Abstract
Ferredoxin-NADP+-oxidoreductase (FNR) mediates electron transfer between ferredoxin (Fd) and NADP+; therefore, it is a key enzyme that provides the reducing power used in the Calvin cycle. Other than FNR, nitrite reductase, sulfite reductase, glutamate synthase, and Fd-thioredoxin reductase also accept electrons from Fd, an electron carrier protein in the stroma. Therefore, the regulation of electron partitioning in the chloroplast is important for photosynthesis and other metabolic pathways. The regulatory mechanism of electron partitioning, however, remains to be elucidated. We found, by taking advantage of a gain-of-function approach, that expression of two rice (Oryza sativa) full-length cDNAs of leaf-type FNRs (OsLFNR1 and OsLFNR2) led to altered chlorophyll fluorescence and growth in Arabidopsis (Arabidopsis thaliana) and rice. We revealed that overexpression of the OsLFNR1 and OsLFNR2 full-length cDNAs resulted in distinct phenotypes despite the high sequence similarity between them. Expression of OsLFNR1 affected the nitrogen assimilation pathway without inhibition of photosynthesis under normal conditions. On the other hand, OsLFNR2 expression led to the impairment of photosynthetic linear electron transport as well as Fd-dependent cyclic electron flow around photosystem I. The endogenous protein level of OsLFNR was found to be suppressed in both OsLFNR1- and OsLFNR2-overexpressing rice plants, leading to changes in the stoichiometry of the two LFNR isoforms within the thylakoid and soluble fractions. Thus, we propose that the stoichiometry of two LFNR isoforms plays an important role in electron partitioning between carbon fixation and nitrogen assimilation.
During photosynthesis, light energy is converted to chemical energy (in the form of ATP) through electron transport, and NADPH, a reducing agent, is also produced. In the final step of the linear electron transport of photosynthesis, ferredoxin-NADP+-oxidoreductase (FNR) catalyzes the reduction of NADP+ by ferredoxin (Fd) and provides the reducing power for CO2 fixation in the Calvin cycle (Arakaki et al., 1997; Carrillo and Ceccarelli, 2003). FNR is supposed to be one of the limiting factors in photosynthetic electron transport. Through the analysis of FNR antisense tobacco (Nicotiana tabacum) plants, the amount of FNR has been shown to correlate with photosynthetic activity (Hajirezaei et al., 2002).
FNR exists in a leaf form (LFNR) and a root form (Hanke et al., 2004). There are two LFNR proteins (LFNR1 and LFNR2) in the Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa) genomes. Both LFNR isoforms have been found in the thylakoid membranes and in the stroma in Arabidopsis (Hanke et al., 2005). It is suggested that Arabidopsis LFNR1 (AtLFNR1) is required for membrane attachment of AtLFNR2, and the formation of an AtLFNR1-AtLFNR2 heterodimer mediates this attachment (Lintala et al., 2007). Arabidopsis lfnr1 showed comparable photosynthetic phenotypes to the LFNR2 Arabidopsis RNA interference plants under normal growth conditions (Lintala et al., 2009), suggesting that LFNR1 and LFNR2 have similar functions in chloroplasts. The two LFNR isoforms, however, have some distinct properties. It is reported that LFNR1 and LFNR2 vary in their pI and affinity for Fd (Hanke et al., 2005). Moreover, only the Atlfnr2 mutant shows resistance to oxidative stress (Lintala et al., 2009). Considering these observations, there remains the possibility that the two LFNR isoforms have distinct functions depending on the growth conditions.
Fd, an electron donor to FNR, also donates an electron to several enzymes: nitrite reductase, Glu synthase, sulfite reductase, and Fd-thioredoxin reductase (Schurmann and Buchanan, 2008). Among them, nitrite reductase and Glu synthase are involved in nitrogen assimilation. Nitrogen is assimilated from nitrate and ammonium, and the nitrate is reduced to nitrite in the cytosol by nitrate reductase (NR). The nitrite is then imported into the chloroplast and reduced by Fd-dependent nitrite reductase to ammonium, which is in turn fixed by Fd-dependent Glu synthase. Thus, the reducing power required in nitrogen assimilation is supplied as a form of reduced Fd. There are several observations that LFNR is also involved in the nitrogen assimilation pathway. The Arabidopsis lfnr1 plants showed greater biomass under low-nitrate conditions than the wild type (Hanke et al., 2008a). In loss-of-function Arabidopsis mutants of LFNR1 and LFNR2, NR genes were up-regulated and nitrate was accumulated compared with the wild type (Lintala et al., 2009). In addition to mutant analysis, LFNR2 mRNA was reported to be increased under high-nitrate conditions, while LFNR1 transcript levels were low (Hanke et al., 2005). FNR activity, therefore, might represent a critical point in electron channeling to determine the form in which the reductant is made available to various chloroplast enzymes, especially those involved in nitrogen assimilation. How two LFNR isoforms are involved in electron partitioning between linear flows to CO2 or nitrogen assimilation in the chloroplast, however, remains poorly understood.
An overexpression approach offers a useful tool for analyzing gene families, especially those with redundant function. Recently, we applied the FOX (for full-length cDNA overexpressor) gene-hunting approach to identify useful genes (Ichikawa et al., 2006). The FOX hunting system is a gain-of-function system that randomly overexpresses a full-length (fl)-cDNA library under the control of the cauliflower mosaic virus (CaMV) 35S promoter. As this system only uses fl-cDNAs to analyze the functions of genes, it can be used with heterologous hosts (i.e. the host plant and the plant supplying the fl-cDNAs can be different species). As an initial model of its use as a heterologous system, more than 33,000 independent Arabidopsis transgenic lines that expressed rice fl-cDNAs (rice FOX Arabidopsis lines) were generated in order to conduct high-throughput screening of rice genes (Kondou et al., 2009). Several useful rice genes have been identified using rice FOX Arabidopsis lines (Yokotani et al., 2008, 2009a, 2009b, 2011; Albinsky et al., 2010; Dubouzet et al., 2011).
One of the most important crops is rice, and it has been utilized as a monocot model plant. However, rice is still not suited for large-scale screening. Many mutants involved in photosynthesis have been isolated by monitoring the chlorophyll fluorescence in cyanobacteria, green algae, and Arabidopsis from loss-of-function populations (Meurer et al., 1996; Niyogi et al., 1997, 1998; Shikanai et al., 1999; Varotto et al., 2000; Ozaki et al., 2007; Higuchi et al., 2009). An approach by chlorophyll fluorescence has not been utilized for the identification of rice genes. Additionally, chlorophyll fluorescence-based mutant screening has not, to our knowledge, been applied using a gain-of-function approach. In this study, we have screened rice FOX Arabidopsis lines using chlorophyll fluorescence to isolate photosynthesis-related mutants. Here, we have isolated two rice FOX Arabidopsis mutants (K15006 and K17234) with altered chlorophyll fluorescence kinetics. The phenotypes of K15006 and K17234 were apparently caused by expression of the rice LFNR1 and LFNR2 genes, respectively. Rice plants overexpressing OsLFNR1 did not show any defects in photosynthesis under normal growth conditions but exhibited growth defects that could be suppressed under nutrient-rich conditions. On the other hand, OsLFNR2 rice overexpression plants were impaired in photosynthetic linear electron transport as well as in Fd-dependent cyclic electron flow around PSI. Thus, we demonstrate that changes in the stoichiometry of the two LFNR isoforms in chloroplasts lead to a disturbance of electron flow either to nitrogen assimilation or to NADP+.
RESULTS
Screening of Rice FOX Arabidopsis Mutants Using Chlorophyll Fluorescence Imaging
We generated a rice fl-cDNA expression library carrying approximately 13,000 rice fl-cDNAs and generated more than 33,000 independent Arabidopsis transgenic plants expressing rice fl-cDNAs under the control of the CaMV 35S promoter (rice FOX Arabidopsis lines; Kondou et al., 2009). About 10,000 rice FOX Arabidopsis lines were screened by monitoring the changes in chlorophyll fluorescence intensities. A total of 10,000 lines correspond to approximately 7,000 fl-cDNAs according to the equation of Clarke and Carbon (1976). Images of chlorophyll fluorescence were captured during illumination at 350 μmol m−2 s−1 for 120 s with a 0.8-s saturating light pulse using a fluorescence imaging system. Since rice FOX Arabidopsis lines are gain-of-function-type transgenic plants, we isolated mutants that showed a dominant phenotype in the T2 generation. Twenty-six candidates showed altered chlorophyll fluorescence kinetics, suggesting that expression of rice fl-cDNAs could affect the photosynthetic metabolism in Arabidopsis plants. To confirm that the observed phenotypes were caused by the expression of the introduced fl-cDNAs, we recovered 17 of them from the rice FOX Arabidopsis lines and generated Arabidopsis plants expressing the respective rice fl-cDNAs. The observed phenotypes were reproduced in 11 transgenic plants. The rice fl-cDNAs recovered are listed in Table I.
Table I. Candidate mutants and their corresponding rice fl-cDNAs in rice FOX Arabidopsis lines.
| Line Name | DNA Data Bank of Japan Accession No. | cDNA Annotation |
| K02013 | AK067378 | Lectin protein kinase domain-containing protein |
| K02143 | AK070779 | Chloroplast 50S ribosomal protein |
| K04425 | AK098990 | Chloroplast 50S ribosomal protein L1 |
| K12734 | AK070198 | U2 snRNP auxiliary factor, small subunit 35b |
| K15006 | AK065309 | Fd NADP+ oxidoreductase 1 |
| K17234 | AK065063 | Fd NADP+ oxidoreductase 2 |
| K19435 | AK101537 | Putative Rubisco chaperonin 60β precursor |
| K20333 | AK067715 | 33-kD oxygen-evolving protein of PSII (PsbO) |
| K20443 | AK072692 | GDA1/CD39 family protein |
| K23449 | AK068585 | Hypothetical protein |
| K28020 | AK065868 | Similar to mitochondrial transcription termination factor |
Chlorophyll fluorescence was also measured by pulse amplitude modulation to analyze the photosynthetic characteristics of the isolated candidates (Supplemental Table S2). In the six retransformed Arabidopsis plants, the chlorophyll fluorescence was indistinguishable from that of the parent line. The phenotypes of these rice FOX Arabidopsis lines might be caused by the insertion of multiple fl-cDNAs and/or by a secondary mutation caused by the T-DNA insertion. We carried out reverse transcription (RT)-PCR analysis using RNA prepared from the retransformed plants to confirm expression of the rice transgenes in Arabidopsis (Supplemental Fig. S1). Expression of the rice fl-cDNAs could be detected in all the retransformed Arabidopsis plants, whereas they could not be found in vector control plants.
Heterologous Expression of Rice LFNR Leads to Altered Chlorophyll Fluorescence Kinetics in Arabidopsis
We previously reported that K17234, one of the isolated rice FOX Arabidopsis lines in this study, showed a pale green phenotype and that this phenotype was caused by the expression of OsLFNR2 (AK065063; Kondou et al., 2009). We also found that one of the rice FOX Arabidopsis lines, K15006, in which the OsLFNR1 fl-cDNA (AK065309) was introduced, showed altered chlorophyll fluorescence. Figure 1A shows the seedling plants and false-color images of nonphotochemical quenching (NPQ) in K15006 and K17234. The false colors show the minimum NPQ in blue and the maximum in red. The NPQs of K15006 and K17234 were lower than in the wild type. K15006 and K17234 exhibited pale green leaves at the seedling stage. As shown in Figure 1B, K15006 and K17234 also exhibited growth defects and had paler green leaves in adult plants compared with the wild type. Chlorophyll content (μg chlorophyll mg−1 fresh weight) was lower in K15006 (1.32 ± 0.32) and K17234 (0.97 ± 0.17) than in the wild type (1.73 ± 0.15). Concomitantly, a higher chlorophyll a/b ratio was observed in K15006 (3.51 ± 0.16) and K17234 (3.39 ± 0.17) than in the wild type (3.13 ± 0.06).
Figure 1.
Isolation of rice FOX Arabidopsis lines with altered chlorophyll fluorescence. A, The left panel shows Arabidopsis wild-type (WT) and T2 seedlings of K15006 and K17234. The right panel shows a pseudocolor image of the NPQ. B, Plants of the wild type and the T2 generation of K15006 and K17234 in the adult stage.
Expression of Rice fl-cDNAs of OsLFNR1 and OsLFNR2 Resulted in the Down-Regulation of Endogenous AtLFNR Genes in Arabidopsis
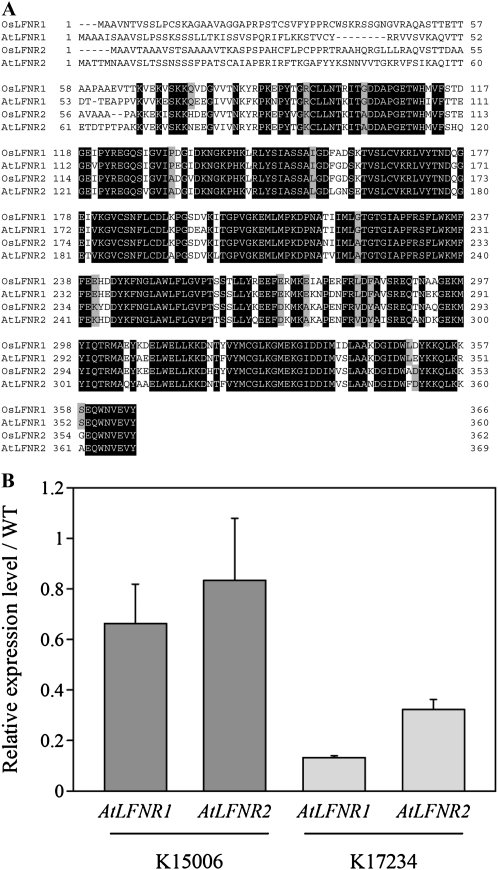
An alignment of LFNR1 and LFNR2 in rice and Arabidopsis is shown in Figure 2A. Both in rice and Arabidopsis, LFNR1 and LFNR2 show high sequence identity at the amino acid level (80% identity between AtLFNR1 and AtLFNR2, 80% identity between OsLFNR1 and OsLFNR2) except for the N-terminal amino acids, corresponding to the chloroplast transit peptide signal sequences. The identity between rice and Arabidopsis is also high in LFNR1 and LFNR2 (80% identity between AtLFNR1 and OsLFNR1, 82% identity between AtLFNR2 and OsLFNR2). Several isoform-specific amino acids were seen as shown in the gray boxes.
Figure 2.
Amino acid sequence alignment of LFNR proteins in rice and Arabidopsis. A, The amino acid sequences of two LFNR proteins in rice and Arabidopsis were compared with each other. AK065309 and AK065063 encode LFNR1 (OsLFNR1) and LFNR2 (OsLFNR2) in rice, respectively. AT5G66190 and AT1G20020 encode LFNR1 (AtLFNR1) and LFNR2 (AtLFNR2) in Arabidopsis, respectively. Black shading indicates conserved amino acids between all proteins, and gray shading indicates conserved amino acids specific to each LFNR isoform. B, Relative expression levels of endogenous FNR genes (AtLFNR1 and AtLFNR2) in K15006 expressing OsLFNR1 and in K172384 expressing OsLFNR2. Expression levels of the LFNR genes were normalized with ACT2 gene expression. Expression levels of each mutant are shown as relative values of the wild type (WT). Real-time PCR experiments were carried out at least four times.
In K15006 and K17234, endogenous expression of AtLFNR1 and AtLFNR2 might be affected by heterologous expression of the OsLFNR genes. As can be seen in Figure 2B, the levels of AtLFNR1 and AtLFNR2 in K15006 decreased to 70% and 80% of those of the wild type, respectively. In K17234, the suppression of AtLFNR1 and AtLFNR2 expression was more severe (i.e. only 10% and 30% of the wild type, respectively). These results indicate that heterologous expression of OsLFNR fl-cDNAs results in a decreased level of AtLFNR mRNA in Arabidopsis.
Overexpression of OsLFNR1 and OsLFNR2 Genes in Rice Affected the Endogenous Transcript and Protein Levels
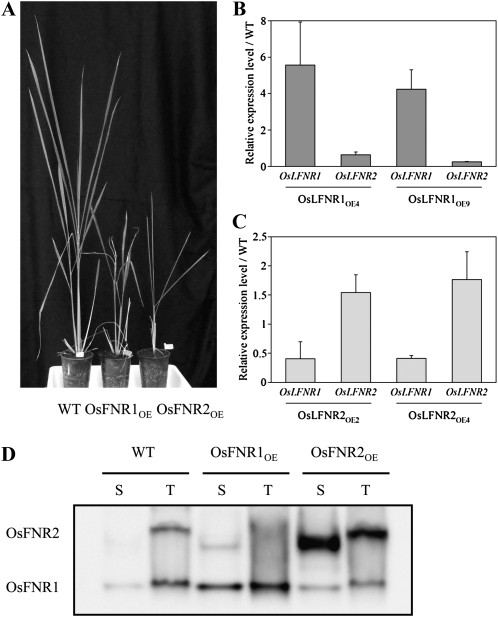
We cloned the fl-cDNAs of OsLFNR1 and OsLFNR2 into pRiceFOX (Nakamura et al., 2007), a monocot expression vector, and they were expressed under the control of the maize (Zea mays) ubiquitin promoter to generate rice plants overexpressing fl-cDNAs of OsLFNR1 and OsLFNR2. We observed 13 T0 transgenic plants of OsLFNR1 (OsLFNR1OE) and found that three of them died and one plant was sterile, all of which showed a severe pale green phenotype. Two of 13 T0 rice overexpression plants of OsLFNR2 (OsLFNR2OE) were sterile, and six plants showed a pale green phenotype. Both rice T1 plants of OsLFNR1OE and OsLFNR2OE showed growth defects (Fig. 3A) when they were grown in soil. Chlorophyll content (μg chlorophyll mg−1 fresh weight), when compared with that of the wild type (4.54 ± 0.62), was low in OsLFNR2OE (2.25 ± 0.47) but similar in OsLFNR1OE (4.34 ± 0.39). The ratio of chlorophyll a/b was slightly higher in OsLFNR2OE (3.83 ± 0.18) than in the wild type (3.52 ± 0.12) or in OsLFNR1OE (3.61 ± 0.10).
Figure 3.
Characterization of rice overexpression plants of OsLFNR1 and OsLFNR2. A, Growth of wild-type (WT) and T1 plants overexpressing OsLFNR1 and OsLFNR2. B, Relative expression levels of LFNR genes in rice plants overexpressing OsLFNR1. Two independent overexpression plants were analyzed by quantitative RT-PCR. C, Relative expression levels of LFNR genes in rice plants overexpressing OsLFNR2. Two independent overexpression plants were analyzed by quantitative RT-PCR. Expression levels of the LFNR genes were normalized with ubiquitin gene expression. Expression levels of each mutant line relative to those of the wild type are shown. Real-time PCR experiments were carried out at least four times. D, Protein levels of OsLFNR1 and OsLFNR2 in soluble (S) and thylakoid (T) fractions. A total of 10 μg of protein was loaded in each lane.
Expression levels of OsLFNR1 and OsLFNR2 in two independent overexpression rice plants were analyzed by quantitative real-time RT-PCR (Fig. 3, B and C). Since fl-cDNAs were used for overexpression, we could not distinguish between the expression of the transgenes and the endogenous genes. Expression of OsLFNR1 was increased 4- to 5-fold in OsLFNR1OE, while an approximate 2-fold increase in OsLFNR2 was found in OsLFNR2OE. We also checked the expression levels of other isoforms in both types of rice overexpression plants. Expression of the OsLFNR2 gene decreased to less than 50% of the wild type in OsLFNR1OE. The OsLFNR1 gene was markedly down-regulated to about 40% of that of the wild type in OsLFNR2OE. These results suggest that overexpression of OsLFNR genes causes the down-regulation of another isoform of the endogenous OsLFNR gene in rice as observed in the rice FOX Arabidopsis lines (K15006 and K17234).
Proteins levels of OsLFNR1 and OsLFNR2 were determined by western blotting using the FNR antibody (Fig. 3D). OsLFNR1 and OsLFNR2 can be distinguished by native PAGE (Hanke et al., 2008a). Since the predicted pI of OsLFNR1 (pI = 5.65) was more acidic than that of OsLFNR2 (pI = 7.69), as in Arabidopsis and wheat (Triticum aestivum; Hanke et al., 2005; Gummadova et al., 2007), migration of OsLFNR1 was faster than OsLFNR2 at around pH 8.0. Subchloroplast distribution of the OsLFNR isoforms was also analyzed by separating the thylakoid membrane fractions and the soluble fractions. In the wild type, the protein levels of OsLFNR1 and OsLFNR2 were comparable in the thylakoid fractions. OsLFNR1 was the major isoform in the soluble fraction and OsLFNR2 was nearly undetectable. OsLFNR1OE contained a higher level of OsLFNR1 both in the thylakoid and soluble fractions compared with the wild type. On the other hand, the level of OsLFNR2 was decreased especially in the thylakoid fraction. Although a slight increase of OsLFNR2 was seen in the stromal fraction, the total amount of OsLFNR2 was decreased, which is consistent with the reduced transcript level of OsLFNR2 (Fig. 3B). In OsLFNR2OE, OsLFNR2 was the major isoform both in the thylakoid and the soluble fractions. OsLFNR1 was decreased in the thylakoid fraction, although it was slightly increased in the stromal fraction compared with the wild type. Total OsLFNR1 was lower, which is also compatible with a decrease in OsLFNR1 mRNA. The degree of increase of OsLFNR2 seemed to be greater than that of the transcript level and less in OsLFNR1. The difference between transcript and protein abundance might be a consequence of posttranscriptional regulation. Apparently, OsLFNR1OE accumulates more OsLFNR1 and less OsLFNR2, while OsLFNR2OE accumulates more OsLFNR2 and less OsLFNR1 compared with the wild type.
Gene Expression Profiles of OsLFNR1- and OsLFNR2-Overexpressing Rice Plants
The gene expression profiles of OsLFNR1OE and OsLFNR2OE were investigated by microarray analysis (Supplemental Table S3); the results are listed in Table II. Many genes encoding nucleus-encoded PSI subunits were down-regulated, and the expression of genes encoding PSII subunits also decreased both in OsLFNR1OE and OsLFNR2OE. Genes other than those encoding the subunits of the photosystems were also down-regulated in both types of overexpression plants, such as genes encoding light-harvesting proteins, chlorophyll biosynthesis-related genes, the Rubisco small subunit, and thylakoid ascorbate peroxidase. Some of these observations are opposite to the results obtained by Lintala et al. (2009), who found that the light-harvesting genes and Rubisco small subunit gene are up-regulated in Arabidopsis lfnr1 and lfnr2 mutants.
Table II. Expression levels of photosynthesis-related genes in OsLFNR1OE and OsLFNR2OE.
Microarray analysis was performed from total RNA isolated from leaf blades of wild-type and rice overexpression plants. Results are shown as averages from two independent experiments.
| Gene | Gene Identifier | OsLFNR1OE/Wild Type | OsLFNR2OE/Wild Type |
| Genes encoding PSI subunits | |||
| PsaD | Os08g0560900 | 0.3413 | 0.3031 |
| PsaE | Os07g0435300 | 0.3718 | 0.2141 |
| PsaF | Os03g0778100 | 0.2528 | 0.2284 |
| PsaG | Os09g0481200 | 0.4093 | 0.3678 |
| PsaH | Os05g0560000 | 0.3287 | 0.1116 |
| PsaK | Os07g0148900 | 0.2799 | 0.2754 |
| PsaL | Os12g0420400 | 0.4006 | 0.2102 |
| PsaN | Os05g0242400 | 0.4884 | 0.5232 |
| Genes encoding PSII subunits | |||
| PsbO | Os01g0501800 | 0.3637 | 0.2387 |
| PsbP | Os07g0141400 | 0.3232 | 0.2273 |
| PsbR | Os07g0147500 | 5.884 | 0.2961 |
| PsbS | Os04g0690800 | 0.7054 | 7.665 |
| PsbX | Os03g0343900 | 0.3267 | 0.1496 |
| PsbY | Os08g0119800 | 0.4404 | 0.2650 |
| PsbW | Os01g0773700 | 0.3717 | 0.3506 |
| Psb28 | Os01g0938100 | 0.4246 | 2.358 |
| Genes encoding light-harvesting proteins | |||
| CP24 | Os04g0457000 | 0.1349 | 0.05311 |
| CP29 | Os07g0558400 | 0.2828 | 0.1130 |
| ASCAB9-A | Os11g0242800 | 0.3066 | 0.2285 |
| LHCII type I CAB-2 | Os01g0600900 | 0.1133 | 0.03197 |
| LHCII type I CAB | Os09g0346500 | 0.4289 | 0.1685 |
| PSI type II chlorophyll a/b-binding protein | Os07g0577600 | 0.3058 | 0.2764 |
| PSI type III chlorophyll a/b-binding protein | Os02g0197600 | 0.5065 | 0.01973 |
| Type I chlorophyll a/b-binding protein | Os01g0720500 | 0.01268 | 0.01973 |
| Type III chlorophyll a/b-binding protein | Os07g0562700 | 0.3778 | 0.1854 |
| Photosynthesis-related genes | |||
| Magnesium-chelatase subunit chlI | Os03g0563300 | 0.1668 | 0.1362 |
| Protochlorophyllide reductase | Os04g0678700 | 0.1161 | 0.01238 |
| Magnesium-chelatase subunit H family protein | Os07g0656500 | 0.2705 | 0.2815 |
| Rubisco small subunit | Os12g0274700 | 0.1114 | 0.2942 |
| Thylakoid-bound ascorbate peroxidase | Os02g0553200 | 0.2028 | 0.216 |
| Early light-inducible protein | Os07g0178700 | 1.847 | 47.68 |
| Early light-inducible protein | Os07g0178800 | 1.271 | 8.231 |
| Rubisco activase | Os11g0707000 | 1.036 | 3.493 |
| Nitrate reductase (NR) | Os08g0468100 | 0.5821 | 3.663 |
| Nitrate reductase (NR) | Os08g0468700 | 0.7400 | 3.106 |
| Ferredoxin I (Fd1) | Os08g0104600 | 0.2843 | 1.216 |
Several genes showed specific expression patterns only in one type of transgenic plant. The gene encoding PsbR, which is one of the PSII extrinsic subunits (Allahverdiyeva et al., 2007), was up-regulated only in OsLFNR1OE. The two genes encoding Psb28, which is reported to function in the assembly of PSII (Sakurai et al., 2007; Dobakova et al., 2009), and PsbS, which is involved in energy dissipation (Li et al., 2000; Zulfugarov et al., 2007), were up-regulated only in OsLFNR2OE. Early light-inducible proteins and Rubisco activase were up-regulated only in OsLFNR2OE.
OsLFNR1-Overexpressing Arabidopsis and Rice Plants Did Not Alter Photosynthetic Activity under Normal Conditions
Table III shows the steady-state chlorophyll fluorescence parameters of Arabidopsis (K15006) and rice (OsLFNR1OE) expressing OsLFNR1 fl-cDNA. K15006 did not show any obvious phenotype in relation to these parameters. This indicates that linear electron transport was not impaired. Similar to K15006, the chlorophyll fluorescence parameters of OsLFNR1OE were also indistinguishable from those of the wild type. However, when its leaves were exposed to higher photon flux densities (1,000 μmol m−2 s−1), photosynthetic electron transport (ΦII), NPQ, and photochemical quenching (qP) showed lower values than those in the wild type, indicating that photosynthetic electron transport was impaired under high-light conditions. The P700 oxidation ratio did not change in OsLFNR1OE, suggesting that Fd-dependent PSI cyclic electron flow was not affected. The defect in NADPH dehydrogenase (NDH)-dependent cyclic electron flow around PSI was reported to result in the absence of a transient increase in chlorophyll fluorescence at the light-to-dark transition (Shikanai et al., 1998). This transient increase was detected in OsLFNR1OE (data not shown), indicating that NDH-mediated PSI cyclic electron flow was intact. These results suggest that an alternative pathway other than cyclic electron flow around PSI might be modified in OsLFNR1OE.
Table III. Photosynthetic characteristics of rice FOX Arabidopsis plants (K15006) and rice OsLFNR1 overexpression plants.
Chlorophyll fluorescence parameters were measured at the indicated intensities of illumination. Rosette leaves and leaf blades were used for measurement. Data are expressed as means of at least three individual leaves. Prior to measurement, leaves were dark adapted for at least 30 min. n.d., Not determined.
| Plant | Fv/Fm | qP | NPQ | Fv′/Fm′ | ΦII | P700 Oxidation Ratio |
| Rice FOX Arabidopsis line (K15006) | ||||||
| Low light (100 μmol m−2 s−1) | ||||||
| Wild type | 0.827 ± 0.004 | 0.906 ± 0.020 | 0.120 ± 0.035 | 0.806 ± 0.005 | 0.730 ± 0.015 | |
| K15006 | 0.819 ± 0.002 | 0.925 ± 0.004 | 0.137 ± 0.026 | 0.795 ± 0.007 | 0.735 ± 0.003 | |
| High light (500 μmol m−2 s−1) | ||||||
| Wild type | 0.452 ± 0.078 | 1.476 ± 0.077 | 0.574 ± 0.016 | 0.259 ± 0.043 | ||
| K15006 | 0.449 ± 0.002 | 1.434 ± 0.032 | 0.567 ± 0.003 | 0.255 ± 0.001 | ||
| Rice OsLFNR1 overexpression plants | ||||||
| Low light (100 μmol m−2 s−1) | ||||||
| Wild type | 0.804 ± 0.007 | 0.906 ± 0.023 | 0.140 ± 0.028 | 0.777 ± 0.008 | 0.704 ± 0.015 | 0.222 ± 0.084 |
| OsLFNR1OE | 0.803 ± 0.008 | 0.935 ± 0.022 | 0.137 ± 0.057 | 0.776 ± 0.004 | 0.726 ± 0.020 | 0.203 ± 0.033 |
| Medium light (500 μmol m−2 s−1) | ||||||
| Wild type | 0.653 ± 0.052 | 0.898 ± 0.139 | 0.640 ± 0.028 | 0.417 ± 0.018 | 0.418 ± 0.028 | |
| OsLFNR1OE | 0.632 ± 0.042 | 0.931 ± 0.308 | 0.633 ± 0.055 | 0.400 ± 0.034 | 0.364 ± 0.097 | |
| High light (1,000 μmol m−2 s−1) | ||||||
| Wild type | 0.317 ± 0.036 | 1.661 ± 0.208 | 0.507 ± 0.063 | 0.167 ± 0.068 | n.d. | |
| OsLFNR1OE | 0.160 ± 0.022 | 1.288 ± 0.064 | 0.557 ± 0.043 | 0.089 ± 0.006 | n.d. |
OsLFNR2-Expressing Arabidopsis and Rice Were Impaired in Linear Electron Transport and Fd-Dependent Cyclic Electron Flow around PSI
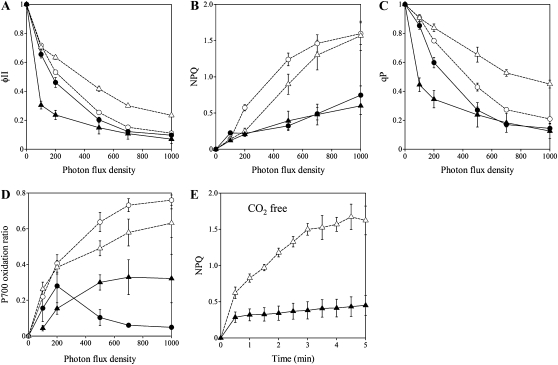
The rice FOX Arabidopsis line K17234 expressing OsLFNR2 showed low ΦII and NPQ compared with the wild type, especially in high-light illumination (Fig. 4, A and B). These results indicate that linear electron transport is impaired in K17234 and that insufficient generation of ΔpH due to the decreased electron transport leads to low NPQ. In addition to ΦII and NPQ, qP was low in K17234 (Fig. 4C), whereas the maximum quantum yield of PSII (Fv/Fm) of K17234 was almost the same as in the wild type (wild type, 0.827 ± 0.004; K17234, 0.820 ± 0.007). These results suggest that the pathway downstream of the plastoquinone pool is impaired in K17234. We also measured the P700 oxidation ratio with different photon flux densities to monitor the redox state of PSI (Fig. 4D). K17234 showed more reduced P700 with increasing illumination compared with the wild type, indicating that the acceptor side of PSI was overreduced in K17234.
Figure 4.
Photosynthetic characterization of K17234 (rice FOX Arabidopsis line) and OsLFNR2 overexpression rice plants. A to D, Photon flux density dependence of ΦII (A), NPQ (B), qP (C), and P700 oxidation ratio (D). E, Time course of NPQ induction in CO2-free air during illumination at 100 μmol m−2 s−1. White circles, Arabidopsis wild type; black circles, K17234; white triangles, rice wild type; black triangles, OsLFNR2-overexpressing rice plants. The measurements were carried out for at least three individual leaves. Prior to measurement, leaves were dark adapted for at least 30 min.
Similar to K17234, rice overexpressing fl-cDNA of OsLFNR2 (OsLFNR2OE) showed lower qP, NPQ, and ΦII and more reduced P700 than the wild type even at low photon flux densities, suggesting that the PSI acceptor side is overreduced. To analyze alternative electron flow in OsLFNR2OE, chlorophyll fluorescence was measured in CO2-free air, under which conditions the photosynthetic alternative electron pathway is enhanced. OsLFNR2OE could not induce NPQ, in contrast to the wild type (Fig. 4E). These results indicate that OsLFNR2OE is impaired in the alternative electron pathway, probably in the Fd-dependent cyclic electron flow around PSI (for details, see “Discussion”). Thus, overexpression of OsLFNR2 leads to the impairment of both the linear electron transport and the Fd-dependent cyclic electron flow around PSI in Arabidopsis and rice.
The Nitrate Reduction Pathway Was Affected in OSLFNR1 Overexpression Rice Plants
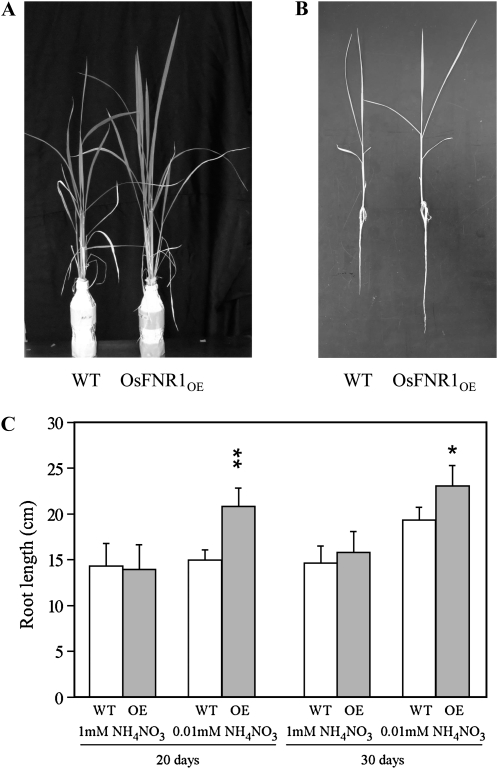
Rice OsLFNR1OE plants showed defects in growth when they were grown in soil, as shown in Figure 3A. Surprisingly, when grown in hydroponic culture in nitrogen-rich medium, OsLFNR1OE did not show any growth defects (Fig. 5A). Chlorophyll fluorescence parameters and CO2 fixation of OsLFNR1OE and the wild type also did not show much difference even under high light at 1,000 μmol m−2 s−1 (Supplemental Table S4). These results suggest that the growth and photosynthetic defects observed in soil-grown rice plants could be due to some kind of nutrient deficiency. On the other hand, the growth defects seen in OsLFNR2OE did not recover in nutrient-rich culture (data not shown).
Figure 5.
Characterization of OsLFNR1 overexpression rice plants. A, Growth of wild-type (WT) and T1 OsLFNR1 overexpression plants. Plants were grown in nutrient-rich hydroponic culture. B, Seedlings of wild-type and T1 OsLFNR1 overexpression plants grown for 20 d in nitrogen-limited (0.01 mm NH4NO3) hydroponic culture. C, Root length of wild-type and OsLFNR1 overexpression plants. Plants were grown in nitrogen-rich (1 mm NH4NO3) or nitrogen-limited (0.01 mm NH4NO3) hydroponic culture. Root lengths of at least three independent plants were measured after 20 and 30 d.
When plants are grown in nitrogen-limited conditions, root elongation is known to be enhanced. We examined the increase in root length of wild-type rice and OsLFNR1OE in nitrogen-rich (1 mm NH4NO3) and nitrogen-limited (0.01 mm NH4NO3) cultures (Fig. 5, B and C). The root length of the wild type continued to increase for 20 to 30 d only under nitrogen-limited conditions. The root length of OsLFNR1OE was almost the same as that of the wild type when grown in nitrogen-rich conditions both at 20 and 30 d. However, OsLFNR1OE showed longer roots compared with the wild type at 20 d as well as at 30 d in nitrogen-limited culture (Fig. 5, B and C). This observation supports the hypothesis that the nitrogen assimilation pathway is affected in OsLFNR1OE. Microarray analysis revealed that transcript levels of nitrate assimilation-related genes such as NR and Gln synthetase did not change in OsLFNR1OE. Moreover, nitrite reduction activity in OsLFNR1OE was almost the same as that in the wild type (data not shown). These results suggest that channeling of electrons by Fd to nitrite reductase and FNR might be the step that is impaired in OsLFNR1OE.
DISCUSSION
Gain-of-Function Screen Is a New Approach for the Identification of Genes Involved in Photosynthesis
In this study, 11 putative mutants with altered chlorophyll fluorescence were isolated from rice FOX Arabidopsis lines (Table I). Apparently, rice fl-cDNAs can affect photosynthetic activity in Arabidopsis at a significant frequency. We revealed that introduction of the rice fl-cDNAs of OsLFNR1 and OsLFNR2 into Arabidopsis and rice leads to similar phenotypes, indicating that OsLFNR1 and OsLFNR2 can serve the same function in both rice and Arabidopsis. These results also indicate that the function of heterologous genes can be analyzed using the FOX hunting system. We applied this system to high-throughput analysis of rice gene function as the first model case of heterologous expression in Arabidopsis. This study thus expands the possibility that the FOX hunting system can be applied to other plant species.
We revealed that one OsLFNR isoform was suppressed when the other was overexpressed in rice (Fig. 3). This result raises two possibilities: (1) the observed phenotype in OsLFNR overexpression plants is caused by the suppression of one LFNR isoform; or (2) the phenotype arises from the combination of overexpression and suppression. The latter interpretation is favored by our observations. Phenotypes of Arabidopsis and rice expressing fl-cDNAs of OsLFNR1 and OsLFNR2 were clearly distinct from AtLFNR1 and AtLFNR2 loss-of-function Arabidopsis mutants. The Arabidopsis Atlfnr1 mutant does not have overreduced acceptors at PSI and has a slightly more oxidized plastoquinone pool than the wild type (Hanke et al., 2008a). Arabidopsis RNA interference plants of AtLFNR2 exhibited defects in CO2 fixation and disturbed PSI cyclic electron flow under normal growth conditions (Lintala et al., 2007, 2009). In this study, OsLFNR1OE rice was affected in nitrogen assimilation and had normal photosynthetic characteristics except under conditions of very high photon flux densities (Table III). OsLFNR2OE rice was altered in linear electron transport and had reduced P700 (Fig. 4). Moreover, Arabidopsis with loss of function of Atfnr1 and Atfnr2 showed up-regulation of the NR gene as well as enhanced NR activity under standard growth conditions (Lintala et al., 2009), whereas up-regulation of the NR gene was only found in OsLFNR2OE. We cannot rule out the possibility that rice LFNR isoforms have different functions from the Arabidopsis ones. However, this is unlikely, because the same phenotypes were observed in Arabidopsis and rice expressing OsLFNR fl-cDNAs. Taking these observations into consideration, we suggest that the phenotype of OsLFNR1OE and OsLFNR2OE rice is a consequence of the combination of overexpression and suppression and not of the suppression of another LFNR isoform. Thus, we demonstrate that novel gene function can be characterized using a gain-of-function approach in this study.
Electron Partitioning to CO2 Fixation and Nitrogen Assimilation Pathways Was Impaired in OsLFNR1OE and OsLFNR2OE Rice Plants
FNR is an enzyme that catalyzes electron transfer between NADP+ and Fd. Reduced Fd is also the source of electrons for nitrite reductase and Glu synthase in the nitrogen assimilation pathway, which is indispensable for plant growth. The concerted regulation of electron partitioning between photosynthetic electron transport and nitrogen assimilation is required to optimize the efficiency of electron flow in the chloroplast.
Our observations demonstrate that overexpression of OsLFNR1 and OsLFNR2 fl-cDNAs in rice results in the modification of photosynthesis and nitrogen assimilation. OsLFNR1OE has some alteration in the nitrogen assimilation pathway (Fig. 5), with less effect on photosynthetic activity (Table III). The growth defects observed in OsLFNR1OE were recovered in nutrient-rich culture (Figs. 3 and 5), suggesting that nitrogen deficiency due to inefficient electron transfer to the nitrogen assimilation pathway might lead to the growth defects seen in the soil-grown plants. Contrary to our observations, tobacco plants expressing a pea (Pisum sativum) LFNR1 did not show any growth defects (Rodriguez et al., 2007). These tobacco plants were grown in soil and nutrient medium was supplied twice a week, whereas the OsLFNR1OE plants were grown in soil without nutrient addition. This suggests that the nutrient supply might be enough for optimal growth in tobacco LFNR1-expressing plants, as observed in the healthy growth of OsLFNR1OE rice in hydroponic culture.
On the other hand, OsLFNR2 overexpression led to the impairment of photosynthetic linear electron transport (Fig. 4). Alteration of linear electron transport and growth defects in OsLFNR2OE might be caused by insufficient electron donation to NADP+.
Thus, we conclude that electron partitioning to nitrogen assimilation or NADP+ is disturbed in OsLFNR1OE and OsLFNR2OE. Rice and Arabidopsis overexpressing OsLFNR1 and OsLFNR2 showed severe growth defects under standard growth conditions, indicating that electron partitioning either to NADP+ or nitrogen assimilation is significant for optimal growth in plants.
Involvement of LFNR Isoforms in Cyclic Electron Flow around PSI
OsLFNR2OE was modified in linear electron transport as well as in alternative electron flow (Fig. 4). PSI cyclic electron transport (Shikanai, 2007), photorespiration (Wingler et al., 2000), and the water-water cycle (Asada, 2000) are proposed as photosynthetic alternative electron pathways. A role for LFNR has also been proposed in cyclic electron flow around PSI based on its association with the cytochrome b6/f complex (Zhang and Cramer, 2004), the NDH complex (Guedeney et al., 1996), and pgr5-like photosynthetic phenotype 1 (PGRL1; DalCorso et al., 2008). In this context, it should be noted that OsLFNR2OE showed a decreasing P700 oxidation ratio with increasing photon flux densities. This phenotype is also found in Arabidopsis pgr5 and pgrl1 mutants, which are reported to have defects in Fd-dependent PSI cyclic electron flow (Munekage et al., 2002; DalCorso et al., 2008). These results suggest that Fd-mediated cyclic electron flow around PSI is impaired in OsLFNR2OE in addition to linear electron transport.
Decreased electron channeling into cyclic electron flow around PSI in OsLFNR2OE may result from insufficient NADP+ reduction. OsLFNR1OE did not show changes in the redox state of P700 (Table III) and NPQ induction in CO2-free air (data not shown). Intact PSI cyclic electron transport in OsLFNR1OE may be a consequence of a normal rate of NADP+ reduction. The redox poise of the chloroplast stroma was supposed to play an important role in the regulation of the rate of PSI cyclic electron flow (Joet et al., 2002; Breyton et al., 2006). Moreover, the activity of PGR5-dependent PSI cyclic electron transport is suggested to be regulated by the redox state of the NADPH pool (Okegawa et al., 2008). Change in the redox state of stroma may result in impairment of the electron flow to PSI cyclic electron flow in OsLFNR2OE.
Changes in the Stoichiometry of OsLFNR1 and OsLFNR2 in Thylakoid and/or Soluble Fractions in Rice Affect Electron Partitioning in Chloroplasts
LFNR1 and LFNR2 are proposed to form a heterodimer complex and regulate electron partitioning in chloroplasts (Lintala et al., 2007). We found that another isoform was suppressed in rice OsLFNR overexpression plants (Fig. 3D), suggesting that the stoichiometry of the LFNR heterodimer greatly changed in the chloroplasts. As described earlier, the phenotype of OsLFNR-overexpressing rice was caused by a combination of overexpression and suppression, suggesting that changes in the stoichiometry of the two OsLFNR isoforms led to the observed phenotypes. From our results here, we propose that the stoichiometry of the OsLFNR isoforms determines electron flow to NADP+ or nitrogen assimilation. Electrons might be transferred preferentially from Fd to the nitrogen assimilation pathway when OsLFNR2 accounts for a larger portion of the total LFNR. Up-regulation of the NR genes only in OsLFNR2OE rice (Table II) might be explained by preferential electron transfer to nitrogen assimilation. We speculate that electrons are selectively transported to NADP+ when OsLFNR1 is predominant in the chloroplasts. Transgenic tobacco plants expressing pea FNR1 have been shown to have high NADP+ photoreduction activity when the PSI electron donor was provided (Rodriguez et al., 2007). LFNR stoichiometry might change in response to environmental cues to regulate the distribution of electrons between the photosynthetic linear electron transfer pathways and nitrogen assimilation.
In OsLFNR1OE, a gene coding an isoform of Fd, Fd1, was down-regulated (Table II). Higher plants possess leaf-type Fd and root-type Fd similar to FNR (Hanke et al., 2004). Fd2 is reported to be a major isoform that is involved in linear electron transport to NADP+ in Arabidopsis (Hanke et al., 2004). Fd1 is a minor isoform, and knockdown plants of the Fd1 gene do not show severe photosynthetic defects, unlike Fd2 knockdown plants (Hanke and Hase, 2008b). It is reported that relative expression of the Fd isoforms is determined by nitrogen status (Hanke et al., 2005). Fd2 mRNA is low under high-nitrate concentrations, although Fd1 is expressed at the same levels under any nitrogen conditions. Fd1 is more abundant than Fd2 in high nitrate, similar to LFNR2. Fd1 and Fd2 are also proposed to have a role in electron portioning (Hanke and Hase, 2008b). In addition to LFNR isoforms, the relative amounts of Fd isoforms might change cooperatively in response to nitrogen status to regulate electron transfer into nitrogen assimilation.
Although LFNR is a soluble protein, it was also found in thylakoid membranes. Early in vitro experiments showed that NADP+ photoreduction activity is higher in thylakoid-bound LFNR than in the soluble one, suggesting that the pool of FNR in the thylakoid is involved in photosynthetic electron transport. There is, however, controversy about the function of the LFNR pool in the thylakoid and stroma in chloroplasts. Tic62 (Benz et al., 2009) and TROL (Juric et al., 2009) have been identified in Arabidopsis as the thylakoid membrane attachments of LFNR. In the tic62 Arabidopsis mutant, photosynthetic activity is not impaired under normal growth conditions. Moreover, LFNR is shown to accumulate in the thylakoid at night and then relocate to the stroma fraction upon illumination (Alte et al., 2010). From these observations, Benz et al. (2010) proposed that the stromal pool of LFNR is active in photosynthetic electron transport rather than in the thylakoid-bound form, and the amount of stromal LFNR might determine Fd-dependent electron partitioning.
As shown in Figure 3D, we found that the soluble LFNR pool was increased both in OsLFNR1OE and OsLFNR2OE (soluble FNR:thylakoid-bound LFNR ratio: wild type, 0.026; OsLFNR1OE, 1.15; OsLFNR2OE, 1.33). On the other hand, the total thylakoid LFNR level was not affected, although the stoichiometry of the two isoforms changed compared with the wild type. An increase in the amount of stromal LFNR might lead to the impairment of electron distribution in OsLFNR-overexpressing rice plants. Furthermore, wild-type rice plants have only a tiny amount of soluble LFNR2, as shown in Figure 3D. This is different from the pattern in Arabidopsis, where LFNR2 was at about the same level as LFNR1 in soluble fractions (Hanke et al., 2008a; Lintala et al., 2009). Rice plants prefer ammonium to nitrate as a major nitrogen source. The difference in stoichiometry between rice and Arabidopsis soluble LFNR isoforms might be explained by the difference in nitrogen responsiveness. It remains unclear even from this study which LFNR pool is active, since the stromal LFNR and thylakoid LFNR ratio changed in both overexpression plants. Further investigation will be needed to clarify this question.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Details of the method for generating the FOX lines are described by Ichikawa et al. (2006). In short, we used a mixture of approximately 13,000 rice (Oryza sativa) full-length cDNAs and made an Agrobacterium tumefaciens fl-cDNA expression library. This Agrobacterium library was used to transform wild-type Arabidopsis (Arabidopsis thaliana; ecotype Columbia) to generate rice FOX Arabidopsis lines. Independent rice fl-cDNAs were expressed under the control of the CaMV 35S promoter. Arabidopsis plants were grown at 22°C under long-day conditions (16 h of light and 8 h of dark). Arabidopsis was transformed using the Agrobacterium in planta transformation method (Clough and Bent, 1998). Rice was transformed as described previously (Toki et al., 2006).
Wild-type rice plants (cv Nipponbare) and transgenic plants were grown in a temperature-controlled growth room at 30°C.
Cloning of fl-cDNAs from Rice FOX Arabidopsis Lines
As we described previously (Ichikawa et al., 2006), fl-cDNA fragments were amplified by PCR using TaKaRa PrimeSTAR HS DNA polymerase with GC buffer (Takara Bio). The following primers were used to amplify the fl-cDNAs: GS17K (5′-GTACGTATTTTTACAACAATTACCAACAAC-3′) and GS18K (5′-GGATTCAATCTTAAGAAACTTTATTGCCAA-3′). Amplified fragments were cloned into the SfiI sites of pBIG2113SF (Ichikawa et al., 2006) and pRiceFOX to retransform into Arabidopsis and rice, respectively. The sequences were analyzed in the Rice Annotation Project database (http://rapdb.dna.affrc.go.jp/) to obtain information about the cDNAs and their annotation.
Chlorophyll Fluorescence Measurements
For screening of photosynthetic mutants, rice FOX Arabidopsis lines were grown on Murashige and Skoog plates for 8 to 10 d. The induction kinetics of chlorophyll fluorescence was measured during illumination (350 μmol m−2 s−1, 120 s) using a chlorophyll fluorescence monitoring system (FluorCam; Photon Systems Instruments). Chlorophyll fluorescence parameters were determined using a pulse-modulated fluorometer (PAM 101/103; Walz) as described by Schreiber et al. (1988). The level of minimum fluorescence (Fo) was recorded after dark adaptation for at least 30 min. The level of maximum fluorescence (Fm) was obtained by applying a 0.8-s saturating light pulse (2,000 μmol m−2 s−1) from a light source (KL 2500; Schott). The variable fluorescence (Fv) was calculated as Fm − Fo. Actinic light was supplied by a KL1500 lamp. The Fv/Fm was calculated as (Fm − Fo)/Fm. The level of maximum fluorescence during illumination (Fm′) was obtained with a saturating pulse during actinic illumination. The level of minimum fluorescence during illumination (Fo′) was obtained after turning off the actinic light. The Fs was the level of fluorescence yield during illumination. Variable fluorescence during illumination (Fv′) was calculated as Fm′ − Fo′. Quantum yield of open PSII in the steady state (Fv′/Fm′) was calculated as (Fm′ − Fo′)/Fm′. Effective quantum yield of ΦII was calculated as (Fm′ − Fs)/Fm′. NPQ was calculated as (Fm − Fm′)/Fm′. qP was calculated as (Fs − Fo′)/(Fm′ − Fo′). The partial pressures of CO2 were controlled with an infrared gas analyzer system (Li-6400; Licor).
Measurement of the P700 Oxidation Ratio
The P700 oxidation ratio was monitored as absorbance changes at 820 nm using a pulse-modulated system (PAM 101/102; Walz). P700 was oxidized by far-red light from a photodiode (FR-102; Walz). The maximum contents of P700+ (ΔAmax) were estimated by applying far-red light (Schreiber et al., 1988). P700+ absorbance changes (ΔA) were determined at indicated photon flux densities. The redox state of P700 was calculated as ΔA/ΔAmax.
Chlorophyll Measurement
Total chlorophyll content and the chlorophyll a/b ratio were determined after extraction with dimethylformamide according to Porra et al. (1989). The relative chlorophyll content per unit of leaf area was estimated using a two-wavelength-type chlorophyll meter (SPAD-502; Minolta).
RT-PCR Analysis
Total RNA was prepared using a NucleoSpin RNA Plant Kit (Macherey-Nagel). cDNA was synthesized using the SuperScript first-strand synthesis system for RT-PCR (Invitrogen Japan) according to the manufacturer’s instructions. PCR was performed with rTaq polymerase (TOYOBO) using the following program: 2 min at 94°C; 30 cycles of 1 min at 95°C, 1 min at 50°C, and 3 min at 72°C; then 10 min at 72°C. The primer sets used in this study are shown in Supplemental Table S1.
Quantitative Real-Time PCR Analysis
Total RNA was isolated from fully expanded leaves as described above. Real-time PCR was performed according to the protocol of the Mx3000P (Agilent Technology). PCR was performed using SYBR Green Real-Time PCR Master Mix (TOYOBO), and the product was analyzed by the Mx3000P multiplex quantitative PCR system. The primer sets used are shown in Supplemental Table S1.
Microarray Analysis
Details of the microarray method were described previously (Kondou et al., 2008). Complementary RNA amplification and fluorescence labeling were performed using the Agilent Low RNA Input Linear Amplification Kit (Agilent Technology). Hybridization was performed using the In Situ Hybridization Kit Plus. We used the Agilent Rice Oligo Microarray for 44K Microarray Analysis (Agilent Technology). Microarrays were scanned using an Agilent G2505 B Microarray Scanner. Microarray images were analyzed and data extracted using the Feature Extraction 9.1 software (Agilent Technology). RNA samples were isolated from aerial parts of rice wild-type and transgenic plants.
Western-Blot Analysis
Thylakoids and the soluble protein fraction were extracted as described by Lintala et al. (2007). Rice leaf blades were homogenized in shock buffer (10 mm HEPES-KOH [pH 7.6], 5 mm Suc, and 5 mm MgCl2). The suspension was filtered through nylon mesh and centrifuged at 2,500g for 4 min at 4°C. After centrifugation, the supernatant was used as the soluble fraction and the pellet was used as the thylakoid fraction. A total of 10 μg of proteins from the thylakoid and soluble fractions were separated in their native form using nondenaturing PAGE. Thylakoid proteins were solubilized in buffer containing 138 mm Tris-HCl (pH 8.0), 22% glycerol, and 1% n-dodecyl-β-d-maltoside and were subjected to native gel electrophoresis (12% acrylamide, 375 mm Tris-HCl, pH 8.8, and 7.6% glycerol). After electrophoresis, proteins were electroblotted to a polyvinylidene difluoride membrane, and immunodetection was performed using the enhanced chemiluminescence western-blotting system. FNR antibodies were purchased from AntiProt.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers NP_001045608 (OsFNR1), NP_001056570 (OsFNR2), AED98174 (AtFNR1), and AEE29923 (AtFNR2).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Expression analysis of rice fl-cDNAs in rice FOX Arabidopsis lines.
Supplemental Table S1. Primer sets used in RT-PCR analysis.
Supplemental Table S2. Steady-state photosynthetic parameters of retransformed Arabidopsis T1 plants.
Supplemental Table S3. The results of microarray analysis for the investigation of gene expression profiles in leaf blades between the wild type and OsLFNR1OE and OsLFNR2OE.
Supplemental Table S4. Steady-state photosynthetic parameters in hydroponically grown plants.
Acknowledgments
We thank Dr. Hiroaki Ichikawa (National Institute of Agrobiological Sciences, Japan) for providing the pRiceFOX vector and Mrs. Miki Ohtake (National Institute of Agrobiological Sciences, Japan) for technical support. We also thank Dr. Takeshi Yoshizumi (RIKEN Plant Science Center, Japan) for valuable help, discussion, and comments.
References
- Albinsky D, Kusano M, Higuchi M, Hayashi N, Kobayashi M, Fukushima A, Mori M, Ichikawa T, Matsui K, Kuroda H, et al. (2010) Metabolomic screening applied to rice FOX Arabidopsis lines leads to the identification of a gene-changing nitrogen metabolism. Mol Plant 3: 125–142 [DOI] [PubMed] [Google Scholar]
- Allahverdiyeva Y, Mamedov F, Suorsa M, Styring S, Vass I, Aro EM. (2007) Insights into the function of PsbR protein in Arabidopsis thaliana. Biochim Biophys Acta 1767: 677–685 [DOI] [PubMed] [Google Scholar]
- Alte F, Stengel A, Benz JP, Petersen E, Soll J, Groll M, Bölter B. (2010) Ferredoxin:NADPH oxidoreductase is recruited to thylakoids by binding to a polyproline type II helix in a pH-dependent manner. Proc Natl Acad Sci USA 107: 19260–19265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arakaki AK, Ceccarelli EA, Carrillo N. (1997) Plant-type ferredoxin-NADP+ reductases: a basal structural framework and a multiplicity of functions. FASEB J 11: 133–140 [DOI] [PubMed] [Google Scholar]
- Asada K. (2000) The water-water cycle as alternative photon and electron sinks. Philos Trans R Soc Lond B Biol Sci 355: 1419–1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benz JP, Lintala M, Soll J, Mulo P, Bölter B. (2010) A new concept for ferredoxin-NADP(H) oxidoreductase binding to plant thylakoids. Trends Plant Sci 15: 608–613 [DOI] [PubMed] [Google Scholar]
- Benz JP, Stengel A, Lintala M, Lee YH, Weber A, Philippar K, Gügel IL, Kaieda S, Ikegami T, Mulo P, et al. (2009) Arabidopsis Tic62 and ferredoxin-NADP(H) oxidoreductase form light-regulated complexes that are integrated into the chloroplast redox poise. Plant Cell 21: 3965–3983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breyton C, Nandha B, Johnson GN, Joliot P, Finazzi G. (2006) Redox modulation of cyclic electron flow around photosystem I in C3 plants. Biochemistry 45: 13465–13475 [DOI] [PubMed] [Google Scholar]
- Carrillo N, Ceccarelli EA. (2003) Open questions in ferredoxin-NADP+ reductase catalytic mechanism. Eur J Biochem 270: 1900–1915 [DOI] [PubMed] [Google Scholar]
- Clarke L, Carbon J. (1976) A colony bank containing synthetic Col E1 hybrid plasmids representative of the entire E. coli genome. Cell 9: 91–99 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- DalCorso G, Pesaresi P, Masiero S, Aseeva E, Schunemann D, Finazzi G, Joliot P, Barbato R, Leister D. (2008) A complex containing PGRL1 and PGR5 is involved in the switch between linear and cyclic electron flow in Arabidopsis. Cell 132: 273–285 [DOI] [PubMed] [Google Scholar]
- Dobakova M, Sobotka R, Tichy M, Komenda J. (2009) Psb28 protein is involved in the biogenesis of the photosystem II inner antenna CP47 (PsbB) in the cyanobacterium Synechocystis sp. PCC 6803. Plant Physiol 149: 1076–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubouzet JG, Maeda S, Sugano S, Ohtake M, Hayashi N, Ichikawa T, Kondou Y, Kuroda H, Horii Y, Matsui M, et al. (2011) Screening for resistance against Pseudomonas syringae in rice-FOX Arabidopsis lines identified a putative receptor-like cytoplasmic kinase gene that confers resistance to major bacterial and fungal pathogens in Arabidopsis and rice. Plant Biotechnol J 9: 466–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guedeney G, Corneille S, Cuine S, Peltier G. (1996) Evidence for an association of ndh B, ndh J gene products and ferredoxin-NADP-reductase as components of a chloroplastic NAD(P)H dehydrogenase complex. FEBS Lett 378: 277–280 [DOI] [PubMed] [Google Scholar]
- Gummadova JO, Fletcher GJ, Moolna A, Hanke GT, Hase T, Bowsher CG. (2007) Expression of multiple forms of ferredoxin NADP+ oxidoreductase in wheat leaves. J Exp Bot 58: 3971–3985 [DOI] [PubMed] [Google Scholar]
- Hajirezaei MR, Peisker M, Tschiersch H, Palatnik JF, Valle EM, Carrillo N, Sonnewald U. (2002) Small changes in the activity of chloroplastic NADP+-dependent ferredoxin oxidoreductase lead to impaired plant growth and restrict photosynthetic activity of transgenic tobacco plants. Plant J 29: 281–293 [DOI] [PubMed] [Google Scholar]
- Hanke GT, Endo T, Satoh F, Hase T. (2008a) Altered photosynthetic electron channelling into cyclic electron flow and nitrite assimilation in a mutant of ferredoxin:NADP(H) reductase. Plant Cell Environ 31: 1017–1028 [DOI] [PubMed] [Google Scholar]
- Hanke GT, Hase T. (2008b) Variable photosynthetic roles of two leaf-type ferredoxins in Arabidopsis, as revealed by RNA interference. Photochem Photobiol 84: 1302–1309 [DOI] [PubMed] [Google Scholar]
- Hanke GT, Kurisu G, Kusunoki M, Hase T. (2004) Fd:FNR electron transfer complexes: evolutionary refinement of structural interactions. Photosynth Res 81: 317–327 [DOI] [PubMed] [Google Scholar]
- Hanke GT, Okutani S, Satomi Y, Takao T, Suzuki A, Hase T. (2005) Multiple iso-proteins of FNR in Arabidopsis: evidence for different contributions to chloroplast function and nitrogen assimilation. Plant Cell Environ 28: 1146–1157 [Google Scholar]
- Higuchi M, Ozaki H, Matsui M, Sonoike K. (2009) A T-DNA insertion mutant of AtHMA1 gene encoding a Cu transporting ATPase in Arabidopsis thaliana has a defect in the water-water cycle of photosynthesis. J Photochem Photobiol B 94: 205–213 [DOI] [PubMed] [Google Scholar]
- Ichikawa T, Nakazawa M, Kawashima M, Iizumi H, Kuroda H, Kondou Y, Tsuhara Y, Suzuki K, Ishikawa A, Seki M, et al. (2006) The FOX hunting system: an alternative gain-of-function gene hunting technique. Plant J 48: 974–985 [DOI] [PubMed] [Google Scholar]
- Joet T, Cournac L, Peltier G, Havaux M. (2002) Cyclic electron flow around photosystem I in C3 plants: in vivo control by the redox state of chloroplasts and involvement of the NADH-dehydrogenase complex. Plant Physiol 128: 760–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juric S, Hazler-Pilepi K, Tomasic A, Lepedus H, Jelicic B, Puthiyaveetil S, Bionda T, Vojta L, Allen JF, Schleiff E, et al. (2009) Tethering of ferredoxin:NADP(+) oxidoreductase to thylakoid membranes is mediated by novel chloroplast protein TROL. Plant J 60: 783–794 [DOI] [PubMed] [Google Scholar]
- Kondou Y, Higuchi M, Takahashi S, Sakurai T, Ichikawa T, Kuroda H, Yoshizumi T, Tsumoto Y, Horii Y, Kawashima M, et al. (2009) Systematic approaches to using the FOX hunting system to identify useful rice genes. Plant J 57: 883–894 [DOI] [PubMed] [Google Scholar]
- Kondou Y, Nakazawa M, Kawashima M, Ichikawa T, Yoshizumi T, Suzuki K, Ishikawa A, Koshi T, Matsui R, Muto S, et al. (2008) RETARDED GROWTH OF EMBRYO1, a new basic helix-loop-helix protein, expresses in endosperm to control embryo growth. Plant Physiol 147: 1924–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XP, Bjorkman O, Shih C, Grossman AR, Rosenquist M, Jansson S, Niyogi KK. (2000) A pigment-binding protein essential for regulation of photosynthetic light harvesting. Nature 403: 391–395 [DOI] [PubMed] [Google Scholar]
- Lintala M, Allahverdiyeva Y, Kangasjarvi S, Lehtimaki N, Keranen M, Rintamaki E, Aro EM, Mulo P. (2009) Comparative analysis of leaf-type ferredoxin-NADP oxidoreductase isoforms in Arabidopsis thaliana. Plant J 57: 1103–1115 [DOI] [PubMed] [Google Scholar]
- Lintala M, Allahverdiyeva Y, Kidron H, Piippo M, Battchikova N, Suorsa M, Rintamaki E, Salminen TA, Aro EM, Mulo P. (2007) Structural and functional characterization of ferredoxin-NADP+-oxidoreductase using knock-out mutants of Arabidopsis. Plant J 49: 1041–1052 [DOI] [PubMed] [Google Scholar]
- Meurer J, Meierhoff K, Westhoff P. (1996) Isolation of high-chlorophyll-fluorescence mutants of Arabidopsis thaliana and their characterisation by spectroscopy, immunoblotting and northern hybridisation. Planta 198: 385–396 [DOI] [PubMed] [Google Scholar]
- Munekage Y, Hojo M, Meurer J, Endo T, Tasaka M, Shikanai T. (2002) PGR5 is involved in cyclic electron flow around photosystem I and is essential for photoprotection in Arabidopsis. Cell 110: 361–371 [DOI] [PubMed] [Google Scholar]
- Nakamura H, Hakata M, Amano K, Miyao A, Toki N, Kajikawa M, Pang J, Higashi N, Ando S, Toki S, et al. (2007) A genome-wide gain-of-function analysis of rice genes using the FOX-hunting system. Plant Mol Biol 65: 357–371 [DOI] [PubMed] [Google Scholar]
- Niyogi KK, Bjorkman O, Grossman AR. (1997) Chlamydomonas xanthophyll cycle mutants identified by video imaging of chlorophyll fluorescence quenching. Plant Cell 9: 1369–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niyogi KK, Grossman AR, Bjorkman O. (1998) Arabidopsis mutants define a central role for the xanthophyll cycle in the regulation of photosynthetic energy conversion. Plant Cell 10: 1121–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okegawa Y, Kagawa Y, Kobayashi Y, Shikanai T. (2008) Characterization of factors affecting the activity of photosystem I cyclic electron transport in chloroplasts. Plant Cell Physiol 49: 825–834 [DOI] [PubMed] [Google Scholar]
- Ozaki H, Ikeuchi M, Ogawa T, Fukuzawa H, Sonoike K. (2007) Large-scale analysis of chlorophyll fluorescence kinetics in Synechocystis sp. PCC 6803: identification of the factors involved in the modulation of photosystem stoichiometry. Plant Cell Physiol 48: 451–458 [DOI] [PubMed] [Google Scholar]
- Porra RJ, Thompson WA, Kriedemann PE. (1989) Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim Biophys Acta 975: 384–394 [Google Scholar]
- Rodriguez RE, Lodeyro A, Poli HO, Zurbriggen M, Peisker M, Palatnik JF, Tognetti VB, Tschiersch H, Hajirezaei MR, Valle EM, et al. (2007) Transgenic tobacco plants overexpressing chloroplastic ferredoxin-NADP(H) reductase display normal rates of photosynthesis and increased tolerance to oxidative stress. Plant Physiol 143: 639–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai I, Mizusawa N, Ohashi S, Kobayashi M, Wada H. (2007) Effects of the lack of phosphatidylglycerol on the donor side of photosystem II. Plant Physiol 144: 1336–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber U, Klughammer C, Neubauer C. (1988) Measuring P-700 absorbance changes around 830 nm with a new type of pulse modulation system. Z Naturforsch Teil C 43: 686–698 [Google Scholar]
- Schurmann P, Buchanan BB. (2008) The ferredoxin/thioredoxin system of oxygenic photosynthesis. Antioxid Redox Signal 10: 1235–1274 [DOI] [PubMed] [Google Scholar]
- Shikanai T. (2007) Cyclic electron transport around photosystem I: genetic approaches. Annu Rev Plant Biol 58: 199–217 [DOI] [PubMed] [Google Scholar]
- Shikanai T, Endo T, Hashimoto T, Yamada Y, Asada K, Yokota A. (1998) Directed disruption of the tobacco ndhB gene impairs cyclic electron flow around photosystem I. Proc Natl Acad Sci USA 95: 9705–9709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikanai T, Munekage Y, Shimizu K, Endo T, Hashimoto T. (1999) Identification and characterization of Arabidopsis mutants with reduced quenching of chlorophyll fluorescence. Plant Cell Physiol 40: 1134–1142 [DOI] [PubMed] [Google Scholar]
- Toki S, Hara N, Ono K, Onodera H, Tagiri A, Oka S, Tanaka H. (2006) Early infection of scutellum tissue with Agrobacterium allows high-speed transformation of rice. Plant J 47: 969–976 [DOI] [PubMed] [Google Scholar]
- Varotto C, Pesaresi P, Meurer J, Oelmuller R, Steiner-Lange S, Salamini F, Leister D. (2000) Disruption of the Arabidopsis photosystem I gene psaE1 affects photosynthesis and impairs growth. Plant J 22: 115–124 [DOI] [PubMed] [Google Scholar]
- Wingler A, Lea PJ, Quick WP, Leegood RC. (2000) Photorespiration: metabolic pathways and their role in stress protection. Philos Trans R Soc Lond B Biol Sci 355: 1517–1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokotani N, Higuchi M, Kondou Y, Ichikawa T, Iwabuchi M, Hirochika H, Matsui M, Oda K. (2011) A novel chloroplast protein, CEST induces tolerance to multiple environmental stresses and reduces photooxidative damage in transgenic Arabidopsis. J Exp Bot 62: 557–569 [DOI] [PubMed] [Google Scholar]
- Yokotani N, Ichikawa T, Kondou Y, Maeda S, Iwabuchi M, Mori M, Hirochika H, Matsui M, Oda K. (2009a) Overexpression of a rice gene encoding a small C2 domain protein OsSMCP1 increases tolerance to abiotic and biotic stresses in transgenic Arabidopsis. Plant Mol Biol 71: 391–402 [DOI] [PubMed] [Google Scholar]
- Yokotani N, Ichikawa T, Kondou Y, Matsui M, Hirochika H, Iwabuchi M, Oda K. (2008) Expression of rice heat stress transcription factor OsHsfA2e enhances tolerance to environmental stresses in transgenic Arabidopsis. Planta 227: 957–967 [DOI] [PubMed] [Google Scholar]
- Yokotani N, Ichikawa T, Kondou Y, Matsui M, Hirochika H, Iwabuchi M, Oda K. (2009b) Tolerance to various environmental stresses conferred by the salt-responsive rice gene ONAC063 in transgenic Arabidopsis. Planta 229: 1065–1075 [DOI] [PubMed] [Google Scholar]
- Zhang H, Cramer WA. (2004) Purification and crystallization of the cytochrome b6f complex in oxygenic photosynthesis. Methods Mol Biol 274: 67–78 [DOI] [PubMed] [Google Scholar]
- Zulfugarov IS, Ham OK, Mishra SR, Kim JY, Nath K, Koo HY, Kim HS, Moon YH, An G, Lee CH. (2007) Dependence of reaction center-type energy-dependent quenching on photosystem II antenna size. Biochim Biophys Acta 1767: 773–780 [DOI] [PubMed] [Google Scholar]