Abstract
Amyloplasts, a subtype of plastid, are found in nonphotosynthetic tissues responsible for starch synthesis and storage. When tobacco (Nicotiana tabacum) Bright Yellow-2 cells are cultured in the presence of cytokinin instead of auxin, their plastids differentiate from proplastids to amyloplasts. In this program, it is well known that the expression of nucleus-encoded starch biosynthesis genes, such as ADP-Glucose Pyrophosphorylase (AgpS) and Granule-Bound Starch Synthase (GBSS), is specifically induced. In this study, we investigated the roles of plastid gene expression in amyloplast differentiation. Microarray analysis of plastid genes revealed that no specific transcripts were induced in amyloplasts. Nevertheless, amyloplast development accompanied with starch biosynthesis was drastically inhibited in the presence of plastid transcription/translation inhibitors. Surprisingly, the expression of nuclear AgpS and GBSS was significantly repressed by the addition of these inhibitors, suggesting that a plastid-derived signal(s) that reflects normal plastid gene expression was essential for nuclear gene expression. A series of experiments was performed to examine the effects of intermediates and inhibitors of tetrapyrrole biosynthesis, since some of the intermediates have been characterized as candidates for plastid-to-nucleus retrograde signals. Addition of levulinic acid, an inhibitor of tetrapyrrole biosynthesis, resulted in the up-regulation of nuclear AgpS and GBSS gene expression as well as starch accumulation, while the addition of heme showed opposite effects. Thus, these results indicate that plastid transcription and/or translation are required for normal amyloplast differentiation, regulating the expression of specific nuclear genes by unknown signaling mechanisms that can be partly mediated by tetrapyrrole intermediates.
Plastids are plant-specific, semiautonomous organelles that have important metabolic functions, such as photosynthesis, carbon and nitrogen assimilation, and biosynthesis of lipids and amino acids. In higher plants, plastids are known to differentiate between different subtypes in response to several developmental and environmental conditions. Each differentiated form of plastid plays a specific role in maintaining a variety of metabolic and physiological functions as a component of plant cells or tissues (for review, see Lopez-Juez and Pyke, 2005). For instance, chloroplasts, the most characterized form of plastid, are found in the majority of green tissues and are involved in photosynthesis. Chromoplasts occur in limited organs, such as fruit and floral petals, or root tissues in carrot (Daucus carota), and are engaged in the synthesis and storage of carotenoid pigments. Leucoplasts are colorless plastids mainly existing in root tissues. Several reactions for terpene biosynthesis take place in leucoplasts as well as in chloroplasts. In meristematic tissues, plastids can be identified as an undifferentiated form, termed proplastids, which are specialized for proliferation instead of any metabolic reactions. Amyloplasts, which are thought to be a specialized form of leucoplasts, exist in sink tissues such as endosperms and tubers and are characterized as plastids for starch synthesis and storage. A series of enzymes related to starch synthesis, such as ADP-Glucose Pyrophosphorylase (AGPase), Granule-Bound Starch Synthase (GBSS), and Starch Branching Enzyme, are specifically enriched in amyloplasts (Martin and Smith, 1995; Fernie et al., 2002). The stored starch can be broken down to free saccharides or sugar derivatives and supplied for germination and plant growth by translocation. In addition, amyloplasts in root cap cells and endodermal cells of inflorescence stems play an essential role in gravity sensing as a kind of statolith, and that specific accumulation of starch granules in the amyloplasts is important in gravity sensing (for review, see Morita and Tasaka, 2004).
It is widely believed that plastids originated from an endosymbiosis between an ancient cyanobacterial cell and a primordial eukaryotic cell. Accordingly, plastids contain their own genomic DNA and gene expression machineries of cyanobacterial origin. However, after the long history of evolution, a large number of genes were lost from the plastid genome or transferred into the nuclear genome (Leister, 2003). The plastid genomes of higher plants are normally double-stranded, circular DNAs containing about 120 genes that are composed of four ribosomal RNAs, 30 transfer RNAs, and 80 proteins required for transcription, translation, and a variety of plastid functions such as photosynthesis (Sugita and Sugiura, 1996). However, the vast majority of plastid proteins are encoded in the nuclear genome. Once expressed and translated in the cytosol, precursors of plastid proteins are imported into plastids via translocating complexes, processed and modified into mature proteins, and function individually or by forming functional complexes with other proteins, whether these are originally plastid encoded or nuclear encoded. In this sense, coordinated expression of the nuclear and plastid genes is considered important for the maintenance of normal plastid functions (Goldschmidt-Clermont, 1998).
Expression of plastid genes is strictly controlled by the nucleus, depending on tissues and environmental conditions. Plastid gene expression is known to be regulated at numerous steps, including transcription. The plastids of higher plants contain at least two types of RNA polymerases, termed nucleus-encoded plastid RNA polymerase (NEP) and plastid-encoded plastid RNA polymerase (PEP; Shiina et al., 2005). NEP is a bacteriophage-type, single-subunit enzyme that is responsible for the transcription of housekeeping genes, including core subunits of PEP. PEP predominantly mediates the transcription of photosynthesis-related genes by combining the holoenzyme with one of the σ factors that are encoded in the nuclear genome. Multiple genes for σ factors have been identified in a variety of higher plants (Lysenko, 2007). An increasing number of genetic and biochemical approaches have revealed the distinct function of each σ factor (SIG1–SIG6) in Arabidopsis (Arabidopsis thaliana). Among these σ factors, it has been shown that both SIG2 and SIG6 were involved in chloroplast development, since lesion of SIG2 as well as SIG6 leads to the phenotype of pale-green leaves (Shirano et al., 2000; Ishizaki et al., 2005; Loschelder et al., 2006). In the case of the sig2 mutant, for instance, abnormal chloroplast development resulted from the decline of glutamyl-tRNA amount, which caused the inhibition of plastid translation as well as chlorophyll biosynthesis (Kanamaru et al., 2001; Hanaoka et al., 2003). Furthermore, glutamyl-tRNA directly bound to and inhibited the transcription activity of NEP in vitro, suggesting that SIG2 mediates the switching of transcriptional machinery from NEP to PEP during chloroplast development (Hanaoka et al., 2005). Thus, recent studies of the transcriptional regulation of plastid genes have focused mainly on the step of differentiation into chloroplasts. On the other hand, little is known about transcriptional regulation during differentiation into other nonphotosynthetic plastids.
As a model system of plastid differentiation in nonphotosynthetic tissues, we chose to use tobacco (Nicotiana tabacum) Bright Yellow-2 (BY-2) cultured cells, which is a model plant cultured cell system with nonphotosynthetic, highly prolific properties (Nagata et al., 1992). This cell line is usually maintained in a medium containing the auxin 2,4-dichlorophenoxyacetic acid (2,4-D), in which plastids are maintained as undifferentiated proplastids. Differentiation into amyloplasts from proplastids is induced by the addition of synthetic cytokinin, 6-benzylaminopurine (BA; Sakai et al., 1992; Miyazawa et al., 1999). During this step, expression of several nucleus-encoded genes related to starch biosynthesis, such as AgpS and GBSS, is known to be specifically induced by the addition of BA (Miyazawa et al., 1999). It was also reported that the transcription activity of several plastid-encoded genes in amyloplasts was lower than that in proplastids and chloroplasts (Sakai et al., 1992; Valkov et al., 2009). However, the importance and requirement of plastid gene expression in amyloplasts have not been clearly elucidated yet.
In this paper, we investigate the roles of plastid gene expression during the course of amyloplast differentiation using a BY-2 cultured cell system. Microarray and northern-blot analyses showed that the profile of plastid transcripts was not changed drastically during amyloplast development, whereas the expression of nucleus-encoded starch biosynthesis genes was specifically regulated by the inhibition of transcription and translation activities in plastids. Induction of nuclear genes was also regulated by tetrapyrrole intermediates, which was hypothesized previously to be a plastid signal. Our results thus highlight the existence of the retrograde signaling pathway from plastid to nucleus, even in the differentiation step into nonphotosynthetic amyloplasts.
RESULTS
Profile of Plastid Gene Expression during Amyloplast Development
Initially, we confirmed the experimental system established in a previous report (Miyazawa et al., 1999). Tobacco BY-2 cultured cells were proliferated in Murashige and Skoog (MS) medium containing 0.2 mg L−1 2,4-D (hereafter designated D medium). When BY-2 cells were inoculated into fresh D medium, transcripts of starch biosynthesis genes (AgpS and GBSS) as well as Elongation Factor1 alpha (EF-1α; control) increased slightly within 24 h (Supplemental Fig. S1). Expression of AgpS and GBSS was further enhanced when cells were grown in the hormone-free MS medium (F medium) or MS medium containing 1 mg L−1 BA (B medium), while expression of EF-1α was similar to that in D medium (Supplemental Fig. S1). Thus, we could reproduce the induction of starch biosynthesis genes as well as starch accumulation within plastids (data not shown), both of which were observed during differentiation into amyloplasts. Since the efficiency of amyloplast differentiation coupled with the induction of starch biosynthesis genes in B medium was similar to, but more drastic than, that in F medium, we considered it sufficient to perform the following analyses in B medium.
To investigate the profile of plastid transcript accumulation during amyloplast development, levels of whole plastid transcripts in BY-2 cells grown in D and B media for 24 h were analyzed using the tobacco plastid microarray established previously (Nakamura et al., 2003). Among 177 DNA regions of the tobacco plastid genome, 74 regions showed no detectable or highly dispersed fluorescent signals, mainly because expression levels of these genes were quite low in both proplastids and amyloplasts. Of the other 103 regions, one of four regions for the ycf2 open reading frame (ORF) showed a marked decrease in signal in amyloplasts, whereas no specific accumulation of plastid transcripts in amyloplasts was observed (Table I). In summary, the expression of almost all plastid genes was constant during amyloplast differentiation.
Table I. Expression levels of plastid genes in BY-2 cells cultured in B and D media.
Signal ratios (Cy5 to Cy3) of six independent spots for appropriate genes on tobacco plastid microarrays from two biological replicates were averaged, and all reliable data sets (coefficient of variation = sd/mean < 0.3) are listed. Spot identification (ID) and corresponding gene names follow Nakamura et al. (2003).
| Spot ID | Gene | Ratio of B to D (Mean ± sd) | Spot ID | Gene | Ratio of B to D (Mean ± sd) | Spot ID | Gene | Ratio of B to D (Mean ± sd) |
| 307 | ycf3(c)exon2 | 1.65 ± 0.28 | 303 | rps16(c)5′exon | 0.99 ± 0.07 | 320 | 3′rps12(c)intron | 0.85 ± 0.12 |
| 314 | clpP(c)exon2 | 1.47 ± 0.14 | 18 | trnG | 0.99 ± 0.17 | 142 | ycf2 | 0.83 ± 0.08 |
| 86 | atpB(c),atpE(c) | 1.37 ± 0.18 | 161 | rrn23 | 0.98 ± 0.19 | 194 | ycf1(c) | 0.81 ± 0.17 |
| 70 | rps4(c) | 1.31 ± 0.24 | 91 | psal | 0.98 ± 0.10 | 129 | rpl16 (c)3′exon | 0.81 ± 0.09 |
| 312 | clpP(c)3′exon | 1.31 ± 0.15 | 72 | trnT(UGU) (c) | 0.98 ± 0.10 | 155 | trnV(GAC) | 0.79 ± 0.20 |
| 164 | rrn5 | 1.26 ± 0.10 | 153 | 3′rps12(c),orf131 | 0.97 ± 0.07 | 323 | trnA intron | 0.78 ± 0.05 |
| 305 | ycf3(c)3′exon | 1.25 ± 0.06 | 89 | accD | 0.96 ± 0.07 | 145 | trnL(CAA) (c) | 0.78 ± 0.13 |
| 139 | ycf2 | 1.24 ± 0.12 | 35 | rpoC1 (c)5′exon | 0.96 ± 0.04 | 173 | rpl32 | 0.78 ± 0.03 |
| 162 | rrn4.5 | 1.24 ± 0.08 | 85 | atpE(c),atpB(c) | 0.95 ± 0.20 | 33 | rpoC1 (c)3′exon | 0.78 ± 0.21 |
| 25 | atpH(c) | 1.22 ± 0.14 | 113 | clpP(c) | 0.95 ± 0.14 | 83 | trnV(UAC) (c) | 0.77 ± 0.15 |
| 75 | trnL(UAA) | 1.20 ± 0.13 | 128 | rpl14(c) | 0.95 ± 0.17 | 135 | rpl2 (c)intron | 0.76 ± 0.07 |
| 62 | psaA (c) | 1.19 ± 0.35 | 15 | psbl | 0.94 ± 0.08 | 313 | clpP(c)intron2 | 0.76 ± 0.06 |
| 319 | rpl16(c)intron | 1.19 ± 0.16 | 61 | psaB(c) | 0.94 ± 0.25 | 140 | ycf2 | 0.73 ± 0.07 |
| 311 | trnV (c)intron | 1.15 ± 0.16 | 34 | rpoC1 (c)intron | 0.94 ± 0.13 | 31 | rpoC2(c) | 0.73 ± 0.11 |
| 110 | rps18 | 1.13 ± 0.05 | 54 | trnS(c) | 0.93 ± 0.06 | 193 | ycf1(c) | 0.71 ± 0.12 |
| 148 | ndhB(c)3′exon | 1.12 ± 0.13 | 97 | psbJ (c),orf99 | 0.93 ± 0.15 | 22 | atpF (c)intron | 0.70 ± 0.13 |
| 158 | trnl(GAU) | 1.12 ± 0.08 | 21 | atpF (c)3′exon | 0.92 ± 0.11 | 4 | trnK(c)3′exon | 0.70 ± 0.12 |
| 124 | rps11(c) | 1.10 ± 0.24 | 125 | rpl36(c) | 0.91 ± 0.13 | 131 | rps3(c),rpl22 | 0.69 ± 0.10 |
| 136 | rpl2(c)5′exon | 1.10 ± 0.13 | 93 | ycf4 | 0.91 ± 0.12 | 68 | trnS(GGA) | 0.68 ± 0.15 |
| 26 | (atpH(c)promoter) | 1.09 ± 0.16 | 308 | ycf3(c) intron 1 | 0.91 ± 0.07 | 130 | rpl16(c)intron,5′exon | 0.67 ± 0.10 |
| 52 | psbD, psbC | 1.07 ± 0.09 | 127 | rps8(c) | 0.91 ± 0.10 | 315 | clpP(c)intron1 | 0.67 ± 0.09 |
| 171 | ndhF,orf350 | 1.06 ± 0.14 | 134 | rpl2 (c)3′exon | 0.90 ± 0.16 | 166 | trnR | 0.67 ± 0.14 |
| 23 | atpF(c)5′exon | 1.04 ± 0.04 | 47 | trnE(c) | 0.90 ± 0.26 | 36 | rpoB(c) | 0.66 ± 0.06 |
| 28 | atpl(c) | 1.04 ± 0.10 | 30 | rpoC2(c) | 0.89 ± 0.10 | 96 | orf99 | 0.66 ± 0.08 |
| 32 | rpoC2(c) | 1.03 ± 0.26 | 112 | 5′rps12(c) | 0.88 ± 0.08 | 121 | petD 5′exon,intron | 0.65 ± 0.17 |
| 317 | petB intron | 1.02 ± 0.15 | 20 | atpA(c) | 0.88 ± 0.23 | 46 | trnY(c) | 0.61 ± 0.14 |
| 102 | petL | 1.01 ± 0.15 | 66 | orf74(c) | 0.88 ± 0.13 | 87 | rbcL | 0.60 ± 0.18 |
| 132 | rpl22,rps3(c) | 1.01 ± 0.21 | 100 | orf103(c) | 0.88 ± 0.17 | 3 | psbA(c) | 0.59 ± 0.09 |
| 65 | ycf3(c),orf74(c) | 1.01 ± 0.11 | 160 | rrn23 | 0.88 ± 0.10 | 322 | trnl intron | 0.58 ± 0.04 |
| 115 | psbB | 1.00 ± 0.17 | 126 | ψ infA(c) | 0.87 ± 0.19 | 103 | petG | 0.53 ± 0.07 |
| 13 | psbK | 1.00 ± 0.24 | 321 | 3′rps12 (c)5′exon | 0.87 ± 0.11 | 45 | trnD(c) | 0.52 ± 0.07 |
| 182 | ndhE(c) | 1.00 ± 0.11 | 191 | rps15(c) | 0.86 ± 0.13 | 56 | ycf9 | 0.51 ± 0.15 |
| 10 | rps16(c) | 1.00 ± 0.08 | 306 | ycf3(c)intron2 | 0.85 ± 0.04 | 156 | rrn16 | 0.51 ± 0.13 |
| 149 | ndhB(c)intron | 1.00 ± 0.20 | 63 | psaA(c) | 0.85 ± 0.02 | 141 | ycf2 | 0.44 ± 0.07 |
| 152 | rps7(c) | 0.99 ± 0.07 |
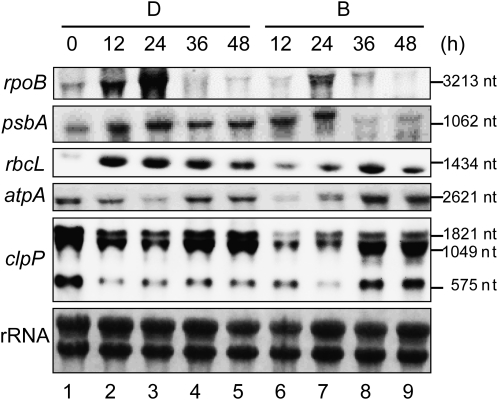
Expression levels of several plastid mRNAs were further confirmed by northern-blot analysis (Fig. 1). Expression of rpoB was transiently up-regulated at 24 h after inoculation into both D and B media, although the expression level was much higher in D medium. Expression patterns of both psbA and rbcL were stable in D medium, while transient expression with different peaks was observed in B medium. No obvious difference in atpA expression was observed between D and B media. The amount of atpA mRNA transiently decreased and gradually increased until 48 h after inoculation. A transient decline of mRNA accumulation was also observed in clpP, although the reduction and reaccumulation level was more extensive in B medium. Thus, the accumulated mRNA of plastid genes examined in this assay remained constant or decreased in B medium after 24 h, consistent with our microarray data. From these results, we concluded that no specific accumulation of plastid genes occurred in the course of differentiation into amyloplasts.
Figure 1.
Northern-blot analysis of plastid gene expression during amyloplast differentiation. Total RNA was extracted from BY-2 cells in D or B medium sampled every 12 h. Five micrograms of total RNA was loaded in each lane. Lane 1, Before inoculation (0 h); lanes 2 to 5, after inoculation in D medium (12, 24, 36, and 48 h after inoculation, from the left); lanes 6 to 9, after inoculation in B medium (12, 24, 36, and 48 h after inoculation, from the left). nt, Nucleotides.
Efficient Inhibition of Plastid Transcription/Translation Repressed Amyloplast Development
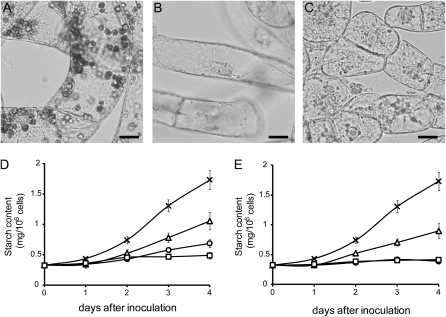
Plastid microarray and northern analyses showed that the expression of plastid genes was constant or slightly decreased during amyloplast differentiation. However, their transcripts were still detected even at a basal level. To investigate whether plastid transcriptional or translational activity is required for amyloplast differentiation, we examined the effect of specific inhibitors of transcription or translation in plastids. When BY-2 cells were cultured in B medium, an apparent pattern of starch staining was detected within amyloplasts (Fig. 2A), as reported previously (Miyazawa et al., 1999). However, the accumulation of starch granules within plastids was not observed when cells were grown in B medium supplemented with 100 mg L−1 rifampicin, a transcriptional inhibitor of bacterial RNA polymerase (Fig. 2B). Repression of starch accumulation was also observed by the addition of several types of translational inhibitors, namely spectinomycin (100 mg L−1), kanamycin (100 mg L−1), streptomycin (200 mg L−1), lincomycin (200 mg L−1), or chloramphenicol (100 mg L−1; Fig. 2C; Supplemental Fig. S2). These observations were confirmed by quantitative analysis. Starch content increased gradually during cultivation in B medium but decreased drastically when cells were grown in B medium in the presence of rifampicin, spectinomycin, or kanamycin, and increasing the concentration of the inhibitor resulted in marked repression of starch accumulation (Fig. 2, D and E; Supplemental Fig. S2F). These results indicated that any specific inhibitor of plastid transcription or translation drastically inhibited starch biosynthesis. So far, these results demonstrated that either transcription or translation activity in plastids was extensively required for amyloplast differentiation.
Figure 2.
Effects of inhibitors of plastid gene expression on starch accumulation within the plastids. A to C, Staining of starch granules in plastids. BY-2 cells were cultured for 48 h in B medium either in the absence (A) or presence of 100 mg L−1 rifampicin (B) or 100 mg L−1 spectinomycin (C) and stained with iodine solution. Bars = 20 μm. D and E, Accumulation of starch in BY-2 cells. Aliquots of cultured cells were sampled every 24 h and subjected to quantitative analysis. D, BY-2 cells were cultured in B medium in the absence (crosses) or presence of 50 mg L−1 (triangles), 100 mg L−1 (circles), or 150 mg L−1 (squares) rifampicin. E, BY-2 cells were cultured in B medium in the absence (crosses) or presence of 100 mg L−1 (triangles), 200 mg L−1 (circles), or 300 mg L−1 (squares) spectinomycin. Values shown are means ± sd (n = 3).
Inhibition of Transcription and Translation Activity in Plastids Caused Specific Reduction of Nucleus-Encoded Starch Synthesis Genes
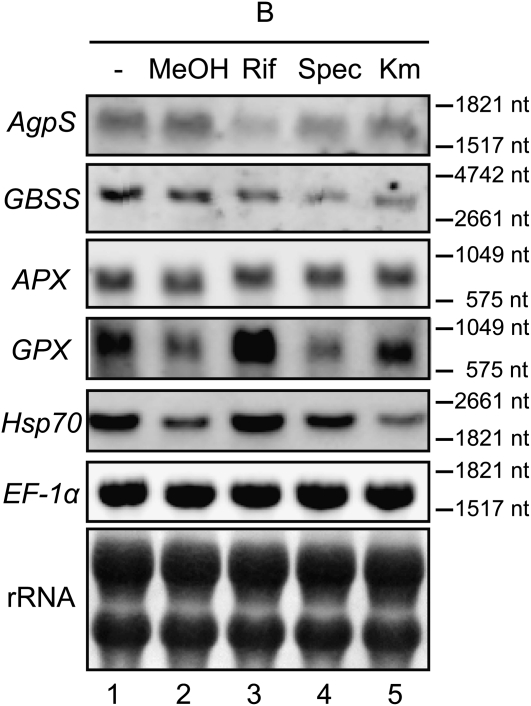
Inhibition of plastid transcription and translation caused the repression of amyloplast differentiation. This result implied that the status of gene expression and/or specific gene products within plastids might play a major role in any step(s) of amyloplast development. In order to check whether plastid gene expression could somehow regulate the transcription of nuclear genes, we performed northern-blot analysis to examine the expression levels of the nucleus-encoded AgpS and GBSS genes in the presence of plastid transcription/translation inhibitors. Surprisingly, the expression of both genes in B medium was significantly and specifically repressed in the presence of rifampicin, spectinomycin, or kanamycin, whereas the expression of EF-1α, another nucleus-encoded gene used as a control, was unchanged in the presence of an inhibitor (Fig. 3). Inhibition of both transcription and translation in plastids globally repressed the expression of AgpS and GBSS, although the inhibition rate was a little different between these inhibitors. In order to test whether supplementation of these plastid transcription/translation inhibitors might affect the expression of a wide variety of nuclear genes by stimulation of any cellular stress response mechanisms, we examined the expression patterns of representative stress response genes, such as the oxidative stress-responsive genes Ascorbate Peroxidase (APX) and Glutathione Peroxidase (GPX; Kadota et al., 2005) and a heat shock-responsive gene, Heat Shock Protein70 (Hsp70; Lara et al., 2005). Whereas the expression level of APX was constitutive, that of GPX and Hsp70 changed considerably in the presence of several inhibitors or even under mock control conditions. Although a certain degree of gene-specific pattern was observed, there seemed to be little correlation of expression profiles between any stress response genes and starch biosynthesis genes. These results suggest that plastid gene expression could modulate amyloplast differentiation through regulating the expression of several nucleus-encoded genes specific for starch synthesis, independent of the general stress response system.
Figure 3.
Effects of plastid transcription and translation inhibitors on the expression of nucleus-encoded starch synthesis genes. Ten micrograms (for AgpS, GBSS, APX, GPX, and Hsp70) or 2 μg (for EF-1α) per lane of total RNAs extracted from 1-d-old BY-2 cells was loaded. Loaded samples were as follows: B medium (lane 1), B medium containing methanol (lane 2), B medium supplemented with 100 mg L−1 rifampicin (in methanol; lane 3), 200 mg L−1 spectinomycin (lane 4), or 10 mg L−1 kanamycin (lane 5). A methylene blue staining image of rRNAs served as a loading control. nt, Nucleotides.
Involvement of Plastid-to-Nucleus Retrograde Signaling in Amyloplast Differentiation
Recently, the existence of regulatory mechanisms for nuclear gene expression in response to stimuli against several processes occurring in plastids, including plastid gene expression, has been noted. One of the candidates for signaling molecules in this signal transduction pathway, termed a plastid signal, was proposed as magnesium-protoporphyrin IX (Mg-proto IX), an intermediate of the tetrapyrrole biosynthesis pathway. To date, almost all studies on plastid-derived signal transduction have focused on communication between the nucleus and chloroplasts in photosynthetic cells. It is unknown whether a similar signaling pathway was also conserved in nonphotosynthetic plastids, such as amyloplasts. As shown above, our results suggested that plastid gene expression could regulate the transcription of nuclear genes, which might be mediated by some plastid-derived signals. To further analyze this, we examined the roles of tetrapyrrole intermediates on amyloplast differentiation.
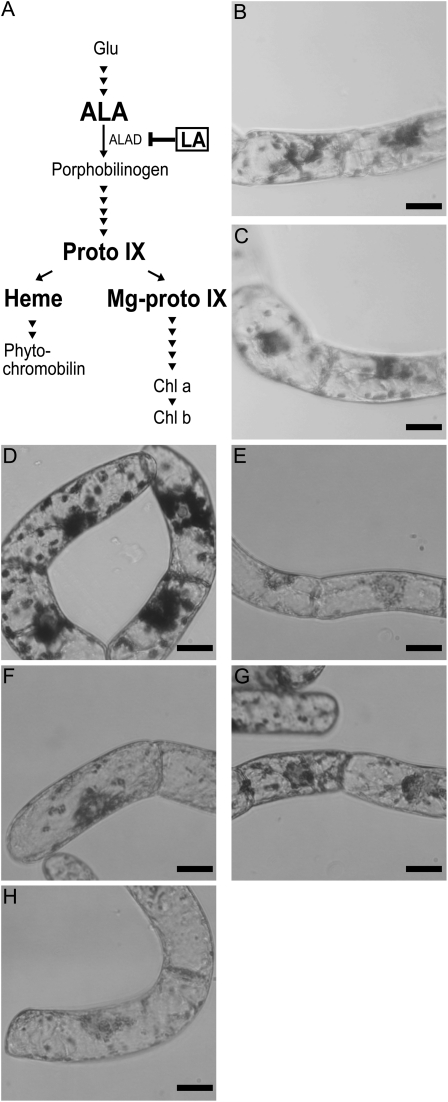
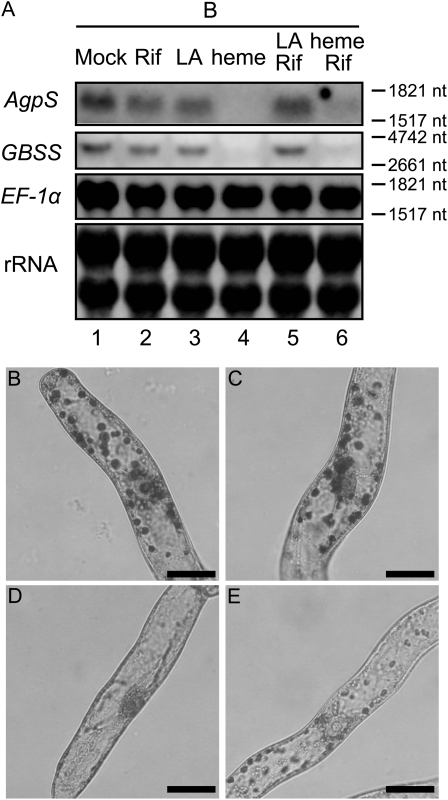
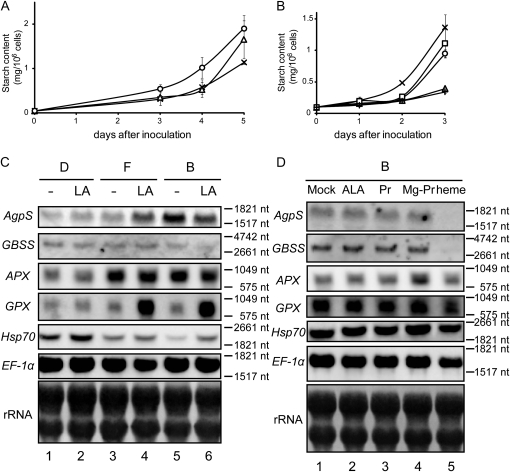
Levulinic acid (LA) is a competitive inhibitor of aminolevulinic acid dehydratase and blocks the metabolic flow of the tetrapyrrole biosynthetic pathway at an early step (Jurgenson et al., 1976), resulting in a drastic decrease in the amount of the downstream intermediate products, including Mg-proto IX (Fig. 4A). Although the addition of 500 μm LA into B medium had little effect compared with the control (data not shown; see Fig. 6, B and C, below), slight enhancement of starch accumulation was observed in response to the addition of the same concentration of LA into F medium (Fig. 4, B and C). These results indicate that inhibition of the biosynthesis of tetrapyrrole intermediates by LA positively affected amyloplast development. It should be noted that a positive effect of LA might be masked by hormonal effects derived from cytokinin supplementation in B medium, consistent with several analyses described below. Since LA could inhibit the accumulation of most tetrapyrrole intermediates, it was highly predictable that one of these might act as a messenger for the signaling pathway from plastids to the nucleus. To specify the candidate molecules required during amyloplast differentiation, we further examined the effects of the addition of several tetrapyrrole intermediates. Among several candidates of downstream intermediates, we examined the effect of protoporphyrin IX (proto IX), Mg-proto IX, and heme in amyloplast differentiation. Observation of starch staining indicated that proto IX and Mg-proto IX caused weak repression of starch accumulation compared with the control (Fig. 4, D, F, and G), whereas the addition of heme resulted in drastic reduction of starch accumulation within plastids (Fig. 4H). When BY-2 cells were grown in B medium containing δ-aminolevulinic acid (ALA), an upstream intermediate in the tetrapyrrole biosynthesis pathway, starch accumulation was also decreased (Fig. 4E).
Figure 4.
Effect of inhibitors or intermediates of the tetrapyrrole biosynthesis pathway on starch accumulation. A, Schematic diagram of the tetrapyrrole biosynthesis pathway. Specific inhibitors and intermediates used in this study are indicated in boldface. Intermediates were as follows: Glu, ALA, proto IX, Mg-proto IX, and chlorophyll a and chlorophyll b (Chl a and Chl b). B to H, Staining of starch granules in plastids. BY-2 cells were grown for 96 h in F medium in the absence (B) or presence of 500 μm LA (C) or in B medium in the absence (D) or presence of 100 μm ALA (E), 40 μm proto IX (F), 10 μm Mg-proto IX (G), or 40 μm heme (H) and stained with iodine solution. Bars = 20 μm.
Figure 6.
Combinational effect of inhibitors targeting different plastid activities on nuclear gene expression. A, Northern-blot analysis was performed using total RNAs isolated from 1-d-old BY-2 cells grown in culture media as follows: B medium containing methanol and dimethyl sulfoxide as a mock control (lane 1), B medium supplemented with 100 mg L−1 rifampicin (Rif; lane 2), 500 μm LA (lane 3), or 40 μm heme (lane 4) alone or B medium supplemented with 500 μm LA (lane 5) or 40 μm heme (lane 6) in the presence of 100 mg L−1 rifampicin simultaneously. Ten micrograms (for AgpS and GBSS) or 2 μg (for EF-1α) of total RNAs was loaded per lane, and a methylene blue staining image of rRNAs served as a loading control. nt, Nucleotides. B to E, Staining of starch granules in plastids. BY-2 cells were grown for 96 h in B medium in the absence (B) or presence of 500 μm LA alone (C), 100 mg L−1 rifampicin alone (D), or LA and rifampicin simultaneously (E) and stained with iodine solution. Bars = 10 μm.
The effects of LA and tetrapyrrole intermediates were further examined by quantification of starch content and northern-blot analysis of starch biosynthesis genes. When 500 μm LA was incorporated into F medium, the starch content rose markedly compared with the control (Fig. 5A). In contrast, the increase in starch content in the control was drastically repressed by the addition of either ALA or heme, whereas a slight reduction of starch accumulation was observed in the presence of proto IX or Mg-proto IX (Fig. 5B). These results were consistent with the microscopic observations. Northern-blot analysis demonstrated that transcription of AgpS and GBSS in F medium was up-regulated in the presence of LA, whereas the addition of LA into D medium slightly enhanced the expression of GBSS (Fig. 5C). On the other hand, obvious enhancement of gene expression by LA addition was not observed in B medium (Fig. 5C), which correlated with the result of starch content in the presence or absence of LA in B medium. These results suggest that the repression of tetrapyrrole biosynthesis by LA enhanced the expression of nuclear starch synthesis genes, although it was dependent on the hormonal condition within cells. In contrast, exclusive repression of AgpS and GBSS expression was observed specifically in the presence of heme (Fig. 5D). These data suggest that heme, not Mg-proto IX, might be involved in plastid-to-nucleus signaling to negatively regulate the expression of nucleus-encoded starch biosynthesis genes. In these conditions, the expression of APX, GPX, and Hsp70 was also examined. Although they were affected to some extent by the addition of a tetrapyrrole biosynthesis inhibitor or intermediates, the expression patterns of these stress response genes differed from each other and were not correlated with those of starch synthesis genes (Fig. 5, C and D). On the other hand, ALA supplementation did not show any influence on the expression of nucleus-encoded genes (Fig. 5D), which might indicate that the decline of starch content in the presence of excess ALA was the result of a posttranscriptional regulatory mechanism.
Figure 5.
Perturbation of tetrapyrrole biogenesis passively affected the expression of starch synthesis genes. A, Effect of LA on starch accumulation. BY-2 cells were cultured in F medium in the absence (crosses) or presence of 200 μm (triangles) or 500 μm (circles) LA. B, Effect of tetrapyrrole intermediates on starch accumulation. BY-2 cells were cultured in B medium either in the absence (crosses) or presence of 100 μm ALA (triangles), 40 μm proto IX (circles), 10 μm Mg-proto IX (squares), or 40 μm heme (plus signs). Values presented are means ± sd (n = 3). C and D, Northern-blot analysis of starch biosynthesis genes. C, Total RNAs were extracted from 1-d-old BY-2 cells grown in D medium (lanes 1 and 2), F medium (lanes 3 and 4), and B medium (lanes 5 and 6) either in the absence (lanes 1, 3, and 5) or presence (lanes 2, 4, and 6) of 500 μm LA. D, Total RNAs were extracted from 1-d-old BY-2 cells grown in B medium in the absence (lane 1) or presence of 100 μm ALA (lane 2), 40 μm proto IX (PR; lane 3), 10 μm Mg-proto IX (Mg-PR; lane 4), or 40 μm heme (lane 5). In C and D, 10 μg (for AgpS, GBSS, APX, GPX, and Hsp70) or 2 μg (for EF-1α) of total RNAs was loaded per lane, and a methylene blue staining image of rRNAs served as a loading control. nt, Nucleotides.
To elucidate whether and to what extent internal levels of tetrapyrrole intermediates were changed by the supplementation of inhibitors or the feeding of excess intermediates, we extracted and quantified net amounts of proto IX, Mg-proto IX, and heme within BY-2 cells (Table II). Generally, levels of these tetrapyrrole intermediates were very low, even in cells cultured in D medium, compared with general plant tissues with photosynthetic chloroplasts (Shalygo et al., 2009; Peter et al., 2010). In our experimental system, proto IX content in BY-2 cells was hardly detected in any growth conditions (data not shown). The internal levels of Mg-proto IX and heme within cells grown in F and B media further declined compared with those in D medium. In particular, Mg-proto IX content was undetectable both in F and B media. When LA was incorporated in D and F media, the cellular level of these intermediates was decreased. However, repression of tetrapyrrole biosynthesis by the addition of LA was not observed in the case of B medium, which was consistent with the results that the expression of starch synthesis genes as well as starch accumulation was not induced by the addition of LA into B medium. This observation indicates that tetrapyrrole synthesis and accumulation in B medium are too low for the addition of LA to be inhibitory. On the other hand, the incorporation of heme and ALA in B medium resulted in a slight accumulation of Mg-proto IX but a similar level of heme compared with those in the fresh B medium. This result may also support the relationship between heme content and the expression level of starch synthesis genes, although it is difficult to estimate the flow of tetrapyrrole biosynthesis in this condition.
Table II. Changes in internal levels of tetrapyrrole intermediates in BY-2.
The data represent means ± sd of three independent experiments. Asterisks, Undetectable; dash, not determined
| Culture | Mg-Proto IX | Heme |
| pmol g−1 fresh wt | ||
| D | 5.06 ± 4.43 | 646.3 ± 74.31 |
| D + LA | 3.97 ± 3.06 | 488.22 ± 179.12 |
| F | * | 367.37 ± 188.63 |
| F + LA | * | 205.84 ± 33.65 |
| B | * | 328.3 ± 138.12 |
| B + LA | * | 310.47 ± 132.16 |
| B + ALA | 1.37 ± 0.8 | 262.25 ± 292.21 |
| B + heme | 1.32 ± 0.54 | – |
Relationship between Pathways for the Modulation of Gene Expression Involved in Amyloplast Transition
The northern-blot analyses demonstrated that the expression of AgpS and GBSS was positively or negatively affected by the effects derived from plastid gene expression or tetrapyrrole biosynthesis, respectively. To assess the relationship between these pathways with opposite effects on nuclear gene expression, we performed another northern-blot analysis with the addition of two drugs simultaneously. As also described above, enhancement of starch biosynthesis gene expression by the addition of LA was not observed in B medium (Fig. 6A), possibly because the tetrapyrrole level in B medium is very low. When rifampicin was incorporated in B medium in combination with 500 μm LA, expression levels of both AgpS and GBSS increased compared with levels in the presence of rifampicin alone, but they were not recovered up to the level detected in the control (Fig. 6A). The relationship between the two regulatory effects was confirmed by further northern-blot analysis using heme instead of LA. Expression of AgpS and GBSS in B medium was repressed moderately in the presence of rifampicin alone and more drastically in the presence of both rifampicin and 40 μm heme (Fig. 6A). This combinational effect of simultaneous supplementation of two inhibitors resulted in modest starch accumulation within plastids (Fig. 6, B–E), consistent with northern-blot results. Similar expression patterns correlated with starch accumulation were also observed when spectinomycin was used instead of rifampicin (data not shown). These results indicate that putative retrograde signal pathways monitoring the condition of tetrapyrrole biosynthesis and gene expression in plastids could be partly related rather than completely independent.
The Retrograde Signaling Pathway Regulating Amyloplast Differentiation Was Independent of Proteasomal Activity
Although molecular details for retrograde signaling from plastid to nucleus were still unknown, it was probable that already-known signal transduction machineries might play an important role in mediating plastid-derived signals. In higher plants, it has been well characterized that protein degradation systems via the ubiquitin-proteasome pathway also contribute to nuclear gene expression in response to several developmental signals and/or environmental stimuli (for review, see Schwechheimer and Schwager, 2004). To test whether the retrograde signal from amyloplast to nucleus was mediated by a proteasomal system, we added epoxomicin, a specific inhibitor of proteasomal activity, into B medium. In microscopic observation of starch staining, no specific differences on starch accumulation were observed in the presence or absence of epoxomicin in both D and B media (Supplemental Fig. S3). This result suggests that proteasome-dependent protein degradation might not be essential for differentiation into amyloplasts.
DISCUSSION
Plastid genes are categorized into three classes depending on their promoter structures. Transcription of class I and III genes is PEP and NEP dependent, respectively, while class II genes are transcribed by both RNA polymerases (Hajdukiewicz et al., 1997). Comprehensive analysis using microarray data indicated that the expression of several genes transcribed by NEP (such as atpB and accD) was unchanged between D and B media. Although only one region for ycf2, which has a comparatively long ORF and is a class III gene, showed decreased accumulation of transcripts in B medium, the signals of the other three regions carrying other portions of the ycf2 ORF were rather constant. These results suggest that NEP activity is not drastically modulated in this condition. Microarray data also demonstrated that plastid gene expression was generally constant or slightly repressed during amyloplast differentiation (Table I). In previous reports of run-on transcription assays using tobacco (Sakai et al., 1992) or sycamore (Acer pseudoplatanus; Ngernprasirtsiri et al., 1990) cultured cell systems, it was reported that the transcription activity of several plastid genes, including rpoB, rbcL, and 16S rRNA, in amyloplasts was reduced compared with that in proplastids or chloroplasts, which is consistent with our data. It should be noted that no plastid genes are extraordinarily up-regulated in B medium, which means that amyloplast development is not caused by specific induction of any plastid genes.
Nevertheless, our inhibitor assays demonstrated that inhibition of either transcription or translation in plastids resulted in significant inhibition of amyloplast differentiation via the repression of specific nuclear gene expression, implying that plastid gene expression was necessary for plastid differentiation. Two possible interpretations of our data are as follows. First, any gene product itself, and/or resulting reaction(s) or product(s) depending on gene expression, is indispensable for amyloplast development. In this case, the amount of putatively essential products was not required to be higher in amyloplasts, but the basal expression level should be essential for differentiation. Second, the activity of transcription and/or translation machinery within plastids itself might serve as the checkpoint for amyloplast differentiation. Numerous studies on a wide variety of higher plants have demonstrated that inhibition of plastid gene expression with several translational inhibitors, such as chloramphenicol and lincomycin, or a transcriptional inhibitor, tagetitoxin, resulted in reduced accumulation of several nuclear transcripts encoding chloroplast proteins (Oelmuller et al., 1986; Rapp and Mullet, 1991; Adamska, 1995; Sullivan and Gray, 1999; Gray et al., 2003; Maclean et al., 2008). Plastid transcriptional/translational inhibitors caused repression of a portion of nucleus-encoded genes, which occurred generally in both photosynthetic and nonphotosynthetic tissues under different light conditions. These observations are highly consistent with our data, demonstrating that drastic repression of specific, but not all, nuclear genes in fact occurred in the presence of any inhibitor of plastid gene expression in BY-2 cells. However, genes examined in previous studies were limited to those encoding proteins for photosynthesis, carbon fixation, or ribosome biogenesis, but not for starch biosynthesis. In addition, although microarray data in Arabidopsis are currently available (NASCArray-336; http://affymetrix.arabidopsis.info/narrays/experimentbrowse.pl) to consider the overall profiles of nuclear gene expression under inhibition of plastid gene expression, transcriptomic information to find the roles of the plastid signals in the course of plastid differentiation is still limited. In this study, we could not specify the essential signal(s) monitoring plastid gene expression, and further experiments, including transcriptomic analyses and genetic assessments, are required to understand signaling mechanisms during plastid differentiation.
The concept of modulation of the transcription of specific nuclear genes by plastid gene expression reminded us of retrograde signaling pathways mediated by plastid signals. Four processes have been proposed as the emerging source of plastid signals: (1) tetrapyrrole biosynthesis, (2) redox state and reactive oxygen species in plastids, (3) organellar gene expression (OGE) status, and (4) protein import into plastids (for review, see Kleine et al., 2009; Inaba, 2010; Pfannschmidt, 2010). Elucidation of molecular mechanisms involving plastid signals has been advanced in this decade by genetic analyses using Arabidopsis mutants. Among five genomes uncoupled (gun) mutants identified in Arabidopsis based on the phenotype of insensitivity to norflurazon, a specific inhibitor of carotenoid biosynthesis (Susek et al., 1993), gun1 was specifically insensitive to lincomycin or erythromycin (Koussevitzky et al., 2007; Ruckle et al., 2007). GUN1 encodes a plastid-localized pentatricopeptide repeat protein and shows putative DNA-binding activity as well as colocalization with other proteins enriched in the plastid transcriptionally active chromosome fraction (Koussevitzky et al., 2007). These data indicate that GUN1 is involved to some extent in the plastid signal pathway related to OGE. On the other hand, fundamental roles of tetrapyrrole intermediates in plastid-to-nucleus signaling pathways have been proposed, according to the fact that gene products of the other four GUN loci (gun2–gun5) have been identified as catalytic enzymes or interaction cofactors for tetrapyrrole biosynthesis in Arabidopsis (Susek et al., 1993; Mochizuki et al., 2001; Larkin et al., 2003). Although several research groups suggested that Mg-proto IX was essential as the signaling molecule of this pathway (Strand et al., 2003; Ankele et al., 2007), recent work found no correlation between steady-state levels of Mg-proto IX and transcription activities of nucleus-encoded genes (Mochizuki et al., 2008; Moulin et al., 2008). Recently, it was demonstrated that heme, another tetrapyrrole intermediate, modulated the expression of nuclear genes in a similar manner to Mg-proto IX in Chlamydomonas reinhardtii (von Gromoff et al., 2008) and Arabidopsis (Woodson et al., 2011). These reports indicate that intermediates of tetrapyrrole biosynthesis might play crucial roles in nuclear gene regulation as one of the plastid signals, although it is still unknown how these molecules mediate the signaling pathway. Considering these reports, our data indicated that heme or its downstream products could be important to modulate nuclear starch biosynthesis gene expression during amyloplast development. However, in this study, we could not identify further mechanisms by which tetrapyrrole intermediates (including heme) might stimulate any signal transduction pathway from plastids to the nucleus or could act as direct signal messengers between these organelles.
In a previous study using a red alga, Cyanidioschyzon merolae, as well as tobacco BY-2 cells, our research group demonstrated that Mg-proto IX is involved in the coordination of nuclear DNA replication with organellar DNA replication (Kobayashi et al., 2009) and that this signaling pathway was mediated by proteasome-dependent protein degradation machinery (Kobayashi et al., 2011). In our inhibitor assay using epoxomicin described here, however, there was little difference in amyloplast differentiation, supporting the notion that the regulatory mechanisms modulating nuclear gene expression and nuclear DNA replication might differ.
Simultaneous inhibition of tetrapyrrole biosynthesis and plastid gene expression activity resulted in the moderated repression of nuclear starch biosynthesis genes. Moreover, simultaneous addition of rifampicin and heme resulted in more significant repression of this specific gene expression. This implies that signals monitoring specific plastid conditions might be modulated by several pathways to regulate nuclear gene expression, although from our experiments it remains uncertain whether these pathways are related. In Arabidopsis, GUN1 is the putative modulator that bundles several plastid signals derived from high-light treatment and defective protein import as well as the inhibition of OGE and tetrapyrrole biogenesis (Koussevitzky et al., 2007; Kakizaki et al., 2009). Although at present there is no information on the existence of a tobacco GUN1 ortholog, it is probable that interplay between each plastid signal is regulated via a GUN1-like protein within plastids, similar to the case in Arabidopsis or, alternatively, that integration of signal transduction pathways might occur in the cytosol or even within the nucleus.
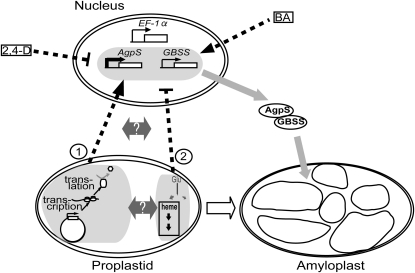
From our data presented here, a probable signaling model modulating amyloplast differentiation via nuclear gene expression is presented in Figure 7. Two possible signal transduction pathways are assumed as follows: induction of nuclear starch biosynthesis genes during amyloplast development is ensured by the normal condition of gene expression systems within plastids, which should be monitored either by checking (1) the activities of plastid transcription and/or translation machinery or (2) the amounts of any products (transcripts, proteins, or their catalytic biomolecules) resulting from plastid gene expression. In contrast, an alternative regulatory mechanism should modulate the expression of nuclear starch biosynthesis genes negatively via a retrograde signal pathway affected by the presence of heme or its downstream intermediates/products of tetrapyrrole biosynthesis. These regulatory systems are presumably active and interact with each other at any step(s) of the signal transduction pathway from plastids to nucleus. Although the detailed molecular mechanism of these plastid-to-nucleus signal transduction pathways is still unknown and remains to be clarified in the future, differentiation into amyloplasts would be strictly controlled by these checkpoints within plastids.
Figure 7.
Hypothetical model for the regulation of nuclear gene expression by retrograde signaling during amyloplast differentiation in BY-2 cells. Putative candidates for the emerging source of retrograde signaling pathways from plastids to the nucleus are indicated as circled numbers. Details of the model are explained in the text.
In conclusion, we demonstrated that the expression of nuclear-encoded starch biosynthesis genes was specifically controlled by plastid-derived signals reflecting both tetrapyrrole biosynthesis and plastid gene expression. This finding further implies the possibility that a retrograde signaling pathway from plastids to the nucleus is widely observed at any developmental stage of plastids and cells.
MATERIALS AND METHODS
Cell Culture
Cultivation and maintenance of tobacco (Nicotiana tabacum) BY-2 cultured cells followed the method of Miyazawa et al. (1999) with a few minor modifications. Briefly, 98 mL of fresh D medium (MS medium containing 0.2 mg L−1 2,4-D) was inoculated with 2 mL of BY-2 suspension cells in the stationary phase every 7 d and cultured at 26°C with shaking at 120 rpm in the dark. To induce amyloplast differentiation, BY-2 cells grown in D medium were transferred into F medium (MS medium lacking 2,4-D) or B medium (MS medium containing 1 mg L−1 BA). For supplementation assays, BY-2 cells were inoculated into medium containing inhibitors or tetrapyrrole intermediates and harvested at the appropriate time points. Details of the supplements and their final concentrations are described in the figure legends.
Measurement of Cell Number and Starch Content
Cell numbers were determined with a counting chamber (Thoma) and a microscope. To prepare BY-2 protoplasts, 3 × 105 cells from the culture were incubated for 90 min at 30°C in an enzyme solution containing 1% (w/v) Cellulase “Onozuka” R-10 (Yakult Pharma Industry) and 0.1% (w/v) Macerozyme R-10 (Yakult Pharma Industry) in 0.4 m mannitol (pH 5.5). Extraction and quantification of starch were performed as described previously (Miyazawa et al., 1999).
Staining of Starch Granules
Suspension cultures containing approximately 3 × 105 BY-2 cells were mixed with a few drops of KI-I solution (1.5 g of KI and 0.5 g of I2 dissolved in 30 mL of distilled water) and observed immediately. Images were recorded with a DP70 digital camera (Olympus) mounted on a light microscope.
RNA Extraction
BY-2 cells were harvested and frozen in liquid nitrogen for grinding with the Multibeads shocker (Yasui Kikai). Total RNA was extracted from cell powder with Trizol reagent (Invitrogen) according to the manufacturer’s protocol. For microarray analysis, total RNA was incubated with 5 units of RQ1 DNase I (Promega), extracted with phenol:chloroform:isoamyl alcohol (25:24:1, v/v), ethanol precipitated, and resolved in RNase-free water.
Northern-Blot Analysis
Tobacco genomic DNA was extracted from ground powder of frozen BY-2 cells as described previously (Murray and Thompson, 1980) and subjected to the following PCR procedure. Gene-specific DNA probes were amplified by PCR with specific primer sets as listed in Supplemental Table S1 and purified by extraction from an agarose gel. For preparation of the probes, an aliquot of 300 ng of each PCR product and antisense strand primer was used. Total RNA was denatured and electrophoresed on a gel containing formaldehyde. The RNAs were vacuum transferred onto a positively charged nylon membrane followed by cross-linking. The rRNA was stained with methylene blue as a loading control. Hybridization with the specific probe in high-SDS hybridization buffer (7% SDS, 50% deionized formamide, 5× SSC, 2% blocking reagent, 50 mm sodium phosphate buffer [pH 7], 0.1% N-lauroylsarcosine sodium salt, and 0.005% yeast RNAs) was performed for 12 h at 48°C as described previously (Kanamaru et al., 2001). Chemiluminescent signals were detected using a LAS-3000 imager (Fujifilm).
Plastid Microarray Analysis
Probe labeling and hybridization with a tobacco chloroplast microarray encompassing whole chloroplast genomic regions (Nakamura et al., 2003) was performed according to the method of Zoschke et al. (2010) with minor modifications. Briefly, 40 μg of total RNA was incubated with specific primer sets utilized for microarray construction (Nakamura et al., 2003). Labeled cDNA fragments were synthesized with Cy3- or Cy5-dUTP (GE Life Science) and ReverTra Ace (Toyobo) and purified with the Qiaquick PCR Purification Kit (Qiagen). Hybridization was performed with two different biological replicates. The ratio of globally normalized signal intensity between Cy3- and Cy5-labeled cDNA derived from D and B media, respectively, was calculated for each microarray spot, and the average of the ratio from six replicated spots was used for quantification analysis.
Quantification of Internal Tetrapyrrole Intermediates
BY-2 cells grown for 1 d after inoculation into appropriate culture media were harvested by centrifugation and weighed. Approximately 1 g of cell pellets was ground in liquid nitrogen with a mortar and pestle, and the resulting cell powders were subjected to the following extraction of tetrapyrrole intermediates. For measurement of proto IX and Mg-proto IX, cell powders were mixed with 1 mL of chilled acetone and centrifuged after vortexing. Pigments extracted in the supernatant were separated and fractionated on a Symmetry C8 column (150 × 4.6 mm; Waters) using a HPLC system (Hitachi) according to a previous report (Zapata et al., 2000). Fluorescent signals were detected with an excitation wavelength of 400 nm and emission at 628 nm for proto IX and with an excitation wavelength of 417 nm and emission at 591 nm for Mg-proto IX, using a LS 50B fluorometer (Perkin-Elmer). The pigments were quantified by comparison with authentic standards (purchased from Frontier Science) eluted in the corresponding fractions. Extraction of internal heme from cell powders was performed according to the method of Weinstein and Beale (1983). The total content of heme was quantified enzymatically using horseradish peroxidase apo-enzyme and ECL-Plus (GE Healthcare) as described previously (Masuda and Takahashi, 2006).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AB018190, AB028026, D63396, U15933, AB041518, and AY253326.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Effect of phytohormones on the expression of starch synthesis genes in tobacco BY-2 cells.
Supplemental Figure S2. Effects of several inhibitors of plastid translation on starch accumulation in plastids.
Supplemental Figure S3. Effect of a proteasome inhibitor on amyloplast differentiation.
Supplemental Table S1. Primer pairs used in this study.
Acknowledgments
We thank Dr. M. Fujimoto, Dr. S. Arimura, Prof. N. Tsutsumi, and Prof. T. Nagata (all from University of Tokyo) for providing tobacco BY-2 cells. We also thank Dr. N. Mochizuki (Kyoto University) and Dr. Y. Kobayashi (Chiba University) for technical suggestions.
References
- Adamska I. (1995) Regulation of early light-inducible protein gene expression by blue and red light in etiolated seedlings involves nuclear and plastid factors. Plant Physiol 107: 1167–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ankele E, Kindgren P, Pesquet E, Strand A. (2007) In vivo visualization of Mg-protoporphyrin IX, a coordinator of photosynthetic gene expression in the nucleus and the chloroplast. Plant Cell 19: 1964–1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernie AR, Willmitzer L, Trethewey RN. (2002) Sucrose to starch: a transition in molecular plant physiology. Trends Plant Sci 7: 35–41 [DOI] [PubMed] [Google Scholar]
- Goldschmidt-Clermont M. (1998) Coordination of nuclear and chloroplast gene expression in plant cells. Int Rev Cytol 177: 115–180 [DOI] [PubMed] [Google Scholar]
- Gray JC, Sullivan JA, Wang JH, Jerome CA, MacLean D. (2003) Coordination of plastid and nuclear gene expression. Philos Trans R Soc Lond B Biol Sci 358: 135–144; discussion 144–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdukiewicz PT, Allison LA, Maliga P. (1997) The two RNA polymerases encoded by the nuclear and the plastid compartments transcribe distinct groups of genes in tobacco plastids. EMBO J 16: 4041–4048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanaoka M, Kanamaru K, Fujiwara M, Takahashi H, Tanaka K. (2005) Glutamyl-tRNA mediates a switch in RNA polymerase use during chloroplast biogenesis. EMBO Rep 6: 545–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanaoka M, Kanamaru K, Takahashi H, Tanaka K. (2003) Molecular genetic analysis of chloroplast gene promoters dependent on SIG2, a nucleus-encoded sigma factor for the plastid-encoded RNA polymerase, in Arabidopsis thaliana. Nucleic Acids Res 31: 7090–7098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba T. (2010) Bilateral communication between plastid and the nucleus: plastid protein import and plastid-to-nucleus retrograde signaling. Biosci Biotechnol Biochem 74: 471–476 [DOI] [PubMed] [Google Scholar]
- Ishizaki Y, Tsunoyama Y, Hatano K, Ando K, Kato K, Shinmyo A, Kobori M, Takeba G, Nakahira Y, Shiina T. (2005) A nuclear-encoded sigma factor, Arabidopsis SIG6, recognizes sigma-70 type chloroplast promoters and regulates early chloroplast development in cotyledons. Plant J 42: 133–144 [DOI] [PubMed] [Google Scholar]
- Jurgenson JE, Beale SI, Troxler RF. (1976) Biosynthesis of delta-aminolevulinic acid in the unicellular rhodophyte, Cyanidium caldarium. Biochem Biophys Res Commun 69: 149–157 [DOI] [PubMed] [Google Scholar]
- Kadota Y, Furuichi T, Sano T, Kaya H, Gunji W, Murakami Y, Muto S, Hasezawa S, Kuchitsu K. (2005) Cell-cycle-dependent regulation of oxidative stress responses and Ca2+ permeable channels NtTPC1A/B in tobacco BY-2 cells. Biochem Biophys Res Commun 336: 1259–1267 [DOI] [PubMed] [Google Scholar]
- Kakizaki T, Matsumura H, Nakayama K, Che FS, Terauchi R, Inaba T. (2009) Coordination of plastid protein import and nuclear gene expression by plastid-to-nucleus retrograde signaling. Plant Physiol 151: 1339–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanamaru K, Nagashima A, Fujiwara M, Shimada H, Shirano Y, Nakabayashi K, Shibata D, Tanaka K, Takahashi H. (2001) An Arabidopsis sigma factor (SIG2)-dependent expression of plastid-encoded tRNAs in chloroplasts. Plant Cell Physiol 42: 1034–1043 [DOI] [PubMed] [Google Scholar]
- Kleine T, Voigt C, Leister D. (2009) Plastid signalling to the nucleus: messengers still lost in the mists? Trends Genet 25: 185–192 [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Imamura S, Hanaoka M, Tanaka K. (2011) A tetrapyrrole-regulated ubiquitin ligase controls algal nuclear DNA replication. Nat Cell Biol 13: 483–487 [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Kanesaki Y, Tanaka A, Kuroiwa H, Kuroiwa T, Tanaka K. (2009) Tetrapyrrole signal as a cell-cycle coordinator from organelle to nuclear DNA replication in plant cells. Proc Natl Acad Sci USA 106: 803–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koussevitzky S, Nott A, Mockler TC, Hong F, Sachetto-Martins G, Surpin M, Lim J, Mittler R, Chory J. (2007) Signals from chloroplasts converge to regulate nuclear gene expression. Science 316: 715–719 [PubMed] [Google Scholar]
- Lara MV, Drincovich MF, Müller GL, Maurino VG, Andreo CS. (2005) NADP-malic enzyme and Hsp70: co-purification of both proteins and modification of NADP-malic enzyme properties by association with Hsp70. Plant Cell Physiol 46: 997–1006 [DOI] [PubMed] [Google Scholar]
- Larkin RM, Alonso JM, Ecker JR, Chory J. (2003) GUN4, a regulator of chlorophyll synthesis and intracellular signaling. Science 299: 902–906 [DOI] [PubMed] [Google Scholar]
- Leister D. (2003) Chloroplast research in the genomic age. Trends Genet 19: 47–56 [DOI] [PubMed] [Google Scholar]
- Lopez-Juez E, Pyke KA. (2005) Plastids unleashed: their development and their integration in plant development. Int J Dev Biol 49: 557–577 [DOI] [PubMed] [Google Scholar]
- Loschelder H, Schweer J, Link B, Link G. (2006) Dual temporal role of plastid sigma factor 6 in Arabidopsis development. Plant Physiol 142: 642–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysenko EA. (2007) Plant sigma factors and their role in plastid transcription. Plant Cell Rep 26: 845–859 [DOI] [PubMed] [Google Scholar]
- Maclean D, Jerome CA, Brown AP, Gray JC. (2008) Co-regulation of nuclear genes encoding plastid ribosomal proteins by light and plastid signals during seedling development in tobacco and Arabidopsis. Plant Mol Biol 66: 475–490 [DOI] [PubMed] [Google Scholar]
- Martin C, Smith AM. (1995) Starch biosynthesis. Plant Cell 7: 971–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda T, Takahashi S. (2006) Chemiluminescent-based method for heme determination by reconstitution with horseradish peroxidase apo-enzyme. Anal Biochem 355: 307–309 [DOI] [PubMed] [Google Scholar]
- Miyazawa Y, Sakai A, Miyagishima S, Takano H, Kawano S, Kuroiwa T. (1999) Auxin and cytokinin have opposite effects on amyloplast development and the expression of starch synthesis genes in cultured Bright Yellow-2 tobacco cells. Plant Physiol 121: 461–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki N, Brusslan JA, Larkin R, Nagatani A, Chory J. (2001) Arabidopsis genomes uncoupled 5 (GUN5) mutant reveals the involvement of Mg-chelatase H subunit in plastid-to-nucleus signal transduction. Proc Natl Acad Sci USA 98: 2053–2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki N, Tanaka R, Tanaka A, Masuda T, Nagatani A. (2008) The steady-state level of Mg-protoporphyrin IX is not a determinant of plastid-to-nucleus signaling in Arabidopsis. Proc Natl Acad Sci USA 105: 15184–15189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita MT, Tasaka M. (2004) Gravity sensing and signaling. Curr Opin Plant Biol 7: 712–718 [DOI] [PubMed] [Google Scholar]
- Moulin M, McCormac AC, Terry MJ, Smith AG. (2008) Tetrapyrrole profiling in Arabidopsis seedlings reveals that retrograde plastid nuclear signaling is not due to Mg-protoporphyrin IX accumulation. Proc Natl Acad Sci USA 105: 15178–15183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray MG, Thompson WF. (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8: 4321–4325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata T, Nemoto Y, Hasezawa S. (1992) Tobacco BY-2 cell line as the “HeLa” cell in the cell biology of higher plants. Int Rev Cytol 132: 1–30 [Google Scholar]
- Nakamura T, Furuhashi Y, Hasegawa K, Hashimoto H, Watanabe K, Obokata J, Sugita M, Sugiura M. (2003) Array-based analysis on tobacco plastid transcripts: preparation of a genomic microarray containing all genes and all intergenic regions. Plant Cell Physiol 44: 861–867 [DOI] [PubMed] [Google Scholar]
- Ngernprasirtsiri J, Kobayashi H, Akazawa T. (1990) DNA methylation is a determinative element of photosynthesis gene expression in amyloplasts from liquid-cultured cells of sycamore (Acer pseudoplatanus L.). Cell Struct Funct 15: 285–293 [DOI] [PubMed] [Google Scholar]
- Oelmuller R, Levitan I, Bergfeld R, Rajasekhar VK, Mohr H. (1986) Expression of nuclear genes as affected by treatments acting on the plastids. Planta 168: 482–492 [DOI] [PubMed] [Google Scholar]
- Peter E, Rothbart M, Oelze ML, Shalygo N, Dietz KJ, Grimm B. (2010) Mg protoporphyrin monomethylester cyclase deficiency and effects on tetrapyrrole metabolism in different light conditions. Plant Cell Physiol 51: 1229–1241 [DOI] [PubMed] [Google Scholar]
- Pfannschmidt T. (2010) Plastidial retrograde signalling: a true “plastid factor” or just metabolite signatures? Trends Plant Sci 15: 427–435 [DOI] [PubMed] [Google Scholar]
- Rapp JC, Mullet JE. (1991) Chloroplast transcription is required to express the nuclear genes rbcS and cab: plastid DNA copy number is regulated independently. Plant Mol Biol 17: 813–823 [DOI] [PubMed] [Google Scholar]
- Ruckle ME, DeMarco SM, Larkin RM. (2007) Plastid signals remodel light signaling networks and are essential for efficient chloroplast biogenesis in Arabidopsis. Plant Cell 19: 3944–3960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai A, Kawano S, Kuroiwa T. (1992) Conversion of proplastids to amyloplasts in tobacco cultured cells is accompanied by changes in the transcriptional activities of plastid genes. Plant Physiol 100: 1062–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwechheimer C, Schwager K. (2004) Regulated proteolysis and plant development. Plant Cell Rep 23: 353–364 [DOI] [PubMed] [Google Scholar]
- Shalygo N, Czarnecki O, Peter E, Grimm B. (2009) Expression of chlorophyll synthase is also involved in feedback-control of chlorophyll biosynthesis. Plant Mol Biol 71: 425–436 [DOI] [PubMed] [Google Scholar]
- Shiina T, Tsunoyama Y, Nakahira Y, Khan MS. (2005) Plastid RNA polymerases, promoters, and transcription regulators in higher plants. Int Rev Cytol 244: 1–68 [DOI] [PubMed] [Google Scholar]
- Shirano Y, Shimada H, Kanamaru K, Fujiwara M, Tanaka K, Takahashi H, Unno K, Sato S, Tabata S, Hayashi H, et al. (2000) Chloroplast development in Arabidopsis thaliana requires the nuclear-encoded transcription factor sigma B. FEBS Lett 485: 178–182 [DOI] [PubMed] [Google Scholar]
- Strand A, Asami T, Alonso J, Ecker JR, Chory J. (2003) Chloroplast to nucleus communication triggered by accumulation of Mg-protoporphyrinIX. Nature 421: 79–83 [DOI] [PubMed] [Google Scholar]
- Sugita M, Sugiura M. (1996) Regulation of gene expression in chloroplasts of higher plants. Plant Mol Biol 32: 315–326 [DOI] [PubMed] [Google Scholar]
- Sullivan JA, Gray JC. (1999) Plastid translation is required for the expression of nuclear photosynthesis genes in the dark and in roots of the pea lip1 mutant. Plant Cell 11: 901–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susek RE, Ausubel FM, Chory J. (1993) Signal transduction mutants of Arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development. Cell 74: 787–799 [DOI] [PubMed] [Google Scholar]
- Valkov VT, Scotti N, Kahlau S, Maclean D, Grillo S, Gray JC, Bock R, Cardi T. (2009) Genome-wide analysis of plastid gene expression in potato leaf chloroplasts and tuber amyloplasts: transcriptional and posttranscriptional control. Plant Physiol 150: 2030–2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Gromoff ED, Alawady A, Meinecke L, Grimm B, Beck CF. (2008) Heme, a plastid-derived regulator of nuclear gene expression in Chlamydomonas. Plant Cell 20: 552–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein JD, Beale SI. (1983) Separate physiological roles and subcellular compartments for two tetrapyrrole biosynthetic pathways in Euglena gracilis. J Biol Chem 258: 6799–6807 [PubMed] [Google Scholar]
- Woodson JD, Perez-Ruiz JM, Chory J. (2011) Heme synthesis by plastid ferrochelatase I regulates nuclear gene expression in plants. Curr Biol 21: 897–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapata M, Rodriguez F, Garrido JL. (2000) Separation of chlorophylls and carotenoids from marine phytoplankton: a new HPLC method using a reversed phase C8 column and pyridine-containing mobile phases. Mar Ecol Prog Ser 195: 29–45 [Google Scholar]
- Zoschke R, Nakamura M, Liere K, Sugiura M, Börner T, Schmitz-Linneweber C. (2010) An organellar maturase associates with multiple group II introns. Proc Natl Acad Sci USA 107: 3245–3250 [DOI] [PMC free article] [PubMed] [Google Scholar]