Abstract
Heterogeneous nuclear RNA-binding proteins, hnRNPs, have been implicated in nuclear export of mRNAs in organisms from yeast to humans. A germ-line mutation in a Drosophila hnRNP, Squid (Sqd)/hrp40, causes female sterility as a result of mislocalization of gurken (grk) mRNA during oogenesis. Alternative splicing produces three isoforms, SqdA, SqdB, and SqdS. Here we show that these isoforms are not equivalent; SqdA and SqdS perform overlapping but nonidentical functions in grk mRNA localization and protein accumulation, whereas SqdB cannot perform these functions. Furthermore, although all three Sqd isoforms are expressed in the germline cells of the ovary, they display distinct intracellular distributions. Both SqdB and SqdS are detected in germ-line nuclei, whereas SqdA is predominantly cytoplasmic. We show that this differential nuclear accumulation is correlated with a differential association with the nuclear import protein Transportin. Finally, we provide evidence that grk mRNA localization and translation are coupled by an interaction between Sqd and the translational repressor protein Bruno. These results demonstrate the isoform-specific contributions of individual hnRNP proteins in the regulation of a specific mRNA. Moreover, these data suggest a novel role for hnRNPs in localization and translational regulation of mRNAs.
Keywords: hnRNP, RNA localization, oogenesis, Gurken, squid, Drosophila
The Drosophila melanogaster gene squid (sqd) encodes a heterogeneous nuclear RNA binding protein (hnRNP), also known as hrp40 (Matunis et al. 1992; Kelley 1993). hnRNPs are a large family of proteins that have been implicated in the processing of nascent mRNA transcripts (for review, see Dreyfuss et al. 1993). Recent studies have demonstrated that a subset of hnRNPs, including human hnRNP A1 and A2, Saccharomyces cerevisiae Nplp3 and Hrp1, and Chironomus tentans Hrp36, rapidly shuttle between the cytoplasm and the nucleus (Pinol-Roma and Dreyfuss 1992; Lee et al. 1996; Visa et al. 1996; Kessler et al. 1997; Hoek et al. 1998). A specific motif, termed M9, has been shown to mediate this nucleocytoplasmic shuttling (Michael et al. 1995; Siomi and Dreyfuss 1995; Weighardt et al. 1995), and this motif is present in Sqd. Nuclear import of M9-containing hnRNPs is achieved by an association with the nuclear import protein Transportin (Pollard et al. 1996). Studies of several of these hnRNPs have indicated that one of their major roles is the nuclear export of mRNAs (Pinol-Roma and Dreyfuss 1992; Lee et al. 1996; Visa et al. 1996), suggesting that Sqd may perform a similar function during Drosophila oogenesis.
Mutations in sqd have been found to disrupt Gurken (Grk)-dependent dorsal-ventral (D-V) patterning during oogenesis (Kelley 1993). grk encodes a TGF-α homolog (Neuman-Silberberg and Schüpbach 1993), which is expressed specifically in the germ line. Grk is a ligand for the epidermal growth factor receptor (Egfr), which is expressed in the somatic follicle cells surrounding the germ line. Activation of Egfr by Grk in mid-oogenesis induces follicle cells to adopt a dorsal cell fate, thus defining the polarity of the egg and the future embryo. Therefore, it is imperative that Grk signaling be restricted to the future dorsal side of the egg chamber. Spatial restriction of Grk protein is achieved, in part through the localization of grk mRNA. Whereas Egfr expression is ubiquitous in all follicle cells, grk mRNA becomes tightly localized to the dorsoanterior corner of the oocyte during mid-oogenesis (for review, see Nilson and Schüpbach 1999). Two female sterile mutations, fs(1)K10 and sqd, consistently cause a mislocalization of grk mRNA along the entire anterior cortex of the oocyte and lead to the production of strongly dorsalized eggs, demonstrating the critical role of grk mRNA localization (Wieschaus et al. 1978; Kelley 1993; Neuman-Silberberg and Schüpbach 1993). Moreover, multiple copies of a grk transgene also result in aberrant grk mRNA localization and a dorsalized egg phenotype, which suggests that the oocyte contains a saturable machinery that is responsible for grk mRNA localization (Neuman-Silberberg and Schüpbach 1994). It is not known whether grk transcription occurs only within the germ-line-derived nurse cell nuclei, or whether grk represents a rare gene that is also transcribed within the oocyte nucleus itself. In either case, grk mRNA must be transported from the nucleus where it is transcribed and somehow anchored or stabilized in the dorso-anterior corner of the oocyte, and Sqd must function at some step in this process.
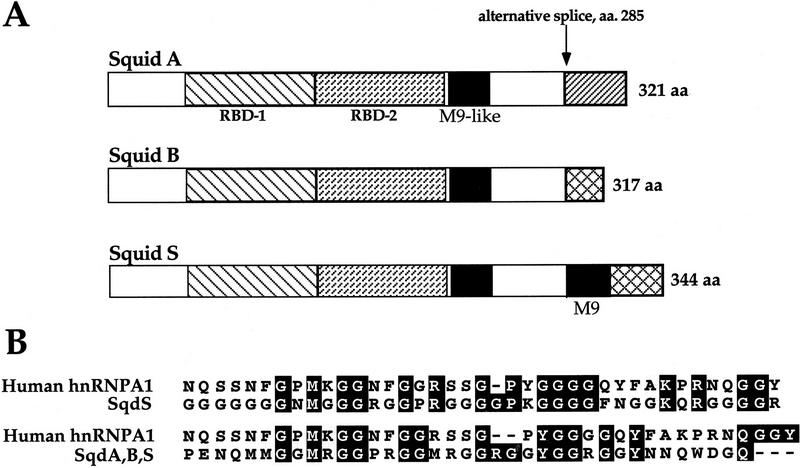
In addition to its requirement for D-V patterning during oogenesis, Sqd is required for viability during larval development (Kelley 1993). The sqd gene is alternatively spliced to produce three protein isoforms: SqdA (hrp40.1), SqdB, and SqdS (hrp40.2). These isoforms are identical over the majority of the protein, including the two RNA-binding domains. Interestingly, although all three isoforms contain an M9-like nuclear import motif within their common coding sequence, only the SqdS isoform contains a strong consensus M9 motif within its alternatively spliced region (Siomi et al. 1998; Fig. 1). Within wild-type ovaries, Sqd protein is expressed ubiquitously and can be detected in the cytoplasm and nuclei of all of the cells in the egg chamber (Matunis et al. 1994). Although null alleles of sqd are lethal because of the somatic requirement for sqd function, the sqd1 allele reveals the germ line requirement for sqd, as it eliminates germ-line Sqd expression during the critical stages of oogenesis when D-V patterning is established (Kelley 1993).
Figure 1.
Schematic diagram of Sqd protein isoforms. The three Sqd isoforms, SqdA, SqdB, and SqdS, are identical to amino acid position 285 (A). Within this conserved region are the two RNA-binding domains RBD-1 and RBD-2 (depicted by the hatched boxes) and an M9-like nuclear-import domain (represented by the black box). At the carboxyl termini of the proteins, all three contain a glycine-rich domain, which in the SqdS and SqdB isoforms continues to the end of the protein. The SqdB and SqdS isoforms use the same 3′ exon; however, the splice site in the SqdB isoform results in a protein with 27 fewer amino acids than SqdS. In addition, SqdS contains another M9 nuclear import motif (amino acids 300–338) that is not present in either SqdA or SqdB. The SqdA isoform uses an alternative 3′ exon and encodes a protein with a unique carboxyl terminus. A comparison of the human hnRNPA1 M9 domain with the M9 domains found in Sqd is shown in B. The M9 motif found only in SqdS shares 16 of 38 amino acid identity with human hnRNPA1 M9 (top), whereas the M9-like motif found in all three Sqd isoforms shares 13 of 38 amino acid identity with hnRNPA1 (bottom). Alignment of the M9 motifs was accomplished using the ClustalW Multiple Sequence Alignment Program.
To gain a better understanding of the mechanism of grk mRNA localization during oogenesis, we have investigated the role of the Sqd protein in this process. More specifically, using isoform-specific transgenes we have been able to analyze the contributions of the individual Sqd isoforms during oogenesis. Despite the extensive protein similarity, we have observed several striking differences among the isoforms. Namely, they do not function equivalently in either the localization of grk mRNA or in the accumulation of Grk protein. Furthermore, because of a differential association with the nuclear import protein Transportin, the Sqd isoforms display distinct intracellular distribution patterns within the ovary. On the basis of its homology to proteins used in the nuclear export of mRNA, it seemed possible that Sqd protein could function in the nuclear export of grk mRNA, although a nuclear export function alone would not fully explain the role of Sqd in grk mRNA localization in the oocyte cytoplasm. Because Sqd is required for grk mRNA localization and our findings that Sqd protein can interact with grk mRNA directly and can associate with cytoplasmic regulators of Grk translation, we propose that Sqd protein is responsible for the regulated nuclear export of grk mRNA.
Results
Rescue of D-V patterning defects by specific Sqd isoforms
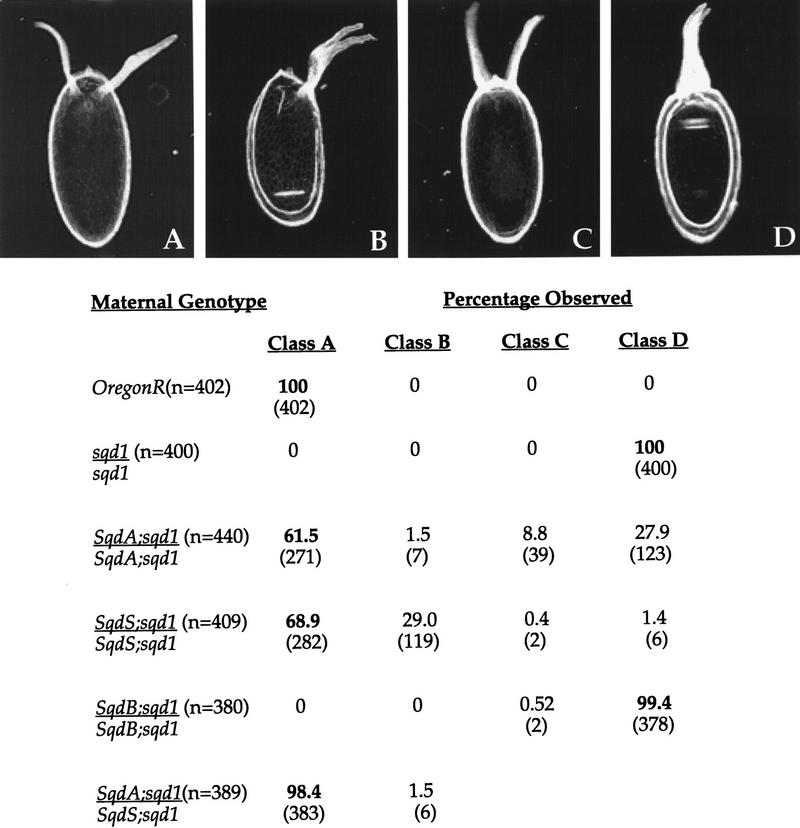
In wild-type ovaries grk mRNA is tightly localized to the dorsoanterior corner of the oocyte during mid-oogenesis in close proximity to the oocyte nucleus. In contrast, in egg chambers from sqd1 mutant females, grk mRNA is localized along the entire anterior cortex of the oocyte. This mislocalized RNA is translated efficiently and leads to a dorsalization of the follicle cells overlying this region of the oocyte (Fig. 2D). The dorsalized eggs produced by sqd1 females have two characteristics. First, the eggs have dorsal appendage material expanding to the ventral side of the eggshell, and second, the dorsal-most cell fate, which is represented by the dorsal midline cells between the dorsal appendages, is missing in sqd1 eggs. This absence of dorsal midline fates is the result of an inability to produce a highpoint of Grk protein expression on the dorsal side of the oocyte (Kelley 1993; Neuman-Silberberg and Schüpbach 1993).
Figure 2.
Rescue of sqd1 D-V patterning defects by individual Sqd isoforms. Females of the indicated genotypes were placed in laying blocks and eggs were collected on days 2 and 3 after blocking. (Top) The series of dorsalized eggshell phenotypes: wild-type egg with two dorsal appendages (A), slightly dorsalized egg with a broad fused dorsal appendage (B), a more dorsalized egg with widely spaced dorsal appendages attributable to the expansion of the dorsal midline fates (C), and a severely dorsalized egg with expanded dorsal appendage material around the anterior circumference of the eggshell (D). For each maternal genotype the percentage of eggs with the indicated phenotypes were calculated (n = total number of eggs examined). In the case of Sqd transgene expressing females, a single female laid the range of eggshell phenotypes shown.
The sqd transcript is alternatively spliced to produce three protein isoforms designated SqdA (hrp40.1), SqdS (hrp40.2), and SqdB (see Fig. 1) (Matunis et al. 1992; Kelley 1993). Previous studies of Sqd within wild-type ovaries have shown that Sqd protein is detected within the somatic follicle cells and the germ-line-derived nurse cells and oocyte (Matunis et al. 1994). The sqd1 mutation affects a germ-line promoter of sqd and eliminates expression of all of the Sqd isoforms at the critical stages of oogenesis when D-V patterning is established. This allowed us to investigate the germ-line requirements for the individual Sqd isoforms using transgenes encoding specific Sqd proteins in a sqd1 homozygous background. Expression of these transgenes is driven by the endogenous Sqd promoter/enhancer region; therefore, their expression should reflect that of the wild-type gene. For brevity, females carrying two copies of a specific transgene in a sqd1 homozygous background will be referred to as SqdA, SqdS, or SqdB females, respectively.
The individual isoforms showed striking differences in their ability to rescue the D-V patterning defects of sqd1 mutant females (Fig. 2). Despite the fact that the transgene is expressed in the germ line (see below), the SqdB isoform cannot restore proper D-V patterning of the eggshell; SqdB females lay 100% strongly dorsalized eggs, which are indistinguishable from those laid by sqd1 homozygotes (Fig. 2D). In contrast, expression of either the SqdS or SqdA isoform can rescue the dorsalized phenotype of sqd1 mutants. Females carrying two copies of either the SqdA or SqdS transgene lay a large percentage of wild-type eggs. Neither transgene alone, however, is capable of entirely restoring the function of the wild-type gene, as SqdA or SqdS females lay a number of dorsalized and partially rescued eggs as well. Furthermore, the classification or phenotype of the partially rescued eggs laid by these females is strikingly different. SqdA females lay a number of very dorsalized eggs (Fig. 2D), whereas in contrast, SqdS females lay eggs with broad, fused dorsal appendages (Fig. 2B). It should be noted, that although SqdA females lay a number of severely dorsalized eggs (Fig. 2D), these eggs are not identical to sqd1 or to SqdB eggs in that the SqdA eggs do not have a uniform crown of dorsal appendage material around the entire anterior circumference of the eggshell. Frequently, the SqdA eggs show a restoration of the dorsal midline fates (indicative of high levels of Grk protein). The differences in the phenotypes of partially rescued eggs are not attributable to differences in transgene expression levels, as decreasing the copy number from two to one does not affect the type of eggs observed (data not shown). Interestingly, females carrying one copy of each of these transgenes (SqdS and SqdA) lay nearly all wild-type eggs (98% as compared with ∼60% in females with two copies of either transgene). Therefore, the two Sqd isoforms, SqdA and SqdS, appear to have nonidentical functions in establishing proper dorsoventral patterning during oogenesis and act together in this process.
In addition to its requirement in oogenesis, Sqd is also required somatically. A number of lethal alleles of sqd have been generated, and we have investigated the ability of the individual isoforms to restore viability of sqd null alleles. Again, as with the ability of the specific isoforms to function during oogenesis, the three Sqd isoforms differ in their ability to rescue the viability of the lethal sqd allelic combination sqdIX50/Df(3R)urd. We observed that both SqdS and SqdB were capable of rescuing the essential somatic Sqd function, in that expression of either of these transgenes allowed recovery of 11% and 19% of the expected number of sqdIX50/Df(3R)urd adults, respectively. In contrast, however, SqdA was incapable of restoring the essential somatic function of Sqd, as <0.2% of the expected number of sqdIX50/Df(3R)urd adults were recovered. These data further demonstrate that the individual Sqd isoforms are not functionally equivalent.
grk mRNA and protein localization in transgenic females
Given the observation that the different Sqd isoforms differ so markedly in their ability to function during oogenesis, we examined their effects on grk mRNA localization and protein accumulation. Ovaries from females carrying two copies of the SqdS or SqdB transgenes showed a grk mRNA and protein localization pattern that was consistent with their effect on patterning of the eggshell. Females carrying the SqdB transgene lay severely dorsalized eggs, and within late stage SqdB egg chambers, grk mRNA and Grk protein are observed in a symmetric ring around the anterior circumference of the oocyte (Fig. 3D,H), much like the grk mRNA and protein distribution in sqd1 egg chambers. In contrast, SqdS expression allows the production of a large number of wild-type eggs, and egg chambers from SqdS females consistently showed grk mRNA and Grk protein tightly localized to the dorsoanterior corner of the oocyte (Fig. 3C,G), similar to wild type (Fig. 3A,E).
Figure 3.
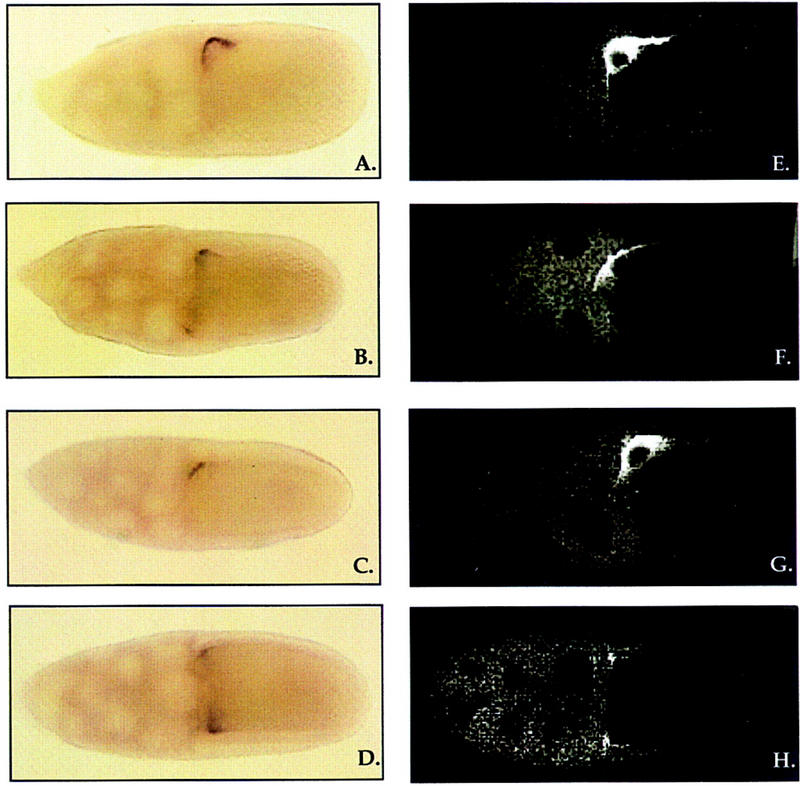
grk mRNA localization and protein distribution within Sqd transgenic egg chambers. In wild-type egg chambers, grk mRNA is tightly localized to the dorso-anterior corner of the oocyte in close proximity to the oocyte nucleus (A). Within egg chambers from SqdA females, grk mRNA is distributed around the anterior circumference of the oocyte, although it is more concentrated on the dorsal side (B). Egg chambers from SqdS females show a grk mRNA localization pattern indistinguishable from wild-type (C), and in contrast, in egg chambers from SqdB females, grk mRNA is mislocalized in an anterior ring in a pattern that is indistinguishable from sqd1 egg chambers (D). Grk protein localization within egg chambers from Sqd transgenic females (E–H). In stage 10 wild-type egg chambers, Grk protein accumulates in the dorso-anterior corner of the oocyte in a location coincident with the grk mRNA (E). Within SqdA and SqdS egg chambers, Grk protein is localized to the dorsoanterior corner of the oocyte (F,G), much like that in wild type, and in SqdB egg chambers Grk protein is mislocalized along the anterior circumference of the oocyte (H). The granular staining observed in the nurse cells in F and H is nonspecific background staining that is seen in grk mutant ovaries as well.
Unexpectedly, the analysis of grk mRNA localization in late stage SqdA egg chambers did not reflect directly the ability of this isoform to rescue the D-V patterning defects of sqd1 mutants. SqdA females lay a large percentage of wild-type eggs, yet <10% of the SqdA stage 10 egg chambers showed tight localization of grk mRNA. In the overwhelming majority of SqdA egg chambers, grk mRNA appeared to be enriched on the dorsal side of the egg chamber but was readily detectable on the ventral side as well. Given the large percentage of wild-type eggs laid by SqdA females, the mislocalization of grk mRNA in these egg chambers is surprising. However, we found that the Grk protein distribution in SqdA egg chambers was much closer to wild type, in that we detected Grk protein localized to the dorsoanterior corner in a large proportion of late stage egg chambers (Fig. 3F). A smaller fraction showed an obvious asymmetric concentration of Grk protein in the dorsoanterior corner and some detectable Grk protein on the ventral side, and a further smaller fraction of egg chambers had symmetrically distributed Grk protein very similar to that observed in sqd1 or SqdB females. Thus, this protein distribution closely mirrors the distribution of eggshell phenotypes produced by SqdA-expressing females. Because the Grk protein distribution was less consistently abnormal than the grk mRNA distribution, these data show that the SqdA isoform is less capable in functioning in grk mRNA localization than SqdS. They also indicate that SqdA affects the production of Grk protein.
In summary, therefore, the Sqd isoforms differ in their ability to localize grk mRNA and in addition, they appear to affect differentially the accumulation of Grk protein. The SqdB isoform seems incapable of restoring normal grk RNA or protein distribution. The SqdS isoform restores the normal pattern of grk RNA and protein distribution, but in some cases does not allow the production of peak levels of Grk protein, as evidenced by the fraction of eggs with a broad fused appendage. Finally, SqdA is much less effective in restoring grk mRNA localization than SqdS, but produces less mislocalized Grk protein than SqdB.
Intracellular distribution of individual Sqd isoforms
Given the striking functional differences (rescue of germ-line and somatic function, as well as grk mRNA and protein distribution) of the individual Sqd isoforms, we were interested in determining whether there were any differences in their subcellular distributions. Previous analysis of Sqd protein expression in wild-type ovaries examined bulk Sqd expression and did not distinguish the distinct isoforms from one another. Recent reports have defined a 38-amino-acid motif, termed M9, which mediates the nuclear import of hnRNPs through an interaction with the nuclear import protein Transportin (Siomi and Dreyfuss 1995; Pollard et al. 1996). Sequence analysis of the Sqd protein isoforms reveals that such an M9 motif is present in Sqd. A comparison of the human hnRNPA1 M9 sequence with the Sqd isoforms shows that M9 is differentially present in the isoforms (see Fig. 1). Specifically, within the alternatively spliced region of SqdS, there is a region (amino acids 300–338) that aligns with the human hnRNPA1 M9 sequence (16 of 38 amino acid identity) (Siomi et al. 1998). Interestingly, this sequence is not present in either the SqdA or the SqdB isoforms. However, we found that there is another M9-like sequence present in all three Sqd isoforms (amino acids 215–254) that aligns with the human hnRNPA1 M9 motif (13 of 38 amino acid identity).
We examined Sqd protein expression in the ovaries of sqd1 females carrying two copies of the individual transgenes (Fig. 4). Both SqdB and SqdS are detected in the nuclei of the germ-line-derived nurse cells (Fig. 4B,C). In addition, SqdS also accumulates in the oocyte nucleus, whereas SqdB does not. Interestingly, examination of SqdA females revealed that this isoform was undetectable in the germ line (Fig. 4A). Given that SqdA rescues efficiently the D-V patterning defect of sqd1 mutants, it seemed unlikely that it is not expressed in the germ line. Therefore, we analyzed Sqd RNA expression in SqdA females using isoform-specific probes and found that the SqdA transcript is readily detectable in the germ line of ovaries from SqdA females (data not shown). It seems, therefore, likely that the intracellular distribution of SqdA is predominantly cytoplasmic and thus, too diffuse to be visualized. Consistent with this possibility, the Sqd protein expression in the somatic follicle cells in SqdA egg chambers appears much more diffuse and cytoplasmic than in SqdS egg chambers (Fig. 4D,E).
Figure 4.
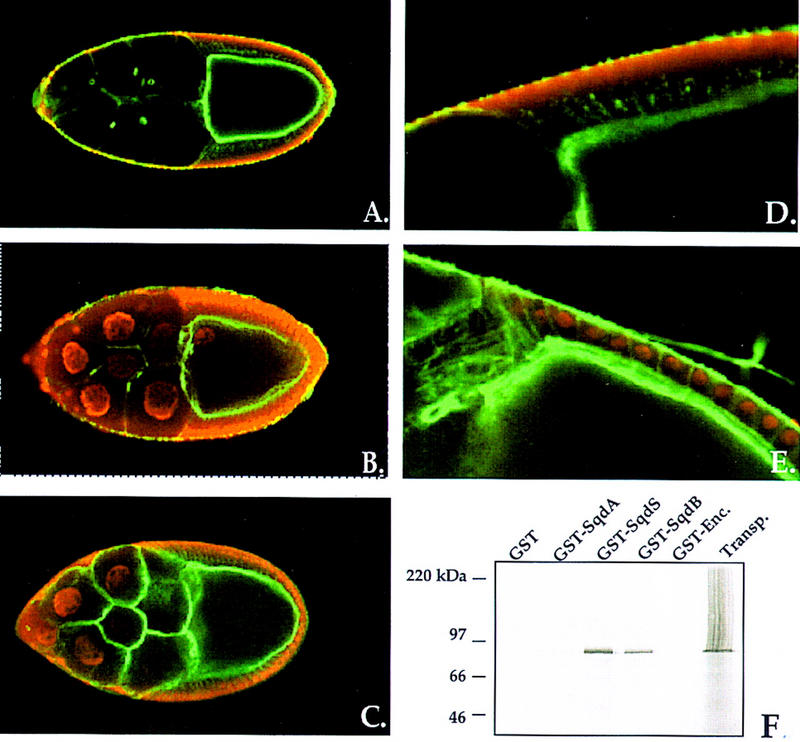
Expression of Sqd isoforms within the ovary. Stage 10 egg chambers from sqd1 females carrying the individual Sqd transgenes were examined for Sqd protein expression. Sqd protein is in red, and cortical actin detected with phalloidin is in green. Within egg chambers from Sqd transgenic females Sqd protein is undetectable in the germ line of SqdA females (A), but is detected within the nurse cell nuclei of both SqdS and SqdB females (B,C). Furthermore, in egg chambers from SqdS females, Sqd protein is detected within the oocyte nucleus (B). Higher magnification view of the somatic follicle cells is shown, in which endogenous protein and the transgene-derived protein, is detected. In SqdS egg chambers the follicle cell Sqd protein is predominantly nuclear (E). However, in SqdA egg chambers diffuse, cytoplasmic Sqd protein is detected (D). The differential nuclear accumulation of SqdB, SqdS, and SqdA is associated with a differential interaction with the nuclear import protein Transportin (F). [35S]methionine-labeled Drosophila Transportin protein was incubated with a series of GST fusion proteins (GST only, GST–SqdA, GST–SqdS, GST–SqdB, and GST–Encore) and precipitated with glutathione-coated-agarose beads. Transportin binding to the GST-fusions was assessed with SDS-PAGE and autoradiography. Transportin protein is brought down by both SqdS and SqdB, but not with GST only, SqdA, or Encore protein.
Nuclear import of M9-containing hnRNPs is mediated through an interaction with the import protein Transportin (Pollard et al. 1996). As the M9 motif is expressed differentially in the Sqd isoforms, it seemed possible that the differences in nuclear accumulation that we observed could be attributable to a differential association with Transportin. To address this issue, we tested the ability of in vitro transcribed and translated Drosophila Transportin protein to bind to GST fusion proteins corresponding to the Sqd isoforms GST alone or a GST–Encore fusion (Fig. 4F). We quantitated the amount of 35S-labeled protein (cpm) associated with each GST fusion using a PhosphorImager. In direct correlation to the intracellular distributions observed, we found that Transportin protein associates strongly with both SqdS (800,901 cpm) and SqdB (269,932 cpm), the two isoforms detected within germ-line nuclei but much less strongly with GST only (213 cpm), GST–Encore (9255 cpm) or SqdA (16,325 cpm), which assumes a cytoplasmic distribution.
At this time it is not clear why SqdA and SqdB, which share the same M9-like motif, behave so differently with respect to nuclear accumulation and association with Transportin. The carboxyl termini of these two isoforms are different from one another, with SqdB having an extensively glycine-rich domain that SqdA does not contain. In addition, the reason for the exclusive accumulation of SqdS, and not SqdB, within the oocyte nucleus is not known. One potential explanation could be that SqdS may be transported preferentially into the oocyte, thus allowing its import into the oocyte nucleus, whereas SqdB may be retained in the nurse cells. In any event, these data demonstrate that the differential nuclear accumulation of the Sqd proteins is associated with a differential ability to interact with Drosophila Transportin.
Sqd protein distribution is altered in egg chambers from fs(1)K10 females
The phenotypes of eggs laid by sqd1 and fs(1)K10 females are similar (Wieschaus et al. 1978; Kelley 1993). In both cases, grk mRNA is mislocalized and Grk protein is produced around the entire anterior circumference of the oocyte. These two female sterile mutations, although similar, are also unique with respect to other known female sterile mutants. In most other cases in which grk mRNA is mislocalized, for example, in orb and spnB mutant egg chambers, the unlocalized RNA is not translated efficiently (for review, see Nilson and Schüpbach 1999). Mutations in cappuccino and spire also result in a mislocalization of grk RNA and translation of the mislocalized message, but these two mutations have more generalized effects on oocyte patterning and do not seem as specific for grk function as K10 or the germ-line forms of Sqd (for review, see Nilson and Schüpbach 1999). In addition, early in oogenesis Grk is necessary for the establishment of anteroposterior patterning. However, eggs laid by both sqd1 and K10 mutant mothers display no anteroposterior defects, but are abnormal along only the D-V axis.
When we examined the expression of K10 protein in sqd1 mutant ovaries we found that the distribution of K10 in sqd1 mutants is unaffected and K10 protein is detected in the oocyte nucleus of late stage egg chambers (data not shown). Conversely, however, the expression of Sqd protein is affected by the K10 mutation (Fig. 5). In K10 mutant ovaries, Sqd is present in the nurse cell nuclei but absent from the oocyte nucleus (Fig. 5C). In addition, in wild-type egg chambers Sqd protein is detected at the posterior pole at late stages and this cytoplasmic Sqd localization is unaffected in K10 egg chambers (data not shown). These data indicate that Sqd protein is, in fact, expressed in K10 mutant egg chambers, but that its accumulation in the oocyte nucleus is specifically lost in the absence of K10 function.
Figure 5.
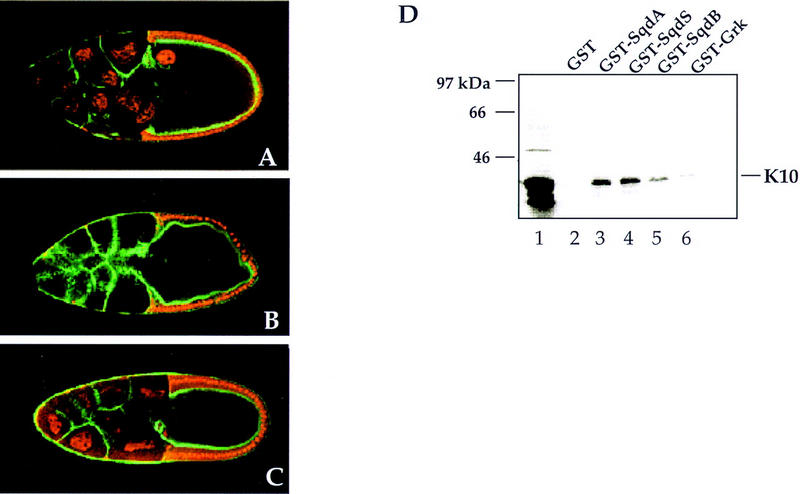
Sqd protein distribution is altered in fs(1)K10 egg chambers and physically interacts with K10 protein. Expression of Sqd protein in wild type, sqd1 and fs(1)K10 stage 10 egg chambers, with Sqd in red and cortical actin detected with phalloidin in green. In wild-type egg chambers, Sqd protein is detected in the follicle cell nuclei, the nurse cell nuclei, and the oocyte nucleus (A). In contrast, in sqd1 egg chambers, Sqd protein is undetectable in the germ line (B). Within fs(1)K10 egg chambers, Sqd protein is detected in the nurse cell nuclei, but is undetectable within the oocyte nucleus (C). To determine whether Sqd protein and K10 protein can physically associate with one another, in vitro binding studies were conducted. 35S-labeled K10 protein was incubated with a series of GST fusion proteins, and after incubation with glutathione-coated–agarose beads, K10 binding was assessed by SDS-PAGE and autoradiography (D). The input 35S-K10 protein is shown in lane 1. Association with the following GST-fusion proteins was tested: (lane 2) GST only; (lane 3) GST–SqdA; (lane 4) GST–SqdS; (lane 5) GST–SqdB; (lane 6) GST–Grk.
To assess whether the role of K10 in Sqd protein localization to the oocyte nucleus is direct, we wanted to determine whether K10 protein and Sqd protein interact with one another physically. The full-length K10 protein (463 amino acids) is comprised of a hydrophilic amino-terminal domain (residues 1–225), a central apolar domain (amino acids 225–337), and a carboxy-terminal domain containing a helix-turn-helix motif (Prost et al. 1988). In vitro-translated labeled K10 protein corresponding to the first two domains of the protein was incubated with purified GST–Sqd fusion proteins, and after incubation with glutathione-coated–agarose beads, K10 association was determined by SDS-PAGE analysis and autoradiography. In a representative experiment (Fig. 5D), K10 protein associates with all three of the Sqd isoforms [SqdA (80,038 cpm), SqdS (80,061 cpm), and SqdB (30,239 cpm)], but interacts much more weakly with either GST alone (11,577 cpm) or with GST–Grk (9552 cpm). These data indicate that Sqd protein and K10 protein can interact directly with one another, even in the absence of other ovarian proteins or RNAs.
Sqd protein can interact with grk mRNA
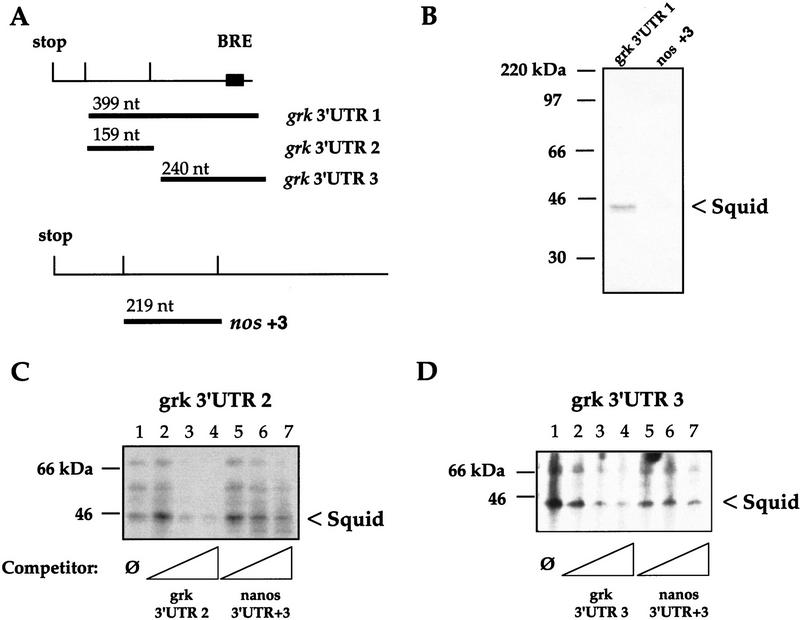
The sqd gene is clearly required for proper grk mRNA localization. As it encodes a known RNA-binding protein, one potential target for Sqd is grk mRNA itself. Therefore, we undertook a UV cross-linking analysis looking for interactions between ovarian proteins and parts of the grk RNA. In these experiments, wild-type ovarian extracts were used as a source of Sqd protein. We used different regions of the grk 3′ untranslated region (UTR) as probes (Fig. 6A), and as a negative control, we used the +3 fragment of the nanos (nos) 3′ UTR (Gavis et al. 1996), because nanos mRNA localization is unaffected by the sqd1 mutation (data not shown).
Figure 6.
Sqd protein binds grk mRNA directly. (A) The regions of the grk 3′ UTR and the nos 3′ UTR that were used as probes are depicted. (B) Wild-type ovarian lysates were subjected to UV cross-linking to the indicated RNA probes, after cross-linking the RNA–protein complexes were precipitated with anti-Sqd and resolved by SDS-PAGE. Sqd protein associates with the grk 3′ UTR (lane 1), whereas it does not interact with the +3 region of the nos 3′ UTR (lane 2). (C,D) To test the specificity of the Sqd–grk interactions, UV cross-linking analyses were repeated in the presence of excess, cold competitor RNAs. The interaction between whole ovarian protein extracts and grk 3′ UTR fragments 2 and 3 were tested. Binding in the presence of no competitor (lane 1), or with increasing amounts (∼20×, 100×, or 200× excess) of specific competitor (lanes 2–4), or with increasing amounts of nonspecific competitor (lanes 5–7) are shown.
To investigate whether Sqd protein can interact with the 3′ UTR of the grk transcript, ovarian extracts were incubated with labeled grk 3′ UTR fragment 1 or the nos +3 fragment. The appropriately treated protein–RNA complexes were subjected to immunoprecipitation with a monoclonal anti-Sqd antibody and visualized after SDS-PAGE (Fig. 6B). The Sqd proteins, which are resolved as a distinct doublet at ∼42 kD, interact well with the 3′ UTR of grk, but interact much less strongly with the nos +3 3′ UTR RNA fragment.
We then further investigated the interaction between Sqd protein and the grk 3′ UTR through competition with excess cold competitor RNAs (Fig. 6C,D). In these experiments, the grk 3′ UTR fragment 1, which comprises nearly all of the grk 3′ UTR, was broken into two smaller fragments: grk 3′ UTR fragment 2 and grk 3′ UTR fragment 3. Incubating either 3′ UTR probe (fragment 2 or 3) with whole ovarian extracts allows detection of several protein RNA complexes, a 42-kD doublet, and two others of higher molecular mass. These complexes are completed away more efficiently by the addition of excess cold self RNA probe (∼20×, 100×, or 200× excess), than by excess nos +3 3′ UTR probe. On the basis of molecular mass, and the immunoprecipitation experiments, it appears that the 42-kD bands are Sqd protein. The largest protein that interacts with the grk 3′ UTR, which runs with an apparent molecular mass of ∼66 kD, appears to correspond to the ovarian protein Bruno. Bruno was identified as a trans-acting factor that represses the translation of unlocalized oskar mRNA (Kim-Ha et al. 1995). A specific binding sequence for Bruno has been identified and the grk 3′ UTR contains one such Bruno response element (BRE). Interestingly, however, both grk 3′ UTR fragments 2 and 3 appear to interact with the 66-kD protein. Although computer searching has revealed a single canonical BRE at the extreme 3′ end of the grk 3′ UTR, it is possible that Bruno can interact with other regions of the grk 3′ UTR, either directly or indirectly through an association with other proteins (in particular, Sqd). Nevertheless, previous studies have reported that Bruno does bind the grk 3′ UTR (Kim-Ha et al. 1995), and our data are consistent with those findings. As of this time, the identity of the 55-kD protein is unknown.
These studies demonstrate that Sqd protein can interact with grk mRNA, although Sqd will most likely interact with a variety of RNAs as well (see Discussion section). In fact, we have observed that excess nos RNA can act as a competitor for Sqd binding to a certain extent. However, these data do indicate that an interaction between Sqd and grk is mediated by sequences located within the 3′ UTR of the grk transcript. Data from our laboratory supports this finding, as studies of the cis-acting sequences required for grk mRNA localization have shown that the 3′ UTR is required for localization to the dorsoanterior corner of the oocyte (G. Thio and T. Schüpbach, in prep.).
Sqd protein can associate with Bruno protein
Because Sqd protein binds grk RNA directly and belongs to the class of hnRNPs implicated in nuclear mRNA export, it seems likely that Sqd functions in the nuclear export of grk mRNA. One complication to this model is that in the sqd1 mutant grk mRNA is still able to leave the nucleus and accumulate in the oocyte cytoplasm but not in the dorso-anterior corner. Therefore, the function of the Sqd protein appears to be in the regulated nuclear export of grk mRNA, such that Sqd is responsible for delivering the grk message to a cytoplasmic protein involved in its anchoring, possibly coupled to translation. Thus, one might expect that Sqd protein should interact with some cytoplasmic ovarian proteins.
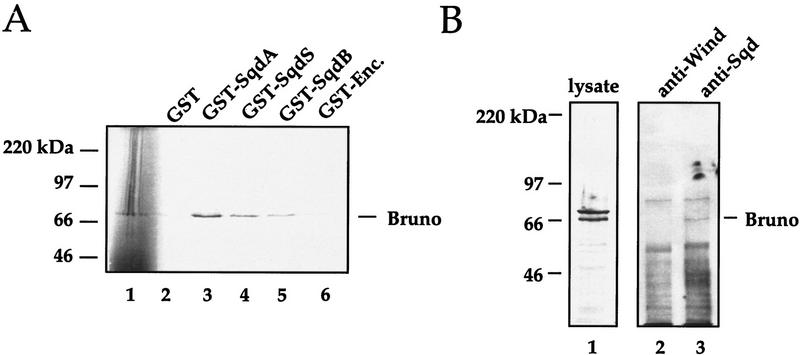
A number of proteins have been implicated in the translational regulation of grk mRNA, both positively (e.g., Encore and Vasa) and negatively (e.g., Bruno) (for review, see Nilson and Schüpbach 1999). Therefore, we decided to investigate whether Sqd protein could directly associate with any of these candidates. Using the in vitro association assay described above, we looked for interactions between the Sqd isoforms and Encore, Vasa, or Bruno. Although we were unable to detect a direct interaction between Sqd and Encore or Vasa (data not shown), Sqd protein did associate with Bruno protein in vitro (Fig. 7A). Although the Sqd–Bruno interaction we have observed in vitro is not extremely strong, it is very consistently observed over multiple experiments. In this representative experiment, the amount of Bruno brought down by the GST fusions was quantitated using a PhosphorImager. The cpm of Bruno associated with each of the fusions was as follows: GST only (10,632 cpm), GST–SqdA (121,092 cpm), GST–SqdS (53,872 cpm), GST–SqdB (32,805 cpm), and GST–Encore (9291 cpm). To further examine the association we performed coimmunoprecipitation experiments looking for an interaction between these two proteins in vivo. As shown in Figure 7B, immunoprecipitation of Sqd protein, but not Windbeutel protein, also brings down a protein of ∼66 kD that reacts with anti-Bruno antisera. Because of the high level of Sqd protein expression in the follicle cells of sqd1 mutant ovaries, it is not possible to use sqd1 lysates as a negative control for the coimmunoprecipitation. However, collectively, our data show that Sqd protein and Bruno protein can associate with one another. Moreover, these data provide evidence for a link between grk mRNA localization and translational regulation.
Figure 7.
Sqd protein associates with the translational repressor protein Bruno. (A) 35S-labeled Bruno protein (lane 1) was incubated with a series of GST fusion proteins, and after incubation with glutathione-coated–agarose beads, Bruno’s association was assessed by SDS-PAGE and autoradiography. The following GST fusions were tested for their ability to bind Bruno: (lane 2) GST only; (lane 3) GST–SqdA; (lane 4) GST–SqdS; (lane 5) GST–SqdB; and (lane 6) GST–Encore. (B) To further investigate the interaction between Sqd protein and Bruno protein, coimmunoprecipitation experiments were done. Whole cell lysates from wild-type ovaries were subjected to immunoprecipitation with a mouse anti-Sqd monoclonal or with a mouse anti-Wind antisera. The blots were probed with a rabbit anti-Bruno antisera: (lane 1) whole cell ovarian extract; (lane 2) anti-Wind immunoprecipitate; (lane 3) anti-Sqd immunoprecipitate.
Discussion
Previous studies have demonstrated that in sqd1 mutant females grk mRNA is mislocalized to the entire anterior circumference of the oocyte in late stage egg chambers. This mislocalized mRNA is translated efficiently and results in a corresponding expansion of Grk protein expression. As a consequence, too many follicle cells receive the Grk signal and adopt a dorsal cell fate that leads to dorsalization of the resulting egg and embryo (Neuman-Silberberg and Schüpbach 1993).
Sqd is a member of the shuttling class of hnRNPs and its homologs include human hnRNPA1, S. cerevisiae Npl3 and Hrp1, as well as C. tentans hrp36 (Pinol-Roma and Dreyfuss 1992; Lee et al. 1996; Visa et al. 1996; Kessler et al. 1997; Hoek et al. 1998). This family of RNA-binding proteins has been implicated in the nuclear export of mRNAs. On the basis of its homology to the shuttling hnRNPs, the nuclear accumulation of some of the Sqd isoforms, and their ability to interact with Transportin, in conjunction with our observation that Sqd protein can interact with sequences within the 3′ UTR of the grk transcript, we suggest that Sqd protein is a key mediator of regulated grk mRNA nuclear export. Because the absence of Sqd protein in the germ line leads to mislocalized grk mRNA rather than the nuclear retention of grk message, we suggest that the association of Sqd with grk message leads to the delivery of the mRNA from the nucleus to appropriately localized anchors in the dorsoanterior corner of the oocyte.
Sqd protein accumulation within the oocyte nucleus is required for grk mRNA localization
At least two lines of evidence indicate that Sqd protein must be present within the oocyte nucleus for grk mRNA to be localized properly during oogenesis. First, of the three Sqd isoforms only SqdS is detected within the oocyte nucleus and among the Sqd transgenic females only those expressing the SqdS isoform show properly localized grk mRNA. Second, the relationship between K10 and the distribution of Sqd protein also demonstrates the importance of Sqd accumulation within the oocyte nucleus. Although the nuclear import of SqdS protein is most likely driven by its association with Transportin, K10 function is required for the stable accumulation of Sqd in the oocyte nucleus. This places K10 function upstream of Sqd in the germ line. Accumulation of Sqd protein in the nurse cells is not affected, and in addition Sqd is detectable within the oocyte cytoplasm of K10 mutants. As the SqdS protein is the only Sqd isoform that is normally detected within the oocyte nucleus, the major effect of K10 must be on the nuclear retention of SqdS.
At this time it is unclear how K10 performs this function mechanistically. We have observed that K10 protein will physically interact with the Sqd isoforms. The only known motif within the K10 protein is a potential helix-turn-helix domain in the carboxyl terminus (Prost et al. 1988), but site-directed mutagenesis of this domain has revealed that this motif is unnecessary for K10 function (Cohen and Serano 1995). K10 could be responsible for the modification of Sqd protein in such a manner as to promote nuclear retention, or alternatively K10 could form a complex with Sqd protein that stabilizes its accumulation within the oocyte nucleus. In either case, the finding that Sqd protein requires the presence of K10 to accumulate in the oocyte nucleus further suggests that the phenotype of K10 mutant eggs is attributable to an effect on Sqd.
These data on Sqd and K10 also have interesting implications for grk transcription. It has been an open question as to whether grk mRNA is transcribed in the nurse cell nuclei, the oocyte nucleus, or both. The observations that Sqd protein can interact with grk transcripts and that Sqd must be present within the oocyte nucleus to localize properly grk mRNA demonstrate the critical importance of the oocyte nucleus in this process. These findings suggest that, at least in mid-to late oogenesis, the oocyte nucleus may be the critical site of grk mRNA transcription and that this is coupled directly to its localization. This interpretation is supported by the recent report of Saunders and Cohen (1999), which also suggests that the oocyte nucleus is the site of grk mRNA transcription.
Regulation of Grk activity is mediated through coupled RNA localization and translational regulation
Despite their extensive protein identity, the three Sqd isoforms behave strikingly differently from one another. As discussed, the three Sqd isoforms assume distinct intracellular distributions within the ovary. Furthermore, the three Sqd isoforms are also unique with respect to function. Sqd is required both somatically and in the germ line, and we have investigated the ability of each of the isoforms to function in these two capacities. Both SqdS and SqdB can provide at least some of the essential somatic functions of Sqd, whereas SqdA cannot. In contrast, the SqdS and SqdA isoforms can function during oogenesis to restore the patterning defects of sqd1 mutant females, whereas SqdB cannot do so. Significantly, although both SqdA and SqdS can rescue the patterning defects of sqd1 mutants, rescue is not complete, and the phenotype of partially rescued eggs laid by these females is qualitatively different. These differences demonstrate that the SqdS and SqdA isoforms perform unique roles in the regulation of Grk activity and that together these proteins cooperate to establish proper D-V patterning during oogenesis.
A comparison of the partially rescued Sqd eggs to those laid by sqd1 and K10 mutant females shows that there are at least two critical aspects of Grk localization that are required for correct D-V patterning: (1) an absolute exclusion of Grk signaling from the ventral side of the egg chamber, and (2) high levels of Grk signaling on the dorsal-most side of the egg chamber to establish the dorsal-most cell fates. The first aspect of Grk localization is normally achieved through the localization of grk mRNA itself. sqd1 and K10 mutant females are both defective in this process, and therefore, these females lay severely dorsalized eggs that have dorsal appendage material on the ventral and lateral sides of the eggshell. This aspect of Grk localization is accomplished efficiently in SqdS females and requires the accumulation of Sqd protein within the oocyte nucleus. In contrast, SqdA is only partially capable of restricting grk mRNA to the dorsal side of the egg chamber. In the overwhelming majority of SqdA egg chambers examined grk mRNA was not excluded from the ventral side of the oocyte. However, 60% of the eggs laid by SqdA females were wild type and Grk protein was often only detected on the dorsal side. Therefore, although SqdA is capable of restricting Grk protein from the ventral side of the egg chamber, this localization is not achieved primarily through the strict localization of grk mRNA itself, but appears to reveal a role for SqdA in translation.
The second aspect of Grk localization is the requirement for high levels of Grk protein on the dorsal-most side of the egg chamber, which produces the dorsal–midline cell fates (Deng and Bownes 1997; Queenan et al. 1997). Eggs from sqd1 and K10 females are not identical in this aspect of Grk function. In the case of K10 eggs, the dorsal–midline cell fates are expanded. Close examination of grk mRNA localization within K10 oocytes has shown that although grk mRNA expression has expanded to the ventral side, a higher concentration of grk mRNA, and consequently Grk protein, remains on the dorsal side of the egg chamber (Roth and Schüpbach 1994). In contrast, sqd1 females produce eggs that lack the dorsal–midline cell fates, and grk mRNA within these egg chambers is deposited symmetrically around the entire anterior circumference of the oocyte. Interestingly, partially rescued SqdS egg chambers do not produce the dorsal-most cell fate, but rather have an expansion of the more lateral dorsal appendage-secreting cell fates. Therefore, SqdS is not as efficient at achieving the peak levels of grk signaling needed for the specification of the dorsal–midline cell fates. Expression of SqdA, however, does allow the correct establishment of the dorsal–midline. Although the partially rescued eggs laid by SqdA females are classified as severely dorsalized, they appear more similar to K10 eggs than sqd1 eggs in that for the most part, the severely dorsalized SqdA eggs do not have the uniform crown of dorsal appendage material that sqd1 eggs have. This suggests that SqdA allows the highest levels of grk signaling to occur properly on the dorsal-most side of the egg chamber. The general similarities in phenotype of K10 mutant egg chambers and egg chambers that express SqdA further supports the conclusion that the major function of K10 is directed at SqdS. Taken together, the ability of SqdA to prevent translation of ventrally localized grk mRNA and its ability to provide peak levels of Grk protein required on the dorsal side of the egg chamber, strongly suggests that SqdA has a role in the accumulation of Grk protein. Moreover, the role of SqdA is both positive and negative, suggesting that SqdA may influence the association of grk mRNA with appropriate translational regulators.
A model for the role of Sqd in the regulation of Grk
The ability to investigate the roles of the individual Sqd isoforms in the regulation of Grk during Drosophila oogenesis has revealed that there are two key aspects of Grk regulation: grk mRNA localization and Grk protein accumulation. Our data demonstrate that both of these critical aspects of Grk regulation are accomplished by Sqd protein; however, these functions are performed differentially by the SqdS and SqdA isoforms. The severity of the sqd1 mutation, therefore, reflects the loss of function of both of these proteins within the germ line, thus causing the mislocalization of grk mRNA and the inappropriate accumulation of ectopic Grk protein. Restoration of either of these levels of regulation allows partial rescue of the D-V patterning defects of sqd1 mutants, but full rescue requires the function of both SqdS and SqdA.
Our data suggest that Sqd protein is a key regulator of both aspects of Grk regulation. The interaction of Sqd with the translational repressor protein Bruno provides a link between grk mRNA localization and its translational regulation. Bruno has been shown directly to have a role in the translational repression of unlocalized oskar (osk) mRNA. The interaction between Bruno and osk mRNA is mediated by a specific sequence within the osk message, which is termed BRE (Kim-Ha et al. 1995). As of yet, the molecular mechanism of Bruno action is not fully understood. However, correctly localized osk mRNA must somehow be relieved from the Bruno-mediated repression by specific trans-acting factors localized to the posterior of the embryo (Gunkel et al. 1998).
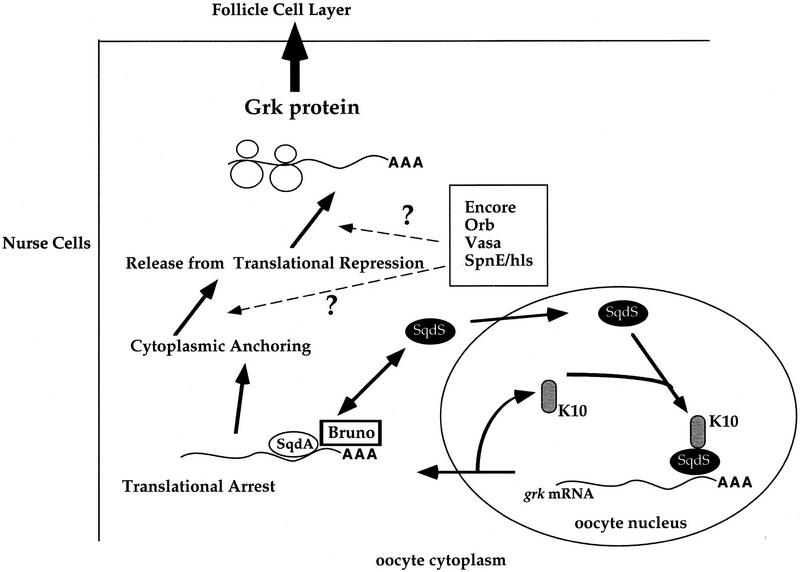
The interaction between Sqd and Bruno suggests that Bruno may play a role in the translational regulation of grk mRNA. In support of this, grk mRNA is known to contain a BRE within its 3′ UTR, and Bruno has been shown to bind the grk message (Kim-Ha et al. 1995; Fig. 7). In addition, it has been demonstrated that during late stages of oogenesis, Bruno protein is concentrated at the anterior end of the oocyte, in a position that is coincident with localized grk mRNA (Webster et al. 1998). On the basis of our protein interaction data, we suggest that it may serve as a translational repressor of unlocalized grk mRNA. As is the case for localized osk mRNA, the appropriately localized grk message would be relieved of its Bruno-mediated repression by other localized trans-acting factors. Moreover, the physical association between Sqd protein and Bruno protein suggests an appealing model to explain the similarity of the sqd and K10 mutant phenotypes. As discussed, these two female sterile mutations represent the only cases in which mislocalized grk mRNA is translated consistently and efficiently in all egg chambers. We propose that the role of Sqd protein is to take grk mRNA from the oocyte nucleus, recruit Bruno in the cytoplasm, and deliver grk mRNA to an anchor (Fig. 8). In the absence of nuclear Sqd, in either sqd or K10 egg chambers, grk RNA exits the nucleus by a generalized export mechanism, but does not associate efficiently with the repressor Bruno nor with the anchor. Because the interaction between grk mRNA and Bruno does not occur, even the unlocalized grk mRNA is translated efficiently. Using this model, Sqd protein provides the physical link between grk mRNA transport, localization, and its appropriately regulated translation.
Figure 8.
Model for the role of Sqd in grk mRNA localization and translation. In the oocyte nucleus SqdS protein binds to grk mRNA and to K10 protein, possibly as a complex. Then SqdS protein in a complex with grk mRNA and potentially other factors, exits the nucleus. Once in the cytoplasm, SqdS delivers grk mRNA to the SqdA isoform and the translational regulatory protein Bruno. The RNA–protein complex is localized and anchored, whereupon positive factors, such as Encore, Vasa, (Orb)oo18 RNA-binding, or (SpnE), Spindle E, relieve the translational repression by Bruno (and possibly interact with Sqd), leading to efficient translation of grk mRNA.
In summary, the analysis of the role of Sqd in Drosophila oogenesis has revealed several novel aspects of hnRNP function. Although the sqd1 mutation has provided the opportunity to study specifically the role of Sqd in oogenesis, the lethality of sqd null alleles demonstrates that it is required for other processes during development. Moreover, as grk itself is not required in other tissues and because Sqd is clearly required at other times during development, grk cannot be the only target of Sqd protein. Although we have observed that Sqd will interact with the grk transcript, it will most certainly bind to other mRNAs as well. Nevertheless, the requirement for Sqd during oogenesis and its specific effect on grk mRNA localization have revealed additional roles for hnRNPs. Specifically, these data have shown that through its direct association with grk mRNA, Sqd protein causes the mRNA to be localized properly to a very specific site within the oocyte cytoplasm. Moreover, through an association with Bruno, a defined translational regulator, Sqd protein provides a link between grk mRNA export and its regulated translation. Finally, one of the most unexpected findings of these studies is the differential functionality of the unique Sqd isoforms, both in terms of Sqd’s somatic function and its germ line requirement. Different isoforms, presumably through differential association with RNA-anchoring proteins, components of the translational machinery, or cellular trafficking molecules, can affect differentially localization and efficient translation of mRNAs.
Materials and methods
Fly stocks, transgenes
The previously described sqd1, sqdIX50, and fs(1)K10 mutant stocks were used (Wieschaus et al. 1978; Kelley 1993). To generate transgenic flies expressing the individual Sqd protein isoforms, a 15-kb genomic fragment containing the complete sqd locus was used. By digestion with EcoR1 and BglII restriction enzymes, the genomic region containing the alternatively spliced exons was removed. This genomic region was replaced with corresponding regions of the appropriate cDNAs, resulting in pseudo-minigene constructs. These constructs were introduced into flies using standard P-element-mediated transformation (Spradling and Rubin 1982). The Sqd transgenes were crossed into sqd1 and sqdIX50 backgrounds to generate stocks that were used to test the ability of each of the Sqd isoforms to rescue the germ-line and somatic requirements for sqd. Somatic function of the isoforms was measured by their ability to rescue the viability of the lethal allelic combination sqdIX50/Df(3R)urd. Similarly, the germ-line function of the isoforms was determined by their ability to rescue the patterning defects of eggs laid by sqd1 homozygous females.
Immunohistochemistry
Immunohistochemistry was performed as previously described (Matunis et al. 1994). Ovaries were fixed in 6% formaldehyde, 16.7 mm KP04, 75 mm KCl, 25 mm NaCl, 3.3 mm MgCl2, and heptane. After fixation, ovaries were washed in PBS + 0.3% Triton X-100, and incubated with appropriate primary and secondary antibodies. The following primary antibodies were used: monoclonal anti-Sqd (8G6) (kindly provided by G. Dreyfuss, University of Pennsylvania) (Matunis et al. 1992), polyclonal rat anti-K10 (a generous gift of R. Cohen, University of Kansas) (Prost et al. 1988), and mouse monoclonal anti-Grk (1D12), which was used at a dilution of 1:40 (Van Buskirk and Schüpbach 1999). After staining, ovaries were mounted in 70% PBS/30% glycerol and analyzed by confocal microscopy (BioRad MRC 600 confocal microscope).
Construction of Sqd–GST fusion proteins
To generate full-length GST fusion proteins of each of the Sqd isoforms, full-length cDNA clones were used as templates for PCR amplification. Primers were designed to produce in-frame fusions, and to produce convenient restriction enzyme sequences for directional cloning. The cDNAs were cloned into the pGex3 bacterial expression vector (Promega, Madison, WI) and transformed into JM109 bacteria. To induce fusion protein expression, cells were grown 1 hr at 37°C under no selection conditions (no antibiotic). After this time, induction was performed by the addition of IPTG to a final concentration of 0.5 mm, grown for an additional 3 hr, and cells were harvested by centrifugation. The cells were lysed in 1× PBS, containing 1% Triton X-100, 0.2 mg/ml lysozyme, 50 mm EDTA, and protease inhibitors. After incubation on ice, the cell lysates were subjected to three rounds of freeze–thawing, and cellular debris was pelleted. Glycerol was to a final concentration of 20%, and the supernatants were stored at −80°C. The GST fusions were purified from the supernatants by incubation with glutathione-coated agarose beads (Sigma, St. Louis, MO). To check the integrity and size of the GST fusions, purified proteins were resolved by SDS-PAGE and Western blotted with an anti-GST monoclonal antibody (SantaCruz Biotechnology, Santa Cruz, CA). Finally, the GST fusions were quantitated using a Bio-Rad Protein Kit.
In vitro association of recombinant proteins
35S-Labeled proteins were made using TNT reticulocyte transcription translation system (Promega, Madison, WI). The following plasmids were used: the K10 cDNA (pKXR; R. Cohen), Bruno cDNA (p4004; P. Macdonald, Stanford University) Vasa cDNA (P. Lasko, McGill University), and Drosophila Transportin (Berkeley Drosophila Genome Project). All binding studies were conducted in 1× PBS + 0.5% BSA. For each sample, the 35S-labeled proteins were precleared by incubation with 50 μl of a 50% slurry glutathione-coated agarose beads (Sigma, St. Louis, MO). Supernatants were transferred to new tubes, 50 μl of fresh glutathione beads and 1 μg GST fusion protein were added. After incubation at room temperature for 30 min, the beads were washed three times with PBS + 0.1% Tween 20, resuspended in 2× reducing SDS-PAGE sample buffer and resolved by SDS-PAGE. After drying, associated proteins were visualized by autoradiography and the amount of radioactivity associated with each GST fusion protein was quantitated using a PhosphorImager and ImageQuant Software (Molecular Dynamics).
Immunoprecipitations
Anti-Sqd immunoprecipitations were performed using the 8G6 mouse monoclonal anti-Sqd antibody (provided by G. Dreyfuss). Briefly, whole cell ovarian lysates were prepared by lysing Oregon R ovaries in lysis buffer [10 mm Tris (pH 7.4), 100 mm NaCl, 2.5 mm MgCl2, and 0.5% Triton X-100]. Lysates were cleared of cellular debris by centrifugation and supernatants were precleared by incubation with protein A-G conjugated–agarose beads (Santa Cruz Biotechnology, Santa Cruz, CA). Supernatants were subjected to immunoprecipitation with the anti-Sqd monoclonal and fresh protein A-G–agarose beads. Immunoprecipitates were washed with lysis buffer, resuspended in 2× SDS-PAGE reducing sample buffer and resolved by SDS-PAGE.
UV cross linking analysis
RNA probes were made by in vitro transcription. Radiolabeled probes were generated by including [32P]UTP in the transcription reaction mix in which 500 ng of linearized DNA template was used (Boehringer Mannheim, Indianapolis, IN). After synthesis, unincorporated nucleotides were removed by purification over Sephadex G-50 microcolumns (Pharmacia Biotech, Piscataway, NJ). Cold, competitor RNA probes were generated in a similar manner, except the amount of template DNA was increased to 2.5 or 30 μg and these probes were purified by ethanol precipitation. UV cross-linking analysis was performed as described (Hedley and Maniatis 1991). Whole cell ovarian lysates were prepared as described above, approximately two to three ovary equivalents were used per reaction. Ovarian proteins were incubated under the following reaction conditions: 1× binding buffer [5 mm HEPES (pH 7.4), 2 mm MgCl2, 150 mm KCl], 3 mg of tRNA, 75 mg of heparin, 1.5 ml of 10 mm DTT, and RNase inhibitor. Water or cold competitor RNAs were added to a final volume of 15 ml and the binding reactions incubated on ice for 10 min. 32 P-Labeled RNAs (5 × 105 to 1 × 106 cpm) were added to the binding reactions and they were further incubated on ice for 10 min. RNAs and proteins were cross-linked on ice in a Stratalinker UV cross-linker (Stratagene, La Jolla, CA). RNase was added, the samples incubated at room temperature for 20 min, and 2× reducing SDS-PAGE sample buffer added. Proteins and associated RNAs were resolved by SDS-PAGE and visualized by autoradiography.
Acknowledgments
We thank Robert Cohen, Gideon Dreyfuss, Elizabeth Gavis, Paul Lasko, and Paul Macdonald for their generous contributions of reagents. We also thank Eric Wieschaus, Elizabeth Gavis, and the members of the Schüpbach laboratory for their critical reading of this manuscript. A.N. was supported by the Howard Hughes Medical Institute and the American Cancer Society postdoctoral fellowship PF-98-011-01 DDC. This work was supported by the Howard Hughes Medical Institute and Public Health Service grant PO1 CA41086 to T.S.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL gschupbach@molbiol.princeton.edu; FAX (609) 258-1547.
References
- Cohen RS, Serano TL. mRNA localization and function of the Drosophila fs(1)K10 gene. In: Lipshitz HD, editor. Localized RNAs. Austin, TX: R.G. Landes Company; 1995. pp. 99–112. [Google Scholar]
- Deng WM, Bownes M. Two signalling pathways specify localised expression of the Broad-Complex in Drosophila eggshell patterning and morphogenesis. Development. 1997;124:4639–4647. doi: 10.1242/dev.124.22.4639. [DOI] [PubMed] [Google Scholar]
- Dreyfuss G, Matunis MJ, Pinol-Roma S, Burd CG. hnRNP proteins and the biogenesis of mRNA. Annu Rev Biochem. 1993;62:289–321. doi: 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- Gavis ER, Curtis D, Lehmann R. Identification of cis-acting sequences that control nanos RNA localization. Dev Biol. 1996;176:36–50. doi: 10.1006/dbio.1996.9996. [DOI] [PubMed] [Google Scholar]
- Gunkel N, Yano T, Markussen F-H, Olsen LC, Ephrussi A. Localization-dependent translation requires a functional interaction between the 5′ and 3′ ends of oskar mRNA. Genes & Dev. 1998;12:1652–1664. doi: 10.1101/gad.12.11.1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedley ML, Maniatis T. Sex-specific splicing and polyadenylation of dsx pre-mRNA requires a sequence that binds specifically to tra-2. Cell. 1991;65:579–586. doi: 10.1016/0092-8674(91)90090-l. [DOI] [PubMed] [Google Scholar]
- Hoek KS, Kidd GJ, Carson JH, Smith R. hnRNP A2 selectively binds the cytoplasmic transport sequence of myelin basic protein mRNA. Biochemistry. 1998;37:7021–7029. doi: 10.1021/bi9800247. [DOI] [PubMed] [Google Scholar]
- Kelley RL. Initial organization of the Drosophila dorsoventral axis depends on an RNA-binding protein encoded by the squid gene. Genes & Dev. 1993;7:948–960. doi: 10.1101/gad.7.6.948. [DOI] [PubMed] [Google Scholar]
- Kessler MM, Henry MF, Shen E, Zhao J, Gross S, Silver PA, Moore CL. Hrp1, a sequence-specific RNA-binding protein that shuttles between the nucleus and the cytoplasm, is required for mRNA 3′-end formation in yeast. Genes & Dev. 1997;11:2545–2556. doi: 10.1101/gad.11.19.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim-Ha J, Kerr K, Macdonald PM. Translational regulation of oskar mRNA by BRUNO, an ovarian RNA-binding protein, is essential. Cell. 1995;81:403–412. doi: 10.1016/0092-8674(95)90393-3. [DOI] [PubMed] [Google Scholar]
- Lee MS, Henry M, Silver PA. A protein that shuttles between the nucleus and the cytoplasm is an important mediator of RNA export. Genes & Dev. 1996;10:1233–1246. doi: 10.1101/gad.10.10.1233. [DOI] [PubMed] [Google Scholar]
- Matunis EL, Matunis MJ, Dreyfuss G. Characterization of the major hnRNP proteins from Drosophila melanogaster. J Cell Biol. 1992;116:257–269. doi: 10.1083/jcb.116.2.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matunis EL, Kelley RL, Dreyfuss G. Essential role for a heterogeneous nuclear ribonucleoprotein (hnRNP) in oogenesis: hrp40 is absent from the germ line in the dorsoventral mutant squid. Proc Natl Acad Sci. 1994;91:2781–2784. doi: 10.1073/pnas.91.7.2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael WM, Choi M, Dreyfuss G. A nuclear export signal in hnRNP A1: A signal-mediated, temperature-dependent nuclear protein export pathway. Cell. 1995;83:415–422. doi: 10.1016/0092-8674(95)90119-1. [DOI] [PubMed] [Google Scholar]
- Neuman-Silberberg FS, Schüpbach T. The Drosophila dorsoventral patterning gene gurken produces a dorsally localized RNA and encodes a TGFα-like protein. Cell. 1993;75:165–174. [PubMed] [Google Scholar]
- ————— Dorsoventral axis formation in Drosophila depends on the correct dosage of the gene gurken. Development. 1994;120:2457–2463. doi: 10.1242/dev.120.9.2457. [DOI] [PubMed] [Google Scholar]
- Nilson LA, Schüpbach T. EGF receptor signaling in Drosophila oogenesis. Curr Top Dev Biol. 1999;44:203–243. doi: 10.1016/s0070-2153(08)60471-8. [DOI] [PubMed] [Google Scholar]
- Pinol-Roma S, Dreyfuss G. Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature. 1992;355:730–732. doi: 10.1038/355730a0. [DOI] [PubMed] [Google Scholar]
- Pollard VW, Michael WM, Nakielny S, Siomi MC, Wang F, Dreyfuss G. A novel receptor-mediated nuclear protein import pathway. Cell. 1996;86:985–994. doi: 10.1016/s0092-8674(00)80173-7. [DOI] [PubMed] [Google Scholar]
- Prost E, Deryckere F, Roos C, Haenlin M, Pantesco V, Mohier E. Role of the oocyte nucleus in determination of the dorsoventral polarity of Drosophila as revealed by molecular analysis of the K10 gene. Genes & Dev. 1988;2:891–900. doi: 10.1101/gad.2.7.891. [DOI] [PubMed] [Google Scholar]
- Queenan AM, Ghabrial A, Schüpbach T. Ectopic activation of torpedo/Egfr, a Drosophila receptor tyrosine kinase, dorsalizes both the eggshell and the embryo. Development. 1997;124:3871–3880. doi: 10.1242/dev.124.19.3871. [DOI] [PubMed] [Google Scholar]
- Roth S, Schüpbach T. The relationship between ovarian and embryonic dorsoventral patterning in Drosophila. Development. 1994;120:2245–2257. doi: 10.1242/dev.120.8.2245. [DOI] [PubMed] [Google Scholar]
- Saunders C, Cohen RS. The role of oocyte transcription, the 5′UTR, and translational repression and derepression in Drosophila gurken mRNA and protein localization. Mol Cell. 1999;3:43–54. doi: 10.1016/s1097-2765(00)80173-2. [DOI] [PubMed] [Google Scholar]
- Siomi H, Dreyfuss G. A nuclear localization domain in the hnRNP A1 protein. J Cell Biol. 1995;129:551–560. doi: 10.1083/jcb.129.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi MC, Fromont M, Rain JC, Wan L, Wang F, Legrain P, Dreyfuss G. Functional conservation of the transportin nuclear import pathway in divergent organisms. Mol Cell Biol. 1998;18:4141–4148. doi: 10.1128/mcb.18.7.4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling A, Rubin GM. Transposition of cloned P elements into Drosophila germ line chromosomes. Science. 1982;218:341–347. doi: 10.1126/science.6289435. [DOI] [PubMed] [Google Scholar]
- Van Buskirk C, Schüpbach T. Versatility in signalling: Multiple responses to EGF receptor activation during Drosophila oogenesis. Trends Cell Biol. 1999;9:1–4. doi: 10.1016/s0962-8924(98)01413-5. [DOI] [PubMed] [Google Scholar]
- Visa N, Alzhanova-Ericsson AT, Sun X, Kiseleva E, Bjorkroth B, Wurtz R, Daneholt B. A pre-mRNA-binding protein accompanies the RNA from the gene through the nuclear pores and into polysomes. Cell. 1996;84:253–264. doi: 10.1016/s0092-8674(00)80980-0. [DOI] [PubMed] [Google Scholar]
- Webster PJ, Liang L, Berg CA, Lasko P, Macdonald PM. Translational repressor bruno plays mulitple roles in development and is widely conserved. Genes & Dev. 1998;11:2510–2521. doi: 10.1101/gad.11.19.2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weighardt F, Biamonti G, Riva S. Nucleocytoplasmic distribution of hnRNP proteins: A search for the targeting domains in hnRNP A1. J Cell Sci. 1995;108:545–555. doi: 10.1242/jcs.108.2.545. [DOI] [PubMed] [Google Scholar]
- Wieschaus E, Marsh JL, Gehring W. fs(1)K10, a germline-dependent female sterile mutation causing abnormal chorion morphology in Drosophila melanogaster. Roux’s Arch Dev Biol. 1978;184:75–82. doi: 10.1007/BF00848670. [DOI] [PubMed] [Google Scholar]