Abstract
Arabidopsis (Arabidopsis thaliana) epidermal seed coat cells follow a complex developmental program where, following fertilization, cells of the ovule outer integument differentiate into a unique cell type. Two hallmarks of these cells are the production of a doughnut-shaped apoplastic pocket filled with pectinaceous mucilage and the columella, a thick secondary cell wall. Cellulose is thought to be a key component of both these secondary cell wall processes. Here, we investigated the role of cellulose synthase (CESA) subunits CESA2, CESA5, and CESA9 in the seed coat epidermis. We characterized the roles of these CESA proteins in the seed coat by analyzing cell wall composition and morphology in cesa mutant lines. Mutations in any one of these three genes resulted in lower cellulose content, a loss of cell shape uniformity, and reduced radial wall integrity. In addition, we found that attachment of the mucilage halo to the parent seed following extrusion is maintained by cellulose-based connections requiring CESA5. Hence, we show that cellulose fulfills an adhesion role between the extracellular mucilage matrix and the parent cell in seed coat epidermal cells. We propose that mucilage remains attached to the seed coat through interactions between components in the seed mucilage and cellulose. Our data suggest that CESA2 and CESA9 serve in radial wall reinforcement, as does CESA5, but CESA5 also functions in mucilage biosynthesis. These data suggest unique roles for different CESA subunits in one cell type and illustrate a complex role for cellulose biosynthesis in plant developmental biology.
In Arabidopsis (Arabidopsis thaliana), the specialized epidermal cells of the seed coat are hexagonally shaped when viewed from the surface and form an interconnected cell layer that protects the embryo from adverse environmental effects such as UV damage, mechanical stress, desiccation, and biotic factors (Windsor et al., 2000; Haughn and Chaudhury, 2005; North et al., 2010; Nowack et al., 2010). Developmentally, epidermal seed coat cells display several unique morphogenic and cell wall characteristics. Between approximately 5 and 9 d after fertilization, seed coat epidermal cells deposit large amounts of pectinaceous mucilage, which accumulates in a mucilage pocket in the apoplast (Haughn and Chaudhury, 2005). A cytoplasmic column is clearly visible in the center of the cell, surrounded by a doughnut-shaped mucilage pocket (Western et al., 2000, 2004; Haughn and Chaudhury, 2005). As maturation proceeds, 9 to 13 d after fertilization, secondary cell wall material is deposited along the outer tangential and radial sides of the plasma membrane, eventually displacing the cytoplasm with a volcano-shaped secondary cell wall called the columella (Western et al., 2000). During the latter stages of seed development, all cells of the seed coat die and desiccate (Haughn and Chaudhury, 2005). Upon imbibition, the pectinaceous mucilage swells and erupts from the pocket by breaking the radial walls above the secondary cell wall thickening. The extruded mucilage forms a halo that surrounds the seed (Western et al., 2000; Arsovski et al., 2009).
Many transcription factors involved in regulating mucilage production and seed coat cell development are known; several genes encoding pectin-modifying and biosynthesis enzymes have also been identified through mucilage hydration screens (for review, see Arsovski et al., 2010). Transcription factors regulate mucilage production directly by regulating mucilage biosynthetic genes or indirectly by effecting mucilage secretory cell development (Arsovski et al., 2010). The transcription factor APETALA2 (AP2) is required for the differentiation of the two outer layers of the seed coat (Jofuku et al., 1994; Western et al., 2001): seeds homozygous for mutations in AP2 show altered cell shape and lack of columellae and mucilage in the seed epidermal layer (Western et al., 2001). Other transcription factors, such as TRANSPARENT TESTA GLABRA1 (TTG1), ENHANCER OF GLABRA3, TRANSPARENT TESTA8 (TT8), and TT2/MYB5, form a WD40-bHLH-MYB complex that appears to regulate mucilage biosynthesis, at least in part by activating the downstream transcription factors TTG2 and GLABRA2 (Western et al., 2001; Gonzalez et al., 2009; Li et al., 2009; Huang et al., 2011). A novel transcription factor, LEUNIG_HOMOLOG/MUCILAGE MODIFIED1 (MUM1), required for the proper extrusion of mucilage, has been identified that positively regulates genes (MUM2, SUBSILIN PROTEASE1.7, and β-XYLOSIDASE1) involved in mucilage extrusion (Huang et al., 2011). Apart from these transcription factors, several pectin biosynthetic/modifying genes involved in mucilage production/extrusion have been identified and characterized (Western et al., 2001; Arsovski et al., 2010). However, the role of cellulose in seed coat development and mucilage biosynthesis remains largely unknown.
Cellulose biosynthesis is an essential process that mediates cell anisotropic growth and morphogenesis in higher plants. Cellulose microfibrils are paracrystalline chains of β-1,4-linked Glc molecules that support the structural framework of the plant cell wall (Brown, 1985). Several lines of evidence suggest that cellulose-synthesizing organisms produce microfibrils from plasma membrane-localized cellulose-synthesizing complexes. In Arabidopsis, there are 10 CELLULOSE SYNTHASE (CESA) genes identified based on sequence homology to bacterial CESAs (Richmond and Somerville, 2000). A combination of biochemical and genetic analyses have shown that at least three CESA subunits directly interact to form a cellulose synthase complex (CSC) in primary (Desprez et al., 2007; Persson et al., 2007) and secondary (Taylor et al., 2003) cell wall formation. It is unclear exactly how all the CESA subunits interact, due to redundancy in the CESA gene family; separate combinations of CESAs, therefore, may be active in different cell types at different times. CESA1 appears to be required for primary wall synthesis because a temperature-sensitive allele, rsw1-1 (Arioli et al., 1998), results in the temperature-dependent disappearance of rosettes from the plasma membrane, and strong alleles are either embryo or male gametophyte lethal (Beeckman et al., 2002; Gillmor et al., 2002). CESA3 is coexpressed with CESA1, and homozygous cesa3 alleles are also male gametophyte lethal (Persson et al., 2007). CESA6 appears to be required for the elongation of hypocotyl cells in etiolated seedlings (Fagard et al., 2000; Desprez et al., 2002) but can be partially complemented by other CESA6-related CESAs, such as CESA2, CESA5, and CESA9 (Desprez et al., 2007; Persson et al., 2007). These genes are thought to encode components of the primary cell wall cellulose biosynthetic machinery, but when mutated individually they do not result in lethality (Desprez et al., 2007). Mutant cesa5 plants have no reported phenotype, but cesa5cesa6 double mutants are seedling lethal (Desprez et al., 2007). Therefore, CESA5 is redundant to CESA6, with at least one of these subunits being required for seedling establishment. Triple cesa2cesa6cesa9 plants are pollen lethal, indicating that they function redundantly in pollen development (Persson et al., 2007). In addition, CESA9 is required for normal secondary wall synthesis in epidermal seed coat cells in Arabidopsis (Stork et al., 2010) and functions redundantly with CESA6 during pollen development (Persson et al., 2007).
Functional specialization of CESAs is thought to be due to both differences in expression and differences in amino acid sequences that could result in chemically distinct microfibrils. Expression of CESA genes in different tissues has been well documented (Beeckman et al., 2002), and transcriptional regulation may be a primary means for defining the CSC stoichiometry (Persson et al., 2005). For example, CESA5 undergoes cell type-specific transcriptional repression by GLABRA2 in Arabidopsis root cells (Tominaga-Wada et al., 2009). CESA5 and CESA6 are coexpressed during seedling establishment, while CESA2, CESA6, and CESA9 are coexpressed during pollen development. During xylem development, all three secondary wall CESAs (CESA4, CESA7, and CESA8) are expressed in the same cells at the same time (Taylor et al., 2003). While the coding sequence of CESA proteins is highly conserved (Richmond and Somerville, 2000), a hypervariable region and numerous subtle amino acid-level differences exist between subunits that could influence CESA function (Fagard et al., 2000). However, no study has shown functional specialization of individual CESA subunits. Nevertheless, immunoprecipitation analyses of differentiating xylem (secondary cell wall biosynthesis) cells of Populus have shown that two different types of CSC exist, which indicates that different CSCs could produce structurally discrete microfibrils that partition during cell wall synthesis (Song et al., 2010). These data suggest that in differentiating cell types, cellulose biosynthesis may be more complicated than a single stoichiometric model for a hexameric CSC, as is proposed in many plant models (Scheible et al., 2001), or that it involves ancillary proteins that function to specialize the CSC. Functional specificity of multiple CESA proteins in a single cell type, particularly one with a well-understood differentiation program, such as that seen in epidermal seed coat cells, has not yet been explored.
During seed development and desiccation, the secondary cell walls in the seed coat protect the embryo. Cellulose has been proposed to have a major role in reinforcing the secondary cell wall in the seed coat epidermal cells (Stork et al., 2010). By way of histochemistry, cellulose is also proposed to be a component of seed mucilage (Willats et al., 2001; Blake et al., 2006; Young et al., 2008). Fluorescently tagged cellulose-binding modules were used to identify cellulose in mucilage, which indicated distinct binding patterns for crystalline and amorphous cellulose microfibrils (Blake et al., 2006; Young et al., 2008). Further studies showed that glycan strands are localized to the internal domain of the inner layer of mucilage (Macquet et al., 2007a). Analysis of mucilage composition using the glycan-binding dye Calcofluor White indicated the presence of glycans in addition to pectins, but no crystalline cellulose could be detected by analytical techniques in extracted mucilage (Windsor et al., 2000; Willats et al., 2001). By genetically dissecting cellulose biosynthesis in seed coat epidermal cell development, we discovered that cellulose production involving the CESA5 subunit is indispensable for mucilage attachment to the seed coat, while CESA2, CESA5, and CESA9 subunits contribute to secondary wall synthesis in epidermal seed coat cells.
RESULTS
Cell Type-Specific Expression of CESA2, CESA5, and CESA9
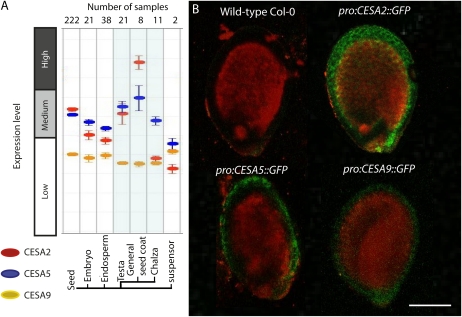
Current models suggest that primary cell wall cellulose biosynthesis requires CESA1, CESA3, and a CESA6-related subunit (Desprez et al., 2007; Persson et al., 2007). We examined the expression of the CESA6-related genes CESA2, CESA5, and CESA9, which were previously shown to be expressed in the Arabidopsis seed (Beeckman et al., 2002), by cross-referencing seed coat gene expression data from GENEVESTIGATOR (Zimmermann et al., 2004). These data showed that gene expression was highest for CESA2, followed by CESA5 and CESA9, in seed coat cells (general seed coat, testa, and chalaza; Fig. 1A). To further explore seed-specific expression, transgenic Arabidopsis plants transformed with promoterCESA2::GFP, promoterCESA5::GFP, or promoterCESA9::GFP fusions were generated, and developing seeds were carefully examined by laser scanning confocal microscopy. Consistent with expression levels shown via GENEVESTIGATOR, seed coat gene expression was observed for promoterCESA2::GFP, promoterCESA9::GFP, and promoterCESA5::GFP (Fig. 1B). Expression patterns of promoterCESA2::GFP and promoterCESA9::GFP were localized through the seed coat, while promoterCESA5::GFP expression patterns were more localized to the outer layer of the seed coat. Observations were made in seeds isolated from whole siliques 10 d post anthesis (DPA) from transgenic plants.
Figure 1.
Expression of CESA2, CESA5, and CESA9 in seeds. A, GENEVESTIGATOR analysis of expression for CESA2, CESA5, and CESA9 in seeds. B, Confirmation of A by localization of promoter::GFP fusions for target CESA genes. Laser scanning confocal microscopy was used to image transgenic plants expressing promoterCESA2::GFP, promoterCESA5::GFP, and promoterCESA9::GFP. These plants were examined at the T2 stage by excising individual seeds from siliques at 10 DPA and mounting them in aqueous solution between two coverslips separated by a thick perimeter of vacuum grease to protect the seeds from being crushed. Seeds were imaged immediately after mounting (within 5 min). Bar = 250 μm. [See online article for color version of this figure.]
Reduction in Secondary Cell Wall Deposition in Seed Coat Cells Is Caused by Mutations in CESA2, CESA5, and CESA9
Cell shape and morphogenesis are partially reliant upon cellulose biosynthesis in various cell types (Brown, 1985; Williamson et al., 2001; Desprez et al., 2002). Based on seed coat preferential gene expression, we next used genetics to investigate whether CESA2, CESA5, and/or CESA9 were required for normal morphogenesis of the seed epidermal cells. To achieve this goal, we obtained T-DNA or point mutants for CESA2 (cesa2-1 [SAIL_400_D01] and cesa2-2 [SALK_096542]), CESA5 (cesa5-1 [SALK_118491] and cesa5-2 [SALK_023353]), CESA6 (cesa6procuste; Fagard et al., 2000), and CESA9 (cesa9-1 [SALK_107750C] and cesa9-2 [SALK_046455]; Persson et al., 2007; Stork et al., 2010). Additionally, cesa2-1cesa9-1 double mutants were obtained (Persson et al., 2007), and a cesa2-1cesa5-1cesa9-1 triple mutant was generated by crossing the cesa2-1cesa9-1 double mutant with cesa5-1 (Supplemental Table S1). Arabidopsis seed coat epidermal cells viewed from the surface appear as an intricate hexameric matrix delineated by the radial cell wall ridge with a pronounced central columella, both of which are consequences of secondary cell wall deposition late in the differentiation process (Haughn and Chaudhury, 2005). Based on this distinctive cell shape, we were able to employ a simple and reliable visual screen for altered morphogenesis using scanning electron microscopy (SEM) of plasma (gold-palladium)-coated seed surfaces. Close examination of multiple generations of cesa2-1 and cesa5-1 seeds revealed distorted and irregular epidermal cell shape as compared with the more uniformly shaped hexagonal cells of wild-type seeds (Fig. 2; Supplemental Fig. S1), similar to a previous report for cesa9 seeds (Stork et al., 2010). The most obvious discriminating feature between wild-type seed coat cells and those observed in the single mutants was a reduction in the radial wall ridge (Fig. 2), suggesting a decrease in secondary cell wall deposition. As observed via SEM, cells could clearly be distinguished by a central columella, but the boundaries between neighboring cells were less visible due to a reduction in the radial wall ridges for both cesa2 and cesa5 (Fig. 2), as was also reported for cesa9 (Stork et al., 2010). Radial wall ridge reduction was even more severe in the cesa2-1cesa9-1 double mutant compared with the single mutants (Fig. 2). Inspection of the cesa2-1cesa5-1cesa9-1 triple mutant revealed a dramatic loss of radial wall ridges between cells, leading to nearly indistinguishable cell boundaries (Fig. 2). However, a mutation in a related subunit, CESA6 (procuste; Supplemental Fig. S2), did not alter the cell shape or cause radial wall ridge reduction.
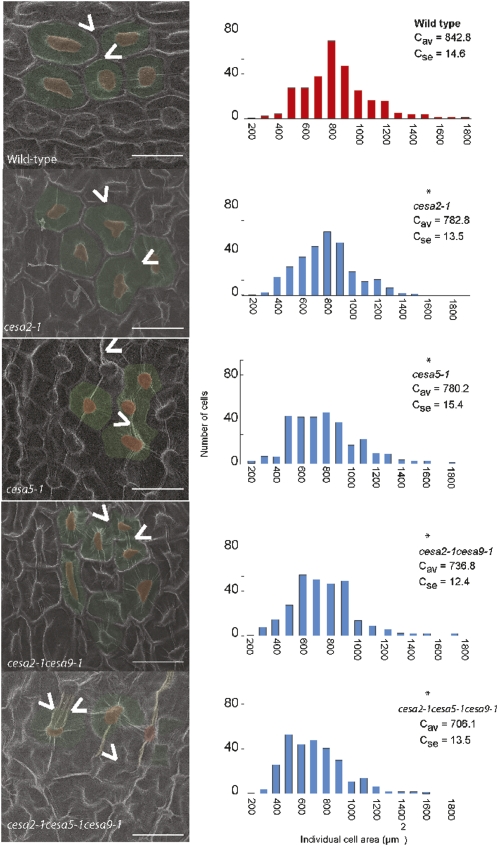
Figure 2.
Quantitative assessment of aberrant cell shape and morphogenesis in cesa mutants via SEM. At left, representative micrographs from mature seeds were false colored with the cell area in green and the columella in red for each genotype. Carats illustrate the presence or absence of radial wall ridges. Bars = 30 μm. At right, individual epidermal cell area in micrographs was measured for 300 cells in 10 different seeds in the wild type, cesa2-1, cesa5-1, cesa9-1, cesa2-1cesa9-1, and cesa2-1cesa5-1cesa9-1. The asterisk indicates a significant difference from the wild type based on results of a Wilcoxon rank-sum test (P < 0.01). [See online article for color version of this figure.]
In addition to reductions in the radial wall ridge, changes in the surface area of individual cells were also observed. To investigate this observation further, the surface area of individual cells was quantified using SEM images. To avoid the interference of the three-dimensional landscape and to promote planar uniformity, a region of cells from the micrograph that did not extend to the border regions of the image was selected for the area measurements. We quantitatively analyzed cell area using frequency distribution comparisons. Cell surface area measurements were made using a minimum population of 10 different seeds from each genetic background and 30 cells from each seed (n = 300; Fig. 2; Supplemental Fig. S1). All cesa single mutant lines showed a decrease in average cell surface area: the wild type (842.8 μm2; se of 14.6 μm2), cesa2-1 (782.8 μm2; se of 13.5 μm2), cesa2-2 (765.4 μm2; se of 15.0 μm2), cesa5-1 (780.2 μm2; se of 15.4 μm2), cesa5-2 (788.8 μm2; se of 17.1 μm2), and cesa9-1 (743.6 μm2; se of 13.1 μm2; Fig. 2; as reported previously by Stork et al. [2010]). The cesa2-1cesa9-1 double mutant showed a slightly stronger reduction compared with the single mutants (736.7 μm2; se of 12.3 μm2), while the cesa2-1cesa5-1cesa9-1 triple mutant showed the most severe reduction (706.0 μm2; se of 13.4 μm2) compared with the wild type. These data demonstrate that a reduction in cellulose synthesis in seed epidermal cells results in smaller overall cell size or smaller seeds.
Histochemical Analysis of Mutant Versus Wild-Type Epidermal Seed Coat Cells Revealed Changes in the Thickness and Height of Radial Wall Secondary Thickening
Calcofluor White has previously been used to effectively explore spatial constraints of cellulose deposition in different plant tissues (Herth and Schnepf, 1980; Albani and Plancke, 1998). Based on the reduced radial wall ridge phenotype, we hypothesized that a deficiency in cellulose should be apparent at the cell borders. To visualize this scenario, we used laser scanning confocal microscopy to create three-dimensional animations (z-plane projections used 500 nm per frame shift for resolution) of seed epidermal cells in mutant and wild-type lines stained with Calcofluor White. Consistent with the surface view of epidermal cells observed by SEM, we could observe aberrant cell shape and regions of reduced and patchy fluorescence across the radial walls separating neighboring cells and the columella in all mutant lines as compared with the wild type (Fig. 3, A and B). To quantify how differences in cellulose biosynthesis correlated with Calcofluor White staining between wild-type and mutant lines, we generated cross-sectional fluorescence intensity plots. These analyses are relative quantifications, since there is no standard available for glycan staining and the cell wall is a complex mixture of glycan-containing biopolymers. Our results revealed a sharp maximum in fluorescence intensity at the radial walls and the central columella of wild-type cells (Fig. 3B). Overall, the fluorescence intensity was quantitatively reduced in the single and double mutants and showed a severe loss of intensity in the triple mutant (Fig. 3B).
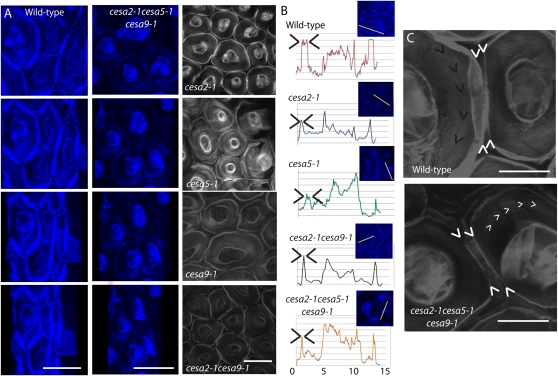
Figure 3.
Radial wall ridge displays decreased thickness in cesa mutants. To carefully examine the structural change of cesa mutant epidermal seed coat cells, Calcofluor White was used to label structural β-glycans and cellular features were visualized via laser scanning confocal microscopy. Imaging was performed approximately 5 min after seeds were immersed in Calcofluor White solution using a 60× objective and a 4′,6-diamino-phenylindole filter. A, Three-dimensional z-plane animation of Calcofluor White-labeled wild-type (column 1) and cesa2-1cesa5-1cesa9-1 (column 2) seed coat cells acquired by merging individual frames taken across a z-plane shift (at 0.5-μm steps) from the top of the columella to the base of the tangential wall. Cells were rotated from surface to side view, illustrating the elevation and fluorescence differences observed between wild-type and triple mutant seed coat radial walls. Bars = 10 μm. Column 3 shows surface views of Calcofluor White-stained seed coat epidermal cells for cesa2-1, cesa5-1, cesa2-1cesa9-1, and cesa2-1cesa5-1cesa9-1. Bar = 30 μm. B, Fluorescence intensity plots generated across a transect that spanned a single cell for the wild type and single, double, and triple mutants. C, High-magnification views of individual cells from Calcofluor White-labeled epidermal cells in the wild type and cesa2-1cesa5-1cesa9-1 visually illustrate the reduced radial wall fluorescence in cesa2-1cesa5-1cesa9-1. Bars = 15 μm. [See online article for color version of this figure.]
The most intriguing comparison was between wild-type cells and the cesa2-1cesa5-1cesa9-1 triple mutant (Fig. 3C; Supplemental Movies S1 and S2). In the wild type, we found that the radial wall appeared elevated and displayed regions of partial fluorescence overlap with the neighboring cells. In cesa2-1cesa5-1cesa9-1 cells, we could not observe fluorescence overlap between neighboring cells. Rather, a thin band of fluorescence was visible surrounding most cells, and some cells showed regions of no fluorescence at the boundary (Fig. 3). When the three-dimensional animations were oriented at a tangential perspective (Fig. 3A), we observed that wall elevation in the triple mutant was reduced compared with the wild type (Supplemental Movies S1 and S2). These data suggest that the secondary cell wall at the boundaries of seed coat cells is primarily cellulosic in nature and is reliant on at least CESA2, CESA5, and CESA9 for its biosynthesis. In wild-type cells, the fluorescence derived from the radial wall appeared to be nearly equal in height as the central columella (Fig. 3, A and B). By contrast, cesa2-1cesa5-1cesa9-1 radial walls barely extended beyond the plane of the trough (Fig. 3, A and B). We interpret the thin band of fluorescence between columella in cesa2-1cesa5-1cesa9-1 as representing the primary radial cell wall, and the loss of fluorescence at this position compared with wild-type cells was due to a reduction in secondary wall thickening.
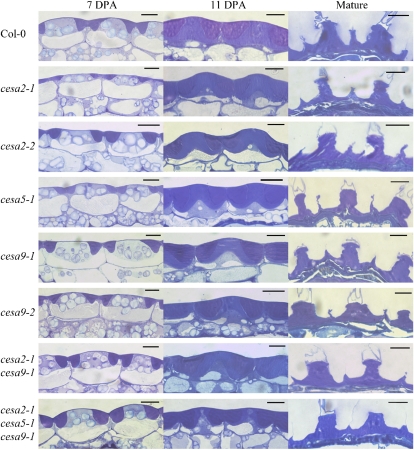
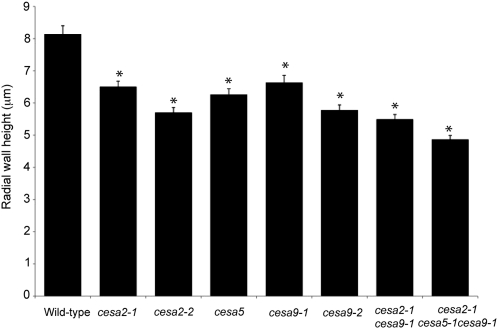
To measure the reduction in secondary wall thickening and to visualize the developmental program of epidermal seed coat cells in CESA mutants, we examined cryofixed sections of wild-type and mutant seeds from 4, 7, and 11 DPA using light microscopy (Fig. 4). Cross-sectional analysis was performed on seeds from cesa2-1, cesa2-2, cesa5-1, cesa9-1, cesa9-2, cesa2-1cesa9-1, and cesa2-1cesa5-1cesa9-1. For the developmental analysis, seeds were excised from at least five siliques for 11 DPA, four siliques for 7 DPA, and three siliques for 4 DPA and sampled randomly from five to six plants for each genotype. Visual examination of embedded seeds revealed relatively minor differences between wild-type and mutant genotypes at 4 DPA (data not shown) and 7 DPA (Fig. 4, column 1). At 11 DPA (Fig. 4, column 2), wild-type ecotype Columbia (Col-0) cells had completed mucilage production and had begun to deposit secondary wall material, filling most of the cytoplasmic column. By contrast, the cesa2-1cesa5-1cesa9-1 triple mutant appeared to be delayed in secondary wall deposition, with cytoplasm still observable through the middle of the columella, extending to the top of the cell. In addition, secondary wall material accumulated in an irregular manner on the sides of the columella. In order to quantify secondary cell wall deposition, mature seeds were fixed with glutaraldehyde, allowing the mucilage pocket to burst, leaving the secondary wall easier to observe (Fig. 4, column 3). Although differences in cell wall thickness between the wild type and mutants were observed, the irregular shape of the secondary cell wall and variability in the plane of the section make quantification of secondary cell wall thickness difficult. To reduce this variability, we chose the height of the secondary cell wall thickening at the radial wall as a determinant for two reasons. First, because it is easy to identify the position of the radial wall in each cell, variability stemming from irregularities in thickness throughout a cell could be minimized. Second, because the secondary cell wall ends at the radial wall, relatively small changes in secondary cell wall thickness can have a pronounced effect on the height of the radial wall thickening. A total of 20 radial walls per resin block were measured for each genotype from a random set of seeds from three different resin blocks (n = 60). Radial walls were measured from the base of the secondary wall depositions to the top of the radial wall thickening (Fig. 5). Radial walls for Col-0 were the highest (8.14 ± 0.27 μm), while all cesa mutant lines were reduced to a similar height [cesa2-1, 6.51 ± 0.17 μm; cesa2-2, 5.71 ± 0.15 μm; cesa5-1, 6.27 ± 0.18 μm; cesa9-1, 6.64 ± 0.22 μm; cesa9-2, 5.78 ± 0.16 μm). The double mutant radial walls were reduced more than any single mutant, and the triple mutant was severely reduced compared with all other mutant lines (cesa2-1cesa9-1, 5.49 ± 0.15 μm; cesa2-1cesa5-1cesa9-1, 4.87 ± 0.12 μm [Fig. 5]). Radial wall thickening was absent in some cells of the triple mutant. Radial wall height measurements for cesa9-1 and cesa9-2 were similar to previous measurements of these genotypes, when differences in the way they were measured were accounted for (measured from the base of the radial wall in this study and from the base of the mucilage pocket in the previous study [Stork et al., 2010]). Based on the wall height measurements, all cesa mutants were significantly different from the wild type (two-tailed t test, P < 0.05; Fig. 5). In addition, cesa2-1cesa5-1cesa9-1 radial walls were significantly smaller than the radial walls of the cesa single mutant lines and the cesa2-1cesa9-1 double mutant (two-tailed t test, P < 0.05). Results of the developmental series indicate that reduced radial wall height is consistent with the decreased radial wall ridges observed by SEM as well as the reduced and patchy regions of fluorescence at the radial wall observed when cells were stained with Calcofluor White (Figs. 2 and 3).
Figure 4.
Developmental analysis of the structure of wild-type and mutant cesa seed coats. Epidermal cell morphology is shown for wild-type, cesa2-1, cesa2-2, cesa5-1, cesa9-1, cesa2-1cesa9-1, and cesa2-1cesa5-1cesa9-1 toluidine blue-stained sections of cryofixed 7-DPA (column 1) and 11-DPA (column 2) seeds. Column 3 shows the epidermal cell morphology of mature wild-type and cesa toluidine blue-stained sections of aqueous (3%, v/v) glutaraldehyde-fixed mature seeds. Mucilage pockets stain dark purple, while secondary wall stains a lighter purple. Bars = 10 μm. [See online article for color version of this figure.]
Figure 5.
Average height of the radial wall of wild-type, cesa2-1, cesa2-2, cesa5-1, cesa9-1, cesa2-1cesa9-1, and cesa2-1cesa5-1cesa9-1 seed coat cells. The asterisks indicate significant differences from the wild type (P < 0.05 by Student’s t test).
Cellulose Content Is Decreased in cesa Mutants
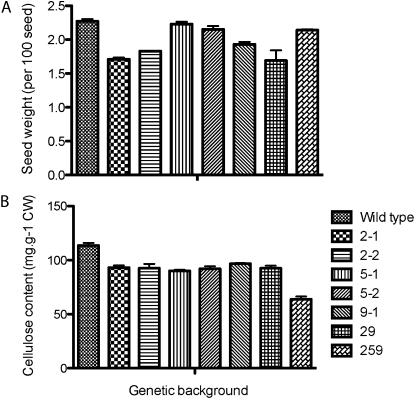
In order to link changes in cell shape, cell structure, and seed coat permeability, we analyzed the cellulose content and cell wall composition of cesa mutant seeds. Due to natural variation, visual examination of seeds was not reliable. To overcome this variability, we measured the average weight of 100 seeds and found that all mutants showed lower seed weight than the wild type, with the exception of cesa5, cesa6 (procuste), and cesa2-1cesa5-1cesa9-1, which were similar in weight to the wild type (Fig. 6A). Seed weights were 2.27 ± 0.06 (wild type), 1.71 ± 0.04 (cesa2-1), 1.83 ± 0.01 (cesa2-2), 2.23 ± 0.06 (cesa5-1), 2.15 ± 0.09 (cesa5-2), 2.22 ± 0.07 (cesa6), 1.81 ± 0.06 (cesa9-1), 1.69 ± 0.04 (cesa2-1cesa9-1), and 2.14 ± 0.02 (cesa2-1cesa5-1cesa9-1) mg per 100 seeds. All of the morphological changes described thus far are presumably due to decreases in cellulose biosynthesis. To investigate this link, we determined the amount of acid-insoluble Glc, a quantitative estimate of crystalline cellulose (Updegraff, 1969), and found a significantly lower cellulose content in the cell walls of single, double, and triple mutants (114 ± 4 [wild type], 93 ± 3 [cesa2-1], 91 ± 2 [cesa2-2], 91 ± 2 [cesa5-1], 92 ± 4 [cesa5-2], 96 ± 1 [cesa9-1], 92 ± 3 [cesa2-1cesa9-1], 63 ± 4 [cesa2-1cesa5-1cesa9-1] mg g−1; P < 0.05 by Student’s t test). No significant difference in cellulose content was observed between cesa2-1, cesa5-1, cesa9-1, and cesa2-1cesa9-1 mutants. These results indicate a noncumulative effect on cellulose biosynthesis between cesa9-1 and cesa2-1; however, a cumulative reduction in cellulose content was identified in the cesa2-1cesa5-1cesa9-1 triple mutant seed compared with single mutant lines and the cesa9-1cesa2-1 double mutant line (Fig. 6B). To address whether CESA2, CESA5, or CESA9 caused a loss of cellulose synthesis in elongating cell types, we measured the acid-insoluble Glc content in the stems of wild-type plants (251 ± 7 mg g−1) and mutant plants (cesa2-1, 230 ± 29; cesa2-2, 232 ± 30; cesa5-1, 241 ± 28; cesa9-1, 250 ± 37; cesa2-1cesa9-1, 226 ± 29; cesa2-1cesa5-1cesa9-1, 237 ± 27 mg g−1), revealing no significant differences (Student’s t test, P < 0.05; Supplemental Fig. S3). These data were consistent with the fact that there was no observable difference in hypocotyl length (4-d-old dark-grown etiolated seedlings grown vertically) or root elongation rate (7-d-old seedlings grown vertically in the light) under the growth conditions used here (Supplemental Fig. S4). Changes in seed cellulose content without observable differences in stem cellulose content further suggest that cellulose biosynthesis is under tissue-specific control with respect to the composition of active CESA subunits.
Figure 6.
Seed size and cellulose content. A, Seed weight was measured as the average of three batches of 100 seeds, and error bars represent the se of three replicates. B, Acid-insoluble crystalline cellulose content of various tissues was determined colorimetrically for mature seeds. Error bars represent the se of three technical replicates from three independent batches of seeds as biological replicates. CW, Cell wall.
Cellulose Biosynthesis via CESA5 Is Required for Mucilage Adherence to the Parent Seed during Hydration
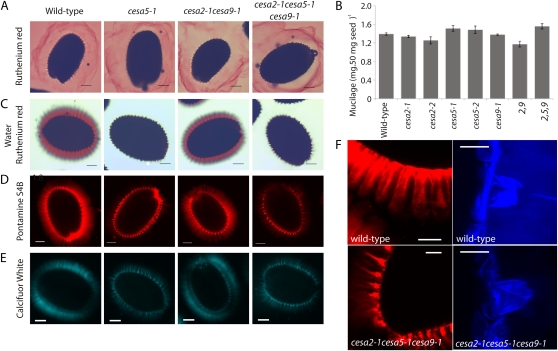
Connections between the mucilage halo and the parent seed during the process of imbibition are poorly understood, but they could involve cellulose present in the mucilage (Willats et al., 2001; Blake et al., 2006; Macquet et al., 2007a; Young et al., 2008). We tested this model by visualizing seed mucilage extrusion and adherence in mutant cesa lines. Our initial experiment was to visually examine for mucilage defects in cesa mutants using Ruthenium Red stain. Seeds were hydrated directly in Ruthenium Red and observed within 5 min with a light microscope (Colombo and Rascio, 1977; Western et al., 2000). All seeds from cesa mutant lines showed equally large mucilage halos (Fig. 7A). These results are consistent with the examination of developing seed coat sections from mutants and the wild type, which showed no differences in the mucilage pocket size (Fig. 4, column 2). We then asked if there was any difference in the amount of mucilage produced in the cesa mutants. We used ammonium oxalate to extract the mucilage (Arsovski et al., 2009) and quantified the amount of mucilage extracted. The total mucilage content of mutants (cesa2-1, 1.34 ± 0.03; cesa2-2, 1.25 ± 0.08; cesa9-1, 1.38 ± 0.02; cesa2-1cesa9-1, 1.17 ± 0.06 mg 50 mg−1 seed) was similar to the wild type (1.39 ± 0.03 mg 50 mg−1 seed). However, cesa5-1 (1.51 ± 0.06 mg 50 mg−1 seed), cesa5-2 (1.48 ± 0.08 mg 50 mg−1 seed), and cesa2-1cesa5-1cesa9-1 (1.55 ± 0.06 mg 50 mg−1 seed) showed increased soluble mucilage recovery compared with the wild type (Fig. 7B). These data suggest that genetic mutation of CESA5 increased the ammonium oxalate-extractable mucilage content. To address the increased mucilage recovery in cesa5-1, hydrated seeds were stained with Ruthenium Red after 2 h of agitation in water (Colombo and Rascio, 1977; Western et al., 2000) and observed with a stereomicroscope. After shaking, the adherent layer of mucilage remained intact in wild-type, cesa2-1, cesa2-2, cesa9-1, and cesa2-1cesa9-1 seeds. In contrast, the adherent layer of mucilage was noticeably absent in cesa5-1, cesa5-2, and cesa2-1cesa5-1cesa9-1 seeds (Fig. 7C; Supplemental Fig. S5). Hence, CESA5 is essential for maintaining mucilage adherence to seeds during hydration.
Figure 7.
Mucilage attachment to the parent seed is mediated by CESA5 in Arabidopsis seeds. A, Ruthenium Red-stained seeds. B, Mucilage weights determined for wild-type and cesa mutant seeds. Error bars represent se of three replicates. C, Water-hydrated Ruthenium Red-stained seeds visualized by light microscopy in the wild type, cesa5-1, cesa2-1cesa9-1, and cesa2-1cesa5-1cesa9-1. Mutants cesa2-1, cesa2-2, cesa9-1, and cesa9-2 are not shown due to no observed mucilage phenotype. D and E, Pontamine S4B-stained (D) and Calcofluor White-stained (E) wild-type, cesa5-1, cesa2-1cesa9-1, and cesa2-1cesa5-1cesa9-1 mature seeds. Bars = 100 μm. F, High-magnification visualization of Pontamine S4B-labeled (whole seeds viewed by light microscopy) and Calcofluor White-labeled (three-dimensional reconstruction of a z-projection; side view via laser scanning confocal microscopy) seed coat cells in the wild type and cesa2-1cesa5-1cesa9-1. Bars = 10 μm. [See online article for color version of this figure.]
Previous studies have shown colocalization of methylesterified pectins through JIM5 antibody labeling, with structural glycans through Calcofluor White staining at the linear rays that extend from the top of the columella (Macquet et al., 2007a). To examine whether cesa5 mucilage had changes in cellulose-staining patterns, we treated imbibed seeds with Pontamine S4B and Calcofluor White and examined them using confocal microscopy. Pontamine S4B shows primary binding affinity to cellulose (Fig. 7D) and xyloglucans to a lesser degree, while Calcofluor White is less specific, showing binding affinity to cellulose, arabinans, and pectic galactans (Herth and Schnepf, 1980; Anderson et al., 2010). A marked loss of fluorescence was observed within the mucilage itself in cesa5-1 and cesa2-1cesa5-1cesa9-1 mutants (Fig. 7E). However, we were still able to see staining associated with rays extending from the columella (Fig. 7, D and E). To examine this phenotype in greater detail, we observed mucilage imbibition after staining with Calcofluor White or Pontamine S4B at high magnification (Fig. 7F). Pontamine S4B staining showed the presence of rays in the cesa5-1 and cesa2-1cesa5-1cesa9-1 mutants but a distinct lack of fluorescence within the mucilage itself (Fig. 7F). Examination of fluorescence after Calcofluor White imbibition (5 min) revealed the presence of brightly stained mucilage surrounding the columella in wild-type seeds, which was absent in the cesa2-1cesa5-1cesa9-1 mutant (Fig. 7F). An additional means to observe this phenotype used consecutive sections from fixed mature seeds that were stained with Pontamine S4B and imaged with the confocal microscope. These images were grouped to form a movie that represented a series of cross-sections from the wild type and cesa2-1cesa5-1cesa9-1 (Supplemental Movies S1 and S2). These data showed the rays, stained with Pontamine S4B, arising from the columella in both wild-type and mutant seeds and a significant loss of fluorescence from the mucilage. Overall, our results support a mechanism of matrix attachment between the mucilage halo and the seed via cellulose that requires CESA5.
Cell Wall Monosaccharide Content Is Altered in cesa5-1 and cesa2-1cesa5-1cesa9-1
Cellulose creates an interconnected network within the plant cell wall via cross-linking with hemicelluloses and matrix associations with pectins (Carpita and Gibeaut, 1993). To determine whether cellulose deficiency would cause feedback changes in the composition of other cell wall sugars, we determined the monosaccharide content of seed preparations using HPLC. Soluble mucilage analysis indicated an increase in the rhamnogalacturonan I backbone sugars, Rha and GalUA, in cesa5-1 and cesa2-1cesa5-1cesa9-1 compared with the wild type and cesa2-1cesa9-1 (Table I). Increases in Rha levels were significant, as determined by a two-tailed t test (P < 0.05). Ara, Gal, Glc, and Xyl levels remained largely unchanged.
Table I. HPLC analysis of mucilage and cell wall monosaccharide composition from wild-type and mutant seeds.
Values shown are averages of three replicates ± se. n.d. Not determined.
| Sugar | Soluble Mucilage |
Whole Seed Enriched Cell Wall Fraction |
||||||
| Col-0 | cesa5-1 | cesa2-1cesa9-1 | cesa2-1cesa5-1cesa9-1 | Col-0 | cesa5-1 | cesa2-1cesa9-1 | cesa2-1cesa5-1cesa9-1 | |
| mg sugar g−1 seed | ||||||||
| Ara | 0.33 ± 0.096 | 0.50 ± 0.10 | 0.244 ± 0.012 | 0.26 ± 0.06 | 53.10 ± 1.73 | 62.87 ± 0.59 | 54.56 ± 1.96 | 62.94 ± 0.59 |
| Rha | 10.31 ± 0.41 | 12.48 ± 0.17a | 11.51 ± 1.52 | 12.56 ± 0.29a | 32.45 ± 2.17 | 22.20 ± 1.49a | 36.45 ± 1.99a | 22.41 ± 0.16 |
| Gal | 0.32 ± 0.11 | 0.29 ± 0.021 | 0.18 ± 0.023 | 0.14 ± 0.012 | 50.62 ± 0.80 | 53.82 ± 0.40 | 52.54 ± 1.18 | 50.04 ± 0.46 |
| Glu | 0.73 ± 0.18 | 0.57 ± 0.037 | 0.37 ± 0.069 | 0.516 ± 0.070 | 10.08 ± 0.30 | 7.65 ± 0.34a | 10.11 ± 0.12 | 7.06 ± 0.34 |
| Xyl | 1.05 ± 0.06 | 1.06 ± 0.040 | 1.06 ± 0.12 | 0.96 ± 0.062 | 27.14 ± 1.93 | 19.47 ± 2.65 | 26.31 ± 1.29 | 19.23 ± 0.92 |
| GalA | 9.21 ± 0.47 | 12.27 ± 2.14 | 9.50 ± 1.57 | 13.0 ± 2.04 | n.d. | n.d. | n.d. | n.d. |
Significantly different from Col-0 (P < 0.05).
To investigate the underlying cause for increases in rhamnogalacturonan I backbone sugars, we next tested monosaccharide levels in whole seeds. We ground whole seeds and performed serial washes with ethanol and acetone, in addition to an amylase treatment to remove soluble mucilage, cytosolic sugars, and starch, leaving an enriched cell wall fraction. Monosaccharide quantification revealed proportional differences among neutral sugars. Rha levels were significantly reduced in cesa5-1 and cesa2-1cesa5-1cesa9-1 compared with the wild type and cesa2-1cesa9-1 (Table I). Ara levels were increased while Xyl levels were decreased in cesa5-1 and cesa2-1cesa5-1cesa9-1 compared with the wild type (Table I). The reductions in Rha levels are consistent with the increases found in soluble mucilage levels, suggesting changes to the adherence of mucilage to the seed in cesa5 mutant lines.
DISCUSSION
Our results illustrate the specific roles of CESA2, CESA5, and CESA9 in seed coat development and differentiation. Specifically, by reverse genetics, we have identified two secondary cell wall processes occurring in the seed coat epidermal cells that require specific cellulose biosynthetic machinery. First, loss of mucilage adherence properties caused by mutations in CESA5 alone link mucilage biosynthesis with CESA5. Second, during seed coat radial wall thickening, mutations in single, double, and triple mutants for cesa2, cesa5, and cesa9 caused decreased radial wall reinforcement (measured as radial wall height; Fig. 5), implicating all three subunits in this process. These data suggest that CESA subunits undergo cell-specific regulation and have functional subspecificity, extending our knowledge of cellulose biosynthetic processes.
Late in the differentiation of seed coat epidermal cells (8–9 DPA), a cellulosic secondary wall is initiated along the membrane of the radial and outer tangential sides of the cell, resulting in a thickening of the radial wall and creation of the volcano-shaped columella. We have shown previously that CESA9 is required for normal synthesis of this secondary cell wall (Stork et al., 2010). In this paper, we show that this process also involves other CESA subunits, CESA2 and CESA5. The additive reduction in radial wall height in the cesa2-1cesa5-1cesa9-1 triple mutant relative to single and double mutants (Figs. 3 and 5) suggests that each subunit plays a similar role. Given the current working models for a hexameric CSC comprising three different CESAs (Taylor et al., 2003; Desprez et al., 2007), a plausible hypothesis is that CESA2, CESA5, and CESA9 combine to form a functional CSC complex, similar to the other such secondary wall complex studied in xylem cells, which requires CESA4, CESA7, and CESA8 (Taylor et al., 2003). An alternative explanation is that a degree of redundancy exists, as illustrated for CESA6-like proteins (Persson et al., 2007). However, reduced radial wall height in single mutants and additive phenotypes in double (cesa2-1cesa9-1) and triple (cesa2-1cesa5-1cesa9-1) mutants support functional significance for each subunit.
Prior to the synthesis of the cellulosic secondary cell wall, seed coat epidermal cells synthesize seed mucilage and deposit it in the apoplast. Upon exposure to an aqueous environment, mature seeds extrude the mucilage, encapsulating the seed (Western et al., 2000; Windsor et al., 2000). The extruded mucilage includes a loosely bound (soluble) layer that can be easily removed through simple agitation and an adherent layer that is difficult to remove completely even with strong alkali solutions (Western et al., 2000, 2001; Macquet et al., 2007a). Mucilage consists primarily of pectin, but xyloglucan and cellulose can also be detected in the adherent layer (Western et al., 2000, 2004; Willats et al., 2001; Blake et al., 2006; Macquet et al., 2007a; Young et al., 2008; Arsovski et al., 2009). It has previously been suggested that cellulose could be involved in maintaining connections to the parent seed (Macquet et al., 2007a), but there has been little experimental evidence to support this hypothesis. We have shown here that CESA5 is required for normal levels of mucilage cellulose and formation of the adherent layer, providing evidence that at least one role of cellulose in seed mucilage is to form a network with pectins such that the mucilage is held to the seed surface following extrusion. Although the nature of the cellulose-pectin interactions has yet to be determined in this tissue, they are likely to be direct, given the tenacity of mucilage adherence. Studies showing direct binding of neutral sugar side chains of rhamnogalacturonan I and cellulose microfibrils (Zykwinska et al., 2005; Dick-Pérez et al., 2011) suggest a mechanism through which such interactions could occur. In support of this hypothesis, several recent studies have shown that increases in neutral side chains of rhamnogalacturonan I inhibit mucilage extrusion (Dean et al., 2007; Macquet et al., 2007b; Arsovski et al., 2009). The discovery that cellulose is integral to the sustained attachment of the mucilage matrix suggests a potential role for cellulose as a molecular scaffold linking the extracellular matrix to the parent cell (Brown, 1985).
Although our data indicate that CESA2, CESA5, and CESA9 are required for secondary cell wall biosynthesis in the seed coat epidermis, these proteins have been associated with primary cell wall synthesis in other tissues. Indeed, current models suggest that primary cell wall cellulose biosynthesis involves CESA1, CESA3, and one of a class of redundant CESA6-related subunits including CESA2, CESA5, CESA6, or CESA9 (Persson et al., 2007). Based on our developmental analysis (Fig. 4), there is little evidence that primary wall cellulose biosynthesis in the seed coat epidermis requires CESA2, CESA5, and CESA9, suggesting that CESA1, CESA3, and CESA6 are sufficient for this purpose. CESA5 could function as the sole biosynthetic unit required for sufficient cellulose production to mediate mucilage attachment. However, given everything that is known about the organization of the CSC and the fact that not all mucilage cellulose is absent in the cesa5 mutant, it is plausible that other CESAs are involved.
Our data are consistent with the speculation that different cell types may require cell-specific cellulose deposition (Carpita and Vergara, 1998). The developing seed coat represents an excellent system to dissect the composition of CSC in two separate processes and an intriguing area for future exploration. Moreover, while it has long been held that cellulose is the structural core of the plant extracellular matrix (Green, 1962; Brown, 1985), its link to mediating adherence between parent cells has been complicated to examine. Our results show that in seed coat epidermal cells, cellulose partially fulfills a matrix attachment role. Finally, we have demonstrated the functional specialization of three CESAs in secondary cell wall deposition. The process of secondary wall cellulose biosynthesis in seed testa cells can be likened to that of cellulosic xylem vessel reinforcement, whereby the epidermal seed coat cells must withstand the immense pressure of desiccation during seed maturation and of rehydration upon imbibition (Windsor et al., 2000), illustrating an intriguing parallel mechanism. Evidence of aberrant cell shape and morphogenesis in cesa2, cesa5, and cesa9 seed coat cells also documents the role for cellulosic secondary cell wall reinforcement in structurally supporting cell shape.
MATERIALS AND METHODS
Plant Material and Growth Conditions
All Arabidopsis (Arabidopsis thaliana) mutants used in this study were of the Col-0 ecotype. We obtained T-DNA insertion or point mutants from the Arabidopsis Biological Resource Center and from Persson et al. (2007) and Stork et al. (2010) as follows: CESA2 (cesa2-1 [SAIL_400_D01] and cesa2-2 [SALK_096542]; Persson et al., 2007), CESA5 (cesa5-1 [SALK_118491] and cesa5-2 [SALK_023353]), CESA6 (procuste-ixr2-1 R1064W; Desprez et al., 2002), and CESA9 (cesa9-1 [SALK_107750C] and cesa9-2 [SALK_046455]; Persson et al., 2007; Stork et al., 2010). Additionally, cesa2-1cesa9-1 double mutants were obtained (Persson et al., 2007) and a cesa2-1cesa5-1cesa9-1 triple mutant was generated by crossing the cesa2-1cesa9-1 double mutant with cesa5-1 (genotyping primers for each allele are documented in Supplemental Table S1). Seeds were surface sterilized using 30% bleach for 20 min. After three sequential washes with sterile water, the seeds were vernalized for 2 d at 4°C in 0.15% agar. For germination, phenotypic analysis, and growth assays, the surface-sterilized seeds were exposed to light for 1 h and grown in either continuous light (200 mmol m–2 s–1) or complete darkness at 22°C on plates containing 0.5× Murashige and Skoog mineral salts (Sigma) and 1% agar.
Staging of Flower Age
Pollination was defined as the time when the flower was beginning to open. At this stage, the stamen is beginning to grow over the gynoecium and the pollen is released (anthesis). Petals can be seen extending beyond the tops of the sepals. Flowers at anthesis were marked using water-soluble paint, and a different color was used for each day of marking.
Promoter::GFP Fusion Constructs
Promoters of CESA2, CESA5, and CESA9 were amplified from Col-0 genomic DNA by PCR using the following primers: CESA2/Prom/SbfI/F (5′-TCCCCCTGCAGGGACAGATTGTGAGTGCGAAACACACATA-3′), CESA2/Prom/KpnI/R (5′-CCGGGGTACCGATGTCTTCTACACCGAATCAAAACGAATT-3′), CESA5/Prom/SbfI/F (5′-TCCCCCTGCAGGACGCTTTCAGTTTGTCGACCCT-3′), CESA5/Prom/KpnI/R (5′-CCGGGGTACCGTACCGACAAAACAGTAGACAGTATGTCAA-3′), CESA9/Prom/SbfI/F (5′-TCCCCCTGCAGGATTAAAGATGGTTTTTAGTGGAGG-3′), and CESA9/Prom/KpnI/R (5′-CCGGGGTACCATTCACTAGTCCGATCAGATCA-3′). Underlined sequence is the promoter sequence and nonunderlined sequence is the restriction site with additional nucleotides. The amplified products were digested with SbfI and KpnI and cloned into the pMDC43 vector, replacing the cauliflower mosaic virus 35S promoter from the original vector, yielding promoterCESA2::GFP, promoterCESA5::GFP, and promoterCESA9::GFP constructs. Individual constructs were sequenced and transformed into Agrobacterium tumefaciens strain GV3101. Wild-type Col-0 plants were transformed with Agrobacterium containing different constructs using the floral dip method (Clough and Bent, 1998). Seeds were collected from the floral-dipped plants and selected on 0.5× Murashige and Skoog mineral salts (Sigma) medium with hygromycin (25 mg L−1), and homozygous T2 plants from three independent lines per construct were used for the expression analysis.
Histochemical Analyses
Ruthenium Red Staining
Seeds incubated in 0.01% (w/v) Ruthenium Red (Sigma) in water for 30 min at 25°C were used to test for aberrant mucilage production (Beeckman et al., 2000). Alternatively, seeds were shaken on an orbital shaker for 2 h, stained with 0.01% (w/v) Ruthenium Red in water for 30 min at 25°C, and imaged with an Olympus MVX-10 stereo fluorescence microscope.
Cellulose Staining Using Pontamine S4B
Seeds were shaken on an orbital shaker for 2 h and then stained with 0.01% (w/v) Pontamine Fast Scarlet S4B (Aldrich Rare Chemical Library; no. S479896) and 50 to 150 mm NaCl2 for 1 h. Seeds were then washed five times with distilled water and imaged using confocal microscopy.
SEM and Image Analysis
Arabidopsis seeds were attached to standard electron microscope stubs and sputter coated with gold-palladium alloy using the Hummer VI sputtering system (Anatech). Specimens were visualized using the Hitachi model S-800 scanning electron microscope, and images were captured using the Evex Nano Analysis digital imaging system. Image analysis was performed using ImageJ software (W. Rasband, National Institutes of Health). Measurement for each cell’s planar area used area measurement output after tracing the polygon via the freehand selection tool (ImageJ). Data were organized in the spreadsheet program Microsoft Excel and transferred to GraphPad Prism-4 for generating histograms and performing statistical comparisons.
Confocal Microscopy
Whole seeds were mounted in water for promoter::GFP fusion analysis and in Calcofluor White for seed histochemical analysis. Seed mounts were created between a slide and a coverslip separated by the perimeter of vacuum grease that acted to keep the seed in contact with Calcofluor White and avoid crushing seed structures. Once mounted, specimens were imaged in darkness after being exposed to Calcofluor White for 5 min or imaged immediately for promoter::GFP fusions. Imaging was performed on an Olympus FV1000 laser scanning confocal microscope using a 63× numerical aperture 1.4 water-immersion objective. The microscope is equipped with lasers for excitation wavelengths ranging from 405 to 633 nm; Calcofluor White was visualized at 405 nm, while promoter::GFP fusions were visualized at 488 nm. All image processing was performed by using Olympus Fluoview software (Olympus) and ImageJ (W. Rasband, National Institutes of Health).
Resin Embedding for Bright-Field Microscopy
Siliques staged at 4, 7, and 11 DPA were removed from the plant and dissected using a razor blade. The seeds were removed and either an insect pin or a razor blade was used to puncture the seed coat. High-pressure freezing, freeze substitution, and resin embedding were performed as described (Rensing et al., 2002). Samples were loaded onto copper hats (Ted Pella) containing 1-hexadecene and frozen under high pressure using a Leica EM HPM 100 high-pressure freezer (Leica). Copper hats were then transferred to frozen cryovials containing freeze substitution medium consisting of 2% (w/v) osmium tetroxide in acetone with 8% (v/v) dimethoxypropane. Freeze substitution was performed for 6 d at −80°C by incubation in a Leica EM AFS chamber, followed by an incubation at −20°C over 20 h to allow for reaction of the fixatives, then the temperature was increased to 4°C. Samples were then removed from the copper hats, rinsed in anhydrous acetone several times, and slowly infiltrated and embedded in Spurr’s epoxy resin (Canemco; Spurr, 1969). Mature seeds were pierced with a razor blade and fixed in 3% glutaraldehyde in 0.1 m PO4 buffer, pH 7.0, then osmicated with 1% (w/v) osmium tetroxide in 0.05 m PO4 buffer, pH 7.0, and washed three times with deionized water followed by dehydration in increasing-percentage ethanol washes. All samples were then transferred to a propylene oxide solution and slowly infiltrated with Spurr’s epoxy resin (Canemco). Resin-embedded samples were then thick sectioned (0.5 mm) using a Reichert Ultracut E microtome and stained with 1% (w/v) toluidine blue O in 1× (w/v) sodium borate (pH 11). Sections were examined by light microscopy and evaluated for intact seed coat cells and proper developmental stage based on the criteria described by Western et al. (2000). Photographs were taken using an Axioskop 2 microscope (Carl Zeiss) and Q Capture pro imaging software (Q Imaging).
Mucilage Precipitation
The mucilage content was determined by using the ammonium-oxalate method (Arsovski et al., 2009; Stork et al., 2010). A total of 50 mg (dry weight) of Arabidopsis seeds was incubated in 0.2% (w/v) aqueous ammonium-oxalate solution at 37°C for 8 h. To facilitate the extraction of mucilage, test tubes were vortexed for 1 min every hour. The seeds were removed by centrifugation after incubation, and the resultant supernatant was transferred to a fresh tube. Five volumes of ethanol were added to the supernatant followed by 30 min of incubation on ice to precipitate the polysaccharides. Samples were centrifuged in a bench-top centrifuge for 30 min at 21,000g. Precipitated mucilage was washed with 70% ethanol, air dried, and weighed. Average weights were calculated based on three mucilage replicates for mutant and wild-type seeds.
Cellulose Content and Monosaccharide Analysis
Cellulose content of seed cell walls was measured colorimetrically (Updegraff, 1969), and neutral sugar composition was determined by HPLC. Briefly, cell wall material from mature seed samples was prepared by sequential washing (five times) with 70% ethanol for 45 min at 70°C followed by five sequential acetone washes at room temperature for 2 min each. To remove the starch, the vacuum-dried samples were redissolved in 1 mL of α-amylase solution (Sigma-Aldrich no. A8220) according to the manufacturer’s instructions and incubated for 15 min at 37°C. The α-amylase-treated samples were washed and vacuum dried. Exactly 4 mg of the air-dried cell wall material was measured into glass tubes, and standards of different neutral sugars (Ara, Fuc, Gal, Glc, Man, Rha, and Xyl) were added to individual glass tubes (5.6 μL of 10 mm stock) and dried at 60°C. A total of 35 μL of 72% (w/v) H2SO4 was added to each sample and standard incubated on ice for 2 h, mixing every 30 min with the vortex. Samples were diluted to 4% (w/v) H2SO4 by adding 965 μL of water and autoclaved at 121°C for 1 h.
For soluble mucilage samples, approximately 20 mg of seeds (exact weight recorded) was hydrated with 1.2 mL of water, 10 μL of 5 mg mL−1 d-erythritol was added as an internal standard, and the samples were shaken on an orbital shaker for 2 h. One milliliter of mucilage was removed and dried at 60°C under a stream of nitrogen. Sulfuric acid (72% [w/v], 17.5 μL) was added to each sample, incubated on ice for 2 h, and shaken every 30 min. Water (480 μL) was then added to give a final concentration of 4% sulfuric acid, and the samples were autoclaved for 60 min at 121°C before being filtered through 0.45-mm nylon syringe filters. Neutral sugar standards (Fuc, Ara, Rha, Gal, Glc, Man, and Xyl) and acid sugar standards (GalUA) were also processed in the same way at the same time.
Neutral and acidic cell wall sugars were identified and quantified by pulsed electrochemical detection using a Dionex ED50 apparatus. Sugar separation was achieved using a BioLC GS50 HPLC device and a CarboPAC-PAl pellicular anion-exchange column with column guard (Dionex). Column and detector temperature was maintained at 30°C using a Dionex LC25 chromatography oven. Samples were introduced to the column from a Dionex AS50 autosampler and neutral sugars were eluted isocratically in 22 mm NaOH over a 30-min period, after which the column was washed in 1 m NaOAc and 200 mm NaOH for 5 min. The column was recharged by passing 200 mm NaOH through it for 10 min and then reequilibrating the column in 22 mm NaOH for 10 min prior to injection of the next sample. This protocol resulted in optimal (near-baseline) resolution of Gal, Glc, Man, and Xyl (Downie and Bewley, 1996). Acidic sugar separation and quantification used the same HPLC procedure but followed the eluent profile of Dean et al. (2007). For soluble mucilage, monosaccharides were quantified with the d-erythritol internal standard after correction of response factors with monosaccharide standards of different concentrations to allow area to be converted to molar amounts. Sugars from mucilage were normalized to the mass of seed used in the extraction. For cell wall monosaccharides, the neutral sugars were normalized to the amount of cell wall material used for the sample preparation.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers NC_003076 for CESA5/AT5G09870, NM_127746 for CESA9/AT2G21770, and NM_120095 for CESA2/AT4G39350.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Quantitative assessment of aberrant cell shape in additional cesa alleles.
Supplemental Figure S2. Mutation in CESA6 (procuste) does not cause seed coat phenotypes.
Supplemental Figure S3. Stem cellulose content in the wild type and cesa mutants.
Supplemental Figure S4. Phenotypic examination of dark- and light-grown seedlings.
Supplemental Figure S5. Phenotypic examination of aberrant mucilage in the wild type, cesa5-1, and cesa5-2.
Supplemental Table S1. T-DNA insertion lines and primers used for analysis.
Supplemental Movie S1. Cell morphology in wild-type seed coats.
Supplemental Movie S2. Cell morphology in cesa2-1cesa5-1cesa9-1 seed coats.
Acknowledgments
We are grateful for the technical assistance of Dr. David McNear, Dr. Michael Goodin, Darby Harris, and Brian Williams, for technical advice from Heather McFarlane and Drs. Robin Young and Tamara Western, and to the University of British Columbia bioimaging facility for equipment use.
References
- Albani JR, Plancke YD. (1998) Interaction between calcofluor white and carbohydrates of alpha 1-acid glycoprotein. Carbohydr Res 314: 169–175 [DOI] [PubMed] [Google Scholar]
- Anderson CT, Carroll A, Akhmetova L, Somerville C. (2010) Real-time imaging of cellulose reorientation during cell wall expansion in Arabidopsis roots. Plant Physiol 152: 787–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arioli T, Peng L, Betzner AS, Burn J, Wittke W, Herth W, Camilleri C, Höfte H, Plazinski J, Birch R, et al. (1998) Molecular analysis of cellulose biosynthesis in Arabidopsis. Science 279: 717–720 [DOI] [PubMed] [Google Scholar]
- Arsovski AA, Haughn GW, Western TL. (2010) Seed coat mucilage cells of Arabidopsis thaliana as a model for plant cell wall research. Plant Signal Behav 5: 796–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arsovski AA, Villota MM, Rowland O, Subramaniam R, Western TL. (2009) MUM ENHANCERS are important for seed coat mucilage production and mucilage secretory cell differentiation in Arabidopsis thaliana. J Exp Bot 60: 2601–2612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beeckman T, De Rycke R, Viane R, Inzé D. (2000) Histological study of seed coat development in Arabidopsis thaliana. J Plant Res 113: 139–148 [Google Scholar]
- Beeckman T, Przemeck GKH, Stamatiou G, Lau R, Terryn N, De Rycke R, Inzé D, Berleth T. (2002) Genetic complexity of cellulose synthase a gene function in Arabidopsis embryogenesis. Plant Physiol 130: 1883–1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake AW, McCartney L, Flint JE, Bolam DN, Boraston AB, Gilbert HJ, Knox JP. (2006) Understanding the biological rationale for the diversity of cellulose-directed carbohydrate-binding modules in prokaryotic enzymes. J Biol Chem 281: 29321–29329 [DOI] [PubMed] [Google Scholar]
- Brown RM., Jr (1985) Cellulose microfibril assembly and orientation: recent developments. J Cell Sci Suppl 2: 13–32 [DOI] [PubMed] [Google Scholar]
- Carpita N, Vergara C. (1998) A recipe for cellulose. Science 279: 672–673 [DOI] [PubMed] [Google Scholar]
- Carpita NC, Gibeaut DM. (1993) Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant J 3: 1–30 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Colombo PM, Rascio N. (1977) Ruthenium red staining for electron microscopy of plant material. J Ultrastruct Res 60: 135–139 [DOI] [PubMed] [Google Scholar]
- Dean GH, Zheng H, Tewari J, Huang J, Young DS, Hwang YT, Western TL, Carpita NC, McCann MC, Mansfield SD, et al. (2007) The Arabidopsis MUM2 gene encodes a β-galactosidase required for the production of seed coat mucilage with correct hydration properties. Plant Cell 19: 4007–4021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desprez T, Juraniec M, Crowell EF, Jouy H, Pochylova Z, Parcy F, Höfte H, Gonneau M, Vernhettes S. (2007) Organization of cellulose synthase complexes involved in primary cell wall synthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA 104: 15572–15577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desprez T, Vernhettes S, Fagard M, Refrégier G, Desnos T, Aletti E, Py N, Pelletier S, Höfte H. (2002) Resistance against herbicide isoxaben and cellulose deficiency caused by distinct mutations in same cellulose synthase isoform CESA6. Plant Physiol 128: 482–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick-Pérez M, Zhang Y, Hayes J, Salazar A, Zabotina OA, Hong M. (2011) Structure and interactions of plant cell-wall polysaccharides by two- and three-dimensional magic-angle-spinning solid-state NMR. Biochemistry 50: 989–1000 [DOI] [PubMed] [Google Scholar]
- Downie B, Bewley JD. (1996) Dormancy in white spruce (Picea glauca [Moench.] Voss.) seeds is imposed by tissues surrounding the embryo. Seed Sci Res 6: 9–15 [Google Scholar]
- Fagard M, Desnos T, Desprez T, Goubet F, Refregier G, Mouille G, McCann M, Rayon C, Vernhettes S, Höfte H. (2000) PROCUSTE1 encodes a cellulose synthase required for normal cell elongation specifically in roots and dark-grown hypocotyls of Arabidopsis. Plant Cell 12: 2409–2424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillmor CS, Poindexter P, Lorieau J, Palcic MM, Somerville C. (2002) α-Glucosidase I is required for cellulose biosynthesis and morphogenesis in Arabidopsis. J Cell Biol 156: 1003–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez A, Mendenhall J, Huo Y, Lloyd A. (2009) TTG1 complex MYBs, MYB5 and TT2, control outer seed coat differentiation. Dev Biol 325: 412–421 [DOI] [PubMed] [Google Scholar]
- Green PB. (1962) Mechanism for plant cellular morphogenesis. Science 138: 1404–1405 [DOI] [PubMed] [Google Scholar]
- Haughn G, Chaudhury A. (2005) Genetic analysis of seed coat development in Arabidopsis. Trends Plant Sci 10: 472–477 [DOI] [PubMed] [Google Scholar]
- Herth W, Schnepf E. (1980) The fluorochrome, calcofluor white, binds oriented to structural polysaccharide fibrils. Protoplasma 105: 129–133 [Google Scholar]
- Huang J, Debowles D, Esfandiari E, Dean G, Carpita NC, Haughn GW. (2011) The Arabidopsis transcription factor LUH/MUM1 is required for extrusion of seed coat mucilage. Plant Physiol 156: 491–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jofuku KD, den Boer BG, Van Montagu M, Okamuro JK. (1994) Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 6: 1211–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SF, Milliken ON, Pham H, Seyit R, Napoli R, Preston J, Koltunow AM, Parish RW. (2009) The Arabidopsis MYB5 transcription factor regulates mucilage synthesis, seed coat development, and trichome morphogenesis. Plant Cell 21: 72–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macquet A, Ralet M-C, Kronenberger J, Marion-Poll A, North HM. (2007a) In situ, chemical and macromolecular study of the composition of Arabidopsis thaliana seed coat mucilage. Plant Cell Physiol 48: 984–999 [DOI] [PubMed] [Google Scholar]
- Macquet A, Ralet M-C, Loudet O, Kronenberger J, Mouille G, Marion-Poll A, North HM. (2007b) A naturally occurring mutation in an Arabidopsis accession affects a β-d-galactosidase that increases the hydrophilic potential of rhamnogalacturonan I in seed mucilage. Plant Cell 19: 3990–4006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- North H, Baud S, Debeaujon I, Dubos C, Dubreucq B, Grappin P, Jullien M, Lepiniec L, Marion-Poll A, Miquel M, et al. (2010) Arabidopsis seed secrets unravelled after a decade of genetic and omics-driven research. Plant J 61: 971–981 [DOI] [PubMed] [Google Scholar]
- Nowack MK, Ungru A, Bjerkan KN, Grini PE, Schnittger A. (2010) Reproductive cross-talk: seed development in flowering plants. Biochem Soc Trans 38: 604–612 [DOI] [PubMed] [Google Scholar]
- Persson S, Paredez A, Carroll A, Palsdottir H, Doblin M, Poindexter P, Khitrov N, Auer M, Somerville CR. (2007) Genetic evidence for three unique components in primary cell-wall cellulose synthase complexes in Arabidopsis. Proc Natl Acad Sci USA 104: 15566–15571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson S, Wei H, Milne J, Page GP, Somerville CR. (2005) Identification of genes required for cellulose synthesis by regression analysis of public microarray data sets. Proc Natl Acad Sci USA 102: 8633–8638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing KH, Samuels AL, Savidge RA. (2002) Ultrastructure of vascular cambial cell cytokinesis in pine seedlings preserved by cryofixation and substitution. Protoplasma 220: 39–49 [DOI] [PubMed] [Google Scholar]
- Richmond TA, Somerville CR. (2000) The cellulose synthase superfamily. Plant Physiol 124: 495–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheible W-R, Eshed R, Richmond T, Delmer D, Somerville C. (2001) Modifications of cellulose synthase confer resistance to isoxaben and thiazolidinone herbicides in Arabidopsis Ixr1 mutants. Proc Natl Acad Sci USA 98: 10079–10084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song D, Shen J, Li L. (2010) Characterization of cellulose synthase complexes in Populus xylem differentiation. New Phytol 187: 777–790 [DOI] [PubMed] [Google Scholar]
- Spurr AR. (1969) A low-viscosity epoxy resin embedding medium for electron microscopy. J Ultrastruct Res 26: 31–43 [DOI] [PubMed] [Google Scholar]
- Stork J, Harris D, Griffiths J, Williams B, Beisson F, Li-Beisson Y, Mendu V, Haughn G, Debolt S. (2010) CELLULOSE SYNTHASE9 serves a nonredundant role in secondary cell wall synthesis in Arabidopsis epidermal testa cells. Plant Physiol 153: 580–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor NG, Howells RM, Huttly AK, Vickers K, Turner SR. (2003) Interactions among three distinct CesA proteins essential for cellulose synthesis. Proc Natl Acad Sci USA 100: 1450–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga-Wada R, Iwata M, Sugiyama J, Kotake T, Ishida T, Yokoyama R, Nishitani K, Okada K, Wada T. (2009) The GLABRA2 homeodomain protein directly regulates CESA5 and XTH17 gene expression in Arabidopsis roots. Plant J 60: 564–574 [DOI] [PubMed] [Google Scholar]
- Updegraff DM. (1969) Semimicro determination of cellulose in biological materials. Anal Biochem 32: 420–424 [DOI] [PubMed] [Google Scholar]
- Western TL, Burn J, Tan WL, Skinner DJ, Martin-McCaffrey L, Moffatt BA, Haughn GW. (2001) Isolation and characterization of mutants defective in seed coat mucilage secretory cell development in Arabidopsis. Plant Physiol 127: 998–1011 [PMC free article] [PubMed] [Google Scholar]
- Western TL, Skinner DJ, Haughn GW. (2000) Differentiation of mucilage secretory cells of the Arabidopsis seed coat. Plant Physiol 122: 345–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Western TL, Young DS, Dean GH, Tan WL, Samuels AL, Haughn GW. (2004) MUCILAGE-MODIFIED4 encodes a putative pectin biosynthetic enzyme developmentally regulated by APETALA2, TRANSPARENT TESTA GLABRA1, and GLABRA2 in the Arabidopsis seed coat. Plant Physiol 134: 296–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willats WGT, McCartney L, Mackie W, Knox JP. (2001) Pectin: cell biology and prospects for functional analysis. Plant Mol Biol 47: 9–27 [PubMed] [Google Scholar]
- Williamson RE, Burn JE, Hocart CH. (2001) Cellulose synthesis: mutational analysis and genomic perspectives using Arabidopsis thaliana. Cell Mol Life Sci 58: 1475–1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windsor JB, Symonds VV, Mendenhall J, Lloyd AM. (2000) Arabidopsis seed coat development: morphological differentiation of the outer integument. Plant J 22: 483–493 [DOI] [PubMed] [Google Scholar]
- Young RE, McFarlane HE, Hahn MG, Western TL, Haughn GW, Samuels AL. (2008) Analysis of the Golgi apparatus in Arabidopsis seed coat cells during polarized secretion of pectin-rich mucilage. Plant Cell 20: 1623–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. (2004) GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zykwinska AW, Ralet M-CJ, Garnier CD, Thibault J-FJ. (2005) Evidence for in vitro binding of pectin side chains to cellulose. Plant Physiol 139: 397–407 [DOI] [PMC free article] [PubMed] [Google Scholar]