TORC1 is a central regulator, controlling cell growth in response to a variety of signals. This study illustrates the crystal structure of the Gtr1p–Gtr2p complex, the yeast homologs of the Rag GTPases that relay amino acid signals to TORC1. The structure provides insight into the domains required for dimerization as well as for Raptor and p18 recognition.
Keywords: Rag GTPases, Gtr1p, Gtr2p, structure, raptor, TORC1
Abstract
The target of rapamycin (TOR) complex 1 (TORC1) is a central cell growth regulator in response to a wide array of signals. The Rag GTPases play an essential role in relaying amino acid signals to TORC1 activation through direct interaction with raptor and recruitment of the TORC1 complex to lysosomes. Here we present the crystal structure of the Gtr1p–Gtr2p complex, the Rag homologs from Saccharomyces cerevisiae, at 2.8 Å resolution. The heterodimeric GTPases reveal a pseudo-twofold symmetric organization. Structure-guided functional analyses of RagA–RagC, the human homologs of Gtr1p–Gtr2p, show that both G domains (N-terminal GTPase domains) and dimerization are important for raptor binding. In particular, the switch regions of the G domain in RagA are indispensible for interaction with raptor, and hence TORC1 activation. The dimerized C-terminal domains of RagA–RagC display a remarkable structural similarity to MP1/p14, which is in a complex with lysosome membrane protein p18, and directly interact with p18, therefore recruiting mTORC1 to the lysosome for activation by Rheb. Our results reveal a structural model for the mechanism of the Rag GTPases in TORC1 activation and amino acid signaling.
The mammalian target of rapamycin (mTOR) is an atypical protein kinase related to ATM and the DNA-PK subfamily. TOR is highly conserved from yeast to mammals and forms two distinct functional complexes: TOC complex 1 (TORC1) and TORC2. mTORC1 is a central cell growth regulator that integrates a wide range of growth stimulatory and inhibitory signals to regulate cell growth (Wullschleger et al. 2006). Key substrates of mTORC1 include S6K and 4EBP1; therefore, mTORC1 activation promotes cell growth by stimulating translation. In addition, mTORC1 plays a critical role in inhibiting catabolic processes, such as autophagy. mTORC1 inhibits autophagy at least in part by phosphorylating and inhibiting the autophagy-initiating kinase ULK1. Uncontrolled TORC1 activation has been observed in human diseases such as cancer (Inoki et al. 2005; Guertin and Sabatini 2007), indicating an important role of tight mTORC1 regulation under physiological conditions. Rapamycin is a specific TORC1 inhibitor, and its analogs are being used for cancer treatment and immunosuppression.
Growth factors act through PI3K, Akt, TSC1/TSC2, and Rheb to stimulate TORC1 (Wullschleger et al. 2006). In addition to growth factors, mTORC1 activation requires energy sufficiency (high ATP levels) and nutrients (amino acids). The AMP-dependent protein kinase AMPK plays a critical role in mTORC1 inhibition in response to cellular energy stress. Amino acids are one of the most important signals for mTORC1 activation. In the absence of amino acids, neither growth factors nor glucose (as a source of energy) can efficiently activate mTORC1. It has been shown that the Rag GTPases, which are distantly related to Ras (Kim et al. 2008; Sancak et al. 2008), play an essential role in TORC1 activation in response to amino acid signals. The requirement of amino acids for TORC1 activation and the involvement of Rag GTPases in amino acid signaling are highly conserved in all eukaryotes. For example, the Saccharomyces cerevisiae Gtr1 and Gtr2, which correspond to human RagA/RagB and RagC/RagD (Binda et al. 2009), respectively, also relay amino acid sufficiency to TORC1 activation in yeast.
Rag GTPases are unique in that they form heterodimers, as RagA or RagB dimerizes with RagC or RagD (Sekiguchi et al. 2001). Similarly, the yeast Gtr1 forms a heterodimer with Gtr2. Another unique property of the Rag GTPases is that the two Rag GTPases in the heterodimer bind guanine nucleotides in an apposing manner; i.e., one subunit binds GTP, and the other binds GDP. Only when RagA or RagB exist in the GTP-bound form, the heterodimer is active to stimulate TORC1 through a direct interaction with raptor (Kim et al. 2008; Sancak et al. 2008). Consistently, addition of amino acids promotes GTP binding of RagA or RagB in the heterodimer. Recently, it has been proposed that the Rag GTPases mainly function to recruit mTORC1 to lysosome, where mTORC1 can be activated by the lysosomal-localized Rheb. Once recruited to the lysosomes by the Rag GTPases, TORC1 is activated by the Rheb GTPase, another Ras family member (Kim et al. 2008; Sancak et al. 2008). The lysosomal localization of Rag is mediated by a direct interaction with the lysosomal membrane protein p18 and its associated protein, p14/MP1 (Sancak et al. 2010).
Recent studies have established an essential role of Rag GTPases in amino acid signaling; however, the molecular mechanism of Rag GTPases in mTORC1 activation is largely unknown. In this study, we report the three-dimensional structure of the Gtr1p–Gtr2p complex. Our structure–function studies have revealed new molecular insights into the molecular basis of mTORC1 regulation by Rag GTPases through interaction with Raptor and the lysosomal protein p18.
Results and Discussion
Overall structure of Gtr1p–Gtr2p, the yeast homolog of RagA–RagC
Previous studies have shown that the function of Rag GTPase complexes are highly conserved across species from yeast to humans (Bun-Ya et al. 1992; Schurmann et al. 1995; Hirose et al. 1998; Nakashima et al. 1999; Sekiguchi et al. 2001; Dubouloz et al. 2005; Binda et al. 2009). The three-dimensional structures are also predicted to be conserved from yeast to humans, with the primary sequence identity of 49% for RagA/Gtr1p and 43% for RagC/Gtr2p (similarity of 75% for RagA/Gtr1p and 76% for RagC/Gtr2p) (Supplemental Fig. S1). To understand the mechanism of Rag GTPase-mediated TORC1 activation, we sought to determine the structure of Rag GTPases from yeast to mammals. After numerous trials, we succeeded in crystallizing the Gtr1p–Gtr2p complex, the yeast homolog of RagA–RagC, and the final model was refined to 2.8 Å resolution (Fig. 1A; Supplemental Fig. S2). The statistics for the structure determination are summarized in Supplemental Table S1. Gtr1p and Gtr2p form a very stable heterodimer in solution, which was copurified from Escherichia coli (Supplemental Fig. S3). The two proteins used for crystallization were both GTP-bound forms, through incorporation of GMPPNP, a nonhydrolyzable GTP analog commonly used in structure studies.
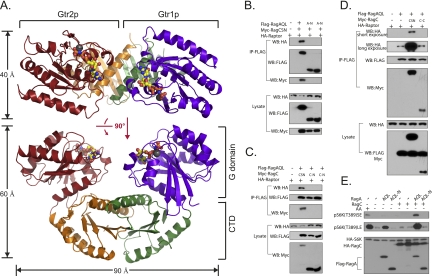
Figure 1.
Overall structure of the Gtr1p–Gtr2p. (A) Overall structure of the Gtr1p–Gtr2p complex as a ribbon representation in two different views. G domains of Gtr1p and Gtr2p bound to GMPPNP are colored in blue and red, respectively, and CTDs are colored in green and orange, respectively. GMPPNP is shown as a ball-and-stick representation, and magnesium atoms are shown as black balls. (B,C) Dimerization is required for RagA–RagC to interact with raptor. Different RagA and RagC constructs were cotransfected with raptor into HEK293 cells. Protein interaction was determined by coimmunoprecipitation. RagAQL and RagCSN are mutants restricted to RagAGTP (Q66L) and RagCGDP(S75N), respectively. (A-N) G domain of RagA; (C-N) G domain of RagC; (IP) immunoprecipitation; (WB) Western blot. (D) The G domain of RagC is required to promote the interaction between RagA and raptor. (CSN) RagCSN (RagCGDP); (C-C) CTD of RagC. (E) The CTD of RagA is required for both basal and RagC-enhanced activity to stimulate TORC1. TORC1 activity was indirectly measured by the level of S6K phosphorylation. RagA and RagC constructs were cotransfected with HA-S6K into HEK293 cells. For amino acid starvation, cells were starved for amino acids for 1 h before harvesting. Amino acid starvation is denoted as AA−. Sample cultured in complete medium is denoted as AA+. Phosphorylation and protein levels were determined by immunoblotting with the indicated antibodies.
As shown in Figure 1A, both Gtr1p and Gtr2p comprise two domains: an N-terminal GTPase domain and a C-terminal domain (designated as the G domain and CTD, respectively). Consistent with their sequence homology, Gtr1p and Gtr2p adopt a similar fold, with a root-mean-squared deviation (RMSD) of 3.92 Å for 216 aligned Cα atoms and, if corresponding domains are superimposed individually, 1.88 Å for 171 Cα atoms of the G domains and 2.68 Å for 99 Cα atoms of the CTDs (Supplemental Fig. S4).
The Gtr1p–Gtr2p heterodimer adopts a pseudo-twofold symmetry and resembles a U-shaped horseshoe. The two G domains of Gtr1p–Gtr2p are located on the same side of the complex, with the corresponding surfaces of two G domains facing opposite directions (Fig. 1A; Supplemental Fig. S5). Interestingly, different from dimerization of G domains in the reported structures (Focia et al. 2004; Low et al. 2009; Chappie et al. 2010; Gao et al. 2010), no direct interaction was found between the two G domains in the Gtr1p–Gtr2p heterodimer structure. The dimerization is mediated by CTDs of both proteins, and the dimer interface is far away from the nucleotide pocket (Fig. 1A). The Gtr1p–Gtr2p heterodimer represents a new architecture among all GTPase structures.
In each monomer, the G domain forms extensive interactions with the CTD, with buried surfaces (G domain and CTD) of 882.2 Å2 for Gtr1p and 878.2 Å2 for Gtr2p, respectively. In the Gtr1pGTP–Gtr2pGTP heterodimer structure, with these intramolecular interactions and extensive interaction between two CTDs (buried surface of 1259 Å2), the Gtr1p–Gtr2p complex adopts a rigid conformation and the two G domains adopt fixed orientation to each other. Nucleotide exchanges in G domains may not change the overall conformation of the complex because the switch regions are far away from both dimer and intramolecular interfaces (Supplemental Fig. S6). Thus, the Gtr1p–Gtr2p heterodimer may keep a rigid overall fold and undergo conformational changes mainly on switch regions upon nucleotide exchanges, through which they recognize raptor and activate mTORC1.
Both G domains are required for raptor interaction
Previous studies showed that the function of Rag/Gtr is highly conserved between yeast and mammals, and the interaction of Rag with raptor in mammalian cells and Gtr with Kog1 in yeast is also conserved (Bun-Ya et al. 1992; Schurmann et al. 1995; Hirose et al. 1998; Nakashima et al. 1999; Sekiguchi et al. 2001; Dubouloz et al. 2005). Compared with yeast TORC1, more extensive biochemical studies have been reported based on human mTORC1 and the Rag complexes (Kim et al. 2008; Sancak et al. 2008, 2010); thus, we used the human RagA–RagC complex to investigate their function in TORC1 binding and activation, guided by structure analyses of the Gtr1p–Gtr2p complex and highly conserved primary sequences (Supplemental Fig. S1). Previous studies have shown a direct interaction between RagA/C and raptor, which was confirmed by our in vitro pull-down assays using purified RagA/C and raptor proteins (Supplemental Fig. S7). Coimmunoprecipitation with raptor and Western blotting for S6K phosphorylation were performed to test the ability of Rag in raptor binding and TORC1 activation, respectively. RagA/C(N) and RagA/C(C) denote the G domain and CTD, respectively. RagAGTP and RagCGDP are mutants restricted to GTP-bound and GDP-bound, respectively.
We first examined whether both G domains are required for proper function of Rag GTPases. RagAGTP–RagCGDP showed strong interaction with raptor, whereas RagA(N)GTP–RagCGDP and RagAGTP–RagC(N)GDP showed little interaction with raptor (Fig. 1B,C). Moreover, expression of RagC(C) did not enhance the weak interaction between RagAGTP and raptor (Fig. 1D). Consistent with these data, RagA(N)GTP could not activate TORC1 in the absence of amino acids (Fig. 1E). These results indicate that both the G domains of RagA and RagC and dimerization are important for raptor binding and mTORC1 activation.
Although the Gtr1p–Gtr2p heterodimer adopts a pseudo-twofold symmetry and both G domains adopt a similar fold, structure comparison shows that the surface features of the G domains from Gtr1p and Gtr2p are rather different. For example, the surface region close to switch I and II of the Gtr1p G domain is more hydrophobic than that of Gtr2p, which is more acidic. Gtr2p and RagC share a similar electrostatic potential distribution in the switch regions of their respective G domains (Supplemental Fig. S8). Together with the finding that both G domains are required for raptor binding, these analyses suggest that Gtr1p/RagA and Gtr2p/RagC may contribute differently to raptor interaction, and together provide the specificity for raptor recognition.
RagA surface for raptor recognition
Next, we mapped the binding interface between raptor and Rag GTPases using coimmunoprecipitation. We first tested whether both G domains are equally important for raptor interaction. As shown in Figure 2A, wild-type RagA, together with either GTP-bound, GDP-bound, or the T90A/L93A/T96A mutant of RagC, strongly bound raptor, while GDP-bound RagA could not. These results indicate that the interaction with raptor was mainly determined by the nucleotide loading status of RagA, although the nucleotide-binding status of RagC modestly influenced the ability of the RagA/C heterodimer to bind raptor. This observation is consistent with previous studies, which have shown that the GTP-bound RagA or RagB interacts with raptor and activates TORC1 when in complex with RagC or RagD; in contrast, the Rag dimer cannot bind mTORC1 if RagA is in the GDP form regardless of the nucleotide-binding status of the associated RagC or RagD (Kim et al. 2008; Sancak et al. 2008). These results suggest that RagA/B play a major role in raptor interaction. Thus, we focus on the RagA G domain to investigate the raptor recognition.
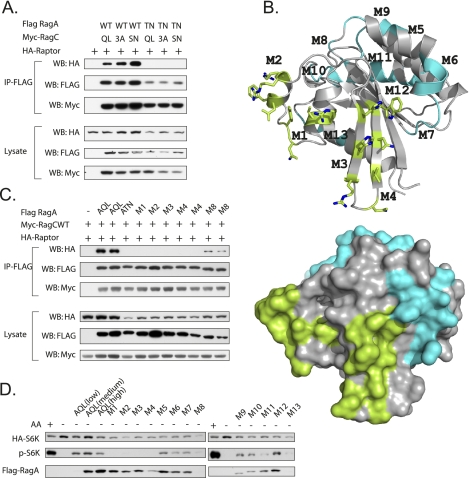
Figure 2.
Mapping the raptor-interacting surface on RagA. (A) RagA plays a major role in raptor binding. The interaction between raptor and cotransfected RagA or RagC was examined by coimmunoprecipitation. 3A is the T90A/L93A/T96A mutation of RagC, in which mutations of the corresponding residues in Ras have been shown to abolish the effector binding. (B) G domain of Gtr1p is shown in a ribbon representation (left panel) and a surface representation (right panel). Corresponding residues involved in composite RagA mutations (M1–M4) are indicated with a stick representation and are colored in green, and residues mutated in M5–M13 are colored in cyan on the surface, as shown in Supplemental Figure S1 and Supplemental Table S2. All mutants were generated based on RagAQL(RagAGTP). (C) The regions close to switch I and II in the RagA G domain are important for raptor interaction. Interaction between raptor and cotransfected RagA mutants was examined by coimmunoprecipitation. (D) The raptor interaction-defective RagA mutants cannot activate TORC1. RagA mutants were cotransfected with HA-S6K into HEK293 cells, and phosphorylation of HA-S6K in the absence of amino acids (indicating the activity of RagA) was determined.
Based on the structural information of Gtr1p, 13 composite mutations were made in RagA-QL, which is a GTP-bound mutant, in order to map the raptor-interacting surface. The involved residues represent small patches of alanine substitutions on the surface area of the RagA G domain (Fig. 2B; Supplemental Table S2). Among the 13 mutants, four (M1–M4) abolished interaction with raptor and lost the ability to activate TORC1 (Fig. 2C,D). Notably, the four mutations are close to the P loop, switch I, and switch II regions. The above data indicate that the surface area (α1, α2 and β2, β3) of the RagA G domain is important for raptor binding and TORC1 activation. This observation is consistent with a notion that nucleotide exchanges alter the surface feature of switch I and II, and thus regulate raptor binding affinity.
Dimerized CTDs of the Rag complexes are required for function
In the structure of Gtr1p–Gtr2p, the CTDs of both proteins contain a central five-stranded anti-parallel β sheet, sandwiched by a long helix on one side of the G domains and two helices on the other side. Gtr1p and Gtr2p form a heterodimer through an edge-to-edge (β9–β9) arrangement of their β sheets (Supplemental Fig. S2). The dimerized CTDs form a compact three-layered structure, with a 10-stranded anti-parallel β sheet sandwiched by two α helices on the concave face and four α helices on the convex face (Fig. 1A; Supplemental Fig. S2). The dimerization is mediated by a network of hydrogen bonds and hydrophobic interactions, and the residues involved in dimerization are highly conserved from yeast to mammals (Fig. 3A,B). Intriguingly, in dimerized CTDs, Gtr2p α8 interacts with α8 and four β strands of Gtr1p, whereas α8 of Gtr1p only interacts with α8 and two β strands of Gtr2p. Structure analyses of Gtr1p–Gtr2p interaction show that more residues in Gtr1p than Gtr2p are involved in dimer formation, suggesting a lower contribution of individual residues in Gtr1p/RagA than Gtr2p/RagC for dimer formation (Supplemental Fig. S9).
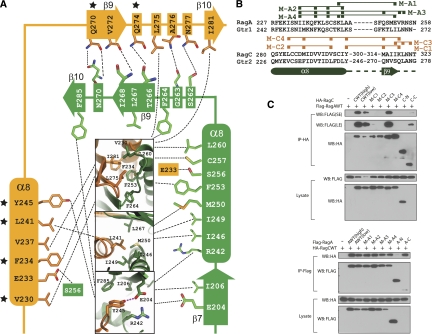
Figure 3.
Mapping the dimerization critical residues. (A) Detailed interaction of Gtr1p–Gtr2p CTDs. Residues involved in hydrogen bond formation are connected with a dashed line directly, and hydrophobic interactions are linked by the central boxes with the detailed interactions shown. Critical residues for dimer formation are indicated as black stars. (B) Sequence alignment of critical regions for dimer formation in the CTDs of Rag GTPases. Composite RagA and RagC mutations are indicated above the sequence and are summarized in Supplemental Table S3. (C) The α8 and β9 of RagC are critical for dimer formation with RagA. Different RagA or RagC mutants were cotransfected as indicated. The interaction was determined by coimmunoprecipitation and Western blot. (SE) Short exposure; (LE) long exposure; (AWT) wild-type RagA; (CWT) wild-type RagC; [AWT(low)] transfection of 100 ng of DNA; [AWT(high)] transfection of 200 ng of DNA.
Since the CTDs’ dimerization is important for the function of Rag GTPases in TORC1 activation, we studied the interaction between the two CTDs. To determine residues critical for dimerization, we generated mutations in RagA and RagC based on the Gtr1p–Gtr2p structure and sequence alignment (Fig. 3B; Supplemental Table S3). Three mutations (M-C1, M-C2, and M-C4) of RagC abolished the interaction with RagA (Fig. 3C). In contrast, similar mutations in RagA did not abolish the interaction with RagC (Fig. 3C), consistent with previous structure analyses (Supplemental Fig. S9).
A Dali search with the Gtr1p–Gtr2p structure indicates that p14/MP1 adopt folds similar to the dimerized CTDs of Gtr1p–Gtr2p (Fig. 4A; Supplemental Fig. S9; Kurzbauer et al. 2004; Lunin et al. 2004). This is a rather surprising finding because these proteins do not share sequence homology. Interestingly, p14/MP1 are essential for lysosomal localization of RagB–RagD and form a complex with p18, which also binds with Rag GTPases (Sancak et al. 2010). However, surface feature differences between the two complexes suggest that they may not interact with p18 in a similar region. Notably, Ego3p, which was identified to interact with Gtr1p–Gtr2p and form an EGO complex, adopts a similar fold to p14/MP1 and was thought to be a potential p14/MP1 functional homolog in yeast, suggesting a conserved mechanism for Rag GTPase localization (Gao and Kaiser 2006; Kogan et al. 2010). Why p14/MP1 adopts a similar fold to the CTDs of Rag GTPases and how p14/MP1 is involved in Rag GTPase-mediated TORC1 activation need to be further investigated.
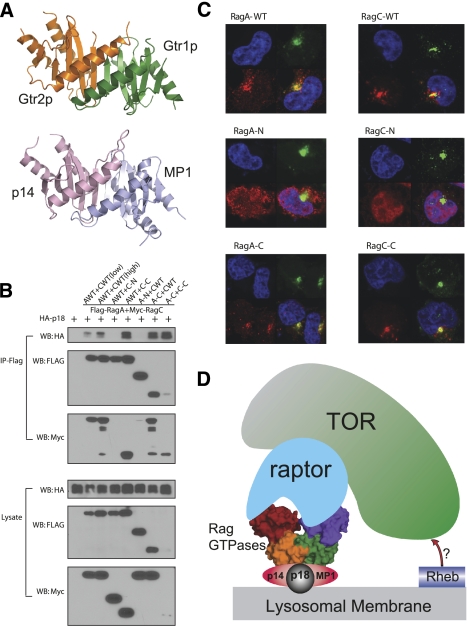
Figure 4.
The CTDs of Rag GTPases share similar structures with p14/MP1 and are responsible for p18 interaction and lysosomal localization. (A) Structure comparison of Gtr1p–Gtr2p CTDs and the p14/MP1 complex. The structures are shown in a ribbon representation, and Gtr1p and Gtr2p CTDs are colored green and orange, respectively, while p14 and MP1 are colored pink and light blue, respectively. (B) The CTD dimer of RagA–RagC interacts with p18. RagA and RagC constructs were cotransfected with the p18 construct and the protein interaction was determined by coimmunoprecipitation. (C) The CTDs of RagA and RagC are necessary and sufficient for lysosomal localization. Different deletion mutants were transfected in 293 cells. The transfected Flag-RagA or HA-RagC was stained (red) along with DNA (blue) and lysosomal marker LAMP2 (green). (D) Working model of Rag GTPases in TORC1 activation. In the cartoon, the Rag CTDs interact with p18 and p14/MP1 to target the GTPases to lysosomes. The G domains of Rag associate with raptor, thus recruiting TORC1 to lysosomes for activation.
It has been shown that p18, together with p14/MP1, interacts with and recruits Rag GTPases to the lysosomal membrane (Sancak et al. 2010). We next studied which regions of Rag GTPase directly interact with p18. The coimmunoprecipitation results show that both CTDs are required for interaction with p18, and dimerization of Rag CTDs is necessary and sufficient for binding to p18 (Fig. 4B; Supplemental Fig. S11A,B). Consistent with the protein interaction results, immunofluorescence experiments showed that the CTDs of both RagA and RagC are necessary and sufficient for colocalization with the lysosomal marker LAMP2 (Fig. 4C). These data indicate that the localization of Rag GTPases is mediated by the interaction with p18 through their dimerized CTDs, which is consistent with the notion that the nucleotide loading status of Rag GTPases does not affect their cellular localization (Sancak et al. 2010).
This study presents the three-dimensional structure of Rag GTPase heterodimers and the structural basis of Rag GTPase-mediated raptor recognition and p18 association. Based on our data, we propose a working model for Rag GTPases in TORC1 recruitment and activation. In this model, the CTDs of Rag GTPase interact with p18, which is permanently anchored to the lysosomal surface (Fig. 4D). p14/MP1 may facilitate the interaction between p18 and Rag GTPases in an unknown mechanism. In a manner depending on GTP-binding status, the Rag heterodimer interacts with raptor mainly via the surfaces close to switch I and II on RagA, although RagC is also required. Through these interactions, the TORC1 complex is recruited to a lysosomal compartment where it is activated, presumably by the lysosome-localized Rheb. Our results provide structural insights into how the Rag GTPases recruit TORC1 to the p18 regulator complex, and thus activation of TORC1 by amino acids. Interestingly, the two p18-interacting complexes, the Gtr1p–Gtr2p CTD domains and p14/MP1, share remarkably similar three-dimensional structures. The function of the RagC/D G domain also needs to be further investigated. Although the G domain of RagC/D plays a minor role in raptor interaction, the nucleotide loading status still regulates the binding affinity. One possibility is that raptor mainly interacts with the RagA G domain, and the GDP-bound RagC G domain may facilitate the interaction and thus provide specific recognition and regulation. A key remaining issue is the activation/nucleotide exchange of Rag GTPases in response to the amino acid signal. Notably, VAM6, also known as VPS39, has been suggested as a guanine nucleotide exchange factor for Gtr1p in yeast (Binda et al. 2009). Future study of amino acids in regulating nucleotide exchange of Rag GTPases will shed new light on this important signaling pathway in cell growth regulation.
Materials and methods
Protein purification and crystallization
Protein expression and purification were performed as described previously (Li et al. 2010). In brief, the ORFs of gtr1p and gtr2p were subcloned into a modified pETDuet-1 vector (Novagen) for bicistronic protein expression in the Escherichia coli strain BL21(DE3). The Gtr1p–Gtr2p complex was purified using Ni-NTA affinity columns, anion exchange, and gel filtration. The crystals were obtained using the hanging-drop, vapor diffusion method with reservoir solution containing 0.1 M HEPES (pH 7.0), 10% PEG monomethyl ether 5000, and 5% v/v Tacsimate (pH 7.0) at 277K. Crystals of the selenomethionine derivative of Gtr1p–Gtr2p were grown under similar conditions.
Data collection and structure determination
Se-SAD (single-wavelength anomalous diffraction) data were collected at a wavelength of 0.97916 Å and the diffraction was extended to 2.8 Å resolution (Hendrickson 1991). Data were indexed, integrated, and scaled using the program XDS (Kabsch 1988). Phases were initially determined by Se-SAD, and automatic model building was performed. All refinements were performed with the restraint of an experimental phase using the refinement module phenix.refine of the PHENIX package (Adams et al. 2002). The model quality was checked with the PROCHECK program (Laskowski et al. 1993).
Other procedures—including antibodies, plasmids, cell culture, transfection, immunofluorescence, and immunoprecipitation—are described in the Supplemental Material.
Accession number
The atomic coordinates of the Gtr1p–Gtr2p has been deposited in the Protein Data Bank with accession code 3R7W.
Acknowledgments
We thank Dr. Jiawei Wang at Tsinghua University for the help in data processing and structure determination. We thank staff members of beamline BL17U at SSRF (China) and other members in the Guan laboratory and the Xu laboratory for technical help. This work was supported by grants (to Y.X.) from the National Basic Research Program of China (2011CB918600 and 2009CB918600), the National Natural Science Foundation of China (31030019, 11079016, and 30870493), and the International Collaboration Program from the Science and Technology Commission of Shanghai Municipality (10430709300), and grants from the NIH (to K.L.G.).
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.16968011.
References
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC 2002. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr 58: 1948–1954 [DOI] [PubMed] [Google Scholar]
- Binda M, Peli-Gulli MP, Bonfils G, Panchaud N, Urban J, Sturgill TW, Loewith R, De Virgilio C 2009. The Vam6 GEF controls TORC1 by activating the EGO complex. Mol Cell 35: 563–573 [DOI] [PubMed] [Google Scholar]
- Bun-Ya M, Harashima S, Oshima Y 1992. Putative GTP-binding protein, Gtr1, associated with the function of the Pho84 inorganic phosphate transporter in Saccharomyces cerevisiae. Mol Cell Biol 12: 2958–2966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappie JS, Acharya S, Leonard M, Schmid SL, Dyda F 2010. G domain dimerization controls dynamin's assembly-stimulated GTPase activity. Nature 465: 435–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubouloz F, Deloche O, Wanke V, Cameroni E, De Virgilio C 2005. The TOR and EGO protein complexes orchestrate microautophagy in yeast. Mol Cell 19: 15–26 [DOI] [PubMed] [Google Scholar]
- Focia PJ, Shepotinovskaya IV, Seidler JA, Freymann DM 2004. Heterodimeric GTPase core of the SRP targeting complex. Science 303: 373–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M, Kaiser CA 2006. A conserved GTPase-containing complex is required for intracellular sorting of the general amino-acid permease in yeast. Nat Cell Biol 8: 657–667 [DOI] [PubMed] [Google Scholar]
- Gao S, von der Malsburg A, Paeschke S, Behlke J, Haller O, Kochs G, Daumke O 2010. Structural basis of oligomerization in the stalk region of dynamin-like MxA. Nature 465: 502–506 [DOI] [PubMed] [Google Scholar]
- Guertin DA, Sabatini DM 2007. Defining the role of mTOR in cancer. Cancer Cell 12: 9–22 [DOI] [PubMed] [Google Scholar]
- Hendrickson WA 1991. Determination of macromolecular structures from anomalous diffraction of synchrotron radiation. Science 254: 51–58 [DOI] [PubMed] [Google Scholar]
- Hirose E, Nakashima N, Sekiguchi T, Nishimoto T 1998. RagA is a functional homologue of S. cerevisiae Gtr1p involved in the Ran/Gsp1–GTPase pathway. J Cell Sci 111: 11–21 [DOI] [PubMed] [Google Scholar]
- Inoki K, Corradetti MN, Guan KL 2005. Dysregulation of the TSC–mTOR pathway in human disease. Nat Genet 37: 19–24 [DOI] [PubMed] [Google Scholar]
- Kabsch W 1988. Evaluation of single-crystal X-ray diffraction data from a position-sensitive detector. J Appl Crystallogr 21: 916–924 [Google Scholar]
- Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL 2008. Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol 10: 935–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogan K, Spear ED, Kaiser CA, Fass D 2010. Structural conservation of components in the amino acid sensing branch of the TOR pathway in yeast and mammals. J Mol Biol 402: 388–398 [DOI] [PubMed] [Google Scholar]
- Kurzbauer R, Teis D, de Araujo ME, Maurer-Stroh S, Eisenhaber F, Bourenkov GP, Bartunik HD, Hekman M, Rapp UR, Huber LA, et al. 2004. Crystal structure of the p14/MP1 scaffolding complex: how a twin couple attaches mitogen-activated protein kinase signaling to late endosomes. Proc Natl Acad Sci 101: 10984–10989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26: 283–291 [Google Scholar]
- Li Z, Zhao B, Wang P, Chen F, Dong Z, Yang H, Guan KL, Xu Y 2010. Structural insights into the YAP and TEAD complex. Genes Dev 24: 235–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low HH, Sachse C, Amos LA, Lowe J 2009. Structure of a bacterial dynamin-like protein lipid tube provides a mechanism for assembly and membrane curving. Cell 139: 1342–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunin VV, Munger C, Wagner J, Ye Z, Cygler M, Sacher M 2004. The structure of the MAPK scaffold, MP1, bound to its partner, p14. A complex with a critical role in endosomal map kinase signaling. J Biol Chem 279: 23422–23430 [DOI] [PubMed] [Google Scholar]
- Nakashima N, Noguchi E, Nishimoto T 1999. Saccharomyces cerevisiae putative G protein, Gtr1p, which forms complexes with itself and a novel protein designated as Gtr2p, negatively regulates the Ran/Gsp1p G protein cycle through Gtr2p. Genetics 152: 853–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM 2008. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 320: 1496–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM 2010. Ragulator–Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 141: 290–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schurmann A, Brauers A, Massmann S, Becker W, Joost HG 1995. Cloning of a novel family of mammalian GTP-binding proteins (RagA, RagBs, RagB1) with remote similarity to the Ras-related GTPases. J Biol Chem 270: 28982–28988 [DOI] [PubMed] [Google Scholar]
- Sekiguchi T, Hirose E, Nakashima N, Ii M, Nishimoto T 2001. Novel G proteins, Rag C and Rag D, interact with GTP-binding proteins, Rag A and Rag B. J Biol Chem 276: 7246–7257 [DOI] [PubMed] [Google Scholar]
- Wullschleger S, Loewith R, Hall MN 2006. TOR signaling in growth and metabolism. Cell 124: 471–484 [DOI] [PubMed] [Google Scholar]