scale alterations in the jasmonate profiles caused by the depletion of the W + OS-induced JA pools (Fig. 4). For each plant, the lamina of one fully expanded leaf close to the rosette base was treated by W + OS on both sides of the midvein and then used as tissue source for the analysis of local responses. Undamaged younger leaves, growing orthostichous to the treated leaf, were selected to examine the systemic responses. Changes in JA metabolism in both treated and systemic leaves were examined after dissection of the leaf into lamina, midvein, and petiole. To get a broad perspective on jasmonate production, we quantified by liquid chromatography (LC)-mass spectrometry (MS/MS/MS) a wide range of the previously reported JA metabolites (Supplemental Table S1).
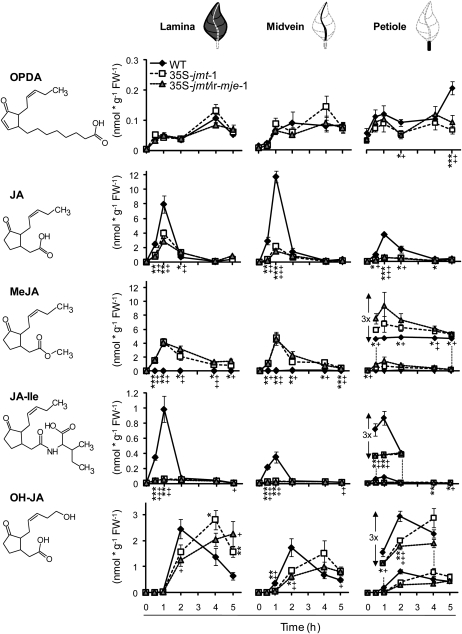
Figure 4.
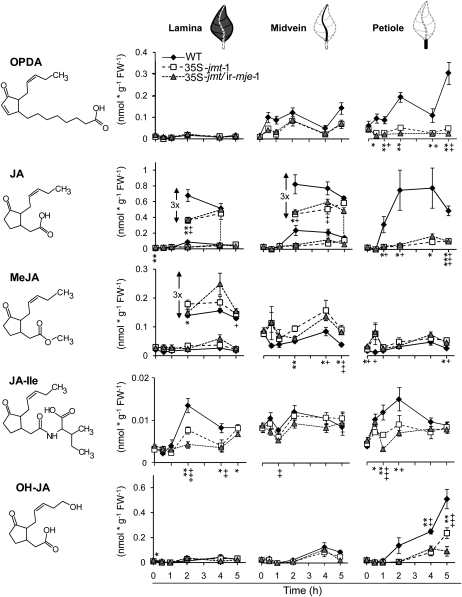
Ectopic expression of AtJMT in 35S-jmt and 35S-jmt/ir-mje has profound, but differential, effects on spatiotemporal accumulation patterns of jasmonates in lamina, midribs, and petioles. Mean (±sd, n = 5) of jasmonate levels—OPDA, JA, MeJA, JA-Ile, and OH-JA (the sum of 12- and 11-OH-JA)—in wild-type, 35S-jmt-1, and 35S-jmt/ir-mje-1 leaves after OS elicitation (W + OS). Leaves were harvested 0 to 5 h after elicitation, dissected into leaf lamina, midvein, and petiole (see schematics above line graphs), and analyzed separately. Different jasmonates attained their maximum levels in different tissues postelicitation. MeJA hyperaccumulation occurred at the expense of JA-Ile and JA accumulation but only retarded the accumulation of OH-JA. AtJMT ectopic expression also had different consequences on the accumulation of JA-Ile and other JA-amino acid conjugates (Supplemental Fig. S4). Asterisks (35S-jmt-1) and plus signs (35S-jmt/ir-mje-1) represent significant differences between wild-type and transgenic lines (unpaired t test; */+ P < 0.05, **/++ < 0.001, ***/+++ < 0.0001).
Formation of OPDA, the JA precursor, increased in a consistent biphasic manner in the leaf laminas of all three genotypes after a single W + OS elicitation (Fig. 4). The second phase in OPDA accumulation occurred in all three genotypes after the waning of the JA burst. Importantly, OPDA levels attained similar levels in wild-type, 35S-jmt, and 35S-jmt/ir-mje-1 plants in response to elicitation, indicating that the manipulation of JA metabolism in these two transformants did not modify upstream components of the JA pathway. As observed above, maximum JA levels, detected 1 h after W + OS elicitation in 35S-jmt-1 and 35S-jmt/ir-mje-1 plants represented only 49% and 35% of those detected in wild type, respectively. The impaired JA burst in both transgenic lines was mirrored by an enhanced and longer-lasting MeJA burst that waned more slowly in 35S-jmt/ir-mje-1. Similar to JA, MeJA reached its highest levels—approximately 4.1 nmol g−1 FW−1 in laminas of both 35S-jmt-1 and 35S-jmt/ir-mje-1 plants compared to less than 0.05 nmol g−1 FW−1 in wild-type plants—1 h post elicitation.
In agreement with the reported high substrate specificity of AtJMT, we did not detect the presence of methylated forms of JA-Ile and OPDA in all tissues examined. Interestingly, decreases in JA-Ile levels were much more pronounced than those detected for JA or other JA-amino acid conjugates including the 12- and 11-hydroxylated derivatives of JA-Ile and 12-carboxy-JA-Ile (12-COOH-JA-Ile; Supplemental Fig. S4). The maximum JA-Ile levels in both 35S-jmt-1 and 35S-jmt/ir-mje-1 laminas were reduced by approximately 95% compared to those in wild type. Formation of 12/11-hydroxy-JA has been proposed to account for the waning of the JA burst. JA hydroxylation after W + OS elicitation was delayed in 35S-jmt-1 and 35S-jmt/ir-mje-1 leaf laminas; 12- and 11-OH-JA levels—here the sum of both is reported as OH-JA—attained wild-type-maximum levels in the two transgenic lines 4 to 5 h after elicitation, values that are attained in wild-type laminas after 2 h.
With the exception of MeJA, the accumulation of all other jasmonates was also strongly reduced in the midveins and petioles of treated 35S-jmt-1 and 35S-jmt/ir-mje-1 leaves. In the case of OPDA, significant reductions were exclusively detected in petioles where OPDA attained its maximal levels. JA levels in midveins of wild type exceeded those measured in leaf laminas and petioles. Levels for JA were those most severely depleted also in the midveins—representing only 19% and 12% respectively, of those detected in wild-type midveins after 1 h. MeJA levels at this sampling time explained approximately half of the reduction in free JA levels. Interestingly, the increase in MeJA levels was less apparent in the petiole of treated leaves of both lines and accounted only for approximately 30% and 20% of the detected reduction in JA. JA-Ile accumulation in vascular tissues followed a different response than that of JA; JA-Ile was most abundant at the wound sites in wild type while strongly reduced in transgenic lines and less abundant in tissues of midveins and especially petioles. As seen before, accumulation of OH-JA in midveins and petioles of the two transgenic lines was also delayed compared to wild-type plants.
Jasmonate Profiles of Systemic Petioles Most Closely Match Those of Elicited Leaves
We examined whether alterations in herbivory-induced jasmonate accumulations also spread to systemic leaves (Fig. 5). For simplicity, we highlight here only the main differences between treated and systemic leaves. Absolute jasmonate levels detected in systemic leaves were except for OPDA, depending on the tissue type, 10- to 60-times lower than in treated leaves and the relative decreases in transgenic lines compared to those in wild type were also less pronounced. While JA and especially JA-Ile were the two jasmonates of elicited leaves most severely affected by the redirection of the JA flux toward MeJA in the transgenic lines, the major alterations in systemic leaves were in decreasing order detected for: JA (88%–92% reductions of peaking levels), OPDA (84%–91%), and OH-JA (53%–82%). Unlike in treated leaves, in systemic leaves the highest levels for each jasmonate were consistently detected in petioles, especially for JA, OH-JA, and OPDA—for this latter jasmonate maximum levels quantified 5 h post elicitation even exceeded those measured in the three tissues of the treated leaves. JA-Ile was the only jasmonate showing the rapidly waxing and waning profile of the typical JA and JA-Ile bursts of elicited leaves, but its accumulation was less affected in the systemic tissues of transgenic lines than previously observed in the elicited tissues. This observation also correlated with a less-pronounced MeJA formation in systemic tissues of transgenic plants.
Figure 5.
Alterations in W + OS elicited jasmonate levels in systemic unelicited leaves of 35S-jmt-1 and 35S-jmt/ir-mje-1 are most pronounced in petioles and differ from those observed in elicited leaves. Mean (±sd, n = 5) levels jasmonates—OPDA, JA, MeJA, JA-Ile, and OH-JA (the sum of 12- and 11-OH-JA)—in wild-type, 35S-jmt-1, and 35S-jmt/ir-mje-1 leaves after OS elicitation (W + OS). Untreated leaves growing on the same plants as in Figure 4 with a minimal angular distance above the treated leaf, and hence orthostichous to the treated leaf, were considered systemic leaves. Leaves were harvested 0 to 5 h after elicitation, dissected into leaf lamina, midvein, and petiole (see schematics above line graphs), and analyzed separately. All jasmonates attained their maximum levels in petioles. Accumulation of JA, OPDA, and OH-JA was largely reduced in systemic petioles, although MeJA formation was less pronounced than in elicited tissues (Fig. 4). Jasmonate levels detected in systemic leaves were, except for OPDA and depending on the tissue type, 10- to 60-times lower than in treated leaves. Asterisks (35S-jmt-1) and plus signs (35S-jmt/ir-mje-1) represent significant differences between wild-type and transgenic lines (unpaired t test; */+ P < 0.05, **/++ < 0.001, ***/+++ < 0.0001).
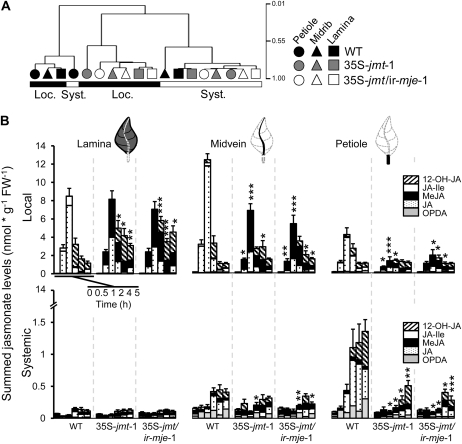
A hierarchical clustering analysis performed on tissue-specific jasmonate profiles further confirmed that although jasmonate accumulation in local and systemic tissues largely behaved independently, only the profile of the systemic petiole wild-type samples clustered together with that of elicited wild-type samples (Fig. 6A). Coexpression networks represent alternative and powerful statistical approaches to visualize large-scale modifications in a metabolic cluster (Han, 2008). The coexpression network drawn from major jasmonates measured here confirmed the pattern observed in the hierarchical clustering analysis: a large uncoupling between jasmonate clusters calculated for elicited and systemic tissues (Supplemental Fig. S5). In wild-type plants, a high degree of connectivity, depicting important correlations among the different jasmonates, was seen for the systemic tissues and interestingly only elicited JA and JA-Ile levels, particularly from petioles displayed significant correlations with the jasmonates measured in systemic tissues. As expected, the topology of this network was strongly distorted in the two transgenic lines (Supplemental Fig. S5). Most importantly, the topology of the network of the two transgenic lines almost perfectly overlapped, suggesting that silencing of NaMJE provided likely little additional effect to the AtJMT-mediated alterations.
Figure 6.
Alterations in jasmonate pools and relative composition are tissue dependent and most pronounced in petioles. A, Hierarchical clustering analysis performed, using the Pearson correlation as metric, on tissue-specific jasmonate signatures using vectors defined by the different levels reached at the different sampling times by a given jasmonate in a given tissue. Jasmonate profiles in systemic petioles relate the most to the signatures elicited after OS elicitation (W + OS) of treated leaves. The structure of locally elicited jasmonate signatures in wild type is not retained in 35S-jmt-1 and 35S-jmt/ir-mje-1 leaves as revealed by the clustering. B, Mean (±sd, n = 5) summed levels of the five most abundant jasmonates (OPDA, JA, MeJA, JA-Ile, and OH-JA) detected at 0 to 5 h after W + OS elicitation in local (top section) and systemic (bottom section) leaf tissues (lamina, midvein, and petiole) of wild-type, 35S-jmt-1, and 35S-jmt/ir-mje-1. Major quantitative changes in jasmonate pools were detected in midveins and petioles (Supplemental Fig. S6). Asterisks represent significant differences between tissues of wild-type and transgenic plants (unpaired t test; * P < 0.05, ** < 0.001, *** < 0.0001).
AtJMT Ectopic Expression Affects Jasmonate Pools in a Tissue-Dependent Manner
To determine whether the observed changes in jasmonate levels and the distortions of the jasmonate coexpression network resulted only from the diversion of JA metabolism toward MeJA or also involved deregulation of the jasmonate biosynthetic capacities, we calculated the sum of the five main abundant jasmonates detected in the individual tissues for each time point (Fig. 6B). As expected, jasmonate pools of elicited leaf laminas of 35S-jmt-1 and 35S-jmt/ir-mje-1 plants clearly differed in their relative composition from that of wild-type counterparts, but the sum of jasmonate levels in transgenic plants attained comparable maximum levels after 1 h as in wild type and vanished more slowly. Important differences in the relative composition and size of the jasmonate pools of local vascular tissues of transgenic lines were also observed when comparing with those of wild-type plants. But more important, for this comparison, maximum jasmonate pools elicited in midveins and petioles of the two transgenic lines were significantly decreased compared to those of wild type, respectively, by 44% (P < 0.001) and 60% (P < 0.001) in 35S-jmt-1 and by 60% (P < 0.001) and 54% (P = 0.014) in 35S-jmt/ir-mje-1.
As expected from the individual kinetics of Figures 4 and 5, the size of the jasmonate pools elicited in systemic lamina tissues was considerably smaller than in local tissues (approximately 0.15 nmol g−1 FW−1 versus more than 8.5 nmol g−1 FW−1 1 h post elicitation) and did not significantly differ between the two transgenic lines and wild type. Compared to wild-type tissues, relative reductions in jasmonate pools of transgenic plants were more pronounced in systemic than in local vascular tissues. These decreases were most severe in petiole samples of systemic leaves—85% reduction in 35S-jmt-1 (P = 0.03) and 91% reduction in 35S-jmt/ir-mje-1 (P = 0.03) after 2 h—and from a quantitative standpoint mainly attributable to decreases in OPDA and JA formation (Fig. 5) that did not directly result from AtJMT-catalyzed MeJA production. This result was best visualized when the relative differences in total jasmonate production in transgenic lines versus wild type were plotted for the different tissue types from the local to the distal lamina (Supplemental Fig. S6).
AtJMT Ectopic Expression Differentially Affects JA Biosynthesis and JA-Dependent Direct Defense Gene Expression
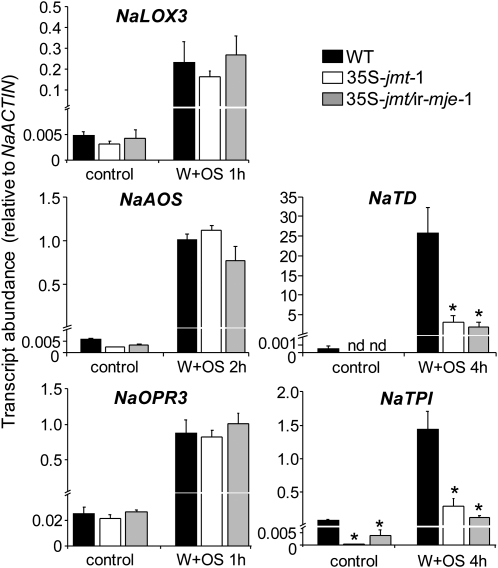
We conclude from the above results that, at least at the elicitation sites, AtJMT ectopic expression caused a redirection of the JA flux toward MeJA without deregulating the entire JA biosynthetic machinery. To evaluate this conclusion, we quantified transcript levels of central JA biosynthesis genes in W + OS elicited leaf laminas. We also measured the expression of two direct defense and JA-inducible genes, NaTD (Kang et al., 2006) and NaTPI (Horn et al., 2005), as references for the output of jasmonate signaling in the two transgenic lines. For both gene categories, transcriptional analyses were performed at times at which maximum transcript levels have been observed in previous studies (Halitschke and Baldwin, 2003; Halitschke et al., 2004). Maximum levels NaLOX3, NaAOS, and NaOPR3 transcripts were similar in all three genotypes (Fig. 7). In contrast, the relative abundance of the two highly JA-induced defense transcripts of NaTD and NaTPI significantly differed before and after W + OS elicitation (Fig. 7), suggesting an uncoupling in the transcriptional regulation of JA biosynthesis and the expression of these two direct defense genes in the transgenic lines.
Figure 7.
Transcript levels of JA biosynthesis and two direct defense genes are differentially affected by AtJMT ectopic expression. Mean (±sd, n = 5) transcript abundance (relative to NaACTIN) of JA biosynthesis genes in leaf lamina—AOS (NaAOS), lipoxygenase3 (NaLOX3), OPDA reductase (NaOPR3)—does not differ between wild-type and 35S-jmt-1 and 35S-jmt/ir-mje-1 plants before and after OS elicitation (W + OS). In contrast, constitutive and W + OS-induced transcript abundance of Thr deaminase (NaTD) and trypsin proteinase inhibitor (NaTPI), two direct defense genes regulated by the JA signaling pathway, were decreased in 35S-jmt-1 and 35S-jmt/ir-mje-1 compared to those in wild-type plants. OS-induced leaf laminas were harvested when peak expression levels have been reported for the selected genes. Asterisks represent significant differences between wild-type and transgenic lines (NaAOS, NaTD, NaLOX3, NaOPR3 unpaired t test, * P < 0.05; NaTPI Welch’s test, * P < 0.05). n.d., Not detected.
DISCUSSION
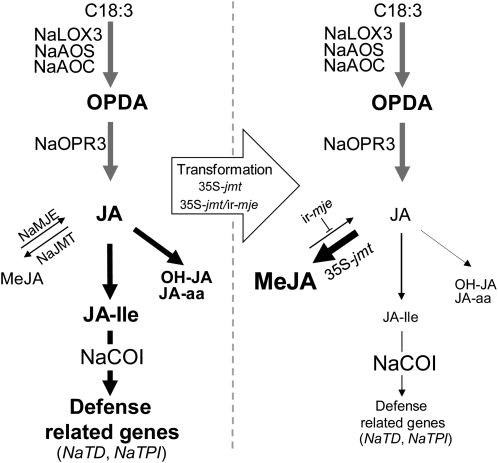
This study demonstrates that ectopically expressing AtJMT in N. attenuata is sufficient to create a strong metabolic sink in the endogenous JA pathway that out competes endogenous catalytic reactions controlling the availability, bioactivity, and catabolism of free JA (Fig. 8). Individual accumulation patterns, especially those of JA-Ile, total jasmonate pools, and networks were altered with different degrees of intensity and in a tissue-dependent manner in transgenic lines. This strong redirection of JA metabolism impaired the accumulation of defense-related transcripts but did not affect transcript levels of JA biosynthesis genes in transformed plants.
Figure 8.
Model of OS-induced JA metabolism and signaling in leaf lamina of 35S-jmt and 35S-jmt/ir-mje plants. AtJMT ectopic expression in concert with NaMJE silencing creates a powerful metabolic sink in the JA pathway that outcompetes herbivory-induced JA and JA-Ile bursts, and in turn, compromises NaCOI1-mediated activation of NaTD and NaTPI expression but not that of JA biosynthesis genes. Font size and arrows’ thickness are proportional to the intensity of metabolite fluxes and activation of gene expression in leaf laminas tissues after simulated M. sexta herbivory.
Creating a Metabolic Sink in the JA Pathway
Methylation is one of the catalytic reactions used by plants to adjust their pools of active hormones to environmental conditions. Conversely, manipulating the scale of this reaction and characterizing its consequences at the metabolic and organismic level has been shown to be a valuable approach to uncover new signaling outputs and to revisit the importance of homeostatic control over metabolite fluxes derived from hormonal signals (Qin et al., 2005; Varbanova et al., 2007; Tieman et al., 2010). By ectopically expressing AtJMT, we engineered a massive redirection of the JA flux toward the formation of MeJA. MeJA hyperaccumulation occurred at the expense of other jasmonates and only after W + OS elicitation, which confirmed that substrate availability, i.e. induced JA formation, was the limiting step for MeJA production in both 35S-jmt and 35S-jmt/ir-mje transformants. Substrate availability exerts an important regulatory function within the JA pathway. For instance, constitutive overexpression of AOS, in tobacco and in Arabidopsis (Laudert et al., 2000), or AOC, in tomato, increased JA and OPDA levels in leaves only after mechanical wounding and in the case of AOC in tomato also dramatically affected the levels and relative ratios of developmentally regulated jasmonates in the different organs of transgenic flowers (Miersch et al., 2004). Our results demonstrate that substrate availability constrains the output of JA metabolism and that its influence varies among different leaf tissues.
The expression metabolic sink has previously been used when diverting or inactivating the metabolic and/or signaling outputs of biosynthetic pathways (Yao et al., 1995; Li and Van Eck, 2007). Decreases in herbivory-induced jasmonate levels reported here that result from the depletion of JA levels by AtJMT, were as large or larger than those of RNAi-based approaches previously used in our group to silence NaLOX3 (Halitschke and Baldwin, 2003) or NaJAR4 and NaJAR6 (Wang et al., 2008) expression. This metabolic sink did not deregulate jasmonate biosynthetic capacities, since no effects were seen in the transcript abundance of JA biosynthesis genes and the accumulation of upstream elements of the JA pathway, such as OPDA. However, we cannot fully rule out that changes had occurred in the activity of enzymes, since in N. attenuata, critical steps within the JA pathway are not transcriptionally regulated (Kallenbach et al., 2010). The strength of the metabolic diversion was sustained in 35S-jmt by the remethylation of deesterified MeJA; this recycling reaction may also explain SA metabolism patterns in salicylic acid O-methyltransferase (SAMT) overexpressing lines (Tieman et al., 2010).
In agreement with the low amounts of MeJA accumulating in wild-type leaves after OS elicitation (Von Dahl and Baldwin, 2004; this study), JA methylation activity measured in vivo was extremely low compared to that of the transgenic lines and the remethylation of deesterified MeJA did not occur in wild-type leaves infiltrated with MeJA. The relative contribution of remethylation to MeJA fate was more important in 35S-jmt than in 35S-jmt/ir-mje leaves, which highlights the importance of considering both JA methylation as well as MeJA stability when analyzing JA and MeJA accumulation patterns.
Jasmonate-Specific Consequences of the Metabolic Sink
Arabidopsis plants overexpressing AtJMT contain elevated MeJA levels and normal levels of JA, however the consequences on other jasmonates were not examined (Seo et al., 2001). Our analysis revealed that MeJA accumulation occurred at the expense of other JA-dependent reactions and that the degree of depletion varied among the different jasmonates. Compartmentalization of the enzymes controlling JA synthesis and metabolism within the different cellular organelles may be responsible for these differences. JMT is predicted, since the amino acid sequence lacks an apparent organ-specific transit signal peptide, to act within the cytosol and therefore to strongly compete with the formation of JA-Ile by JAR enzymes that have been shown to be localized in the cytosol (Hsieh et al., 2000). Additionally, the almost complete depletion of the JA-Ile pool may in part have resulted from the reduced NaTD expression that mediates Ile formation (Kang et al., 2006). The contribution of NaTD down-regulation to the depletion of JA-Ile pools may also explain why less-pronounced alterations were seen for other JA-amino acid conjugates than for JA-Ile (Supplemental Fig. S4). An explanation for not having observed developmental alterations resembling those of N. attenuata transgenic lines strongly silenced for NaTD expression (Kang et al., 2006) could likely be that the metabolic sink created in the two transgenic backgrounds affected only inducible levels of expression of this gene and not its developmental regulation.
Tissue-Specific Spread of Jasmonate Bursts and the Role of Petioles
Different jasmonates reach their maximum levels in different tissues (Hause et al., 2000; Glauser et al., 2008) and importantly JA-Ile, the bioactive jasmonate, accumulates preferentially at wound sites and was the only jasmonate with a clear burst character in distal laminas, as previously shown (Koo et al., 2009). As previously reported in Arabidopsis (Koo et al., 2009) and N. attenuata (Wang et al., 2008), JA and JA-Ile in systemic leaves were, depending on the tissue type, 10- to 60-fold lower than in treated tissues. Our results are consistent with a central role played by vascular tissues in JA biosynthesis and the spread of jasmonate bursts. Clustering and network analyses highlighted the particularity of jasmonate profiles of petioles. The jasmonate profiles in petioles resembled those detected in treated leaves and the gradual increases in certain jasmonates preceding JA-Ile accumulation in distal laminas were only observed in unelicited petioles.
Calculation of jasmonate pools at each time point and for the distinct tissues revealed that AtJMT ectopic expression translated, only in vascular tissues, into significant reductions of total jasmonate pools, with the largest effects in petioles. We did not find any evidence for significantly higher AtJMT expression in vascular that may have directly contributed to the larger depletions of jasmonate pools seen in petioles and midveins. We rather think that these decreases of total jasmonate pools arose both from alterations in jasmonate translocation as revealed from petiole exudates (PEX) analysis (Supplemental Fig. S7) and from alterations in de novo synthesis. This in turn implies that petioles of both local and systemic leaves may play an important role in the amplification of jasmonate synthesis and in sustaining the spread of jasmonate bursts toward distal tissues. Recent studies have reopened the discussion about jasmonate formation and translocation in vascular tissues (Mielke et al., 2011). Testing this interesting hypothesis would demand an analysis of JA biosynthesis at the transcriptional and enzymatic levels in midvein and petiole tissues of 35S-jmt-1 and 35S-jmt/ir-mje-1 plants.
JA Biosynthesis and Direct Defense Transcript Levels Are Uncoupled in Transgenic Lines
JA biosynthesis, at least at the elicitation sites, was not repressed by the sink created within JA metabolism. This hypothesis was verified at the transcriptional level by the absence of differences, before and after induction, for central genes of the JA biosynthesis pathway (Fig. 7). In other plants species, overexpression of AOS or AOC has also been shown to not affect significantly the expression of other genes of the JA pathway (Harms et al., 1995). However, these results contrast with studies in Arabidopsis 35S-jmt plants reporting constitutively higher transcript levels for JA biosynthesis genes (Seo et al., 2001; Jung et al., 2003). Similarly, a positive feedback effect on SA biosynthesis mediated either by the increase of methyl SA itself or by a disruption in the control of the SA-related pool or flux has been observed in SAMT overexpressing tomato plants (Tieman et al., 2010). We cannot therefore completely exclude the possibility that the flux of compounds through the JA pathway may have been accelerated in N. attenuata in a transcription-independent manner that could not be captured by quantifying individual metabolites. We indeed may have underestimated MeJA levels, which according to the PEX measurements (Supplemental Fig. S7) and literature reports, may likely be quickly translocated within the plant’s phloem (Thorpe et al., 2007).
The absence of deregulation of JA biosynthesis gene expression, despite strong reduced JA-Ile levels suggests that the transcriptional positive feedback of the JA pathway may be independent of the JA-Ile gradient established after simulated herbivory. Wang et al. (2008) suggested from microarray analyses comparing N. attenuata lines silenced for NaLOX3 and JA-Ile forming genes (NaJAR4 and NaJAR6) that NaAOS, NaAOC, and NaOPR3 expression was not directly linked to JA-Ile synthesis. In contrast, the abundance of NaTD and NaTPI transcripts, which have been consistently shown by microarray analysis to be among the most highly JA and JA-Ile inducible genes in N. attenuata, was strongly impaired in both transformants and this effect was also observed in systemic lamina tissues of 35S-jmt-1 plants (Supplemental Fig. S8). Such uncoupling between the expression of JA biosynthesis genes from that of late-induced defense-related genes has been demonstrated and extensively studied at the temporal and spatial levels in tomato (Howe et al., 2000; Ryan, 2000; Strassner et al., 2002). Moreover, tomato mutants differentially affected in these two classes of genes, as were our transgenic lines, have been reported. For instance, in the def-1 mutant that is deficient in wound-induced JA accumulation and in turn impaired in PI expression, AOS expression, as well as that of LOX-D involved in JA biosynthesis, is not affected (Howe et al., 2000). Tomato plants silenced for Leu Aminopeptidase A also show specific alterations in late wound/JA-responsive gene expression, although JA biosynthesis and perception appears to be intact in these lines (Fowler et al., 2009). Such uncoupling may originate in the aforementioned tomato and N. attenuata transformants from alterations downstream of COI1 signaling, for instance from specific deregulations of JAZ levels, since activation of the expression of these transcriptional repressors, like that of the two aforementioned gene classes, involves COI1 (Chung et al., 2008; Paschold et al., 2008).
To summarize, ectopically expressing AtJMT in N. attenuata dramatically depletes herbivory-induced jasmonate metabolism in elicited and systemic tissues. These results support the need for future work analyzing more deeply alterations in defense-related metabolic processes in AtJMT-expressing lines and their consequences on the plant’s ecological performance.
MATERIALS AND METHODS
Plant Material and Growing Conditions
Wild-type Nicotiana attenuata Torr. ex Watson from an inbred line in its 30th generation were used for all experiments. The original seeds were collected in 1988 from an isolated population at the Desert Inn ranch in southwestern Utah. Before germination on agar plates containing Gamborg B5 media, all seeds were sterilized and incubated with diluted smoke and 0.1 m GA3, as described in Kruegel et al. (2002). Plants were grown with a day/night cycle of 16 h (26°C–28°C)/8 h (22°C–24°C) under supplemental light from Master Sun-T PIA Agro 400 or Master Sun-TPIA Plus 600-W sodium lights (Philips).
For the generation of 35S-jmt plants, the complete cDNA sequences of AtJMT and of the hptII hygromycin resistance genes were inserted into the pRESC2 transformation vector, both in sense orientation under the control of the cauliflower mosaic virus 35S promoter. To generate 35S-jmt/ir-mje transgenic lines, plants were transformed with a vector that contained additionally to the cDNA sense constructs of AtJMT and hptII, a cDNA fragment in inverted orientation of NaMJE (accession no. EU196055; Wu et al., 2008). Vectors were inserted into N. attenuata wild-type plants’ genome using Agrobacterium tumefaciens-mediated transformation (Kruegel et al., 2002). Homozygosity of the resulting T2 plants was determined by screening for the resistance to the antibiotic hygromycin and the number of insertions was determined as described in Gase et al. (2010), by Southern-blot hybridization of genomic DNA using a PCR fragment of the hptII gene as a probe (Supplemental Fig. S1).
Plant Treatments
For all experiments, plant treatments were randomly assigned among rosette stage plants and the first fully elongated leaf (+1 position) was treated. Manduca sexta feeding was simulated by wounding the leaf lamina with a fabric pattern wheel on both sides of the midrib and immediately applying 20 μL of M. sexta OS (diluted 1:10 in water) to the fresh wounds (W + OS); this procedure that is referred to as OS elicitation, provides a convenient means of accurately standardizing herbivore elicitation of N. attenuata plants and allowing for detailed kinetic analyses of the elicitation process. M. sexta OS were collected from third- to fourth-instar larvae reared on N. attenuata wild-type leaves as described in Roda et al. (2004). Eggs of the tobacco (Nicotiana tabacum) hornworm M. sexta were obtained from North Carolina State University (Raleigh, NC).
For jasmonate profiling, leaves of similar size and at the same developmental stage were harvested after W + OS treatment and, before being flash frozen, rapidly dissected with a scalpel into three distinct tissue types: petioles (the vascular tissue connecting leaf laminas to the plant’s shoot), midveins (the leaf’s major vein acting as a vascular manifold), and the lamina (expanding right and left of the midvein). Petioles were flash frozen immediately after being detached (less than 10 s). The midvein and the right and left sections of the leaf lamina were dissected and flash frozen within approximately 10 s. The treated leaf was analyzed for local responses. The untreated leaf growing on the same plant with a minimal angular distance above the treated leaf and therefore orthostichous to the treated leaf was considered the systemic leaf.
Phytohormone Analysis
Approximately 150 mg of frozen tissues were homogenized in a mortar and further pulverized to a fine powder by shaking with two steel beads (5 mm) in 2-mL reaction tubes using a Genogrinder (SPEX Certi Prep) at a frequency of 1,200 strokes/min for 40 s. Jasmonates were extracted by shaking for 3 min with 1 mL ethylacetate containing internal standards (IS) for JA (9,10-2H2-dihydro-JA), JA-Ile (jasmonyl-[13C6]Ile), and MeJA ([1, 2, 13-13C]MeJA, synthesized by esterification of [1, 2-13C]JA with 13C methanol as previously described in Zhang and Baldwin, 1997). Samples were analyzed as previously described by Wang et al. (2007). Briefly, 15 μL of the resulting extracts were analyzed for jasmonates using a Varian 1200L LC-MS/MS/MS system (Varian) working with an electrospray ionization source. Negative or positive ionization mode was used depending on the jasmonate structure (Supplemental Table S1). JA IS was used as IS for the quantification of 12- and 11-hydroxy-jasmonic-acid (OH-JA) and OPDA and concentrations were adjusted with response factors: Peak response factors for OPDA (1.28) and OH-JA (1.06) calculated versus the JA IS (Supplemental Table S1) were obtained by measurement of dilution series of pure OPDA, OH-JA, and JA IS dissolved into a leaf matrix. The JA-Ile IS was used to perform relative quantification of 12/11-hydroxy-JA-Ile, 12-COOH-JA-Ile, JA-Gln, and JA-Val, and their levels expressed in relative amounts (Supplemental Fig. S4) since no response factors could be calculated for these compounds for which no authentic standard was available.
Elemental formulas of [1, 2-13C]MeJA and 12-COOH-JA-Ile were verified by ultra-high-pressure LC time-of-flight MS using the method and calculation settings described in Gaquerel et al. (2010).
Collection of PEX
The PEX collection protocol was modified from previously published methods (King and Zeevaart, 1974; Maldonado et al., 2002; Park et al., 2007). Wild type, 35S-jmt-1, and 35S-jmt-2 rosette stage plants of similar size were used for PEX collection. For one replicate, three leaves of similar developmental stage were gently detached with a scalpel at the very base of their petioles and then subsequently recut in 1 mm EDTA solution (pH 7.5) to prevent callose deposition and closure of phloem vessels. Leaf laminas were mechanically wounded with a fabric pattern wheel and M. sexta OS were applied to the wound sites (W + OS). Petioles of three induced leaves of one genotype were then immersed into a fresh 1.5 mL 1 mm EDTA solution (pH 7.5). After 2.5 h the collection solution was renewed and PEX collected for another 2.5 h combined with the first fraction. PEX were freeze dried and dissolved in 70% methanol prior to the analysis for jasmonates.
In Vivo JMT and MJE Enzyme Activity Assays
To examine AtJMT and NaMJE activities, we infiltrated with a syringe JA, MeJA, or [1, 2, 13-13C]MeJA (labeled with 13C on the three first carbons counting from the methylester group) dissolved in 1% dimethyl sulfoxide in distilled water into leaves of rosette stage plants. Concentrations of the solutions were for JA of 12.5 μg/mL, for MeJA of 12.5 μg/mL, and for 13C3-MeJA of 6.25 μg/mL. The amounts of infiltrated substrate per leaf discs were of 0.5 μg for JA and MeJA and of 0.25 μg for 13C3-MeJA. To evaluate the effect of the infiltration on endogenous jasmonate production, similar volumes of 1% dimethyl sulfoxide solution (mock infiltration) were infiltrated with a syringe into leaves of equal size. After 0.5, 10, and 45 min, a leaf disc of 0.4 cm2 was removed from the infiltrated leaf area with a cork borer and immediately flash frozen in liquid nitrogen until analyses for jasmonates.
Quantitative Real-Time PCR Analysis
Total RNA from five biological replicates per line was extracted as described in Linke at al. (2002). RNA extracts were treated with DNAse using the DNA-free kit from Ambion (Applied Biosystems/Ambion). cDNA was synthesized from 500 ng RNA using SuperScript II reverse transcriptase (Invitrogen) and a poly-T primer. Quantitative real-time PCR (Stratagene 500 Mx3005P) was conducted with 30 ng cDNA using the core reagent kit (Eurogentec, http://www.eurogentec.be) and pairs of gene-specific primers (Supplemental Table S2). qPCR products were either detected by gene-specific double fluorescent dye-labeled TaqMan probes or after reaction with SYBR green (qPCR core kit for SYBR green I; Eurogentec). Relative gene expression was calculated using a 200-fold dilution series of cDNAs synthesized from RNA samples of the same experiment and normalized, according to Pfaffl et al. (2002), by the expression value of the N. attenuata ACTIN gene.
Statistical Analysis
Most statistics were performed with StatView (Abacus Concepts Inc., http://www.statview.com). Jasmonate pools were calculated in each tissues of local and systemic leaves after W + OS elicitation by summing, for each time point, the average (n = 5) OPDA, JA, MeJA, OH-JA, and JA-Ile levels of individual leaf samples. Jasmonate pools were then presented as bar charts and analyzed by Student’s t test.
The hierarchical clustering analysis was performed with the TIGR MeV 3.1 software (http://www.tm4.org/mev.html) on autoscaled data using as expression vector, the average levels of each jasmonate (OPDA, JA, MeJA, OH-JA, and JA-Ile) at the different time points in a particular tissue of local or systemic leaves (20 variables). The data matrix consisted of 20 variables × 18 samples (three tissue types × two leaf positions × three genotypes). The Pearson correlation was used as clustering metric and the complete linkage aggregation algorithm as clustering method.
For the network analysis, Pearson correlation coefficients were calculated among the different jasmonates using the mean of all biological replicates at each time point and for each tissue harvested after dissection from treated and systemic leaves independently. Significance levels for correlation coefficients (r) were determined following the number of metabolite pairs (n) using the equation t = r × (n − 2)0.5/(1 − r2)0.5. Correlations corresponding to a coefficient with P < 0.05 from a distance matrix calculations were transposed into a pairwise format and visualized using Networks cartography and Pajek software version 1.22 (http://vlado.fmf.uni-lj.si/pub/networks/pajek/). The distance used to rebuild the network was 1 minus the absolute value of the correlation coefficient with the Fruchtermann-Reingold three-dimensional algorithm.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Southern-blot analysis of 35S-jmt and 35S-jmt/ir-mje lines.
Supplemental Figure S2. Conversion of infiltrated JA and MeJA into JA-Ile is outcompeted by AtJMT activity in 35S-jmt-1 and 35S-jmt/ir-mje-1.
Supplemental Figure S3. Measurement of volatile MeJA in the headspace of W + OS-induced leaves.
Supplemental Figure S4. Levels of JA-Ile metabolites, JA-Gln, and JA-Val are differentially affected by AtJMT activity in different tissues of W + OS-treated leaves.
Supplemental Figure S5. The topology of the jasmonate metabolic network is largely and similarly altered in 35S-jmt and 35S-jmt/ir-mje leaves.
Supplemental Figure S6. Decreases in W + OS-induced total jasmonate pools of transgenic plants are more severe in petiole tissues.
Supplemental Figure S7. MeJA is the most abundant jasmonate in the PEX of W + OS-treated 35S-jmt-1 and 35S-jmt-2 leaves.
Supplemental Figure S8. Transcript levels of direct defense genes NaTPI and NaTD are strongly decreased locally and systemically by AtJMT ectopic expression.
Supplemental Table S1. Multiple reaction monitoring conditions for jasmonate profiling in N. attenuata.
Supplemental Table S2. Sequences of gene-specific primers used for quantitative real-time PCR.
Acknowledgments
We thank Dr. Matthias Schoettner, Mario Kallenbach, and Christian Hettenhausen for help with analytical equipments, Dr. Markus Hartl for fruitful discussions, and Prof. Claus Wasternack for providing the 11/12-OH-JA for the determination of the OH-JA/JA response factor. We thank the two reviewers and the handling editor (Gregg Howe) for insightful comments. Dedicated to the memory of Dr. Jean-Pierre Salaün (1944–2011).
References
- Allmann S, Baldwin IT. (2010) Insects betray themselves in nature to predators by rapid isomerization of green leaf volatiles. Science 329: 1075–1078 [DOI] [PubMed] [Google Scholar]
- Baldwin IT, Schmelz EA, Ohnmeiss TE. (1994) Wound-induced changes in root and shoot jasmonic acid pools correlate with induced nicotine synthesis in Nicotiana sylvestris. J Chem Ecol 20: 2139–2157 [DOI] [PubMed] [Google Scholar]
- Browse J. (2005) Jasmonate: an oxylipin signal with many roles in plants. Vitam Horm 72: 431–456 [DOI] [PubMed] [Google Scholar]
- Browse J, Howe GA. (2008) New weapons and a rapid response against insect attack. Plant Physiol 146: 832–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini A, Fonseca S, Fernández G, Adie B, Chico JM, Lorenzo O, García-Casado G, López-Vidriero I, Lozano FM, Ponce MR, et al. (2007) The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671 [DOI] [PubMed] [Google Scholar]
- Chung HS, Koo AJ, Gao X, Jayanty S, Thines B, Jones AD, Howe GA. (2008) Regulation and function of Arabidopsis JASMONATE ZIM-domain genes in response to wounding and herbivory. Plant Physiol 146: 952–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creelman RA, Mullet JE. (1995) Jasmonic acid distribution and action in plants: regulation during development and response to biotic and abiotic stress. Proc Natl Acad Sci USA 92: 4114–4119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devoto A, Turner JG. (2005) Jasmonate-regulated Arabidopsis stress signalling network. Physiol Plant 123: 161–172 [Google Scholar]
- Fowler JH, Narváez-Vásquez J, Aromdee DN, Pautot V, Holzer FM, Walling LL. (2009) Leucine aminopeptidase regulates defense and wound signaling in tomato downstream of jasmonic acid. Plant Cell 21: 1239–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaquerel E, Heiling S, Schoettner M, Zurek G, Baldwin IT. (2010) Development and validation of a liquid chromatography-electrospray ionization-time-of-flight mass spectrometry method for induced changes in Nicotiana attenuata leaves during simulated herbivory. J Agric Food Chem 58: 9418–9427 [DOI] [PubMed] [Google Scholar]
- Gase K, Weinhold A, Bozorov T, Schuck S, Baldwin IT. (2011) Efficient screening of transgenic plant lines for ecological research. Mol Ecol Resour (in press) [DOI] [PubMed] [Google Scholar]
- Gfeller A, Dubugnon L, Liechti R, Farmer EE. (2010a) Jasmonate biochemical pathway. Sci Signal 3: cm3. [DOI] [PubMed] [Google Scholar]
- Gfeller A, Liechti R, Farmer EE. (2010b) Arabidopsis jasmonate signaling pathway. Sci Signal 3: cm4. [DOI] [PubMed] [Google Scholar]
- Glauser G, Grata E, Dubugnon L, Rudaz S, Farmer EE, Wolfender JL. (2008) Spatial and temporal dynamics of jasmonate synthesis and accumulation in Arabidopsis in response to wounding. J Biol Chem 283: 16400–16407 [DOI] [PubMed] [Google Scholar]
- Halitschke R, Baldwin IT. (2003) Antisense LOX expression increases herbivore performance by decreasing defense responses and inhibiting growth-related transcriptional reorganization in Nicotiana attenuata. Plant J 36: 794–807 [DOI] [PubMed] [Google Scholar]
- Halitschke R, Ziegler J, Keinänen M, Baldwin IT. (2004) Silencing of hydroperoxide lyase and allene oxide synthase reveals substrate and defense signaling crosstalk in Nicotiana attenuata. Plant J 40: 35–46 [DOI] [PubMed] [Google Scholar]
- Han JDJ. (2008) Understanding biological functions through molecular networks. Cell Res 18: 224–237 [DOI] [PubMed] [Google Scholar]
- Harms K, Atzorn R, Brash A, Kuhn H, Wasternack C, Willmitzer L, Pena-Cortes H. (1995) Expression of a flax allene oxide synthase cDNA leads to increased endogenous jasmonic acid (JA) levels in transgenic potato plants but not to a corresponding activation of JA-responding genes. Plant Cell 7: 1645–1654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hause B, Hause G, Kutter C, Miersch O, Wasternack C. (2003) Enzymes of jasmonate biosynthesis occur in tomato sieve elements. Plant Cell Physiol 44: 643–648 [DOI] [PubMed] [Google Scholar]
- Hause B, Stenzel I, Miersch O, Maucher H, Kramell R, Ziegler J, Wasternack C. (2000) Tissue-specific oxylipin signature of tomato flowers: allene oxide cyclase is highly expressed in distinct flower organs and vascular bundles. Plant J 24: 113–126 [DOI] [PubMed] [Google Scholar]
- Horn M, Patankar AG, Zavala JA, Wu JQ, Dolecková-Maresová L, Vujtechová M, Mares M, Baldwin IT. (2005) Differential elicitation of two processing proteases controls the processing pattern of the trypsin proteinase inhibitor precursor in Nicotiana attenuata. Plant Physiol 139: 375–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe GA, Lee GI, Itoh A, Li L, DeRocher AE. (2000) Cytochrome P450-dependent metabolism of oxylipins in tomato: cloning and expression of allene oxide synthase and fatty acid hydroperoxide lyase. Plant Physiol 123: 711–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe GA, Schilmiller AL. (2002) Oxylipin metabolism in response to stress. Curr Opin Plant Biol 5: 230–236 [DOI] [PubMed] [Google Scholar]
- Hsieh HL, Okamoto H, Wang ML, Ang LH, Matsui M, Goodman H, Deng XW. (2000) FIN219, an auxin-regulated gene, defines a link between phytochrome A and the downstream regulator COP1 in light control of Arabidopsis development. Genes Dev 14: 1958–1970 [PMC free article] [PubMed] [Google Scholar]
- Jung CK, Lyou SH, Koo YJ, Song JT. (2003) Constitutive expression of defense genes in transgenic Arabidopsis overproducing methyl jasmonate. Agricultural Chemistry and Biotechnology 46: 52–57 [Google Scholar]
- Kallenbach M, Alagna F, Baldwin IT, Bonaventure G. (2010) Nicotiana attenuata SIPK, WIPK, NPR1, and fatty acid-amino acid conjugates participate in the induction of jasmonic acid biosynthesis by affecting early enzymatic steps in the pathway. Plant Physiol 152: 96–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JH, Wang L, Giri A, Baldwin IT. (2006) Silencing threonine deaminase and JAR4 in Nicotiana attenuata impairs jasmonic acid-isoleucine-mediated defenses against Manduca sexta. Plant Cell 18: 3303–3320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RW, Zeevaart JA. (1974) Enhancement of phloem exudation from cut petioles by chelating agents. Plant Physiol 53: 96–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo AJ, Howe GA. (2009) The wound hormone jasmonate. Phytochemistry 70: 1571–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo AJK, Gao XL, Jones AD, Howe GA. (2009) A rapid wound signal activates the systemic synthesis of bioactive jasmonates in Arabidopsis. Plant J 59: 974–986 [DOI] [PubMed] [Google Scholar]
- Kruegel T, Lim M, Gase K, Halitschke R, Baldwin IT. (2002) Agrobacterium-mediated transformation of Nicotiana attenuata, a model ecological expression system. Chemoecology 12: 177–183 [Google Scholar]
- Laudert D, Schaller F, Weiler EW. (2000) Transgenic Nicotiana tabacum and Arabidopsis thaliana plants overexpressing allene oxide synthase. Planta 211: 163–165 [DOI] [PubMed] [Google Scholar]
- Li L, Van Eck J. (2007) Metabolic engineering of carotenoid accumulation by creating a metabolic sink. Transgenic Res 16: 581–585 [DOI] [PubMed] [Google Scholar]
- Li L, Zhao Y, McCaig BC, Wingerd BA, Wang J, Whalon ME, Pichersky E, Howe GA. (2004) The tomato homolog of CORONATINE-INSENSITIVE1 is required for the maternal control of seed maturation, jasmonate-signaled defense responses, and glandular trichome development. Plant Cell 16: 126–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linke C, Conrath U, Jeblick W, Betsche T, Mahn A, Düring K, Neuhaus HE. (2002) Inhibition of the plastidic ATP/ADP transporter protein primes potato tubers for augmented elicitation of defense responses and enhances their resistance against Erwinia carotovora. Plant Physiol 129: 1607–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado AM, Doerner P, Dixon RA, Lamb CJ, Cameron RK. (2002) A putative lipid transfer protein involved in systemic resistance signalling in Arabidopsis. Nature 419: 399–403 [DOI] [PubMed] [Google Scholar]
- Mielke K, Forner S, Kramell R, Conrad U, Hause B. (2011) Cell-specific visualization of jasmonates in wounded tomato and Arabidopsis leaves using jasmonate-specific antibodies. New Phytol 190: 1069–1080 [DOI] [PubMed] [Google Scholar]
- Miersch O, Weichert H, Stenzel I, Hause B, Maucher H, Feussner I, Wasternack C. (2004) Constitutive overexpression of allene oxide cyclase in tomato (Lycopersicon esculentum cv. Lukullus) elevates levels of some jasmonates and octadecanoids in flower organs but not in leaves. Phytochemistry 65: 847–856 [DOI] [PubMed] [Google Scholar]
- Park SW, Kaimoyo E, Kumar D, Mosher S, Klessig DF. (2007) Methyl salicylate is a critical mobile signal for plant systemic acquired resistance. Science 318: 113–116 [DOI] [PubMed] [Google Scholar]
- Paschold A, Bonaventure G, Kant MR, Baldwin IT. (2008) Jasmonate perception regulates jasmonate biosynthesis and JA-Ile metabolism: the case of COI1 in Nicotiana attenuata. Plant Cell Physiol 49: 1165–1175 [DOI] [PubMed] [Google Scholar]
- Pfaffl MW, Horgan GW, Dempfle L. (2002) Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res 30: e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin GJ, Gu HY, Zhao YD, Ma ZQ, Shi GL, Yang Y, Pichersky E, Chen HD, Liu MH, Chen ZL, et al. (2005) An indole-3-acetic acid carboxyl methyltransferase regulates Arabidopsis leaf development. Plant Cell 17: 2693–2704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond P, Weber H, Damond M, Farmer EE. (2000) Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis. Plant Cell 12: 707–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roda A, Halitschke R, Steppuhn A, Baldwin IT. (2004) Individual variability in herbivore-specific elicitors from the plant’s perspective. Mol Ecol 13: 2421–2433 [DOI] [PubMed] [Google Scholar]
- Ryan CA. (2000) The systemin signaling pathway: differential activation of plant defensive genes. Biochim Biophys Acta 1477: 112–121 [DOI] [PubMed] [Google Scholar]
- Seo HS, Song JT, Cheong JJ, Lee YH, Lee YW, Hwang I, Lee JS, Choi YD. (2001) Jasmonic acid carboxyl methyltransferase: a key enzyme for jasmonate-regulated plant responses. Proc Natl Acad Sci USA 98: 4788–4793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibbe M, Qu N, Galis I, Baldwin IT. (2008) Induced plant defenses in the natural environment: Nicotiana attenuata WRKY3 and WRKY6 coordinate responses to herbivory. Plant Cell 20: 1984–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE, Tiryaki I. (2004) The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis. Plant Cell 16: 2117–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE, Tiryaki I, Rowe ML. (2002) Jasmonate response locus JAR1 and several related Arabidopsis genes encode enzymes of the firefly luciferase superfamily that show activity on jasmonic, salicylic, and indole-3-acetic acids in an assay for adenylation. Plant Cell 14: 1405–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenzel I, Hause B, Maucher H, Pitzschke A, Miersch O, Ziegler J, Ryan CA, Wasternack C. (2003) Allene oxide cyclase dependence of the wound response and vascular bundle-specific generation of jasmonates in tomato—amplification in wound signalling. Plant J 33: 577–589 [DOI] [PubMed] [Google Scholar]
- Stintzi A, Browse J. (2000) The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc Natl Acad Sci USA 97: 10625–10630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strassner J, Schaller F, Frick UB, Howe GA, Weiler EW, Amrhein N, Macheroux P, Schaller A. (2002) Characterization and cDNA-microarray expression analysis of 12-oxophytodienoate reductases reveals differential roles for octadecanoid biosynthesis in the local versus the systemic wound response. Plant J 32: 585–601 [DOI] [PubMed] [Google Scholar]
- Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu GH, Nomura K, He SY, Howe GA, Browse J. (2007) JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448: 661–665 [DOI] [PubMed] [Google Scholar]
- Thorpe MR, Ferrieri AP, Herth MM, Ferrieri RA. (2007) 11C-imaging: methyl jasmonate moves in both phloem and xylem, promotes transport of jasmonate, and of photoassimilate even after proton transport is decoupled. Planta 226: 541–551 [DOI] [PubMed] [Google Scholar]
- Tieman D, Zeigler M, Schmelz E, Taylor MG, Rushing S, Jones JB, Klee HJ. (2010) Functional analysis of a tomato salicylic acid methyl transferase and its role in synthesis of the flavor volatile methyl salicylate. Plant J 62: 113–123 [DOI] [PubMed] [Google Scholar]
- Varbanova M, Yamaguchi S, Yang Y, McKelvey K, Hanada A, Borochov R, Yu F, Jikumaru Y, Ross J, Cortes D, et al. (2007) Methylation of gibberellins by Arabidopsis GAMT1 and GAMT2. Plant Cell 19: 32–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Dahl CC, Baldwin IT. (2004) Methyl jasmonate and cis-jasmone do not dispose of the herbivore-induced jasmonate burst in Nicotiana attenuata. Physiol Plant 120: 474–481 [DOI] [PubMed] [Google Scholar]
- Wang L, Allmann S, Wu J, Baldwin IT. (2008) Comparisons of LIPOXYGENASE3- and JASMONATE-RESISTANT4/6-silenced plants reveal that jasmonic acid and jasmonic acid-amino acid conjugates play different roles in herbivore resistance of Nicotiana attenuata. Plant Physiol 146: 904–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Halitschke R, Kang J-H, Berg A, Harnisch F, Baldwin IT. (2007) Independently silencing two JAR family members impairs levels of trypsin proteinase inhibitors but not nicotine. Planta 226: 159–167 [DOI] [PubMed] [Google Scholar]
- Wasternack C. (2007) Jasmonates: an update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann Bot (Lond) 100: 681–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber H, Chételat A, Caldelari D, Farmer EE. (1999) Divinyl ether fatty acid synthesis in late blight-diseased potato leaves. Plant Cell 11: 485–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JS, Wang L, Baldwin IT. (2008) Methyl jasmonate-elicited herbivore resistance: does MeJA function as a signal without being hydrolyzed to JA? Planta 227: 1161–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan YX, Stolz S, Chételat A, Reymond P, Pagni M, Dubugnon L, Farmer EE. (2007) A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell 19: 2470–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao KN, De Luca V, Brisson N. (1995) Creation of a metabolic sink for tryptophan alters the phenylpropanoid pathway and the susceptibility of potato to Phytophthora infestans. Plant Cell 7: 1787–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZP, Baldwin IT. (1997) Transport of [2-C-14]jasmonic acid from leaves to roots mimics wound-induced changes in endogenous jasmonic acid pools in Nicotiana sylvestris. Planta 203: 436–441 [Google Scholar]