Abstract
Bud endodormancy in woody plants plays an important role in their perennial growth cycles. We previously identified a MADS box gene, DORMANCY-ASSOCIATED MADS box6 (PmDAM6), expressed in the endodormant lateral buds of Japanese apricot (Prunus mume), as a candidate for the dormancy-controlling gene. In this study, we demonstrate the growth inhibitory functions of PmDAM6 by overexpressing it in transgenic poplar (Populus tremula × Populus tremuloides). Transgenic poplar plants constitutively expressing PmDAM6 showed growth cessation and terminal bud set under environmental conditions in which control transformants continued shoot tip growth, suggesting the growth inhibitory functions of PmDAM6. In the Japanese apricot genome, we identified six tandemly arrayed PmDAM genes (PmDAM1–PmDAM6) that conserve an amphiphilic repression motif, known to act as a repression domain, at the carboxyl-terminal end, suggesting that they all may act as transcriptional repressors. Seasonal expression analysis and cold treatment in autumn indicated that all PmDAMs were repressed during prolonged cold exposure and maintained at low levels until endodormancy release. Furthermore, PmDAM4 to PmDAM6 responses to a short period of cold exposure appeared to vary between low- and high-chill genotypes. In the high-chill genotype, a short period of cold exposure slightly increased PmDAM4 to PmDAM6 expression, while in the low-chill genotype, the same treatment repressed PmDAM4 to PmDAM6 expression. Furthermore, PmDAM4 to PmDAM6 expression was negatively correlated with endodormancy release. We here discuss the genotype-dependent seasonal expression patterns of PmDAMs in relation to their involvement in endodormancy and variation in chilling requirements.
Perennial plants in temperate and boreal zones have an annual growth cycle consisting of dormant and active growth phases. After shoot growth cessation and bud set, apical buds enter a dormant state called endodormancy. At the same time, lateral buds shift to the endodormant state from the paradormant state, in which the major growth inhibitory effects are imposed by apical dominance (Lang, 1987). Endodormant buds are incapable of initiating growth under favorable conditions without prior chilling (Crabbe and Barnola, 1996; Faust et al., 1997). These buds shift to the ecodormant state after a specific amount of chilling. In contrast to endodormancy, ecodormancy is imposed by external environmental factors such as cold or drought stress that induce critical signals and prevent bud growth (Lang, 1987; Crabbe and Barnola, 1996; Horvath et al., 2003).
Although internal physiological changes related to endodormancy in perennial plants, such as alterations in plant hormone contents, carbohydrate metabolism, and cell-to-cell communication (Rohde et al., 2002; Ruonala et al., 2006; Rohde and Bhalerao, 2007), have been extensively studied, the molecular mechanism of bud endodormancy is not yet well understood. In poplar trees (Populus species), the CONSTANS (CO)/FLOWERING LOCUS T (FT) module was involved in short day-triggered growth cessation and bud set (Böhlenius et al., 2006). Down-regulation of CENTRORADIALIS-LIKE1 (CENL1) correlates with short day-induced growth cessation (Ruonala et al., 2008). On the other hand, Mohamed et al. (2010) reported that CENTRORADIALIS (CEN)/TERMINAL FLOWER1 (TFL1) affects dormancy release and growth after dormancy release. These recent results suggest that flowering regulators found in poplar also play significant roles in growth cessation and possibly in dormancy. However, the genes involved in the induction and release of lateral bud endodormancy in temperate fruit tree species are not well understood. In addition, mechanisms of cultivar-dependent chilling requirements for endodormancy release are unknown.
Endodormancy is regulated by internal factors, suggesting that internal growth inhibitors, if any, are specifically localized in buds during endodormancy and prevent the buds from resuming growth. We previously performed suppression subtractive hybridization combined with mirror orientation selection to identify candidates for internal factors. We also performed differential screening to identify genes that are expressed preferentially in the endodormant buds of temperate fruit tree species such as Japanese apricot (Prunus mume; Yamane et al., 2008). We identified a MADS box gene with endodormancy-associated expression. Seasonal expression analysis suggested that the gene was up-regulated during endodormancy induction and down-regulated during endodormancy release. Full-length cDNA cloning of the MADS box gene and phylogenetic analysis revealed that the gene was similar to the StMADS11-clade MADS box genes such as SHORT VEGETATIVE PHASE (SVP) and AGAMOUS-LIKE24 (AGL24) of Arabidopsis (Arabidopsis thaliana; Yamane et al., 2008). Bielenberg et al. (2008) independently identified six StMADS11-clade MADS box genes as candidate genes associated with terminal bud formation in peach (Prunus persica) and named them DORMANCY-ASSOCIATED MADS1 to -6 (DAM1–6) genes. The gene we found in Japanese apricot appeared to be an ortholog of peach DAM6, and we named it PmDAM6 (Yamane et al., 2008). Six peach PpDAM genes showed distinct seasonal expression changes in the shoot apex of peach. Furthermore, PpDAM1, PpDAM2, and PpDAM4 were more closely associated with terminal bud formation (Li et al., 2009). Horvath et al. (2008) found that DAM homologs in leafy spurge (Euphorbia esula), EeDAM1 and EeDAM2, were associated with endodormancy induction. Although the StMADS11-clade MADS box genes were highly repeated in the poplar genome (Leseberg et al., 2006), ESTs similar to poplar DAM-like genes such as Populus trichocarpa MADS9 (accession no. XM_002301057) were up-regulated in the vascular tissue on seasonal dormancy induction (Druart et al., 2007) and bud set after short-day perception (Ruttink et al., 2007). Recently, a strong quantitative trait locus for chilling requirement and blooming time was found to be localized near PpDAM6 in peach (Fan et al., 2010). Furthermore, peach PpDAM5 and PpDAM6 expression was negatively correlated with the time required for terminal bud break in peach (Jiménez et al., 2010). The same trend of PpDAM5 and PpDAM6 expression was reported for lateral vegetative (Yamane et al., 2011a) and flower (Yamane et al., 2011b) buds. These results suggest that the StMADS11-clade MADS box genes are candidates for internal factors controlling endodormancy in perennial plants. However, no direct evidence is available showing that DAM functions as a growth inhibitor and is directly involved in bud dormancy.
In this study, we used transgenic approaches with poplar (Populus tremula × Populus tremuloides), a model plant species (Jansson and Douglas, 2007), to show the growth inhibitory function of PmDAM6 in Japanese apricot, one of the tree species recalcitrant to transformation. Furthermore, our survey of the Japanese apricot genome revealed the presence of tandemly arrayed PmDAM6 homologs, PmDAM1 to PmDAM5, in the genome region where PmDAM6 was present. A series of expression analyses strongly indicated the association of PmDAMs with endodormancy induction, maintenance, and release in Japanese apricot.
RESULTS
Constitutive PmDAM6 Expression Induced Growth Cessation and Bud Set in Transgenic Poplar
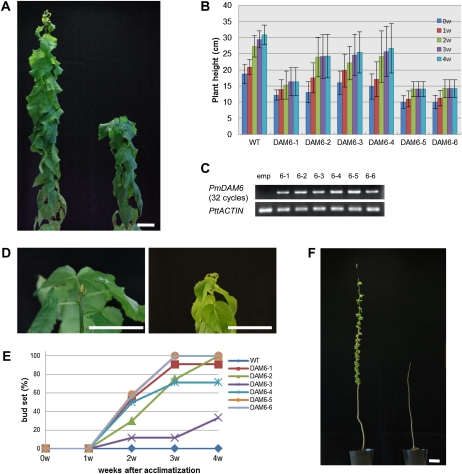
To elucidate the biological functions of PmDAM6, we produced transgenic poplar plants that constitutively expressed PmDAM6. These plants constitutively expressing PmDAM6 under the control of the cauliflower mosaic virus 35S promoter (35S:PmDAM6) were more difficult to obtain than control transformants with the empty vector (35S:empty) because the regeneration rate was much lower with 35S:PmDAM6 than with 35S:empty (data not shown). However, we successfully obtained six independently transformed lines (35S:PmDAM6-1 to -6). Although shoot growth was repressed in all six 35S:PmDAM6 lines, 35S:PmDAM6-1, -5, and -6 produced much shorter shoots (Fig. 1, A–C). In specific poplar genotypes, short days (less than approximately 14 h) are known to be an environmental signal that triggers growth cessation, bud set, and endodormancy (Böhlenius et al., 2006; Ruttink et al., 2007). Under long-day (LD) conditions (16/8 h of light/dark, 22°C), growth cessation was promoted and terminal bud set was induced in transgenic poplar plants with 35S:PmDAM6, whereas control plants, both wild-type and 35S:empty plants, showed continuous shoot growth (Fig. 1, D and E). To investigate the dormancy status of the terminal and lateral buds of the transgenic poplar plants, we excluded the inhibitory effects of leaves on bud burst by removing all leaves and observed bud growth under LD conditions. 35S:PmDAM6 poplar did not resume their growth, whereas the control plants showed bud burst (Fig. 1F).
Figure 1.
Constitutive expression of PmDAM6 induces growth cessation, bud set, and lateral bud endodormancy in poplar. A, PmDAM6 overexpression inhibited growth. Growth in the control plant (35S:empty; left) continued, whereas that in 35S:PmDAM6-1 (right) was arrested 1 month after acclimatization under LD conditions (16/8 h of light/dark, 22°C). B, Plant heights of control (wild type [WT]) and six independent 35S:PmDAM6 transgenic plants for 4 weeks immediately after acclimatization. C, Constitutive expression of PmDAM6 in six transgenic lines (RT-PCR with 32 cycles). PttACTIN was used as a control. D, Terminal bud set was induced in 35S:PmDAM6-1 (left), whereas growth in the control plant (35S:empty) continued under LD conditions. E, Bud set was induced earlier in six independent 35S:PmDAM6 transgenic plants. Percentages of bud set in seven to 14 plants of each line are shown. F, Burst of terminal and lateral buds was not observed in 35S:PmDAM6-2 under LD conditions. At approximately 6 weeks after acclimatization, leaves were removed and plants were placed under LD conditions for 2 weeks. Bud burst was observed in the control plant (35S:empty; left), whereas 35S:PmDAM6 buds (right) entered the dormant stage. Bars = 5 cm. [See online article for color version of this figure.]
Presence of Six Tandemly Arrayed PmDAM Genes in the Japanese Apricot Genome
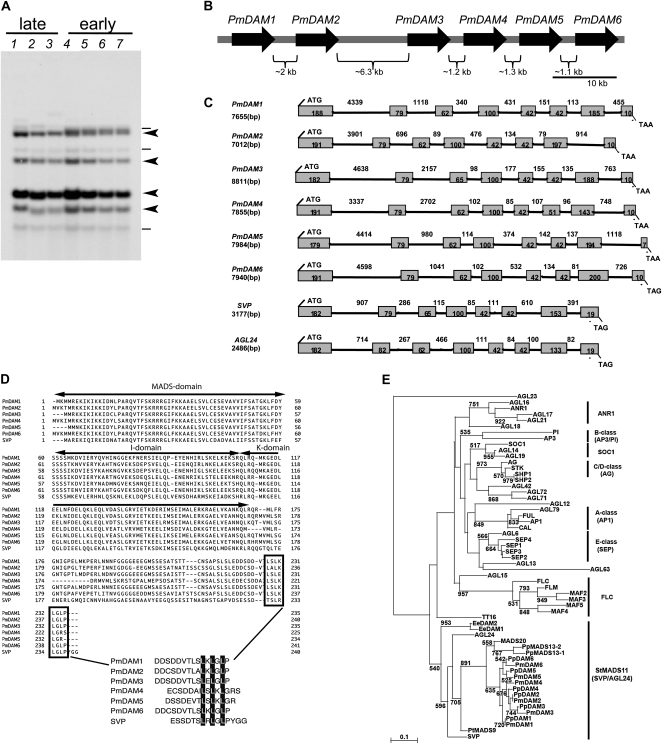
Genomic DNA-blot analysis with the PmDAM6 probe yielded plural hybridization signals, suggesting that the Japanese apricot genome has several sequences similar to PmDAM6 (Fig. 2A). Four strong and three faint bands were conserved in all cultivars tested, although band sizes differed slightly depending on the cultivars. Genomic library screening and shotgun sequencing revealed that Japanese apricot contains six tandemly arrayed MADS box genes that are putative homologs of PpDAM1 to PpDAM6 genes in peach (Bielenberg et al., 2008; Fig. 2B). We named these six genes in Japanese apricot PmDAM1 to PmDAM6 (Fig. 2B). All six PmDAMs have similar genomic structures consisting of eight exons and seven introns flanked by translation initiation and stop codons (Fig. 2C). The deduced amino acid sequences of all six PmDAMs contained the MADS box domain at the N-terminal end and the putative I region and K box domain at the middle position, similar to those observed in other MIKCc-type MADS box genes, suggesting that PmDAM genes encode a MIKCc-type MADS box (Fig. 2D). At the C-terminal end, PmDAMs have an ethylene-responsive element-binding factor-associated amphiphilic repression motif, SRDX, that acts as a repression domain and converts transcriptional activators to strong repressors when fused with DNA-binding proteins (Ohta et al., 2001). Phylogenetic analysis showed that the six PmDAMs belong to the StMADS11 (SVP/AGL24) clade of angiosperm MADS box genes, similar to PpDAMs of peach, EeDAMs of leafy spurge, and database-registered DAM-like genes of other species of Rosaceae (Fig. 2E). Each pair of DAM orthologs of Prunus, such as PmDAM1 and PpDAM1, was placed closely together in the phylogenetic tree.
Figure 2.
Six tandemly arrayed DAM genes in Japanese apricot. A, DNA gel-blot analysis using PmDAM6 as the probe. Cv Nanko (lane 1), cv Shirakaga (lane 2), and cv Oushuku (lane 3) are late-flowering cultivars, and cv Ellching (lane 4), cv Nisei (lane 5), selection SC (lane 6), and selection ST (lane 7) are early-flowering cultivars. Genomic DNAs digested with HindIII were hybridized with the PmDAM6 probe. Strong and faint bands are shown by arrowheads and horizontal lines, respectively. B, Overview of the PmDAM locus in the Japanese apricot genome. Six PmDAM genes are located as tandem repeats. C, Structures of PmDAM genes in Japanese apricot. Boxes and lines represent exons and introns, respectively. The number of nucleotide base pairs of each exon and intron are indicated. D, Alignment of the deduced amino acid sequences of PmDAMs of Japanese apricot and SVP of Arabidopsis. MADS box, K box, and I region domains are indicated by arrows. The boxes indicate the conserved ethylene-responsive element-binding factor-associated amphiphilic repression motif. E, Phylogenetic relationship among 40 Arabidopsis MADS box proteins and six Japanese apricot, six peach, two leafy spurge, two pear, one apple, and one poplar DAM-like proteins (Supplemental Table S2). The number at each branch indicates the bootstrap value of 1,000 replicates, and branches with more than 50% bootstrap values are shown.
Seasonal Endodormancy Status and PmDAM Expression Patterns in Two Japanese Apricot Cultivars with Different Chilling Requirements for Bud Break
The chilling requirements for bud break and seasonal changes in endodormancy depth are known to vary depending on the Japanese apricot cultivar. We previously investigated the chilling requirements of Japanese apricot cultivars that showed different blooming times in the field: a late-blooming cultivar, Nanko, and an early-blooming cultivar, Ellching, and found that the chilling requirement of cv Ellching flower buds was much lower than that of cv Nanko (Yamane et al., 2006). In this study, the seasonal endodormancy status of lateral vegetative buds was investigated in using these two Japanese apricot cultivars. Nanko vegetative buds did not open from July to January under forcing conditions and were considered to be endodormant during this period (Table I). Endodormancy in cv Nanko was released in February, since 72.2% of vegetative buds opened. Endodormancy in cv Ellching was shorter and lasted for 3 months (from August to October). In November, 25% of the vegetative buds of cv Ellching opened, and the percentages of bud burst in this cultivar from December to February were higher than those in cv Nanko. When single-node cuttings were used instead of whole branches to examine the dormancy status, cv Ellching buds opened within 1 month in all the months tested, although the buds in September and October took more days to open than in other months. No cv Nanko bud opened from August to November. These results confirmed that the Taiwanese early-blooming cv Ellching is a low-chill type, while cv Nanko is a high-chill type. Although the first bud bursts in cv Nanko and cv Ellching were observed in April and March, respectively, under field conditions, bud flush peaked almost at the same time in April, probably because of the lack of high-temperature days in March (data not shown).
Table I. Seasonal endodormancy status of lateral vegetative buds of two Japanese apricot cultivars tested under forcing conditions (16 h of light/8 h of dark, 22°C).
Long one-year-old branches and single-node cuttings from the long 1-year-old branches of field-grown trees were used for tests. For days to bud burst, first bud burst was observed within 10 d (++), within 30 d (+), or not observed within 30 d (−).
| Cultivar | Month |
||||||||
| June | July | August | September | October | November | December | January | February | |
| days to bud burst | |||||||||
| Long 1-year-old branches | |||||||||
| Nanko | ++ | − | − | − | − | − | − | − | ++ |
| Ellching | ++ | + | − | − | − | + | ++ | ++ | ++ |
| % bud burst | |||||||||
| Nanko | 33.3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 72.2 |
| Ellching | 70 | 5 | 0 | 0 | 0 | 25 | 85 | 100 | 100 |
| days to bud burst | |||||||||
| Single-node cuttings | |||||||||
| Nanko | ++ | ++ | − | − | − | − | + | ++ | ++ |
| Ellching | ++ | ++ | ++ | + | + | ++ | ++ | ++ | ++ |
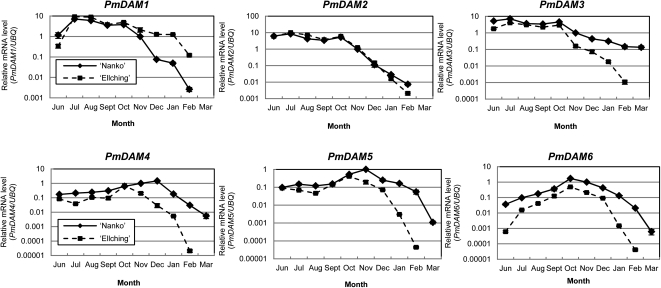
We investigated seasonal expression changes in PmDAM genes in the vegetative buds of two Japanese apricot cultivars by real-time reverse transcription (RT)-PCR using gene-specific TaqMan probes and primers (Fig. 3). Sequencing of genomic fragments and cDNAs cloned from cv Nanko and cv Ellching revealed that the designed probe and primer sequences were conserved in these cultivars (data not shown). PmDAM6 transcript levels significantly increased from June to October in cv Nanko and cv Ellching. PmDAM1 was up-regulated from June to July in both cultivars, while PmDAM2, PmDAM3, PmDAM4, and PmDAM5 transcript levels remained relatively constant during this period. Although transcript levels of all six PmDAMs decreased toward spring, the down-regulation patterns were different depending on the cultivars and type of PmDAMs. Namely, PmDAM3 to PmDAM6 transcript levels decreased earlier and faster in the low-chill cultivar, and PmDAM3 to PmDAM6 transcript levels were higher in cv Nanko than in cv Ellching throughout the period analyzed, while the PmDAM1 transcript level was higher in cv Ellching than in cv Nanko. The accumulation of PmDAM2 transcripts was similar in both cultivars. This trend was also found in another biological replicate, although the relative values of mRNA levels were not exactly the same between the two, especially when the transcript levels were low (Fig. 3; Supplemental Fig. S1).
Figure 3.
Seasonal expression changes in PmDAMs in the lateral vegetative buds of two Japanese apricot cultivars. Gene expression in the lateral vegetative buds of cv Nanko and cv Ellching grown in the field was assessed at monthly intervals by real-time PCR using TaqMan probes from June to March. Transcript levels of each gene were normalized by PmUBQ. The means of three technical replicates are shown, with error bars representing sd. The means of another biological replicate are shown in Supplemental Figure S1. Changes in expression are shown as logarithmic graphs. PmDAM1 and PmDAM2 transcripts in cv Nanko in March were present at undetectable levels under our experimental conditions; thus, we did not plot them in the figures.
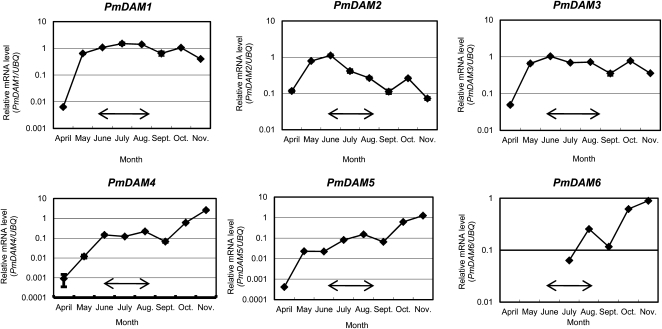
We also performed seasonal expression analysis of PmDAMs in cv Nanko leaves. Vegetative bud burst and growth in cv Nanko occurred in April under field conditions. Shoot growth cessation was first observed in late June, and the majority of shoots stopped their active growth in August; trees shed their leaves by early December. The seasonal expression change in PmDAMs in leaves was investigated from April to November. PmDAMs showed roughly two distinct seasonal expression trends (Fig. 4). This trend was confirmed by an experiment using another biological replicate (Supplemental Fig. S2). PmDAM1 to PmDAM3 were rapidly up-regulated in spring until the beginning of summer and were gradually down-regulated toward autumn. In contrast, PmDAM4 to PmDAM6 transcript levels gradually increased until the peak in November immediately before leaf fall. Seasonal changes in the accumulation of PmDAM transcripts were similar in leaves and buds. Thus, in buds, the accumulation of PmDAM1 to PmDAM3 transcripts peaked in early summer, while that of PmDAM4 to PmDAM6 transcripts peaked in autumn (Figs. 3 and 4; Supplemental Figs. S1 and S2).
Figure 4.
Seasonal expression changes in PmDAMs in the leaves of the Japanese apricot cv Nanko. Gene expression in cv Nanko leaves grown in the field from April to November was assessed at monthly intervals by real-time PCR as described in Figure 3. Transcript levels of each gene were normalized by PmUBQ. The means of three technical replicates are shown, with error bars representing sd. The means of another biological replicate are shown in Supplemental Figure S2. Changes in expression are shown as logarithmic graphs. PmDAM6 transcripts in April, May, and June were present at undetectable levels under our experimental conditions; thus, we did not plot them in the figures. The period when shoot growth cessation was first observed until the majority of shoots stopped growing in the field is shown by arrows.
PmDAM Down-Regulation by Prolonged Cold Exposure (5°C–9°C)
Buds are released from endodormancy by prolonged cold exposure. To investigate the effect of prolonged cold exposure on PmDAM expression, we placed branches collected in October under prolonged cold [cold(+)] or noncold [cold(−)] temperature conditions. As shown in Table II and Figure 5, the low-chill cv Ellching required less chilling exposure for dormancy release than the high-chill cv Nanko.
Table II. Effects of a prolonged cold temperature (5°C–9°C) on endodormancy release of Japanese apricot.
First bud burst was observed within 10 d (++), within 30 d (+), or not observed within 60 d (2).
| Days for Cold Temperature Treatment | Days to Bud Burst after Transferring to Forcing Conditions |
|
| cv Nanko | cv Ellching | |
| 0 | − | 2 |
| 16 | 2 | 2 |
| 32 | 2 | + |
| 64 | + | ++ |
Figure 5.
Difference in the endodormancy status of two Japanese apricot cultivars. Endodormancy in cv Ellching (left) was released with a relatively short period of cold exposure, while cv Nanko (right) remained endodormant. Each plant was transferred to forcing conditions (greenhouse controlled at 25°C ± 3°C under natural daylength) after 32 d of cold exposure (7°C ± 2°C). Only cv Ellching resumed growth and produced new leaves. [See online article for color version of this figure.]
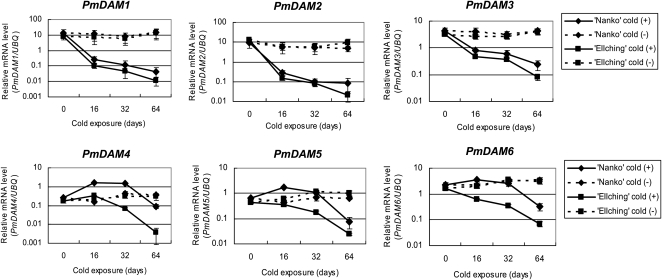
PmDAM expression can be approximately classified into two distinct patterns (Fig. 6). PmDAM1 to PmDAM3 expression levels decreased by a relatively short period of cold exposure and were steadily repressed by a prolonged period of cold exposure in both cv Nanko and cv Ellching, with both cultivars showing similar expression levels of these genes. PmDAM1 to PmDAM3 expression remained constant during cold(–) treatment in both cultivars (Fig. 6). However, PmDAM4 to PmDAM6 showed distinct expression patterns depending on the cultivar. After a short period of cold(+) treatment, PmDAM4 to PmDAM6 expression slightly increased or remained constant in cv Nanko, while it decreased in cv Ellching. However, prolonged cold exposure repressed PmDAM4 to PmDAM6 in both cultivars, although expression levels in cv Ellching were significantly lower than those in cv Nanko. In both cultivars, PmDAM4 to PmDAM6 were slightly up-regulated or remained constant during cold(−) treatment. Another biological replicate also showed the same trends in expression patterns (Supplemental Fig. S3).
Figure 6.
The effect of prolonged cold exposure on PmDAM expression in October. One-year-old branches were cut from cv Nanko and cv Ellching trees in October, artificially defoliated, and placed in a growth chamber for cold(+) treatment (7°C ± 2°C) or cold(−) treatment (25°C ± 3°C) under dark conditions. Vegetative buds were collected from the middle portions of branches at 0, 16, 32, or 64 d of cold(+) or cold(−) treatment. Gene expression was measured by real-time PCR as described in Figure 3. Transcript levels of each gene were normalized by PmUBQ. The means of three technical replicates are shown, with error bars representing sd. The means of another biological replicate are shown in Supplemental Figure S3. Changes in expression are shown as logarithmic graphs.
DISCUSSION
Transgenic studies have been used effectively to show the involvement of the poplar homolog of FT (Böhlenius et al., 2006), birch (Betula pendula) homolog of FRUITFULL (Hoenicka et al., 2008), oat (Avena sativa) homolog of PHYTOCHROME A (Olsen et al., 1997), and poplar homolog of the CEN/TFL1 subfamilies (Mohamed et al., 2010) in seasonal growth cycles of shoots and buds in higher plants. In addition to these genes, transgenic experiments in this study indicated the possible involvement of PmDAM6, a Japanese apricot homolog of SVP/AGL24, in perennial growth cycles of plants. To our knowledge, this is the first report to use a transgenic technique to functionally characterize SVP/AGL24 homologs in woody plant species.
We successfully obtained transgenic poplar lines constitutively expressing PmDAM6; however, the regeneration rate was much lower with 35S:PmDAM6 than with 35S:empty. The same trend was observed in transformation experiments with Japanese apricot and apple (Malus × domestica; H. Yamane, M. Wada, and R. Tao, unpublished data). The regeneration rate of 35S:PmDAM6 apple was approximately 10-fold lower than that of control apple transformants. In addition, during root initiation and acclimatization, shoot growth was repressed in some 35S:PmDAM6 lines, resulting in shorter plant heights in 35S:PmDAM6 than in control lines (Fig. 1B). It is obvious that PmDAM6 could affect adventitious shoot regeneration processes and shoot growth during root initiation and acclimatization as well as after acclimatization. Because several growth cessation-related genes have been reported in poplar, such as CENL1, CO, and FT (Böhlenius et al., 2006; Ruonala et al., 2008), and because DAM genes of leafy spurge have been hypothesized to regulate FT (Horvath et al., 2008, 2010), it would be interesting to determine if ectopic expression of PmDAM6 has any effect on the expression of the above-mentioned growth cessation-related genes in transgenic poplar. No significant difference was observed in CO2 expression in leaves and CENL1 expression in shoot apices between 35S:PmDAM6 and wild-type poplar (data not shown). FT1 was present at undetectable levels in both 35S:PmDAM6 and wild-type poplar (data not shown). FT2 expression varies with transgenic lines. FT2 expression was repressed in transgenic lines whose growth was strongly inhibited (35S:PmDAM-1, -5, and -6), whereas it was not affected or up-regulated in transgenic lines whose growth was not strongly inhibited immediately after acclimatization (35S:PmDAM-2, -3, and -4; Fig. 1B; Supplemental Fig. S4A). Since a distinct difference in PmDAM6 expression level was not found among 35S:PmDAM6 poplar, we are unable to conclusively state anything about the correlation between PmDAM6 and FT2 transcript levels. Furthermore, no direct correlation between PmDAM6 and PmFT (Esumi et al., 2009) expression levels was indicated from the seasonal changes in PmFT expression patterns in Japanese apricot (Supplemental Fig. S4, B and C). Thus, the involvement of PmDAM6 in CO-, CENL1-, or FT-mediated growth cessation is currently unclear. A comprehensive expression survey such as microarray analysis would be required to evaluate the biological role of PmDAM6 in 35S:PmDAM6 poplar. Although this study does not provide conclusive information on how PmDAM6 functions in transgenic lines, it is noteworthy that overexpressed PmDAM6 had a growth inhibitory function in transgenic poplar.
Endodormancy is presumed to be regulated by putative internal growth inhibitors that may be localized in endodormant buds to prevent the resumption of growth. Because PmDAM6 was originally identified as the gene up-regulated during the endodormancy period (Yamane et al., 2008) and expressed in the area containing shoot apical meristem and rib meristem regions within Japanese apricot buds (Supplemental Fig. S5), we assumed a growth inhibitory function of PmDAM6 in buds. Because not only growth cessation but also terminal bud set was observed in 35S:PmDAM6 poplar under environmental conditions in which the control plants continued shoot tip growth, our hypothesis is that PmDAM6 may function through its growth inhibitory effect to control endodormancy. We are now transforming Japanese apricot, one of the tree species resistant to transformation, to test our hypothesis in a homologous plant system.
In this study, we found six PmDAM genes, including PmDAM6, in a tandem arrangement in the Japanese apricot genome. The presence of tandemly arrayed PmDAM6 homologs in the genome was expected because the six PmDAM6 homologs in peach, a close relative of Japanese apricot, have also been reported to be tandemly arrayed in its genome. However, more than six bands found on the DNA blot could suggest the possible presence of additional DAM-like genes in the Japanese apricot genome. When the whole genome sequences of peach were searched on the Genome Database for Rosaceae Web site (http://www.rosaceae.org/), we found at least two more SVP-like genes (peach gene accession nos. ppa011063 and ppa022274). Although these two SVP-like genes may have some function in dormancy, we focused on tandemly arrayed PmDAM6 homologs in Japanese apricot in this study. Based on their homology to PpDAMs and results obtained with phylogenetic analysis, we named these tandemly arrayed genes with PmDAM6 as PmDAM1, PmDAM2, PmDAM3, PmDAM4, and PmDAM5.
Along with PmDAM6 (Yamane et al., 2008) and other PpDAMs of peach (Bielenberg et al., 2008) and EeDAMs of leafy spurge (Horvath et al., 2010), PmDAM1 to PmDAM5 belong to the StMADS11 (SVP/AGL24) clade of MIKCc-type MADS box genes. Interestingly, the deduced amino acid sequences of PmDAMs are similar to each other, and all PmDAMs contain the SRDX repressor motif at the C-terminal end (Ohta et al., 2001). This suggests that all six PmDAMs may act as transcriptional repressors and have functional redundancy. Although PmDAMs showed distinct seasonal expression changes as discussed below, all PmDAMs were up-regulated after growth cessation and down-regulated in February and March, when buds have the ability to resume growth under favorable conditions. Furthermore, all six PmDAMs were repressed by prolonged cold exposure. Prolonged cold exposure in winter is known to induce endodormancy release in perennial plants (Rohde and Bhalerao, 2007). These results may indicate that PmDAMs have functional redundancy and that unknown target genes of PmDAMs are up-regulated when buds are released from endodormancy. However, PmDAMs may have been subfunctionalized during evolution. Despite the similarity in peach PpDAM coding sequences, Jiménez et al. (2009) detected strong purifying selection in all six PpDAM genes. Furthermore, a peach evergrowing mutant lacking PpDAM1 to PpDAM6 expression showed dimorphism with an inability to set terminal buds and enter lateral bud dormancy (Rodriguez et al., 1994; Bielenberg et al., 2004, 2008; Li et al., 2009). Based on these facts, Jiménez et al. (2009) proposed the possibility of peach PpDAM subfunctionalization. Although it is unclear whether terminal bud set and lateral bud endodormancy are under the control of the same regulatory pathway, our transformation study showed that constitutive expression of PmDAM6 alone could modify both terminal bud set and dormancy induction. Further transformation studies using the five PmDAMs other than PmDAM6 would help address questions regarding the functional redundancy or subfunctionalization of PmDAMs.
After flushing in April and subsequent active shoot growth, the majority of cv Nanko shoots had stopped growing by August and all six PmDAMs in leaves were up-regulated compared with their expression in April. Using branch cuttings, we determined that endodormancy periods in cv Nanko and cv Ellching trees under our experimental conditions were from July to January and from August to October, respectively. During these months, the deep dormant periods of cv Nanko and cv Ellching can be estimated to be from August to November and from September to October, respectively, on the basis of the results obtained using single-node cuttings. PmDAM6 expression levels showed positive correlations with induction of lateral bud endodormancy. On the other hand, in both cultivars, negative correlations between changes in expression and endodormancy release were found in PmDAM3 to PmDAM6. These observations suggested that the transcriptional control of PmDAMs is different, although they may have functional redundancy (i.e. they could act as internal growth inhibitors), as suggested for PmDAM6 from the transgenic experiment in this study. Seasonal expression patterns of PmDAMs in buds and leaves were roughly classified into two distinct patterns. PmDAM1 to PmDAM3 showed earlier expression peaks (in summer), while PmDAM4 to PmDAM6 showed later expression peaks (in autumn). Because Japanese apricot buds enter deep dormancy in autumn (September and October), PmDAM4 to PmDAM6 expression appeared to be more closely correlated with endodormancy depth than PmDAM1 to PmDAM3 expression. We assumed that this difference could be at least partly due to variation in PmDAM1 to PmDAM3 and PmDAM4 to PmDAM6 responses to an ambient cool temperature in September (15°C–18°C). The artificial cool-temperature treatment in September significantly increased the accumulation of PmDAM4 to PmDAM6 transcripts in buds (Supplemental Fig. S6). Peach DAMs, PpDAM5 and PpDAM6, were also up-regulated by cool temperature in September (Yamane et al., 2011a).
As suggested by the phylogenetic similarity of DAM orthologs between Japanese apricot and peach (Fig. 2E), all PmDAMs, except PmDAM4, showed seasonal expression changes similar to the respective peach orthologs, in that DAM1 and DAM2 peaked in summer and decreased before winter, whereas DAM3, DAM5, and DAM6 showed negative correlation with endodormancy release (Li et al., 2009; Jiménez et al., 2010; Yamane et al., 2011a). Although PmDAM4 and PpDAM4 expression was negatively correlated with endodormancy release, as shown in this study and as reported by Leida et al. (2010), PmDAM4 peaked in autumn (this study) whereas PpDAM4 peaked in summer (Li et al., 2009). Nevertheless, all DAMs of Japanese apricot and peach reported so far are down-regulated when buds are able to resume their growth (Bielenberg et al., 2008; Yamane et al., 2008, 2011a, 2011b; Li et al., 2009; Jiménez et al., 2010; Leida et al., 2010). Because prolonged cold exposure down-regulated DAM5 and DAM6 in both species (Jiménez et al., 2010; Yamane et al., 2011a, 2011b; this study), these genes could function in endodormancy release in Prunus.
PpDAM4 to PpDAM6 were shown to be responsive to a reduction in daylength under controlled environmental conditions (Li et al., 2009). Although we did not analyze short-day effects on PmDAM expression, reduction in daylength could be one of the triggers for PmDAM4 to PmDAM6 because they were up-regulated toward autumn (Fig. 3). In this study, instead, we found that PmDAM4 to PmDAM6 were up-regulated by low temperature (Supplemental Fig. S6). Horvath et al. (2010) found that EeDAM1 was cold stress (11°C) responsive and contained putative C-repeat/DRE-Binding Factor (CBF) sites, which are cis-regulating motifs targeted by the cold/drought stress CBF regulon found within the 2,000-bp region upstream of the EeDAM1 translation initiation codon. This finding suggested that the cold-responsive EeDAM1 gene was controlled by the CBF protein. Similar to EeDAM1 (Horvath et al., 2010), conserved CBF sites were found within the 1,000-bp region upstream of DAM4 to DAM6 translation initiation codons of both peach and Japanese apricot. In particular, the positions of CBF sites were highly conserved in DAM5 and DAM6 of peach and Japanese apricot. CBF sites were found at 527 and 536 bp upstream of the translation initiation codon of peach and Japanese apricot DAM5, respectively, while they were at 692 and 652 bp upstream of the translation initiation codon of peach and Japanese apricot DAM6, respectively. In contrast, putative CBF sites were not found within 1,000 bp upstream of DAM1, DAM2, and DAM3 of peach and Japanese apricot. These results could suggest that the CBF-mediated cold response may be conserved in DAM4 to DAM6 of Japanese apricot and peach.
Cold treatment in October induced endodormancy release in cv Nanko and cv Ellching at 64 and 32 d, respectively, coinciding very well with a prominent decrease in PmDAM4 to PmDAM6 transcript levels in buds. In particular, it is notable that dormancy release was observed when PmDAM4 to PmDAM6 transcript levels were down-regulated to an approximately 10-fold decrease from their peak levels. Taking these results into consideration, along with the seasonal expression changes in PmDAM4 to PmDAM6, it is suggested that PmDAM4 to PmDAM6 expression could be associated with endodormancy release by chilling accumulation. In contrast, although PmDAM1 to PmDAM3 were responsive to cold temperature, their transcript levels decreased well before endodormancy release at a similar rate in both cv Nanko and cv Ellching. This could indicate that PmDAM1 to PmDAM3 cannot be considered as determinants of endodormancy release, even though they could still serve as internal growth inhibitors.
The chilling requirements for dormancy release vary widely depending on the genotypes of a given species. However, the molecular basis for differences in chilling requirements has yet to be elucidated. This study demonstrated the association of seasonal expression changes in PmDAMs with temperature-mediated phenological dormancy events in two Japanese apricot cultivars differing in chilling requirements. If we closely observe the seasonal expression patterns of PmDAM4 to PmDAM6 and their cold temperature response in October, genotype-dependent regulation patterns can be found. PmDAM4 to PmDAM6 expression was up-regulated until late autumn or early winter, after which it was down-regulated toward spring. Although no substantial difference was observed in the initial up-regulation patterns of PmDAM4 to PmDAM6 expression in cv Nanko and cv Ellching, PmDAM4 to PmDAM6 expression levels in low-chill cv Ellching started to decrease earlier and faster than those in high-chill cv Nanko. From October to December, the difference between the two cultivars was prominent. Namely, PmDAM4 to PmDAM6 in low-chill cv Ellching were down-regulated while those in high-chill cv Nanko were up-regulated or remained constant. This difference could be attributed to variation in the response to cold temperature in PmDAM4 to PmDAM6 between the two cultivars in October. Cold treatment in October readily induced PmDAM4 to PmDAM6 down-regulation in low-chill cv Ellching, while PmDAM4 to PmDAM6 in high-chill cv Nanko remained constant or were up-regulated during the first 32 d of treatment. Although the cause of the distinct responses of PmDAM4 to PmDAM6 to cold temperature in these two cultivars is unknown, these results may indicate that low-chill cv Ellching reacts to cold temperature in October as chilling but high-chill cv Nanko does not. Alternatively, a certain amount of chilling accumulation may be necessary for PmDAM4 to PmDAM6 down-regulation in high-chill cv Nanko. In any case, the distinct changes in PmDAM4 to PmDAM6 expression may possibly contribute to the different amounts of chilling requirements for dormancy release in cv Nanko and cv Ellching.
CONCLUSION
In this study, we demonstrated the growth inhibitory functions of PmDAM6 in transgenic poplar overexpressing it. We identified six tandemly arrayed PmDAM genes (PmDAM1–PmDAM6) and found that all PmDAMs were repressed during prolonged cold exposure and maintained at low levels until endodormancy release, suggesting that all PmDAMs, similar to PmDAM6, act as growth inhibitors. Our study, along with other reported studies, strongly suggests that DAM genes play significant roles in the regulation of bud dormancy in perennial plants. Thus, it is apparent that DAM genes could be one of the most promising key bud dormancy factors. Hence, elucidation of the genetic, molecular, and biochemical aspects of DAM genes would be of great interest in a wide range of studies of environmental adaptation.
Our study also suggested the association of PmDAMs with the genetic control of chilling requirements for dormancy release, because PmDAM4 to PmDAM6 down-regulation was correlated with cold temperature-mediated phenological events of dormancy release in two Japanese apricot cultivars differing in chilling requirements for dormancy release. The genotype-dependent changes in PmDAM4 to PmDAM6 expression may possibly contribute to the different levels of chilling requirements, providing new insights in the understanding of the molecular basis of chilling requirements for dormancy release in temperate fruit trees.
MATERIALS AND METHODS
Transformation of Poplar and Growth Conditions
Hybrid poplar (Populus tremula × Populus tremuloides; clone T89) was transformed with a chimeric gene construct containing PmDAM6. PmDAM6 was introduced into wild-type plants for constitutive expression under the control of the cauliflower mosaic virus 35S promoter. To construct the binary vector p35S:PmDAM6, PmDAM6 cDNA (National Center for Biotechnology Information accession no. AB437345; Yamane et al., 2008) was blunt-end ligated in the sense orientation at the BamHI site located between the cauliflower mosaic virus 35S promoter and terminator sequences in the T-DNA region of the binary vector pDU92.3103 (Tao et al., 1995). pDU92.3103 was used for control transformation. p35S:PmDAM6 and pDU92.3103 vectors were introduced in the disarmed Agrobacterium tumefaciens strain EHA105 and used to transform hybrid poplar by the conventional method (Nilsson et al., 1996). Six independent transformed lines and a single control transformed line were obtained. PmDAM6 expression was confirmed by RT-PCR using PmDAM6-F2 and PmDAM6-R2 primers (Supplemental Table S1). RT-PCR was performed with cDNAs synthesized from total RNAs extracted from the leaves of each transgenic plant immediately after acclimatization using the RNeasy Plant Mini Kit (Qiagen). PCR conditions were as follows: 32 cycles at 98°C for 10 s, 57°C for 30 s, and 72°C for 20 s, with initial denaturation at 98°C for 3 min and final extension at 72°C for 7 min. Transgenic shoots were transplanted to half-strength Murashige and Skoog (1962) medium for root initiation. As controls, the control transformed line (35S:empty) and wild-type plants were simultaneously grown under the same conditions. When the plants rooted, they were planted in plastic pots covered with plastic bags containing vermiculite that had been autoclaved and wetted with 1:1,000 Hyponex (Hyponex Japan). They were grown under LD conditions (16/8 h of light/dark, 22°C) with cool-white fluorescent light (60 μmol m−2 s−1; FL 40SS W/37 lamps; Matsushita Electronics) for 4 weeks of acclimatization. At 4 weeks, the plastic bags were removed and the plants were transplanted to larger pots.
Phenotypic Assessment of Transgenic Poplar
Transgenic and control poplar plants were grown under LD conditions for 4 weeks after acclimatization (namely for 4 weeks after removing the plastic bags). Plant height and the timing of terminal bud set were investigated. Growth cessation and terminal bud set in transgenic plants were observed within 1 month after removing the plastic bags. Six weeks after acclimatization, or approximately 2 to 3 weeks after growth cessation and terminal bud set in transgenic lines, both transgenic and control plants were defoliated and grown under LD conditions for 2 weeks to observe bud burst.
Plant Materials for Genomic DNA-Blot and Expression Analyses of Japanese Apricot
The two Japanese apricot (Prunus mume) cultivars used in this study were the early-blooming Taiwanese cultivar, Ellching (19 years old, seed grafted), and a Japanese cultivar with an average blooming time in Japan, Nanko (18 years old, seed grafted). Both cultivars were grown at the Horticultural Experiment Center of the Wakayama Research Center of Agriculture, Forestry, and Fisheries in Gobo, Japan (34°N, 135°E). Two-year-old pot-grown cv Nanko and cv Ellching self-rooted plants were also used. For genomic DNA-blot analysis, five other cultivars showing blooming times similar to cv Nanko (cv Shirokaga and cv Oushuku) or cv Ellching (cv Nisei, selection SC, selection ST) were used. These cultivars and selections were grown at the Kyoto University Experimental Farm in Kyoto, Japan (34°N, 135°E).
Genomic DNA-Blot Analysis
Genomic DNAs were isolated from young leaves of seven cultivars and selections using the Nucleon PhytoPure Plant and Fungal DNA Extraction Kit (GE Healthcare), with some modifications (Yamane et al., 2009). In brief, 1.5 g of frozen leaves was ground to a powder using the Multi-Beads Shocker (Yasui kikai), suspended in washing buffer (10 mm Tris-HCl [pH 9.0], 0.5 m Suc, 10 mm EDTA [pH 8.0], and 80 mm KCl), and mixed thoroughly. The mixture was centrifuged (6,500g at 4°C for 15 min) to collect the pellet; the pellet was resuspended in washing buffer and centrifuged again. Genomic DNA was isolated from the pellet using the above-mentioned plant and fungal DNA extraction kit and further purified by phenol/chloroform extraction.
Genomic DNA (5 μg) was digested with HindIII, run on a 0.8% (w/v) agarose gel, and transferred to a Biodyne Plus nylon filter (Pall). The membrane was hybridized with a digoxigenin-labeled PmDAM6 probe containing nucleotide sequences corresponding to the MADS domain of PmDAM6 to detect all PmDAMs. After hybridization at 60°C, the membrane was washed under low-stringency conditions (Watari et al., 2007). The hybridized signals were visualized using LAS-3000 mini (Fujifilm).
Genomic DNA Library Construction, Screening, and Sequencing
Fosmid libraries were constructed from the genomic DNAs of the Japanese apricot cv Nanko and cv Ellching using the CopyControl Fosmid Library Production Kit (Epicentre). The libraries were screened using a digoxigenin-dUTP-labeled probe synthesized from a PmDAM6 partial fragment corresponding to the K box region. The cv Nanko library was also screened with probes synthesized from PmDAM2 cDNAs cloned by RT-PCR (data not shown), and the cv Ellching library was screened with probes from the PmDAM3 cDNA of cv Nanko. Positive clones were subjected to gene-specific PCR to confirm the presence of PmDAMs. Nucleotide sequences of the selected fosmid clone were determined by partial digestion and shotgun sequencing using the BigDye Terminator version 3.1 Cycle Sequencing Kit (Applied Biosystems) and an ABI 3730xl capillary sequencer (Applied Biosystems).
Phylogenetic Analysis
PmDAMs of Japanese apricot (this study), DAMs reported in other plant species (leafy spurge [Euphorbia esula] and peach [Prunus persica]), database-registered DAM-like genes of poplar, apple (Malus × domestica), pear (Pyrus communis), rosaceous fruit tree species other than Japanese apricot and peach, and 39 MIKCc-type MADS box genes of Arabidopsis (Arabidopsis thaliana) were used to construct the phylogenetic tree. AGL23 of Arabidopsis, which belongs to type I MADS box genes, was used as an outgroup. Accession numbers of the genes used are shown in Supplemental Table S2. Phylogenetic analysis was performed using the ClustalW program at the DNA Data Bank of Japan (http://clustalw.ddbj.nig.ac.jp/top-j.html). The tree was displayed using NJplot software.
Seasonal Endodormancy Status and Expression Analysis of Japanese Apricot
For each cultivar, 1-year-old branches (current season’s growth; n = 3) with a length of approximately 40 cm (containing approximately 25 buds) were cut at monthly intervals from trees in the field from June to March, 2005 and 2006, for cv Nanko or from June to February, 2005 and 2006, for cv Ellching. Lateral vegetative buds on the middle portion of each branch were used to calculate the percentage of bud burst. When the branches were collected before the trees shed their leaves in the field, they were artificially defoliated. The branches were placed in water containing Misakifarm (Otsuka kagaku; containing nutrients and fungicides). At the same time, the basal parts of 10 single-node cuttings obtained from the middle portion of each of the three branches were placed in water containing Misakifarm. The branches and single-node cuttings were maintained at 22°C under cool-white fluorescent light (60 μmol m−2 s−1) for a 16-h-light/8-h-dark photoperiod. The water containing Misakifarm was replaced every 2 weeks. After 1 month in the growth chamber, buds showing green leaves were considered to have burst.
The lateral vegetative buds excised from the middle portions of branches of both cv Nanko and cv Ellching and cv Nanko leaves were collected at monthly intervals, immediately frozen in liquid nitrogen, and stored at −80°C until use. Total RNA was isolated from the buds and leaves as described by Yamane et al. (2008). After DNaseI treatment (Takara BIO), 1 μg of total RNA was used for cDNA synthesis with SuperScript III reverse transcriptase (Invitrogen). Based on the genomic DNA and cDNA sequences for PmDAM1 to PmDAM6, gene-specific TaqMan probes and primers (Supplemental Table S1) for detecting each gene were synthesized. Real-time PCR analysis using a TaqMan probe was performed using LightCycler 480 (Roche) and a probe master mix (Roche). The reaction mixture consisted of 1× probe master mix, 500 nm each of forward and reverse primers, 200 nm TaqMan probe, and cDNA equivalent to 4 ng of total RNA in 20-μL reaction volumes. As a reference, the accumulation of the Japanese apricot UBIQUITIN (PmUBQ) transcript was monitored by real-time PCR using SYBR Green Master Mix (Roche) and gene-specific primers (Supplemental Table S1). PCR was performed using a program of 45 cycles at 95°C for 10 s and 60°C for 20 s, with initial heating at 95°C for 5 min. For PmUBQ gene-specific real-time PCR, dissociation curve analysis was performed to confirm that the fluorescence was only derived from gene-specific amplification. Two biological replicates each with three technical replicates were performed for each gene. Quantities of PmDAM1 to PmDAM6 transcripts in each sample were normalized using PmUBQ transcripts.
Prolonged Cold Exposure under Controlled Environmental Conditions
For prolonged cold treatment in October, 1-year-old long branches of cv Nanko and cv Ellching trees were collected in October 2007. In addition, 12 pot-grown trees of each cultivar were used for estimating the endodormancy status of vegetative buds. Collected branches and pot-grown trees were artificially defoliated and transferred to a growth chamber at 7°C ± 2°C [cold(+)] or 25°C ± 3°C [cold(−)] under dark conditions. The branches were placed in water containing Misakifarm, and the water was replaced every 2 weeks. The pots were watered once a week. After 16, 32, or 64 d, three pot-grown trees of each cultivar were transferred to a greenhouse controlled at 25°C ± 3°C under natural daylength to force growth. Lateral vegetative buds were excised from the middle portions of branches at 0, 16, 32, or 64 d of cold(+) or cold(−) treatment, immediately frozen in liquid nitrogen, and stored at −80°C until further use. Total RNA was isolated from the buds stored at −80°C and used for the analysis of PmDAM1 to PmDAM6 expression as described above. Two biological replicates each with three technical replicates were performed for each gene.
Sequence data from the article can be found in the GenBank/EMBL/DNA Data Bank of Japan data libraries under the following accession numbers: PmDAM1 (AB576350), PmDAM2 (AB576351), PmDAM3 (AB576352), PmDAM4 (AB576353), and PmDAM5 (AB576349).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Biological replicate of seasonal expression changes in PmDAMs in the lateral vegetative buds of two Japanese apricot cultivars.
Supplemental Figure S2. Biological replicate of seasonal expression changes in PmDAMs in the leaves of the Japanese apricot cv Nanko.
Supplemental Figure S3. Biological replicate of changes in PmDAM expression affected by prolonged cold exposure.
Supplemental Figure S4. Expression of FT orthologs in transgenic poplar and Japanese apricot.
Supplemental Figure S5. Expression of PmDAM6 in shoot apex of lateral vegetative buds of Japanese apricot.
Supplemental Figure S6. Effect of ambient cool temperature in autumn on PmDAM expression in Japanese apricot.
Supplemental Table S1. Sequences of primers and TaqMan probes used in this study.
Supplemental Table S2. Genes used for constructing the phylogenetic tree and their accession numbers.
Acknowledgments
We thank Dr. Yoshikazu Ozawa (former director of the Horticultural Experiment Center of Wakayama Research Center of Agriculture, Forestry, and Fisheries, Japan) and Dr. Kyohei Hayashi (Wakayama Research Center of Agriculture, Forestry, and Fisheries, Japan) for helping us to collect experimental materials. P. tremula × P. tremuloides T89 clones were kindly provided by Dr. Nobuyuki Nishikubo (Oji Paper Group, Japan) by courtesy of Prof. Björn Sundberg (Umeå Plant Science Centre, Swedish University of Agricultural Sciences).
References
- Bielenberg DG, Li Z, Zhebentyayeva T, Fan S, Reighard GL, Scorza R, Abbott AG. (2008) Sequencing and annotation of the evergrowing locus in peach [Prunus persica (L.) Batsch] reveals a cluster of six MADS-box transcription factors as candidate genes for regulation of terminal bud formation. Tree Genet Genomes 4: 495–507 [Google Scholar]
- Bielenberg DG, Wang Y, Fan S, Reighard GL, Scorza R, Abbott AG. (2004) A deletion affecting several gene candidates is present in the Evergrowing peach mutant. J Hered 95: 436–444 [DOI] [PubMed] [Google Scholar]
- Böhlenius H, Huang T, Charbonnel-Campaa L, Brunner AM, Jansson S, Strauss SH, Nilsson O. (2006) CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science 312: 1040–1043 [DOI] [PubMed] [Google Scholar]
- Crabbe J, Barnola P. (1996) A new conceptual approach to bud dormancy in woody plants. Lang GA, , Plant Dormancy: Physiology, Biochemistry and Molecular Biology. CAB International, Wallingford, UK, pp 83–113 [Google Scholar]
- Druart N, Johansson A, Baba K, Schrader J, Sjödin A, Bhalerao RR, Resman L, Trygg J, Moritz T, Bhalerao RP. (2007) Environmental and hormonal regulation of the activity-dormancy cycle in the cambial meristem involves stage-specific modulation of transcriptional and metabolic networks. Plant J 50: 557–573 [DOI] [PubMed] [Google Scholar]
- Esumi T, Hagihara C, Kitamura Y, Yamane H, Tao R. (2009) Identification of an FT ortholog in Japanese apricot (Prunus mume Sieb. Et Zucc.). J Hortic Sci Biotechnol 84: 149–154 [Google Scholar]
- Fan S, Bielenberg DG, Zhebentyayeva TN, Reighard GL, Okie WR, Holland D, Abbott AG. (2010) Mapping quantitative trait loci associated with chilling requirement, heat requirement and bloom date in peach (Prunus persica). New Phytol 185: 917–930 [DOI] [PubMed] [Google Scholar]
- Faust M, Erez A, Rowland LJ, Wang SY, Norman HA. (1997) Bud dormancy in perennial fruit trees: physiological basis for dormancy induction, maintenance, and release. HortScience 32: 623–629 [Google Scholar]
- Hoenicka H, Nowitzki O, Hanelt D, Fladung M. (2008) Heterologous overexpression of the birch FRUITFULL-like MADS-box gene BpMADS4 prevents normal senescence and winter dormancy in Populus tremula L. Planta 227: 1001–1011 [DOI] [PubMed] [Google Scholar]
- Horvath DP, Anderson JV, Chao WS, Foley ME. (2003) Knowing when to grow: signals regulating bud dormancy. Trends Plant Sci 8: 534–540 [DOI] [PubMed] [Google Scholar]
- Horvath DP, Chao WS, Suttle JC, Thimmapuram J, Anderson JV. (2008) Transcriptome analysis identifies novel responses and potential regulatory genes involved in seasonal dormancy transitions of leafy spurge (Euphorbia esula L.). BMC Genomics 9: 536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath DP, Sung S, Kim D, Chao WS, Anderson JV. (2010) Characterization, expression and function of DORMANCY ASSOCIATED MADS-BOX genes from leafy spurge. Plant Mol Biol 73: 169–179 [DOI] [PubMed] [Google Scholar]
- Jansson S, Douglas CJ. (2007) Populus: a model system for plant biology. Annu Rev Plant Biol 58: 435–458 [DOI] [PubMed] [Google Scholar]
- Jiménez S, Lawton-Rauh AL, Reighard GL, Abbott AG, Bielenberg DG. (2009) Phylogenetic analysis and molecular evolution of the dormancy associated MADS-box genes from peach. BMC Plant Biol 9: 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez S, Reighard GL, Bielenberg DG. (2010) Gene expression of DAM5 and DAM6 is suppressed by chilling temperatures and inversely correlated with bud break rate. Plant Mol Biol 73: 157–167 [DOI] [PubMed] [Google Scholar]
- Lang GA. (1987) Dormancy: a new universal terminology. HortScience 22: 817–820 [Google Scholar]
- Leida C, Terol J, Martí G, Agustí M, Llácer G, Badenes ML, Ríos G. (2010) Identification of genes associated with bud dormancy release in Prunus persica by suppression subtractive hybridization. Tree Physiol 30: 655–666 [DOI] [PubMed] [Google Scholar]
- Leseberg CH, Li A, Kang H, Duvall M, Mao L. (2006) Genome-wide analysis of the MADS-box gene family in Populus trichocarpa. Gene 378: 84–94 [DOI] [PubMed] [Google Scholar]
- Li Z, Reighard GL, Abbott AG, Bielenberg DG. (2009) Dormancy-associated MADS genes from the EVG locus of peach [Prunus persica (L.) Batsch] have distinct seasonal and photoperiodic expression patterns. J Exp Bot 60: 3521–3530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed R, Wang CT, Ma C, Shevchenko O, Dye SJ, Puzey JR, Etherington E, Sheng X, Meilan R, Strauss SH, et al. (2010) Populus CEN/TFL1 regulates first onset of flowering, axillary meristem identity and dormancy release in Populus. Plant J 62: 674–688 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Nilsson O, Little CHA, Sandberg G, Olsson O. (1996) Expression of two heterologous promoters, Agrobacterium rhizogenes rolC and cauliflower mosaic virus 35S, in the stem of transgenic hybrid aspen plants during the annual cycle of growth and dormancy. Plant Mol Biol 31: 887–895 [DOI] [PubMed] [Google Scholar]
- Ohta M, Matsui K, Hiratsu K, Shinshi H, Ohme-Takagi M. (2001) Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13: 1959–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen JE, Junttila O, Nilsen J, Eriksson ME, Martinussen I, Olsson O, Sandberg G, Moritz T. (1997) Ectopic expression of oat phytochrome A in hybrid aspen changes critical daylength for growth and prevents cold acclimatization. Plant J 12: 1339–1350 [Google Scholar]
- Rodriguez AJ, Sherman WB, Scorza R, Wisniewski M, Okie WR. (1994) ‘Evergreen’ peach, its inheritance and dormant behavior. J Am Soc Hortic Sci 119: 789–792 [Google Scholar]
- Rohde A, Bhalerao RP. (2007) Plant dormancy in the perennial context. Trends Plant Sci 12: 217–223 [DOI] [PubMed] [Google Scholar]
- Rohde A, Prinsen E, De Rycke R, Engler G, Van Montagu M, Boerjan W. (2002) PtABI3 impinges on the growth and differentiation of embryonic leaves during bud set in poplar. Plant Cell 14: 1885–1901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruonala R, Rinne PL, Baghour M, Moritz T, Tuominen H, Kangasjärvi J. (2006) Transitions in the functioning of the shoot apical meristem in birch (Betula pendula) involve ethylene. Plant J 46: 628–640 [DOI] [PubMed] [Google Scholar]
- Ruonala R, Rinne PL, Kangasjärvi J, van der Schoot C. (2008) CENL1 expression in the rib meristem affects stem elongation and the transition to dormancy in Populus. Plant Cell 20: 59–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruttink T, Arend M, Morreel K, Storme V, Rombauts S, Fromm J, Bhalerao RP, Boerjan W, Rohde A. (2007) A molecular timetable for apical bud formation and dormancy induction in poplar. Plant Cell 19: 2370–2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao R, Uratsu SL, Dandekar AM. (1995) Sorbitol synthesis in transgenic tobacco with apple cDNA encoding NADP-dependent sorbitol-6-phosphate dehydrogenase. Plant Cell Physiol 36: 525–532 [DOI] [PubMed] [Google Scholar]
- Watari A, Hanada T, Yamane H, Esumi T, Tao R, Yaegaki H, Yamaguchi M, Beppu K, Kataoka I. (2007) A low transcriptional level of Se-RNase in the Se-haplotype confers self-compatibility in Japanese plum. J Am Soc Hortic Sci 132: 396–406 [Google Scholar]
- Yamane H, Fukuta K, Matsumoto D, Hanada T, Gao M, Habu T, Fuyuhiro Y, Ogawa S, Yaegaki H, Yamaguchi M, et al. (2009) Characterization of a novel self-compatible S3′ haplotype leads to the development of a universal PCR marker for two distinctly originated self-compatible S haplotypes in Japanese apricot (Prunus mume Sieb. et Zucc.). J Jpn Soc Hortic Sci 78: 40–48 [Google Scholar]
- Yamane H, Kashiwa Y, Kakehi E, Yonemori K, Mori H, Hayashi K, Iwamoto K, Tao R, Kataoka I. (2006) Differential expression of dehydrin in flower buds of two Japanese apricot cultivars requiring different chilling accumulation for bud break. Tree Physiol 26: 1559–1563 [DOI] [PubMed] [Google Scholar]
- Yamane H, Kashiwa Y, Ooka T, Tao R, Yonemori K. (2008) Suppression subtractive hybridization and differential screening reveals endodormancy-associated expression of an SVP/AGL24-type MADS-box gene in lateral vegetative buds of Japanese apricot. J Am Soc Hortic Sci 133: 708–716 [Google Scholar]
- Yamane H, Ooka T, Jotatsu H, Hosaka Y, Sasaki R, Tao R. (2011a) Expressional regulation of PpDAM5 and PpDAM6, peach (Prunus persica) dormancy-associated MADS-box genes, by low temperature and dormancy-breaking reagent treatment. J Exp Bot 62: 3481–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane H, Ooka T, Jotatsu H, Sasaki R, Tao R. (2011b) Expression analysis of PpDAM5 and PpDAM6 during flower bud development in peach (Prunus persica). Sci Hortic (Amsterdam) 129: 844–848 [Google Scholar]