Abstract
While oat (Avena sativa) has long been known to produce epoxy fatty acids in seeds, synthesized by a peroxygenase pathway, the gene encoding the peroxygenase remains to be determined. Here we report identification of a peroxygenase cDNA AsPXG1 from developing seeds of oat. AsPXG1 is a small protein with 249 amino acids in length and contains conserved heme-binding residues and a calcium-binding motif. When expressed in Pichia pastoris and Escherichia coli, AsPXG1 catalyzes the strictly hydroperoxide-dependent epoxidation of unsaturated fatty acids. It prefers hydroperoxy-trienoic acids over hydroperoxy-dienoic acids as oxygen donors to oxidize a wide range of unsaturated fatty acids with cis double bonds. Oleic acid is the most preferred substrate. The acyl carrier substrate specificity assay showed phospholipid and acyl-CoA were not effective substrate forms for AsPXG1 and it could only use free fatty acid or fatty acid methyl esters as substrates. A second gene, AsLOX2, cloned from oat codes for a 9-lipoxygenase catalyzing the synthesis of 9-hydroperoxy-dienoic and 9-hydroperoxy-trienoic acids, respectively, when linoleic (18:2-9c,12c) and linolenic (18:3-9c,12c,15c) acids were used as substrates. The peroxygenase pathway was reconstituted in vitro using a mixture of AsPXG1 and AsLOX2 extracts from E. coli. Incubation of methyl oleate and linoleic acid or linolenic acid with the enzyme mixture produced methyl 9,10-epoxy stearate. Incubation of linoleic acid alone with a mixture of AsPXG1 and AsLOX2 produced two major epoxy fatty acids, 9,10-epoxy-12-cis-octadecenoic acid and 12,13-epoxy-9-cis-octadecenoic acid, and a minor epoxy fatty acid, probably 12,13-epoxy-9-hydroxy-10-transoctadecenoic acid. AsPXG1 predominately catalyzes intermolecular peroxygenation.
Peroxygenase is a hydroperoxide-dependent oxygenase that catalyzes the transfer of one oxygen atom from a hydroperoxide to a substrate that is oxidized (Blée et al., 1993). Unlike cytochrome P450 epoxygenase, peroxygenase does not require cofactors such as NAD(P)H and molecular oxygen for substrate epoxidation (Blée et al., 1993; Matsunaga and Shiro, 2004).
Peroxygenase activity was first detected in oat (Avena sativa) when 9- and 13-hydroperoxy-octadecadienoic acids were reduced to the corresponding alcohols and 9,10-epoxy-13-hydroxyoctadecenoic acids or 12,13-epoxy-9-hydroxyoctadecenoic acids was formed (Heimann and Schreier, 1970; Heimann and Dresen, 1973). However, this reaction was then believed to be catalyzed by a lipoperoxidase activity or hydroperoxide isomerase. In 1977, peroxygenase was first defined by a labeling study using pea (Pisum sativum) microsomes (Ishimaru and Yamazaki, 1977) as hydroperoxide-dependent monooxygenase, which differs from typical cytochrome P450 monooxygenase in the oxygen donor. Since then, this type of enzyme activity has been detected in various microorganisms and plants species (Blée et al., 1993; Matsunaga and Shiro, 2004). In oat, peroxygenase-catalyzed fatty acid epoxidation/hydroxylation was confirmed using a seed microsomal fraction (Hamberg and Hamberg, 1996). In 2006, to our knowledge, the first peroxgenase gene was cloned from Arabidopsis (Arabidopsis thaliana) using the sequence information of a partially purified peroxygenase from oat (Hanano et al., 2006). The encoded Arabidopsis peroxygenase shares no sequence similarity with either peroxidase or cytochrome P450, but rather with caleosin, a small oil body-associated protein with both heme- and calcium-binding motifs in plant seeds. The peroxygenase activity is strictly dependent on the binding of calcium and ferric-heme (Hanano et al., 2006).
Association of the peroxygenase activity with caleosin is surprising. Caleosin was first described as an oil body protein which, like oleosin, was believed to be imbedded in the phospholipid monolayer of lipid particles playing solely a structural role in maintaining the integrity of oil bodies (Chen et al., 1999). However, with the discovery of the biochemical activity, this type of protein has been widely identified in fungi and other plant species and is localized not only in oil bodies, but also in nonlipid storage organelles and tissues (Murphy, 2005; Partridge and Murphy, 2009).
Oat has been known to produce several epoxy fatty acids in seeds, such as 9,10-epoxy-18:0, 9,10-epoxy-18:1-12c, and 12,13-epoxy-18:1-9c together accounting for approximately 0.5% of the total fatty acids. These fatty acids are mainly present in neutral lipids such as triacylglycerols and diacylglycerols (Leonova et al., 2008; Doehlert et al., 2010). Oat has also been extensively used as an enzyme source to study their biosynthesis, catalyzed by peroxygenase activity (Heimann and Schreier, 1970; Heimann and Dresen, 1973; Hamberg and Hamberg, 1996; Leonova et al., 2008). Although the exact biological function of these fatty acids is not defined, they are perceived to act as precursors for the synthesis of oxylipins, a group of oxygenated fatty acids, in response to biotic and abiotic stress (Feussner and Wasternack, 2002; Andreou et al., 2009) or as monomers for the biosynthesis of cutin polymers to cover aerial parts of the plant surface providing the hydrophobic structural barrier to the environment (Lequeu et al., 2003). While the activity of peroxygenase has been well defined in oat in planta, the gene encoding the enzyme remains unknown. Thus, the primary goal of this research is to clone the peroxygenase gene and characterize the peroxygenase pathway in vitro that is involved in the biosynthesis of epoxy fatty acids in oat.
RESULTS
Identification of cDNAs Encoding Fatty Acid Peroxygenase and Lipoxygenases from Oat
To identify a cDNA encoding fatty acid peroxygenase from oat, oat EST databases prepared from leaves, roots, and developing seeds at different stages that are publically available from the National Center for Biotechnology Information were searched using Arabidopsis ATS1, an embryo-specific caleosin (Nuccio and Thomas, 1999) as a query sequence. The search resulted in identification of ESTs representing six distinct caleosin-like sequences in oat. All these sequences appeared only in EST libraries prepared from developing seeds and were represented by only one or two ESTs. An exception was a single sequence in an EST collection from developing seeds at the watery stage that was represented by 34 ESTs. The sequence assembly using these highly abundant ESTs gave rise to a contig with 1,073 bp in length (AsPXG1). The sequence analysis of this contig revealed an open reading frame (ORF) with the putative start and stop codons at positions 63 and 810, respectively, and an in-frame stop codon at 30 nucleotides upstream the start codon. To clone the full-length cDNA, total RNA isolated from developing seeds of oat was used as template for reverse transcriptase (RT)-PCR amplification with two specific primers containing the putative start and stop codons. Sequencing of amplified PCR products revealed three types of isomers that differed in length at the 5′ ends, giving three different ORFs with 750, 735, and 717 bp in length. The reason why three different isoforms were amplified by a single pair of specific primers was not clear. Possibly they were spliced alternatively from the same transcript or transcribed from different loci/alleles that are highly homologous, as oat is a hexaploid. The longest ORF corresponded to the contig from the EST database and coded for a polypeptide of 249 amino acids with a predicted molecular mass of 28.1 kD (AsPXG1). Two conserved His residues (His-76 and His-144) were found in AsPXG1 that were believed to be involved in heme binding. The EF-hand motif for calcium binding was located between position 80 and 93. The sequence of 19 amino acids between position 10 and 29 (AVVVSDAMSSVAKGAPVTAQ) of AsPXG1 was identical to the sequenced fragment of the protein purified from oat lipid particles previously that was used to identify the first peroxygenase gene in Arabidopsis (Hanano et al., 2006; Fig. 1). Sequence comparison showed that AsPXG1 shares high amino acid sequence identity to abscisic acid-induced EF-hand, abscisic acid responsive27 (EFA27) (66%) from rice (Oryza sativa), AtPXG1 (At4g26740; 60%), and AtPXG2 (At5g55240; 58%) from Arabidopsis (Supplemental Fig. S1). The hydropathy analysis indicated that AsPXG1, similar to group I caleosins, has a hydrophobic membrane-associated domain in the middle of the protein sequence (Fig. 1; Hanano et al., 2006).
Figure 1.
A diagram illustrating the primary structure of the putative peroxygenase AsPXG1 from oat. The black box indicates 19 amino acids between residue 10 and 29 identical to the sequenced fragment of the peroxygenase protein purified from oat lipid particles previously. Two conserved His residues are located at position 76 and 144 that are involved in heme binding. The gray box indicates the EF-hand motif that is involved in calcium binding. The hatched box indicates a hydrophobic membrane-associated domain.
To identify cDNAs encoding fatty acid lipoxygenase from oat, the same oat EST databases were searched using Arabidopsis lipoxygenase 9-LOX (AtLOX5, Q9LUW0) and 13-LOX (AtLOX2, P38418; Bell and Mullet, 1993; Vellosillo et al., 2007) as query sequences. This resulted in identification of two ESTs from developing seed libraries. One was 700 bp long, while the other was 287 bp in length. To clone the full-length cDNAs, 5′RACE and 3′RACE methods were employed using mRNA isolated from developing seeds as templates. RACE amplifications identified two full-length cDNAs, AsLOX1 and AsLOX2, encoding two distinct putative lipoxygenases. Both cDNAs had ORFs of 2,586 bp long coding for a polypeptide of 862 amino acids with predicted molecular mass of approximately 97 kD. AsLOX1 and AsLOX2 shared amino acid sequence identity at 74% and exhibited high sequence identity to lipoxygenases from rice (P29250; 77% and 78%), Hordeum vulgare (P29114; 73% and 90%), and Zea mays (Q9LKL4; 75% and 74%, respectively; Supplemental Fig. S2). Phylogenic analysis indicated both AsLOX1 and AsLOX2 belong to type I lipoxygenase that does not possess any chloroplast transit peptide. Unlike type II lipoxygenase that is associated with chloroplasts and usually introduces a molecular oxygen at the C13 position of polyunsaturated acids, type I lipoxygenase is believed to be extraplastidic and introduce a peroxide group at the C9 position of linoleic and linolenic acids (Feussner and Wasternack, 2002).
Functional Characterization of the Putative Peroxygenase in Yeast
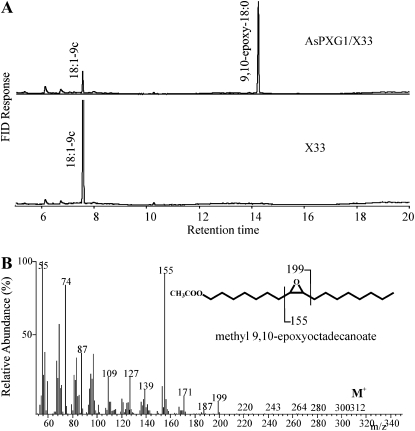
To functionally characterize the putative peroxygenase, the coding region of AsPXG1 was cloned into the yeast (Saccharomyces cerevisiae) expression vector pPICZ-B under control of the AOX1 promoter. The recombinant plasmid was introduced into Pichia pastoris X33. In presence of cumene hydroperoxide as oxygen donor and oleic acid as a substrate, cell-free extracts of AsPXG1/X33 expressing the putative peroxygenase produced a new fatty acid, compared with the control, with the same chromatographic retention time as 9,10-epoxy stearate (9,10-epoxy-18:0; Fig. 2A). Gas chromatography-mass spectrometry (GC-MS) analysis confirmed that the mass spectrum of this fatty acid is identical to that of authentic cis-9,10-epoxyoctadecanoic acid (Fig. 2B) with a molecular ion at mass-to-charge ratio (m/z) 312 and two prominent diagnostic fragments at m/z 199 and m/z 155. The m/z 199 fragment was assigned to the carboxyl-containing ion formed by the cleavage between carbon 10 and 11, while the m/z 155 fragment was the methyl terminal ion formed by the cleavage between carbon 8 and 9. In addition, it was found that formation of the epoxy fatty acid catalyzed by AsPXG1 was strictly hydroperoxide dependent and sensitive to β-mercaptoethanol, a known peroxygenase inhibitor (Supplemental Fig. S3). These results indicated AsPXG1 is the long-sought gene encoding a peroxygenase in oat that catalyzes strictly hydroperoxide-dependent monooxygenation of unsaturated fatty acids, giving rise to corresponding epoxy fatty acids.
Figure 2.
Functional characterization of oat putative peroxygenase AsPXG1 in P. pastoris in presence of cumene hydroperoxide as oxygen donor and oleic acid as a substrate. A, GC analysis of fatty acid methyl esters prepared from enzymatic reactions using extracts of Pichia AsPXG1/X33 and the control X33 cells (the empty vector). B, The mass spectrum of methyl 9,10-epoxy stearate produced by AsPXG1. The fatty acid derivative was prepared from enzymatic reactions using extracts of Pichia AsPXG1/X33.
Enzymatic Properties of AsPXG1
Oat peroxygenase is considered to be a membrane-bound monooxygenase associated with either microsomes or lipid particle membranes (Hanano et al., 2006). To confirm whether AsPXG1 is associated with microsomal membrane or lipid particles, the yeast crude extracts of AsPXG1/X33 were fractionated into soluble, lipid particle, and microsomal portions by centrifugation and the three subfractions were assayed. The result showed that the lipid particle and microsomal fractions possessed high levels of peroxygenase activities (415.4 ± 30.6 and 50.6 ± 3.2 nmol min−2 mg−1, respectively) that were approximately 20 and 2.5 times that of the soluble fraction (19.5 ± 1.0 nmol min−2 mg−1). In addition, no activity was observed in the microsomal fraction when emulphogene, a detergent that could effectively solubilize the peroxgenase from membrane (Hanano et al., 2006), was added to the lysis buffer. Rather, all the peroxygenase activity was observed in the detergent-solubilized supernatant fraction. These results indicate that, consistent with the result from the hydropathy analysis, AsPXG1 is a membrane-associated enzyme that might be located at both lipid particles and endoplasmic reticulum. However, it could be readily solubilized from the membrane by a detergent and the solubilized protein retained enzyme activity.
To further characterize the enzymatic properties of AsPXG1, the crude extract of AsPXG1/X33 was subjected to the activity assay under different temperature and pH buffers. The results showed that AsPXG1 exhibited activity at a range of pH (5–9) and temperature (15°C–60°C) with an optimal pH value of 7 and an optimal temperature of 45°C (Supplemental Figs. S4 and S5, respectively).
To determine the substrate specificity of AsPXG1, a variety of free fatty acids including saturated, unsaturated, and unusual ones ranging in the chain length from C14 to C24 were provided in the in vitro assays using cumene hydroperoxide as oxidant. Results showed that AsPXG1 possessed high substrate selectivity. It could only use unsaturated fatty acids with double bonds in the cis configuration as substrates, whereas saturated fatty acids or unsaturated fatty acids with double bonds in the trans configuration were not accepted by the enzyme. In addition, the enzyme was inactive toward ricinoleic acid (12-OH-18:1-9c), although this fatty acid contains a cis double bond at position 9 (Table I). Among fatty acids examined, 9-cis-octadecenoic acid (18:1-9c, oleic acid) is the most preferred substrate for AsPXG1, which was followed by 10-cis-pentadecenoic acid (15:1-10c), 10-cis-heptadecenoic acid (17:1-10c), 9-cis-hexadecenoic acid (16:1-9c), and 6-cis-octadecenoic acid (18:1-6c; Table I). Polyunsaturated fatty acids with 18 carbon such as linoleic acid (18:2-9c,12c) and linolenic acid (18:3-9c,12c,15c), as well as very-long-chain monounsatutated fatty acids such as erucic acid (22:1-13C) and nervonic acid (24:1-15c) could be epoxidized, but with less efficiency.
Table I. The substrate specificity of AsPXG1 in P. pastoris.
The in vitro assays with different substrates were performed using cumene hydroperoxide as oxidant as described in “Materials and Methods.” The substrates for which no activity was detected are 18:0, 16:1-9t, 18:1-9t, and 12-OH-18:1-12c.
| Substrate | Relative Activity |
| % | |
| 14:1-9c | 76.3 |
| 15:1-10c | 98.8 |
| 16:1-9c | 88.9 |
| 17:1-10c | 94.1 |
| 18:1-6c | 87.3 |
| 18:1-9c | 100 |
| 18:1-11C | 71.9 |
| 19:1-10c | 77.0 |
| 20:1-11C | 74.8 |
| 22:1-13C | 54.8 |
| 24:1-15c | 52.6 |
| 18:2-9c,12c | 46.7 |
| 18:3-9c,12c,15c | 54.9 |
To identify the acyl carrier substrate specificity of AsPXG1, different types of oleic derivatives such as free fatty acid, fatty acid methyl ester, fatty acyl-CoA, and phosphatidylcholine were examined by the in vitro assay. The results showed that AsPXG1 could only use free fatty acid and fatty acid methyl esters as substrates; whereas phospholipid and acyl-CoA were not effective substrate forms (see Supplemental Fig. S6). The ability to use fatty acid methyl ester as substrate by AsPXG1 provides a simple and sensitive way to evaluate the activity by direct chromatographic analysis of reaction products without fatty acid methyl ester derivatization.
To investigate the cosubstrate specificity of AsPXG1, common types of hydroperoxy-dienoic (HPOD) and hydroperoxy-trienoic (HPOT) acids such as 9-OOH-18:2-10t,12c, 13-OOH-18:2-9c,11t, 9-OOH-18:3-10t,12c,15c, and 13-OOH-18:3-9c,11t,15c were examined by in vitro assays using methyl oleate as a substrate to be epoxidized. The result showed that AsPXG1 could effectively utilize all these hydroperoxy fatty acids as oxygen donors. However, HPOT acids (13-HPOT and 9-HPOT) were more efficiently reduced by the peroxygenase as they had significantly lower Km values compared with those of HPOD acids (13-HPOD and 9-HPOD) although HPOD acids possessed slightly higher, but not statistically significantly different, Vmax values (Table II). These results indicated AsPXG1 preferred HPOT acid over HPOD acid as oxidants to epoxidize oleic acid.
Table II. The kinetic parameters of oat AsPXG1 in P. pastoris on cooxidants.
The in vitro assays were performed using methyl oleate as substrate as described in “Materials and Methods.” Means with the same letters are not significantly different according to statistical analysis (Duncan test at P = 0.01, n = 3).
| Substrate | Kmapp | Vmaxapp | Vmax/Kmapp |
| μm | pkat·mg−1 | pkat·mg−1·μm | |
| 9-HPOD | 16.07 ± 0.78 a | 214.04 ± 11.84 e | 13.33 ± 0.92 g |
| 9-HPOT | 10.42 ± 0.42 b | 188.69 ± 22.88 e | 18.06 ± 1.46 h |
| 13-HPOD | 25.18 ± 1.80 c | 306.57 ± 22.02 e | 12.20 ± 0.92 g |
| 13-HPOT | 7.25 ± 1.52 b | 173.49 ± 23.51 e | 24.43 ± 4.54 i |
| Cumene hydroperoxide | 215.76 ± 1.23 d | 883.34 ± 118.79 f | 4.10 ± 0.57 j |
To examine whether AsPXG1 is also functional in Escherichia coli, the coding region of AsPXG1 was cloned into the bacterial vector pET15b. The recombinant plasmid was introduced into the Rosetta2(DE3)pLysS strain. The in vitro assay was conducted under the same condition used in the Pichia system. The result showed that, like AsPXG1 expressed in Pichia, AsPXG1 expressed in E. coli could also efficiently epoxidize oleic acid when cumin hydroperoxide was used as oxygen donor (Supplemental Fig. S7). Functional expression of AsPXG1 in E. coli provides us with the opportunity to reconstitute the two-gene peroxygenase pathway in the bacterium (see below).
Functional Characterization of the Oat Putative Lipoxygenases
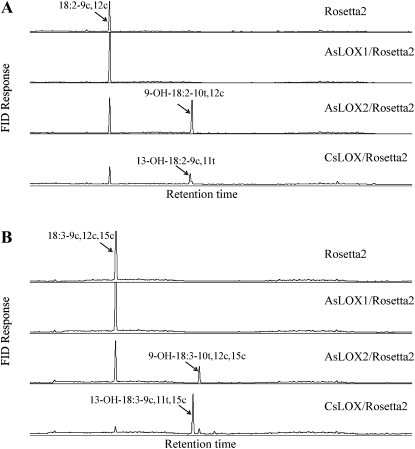
To functionally characterize putative oat lipoxygenases genes, the coding regions of AsLOX1 and AsLOX2 were cloned into the bacterial vector pET28a. The recombinant plasmids were transformed into the E. coli strain Rosetta2(DE3)pLysS. Three common fatty acids found in oat seeds, i.e. oleic acid (18:1-9c), linoleic acid (18:2-9c,12c), and linolenic acid (18:3-9c,12c,15c; Leonova et al., 2008) in both free acid and methyl ester forms were used for the activity assay using a previously well-characterized 13-lipoxygenase CsLOX from cucumber (Cucumis sativus) as a positive control (Hornung et al., 1999). The results showed that, similar to the positive control CsLOX13/Rosetta2, AsLOX2/Rosetta2 had high activity toward both linoleic and linolenic free fatty acids, and no activity was observed on either form of oleic acid or methyl ester forms of linoleic acid and linolenic acid. However, unlike cucumber CsLOX that produced 13-HPOD acid (13-OOH-18:2-9c,11t) and 13-HPOT acid (13-OOH-18:3-9c,11t,15c) predominately with a very small amount of 9-hydroperoxides, oat AsLOX2 only produced 9-HPOD acid (9-OOH-18:2-10t,12c) and 9-HPOT acid (9-OOH-18:3-10t,12c,15c), respectively, when linoleic acid and linolenic acid were used as substrates (Fig. 3). The identity of these hydroperoxydienoic and hydroperoxytrienoic acid products was confirmed by GC-MS analysis of the reduced derivatives in comparison with appropriate standards (Supplemental Fig. S8). AsLOX1, unlike AsLOX2, showed no activity toward any of the substrates tested when expressed in E. coli.
Figure 3.
Functional characterization of oat putative lipoxygenases in E. coli. Hydroperoxy fatty acids were reduced to hydroxyl fatty acids by TnCl, followed by derivatization by trimethylsilyl as described in “Materials and Methods.” A, 18:2-9c,12c as substrate. B, 18:3-9c,12c,15c as substrate. The control is Rosetta2 cells transformed with the empty vector.
Cooxidation of Unsaturated Fatty Acids by Oat Lipoxygenase and Peroxygenase
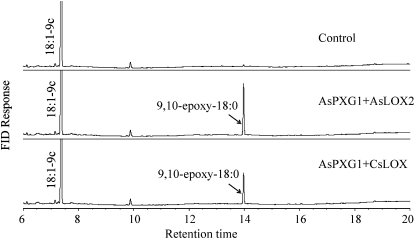
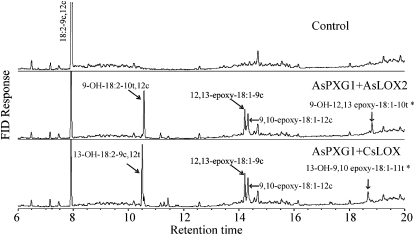
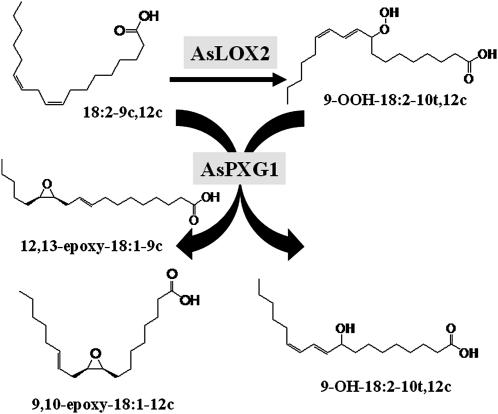
To determine whether fatty acid hydroperoxides produced by oat lipoxygenase could be used by the peroxygenase as oxygen donor to oxidize unsaturated fatty acids, an in vitro assay was established using AsPXG1 and AsLOX2 expressed separately in E. coli Rosetta2(DE3)pLysS. A mixture of the equal amount of the two enzyme extracts was incubated with methyl oleate and free linoleic acid (Fig. 4) or free linolenic acid (Supplemental Fig. S9). Direct GC analysis of fatty acid methyl esters in the reaction products showed that the combination of AsPXG1 and AsLOX2 could effectively cooxidize the substrates provided, resulting in production of methyl epoxy stearate. Similar results were obtained using a mixture of E. coli AsPXG1 and CsLOX extracts. These results indicated AsPXG1 could use both 9-HPOD/HPOT acids and 13-HPOD/HPOT acids synthesized by the lipoxygenases as oxygen donors to oxidize the monounsaturated fatty acid, producing the corresponding epoxy fatty acid. When a mixture of the two enzyme extracts was incubated with linoleic acid alone, the reaction produced four new products detected by GC analysis of trimethylsilyl derivatives of fatty acid methyl esters: 9-OH-18:2-10t,12c (15.4%), 12,13-epoxy-18:1-9c (7.0%), 9,10-epoxy-18:1-12c (5.7%), and probably 9-OH-12,13-epoxy-18:1-10t (3.4%) in order of abundance (mol % of the total fatty acids; Fig. 5). The identity of these products, except for 9-OH-12,13-epoxy-18:1-10t, was confirmed by GC-MS in comparison with relevant standards (Supplemental Fig. S10). Although there is no standard available for 9-OH-12,13-epoxy-18:1-10t to confirm the structure, this fatty acid has been previously observed as one of the products produced by oat peroxygenase (Hamberg and Hamberg, 1996). 9-OH-18:2-10t,12c was presumably derived from reduction of 9-OOH-18:2-10t,12c synthesized by AsLOX2 from linoleic acid. 12,13-Epoxy-18:1-9c was presumably derived from epoxidation of the Δ12 double bond of linoleic acid using 9-OOH-18:2-10t,12c as cooxidant. 9,10-Epoxy-18:1-12c was presumably derived from epoxidation of the Δ9 double bond of linoleic acid using 9-OOH-18:2-10t,12c as cooxidant. 9-OH-12,13-epoxy-18:1-10t was presumably derived from epoxidation of the Δ12 double bond of 9-OH-18:2-10t,12c (Fig. 6). A higher level of 12,13-epoxy-18:1-9c than 9,10-epoxy-18:1-12c produced from the reconstitution indicated that the epoxidation occurred preferentially at the Δ12 double bond over the Δ9 double bond of linoleic acids.
Figure 4.
Cooxidation of methyl oleate and linoleic acid by AsPXG1 and AsLOX2 or AsPXG1 and CsLOX in E. coli. Direct GC analysis of fatty acid methyl esters in the reaction products was performed as described in “Materials and Methods.” The control is Rosetta2 cells transformed with the empty vector.
Figure 5.
Cooxidation of linoleic acid alone by AsPXG1 and AsLOX2 or AsPXG1 and CsLOX in E. coli. GC analysis of trimethylsilyl derivatives of fatty acid methyl esters was performed as described in “Materials and Methods.” The control is Rosetta2 cells transformed with the empty vector. *, The structure was not confirmed.
Figure 6.
A diagram illustrating the reconstituted peroxygenase pathway in E. coli in presence of linoleic acid.
To confirm hydroperoxides produced by a cucumber 13-lipoxygenase could also be used by the oat peroxygenase as oxygen donor to oxidize unsaturated fatty acids, a similar coexpression experiment was conducted using AsPXG1 and CsLOX2 expressed separately in E. coli Rosetta2(DE3)pLysS. A mixture of the equal amounts of the two enzyme extracts incubated with linoleic acid (18:2-9c,12c) alone also produced four new products detected by GC analysis of trimethylsilyl derivatives of fatty acid methyl esters: 13-OH-18:2-9c,11t (20.3%), 12,13-epoxy-18:1-9c (7.6%), 9,10-epoxy-18:1-12c (6.3%), and probably 13-OH-9,10-epoxy-18:1-11t (4.1%) in order of abundance (Fig. 5). 13-OH-18:2-9c,11t was presumably derived reduction of 13-OOH-18:2-9c,11t synthesized by the cucumber lipoxygenase from linoleic acid. 12,13-Epoxy-18:1-9c was presumably derived from epoxidation of the Δ12 double bond of linoleic acid using 13-OOH-18:2-9c,11t as cooxidant. 9,10-Epoxy-18:1-12c was derived from epoxidation of the Δ9 double bond of linoleic acid using 13-OOH-18:2-9c,11t as cooxidant. 13-OH-9,10-epoxy-18:1-11t was derived from epoxidation of the Δ9 double bond of 13-OH-18:2-9c,11t. Again, these results confirmed that the epoxidation occurred preferentially at the Δ12 double bond of linoleic acids.
Tissue-Specific Expression of Oat Lipoxygenase and Peroxygenase Genes
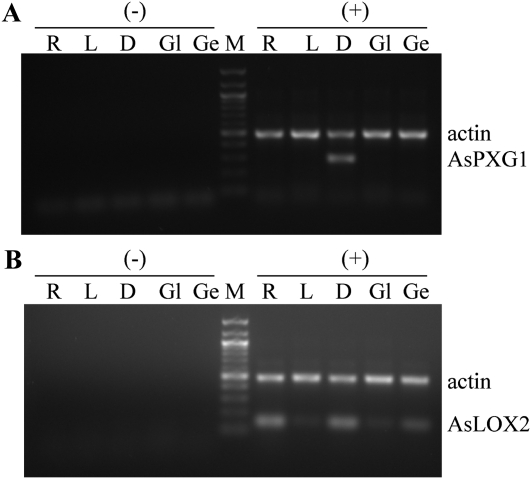
As shown in Figure 7, AsPXG1 was exclusively expressed in developing seeds, not in other tissues examined such as roots, leaves, glumes, and germinating seeds, whereas AsLOX2 was expressed in all tissues examined with relatively higher expression in developing seeds, roots, and germinating seeds.
Figure 7.
Tissue-specific expression of AsPXG1 (A) and AsLOX2 (B) in oat. RT-PCR amplification was conducted using total RNA isolated from roots (R), leaves (L), developing seeds (D), glumes (Gl), and germinating seeds (Ge) with (+) or without (−) RT.
DISCUSSION
There are three mechanisms underlying the biosynthesis of epoxy fatty acids in plants. The first one was identified by Stymne and colleagues that involves a Δ12 desaturase-like oxygenase epoxidizing the Δ12 double bond of linoleic acid linked to phosphatidylcholine, giving rise to 12,13-epoxy-18:1-9c (vernolic acid) in Crepis palaestina (Lee et al., 1998). The second one was elucidated by Cahoon and colleagues that involves a cytochrome P450-like oxygenase epoxidizing the same substrate, giving the same product in Euphorbia lagascae (Cahoon et al., 2002). The peroxygenase system is the third mechanism involved in the biosynthesis of epoxy fatty acids in plants. Unlike the first two mechanisms that use molecular oxygen as oxidant and phospholipid as acyl carrier substrate, the peroxygenase uses an oxygen atom from fatty acid hydroperoxide as oxidant to epoxidize substrates. Our in vitro study on the acyl carrier substrate specificity showed that AsPXG1 could only use free fatty acid or fatty acid methyl esters as substrates, while phospholipid and acyl-CoA were not effective substrate forms. Free fatty acid is likely the biological substrate for AsPXG1 as fatty acid methyl ester is not a viable form in the cells.
Oat has been known to produce several epoxy and hydroxyl fatty acids in seeds synthesized by a peroxygenase pathway, and has thus served as a model system to study enzymatic properties of the peroxygenase (Hamberg and Hamberg, 1996). While the oat peroxygenase activity that catalyzes the biosynthesis of these fatty acids has been well characterized in planta, the gene encoding the peroxygenase has not been cloned and characterized in vitro in detail. This study identified a peroxygenase cDNA AsPXG1 from oat developing seeds using the sequence similarity search of EST databases prepared from different tissues of oat plants with an Arabidopsis caleosin as query sequence. AsPXG1 is a small protein with 249 amino acids in length and contains conserved heme-binding residues and calcium-binding motif. Like type I caleosin, AsPXG1 has a hydrophobic domain in the middle of the sequence (Hanano et al., 2006). Consistent with this observation, activity of the recombinant enzyme AsPXG1 in Pichia was found to be primarily associated with the lipid particle and microsomal fraction. However, a substantial amount of peroxygenase activity was also found in the soluble fraction. This proportion of activity might result from the process of enzyme preparation, as AsPXG1 contains only a single 20-amino acid membrane-associated domain and the weak association of AsPXG1 with membrane might render it being easily solubilized into the supernatant fraction. This assumption is supported by observation that AsPXG1 is to be readily solubilized from the microsomal membrane by the emulphogene detergent.
When expressed in either Pichia or E. coli, AsPXG1 catalyzes strictly hydroperoxide-dependent epoxidation of unsaturated fatty acids giving rise to corresponding epoxy fatty acids with high selectivity for the substrate to be oxidized as well as the cosubstrate to be reduced. Under the assay condition used in this study, AsPXG1 significantly prefers HPOT acids over HPOD acids as oxygen donors to oxidize a very wide range of unsaturated fatty acids with cis-double bonds. Oleic acid is the most preferred substrate to be epoxidized by the peroxygenase. With the broad substrate selectivity, high stereospecificity, and low regioselectivity, AsPXG1 can account for the synthesis of all the epoxy fatty acids in oat seeds.
In this report, we have also cloned two lipoxygenase-like genes, AsLOX1 and AsLOX2 from oat seeds. AsLOX1 showed no lipoxygenase activity when expressed in E. coli. Nonfunction of AsLOX1 might be due to potential mutations already existing in the native sequence or accidently introduced during cloning process. However, other possibility such as a low level of expression or misfolding of the expressed protein in E. coli cannot be excluded. AsLOX2, when expressed in E. coli, catalyzes the synthesis of 9-HPOD and 9-HPOT acids when linoleic and linolenic acids were used as substrates, indicating it codes for 9-lipoxygenase. Unlike 13-lipoxygenase, AsLOX2 does not contain a transit peptide for chloroplast targeting; thus, it is presumably extraplastidic. Previous enzymatic assays in planta showed that the peroxygenation in oat seeds is almost exclusively derived from 9-lipoxygenase (Hamberg and Hamberg, 1996). Thus, AsLOX2 identified in this report might be the lipoxygenase in oat seeds.
The peroxygenase pathway constitutes one branch of the lipoxygenase pathway where oxygenation of a polyunsaturated fatty acid by lipoxygenase gives rise to corresponding fatty acid hydroperoxide, which is then used by peroxygenase as oxygen donor to oxidize an unsaturated fatty acid (Hanano et al., 2006). This study reconstituted the peroxygenase pathway in vitro using a mixture of AsPXG1 and AsLOX2 extracts from E. coli. Incubation of the mixture of the enzymes with methyl oleate and linoleic acid or linolenic acid produced a high level of methyl 9,10-epoxy stearate. The initial reaction in this reconstituted peroxygenase pathway catalyzes the formation of 9-HPOD acid by AsLOX2, which is then followed by the second reaction that catalyzes epoxidation of oleic acid by AsPXG1 using a 9-HPOD acid or 9-HPOT acid as the cooxidant. Incubation of a mixture of AsPXG1 and AsLOX2 with linoleic acid alone produced two major epoxy fatty acids, 9,10-epoxy-18:1-12c and 12,13-epoxy-18:1-9c, while structurally unconfirmed 9-OH-12,13-epoxy-18:1-10t is only a minor product of the cooxidation (Fig. 5). 9,10-Epoxy-18:1-12c and 12,13-epoxy-18:1-9c are derived from intermolecular peroxygenation, i.e. epoxidation of Δ9 and Δ12 double bonds of linoleic acid, respectively, using 9-HPOD acid as oxygen donor, while 9-OH-12,13-epoxy-18:1-10t can be derived from either intramolecular or intermolecular peroxygenation (Fig. 6). Intramolecular peroxygenation occurs when 9-HPOD acid is epoxidized using oxygen from the hydroperoxy group of the same molecule, while intermolecular peroxygenation occurs when 9-hydroxy-dienoic acid reduced from a 9-HPOD acid is epoxidized using oxygen atom from another 9-HPOD acid molecule. Based on this analysis, we concluded that intermolecular peroxygenation of polyunsaturated fatty acids occurs predominately, if not exclusively, in the oat peroxygenase pathway involved in the biosynthesis of epoxy fatty acids.
The peroxygenase pathway has been perceived to be involved in the biosynthesis of oxylipins in response to environmental stress or in the biosynthesis of cutin polymers in the plant surface area (Hanano et al., 2006). In oat, AsPXG1 is exclusively expressed in developing seeds, not in any other tissues such as leaves, roots, and germinating seeds. It is mainly associated with endoplasmic reticulum and lipid particles. AsLOX2, on the other hand, is widely expressed in oat tissues including developing seeds. It has been documented that lipoxylgenase can also be associated with lipid particles (Feussner et al., 1997). Thus, coordinately temporal and spatial expression of the two enzymes in the peroxygenase pathway would result in accumulation of a small amount of epoxy fatty acids in seed triacylglycerols. These stored oxygenated fatty acids could be used as precursors for synthesis of active oxylipins in germinating seeds, whereby inducing defensive systems against abiotic and biotic stress during seed germination and seedling establishment.
MATERIALS AND METHODS
Oat seeds (Avena sativa ‘CDC Dancer’) were kindly provided by Dr. Aaron Beattie, Department of Plant Sciences, University of Saskatchewan. All common fatty acids used in this study were purchased from Nuchek. Fatty acid 9-hydroperoxides (9-HPOD and 9-HPOT), 13-hydroperoxides (13-HPOD and 13-HPOT), such as 9-OOH-18:2-10t,12c, 9-OOH-18:3-10t,12c,15c, 13-OOH-18:2-9c,11t, and 13-OOH-18:3-9c,11t,15c as well as epoxy fatty acids such as 12,13-epoxy-18:1-9c, 9,10-epoxy-18:1-12c were purchased from Larodan Fine chemicals (Handelsbanken SE-205 40). 9-OH-18:2-10t,12c, 9-OH-18:3-10t,12c,15c, 13-OH-18:2-9c,11t, and 13-OH-18:3-9c,11t,15c were purchased from Cayman Chemical Company. The cumene hydroperoxide was obtained from Sigma-Aldrich.
Cloning of AsPXG1 and AsLOX cDNAs from Oat
To clone genes encoding fatty acid peroxygenase (AsPXG1) and lipoxygenases (AsLOX1 and AsLOX2), total RNA was isolated from developing seeds of oat using TRIzol reagent (Invitrogen). Five micrograms of the total RNA was used to synthesize first-strand cDNA using the SuperScript III first-strand synthesis system (Invitrogen). Two microliters of the cDNA was used as a template for PCR amplification for AsPXG1 cDNA with two specific primers, DM203, 5′-ATGGCGGAGGACGCGGT-3′ and DM204, 5′-CTAGTGCTGCTTCCCGTGTGC-3′ using Pfx50 DNA polymerase (Invitrogen). The amplified products having the expected size of approximately 750 bp were gel purified, then cloned into pYES2.1-TOPO vector (Invitrogen), producing construct pDM60. The 5′ and 3′ ends of AsLOX1 and AsLOX2 cDNAs encoding putative 9-lipoxygenases were obtained using the Marathon cDNA amplification kit (BD Biosciences, CLONTECH) following the manufacturer’s instruction. Primers DM172 (5′-CCGCACCGCCATGCCCCTCTTGATTAG-3′), DM173 (5′-CCCGCCGGCATTGACGAGCAACTC-3′), DM183 (5′-GGCGTGGTTGTTCTTGACGATGATGGCG-3′), and primer DM174 (5′-CGCCGTGATGGAGCCCTTCATTATCGC-3′) were used to obtain the 5′ and 3′ ends of AsLOX1 and primers DM176 (5′-GCCCACCACCTTGCTCTCGATGTCCACG-3′), DM177 (5′-TCTGGCTGGTGATGGTGTGGATGAACGC-3′), DM196 (5′-GCCGTCGCCGCGCAGGAAGAAGAG-3′), DM197 (5′-CGCCGCCGTGATGGTGCTGGTGTG-3′), and primer DM178 (5′-GGCCAGTACCCGTACGCCGGGGACCTC-3′) were used to obtain the 5′ and 3′ ends of AsLOX2, respectively.
Construction of Yeast Expression Plasmid of AsPXG1
The ORF of AsPXG1 was amplified by PCR using a forward primer DM194 containing an EcoRI restriction site and a Kozak consensus sequence (5′-GCGAATTCATCATGGCGGAGGACGC-3′), and a reverse primer containing a XbaI site at the 5′ end (5′-TGCTCTAGAAAGTGCTGCTTCCCGTGTG-3′). The PCR product with expected size was purified, digested, and cloned into pPICZ-B vector (Invitrogen) using EcoRI and XbaI sites. The resulting plasmid (pDM71) contained an additional sequence encoding 23 amino acid residues (FLEQKLISEEDLNSAVDHHHHHH) at the 3′ end of the cloned gene.
Expression of the AsPXG1 cDNA in Pichia pastoris
Ten micrograms of pDM71 containing AsPXG1 was linearized with DraI and transformed into Pichia pastoris X-33 (Invitrogen) by electroporation using an electroporator 2510 (Eppendorf). The transformed cells were grown at 28°C for 24 h with shaking in buffered minimal glycerol medium containing 100 mm potassium phosphate, pH 6.0, 1.34% yeast (Saccharomyces cerevisiae) nitrogen base without amino acids, 4 × 10−5% biotin, and 1% glycerol. The cells were then centrifuged and resuspended in a buffered minimal methanol medium containing 100 mm potassium phosphate, pH 6.0, 1.34% yeast nitrogen base without amino acids, 4 × 10−5% biotin, and 1% methanol at an OD600 of 1.0 to induce the expression. After 60 h, the resulting cultures were harvested and washed once with Tris buffer (50 mm Tris-HCl pH 7.5). The pellet was resuspended in a homogenizing buffer containing 50 mm Tris-HCl (pH 7.5) and 0.6 m sorbitol, and the cells were disrupted with glass beads using a Mini-Beadbeater (Biospect; three cycles of 1 min each). The homogenate was subfractionated into supernatant, microsome, and lipid articles (Kamisaka and Nakahara, 1994), and these subfractions were then used as the enzyme sources for the activity assay. Protein concentration was determined according to Bradford using bovine serum albumin as the standard (Bradford, 1976).
Peroxygenase Activity Assays
Unless otherwise stated, peroxygenase assays were carried out in a volume of 200 μL containing 0.1 mg protein, 0.5 mm cumene hydroperoxide, 0.5 mm oleic acid, 50 mm Tris-HCl (pH 7.0), and 10% glycerol. The reaction mixture was incubated at 45°C for 15 min with 500 rpm shaking and stopped by the addition of 500 μL ethyl acetate containing 10 μL of glacial acetic acid. The fatty acid products were extracted twice with ethyl acetate and methylated with diazomethane. The resulting fatty acid methyl esters were analyzed by GC and/or GC-MS. Identity of the products was confirmed by comparing GC retention time and MS data with those of standards. For quantifying the products, 3 μg of 18:0 was used as an internal standard. The relative activity of substrates was determined by GC(flame ionization detector) quantification of products versus substrate using the internal standard for each substrate and then comparison of its value to that of oleic acid substrate. The fatty acid methyl ester samples were analyzed on an Agilent 6890N GC equipped with a DB-23 column (30-m × 0.25-mm) with 0.25-μm film thickness (J&W Scientific). The column temperature was maintained at 160°C for 1 min, and then raised to 240°C at a rate of 4°C/min.
The pH optimum of AsPXG1 was evaluated using 0.5 mm cumene hydroperoxide and 0.5 mm methyl oleate from pH 5.0 to 9.0 using one of three 50 mm buffers (MES, HEPES, and Tris-HCl) in 0.5-unit intervals. The optimal temperature was similarly evaluated from 15°C to 60°C in 5°C intervals at pH 7.0.
Apparent kinetic parameters were determined under optimum conditions. The range of substrate concentrations used to determine the kinetic parameters of 9-HPOD and 9-HPOT was 4 to 128 μm, while those for 13-HPOD and 13-HPOT were 4 to 130 μm and 3 to 107 μm, respectively. The kinetic constants were estimated from Lineweaver-Burke plots using the average of triplicate measurements.
Expression of Peroxygenase and Lipoxygenase in Escherichia coli
For activity assay of AsPXG in Escherichia coli, coding region was reamplified with two primers containing NdeI restriction sites DM200 5′-CCATATGATGGCGGAGGACGCGGTG-3′ and DM201 5′-CCATATGCTAGTGCTGCTTCCCGTGTG-3′ using pDM60 as template. The amplified fragment was subcloned into intermediate vector pCR4-TOPO TA (Invitrogen) and then transferred into E. coli expression vector, pET15b (Novagen) to yield pDM70. The activity assay of peroxygenase expressed in E. coli was carried out with crude extracts of AsPXG1/Rosetta2(DE3)pLysS following the protocol as described above.
The bacterial expression vector pET28a (Novagen) was used to express three lipoxygenase cDNAs in E. coli. Two full-length lipoxygenase cDNAs from oat (AsLOX1 and AsLOX2) were amplified by RT-PCR using total RNA from developing seeds as template with two primers containing BamHI restriction sites DM188 (5′-CGAGGATCCAAGATGTTCGGCGGCCTGG-3′) and DM189 (5′-CGCGGATCCTTAGATGGAGATACTGTTGGGG-3′) for AsLOX1 and with two primers containing EcoRI restriction sites DM207 (5′-GCGAATTCAGGATGCTGCTGGGCGG-3′) and DM208 (5′-GCGAATTCTCAGATGGAGATGCTGTTGGG-3′) for AsLOX2. The third full-length lipoxygenase cDNA encoding 13-lipoxygenase from cucumber (Cucumis sativa; CsLOX13; Hornung et al., 1999) was amplified by RT-PCR using the RNA from cucumber germinating seeds (7 d) as template and two primers containing BamHI restriction sites DM184 (5′-CGCGGATCCAAAATGTTTGGAATTGGGAAGAAC-3′) and DM185 (5′-CGCGGATCCTTAGATAGAAATACTATTAGGAATTCC-3′). PCR fragments were digested with appropriate restriction enzymes and gel purified. Both AsLOX1 and CsLOX13 were cloned into the BamHI site of pET28a to yield pDM80 and pDM82, respectively. The AsLOX2 were cloned into the EcoRI site of pET28a to yield pDM81. All constructs were checked by sequencing prior to the expression studies.
For functional analysis of the lypoxygenases in E. coli, the constructs were transformed into the Rosetta2(DE3)pLysS strain. The E.coli transformant, Rosetta2(DE3)pLysS/pDM70, Rosetta2(DE3)pLysS/pDM80, Rosetta2(DE3)pLysS/pDM81, or Rosetta2(DE3)pLysS/pDM82 was first grown at 37°C overnight in 10 mL Luria-Bertani medium containing either kanamycin (50 μg/mL) or ampicillin (100 μg/mL) along with chloramphenicol (34 μg/mL). The fresh culture was then inoculated into 50 volumes of Luria-Bertani medium containing the same antibiotics. When the culture was grown at 37°C till OD600 = 0.5 to 1.0, isopropyl-β-d-thiogalactopyranoside was added to a final concentration of 1 mm to induce the expression at 30°C for 4 h. After induction, the cells were harvested by centrifugation (5,000 rpm, 15 min). The cell pellets were first resuspended in a homogenizing buffer containing 50 mm Tris-HCl (pH 7.5), 0.6 m sorbitol, 50 μg/mL RNaseA, and 20 μg/mL DNaseI with sonication. Lipoxygenase activity was determined by incubation of 20 μL of lysate with 250 μm of fatty acid in the reaction buffer containing 50 mm Tris-HCl, pH 7.0, and 10% glycerol. The reaction was incubated at 30°C for 30 min with 500 rpm shaking. After the reaction, the resulting hydroperoxide products were reduced to their corresponding hydroxy derivatives with 25 mg/mL of Tin (II) Chloride (SnCl2) in ethanol. The mixture was then extracted twice with 2 mL ethyl acetate. The pooled organic phases were evaporated under nitrogen gas and the residue was derivatized with 200 μL of bovine serum albumin (Aldrich)/pyridine (1:1) at 80°C for 30 min. The trimethylsilyl esters were analyzed by GC and/or GC-MS.
Tissue-Specific Expression of AsPXG1 and AsLOX2
Total RNAs were prepared from roots, leaves, developing seeds, glumes, and germinating seeds at 5 d using TRIzol reagent (Invitrogen). One microgram of total RNA was treated with DNase I (Invitrogen) and used for cDNA synthesis using SuperScript III RT-PCR system in 20 μL reaction with Oligo(dT)20 primer according to the manufacturer’s instructions. One microliter of the first-strand reaction was then used as a template for 25-μL multiplex PCR reaction using Taq DNA polymerase (UBI Life sciences). The specific primers DM218 (5′-TGATGACCAATGACCACAGGC-3′) and DM219 (5′-GGCGGTAGCTTGTCCTCCTC-3′) were used to amplify a fragment of AsPXG1 cDNA with 282 bp. The primers DM178 (5′-GGCCAGTACCCGTACGCCGGGGACCTC-3′) and DM177 (5′-TCTGGCTGGTGATGGTGTGGATGAACGC-3′) were used to amplify a fragment of AsLOX2 cDNA with 139 bp. Primers DM216 (5′-GGTGGGGATGGGGCAGAA-3′) and DM217 (5′-CCCGCTCGGCAGTGGTG-3′) were used to generate a 488-bp internal control for a housekeeping gene, actin2. The PCR conditions for both multiplex PCR reaction were 30 cycles of 95°C for 30 s, 62°C for 30 s, and 72°C for 30 s. A 10-μL aliquot of both reactions was used for agarose gel analysis.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers JN390966 for AsPXG1 and JN390967 for AsLOX2.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Alignment of AsPXG1 with related sequences.
Supplemental Figure S2. Alignment of AsLOX1 and AsLOX2 with related sequences.
Supplemental Figure S3. AsPXG1 activity in presence of beta-mercaptoethanol.
Supplemental Figure S4. The optimal pH value of AsPXG1.
Supplemental Figure S5. The optimal temperature of AsPXG1.
Supplemental Figure S6. The acyl carrier substrate specificity of AsPXG1.
Supplemental Figure S7. Functional characterization of oat putative peroxygenase AsPXG1 in E. coli.
Supplemental Figure S8. The mass spectra of reduced derivatives of hydroperoxides produced by AsLOX2 and CsLOX when linoleic or linolenic acid was used as substrate.
Supplemental Figure S9. Co-oxidation of methyl oleate and linolenic acid by AsPXG1 and AsLOX2 or AsPXG1 and CsLOX in E. coli.
Supplemental Figure S10. The mass spectra of methyl 12,13-epoxy-18:1-9c and 9,10-epoxy-18:1-12c.
Acknowledgments
We thank Dr. Aaron Beattie (Plant Science Department, University of Saskatchewan) for providing oat seeds, Jack Chen (University of Saskatchewan) for technical help, Sangjie Jiang (University of Saskatchewan) for statistical analysis, Darwin Reed (National Research Council of Canada, Plant Biotechnology Institute) for technical discussions, and Drs. Pat Covello and Mark Smith (National Research Council of Canada, Plant Biotechnology Institute) for critical review of the manuscript.
References
- Andreou A, Brodhun F, Feussner I. (2009) Biosynthesis of oxylipins in non-mammals. Prog Lipid Res 48: 148–170 [DOI] [PubMed] [Google Scholar]
- Bell E, Mullet JE. (1993) Characterization of an Arabidopsis lipoxygenase gene responsive to methyl jasmonate and wounding. Plant Physiol 103: 1133–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blée E, Wilcox AL, Marnett LJ, Schuber F. (1993) Mechanism of reaction of fatty acid hydroperoxides with soybean peroxygenase. J Biol Chem 268: 1708–1715 [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Cahoon EB, Ripp KG, Hall SE, McGonigle B. (2002) Transgenic production of epoxy fatty acids by expression of a cytochrome P450 enzyme from Euphorbia lagascae seed. Plant Physiol 128: 615–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JC, Tsai CC, Tzen JT. (1999) Cloning and secondary structure analysis of caleosin, a unique calcium-binding protein in oil bodies of plant seeds. Plant Cell Physiol 40: 1079–1086 [DOI] [PubMed] [Google Scholar]
- Doehlert DC, Angelikousis S, Vick B. (2010) Accumulation of oxygenated fatty acids in oat lipids during storage. Cereal Chemistry 87: 532–537 [Google Scholar]
- Feussner I, Balkenhohl TJ, Porzel A, Kühn H, Wasternack C. (1997) Structural elucidation of oxygenated storage lipids in cucumber cotyledons: implication of lipid body lipoxygenase in lipid mobilization during germination. J Biol Chem 272: 21635–21641 [DOI] [PubMed] [Google Scholar]
- Feussner I, Wasternack C. (2002) The lipoxygenase pathway. Annu Rev Plant Biol 53: 275–297 [DOI] [PubMed] [Google Scholar]
- Hamberg M, Hamberg G. (1996) Peroxygenase-catalyzed fatty acid epoxidation in cereal seeds (sequential oxidation of linoleic acid into 9(S),12(S),13(S)-trihydroxy-10(E)-octadecenoic acid). Plant Physiol 110: 807–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanano A, Burcklen M, Flenet M, Ivancich A, Louwagie M, Garin J, Blée E. (2006) Plant seed peroxygenase is an original heme-oxygenase with an EF-hand calcium binding motif. J Biol Chem 281: 33140–33151 [DOI] [PubMed] [Google Scholar]
- Heimann W, Dresen P. (1973) Enzymatic hydroperoxide degradation in cereals, enzyme characterization and reaction products. Helv Chim Acta 56: 463–469 [Google Scholar]
- Heimann W, Schreier P. (1970) The formation of 9-hydroxy-10,12- and 13-hydroxy-9,11-octadecadienoic acid through “lipoperoxidase” of cereals. Helv Chim Acta 53: 2296–2297 [Google Scholar]
- Hornung E, Walther M, Kühn H, Feussner I. (1999) Conversion of cucumber linoleate 13-lipoxygenase to a 9-lipoxygenating species by site-directed mutagenesis. Proc Natl Acad Sci USA 96: 4192–4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimaru A, Yamazaki I. (1977) Hydroperoxide-dependent hydroxylation involving “H2O2-reducible hemoprotein” in microsomes of pea seeds: a new type enzyme acting on hydroperoxide and a physiological role of seed lipoxygenase. J Biol Chem 252: 6118–6124 [PubMed] [Google Scholar]
- Kamisaka Y, Nakahara T. (1994) Characterization of the diacylglycerol acyltransferase activity in the lipid body fraction from an oleaginous fungus. J Biochem 116: 1295–1301 [DOI] [PubMed] [Google Scholar]
- Lee M, Lenman M, Banaś A, Bafor M, Singh S, Schweizer M, Nilsson R, Liljenberg C, Dahlqvist A, Gummeson PO, et al. (1998) Identification of non-heme diiron proteins that catalyze triple bond and epoxy group formation. Science 280: 915–918 [DOI] [PubMed] [Google Scholar]
- Leonova S, Shelenga T, Hamberg M, Konarev AV, Loskutov I, Carlsson AS. (2008) Analysis of oil composition in cultivars and wild species of oat (Avena sp.). J Agric Food Chem 56: 7983–7991 [DOI] [PubMed] [Google Scholar]
- Lequeu J, Fauconnier ML, Chammaï A, Bronner R, Blée E. (2003) Formation of plant cuticle: evidence for the occurrence of the peroxygenase pathway. Plant J 36: 155–164 [DOI] [PubMed] [Google Scholar]
- Matsunaga I, Shiro Y. (2004) Peroxide-utilizing biocatalysts: structural and functional diversity of heme-containing enzymes. Curr Opin Chem Biol 8: 127–132 [DOI] [PubMed] [Google Scholar]
- Murphy DJ. (2005) Lipid-associated proteins. Murphy DJ, , Plant Lipids: Biology, Utilisation and Manipulation. Blackwell, Oxford, pp 226–269 [Google Scholar]
- Nuccio ML, Thomas TL. (1999) ATS1 and ATS3: two novel embryo-specific genes in Arabidopsis thaliana. Plant Mol Biol 39: 1153–1163 [DOI] [PubMed] [Google Scholar]
- Partridge M, Murphy DJ. (2009) Roles of a membrane-bound caleosin and putative peroxygenase in biotic and abiotic stress responses in Arabidopsis. Plant Physiol Biochem 47: 796–806 [DOI] [PubMed] [Google Scholar]
- Vellosillo T, Martínez M, López MA, Vicente J, Cascón T, Dolan L, Hamberg M, Castresana C. (2007) Oxylipins produced by the 9-lipoxygenase pathway in Arabidopsis regulate lateral root development and defense responses through a specific signaling cascade. Plant Cell 19: 831–846 [DOI] [PMC free article] [PubMed] [Google Scholar]