Abstract
Sucrose (Suc) synthase (Sus) is the major enzyme of Suc breakdown for cellulose biosynthesis in cotton (Gossypium hirsutum) fiber, an important source of fiber for the textile industry. This study examines the tissue-specific expression, relative abundance, and temporal expression of various Sus transcripts and proteins present in cotton. A novel isoform of Sus (SusC) is identified that is expressed at high levels during secondary cell wall synthesis in fiber and is present in the cell wall fraction. The phylogenetic relationships of the deduced amino acid sequences indicate two ancestral groups of Sus proteins predating the divergence of monocots and dicots and that SusC sequences form a distinct branch in the phylogeny within the dicot-specific clade. The subcellular location of the Sus isoforms is determined, and it is proposed that cell wall-localized SusC may provide UDP-glucose for cellulose and callose synthesis from extracellular sugars.
In most higher plants, Suc is the major translocated sugar, requiring cleavage to its component hexoses before further biochemical conversion can occur. This cleavage can occur via two enzymes: invertase or Suc synthase (Sus; for review, see Winter and Huber, 2000; Koch, 2004). Sus is often regarded as the more energy conservative of the two enzyme reactions, as it produces Fru and UDP-Glc, the latter being used as a phosphorylated sugar in biosynthetic processes. Sus is believed to play a major role in cellulose biosynthesis, where UDP-Glc produced by a membrane-associated form of Sus is thought to be used directly as a substrate for the cellulose synthase complex (Amor et al., 1995). This so-called “PM” (for plasma membrane) association of Sus has been studied in a number of species (Winter and Huber, 2000; Haigler et al., 2001; Komina et al., 2002). Conclusive evidence for substantial partitioning of cellular Sus to the plasma membrane is difficult to find in the literature for any species; however, recent evidence that Sus is an integral component of the cellulose synthase rosette in bean (Phaseolus vulgaris) hypocotyls has been published (Fujii et al., 2010). It has been shown that suppression of Sus expression in transgenic cotton (Gossypium hirsutum) ovules decreased fiber elongation (Ruan et al., 2003) and affected cellulose deposition (Ruan, 2007), reinforcing the importance of this enzyme in plant organs where cellulose is synthesized at high rates. Sus has also been implicated as an important gene in wood formation, with expression levels correlating with secondary thickening of xylem and wood strength (Hauch and Magel, 1998; Salnikov et al., 2001; Andersson-Gunnerås et al., 2006; Nilsson et al., 2010). Sus has been shown to be important in the pathway of starch biosynthesis in legumes such as Vicia faba (Heim et al., 1993) and canola (Brassica napus; King et al., 1997) embryos, in the cellularization of cotton seed endosperm (Ruan et al., 2008), and in starch and cellulose synthesis in maize (Zea mays; for review, see Koch, 2004). In Arabidopsis (Arabidopsis thaliana), six isoforms of Sus are encoded in the genome, and extensive studies have been made of the tissue-specific expression patterns of all six isoforms (Baud et al., 2004; Bieniawska et al., 2007). However, despite reverse genetic and biochemical evidence for crucial roles of Sus in the crop species mentioned above, it has recently been reported that Arabidopsis Sus gene knockout lines have little or no obvious phenotypes (Bieniawska et al., 2007). This includes analysis of a knockout in the isoform involved in Suc cleavage in the embryo and the isoforms present in stems and root, major sites of secondary cell wall synthesis in this species (Bieniawska et al., 2007). It appears that inactivation of multiple isoforms of Sus and, in particular, invertase is necessary to impair Suc breakdown in this model species and that invertase may work together with UDPG pyrophosphorylase to supply carbon skeletons to cellulose synthase (Barratt et al., 2009). In tobacco (Nicotiana tabacum), overexpression of both Sus and invertase had little impact on cellulose content (Coleman et al., 2006), while in the woody species Populus alba, overexpression of Sus produced a small but reproducible increase in cellulose content and had an impact on wood properties (Coleman et al., 2009). Thus, despite decades of research, the roles and importance of Sus in plant metabolism, and in particular in cellulose biosynthesis, still remain contentious.
In cotton, a major source of fiber for the textile industry, cellulose synthesis is of particular interest. Cotton fiber is composed of single cells initiated from the surface of the ovule epidermis around the time of flowering. Over the next 20 d, these cells elongate up to approximately 3 cm before laying down massive amounts of secondary cell wall (Basra and Malik, 1984), and they contain up to 98% cellulose when desiccated. The flux of Glc moieties into cellulose at this secondary cell wall synthesis stage is extremely high and comparable in magnitude only to the process of secondary wall formation in xylem tracheids of woody species. To date, most research at the biochemical and molecular levels in cotton has focused on a single isoform of Sus expressed at high levels in fiber and seed (Ruan et al., 1997). This Sus has been claimed to represent the predominant isoform in cotton fiber, and the transcript corresponding to this Sus is highly expressed in microarray (Arpat et al., 2004) and subtractive hybridization experiments on cotton fiber (Haigler et al., 2005). A major focus for research on Sus has been the study of the membrane association of this protein and its proposed role in channeling UDP-Glc to the cellulose synthase complex. The hypothesis proposed by Amor et al. (1995) and subsequently explored in many other laboratories (for review, see Winter and Huber, 2000; Haigler et al., 2001; Koch, 2004) has been that a single, highly expressed Sus isoform can exist either in a free cytosolic state or as a membrane-bound form and that this equilibrium can be regulated by protein phosphorylation and/or other posttranslational processes. The exact mechanism responsible for this membrane association and the biochemical control of the proportion of membrane-bound versus soluble protein has thus far remained elusive.
In this study, we examine the abundance and temporal expression of various Sus transcripts present during fiber development and identify a novel isoform that is expressed at high levels during secondary cell wall synthesis. The gene sequences and tissue-specific expression patterns of this Sus isoform and other genes in the cotton Sus family are described, along with subcellular location, predicted protein structure, and proposed role of this novel Sus isoform.
RESULTS
Identification of the Sus Gene Family in Cotton
It has previously been suggested that a single isoform of Sus is expressed during both cotton fiber elongation and secondary cell wall synthesis, and it is widely believed that posttranslational modification of Sus is important in the secondary cell wall synthesis stage to direct UDP-Glc to the cellulose synthase complex (Amor et al., 1995; for review, see Winter and Huber, 2000). However, very little work has been done on elucidation of the Sus genes expressed over fiber development. We examined this issue by creating a small library of Sus partial cDNA sequences derived by reverse transcription (RT)-PCR of fiber RNA extracted from fibers undergoing rapid elongation (approximately 10 d after flowering [DAF]) and in the secondary cell wall synthesis stages (approximately 20 DAF). Primers were designed to hybridize to a region of the Sus sequence highly conserved across all higher plants (see “Materials and Methods”). Full-length cDNAs of representatives of the classes of sequences were then obtained by 5′ RACE. The sequences of partial cDNAs obtained from the fiber RNA pools fell into four classes, termed SusA, SusB, SusC, and SusD. Full-length deduced amino acid sequences of these are shown in Figure 1. Deduced amino acid sequences for SusB and -D were 93% and 95.5% similar to SusA, respectively. Sequence A is identical to the sequence reported in the literature by Amor et al. (1995) and Ruan et al. (1997) and is the isoform commonly studied in the literature (GenBank accession no. U73588). As cotton is tetraploid, it is possible that SusA, -B, and -D are homeologous, but this cannot be definitively determined from these data. In contrast, the deduced amino acid sequence of SusC was only 76% identical to that of the SusA protein, with regions of marked sequence divergence at both the N and C termini.
Figure 1.
Deduced amino acid sequence comparison of cotton SusA, -B, -D, and -C isoforms (generated using Genetics Computer Group Wisconsin Package Pileup software). Note the sequence divergence at the N and C termini of SusC compared with the A,B,D sequences. The line below the SusC sequence indicates the oligopeptide synthesized to raise antiserum specific to the SusC isoform.
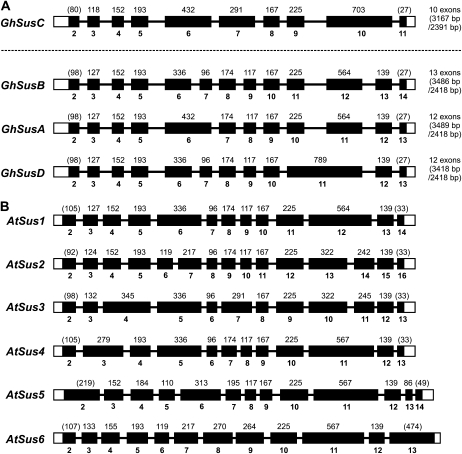
To examine the likely genomic origins and relatedness of the four classes of Sus genes, genomic clones of each class were isolated and the gene structures were compared. Figure 2 shows the intron-exon distribution of the Sus genes isolated from cotton. All four classes of Sus have complex gene structures, 10 exons in the case of SusC, 12 in SusA and -D, and 13 in the case of SusB. This is broadly similar to the gene structure of the Arabidopsis Sus genes, the only other dicot Sus gene family sequenced, which ranges from 12 to 15 exons (Baud et al., 2004).
Figure 2.
Intron/exon distributions in the genomic sequences of the four Sus isoforms present in the cotton genome (A) compared with the gene structures of the six isoforms present in the Arabidopsis genome (B; Baud et al., 2004).
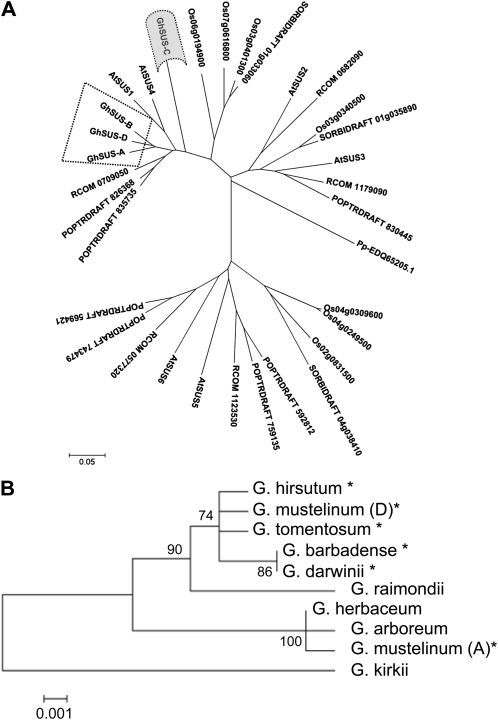
The phylogenetic relationship between the deduced amino acid sequences of the SusA, -B, -C, and -D proteins from cotton, SusC proteins from a range of diploid and tetraploid Gossypium species, and Sus sequences from other dicot and monocot plant species with fully sequenced genomes are shown in Figure 3 (the alignments are provided as Supplemental Fig. S1). There are clearly two ancestral groups of Sus proteins that predate the divergence of monocots and dicots but also more specialized forms of Sus that are specific to either dicots or monocots. SusC sequences form a distinct branch in the phylogeny within the dicot-specific clade and have evolved away from the other members of the cotton Sus gene family. The major regions of divergence are in the N and C termini of the SusC protein rather than in regions responsible for enzymatic activity. The SusA, -D, and -B isoforms are most closely related to each other than to SusC. It is notable that there seems to be no direct homolog of SusC in the Arabidopsis or Populus genomes. Within the Gossypium species, SusC appears to be quite ancestral, with distinct clades in the A and D genome diploids (Fig. 3B). In the AD tetraploids, the A genome form of SusC appears to have been lost at some time after polyploidization, except for Gossypium mustelinum, which has retained both homeologues. This was confirmed by Southern blotting using SusC-specific probes (data not shown).
Figure 3.
Phylogenetic tree of Sus protein sequences generated using MEGA 4.0 and the neighbor-joining method (see “Materials and Methods”). A, Unrooted tree of 28 plant Sus proteins from plants with sequenced genomes: the dicots Arabidopsis (AtSUS1 to -6), Populus trichocarpa (POPTRDRAFT), and Ricinus communis (RCOM_xxxxxxx), the monocots Oryza sativa japonica group (Osxxxxxxxx) and Sorghum bicolor (SORBIDRAFTxxxxxxxx), together with the four cotton Sus proteins reported in this paper, and with the moss (Physcomitrella patens subspecies patens) Sus protein (Pp-EDQ65205.1) as an outgroup. The cotton Sus proteins are surrounded by dotted lines, and the SusC protein is shaded in gray (alignment files are provided as Supplemental Fig. S1). B, Phylogenetic tree of the SusC proteins from selected diploid and tetraploid Gossypium species (alignment files shown in Supplemental Fig. S2). The G. kirkii SusC sequence was used as the outgroup. Tetraploid cotton species names are indicated by asterisks. The numbers on the interior branches refer to the bootstrap values for 1,000 replications. Bootstrap values less than 50% are not given. The scale at the bottom is in units of amino acid substitutions per site.
Expression of Sus Genes in Cotton Fiber and Other Tissues
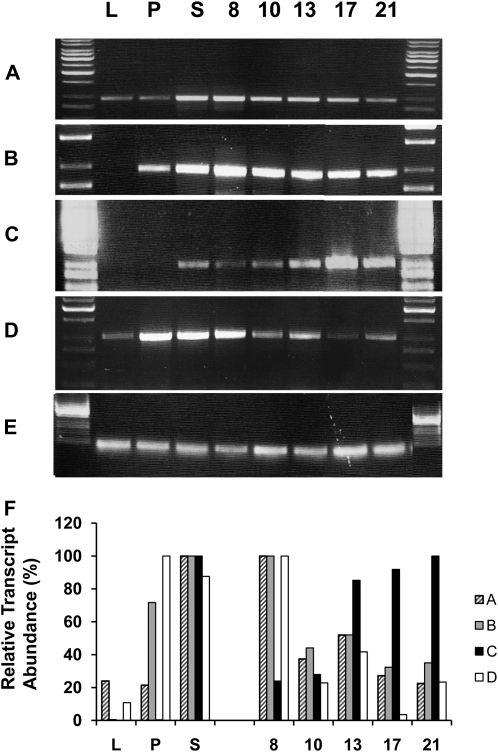
The expression levels of the Sus isoforms in various organs of cotton plants and across fiber development were examined using semiquantitative PCR. Gene-specific primers for SusA, SusB, SusC, and SusD transcripts were used (Fig. 4, A–D), along with ubiquitin as a normalization control (Fig. 4E; see “Materials and Methods”). All four transcripts were detected at high levels in stem and fiber RNA pools (Fig. 4). Transcripts for SusA, -B, and -D were also detected at low levels in leaf and at higher levels in petal samples. During fiber development, transcripts encoding SusA, -B, and -D were detected at all six stages of fiber development, with transcript levels generally falling from 8 to 21 DAF (Fig. 4). In contrast, SusC transcript abundance increased with developmental age, declining slightly at 21 DAF. Public G. hirsutum EST collections were also searched using the unique peptides from the N and C termini of the SusC protein sequence; no SusC ESTs were found in libraries isolated from the fiber initiation, elongation, and primary cell wall biosynthesis stages of development (0–15 DAF). In contrast, in EST libraries constructed from fibers at 20 to 40 DAF, 30 SusC EST sequences were identified using the C-terminal region as a query sequence and 35 using the N-terminal region as a query. Consistent with the phylogenetic analysis of the SusC amino acid sequence, no ESTs were identified from genera other than Gossypium using the N- or C-terminal region of the SusC protein as query sequence (data not shown).
Figure 4.
Semiquantitative PCR of Sus RNA prepared from different plant organs and over fiber development: young fully expanded leaf (L), petal (P), and stem (S) and fiber RNA from bolls at 8, 10, 13, 17, and 21 DAF. Primers specific to SusA, -B, -C, and -D were used (A–D, respectively) and to a member of the ubiquitin family as a quantitative control (E). F compares relative expression levels of Sus isoforms shown in A to E by normalizing the fluorescence intensity of each PCR product on the gel to the respective Ubiquitin control and presenting this as a percentage of the maximum expression level for the Sus isoform in that tissue sample and developmental stage.
Immunological Detection and Subcellular Targeting of Sus Isoforms
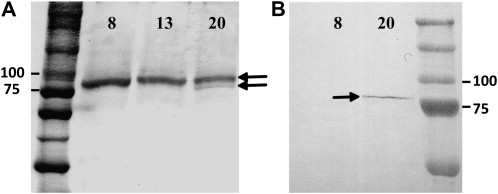
As discussed above, it has previously been proposed that during secondary cell wall synthesis, Sus binds to the plasma membrane, channeling UDPG directly to the cellulose synthase complex (for review, see Huber and Winter, 2000; Haigler et al., 2001). Sus has also been proposed to associate with the actin cytoskeleton (Winter et al., 1998), mitochondria (Subbaiah et al., 2006), and the tonoplast (Etxeberria and Gonzalez, 2003). It has been proposed that a single Sus isoform can be targeted to these diverse subcellular compartments, although no overarching mechanism for controlling this targeting and association has been resolved. Protein phosphorylation has been implicated in partitioning the protein between the plasma membrane and the cytosol and between the cytosol and the cytoskeleton (for review, see Winter and Huber, 2000), and regions of both the C and N terminus and the presence of a pleckstrin-like domain have been implicated in membrane binding in maize Sus (Hardin et al., 2006). The data above show that Sus gene family members have distinctly different expression profiles during fiber development and that a single protein does not dominate. This presents the possibility that the individual isoforms may play distinct roles in fiber metabolism, possibly in separate subcellular compartments. To explore this possibility, antibodies were raised against oligopeptides specific to the SusC protein to produce antiserum that would not cross-react with the A/D or B isoform. Separation of the other protein isoforms immunologically was not possible due to their high sequence similarity. To test the specificity of the SusC antibody, western blotting was carried out using protein prepared from fiber samples in the elongation phase (8–13 DAF) and the secondary cell wall stage (20 DAF; Fig. 5). Western blots probed with an antiserum raised against purified rice Sus protein, which detects multiple Sus isoforms (Fallahi et al., 2008), detected a single band at 92 kD in fiber sampled at 8 to 13 DAF (Fig. 5A). In 20-DAF fiber, two bands were detected at approximately 90 and 92 kD (Fig. 5A). When probed with the specific SusC antibody, a single band at 90 kD was detected in 20-DAF fiber, corresponding to the lower molecular mass band detected by the rice Sus antiserum, which is consistent with the predicted molecular mass of the SusC protein being smaller than the A, B, and D isoforms (Fig. 1). No band was detected with specific SusC antiserum in fiber at the elongation stage at 8 DAF (Fig. 5B). To confirm the identity of the proteins recognized by these antibodies, a Coomassie blue-stained gel slice corresponding to these two protein bands was excised from duplicate SDS-PAGE gels and subjected to tryptic digestion and subsequent liquid chromatography-tandem mass spectrometry (LC-MS/MS). A mixture of peptides from the A, B, and D isoforms was detected in both the 8- to 13-DAF and 20-DAF fiber samples, while the specific lower molecular mass protein recognized by the SusC antiserum in the 20-DAF fiber sample also contained peptides unique to the SusC isoforms (data not shown). This LC-MS/MS result is also confirmed in the apoplast and microsomal protein fractions from 8-DAF and 20- to 25-DAF fiber, showing no SusC peptides in 8-DAF fiber samples and numerous unique SusC peptides in 20-DAF fiber (Fig. 6; Supplemental Table S1).
Figure 5.
Western analysis of cotton fiber proteins extracted from bolls at 8 and 13 DAF (elongation stage) and 20 DAF (secondary cell wall stage). A, Protein from these three stages probed with the nonselective antiserum, with the Sus protein bands indicated by arrows. Note the appearance of labeling of a lower molecular mass band at 20 DAF shown by MS/MS to be SusC. B, Western blot of cotton fiber proteins at 8 and 20 DAF probed with the SusC-specific antiserum, detecting a single protein band shown by MS/MS to be SusC.
Figure 6.
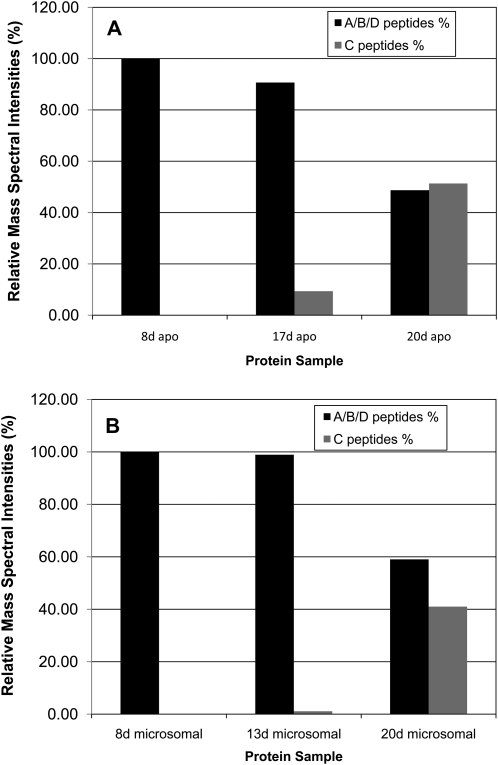
Relative abundance of Sus protein isoforms quantified by relative mass spectral intensities of diagnostic peptides for SusC and the combined A, B, and D isoforms. A, Relative spectral intensities for the apoplast protein pool of fiber at 8, 17, and 20 DAF. B, These same data for the microsomal protein pool at 8, 13, and 20 DAF.
The SusC-specific antiserum was used to investigate the subcellular location of SusC both through western blotting and LC-MS/MS of tryptic peptide from proteins prepared from fiber-cell fractions (Figs. 6–8) or by immunohistochemistry (Fig. 9). Cotton fibers harvested at 8 to 13 DAF (elongation phase) and 17 to 25 DAF (secondary cell wall stage) were frozen in liquid nitrogen, ground to a fine powder, and then extracted in buffer (see “Materials and Methods”) intended to extract cytosolic proteins plus proteins loosely bound to cell wall or membrane components. This extract was subjected to centrifugation to remove particulate material, mostly cell wall debris (“cell wall fraction”). The supernatant fraction from this first centrifugation also contained suspended membrane components that were removed by ultracentrifugation. The supernatant from this ultracentrifugation was termed “soluble fraction” and the pellet was termed “microsomal fraction,” the latter being enriched in plasma membrane and other membranous cell fractions (Carlson and Chourey, 1996). The microsomal fraction was also further purified by density gradient ultracentrifugation to enrich the proportion of plasma membranes (“PM fraction”; Carlson and Chourey, 1996). The cell wall pellet was either further extracted or solubilized for SDS-PAGE, western blotting, and subsequent LC-MS/MS. The supernatant, microsomal, and PM fractions were subjected to SDS-PAGE and subsequent LC-MS/MS. Consistent with previous experiments from total soluble fiber fractions (Fig. 5), SusC protein was only detected in fiber microsomal extracts from approximately 13 to 17 DAF onward, increasing in the secondary cell wall synthesis stage (20–25 DAF) both in western blotting experiments (data not shown) and LC-MS/MS (Fig. 6).
Figure 8.
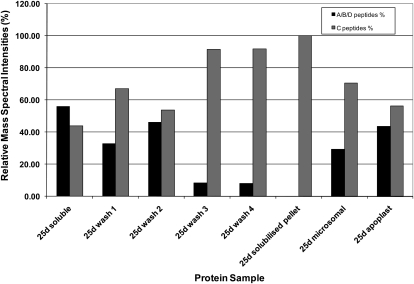
MS/MS of Sus proteins quantified by relative mass spectral intensity of SusC and SusA, -B, and -D diagnostic peptides (as in Fig 6) but derived from the 25-DAF fiber protein samples (described in Fig 7). Note the persistence of SusC peptides in the protein pool in later washes and the dominance of this isoform in the final extensively washed and solubilized pellet.
Figure 9.
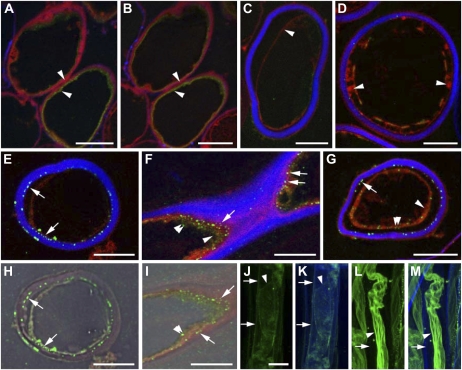
Confocal laser scanning micrographs of cotton fiber cross sections (A–I) or whole fibers (J–M) labeled with SusC affinity-purified antibodies (A and E–K) or preimmune serum (B and D) and then Alexa-488 secondary antibody, secondary antibody only (C), or monoclonal anti-a-tubulin followed by Alexa-488 secondary antibody (L and M). A and B, Sections of 10-DAF fibers showing nonspecific autofluorescence (green, red; arrowheads) and weak cellulose fluorescence (blue) after incubation in SusC antibody (A) or preimmune serum (B). C to I, Sections of 18-DAF fibers showing background autofluorescence (mostly red; arrowheads in C, D, and G) and strong cellulose fluorescence (blue). Very little background label was seen in sections treated with secondary antibody only (C) or preimmune serum and then secondary antibody (D). SusC (punctate green fluorescence) was detected primarily on the inner surface of the cell wall (E–I; arrows) and was occasionally detected in the cytoplasm, where the plasma membrane and cytoplasm have retracted from the cell wall (F, G, and I; double arrowheads). A glancing section (F and I) shows diffuse autofluorescence in the cytoplasm (F; arrowhead) as well as SusC label. H and I, SusC fluorescence (green) from E and F, respectively, overlaid on bright-field images. J, Intact 18-DAF fibers treated with 0.3 m mannitol before SusC labeling (green) show fluorescence associated with contracted cytoplasm (arrowhead) and with the cell walls (arrows). K, Overlay of SusC label in J with cellulose fluorescence (blue) showing colocalization with the cell wall (arrows). L, Microtubules (green) in 18-DAF fibers treated with 0.6 m mannitol are contracted away from the cell walls (arrowhead), with no detectable wall-associated background fluorescence (arrow). M, Overlay of microtubule label in L with cellulose fluorescence (blue) showing no colocalization with the cell wall (arrow). Bars = 10 μm (A–I) or 20 μm (in J for J–M).
Over the whole experiment and considering all the observed peptides identified to Sus isoforms A, B, C, and D, 19 distinct peptides were unique to SusC, whereas the remaining peptides could have been derived from SusA, -B, or -D or subsets of that group (Supplemental Table S1; Supplemental Fig. S3). Only one peptide was observed that could have been derived from SusC and also could have been derived from SusB and -D. For an estimate of the relative abundance of Sus isoforms in each sample, we compared the total mass spectrum intensities of peptides unique to SusC with the total intensities for peptides indicating SusA, -B, or -D expressed as a proportion of the total spectral intensity for all peptides indicating Sus isoforms. For the purposes of this estimation, the one peptide observed that could have been derived by SusB, -C, or -D was excluded. The high similarity of the SusA, -B, and -D amino acid sequences meant that these proteins could not be estimated individually. This approach does not allow absolute amounts of the proteins to be estimated without the use of standards (Perkins et al., 2005), but it was sufficient for our purposes here to indicate where and when SusA/B/D dominated relative to SusC and vice versa.
Relative abundance of the Sus isoforms in cellular fractions at 25 DAF was estimated by quantification of the spectral intensities of unique, diagnostic peptides for SusC found in each sample, relative to those of isoforms A/B/D and normalized to the total spectral intensity of all Sus peptides in that sample (Fig. 6). Because of the high similarity of the SusA, -B, and -D amino acid sequences, these proteins could not be individually quantified using this method. SusA,B,D and SusC were all present in the soluble samples and the microsomal fractions at similar abundance in 25-DAF fiber (Fig. 6); however, SusC was barely detectable in the PM fraction (data not shown).
Two approaches were used to examine the presence of Sus isoforms in the cell wall fraction. First, ovules were excised from cotton fruits with attached fibers intact, washed briefly in cold isotonic buffer, and then proteins that were loosely bound to the cell walls were extracted by gentle washing in isotonic buffer containing a variety of protease inhibitors and stabilizing agents. Proteins recovered by concentrating this wash were subjected to SDS-PAGE, the region of the gel containing Sus proteins was excised, and the tryptic peptides were identified by MS/MS. While SusA/B/D peptides were present and dominant in the early stages of fiber development, SusC diagnostic peptides only began to appear in the Sus tryptic peptide profile by 17 DAF and dominated the profile from 20 DAF onward in the cell wall fraction (Fig. 6B). This was confirmed by western blotting of duplicate gels with the SusC-specific antibody (data not shown).
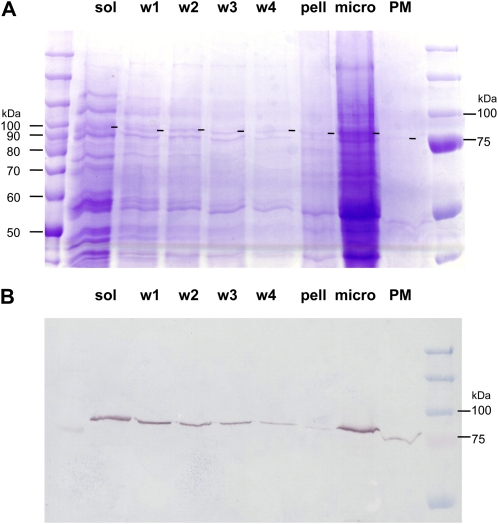
The second approach to extracting cell wall-associated proteins involved sequential washing of the “cell wall pellet” fraction from 25-DAF fibers following centrifugation and subjecting these washes to SDS-PAGE (Fig. 7A), western blotting (Fig. 7B), and MS/MS peptide quantification, as described above (Fig. 8). Interestingly, the proportion of SusA/B/D present in the washes declined substantially with each wash, and in the solubilized pellet after the fourth wash, only SusC peptides were present (Fig. 8). However, it should be noted that the total amount of all Sus proteins remaining was low (Fig. 7), with 20 Sus peptides identified in wash 1, decreasing to only five peptides in the solubilized pellet after four washes. At this stage of development, microsomal and apoplast fractions also contained more SusC than the other isoforms combined.
Figure 7.
SDS-PAGE and western blotting of 25-DAF cotton fiber protein with the SusC-specific antiserum. Crude extract was fractionated into soluble (sol), microsomal (micro), and plasma membrane (PM) fractions (see “Materials and Methods”), and the pellet (pell) remaining from the crude extract was washed extensively four more times in extraction buffer (w1, w2, w3, and w4; see “Materials and Methods”) in order to remove all residual loosely bound proteins. [See online article for color version of this figure.]
Immunolocalization of SusC in Cotton Fiber
Sections of fixed and resin-embedded cotton fibers from ovules at 8 to 10 DAF and 18 to 20 DAF were labeled with SusC-specific antiserum (Fig. 9, A and E–K) or preimmune serum (Fig. 9, B and D) or secondary antibody only (Fig. 9C). Cell walls were visualized by the fluorescent dye Calcofluor white, and antiserum binding was visualized using an Alexa-488-conjugated secondary antibody. Fluorescence was detected using laser confocal scanning microscopy. Nonspecific binding by the preimmune serum was problematic in these experiments and required extensive trials of blocking agents and hybridization conditions and reverse affinity purification of antisera (see “Materials and Methods”). As predicted from western blotting and RT-PCR of fiber samples, no SusC could be detected in transverse sections of cotton fibers at 8 to 10 DAF (Fig. 9A). In contrast, at 18 to 20 DAF, the inner part of the cell wall was strongly labeled by the SusC antiserum, with very weak fluorescence in the cytoplasm (Fig. 9, E–I). It was not possible to detect whether the cell wall signal was present in the inner secondary cell wall or on the outer surface of the plasma membrane, as the latter could not be easily seen in these sections. In glancing sections, there was weak labeling of cytoplasm close to the cell wall, which could be associated with the plasma membrane (Fig. 9, F and I). However, examination of fibers in which the protoplast had pulled away from the cell wall during processing would suggest that the inner cell wall is most strongly labeled (Fig. 9, E, G, and H). Intact fibers that were deliberately plasmolyzed before whole-mount immunofluorescence showed weak labeling of both the cell wall and plasma membrane (Fig. 9, J and K), whereas similar fluorescence localization of microtubules, which are located just inside the plasma membrane, showed a very bright cytoplasmic signal with no cell wall labeling (Fig. 9, L and M).
DISCUSSION
SusC Is a Novel Sus Isoform Abundant in Cotton Fiber during Secondary Cell Wall Synthesis
The highly regulated expression of the Sus gene family during fiber development and the abundance at the transcript and protein levels (Figs. 4 and 5) during the later phases of fiber development lead us to postulate that this isoform of Sus plays an important role in secondary cellulose synthesis. To understand this role, it is important to place the observations above into the context of the complex hypotheses and experiments reported in the literature on “PM-Sus.”
It has been a long-held view that the Sus protein pool in cotton fiber is dominated by a single gene product (or proteins so similar in amino acid composition as to be considered functionally identical) and that this single Sus protein is targeted either to the cytosol or the plasma membrane by posttranslational modification (Amor et al., 1995; Winter and Huber, 2000). Evidence for the dominance of a single dominant isoform of Sus is in fact scant (Amor et al., 1995; for review, see Haigler et al., 2001), and until our study, a comprehensive analysis of the Sus gene family and expression in cotton fiber had not been performed, apart from analyses of EST collections (Arpat et al., 2004; Haigler et al., 2005; Wilkins and Arpat, 2005). The sequence information in this study now allows a bioinformatic examination of SusC transcript abundance in cDNA collections in publicly available databases. SusC is highly represented in these EST collections during secondary cell wall synthesis stages but not early in fiber elongation phases. Careful examination of the subtractive hybridization analysis of ESTs differentially represented between cotton fiber secondary cell wall synthesis and elongation stages (Haigler et al., 2005) reveals clear differential expression of a SusC-like transcript.
Part of the reasoning behind the hypothesis that a “single” Sus protein, of the SusA type, is targeted either to the PM or other parts of the cell was based on the sequence similarity of known full-length Sus proteins and the conservative nature of the sequence divergence at the amino acid level (Amor et al., 1995; Ruan et al., 1997; Haigler et al., 2001, 2005). Figure 1 clearly shows that there is significant amino acid sequence divergence both at the N and C termini of the SusC protein when compared with the A,B,D isoforms, which are highly similar. The functional significance of this amino acid divergence is at present unknown and requires considerable further work to elucidate. At the N terminus, there are several interesting differences between the SusC and A,B,D isoforms. First, the SusC predicted protein is truncated at the N terminus when compared with the other isoforms. This means that a putative phosphorylation site (8-RVHS-11) present in the A,B,D proteins and conserved in most other Sus sequences from other species is absent in SusC. Ser-11 in the SusA sequence is orthologous to Ser-15 in maize, implicated in PM binding (Winter et al., 1997; Hardin et al., 2004). Phosphorylation of this Ser has also been implicated in the reversal of PM binding in soybean (Glycine max) nodules (Zhang et al., 1999) and with determining kinetic properties of the enzyme in mung bean (Vigna radiata; Nakai et al., 1998). Truncation of the N terminus in SusC also results in a considerable increase in predicted hydrophobicity of this region of the protein compared with the SusA, -B, and -D proteins (data not shown).
To determine whether the amino acid sequence divergence in SusC might be significant for protein structure and function, a protein structural prediction server (JPRED) was used to compare SusC with SusA, -B, and -D predicted structures in these regions. Of particular note is the region from amino acids 36 to 59 of the SusC sequence, which are unique to this protein compared with A,B,D and most other predicted Sus sequences in the database. In the case of SusC, a predicted α-helix in the SusA protein becomes a β-sheet, followed in the structure by the converse change. It is not known how these changes in structure would affect protein function, as they are distant from the predicted substrate-binding sites (likely to be between amino acids 270 and 329), a region highly conserved across Sus gene sequences and showing homology to the UDPG-binding site in the related enzyme Suc phosphate synthase (McMichael et al., 1993; Lunn and MacRae, 2003).
At the C terminus, considerable sequence divergence is seen in the SusC sequence from amino acids 765 to 790. Once again, structural prediction suggests that compared with SusA, -B, and -D, SusC would contain an α-helix in place of a β-sheet. Once again, the functional significance of this region of the protein is unclear, although it is close to the predicted glycosyl transferase domain (amino acids 549–737; identified with InterProScan). There are no predicted phosphorylation sites in this region of the Sus protein sequence. It is not clear how these regions relate to the predicted structures identified in maize Sus by Hardin et al. (2006).
Targeting of Sus during Secondary Cell Wall Synthesis
Despite the large effort that has been made to study the so-called PM association of Sus in a number of species (Winter and Huber, 2000; Haigler et al., 2001), firm evidence for partitioning of a quantitatively significant portion of Sus to the PM of cotton fiber is difficult to find in the literature. The standard technique of extraction of a “pellet” fraction with EGTA following centrifugation varies in the efficacy of extraction of Sus in the literature, even from the same groups working in cotton (Amor et al., 1995; Haigler et al., 2001, Komina et al., 2002). There is little conclusive evidence of “docking” of Sus with the cellulose synthase rosette on the plasma membrane using either the standard techniques of immunoprecipitation or techniques such as fluorescence resonance energy transfer (Dixit et al., 2006); however, recently, evidence has been published that Sus is an integral component of the cellulose synthase rosette using detergent-soluble granular particles from the plasma membrane of bean epicotyls (Fujii et al., 2010). Localization of a significant proportion of SusC to the cell wall, suggested from both the protein mass spectrometry of subcellular fractions and the immunolocalization experiments shown here may shed light on some of these earlier observations. It is not clear why EGTA should specifically release Sus from the plasma membrane. It has been suggested that the interaction with the membrane may involve calcium, although evidence for this is circumstantial and the membrane binding of Sus varies in reversibility markedly between species (Winter and Huber 2000; Komina et al., 2002). It is well known that EGTA loosens the interactions between pectins and other cell wall components and as such is a standard reagent in buffers used to elute proteins from cell walls (Zhao et al., 2008, and refs. therein). Clearly, from Figure 5, a simple buffer excluding EGTA eluted SusA and -C from the cell wall fraction of cotton fiber, but it is difficult to elute all the SusC protein in the cell wall pellets, suggesting that this isoform is more tightly bound to the cell wall fraction. Little or no SusA, -B, or -D is present in successive washes of cell wall material, suggesting that these proteins are likely to be either loosely bound or even cytosolic isoforms binding nonspecifically to this fraction (Figs. 7 and 8). It would be difficult to design definitive experiments to determine which of these options is correct. However, irrespective of this uncertainty, the presence of significant amounts of Sus in the cell wall fraction would certainly call into question the standard method of estimating PM-Sus levels by comparing soluble Sus levels with EGTA washes of the pellet fraction of crude cotton fiber extracts, or other tissues for that matter.
The immunolocalization of SusC using the isozyme-specific antiserum (Fig. 9) also indicated that a large proportion of SusC is present in the cell wall of fiber at 18 to 20 DAF. This is entirely consistent with the cell wall pellet washing experiments described above. The other isoforms of Sus, in particular SusA, were also shown to be present in this compartment at all stages of development using a nondiscriminatory anitiserum (data not shown). Localization of Sus to the fiber cell wall of cultured ovules has been reported several times (Amor et al., 1995; Haigler et al., 2001; Salnikov et al., 2003); however, the significance of this observation has not been emphasized, seemingly because the observations were interpreted to be artifacts of fixation with the cell wall label, indicating plasma membrane-bound Sus (Amor et al., 1995; Haigler et al., 2001). Recent work on immunolocalization of Sus in pollen tubes has also suggested that Sus is present in cell walls (Persia et al., 2008). This evidence, however, is not compelling, based on the presence of the protein in the postnuclear supernatant fraction of cell extracts and antibody labeling of detergent-treated pollen tubes, in which the site of labeling could be in the cell wall, plasmalemma, or cytosol. (Persia et al., 2008). In cotton fiber, immunolocalization experiments at later stages of fiber development reported here and in the literature (Salnikov et al., 2003) are much clearer, consistently showing almost no Sus labeling in the cytoplasm, despite reasonable preservation of this compartment. In our experiments and in those of Salnikov et al. (2003), greater than 90% of Sus labeling is in the “exoplasmic zone” (i.e. cell wall apoplast). Quantification of this labeling at the plasma membrane surface (internally or externally) is difficult; however, both the fluorescent immunolocalization carried out here and the previous immunogold work (Salnikov et al., 2003; Ruan, 2007) show that a large percentage of Sus is external to the plasma membrane “zone” as defined by Salnikov et al. (2003).
There is convincing evidence that “particulate” or microsomal Sus is also strongly associated with microtubules and F-actin filaments (for review, see Winter and Huber, 2000). The region of SusC corresponding to the putative actin-binding site identified in other species (Winter and Huber 2000) has the following consensus amino acid sequence: [*,E,D]-D-[V,A]-[A,G,S,T]-X-E-[L,V,I]-[T,S,A,M]-[K,R,G,M,L]-E-[*,L,M]-[Q,N], where boldface amino acids are polar-hydrophylic and italic amino acids are nonpolar-hydrophobic at neutral pH. The sequence motif corresponding to this F-actin-binding domain in SusC is 383-KDVAAEITKEFQ-394. The amino acids highlighted in boldface are unique to SusC. Of interest is that 383-K (Lys) has a basic side chain while E (Glu) and D (Asp) have acidic side chains. The unique K substitution in the SusC sequence may indicate that this protein may no longer bind to the F-actin filaments. This is also compounded by the presence of F (Phe) in this motif, also unique to SusC, containing a side chain more voluminous than L and M, which are present in SusA, -B, and -D and their counterparts in other species, potentially leading to added instability in any actin-SusC interaction.
The Roles of SusA and -C in Cotton Fiber Cell Wall Synthesis
Conclusive elucidation of the role of SusC in fiber cellulose biosynthesis awaits the results of isoform-specific gene suppression and overexpression experiments currently underway for cotton; however, the data presented here may support some informed speculation. Previous cotton Sus gene suppression experiments in our laboratory, using a construct targeted to SusA, showed a clear disruption of fiber and seed development, producing shorter fibers (Ruan et al., 2003) containing less cellulose (Ruan, 2007). Due to the severity of phenotypic effects, it was difficult to isolate the impact of low SusA on secondary cell wall synthesis from pleiotropic developmental effects on fiber growth and primary wall synthesis, and it is also unknown to what degree this gene construct cross-silenced SusC gene expression. The transcriptional analysis of Sus expression during fiber development shown here and the analysis of SusA/B/D and SusC abundance in cDNA libraries from cotton fiber suggest that the SusA/B/D isoforms are expressed highly during fiber elongation, falling off during secondary cell wall synthesis (Fig. 4). In contrast, we have shown that SusC is absent at both the transcript and protein levels in early fiber development but highly expressed in later fiber development (Figs. 4, 5, and 9). This high-level expression of SusC specifically during secondary cell wall synthesis in cotton fibers, its targeted expression in other tissues that undergo secondary cell wall synthesis (such as stem; Fig. 4), and its distant phylogenetic relationship to Sus in most herbaceous species (Fig. 3) provide strong circumstantial evidence for its specific involvement in secondary cell wall cellulose synthesis.
If we accept that the data shown here cast doubt on the precise location of SusC (and SusA) but accept that particulate or “P-Sus” is important for secondary cell wall synthesis, we can calculate how much P-Sus might be required in cotton fiber to support flux. Haigler et al. (2005) calculated that 80% of sugar flux to cotton fibers is used for cellulose synthesis during the peak secondary wall synthesis stage. Their model predicts that greater than 80% of Sus should be “membrane associated.” Extraction of cell wall/membrane fractions isolated with EGTA washing from a variety of species commonly yields figures of only 10% to 50% of total Sus protein associated with the particulate fraction, with microsomal and more purified preparations yielding estimates at the lower end of this range (Amor et al., 1995; Winter et al., 1997; Haigler et al., 2001, and refs. therein). This disparity between standard biochemical methods for determining Sus subcellular targeting and cell biological techniques may be explained by the data shown here. It is apparent from Figure 9 that the major Sus isoforms present in cotton fiber bind differentially to cell walls. The soluble fraction, described by Amor et al. (1995) and subsequent authors, may in fact be contaminated with a large proportion of loosely bound SusA and SusC. A significant proportion of SusC will also be retained in the pellet fraction and discarded when microsomal fractions are prepared. Thus, membrane-bound Sus, on which a great deal of research has been conducted, may be a very minor proportion of particulate Sus, the majority of which is bound to varying degrees within the cell wall apoplast. The role of SusC would be clearer if this isoform alone were targeted to the cell wall. This is not the case, however, and SusA/B/D are also found in this fraction to varying degrees, shown in both destructive and in situ (data not shown) immunological approaches and by diagnostic protein mass spectrometry carried out here.
The implications of a large pool of cell wall-associated, extracellular Sus are manifold, regardless of the identity of the Sus isoforms involved. An active apoplastic Sus could function in much the same way as cell wall-associated invertase (for review, see Koch, 2004); however, it would require apoplastic UDP as a substrate in addition to Suc. The sugar composition of the apoplastic fluid of cotton locules 18 to 20 was determined here using a protocol developed for leaves (Nadwodnik and Lohaus, 2008; data not shown) and HPLC-pulsed-amperometric detection (Ruuska et al., 2006). While UDP could not be measured in this system, Suc, Glc, and Fru were abundant (data not shown). High sugar levels are not unusual in the apoplast of other tissues, particularly leaves (Canny, 1995), and observations of high extracellular Suc levels in leaf apoplast gave rise to the theory of apoplastic loading of leaf phloem (for review, see Giaquinta, 1983; Van Bel, 1993; Turgeon and Wolf, 2009). In cotton fiber, the detailed postphloem pathway of sugar import into the fiber over development has only recently been elucidated (Ruan et al., 2000, 2001). Suc is proposed to move from the phloem through the outer seed coat symplastically to the fiber foot, where it either enters the fiber cell through plasmodesmata or via active transport, depending on developmental stage (Ruan et al., 2001). The origin of sugars in the fiber apoplast, contiguous with the boll cavity, however, is less certain. When cotton bolls are dissected for fiber removal, liquid is obviously present in this space, declining with fiber maturity (data not shown). Further work is required to determine if apoplastic Suc can be used by a cell wall-localized Sus and whether this is a major alternative pathway for the generation of UDP-Glc for cellulose synthesis.
An alternative role for Sus in the apoplasm could also be the synthesis of callose, and this has been suggested for the Sus5 and Sus6 isoforms in Arabidopsis (Barratt et al., 2009). Callose synthesis begins near the onset of secondary wall deposition in cotton, and the amount of callose remains high throughout secondary wall deposition (Maltby et al., 1979; Waterkeyn, 1981). Experiments with developmentally similar Gossypium arboreum showed that callose synthesis persists throughout secondary wall deposition in cotton fiber (Pillonel et al., 1980). However, callose flux is comparatively low compared with cellulose, and the reported timing of the onset of synthesis does not coincide with the period when SusC dominates the fiber Sus protein profile (Figs. 5–9).
What triggers cell wall targeting of Sus remains unresolved. One possibility is that heterotetrameric SusA-SusC protein is required for secretion to the cell wall or for the activity of Sus in the apoplasm. Sus is known to form heterotetramers in maize (Duncan et al., 2006), and the unique nature of the N and C termini of the SusC protein may provide targeting information, although no secretion signals or obvious targeting domains are present. Differential targeting of the heterotetramer would explain observations in the literature that particulate Sus is only a major isoform of total fiber Sus during secondary wall synthesis, and this hypothesis would not require developmentally controlled phosphorylation. While we observed some SusA, -B, and -D in the apoplasm at all developmental stages, the MS measurements made here only allow relative, not absolute, comparisons to be made. Regardless of mechanism, the strict developmental regulation of SusC expression would suggest a pivotal role at the secondary cell wall synthesis stage.
CONCLUSION
The role of Sus in secondary cell wall cellulose synthesis has been a complex area of research, and mechanisms controlling the partitioning of Sus between soluble and particulate or membrane-associated Sus have remained elusive. Despite assertions that one Sus family, more than 93% identical in amino acid sequence between its members, dominates the Sus isoforms found in developing cotton fiber (Haigler et al., 2001), we have isolated a novel Sus from cotton fiber (termed SusC) that is present at high levels in the fiber cell wall apoplast during secondary cell wall synthesis. While this isoform dominates the Sus protein pool during this stage, it is not exclusive in its partitioning between cellular compartments. The precise role of this Sus in cell wall biosynthesis remains to be elucidated.
MATERIALS AND METHODS
Plant Material
Cotton (Gossypium hirsutum ‘Coker 315’) seeds were grown under naturally lit greenhouse conditions with partial temperature control (25°C–30°C during the day and 18°C–22°C during the night). About 100 g per pot of controlled-release fertilizer (Osmocote; Scotts) was applied once every 14 d. The plants were watered once per day. Standard pest- and disease-control practices were used. Cotton boll age was determined by tagging the flowering truss when the flower was fully opened.
RNA and Protein Extraction
Cotton tissues were harvested from glasshouse-grown plants, snap frozen in liquid nitrogen, and stored at −80°C until needed. The cotton fibers were separated from seed under liquid nitrogen using fine-grinding techniques and forceps. The remaining fiber tissue was then ground to a fine powder under liquid nitrogen with a mortar and pestle.
Protein was extracted by adding frozen ground powder to 4 volumes of extraction buffer (25 mm HEPES-KOH, pH 7.3, 0.1% polyethylene glycol-6000, 250 mm Suc, 10 mm leupeptin, 100 mm phenylmethylsulfonyl fluoride, and 1 mm dithiothreitol) and grinding for a further 1 min. This extract was then centrifuged for 15 min at 4,000 rpm at 4°C. The supernatant was filtered through one layer of Miracloth (Calbiochem), and the resultant filtrate is the crude protein extract. The remaining pellet was subsequently washed four times with 2 volumes of extraction buffer to remove any remaining soluble proteins, with centrifugation for 15 min at 4,000 rpm at 4°C after each wash. After four washes, the pellet was further solubilized using solubilization buffer containing 0.1% Triton X-100, 5 mm EDTA, 1 mm phenylmethylsulfonyl fluoride, 25 mm HEPES, pH 7.5, 1 m NaCl, and 1 mm dithiothreitol. Microsomal vesicles and plasma membrane were extracted from the prepared crude extract by published methods (Carlson and Chourey, 1996) using a Beckman TL-100 tabletop ultracentrifuge and a TLS-55 swing-out rotor at 55,000 rpm (201,078 relative centrifugal force average) for 2 h. Apoplastic protein was isolated by placing freshly harvested, intact cotton locules into a tube containing 4 volumes of extraction buffer. This was gently shaken on a platform rocker (Seoulin Mylab shaker SLS4) set at 20 rpm at 4°C overnight. The locules and buffer were filtered through one layer of Miracloth, and the resultant supernatant was the putative apoplastic fraction. Contamination of the apoplastic fraction with cytoplasmic components was assessed in two ways. First, Glc-6-P dehydrogenase activity was assayed in the apoplastic washes and in the intact fiber; apoplast activities were less than 1% of whole fiber values. Second, fibers were labeled with the membrane-permeable dye 6-carboxyfluorescein diacetate (Ruan et al., 2003), subjected to the apoplastic wash, and then the appearance of dye in the wash was monitored by fluorescence microscopy over time. This dye was cleaved intracellularly to become fluorescent and membrane impermeable. Dye passing out of fibers into the wash medium can only do so if fiber plasma membranes are compromised. Fluorescence due to dye leakage was undetectable in the apoplastic wash.
RNA was extracted by previously published methods (Wu et al., 2002) and treated with DNaseI (Qiagen) to remove any remaining contaminating genomic DNA.
cDNA Cloning and Sequence Analysis
Sus amino acid and deduced amino acid sequences from soybean (Glycine max; accession no. AF030231), mung bean (Vigna radiata; D10266), bean (Phaseolus vulgaris; AF315375), cotton (U73588), satsuma mandarin (Citrus unshiu; AB022092), Arabidopsis (Arabidopsis thaliana; NM_122090), potato (Solanum tuberosum; M18745), and maize (Zea mays; X02382, X02400, L22296, and AY124703) were aligned using the Pileup program from Genetics Computer Group Wisconsin Package to find any possible regions of good amino acid homology between all the sequences. An area of homology (FDPKFNI) was identified that corresponded to amino acid 516 in the cotton sequence. This region was then compared at the nucleotide level across all the sequences, and good homology was also found at this level. Two oligonucleotides were designed based on this homology: primer A (5′-TTTGATCCCAAATTCAACAT-3′) and primer B (5′-TTTGATCCTAAATTCAACAT-3′). These oligonucleotides were used in combination with an oligo(dT)30 primer that included an EcoRI restriction site (underlined; 5′-CGunderline>GAATTC/underline>T30N-3′) to isolate partial Sus cDNA sequences.
Five micrograms of total RNA from 10-DAF fiber and 20-DAF fiber was reverse transcribed to make first-strand cDNA using ThermoScript reverse transcriptase (Invitrogen) and 50 μmol of oligo(dT)30 primer and incubating at 50°C for 1 h. Aliquots of cDNA, equivalent to 0.1 μg of total RNA, were PCR amplified with 20 pmol of either primer A or primer B in combination with the oligo(dT)30 primer using HotStarTaq DNA polymerase (Qiagen). Low-stringency PCR amplification was performed with a Corbett FTS 4000 thermal sequencer using the following program: 95°C for 15 min (one cycle); 94°C for 45 s, 48°C for 45 s, and 72°C for 1.5 min (30 cycles); and then 72°C for 10 min and 25°C for 1 min (one cycle). The products obtained from these reactions were purified using the Wizard PCR Preps DNA Purification System (Promega). The products were subcloned into pGEMT-Easy (Promega) and transformed into Escherichia coli TOP10F′ competent cells (Invitrogen) following the manufacturer’s protocols and sequenced using Big Dye Terminator sequencing (Applied Biosystems).
Three novel Sus cDNA sequences (Sus B, C, and D) were identified, and specific primers complementary to these sequences were designed in order to isolate the full-length cDNA clones. The primers used were as follows: SusB, 5′-GGAAATCACAATCTTTTGTTGGAATCCAGG-3′; SusC, 5′-GCAATCAATGGGACCAAACCCAGAGTTC-3′; SusD, 5′-CAGATGTTGAAACAATGCCCAAAACATGAAC-3′. Full-length clones of SusB, -C, and -D were isolated from 20-DAF total RNA by 5′-RACE using the gene-specific primers and BD SMART RACE technology (BD Biosciences Clontech). The products were subcloned into pGEMT-Easy (Promega) and transformed into E. coli TOP10F′ competent cells (Invitrogen) following the manufacturer’s protocols and sequenced using Big Dye Terminator sequencing (ABI). Genomic sequences for SusC were amplified from various diploid and tetraploid Gossypium species, including G. arboreum, G. raimondii, G. herbaceum, G. kirkii, G. hirsutum (FM966), G. barbadense (Pima S7), G. darwinii, G. tomentosum, and G. mustelinum, using the primers C-type-5′UTR (5′-CCCTTCTGCCATTTCAGGAACC-3′) and C-type-3′UTR (5′-AATGGGACCAAACCCAGAGTTC-3′), and PCR products cloned into the pCR4-TOPO vector (Invitrogen) and four independent clone inserts were sequenced to make a consensus sequence. In all the tetraploids except G. mustelinum, which had two different SusC genes (one more similar to the diploid D genome SusC and the other to the A genome SusC), only one version (more similar to the D genome form) of SusC was recovered.
A total of 29 putative plant Sus protein sequences from dicots, monocots, and mosses whose complete genomes have been sequenced (GenBank RefSeq genomes) were retrieved from the National Center for Biotechnology Information and aligned with the cotton SusA, -B, -C, and -D sequences using ClustalW within the MEGA 4.0 software package (Molecular Evolutionary Genetics Analysis; Kumar et al., 2001). A phylogenetic tree was drawn with the same package using the neighbor-joining method with complete deletion; 1,000 replicates were used for bootstrap analysis, and the cutoff value was 50%. Phylogenetic comparisons between the different SusC proteins from tetraploid and diploid cotton species were treated in the same way.
Semiquantitative RT-PCR
To determine relative transcript levels of each of the Sus genes, specific primers were designed to amplify fragments specific for each gene. The sequences of the primer pairs for each Sus gene were as follows: Sus A (62 bp of 3′-untranslated region [UTR]), 5′-GACAAGATGAAATACAAAGGAGC-3′ and 5′-CATTGGGCCGGTTTTTCTTGGAG-3′; Sus B (105 bp of 3′-UTR), 5′-GGCTTTTTCTTGTCCGACCATA-3′ and 5′-AAAGGAAGAGGCGGGTTTTCC-3′; Sus C (150 bp of 3′-UTR), 5′-CAATGGGACCAAACCCAGAGTTC-3′ and 5′-AGCAAAAGGCTGCTTGGAAAC-3′; Sus D (1,030 bp), 5′-TTTGATCCTAAATTCAACAT-3′ and 5′-CAGATGTTGAAACAATGCCCAAAACATGAAC-3′.
Five micrograms of total RNA from leaf, petal, stem, and fiber from 8, 10, and 13 DAF (spanning the elongation and primary cell wall synthesis stages) and from 17 and 21 DAF (representing secondary cell wall synthesis stages) was reverse transcribed to make first-strand cDNA using 50 mmol of oligo(dT)30 primer and ThermoScript reverse transcriptase (Invitrogen) and incubating at 50°C for 1 h. Aliquots of cDNA, equivalent to 0.1 μg of total RNA, were PCR amplified using 20 pmol of both primers from each primer set and HotStarTaq DNA polymerase (Qiagen). PCR amplification was performed with a Corbett FTS 4000 thermal sequencer using the following program: 95°C for 15 min (one cycle); 94°C for 30 s, 48°C for 30 s, and 72°C for 1.5 min (30 cycles); and then 72°C for 10 min and 25°C for 1 min (one cycle). Normalization of all the templates was done using primers for a cotton ubiquitin gene (CK738219), producing a 200-bp fragment: 5′-CAAGACAAGGAAGGCATCCCAC-3′ and 5′-TCGGAACTCTCCACCTCCAAAG-3′. Conditions for normalizing PCR were the same as for the Sus genes, but using only 10 pmol of primers, an annealing temperature of 65°C, a 1-min extension time, and only 20 cycles.
Protein Separation and Identification by Mass Spectrometry
One-dimensional SDS-PAGE of standards and samples (as above) was performed on precast gradient gels (Invitrogen: MultiMark Multi-Colored Standard [LC5725], NuPAGE 4–12% Bis-Tris Gel, 1-mm × 10 well [NP0321Box], and NuPAGE MOPS running buffer [NP0001]) according to the manufacturer’s instructions for about 50 min at 200 V; then they were transferred to nitrocellulose membranes (Amersham Biosciences) by western blotting. The membranes were probed with either a nonspecific antibody raised against a rice SUS2 peptide (provided by T. Hirose, National Agricultural Research Center, Japan), used at a dilution of 1:10,000 in Tris-buffered saline buffer, or an antibody raised against oligopeptides specific to the SusC protein, used at a dilution of 1:5,000 in Tris-buffered saline buffer.
Sus protein bands, identified by western blotting of duplicate gels, were excised from Coomassie blue-stained gels. Proteins were digested in-gel with trypsin, and the resultant peptides were analyzed using an Agilent 1100 capillary liquid chromatography system and an Agilent XCT ion-trap mass spectrometer as described by Campbell et al. (2008).
Mass spectral data sets were used to search sequence databases using Agilent’s Spectrum Mill software (Revision A.03.02.060). False-positive identifications were avoided by using the software’s stringent “autovalidation” default settings. This includes a requirement for the peptide matches to be considerably better than the best match against the reversed database and various weightings favoring more probable ionization and fragmentation patterns (“proton mobility scoring”). The data were first used to search a small database of possible contaminants such as keratins and trypsin. Autovalidation with this data set removed from consideration spectra of contaminant peptides that might otherwise have produced low-quality matches to cotton sequences. The unmatched data were then used to search an in-house database of cotton cDNA sequences combined with public domain sequences from cotton and its near relatives, again with a round of autovalidation. Sus isoforms were identified with autovalidation by at least two distinct peptides in all samples reported here. Only tryptic peptides were considered, allowing for the possibility of one missed cleavage and oxidized Met.
Immunolocalization
For immunolocalization of Sus in resin sections, cotton ovules were dissected into a drop of the fixative, comprising 2% (v/v) paraformaldehyde and 0.1% (v/v) glutaraldehyde in 25 mm phosphate buffer, pH 7.2, and fixed for 2 h at room temperature. After washing in buffer, the ovules were dehydrated in an ethanol series and embedded in LR White resin (medium grade; Alltech). Semithin sections were incubated with Sus antibody or preimmune serum in phosphate-buffered saline (PBS) that had been reverse affinity purified by filtering through cotton wool, which removes background labeling caused by serum components binding to cellulose. After incubation in Sus antibody for 2 h, sections were rinsed in PBS and incubated in Alexa-488-tagged anti-mouse antibody (Invitrogen). Cell walls were highlighted by staining in 0.05% Calcofluor white for 2 min prior to imaging on a SP2 confocal laser scanning microscope (Leica Microsystems)
For analysis of plasmolyzed fibers, whole locules, containing several developing seeds, were dissected from 10-DAF or 18- to 20-DAF cotton bolls and processed as described by Preuss et al. (2003). Briefly, the locules were fixed in 4% formaldehyde and 0.1% glutaraldehyde in 50 mm PIPES, 1 mm MgSO4, and 5 mm EGTA, pH 6.9, containing 0.1% Tween 20 and 0.3 m mannitol for 1 h at room temperature. Some replicates contained 0.6 m mannitol to induce plasmolysis. After three washes in appropriate buffer, individual ovules were carefully dissected into individual wells of a multiwell plate for further processing. Ovules were then incubated in 2% cellulase (Onozuka) R-10 (Yakult Pharmaceutical) and 0.1% macerozyme R-10 (Yakult Pharmaceutical) in buffer for 20 to 30 min for 10-DAF fibers and for 45 to 60 min for 18- to 20-DAF fibers. After washing in buffer, tissue was incubated for 1 h in 1% Triton X-100 in buffer at room temperature. After three more washes in buffer, the tissues were plunged into methanol at −20C for 10 min. After rehydration in PBS, ovules were incubated in Sus antibody diluted 1:50 in PBS for 2 h, rinsed in PBS, incubated in Alexa-488 anti-mouse secondary antibody for 2 h, rinsed in PBS, and then immediately imaged on Leica SP2 confocal laser scanning microscope.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers JN248431 to JN248440 and JN376125 to JN376127.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Sequence alignments of Sus protein sequences used to generate Figure 3A.
Supplemental Figure S2. Sequence alignments of cotton Sus protein sequences used to generate Figure 3B.
Supplemental Figure S3. Sequence alignments of SusA, -B, -C, and -D proteins showing sequence coverage of diagnostic peptides used for mass spectrometry.
Supplemental Table S1. All known Sus peptides identified in individual experiments identifying which isoform(s) they are present in; spectral intensities shown were used to calculate the total spectral intensity for SusC versus SusA, -B, and -D in each experiment for Figures 6 and 8.
Acknowledgments
We thank Colin Jenkins (CSIRO Plant Industry, Australia) for assistance with sugar analysis and Mark Talbot (CSIRO Plant Industry, Australia) for microscopy work. We thank Dr. Tatsuro Hirose (Department of Rice Research, National Agricultural Research Center, Japan) for providing the anti-rice SUS2 antibody.
References
- Amor Y, Haigler CH, Johnson S, Wainscott M, Delmer DP. (1995) A membrane-associated form of sucrose synthase and its potential role in synthesis of cellulose and callose in plants. Proc Natl Acad Sci USA 92: 9353–9357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson-Gunnerås S, Mellerowicz EJ, Love J, Segerman B, Ohmiya Y, Coutinho PM, Nilsson P, Henrissat B, Moritz T, Sundberg B. (2006) Biosynthesis of cellulose-enriched tension wood in Populus: global analysis of transcripts and metabolites identifies biochemical and developmental regulators in secondary wall biosynthesis. Plant J 45: 144–165 [DOI] [PubMed] [Google Scholar]
- Arpat AB, Waugh M, Sullivan JP, Gonzales M, Frisch D, Main D, Wood T, Leslie A, Wing RA, Wilkins TA. (2004) Functional genomics of cell elongation in developing cotton fibers. Plant Mol Biol 54: 911–929 [DOI] [PubMed] [Google Scholar]
- Barratt DH, Derbyshire P, Findlay K, Pike M, Wellner N, Lunn J, Feil R, Simpson C, Maule AJ, Smith AM. (2009) Normal growth of Arabidopsis requires cytosolic invertase but not sucrose synthase. Proc Natl Acad Sci USA 106: 13124–13129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basra AS, Malik CP. (1984) Development of the cotton fiber. Int Rev Cytol 89: 65–113 [Google Scholar]
- Baud S, Vaultier MN, Rochat C. (2004) Structure and expression profile of the sucrose synthase multigene family in Arabidopsis. J Exp Bot 55: 397–409 [DOI] [PubMed] [Google Scholar]
- Bieniawska Z, Paul Barratt DH, Garlick AP, Thole V, Kruger NJ, Martin C, Zrenner R, Smith AM. (2007) Analysis of the sucrose synthase gene family in Arabidopsis. Plant J 49: 810–828 [DOI] [PubMed] [Google Scholar]
- Campbell PM, Cao AT, Hines ER, East PD, Gordon KHJ. (2008) Proteomic analysis of the peritrophic matrix from the gut of the caterpillar, Helicoverpa armigera. Insect Biochem Mol Biol 38: 950–958 [DOI] [PubMed] [Google Scholar]
- Canny MJ. (1995) Apoplastic water and solute movement: new rules for an old space. Annu Rev Plant Physiol Plant Mol Biol 46: 215–236 [Google Scholar]
- Carlson SJ, Chourey PS. (1996) Evidence for plasma membrane-associated forms of sucrose synthase in maize. Mol Gen Genet 252: 303–310 [DOI] [PubMed] [Google Scholar]
- Coleman HD, Ellis DD, Gilbert M, Mansfield SD. (2006) Up-regulation of sucrose synthase and UDP-glucose pyrophosphorylase impacts plant growth and metabolism. Plant Biotechnol J 4: 87–101 [DOI] [PubMed] [Google Scholar]
- Coleman HD, Yan J, Mansfield SD. (2009) Sucrose synthase affects carbon partitioning to increase cellulose production and altered cell wall ultrastructure. Proc Natl Acad Sci USA 106: 13118–13123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit R, Cyr R, Gilroy S. (2006) Using intrinsically fluorescent proteins for plant cell imaging. Plant J 45: 599–615 [DOI] [PubMed] [Google Scholar]
- Duncan KA, Hardin SC, Huber SC. (2006) The three maize sucrose synthase isoforms differ in distribution, localization, and phosphorylation. Plant Cell Physiol 47: 959–971 [DOI] [PubMed] [Google Scholar]
- Etxeberria E, Gonzalez P. (2003) Evidence for a tonoplast-associated form of sucrose synthase and its potential involvement in sucrose mobilization from the vacuole. J Exp Bot 54: 1407–1414 [DOI] [PubMed] [Google Scholar]
- Fallahi H, Scofield GN, Badger MR, Chow WS, Furbank RT, Ruan Y-L. (2008) Localization of sucrose synthase in developing seed and siliques of Arabidopsis thaliana reveals diverse roles for SUS during development. J Exp Bot 59: 3283–3295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii S, Hayashi T, Mizuno K. (2010) Sucrose synthase is an integral component of the cellulose synthesis machinery. Plant Cell Physiol 51: 294–301 [DOI] [PubMed] [Google Scholar]
- Giaquinta RT. (1983) Phloem loading of sucrose. Annu Rev Plant Physiol 34: 347–387 [Google Scholar]
- Haigler CH, Ivanova-Datcheva M, Hogan PS, Salnikov VV, Hwang S, Martin K, Delmer DP. (2001) Carbon partitioning to cellulose synthesis. Plant Mol Biol 47: 29–51 [PubMed] [Google Scholar]
- Haigler CH, Zhang D, Wilkerson CG. (2005) Biotechnological improvement of cotton fibre maturity. Physiol Plant 124: 285–294 [Google Scholar]
- Hardin SC, Duncan KA, Huber SC. (2006) Determination of structural requirements and probable regulatory effectors for membrane association of maize sucrose synthase1. Plant Physiol 141: 1106–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardin SC, Winter H, Huber SC. (2004) Phosphorylation of the amino terminus of maize sucrose synthase in relation to membrane association and enzyme activity. Plant Physiol 134: 1427–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauch S, Magel E. (1998) Extractable activities and protein content of sucrose-phosphate synthase, sucrose synthase and neutral invertase in trunk tissues of Robinia pseudoacacia L. are related to cambial wood production and heartwood formation. Planta 207: 266–274 [Google Scholar]
- Heim U, Weber H, Bäumlein H, Wobus U. (1993) A sucrose-synthase gene of Vicia faba L.: expression pattern in developing seeds in relation to starch synthesis and metabolic regulation. Planta 191: 394–401 [DOI] [PubMed] [Google Scholar]
- King SP, Lunn JE, Furbank RT. (1997) Carbohydrate content and enzyme metabolism in developing canola (Brassica napus L.) siliques. Plant Physiol 114: 153–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch KE. (2004) Sucrose metabolism: regulatory mechanisms and pivotal roles in sugar sensing and plant development. Curr Opin Plant Biol 7: 235–246 [DOI] [PubMed] [Google Scholar]
- Komina O, Zhou Y, Sarath G, Chollet R. (2002) In vivo and in vitro phosphorylation of membrane and soluble forms of soybean nodule sucrose synthase. Plant Physiol 129: 1664–1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Jakobsen IB, Nei M. (2001) MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17: 1244–1245 [DOI] [PubMed] [Google Scholar]
- Lunn JE, MacRae E. (2003) New complexities in the synthesis of sucrose. Curr Opin Plant Biol 6: 208–214 [DOI] [PubMed] [Google Scholar]
- Maltby D, Carpita NC, Montezinos D, Kulow C, Delmer DP. (1979) β-1,3-Glucan in developing cotton fibers: structure, localization and relationship of synthesis to that of secondary wall cellulose. Plant Physiol 63: 1158–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMichael RW Jr, Klein RR, Salvucci ME, Huber SC. (1993) Identification of the major regulatory phosphorylation site in sucrose-phosphate synthase. Arch Biochem Biophys 307: 248–252 [DOI] [PubMed] [Google Scholar]
- Nadwodnik J, Lohaus G. (2008) Subcellular concentrations of sugar alcohols and sugars in relation to phloem translocation in Plantago major, Plantago maritima, Prunus persica, and Apium graveolens. Planta 227: 1079–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai T, Konishi T, Zhang XQ, Chollet R, Tonouchi N, Tsuchida T, Yoshinaga F, Mori H, Sakai F, Hayashi T. (1998) An increase in apparent affinity for sucrose of mung bean sucrose synthase is caused by in vitro phosphorylation or directed mutagenesis of Ser-11. Plant Cell Physiol 39: 1337–1341 [DOI] [PubMed] [Google Scholar]
- Nilsson R, Bernfur K, Gustavsson N, Bygdell J, Wingsle G, Larsson C. (2010) Proteomics of plasma membranes from poplar trees reveals tissue distribution of transporters, receptors, and proteins in cell wall formation. Mol Cell Proteomics 9: 368–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins PD, Miller CA, Kuhlmann FE. (2005) Comparison of different approaches for the label-free relative quantitation of proteins. Application note, Agilent Technologies; www.agilent.com/chem (August 3, 2011) [Google Scholar]
- Persia D, Cai G, Del Casino C, Faleri C, Willemse MT, Cresti M. (2008) Sucrose synthase is associated with the cell wall of tobacco pollen tubes. Plant Physiol 147: 1603–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillonel C, Buchala AJ, Meier H. (1980) Glucan synthesis by intact cotton fibres fed with different precursors at the stages of primary and secondary cell wall formation. Planta 149: 306–312 [DOI] [PubMed] [Google Scholar]
- Preuss ML, Delmer DP, Liu B. (2003) The cotton kinesin-like calmodulin-binding protein associates with cortical microtubules in cotton fibers. Plant Physiol 132: 154–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Y-L. (2007) Rapid cell expansion and cellulose synthesis regulated by plasmodesmata and sugar: insights from the single-celled cotton fibre. Funct Plant Biol 34: 1–10 [DOI] [PubMed] [Google Scholar]
- Ruan Y-L, Chourey PS, Delmer DP, Perez-Grau L. (1997) The differential expression of sucrose synthase in relation to diverse patterns of carbon partitioning in developing cotton seed. Plant Physiol 115: 375–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Y-L, Llewellyn DJ, Furbank RT. (2000) Pathway and control of sucrose import into initiating cotton fibre cells. Aust J Plant Physiol 27: 795–800 [Google Scholar]
- Ruan Y-L, Llewellyn DJ, Furbank RT. (2001) The control of single-celled cotton fiber elongation by developmentally reversible gating of plasmodesmata and coordinated expression of sucrose and K+ transporters and expansin. Plant Cell 13: 47–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Y-L, Llewellyn DJ, Furbank RT. (2003) Suppression of sucrose synthase gene expression represses cotton fiber cell initiation, elongation, and seed development. Plant Cell 15: 952–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Y-L, Llewellyn DJ, Liu Q, Xu X-M, Wu L-M, Wang L, Furbank RT. (2008) Expression of sucrose synthase in developing endosperm is essential for early seed development in cotton. Funct Plant Biol 35: 382–393 [DOI] [PubMed] [Google Scholar]
- Ruuska SA, Rebetzke GJ, van Herwaarden AF, Richards RA, Fettell NA, Tabe L, Jenkins CLD. (2006) Genotypic variation in water-soluble carbohydrate accumulation in wheat. Funct Plant Biol 33: 799–809 [DOI] [PubMed] [Google Scholar]
- Salnikov VV, Grimson MJ, Delmer DP, Haigler CH. (2001) Sucrose synthase localizes to cellulose synthesis sites in tracheary elements. Phytochemistry 57: 823–833 [DOI] [PubMed] [Google Scholar]
- Salnikov VV, Grimson MJ, Seagull RW, Haigler CH. (2003) Localization of sucrose synthase and callose in freeze-substituted secondary-wall-stage cotton fibers. Protoplasma 221: 175–184 [DOI] [PubMed] [Google Scholar]
- Subbaiah CC, Palaiappan A, Duncan DM, Rhoads DM, Huber SC, Sachs MM. (2006) Mitochondrial localization and putative signaling function of sucrose synthase in maize. J Biol Chem 281: 15625–15635 [DOI] [PubMed] [Google Scholar]
- Turgeon R, Wolf S. (2009) Phloem transport: cellular pathways and molecular trafficking. Annu Rev Plant Biol 60: 207–221 [DOI] [PubMed] [Google Scholar]
- Van Bel AJE. (1993) Strategies of phloem loading. Annu Rev Plant Physiol Plant Mol Biol 44: 253–281 [Google Scholar]
- Waterkeyn L. (1981) Cytochemical localisation and function of the 3-linked glucan callose in the developing cotton fibre cell wall. Protoplasma 106: 49–67 [Google Scholar]
- Wilkins TA, Arpat AB. (2005) The cotton fiber transcriptome. Physiol Plant 124: 295–300 [Google Scholar]
- Winter H, Huber S. (2000) Regulation of sucrose metabolism in higher plants: localisation and regulation of activity of key enzymes. Crit Rev Plant Sci 19: 31–67 [DOI] [PubMed] [Google Scholar]
- Wu Y, Llewellyn DJ, Dennis ES. (2002) A quick and easy method for isolating good quality RNA from cotton (Gossypium hirsutum L.) tissues. Plant Mol Biol Rep 20: 213–218 [Google Scholar]
- Zhang XQ, Lund AA, Sarath G, Cerny RL, Roberts DM, Chollet R. (1999) Soybean nodule sucrose synthase (nodulin-100): further analysis of its phosphorylation using recombinant and authentic root-nodule enzymes. Arch Biochem Biophys 371: 70–82 [DOI] [PubMed] [Google Scholar]
- Zhao Q, Yuan S, Wang X, Zhang Y, Zhu H, Lu C. (2008) Restoration of mature etiolated cucumber hypocotyl cell wall susceptibility to expansin by pretreatment with fungal pectinases and EGTA in vitro. Plant Physiol 147: 1874–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]