ChIP-seq and RNA-seq technologies were combined with analysis of LRH-1-null mice to describe the first complete analysis of the cistrome and transcriptome for the nuclear receptor LRH-1 in an adult animal. The authors found that LRH-1 interaction with the pancreas pioneering factor PTF1-L is required for the expression and secretion of exocrine pancreas proteins that aid in digestion.
Keywords: LRH-1, PTF1-L, exocrine pancreas, digestive enzymes, ChIP-seq, RNA-seq
Abstract
We have determined the cistrome and transcriptome for the nuclear receptor liver receptor homolog-1 (LRH-1) in exocrine pancreas. Chromatin immunoprecipitation (ChIP)-seq and RNA-seq analyses reveal that LRH-1 directly induces expression of genes encoding digestive enzymes and secretory and mitochondrial proteins. LRH-1 cooperates with the pancreas transcription factor 1-L complex (PTF1-L) in regulating exocrine pancreas-specific gene expression. Elimination of LRH-1 in adult mice reduced the concentration of several lipases and proteases in pancreatic fluid and impaired pancreatic fluid secretion in response to cholecystokinin. Thus, LRH-1 is a key regulator of the exocrine pancreas-specific transcriptional network required for the production and secretion of pancreatic fluid.
The exocrine pancreas constitutes >98% of the pancreatic gland and is composed of acinar cells, which produce hydrolytic digestive enzymes, and ductal cells, which produce a bicarbonate-rich fluid. Within acini, the majority of the protein synthetic capacity is used to synthesize secretory proteins and is supported by more ribosomes than any other cell type in the body and by numerous mitochondria to provide energy (Harding et al. 1977; Van Nest et al. 1980). In response to the post-prandial hormones cholecystokinin (CCK), secretin, and gastrin, the mixture of enzymes and fluid that compose the pancreatic fluid is released via the pancreatic duct into the duodenum to facilitate digestion.
The pancreas transcription factor 1-L complex (PTF1-L) plays a prominent role in regulating exocrine pancreas-specific gene expression (Cockell et al. 1989; Rose et al. 2001; Masui et al. 2010). It does so by binding to bipartite response elements composed of an E-box and a TC-box spaced by one or two DNA turns (Cockell et al. 1989; Beres et al. 2006). The PTF1-L complex is comprised of three proteins, each of which is required for the complex to bind to DNA (Roux et al. 1989; Beres et al. 2006): PTF1A, which is a class B basic helix–loop–helix (bHLH) factor (Krapp et al. 1996; Rose et al. 2001; Beres et al. 2006); RBPJL, which is closely related to RBPJ but (unlike RBPJ) does not mediate Notch signaling (Minoguchi et al. 1997); and one of several class A bHLH family members. PTF1A and class A bHLH proteins form a heterodimer that binds to the E-box. RBPJL interacts with the C-terminal domain of PTF1A and binds to the TC-box. Elimination of RBPJL in mice results in a smaller pancreas size and defective acinar cell gene expression, including marked reductions in the expression of genes encoding digestive enzymes, and proteins involved with secretory and mitochondrial function (Masui et al. 2010).
Liver receptor homolog-1 (LRH-1; also called NR5A2) is a member of the nuclear receptor family of ligand-activated transcription factors (Fayard et al. 2004; Lee and Moore 2008). LRH-1 regulates gene expression by binding as a monomer to DNA sequence elements with the consensus sequence 5′-Py-CAAGGPyCPu-3′. LRH-1 has high constitutive transcriptional activity, and while it can bind to various phospholipids (Lee and Moore 2008), it remains unclear whether these are physiologic ligands. LRH-1 is present during the earliest stages of development and in embryonic stem cells, where it maintains pluripotency (Pare et al. 2004; Gu et al. 2005; Heng et al. 2010). In adults, LRH-1 is abundantly expressed in endoderm-derived enterohepatic tissues (Rausa et al. 1999), where it regulates genes involved in cholesterol, fatty acid, and bile acid homeostasis, and intestinal epithelial cell renewal (Fayard et al. 2004; Lee and Moore 2008). LRH-1 is also expressed in both the acinar and ductal cells of the exocrine pancreas (Rausa et al. 1999). While LRH-1 regulates expression of carboxyl ester lipase in acinar cells (Fayard et al. 2003), little else is known about its function in this tissue. Here, we investigated the function of LRH-1 in exocrine pancreas.
Results and Discussion
Since conventional knockout of LRH-1 is embryonic-lethal (Pare et al. 2004; Gu et al. 2005), we used a tamoxifen-inducible estrogen receptor-cre system to eliminate LRH-1 in adult tissues, including the pancreas. Mice homozygous for the floxed Lrh1 allele and heterozygous for the ROSA26-ER-cre allele showed no overt abnormalities. At 3–5 mo of age, Lrh1flox/flox male mice that were either positive or negative for the Cre allele were treated with tamoxifen for two consecutive days to generate LRH-1-null (Lrh1t−/−) or control floxed (Flox) mice. In initial experiments, all Lrh1t−/− mice became moribund 7–12 d after tamoxifen treatment and died ∼2 d later. Thus, all subsequent experiments were performed in mice 5 d after tamoxifen treatment. At this time point, Lrh1 mRNA was reduced ∼20-fold in the pancreas (Fig. 1A, bottom panel) and was also virtually absent in the liver and intestine (data not shown). Western blot analysis confirmed the absence of the LRH-1 protein in the pancreas of the Lrh1t−/− mice (Fig. 1A, top panel). There were no gross morphological differences in pancreatic acini, ducts, or islets as assessed by hemotoxylin-eosin staining, nor were there differences in serum insulin concentrations measured at the beginning of the light cycle in mice 5 d after tamoxifen treatment (data not shown).
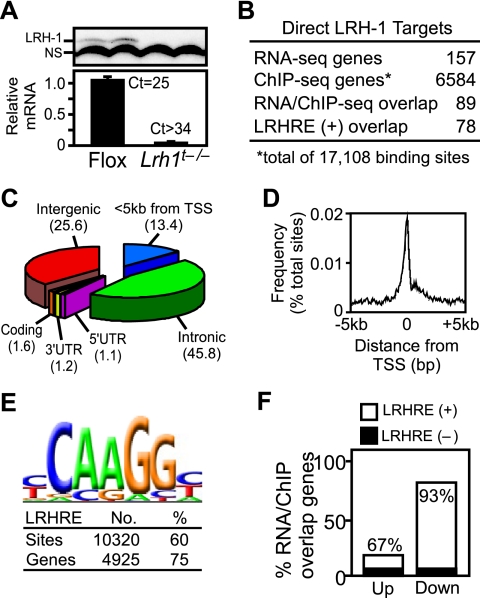
Figure 1.
Identification of LRH-1 target genes in exocrine pancreas by RNA-seq and ChIP-seq analyses. (A) Immunoblots of LRH-1 protein (top panel, n = 2 mice per genotype) and qPCR of LRH-1 mRNA (bottom panel, n = 4 per group) from pancreas of Flox and Lrh1t−/− mice. (NS) Nonspecific band. (B) Comparison and overlap of LRH-1 target genes. RNA-seq identified genes up-regulated or down-regulated twofold or more in Lrh1t−/− pancreas compared with Flox pancreas. ChIP-seq analysis from pancreas of wild-type mice identified genes with at least one LRH-1-binding site within 50 kb of a gene. The overlap of genes found in both data sets containing a consensus LRHRE are shown. (C,D) Distribution of LRH-1 ChIP-seq peaks across the genome (C) and relative to gene transcriptional start sites (TSSs) (D). (E) Analysis of LRH-1 ChIP-seq peak sequences revealed the consensus LRHRE as an overrepresented motif. Numbers of LRHRE-positive peaks and associated genes are shown. (F) Direct LRH-1 target genes identified by RNA-seq and ChIP-seq that were up-regulated or down-regulated in the Lrh1t−/− pancreas are shown.
To identify LRH-1 target genes in the pancreas, RNA-seq and chromatin immunoprecipitation (ChIP)-seq experiments were done. RNA-seq revealed 157 mRNAs that were either increased or decreased twofold or more in Lrh1t−/− compared with the Flox mouse pancreas (Fig. 1B; Supplemental Table S1). The conservative twofold cutoff was chosen to identify genes and pathways that are robustly regulated by LRH-1. Of these genes, 31% were up-regulated and 69% were down-regulated. ChIP-seq with an LRH-1 antibody identified 17,108 binding sites mapping within 50 kb of 6584 genes in the pancreas from wild-type mice (Fig. 1B). The breakdown of binding sites across the genome showed that 13% are located within 5-kb upstream promoter regions and 46% are in intronic regions (Fig. 1C). For those bound sites within upstream promoter sequences, most localized near the transcriptional start site (Fig. 1D). An unbiased motif search of the DNA sequences under the LRH-1-binding sites revealed that 60% contain the LRHRE consensus core CAAGG (Fig. 1E). Eighty-nine out of 157 genes from the RNA-seq data set had an LRH-1-binding site located by ChIP-seq within 50 kb of the gene, with 78 of these genes harboring a consensus LRHRE core (Fig. 1B). Of these 78 genes, 14 were up-regulated and 64 were down-regulated in the Lrh1t−/− pancreas, and 67% of the up-regulated genes and 93% of the down-regulated genes had LRHREs (Fig. 1F). Comparison of the RNA-seq and ChIP-seq data showed that only ∼1% of genes with LRH-1 bound within 50 kb showed a twofold or more change in expression. Thus, ChIP-seq data alone are insufficient to predict the LRH-1-regulated targets in a tissue. Similar comparisons of mRNA regulation and ChIP-seq for other transcription factors, including estrogen receptor α, peroxisome proliferator-activated receptor γ, and RBPJL, have also revealed that only a small fraction of binding sites contributes to gene regulation in the context of a particular tissue (Carroll et al. 2006; Nielsen et al. 2008; Lefterova et al. 2010; Masui et al. 2010).
DAVID/PANTHER gene ontology analysis was performed for those genes present in both the RNA-seq and ChIP-seq data sets that harbored LRHREs. These LRH-1 targets were enriched for genes involved in amino acid metabolism, proteolysis, and lipid metabolism (Supplemental Table S2A). Further analysis of these genes revealed enrichment for exocrine pancreas-related processes, including digestion, secretion, and mitochondrial metabolism (Supplemental Table S2B). The altered expression of a subset of these genes in Lrh1t−/− pancreas was confirmed by quantitative PCR (qPCR) (Fig. 2A). Loss of LRH-1 binding to the promoters of several of these genes in Lrh1t−/− pancreas was confirmed by ChIP (Fig. 2B). Thus, LRH-1 regulates exocrine pancreas-specific gene expression.
Figure 2.
LRH-1 regulates exocrine pancreas-specific genes. (A) LRH-1-regulated genes identified by RNA-seq were confirmed by qPCR by comparing expression in Flox and Lrh1t−/− (KO) pancreas (n = 5 per group). (B) ChIP-seq data were confirmed by conventional ChIP using an LRH-1 antibody and Flox and Lrh1t−/− pancreas (n = 3 per group). (Cel) Carboxyl ester lipase; (Pnlip) pancreatic lipase; (Ela1) elastase 1; (Ctrl) chymotrypsin-like; (Cpa1) carboxypeptidase A1; (Shp) small heterodimer partner; (Aqp12) aquaporin 12; (Sycn) syncollin; (Gatm) glycine amidinotransferase; (Gls2) glutaminase 2.
Among the genes down-regulated in Lrh1t−/− pancreas was Rbpjl (Fig. 2A; Supplemental Table S2B), which encodes a component of the PTF1-L complex (Cockell et al. 1989; Minoguchi et al. 1997; Rose et al. 2001; Masui et al. 2010). ChIP-seq analysis revealed an LRHRE within 100 base pairs (bp) of the known PTF1-L-binding site in the Rbpjl promoter. ChIP-seq data also showed that RBPJL and PTF1A bind to this region (Supplemental Fig. S1A). We next examined whether LRH-1 cooperates with PTF1-L to regulate Rbpjl and other acinar cell-specific genes. ChIP-seq for RPBJL and PTF1A in the adult pancreas was used to identify PTF1-L-binding sites (Masui et al. 2010). Colocalized sequences under PTF1A and RPBJL ChIP-seq peaks that shared ≥1-bp overlap with each other were assumed to represent PTF1-L-binding sites. A genome-wide comparison of LRH-1- and PTF1-L-binding sites in the pancreas revealed colocalization at 1533 sites mapping within 50 kb of 1063 genes (Supplemental Fig. S1B). These binding sites generally lay in close proximity to one another (Supplemental Fig. S1C). The coincident binding of LRH-1 and PTF1-L was highly significant (P < 1 × 10−300). RNA-seq from the pancreas of embryonic day 17.5 (E17.5) Rbpjl−/− mice revealed 423 genes with twofold or more change in mRNA levels that also harbored at least one PTF1-L-binding site within 50 kb of the gene (Masui et al. 2010). Comparison of LRH-1 and PTF1-L target genes revealed 18 genes that were regulated twofold or more in the same direction in Lrh1t−/− and Rbpjl−/− mice (Fig. 3A). Of these 18 coregulated genes, 17 had colocalized LRH-1 and PTF1-L binding (Fig. 3A). The concurrence of coregulated and colocalized genes among the pool of genes bound by both LRH-1 and PTF1-L was statistically significant (P < 0.001). Sixteen of the colocalized/coregulated genes were down-regulated in the pancreas from Lrh1t−/− and Rbpjl−/− mice (Fig. 3A). Notably, 11 of the 17 colocalized genes belonged to the exocrine pancreas-enriched gene list in Supplemental Table S2B.
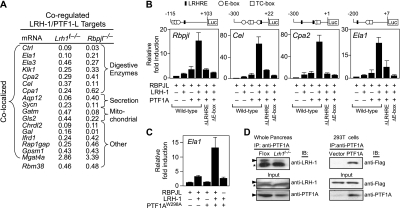
Figure 3.
LRH-1 and PTF1-L coregulate exocrine pancreas-specific genes. (A) ChIP-seq and RNA-seq data from Lrh1t−/−, E17.5 Rbpjl−/−, and control mouse pancreas were used to identify genes that contained overlapping LRH-1 and PTF-1L ChIP-seq peaks (within 50 kb) and that were dysregulated twofold or more in the same direction (either up or down) in the absence of LRH-1 or PTF1-L. The relative mRNA levels in the Lrh1t−/− and Rbpjl−/− mice compared with control mice are shown. Common LRH-1/PTF1-L target genes are listed according to function. (B) 293T cells were cotransfected with expression plasmids for RBPJL, LRH-1, and PTF1A as indicated together with luciferase reporter plasmids containing the mouse Rbpjl, Cel, Cpa2, and Ela1 promoters, or these same promoters in which either the LRHRE or E-boxes were mutated. Schematics showing the relative positions of LRHREs, E-boxes, and TC-boxes in each promoter are shown. (C) 293T cells were cotransfected with the Ela1-luc reporter plasmid and expression plasmids for RBPJL, LRH-1, and PTF1AW298A as indicated. In B and C, n = 3 ± SEM. Luciferase activity is normalized to vector alone. (D) Coimmunoprecipitation experiments were performed with either whole pancreas extracts from Flox and Lrh1t−/− mice (left panels) or 293T cells transfected with Flag-tagged LRH-1 in the presence or absence of PTF1A (right panel). Immunoprecipitations were done with a PTF1A antibody, and immunoblotting was done with an LRH-1 antibody (top left panel) or an anti-Flag antibody (top right panel). Immunoblots for total input LRH-1 and PTF1A are shown at the bottom.
We tested whether LRH-1 and PTF1-L cooperate in regulating the promoters of Rbpjl and three additional genes that had decreased expression in both Lrh1t−/− and Rbpjl−/− pancreas: carboxyl ester lipase (Cel), carboxypeptidase A2 (Cpa2), and elastase 1 (Ela1). Cell-based reporter assays were done in nonpancreatic HEK293T cells, which represent a null background for PTF1A, RBPJL, or LRH-1 expression. Introduction of RBPJL with either LRH-1 or PTF1A alone resulted in a modest induction of all three promoters (Fig. 3B). However, cointroduction of RBPJL, LRH-1, and PTF1A caused a synergistic induction of reporter activity. Comparison of the mouse and human promoters of Ela1, Cel, Cpa2, and Rbpjl showed conserved LRHREs and E-boxes coincident with the binding sites identified by ChIP (Supplemental Fig. S2). Mutating either the LRHREs or E-boxes within the PTF1A- and LRH-1-binding regions in all four reporters eliminated the synergy (Fig. 3B).
Although RPBJL is absent in HEK293T cells, the related ubiquitous central Notch transcriptional mediator RBPJ is expressed (Beres et al. 2006). RBPJL and RBPJ both interact with PTF1A (Beres et al. 2006). However, whereas RBPJL binds to PTF1A via two interaction surfaces involving tryptophan residues at positions 298 and 280, RBPJ binding to PTF1A depends only on W298 (Masui et al. 2007). Thus, a W298A mutation in PTF1A blocks interaction with RBPJ but retains its interaction with RBPJL. To confirm that the synergy observed with LRH-1, PTF1A, and RBPJL requires all three of these proteins, we took advantage of the PTF1AW298A mutant in transfection assays. Using the Ela1-luc reporter, transfection assays were performed as above, except that wild-type PTF1A was replaced with the W298A mutant. A comparable high level of induction of reporter activity was observed when LRH-1 and mutant PTF1A were introduced together with RBPJL (Fig. 3C). This induction was absent when RBPJL was excluded from the assay. These data show that LRH-1 functions with the entire PTF1-L complex to activate pancreatic gene targets.
To determine whether the cooperativity between LRH-1 and PTF1-L involves direct interactions, coimmunoprecipitation experiments were performed. LRH-1 was immunoprecipitated with a PTF1A antibody from murine pancreatic extracts (Fig. 3D) and HEK293T cells in which PTF1A and Flag-tagged LRH-1 were coexpressed (Fig. 3D). In complementary ChIP studies, there was no significant difference in PTF1A binding to various promoters in Lrh1t−/− and Flox mouse pancreas (Supplemental Fig. S3). Thus, LRH-1 and the PTF1-L complex cooperate in activating acinar-specific gene transcription through a mechanism that likely involves physical interactions but does not involve cooperative DNA binding.
A previous genome-wide ChIP-seq analysis in stem cells showed that LRH-1 shares many common gene targets with SOX2 and KLF4 and that each of these transcription factors is required for stem cell pluripotency (Heng et al. 2010). Comparison of the pancreatic and stem cell ChIP-seq data sets revealed only eight common LRH-1-binding sites with <1-bp overlap (data not shown). Furthermore, stem cell LRH-1-binding sites overlapped with PTF1-L sites at seven regions, in contrast to the 1533 observed with LRH-1 and PTF1-L in the pancreas. This indicates that the genome-wide binding pattern of LRH-1 is strongly tissue-dependent and that colocalization of LRH-1 with PTF1-L is a pancreas-specific event.
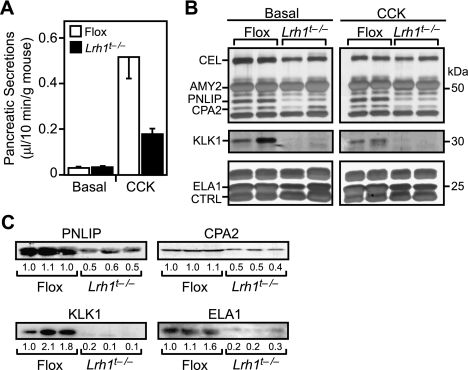
Last, we compared pancreatic fluid between Flox and Lrh1t−/− mice in both the presence and absence of CCK, which induces pancreatic fluid secretion. Basal secretion rates were similar in Flox and Lrh1t−/− mice (Fig. 4A). CCK injection resulted in a 20-fold increase in pancreatic secretion volume in the Flox mice. In contrast, Lrh1t−/− mice showed only a fivefold induction. Since acinar cells do not contribute significantly to bicarbonate production and fluid volume, this likely is due to a defective ductal cell response. Silver staining of both basal and CCK-induced secretions showed that several digestive enzymes, including pancreatic lipase (PNLIP), CPA2, kallikrein-1 (KLK1), and ELA1, were selectively decreased in the Lrh1t−/− pancreatic fluid (Fig. 4B). Western blot analysis confirmed that these enzymes were less abundant in the pancreatic secretions of Lrh1t−/− mice (Fig. 4C).
Figure 4.
LRH-1 regulates the secretion and composition of pancreatic fluid. (A) Pancreatic fluid volumes from basal and CCK-stimulated secretions were collected for 10 min each, and volumes were normalized to body weight (n = 5–7 mice per group). (B) Pancreatic secretions from A were resolved by SDS-PAGE and visualized by silver staining. Each lane represents pancreatic fluid pooled from two mice. Expected positions of abundant digestive enzymes are indicated. (CEL) Carboxyl ester lipase; (AMY2) amylase 2; (PNLIP) pancreatic lipase; (CPA2) carboxypeptidase A2; (KLK1) kallikrein 1; (ELA1) elastase 1; (CTRL) chymotrypsin-like. (C) CCK-stimulated pancreatic secretions from three Flox and three Lrh1t−/− mice were analyzed by Western analysis for PNLIP, CPA2, KLK1, and ELA1.
In summary, we show that LRH-1 coordinately regulates a program of genes involved in the synthesis and secretion of pancreatic fluid. LRH-1 was previously shown to regulate fatty acid, cholesterol, and bile acid homeostasis in the intestine and liver (Fayard et al. 2004; Lee and Moore 2008). Thus, LRH-1 plays an overarching role in regulating digestive processes. Since the transcriptional activity of LRH-1 can be modulated by small molecules (Whitby et al. 2011), LRH-1 may serve as a pharmacologic target for treating diseases such as pancreatitis and cystic fibrosis that are associated with either the overproduction or underproduction of pancreatic enzymes.
Materials and methods
Tamoxifen-inducible Lrh1−/− mice
To generate the tamoxifen-inducible cre Lrh1 deletion, mice with floxed Lrh1 alleles (Lee et al. 2008) were crossed with a ROSA26 Tam-ERCre strain (no. 004847, Jackson Laboratories). This line was backcrossed to generate a mixed strain, homozygous Lrh1 floxed, and heterozygous ROSA26 Tam-ERCre line. From this line, adult male mice with or without ROSA26 Tam-ERCre (3–5 mo old) were treated with 0.25 mg/g tamoxifen (T5648, Sigma-Aldrich) in corn oil by oral gavage for two consecutive days followed by sacrifice after 5 d. Mice were kept on a 12-h light–dark cycle from 6:00 am to 6:00 pm with free access to water and standard chow diet (TD.2016, Harland-Teklad Laboratories). All animal procedures were performed according to institutional and NIH guidelines with the approval of the Institutional Animal Care and Use Committee at the University of Texas Southwestern Medical Center.
Reporter constructs and assays
Reporter plasmids were generated using a pGL3-basic (Promega) vector. Genomic DNA regions from mouse Ela1 (−200 to +7), Cpa2 (−300 to +22), Rbpjl (−115 to +103), and Cel (−300 to +1) were amplified by PCR. Mutagenesis was performed as described (Sawano and Miyawaki 2000). pCMX-βgal, pCDNA3.1-hPTF1A, pCDNA3.1-mRBPJL, and pCDNA3.1-hPTF1A-W298A were as described (Masui et al. 2007). Full-length mouse Lrh1 was cloned into pCDNA3.1. pCMV-3xFlag(His6)-mLRH-1 was constructed by inserting a 3xFlag (His6) tag upstream of mouse Lrh1. 293T cells were transfected using calcium phosphate coprecipitation as described (Makishima et al. 1999). Each 96-well was transfected with 40 ng of pGL3-basic reporter plasmid, 20 ng of pCMX-βgalactosidase, and 20 ng each of pCDNA3.1-based mLRH-1, RBPJL, or PTF1A expression plasmids. Total DNA was brought up to 140 ng with Bluescript II.
Pancreatic fluid collection and analysis
Five days after tamoxifen treatment, male Lrh1t−/− or Flox mice (n = 5–11 per group) were fasted for 16 h and anesthetized with 2.5% avertin solution (350 μL/25 g mouse), and a 2-cm incision was made at the midline of the abdomen to expose the common duct. A 10-0 suture was used to ligate the common bile duct upstream of the pancreatic duct to exclude bile flow. A PE-10 cannula was inserted 1 mm into the bile duct and secured to the opening of the ampulla of vater with an 8-0 suture. Pancreatic secretion flow was allowed to equilibrate for 5 min, followed by collection of basal flow for 10 min. The cannula was replaced and mice were injected subcutaneously with CCK-8 (2 nmol/kg; Pheonix Pharmacueticals). After CCK treatment, the secretions were collected for an additional 10 min and the volume was measured. The pancreatic secretions were then diluted 10-fold in HEPES-Ringer buffer (15 mM HEPEPS, 120 mM NaCl, 5 mM KCl, 2.2 mM MgCl2, 1.28 mM CaCl2 at pH 7.4) containing EDTA-free protease inhibitors (Roche) and stored at −80°C until use. For visualizing digestive enzymes, 3 μL of 10× diluted pancreatic secretions were resolved by SDS-PAGE and probed with anti-CPA2 (3816, Biovision), anti-KLK1 (kind gift from Quan Li, University of Texas Southwestern Medical Center), anti-PNLIP (98992, Santa Cruz Biotechnology), or anti-ELA1 (21593, Abcam). Densitometry was done with ImageJ (Bearer 2003).
Coimmunoprecipitation experiments
Whole pancreas (∼300 mg) from two Flox or Lrh1t−/− mice were homogenized in 2 mL of lysis buffer (20 mM HEPES, 1% Triton X-100, 200 mM NaCl, 5 mM EDTA, 1 mM EGTA, 5% glycerol at pH 7.4) containing protease inhibitors. Lysate was cleared with a 12,000 rpm spin for 5 min, followed by preclearing with 40 μL of protein A beads (Roche) for 30 min at 4°C. Precleared lysates were immunoprecipitated with 3 μg mouse anti-rabbit PTF1A antibody (Beres et al. 2006) for 1 h followed by addition of 50 μL of equilibrated 50% protein A beads. After three washes, beads were boiled in 1× Lamelli buffer, and 20 μL of extract was resolved by SDS-PAGE and blotted with anti-LRH-1 antibody (18293, Abcam) or anti-PTF1A antibody (EB07839, Everest). In a separate experiment, homogenized extracts were pretreated with 50 μg/mL ethidium bromide for 30 min at 4°C before coimmunoprecipitation to test the dependence of the interactions on DNA. Ethidium bromide had no effect on the coimmunoprecipitation (data not shown).
For in vitro coimmunoprecipitation experiments, 293T cells were transfected with vector or pCMV-3xFlag(His6)-mLRH-1 expression plasmids in combination with vector alone or pCDNA3.1-hPtf1a. Cells were harvested with 1% Triton X-100 lysis buffer, and coimmunoprecipitations and SDS-PAGE were performed as described above, then blotted for Flag epitope using anti-M2 Flag-HRP (Sigma-Aldrich) and anti-PTF1A antibody (EB07839, Everest).
RNA isolation, qPCR, and RNA-seq
Total RNA was isolated from the pancreas of individual, 3- to 5-mo-old Flox or Lrh1t−/− mice (n = 4 knockout and 6 Flox) 5 d after tamoxifen treatment using Trizol (Invitrogen). cDNA was synthesized using Invitrogen reverse transcriptase system, and 25 ng was used for qPCR analysis with the ABI7700 Fast instrument (Applied Biosystems) and normalized to U36b4 as described (Bookout et al. 2006). Primer sequences are listed in Supplemental Figure S4. Ten micrograms of total RNA from two separate pools of three Flox or two Lrh1t−/− pancreas RNA preparations was subjected to the Illumina RNA-seq protocol (Illumina). DNA (0.7 fmol) from each amplified library was analyzed with an Illumina Genome analyzer. The sequence tags were aligned with Bowtie 3, and normalized gene expression levels were calculated with ERANGE 4. The two Lrh1t−/− pancreas libraries had 13.0 million and 13.3 million tags that aligned to the mouse, and the two Flox pancreas libraries had 15.5 million and 16.3 million with RNA from Lrh1t−/−. The fold change of mRNA levels was calculated from RNA-seq data as the ratio of the rpkm values (rpkm is a measure of the relative level of an RNA normalized to 1 million total reads) for Flox and Lrh1t−/− pancreas. We chose a minimum of 1 rpkm in either the normal or Lrh1t−/− pancreatic RNA, which is equivalent to ∼0.1 mRNA molecules per cell, based on a RNA:DNA ratio of 8 for wild-type pancreas 5 and 6 pg of DNA per cell. Eight-thousand-five-hundred-fifty-eight genes had mRNA levels of ≥1 rpkm. All mRNA values from the RNA-seq data were normalized internally to the mean of the following reference gene mRNA levels: Gusb, Hmbs, Ipo8, Rpl19, Rpl23, Sfrs9, Tbp, Tfrc, Ubc, and Ywhaz.
ChIP and ChIP-seq
ChIP-seq was performed as described (Masui et al. 2010) using antibodies against LRH-1 (R&D Systems), PTF1A (Beres et al. 2006), or RBPJL (Beres et al. 2006). Primer sequences are listed in Supplemental Figure S4. An amplified library was prepared using the manufacturer's protocol (Illumina). We analyzed 0.7 fmol of DNA with an Illumina/Solexa Genome analyzer. Final data compilation includes a total of 11.2 million, 17.3 million, and 8.6 million aligned sequence reads (36-nucleotide tags) for LRH-1, PTF1A, and RBPJL, respectively. We identified 17,108 LRH-1-, 12,821 PTF1A-, and 10,294 RBPJL-bound regions (peaks) with CisGenome 8, restricting the false discovery rate to a maximum of 1 × 10−5.
Data analysis and visualization
Motif analysis was performed using MEME version 4.3.0 using both strands, a minimum occurrence of 50 sites in the data set, and minimum motif size of 8 bases. Aligned tags were converted to WIG files by MACs software and were graphed using MochiViewer. Degree of peak overlap was performed using the Galaxy Web tools, and peaks with ≥1-bp overlap were determined. E-box- and LRH-1-binding motifs determined by MEME were used to identify potential binding sites in each peak using the RSAT Tools (http://embnet.ccg.unam.mx/rsa-tools) pattern matching tool. Spacing between each potential binding site was then measured and graphed. Gene ontology analysis of data was performed using DAVID software (Huang et al. 2007).
Statistical analysis
Significance of the number of observed overlaps between LRH-1 and PTF1-L was established using the binomial distribution describing the random occurrences of overlap in the genome as a null model. Significance of LRH-1/PTF1-L coregulated genes that were also colocalized was established by using a log linear analysis for a 2 × 2 × 2 contingency table (http://faculty.vassar.edu/lowry/log.html). The three categorical variables were colocalized genes, LRH-1 target genes, and PTF1-L target genes. Significance was identified in the number of coregulated genes. Within the colocalized genes, the number of coregulated genes was significant (P < 0.001), but not in the case of the non-colocalized genes.
Acknowledgments
We thank Dr. Shmuel Muallem for useful discussions, Dr. Quan-Zhen Li for the kallikrein-1 antibody, Chengcheng Shen for RNA-seq and ChIP-seq bioinformatics expertise, and Varun Sondhi for help with pancreatic secretion collection. This work was supported by the Howard Hughes Medical Institute (to D.J.M.); National Institutes of Health grants F32-DK079544 (to S.R.H.), U19-DK06243409 (to D.J.M.), and R01-DK061220 (to R.J.M.); and Robert A. Welch Foundation grants I-1275 (to D.J.M.) and I-1558 (to S.A.K.). D.J.M. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.16860911.
References
- Bearer EL 2003. Overview of image analysis, image importing, and image processing using freeware. Curr Protoc Mol Biol 14.15.1–14.15.9 doi: 10.1002/0471142727.mb1415s63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beres TM, Masui T, Swift GH, Shi L, Henke RM, MacDonald RJ 2006. PTF1 is an organ-specific and Notch-independent basic helix–loop–helix complex containing the mammalian Suppressor of Hairless (RBP-J) or its paralogue, RBP-L. Mol Cell Biol 26: 117–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bookout AL, Cummins CL, Mangelsdorf DJ, Pesola JM, Kramer MF 2006. High-throughput real-time quantitative reverse transcription PCR. Curr Protoc Mol Biol 15.8.1–15.8.28 doi: 10.1002/0471142727.mb1508s73 [DOI] [PubMed] [Google Scholar]
- Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, Hall GF, et al. 2006. Genome-wide analysis of estrogen receptor binding sites. Nat Genet 38: 1289–1297 [DOI] [PubMed] [Google Scholar]
- Cockell M, Stevenson BJ, Strubin M, Hagenbuchle O, Wellauer PK 1989. Identification of a cell-specific DNA-binding activity that interacts with a transcriptional activator of genes expressed in the acinar pancreas. Mol Cell Biol 9: 2464–2476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fayard E, Schoonjans K, Annicotte JS, Auwerx J 2003. Liver receptor homolog 1 controls the expression of carboxyl ester lipase. J Biol Chem 278: 35725–35731 [DOI] [PubMed] [Google Scholar]
- Fayard E, Auwerx J, Schoonjans K 2004. LRH-1: an orphan nuclear receptor involved in development, metabolism and steroidogenesis. Trends Cell Biol 14: 250–260 [DOI] [PubMed] [Google Scholar]
- Gu P, Goodwin B, Chung AC, Xu X, Wheeler DA, Price RR, Galardi C, Peng L, Latour AM, Koller BH, et al. 2005. Orphan nuclear receptor LRH-1 is required to maintain Oct4 expression at the epiblast stage of embryonic development. Mol Cell Biol 25: 3492–3505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding JD, MacDonald RJ, Przybyla AE, Chirgwin JM, Pictet RL, Rutter WJ 1977. Changes in the frequency of specific transcripts during development of the pancreas. J Biol Chem 252: 7391–7397 [PubMed] [Google Scholar]
- Heng JC, Feng B, Han J, Jiang J, Kraus P, Ng JH, Orlov YL, Huss M, Yang L, Lufkin T, et al. 2010. The nuclear receptor Nr5a2 can replace Oct4 in the reprogramming of murine somatic cells to pluripotent cells. Cell Stem Cell 6: 167–174 [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Tan Q, Kir J, Liu D, Bryant D, Guo Y, Stephens R, Baseler MW, Lane HC et al. 2007. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res 35: W169–W175 doi: 10.1093/nar/gkm415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krapp A, Knofler M, Frutiger S, Hughes GJ, Hagenbuchle O, Wellauer PK 1996. The p48 DNA-binding subunit of transcription factor PTF1 is a new exocrine pancreas-specific basic helix–loop–helix protein. EMBO J 15: 4317–4329 [PMC free article] [PubMed] [Google Scholar]
- Lee YK, Moore DD 2008. Liver receptor homolog-1, an emerging metabolic modulator. Front Biosci 13: 5950–5958 [DOI] [PubMed] [Google Scholar]
- Lee YK, Schmidt DR, Cummins CL, Choi M, Peng L, Zhang Y, Goodwin B, Hammer RE, Mangelsdorf DJ, Kliewer SA 2008. Liver receptor homolog-1 regulates bile acid homeostasis but is not essential for feedback regulation of bile acid synthesis. Mol Endocrinol 22: 1345–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefterova MI, Steger DJ, Zhuo D, Qatanani M, Mullican SE, Tuteja G, Manduchi E, Grant GR, Lazar MA 2010. Cell-specific determinants of peroxisome proliferator-activated receptor gamma function in adipocytes and macrophages. Mol Cell Biol 30: 2078–2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makishima M, Okamoto AY, Repa JJ, Tu H, Learned RM, Luk A, Hull MV, Lustig KD, Mangelsdorf DJ, Shan B 1999. Identification of a nuclear receptor for bile acids. Science 284: 1362–1365 [DOI] [PubMed] [Google Scholar]
- Masui T, Long Q, Beres TM, Magnuson MA, MacDonald RJ 2007. Early pancreatic development requires the vertebrate Suppressor of Hairless (RBPJ) in the PTF1 bHLH complex. Genes Dev 21: 2629–2643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masui T, Swift GH, Deering T, Shen C, Coats WS, Long Q, Elsasser HP, Magnuson MA, MacDonald RJ 2010. Replacement of Rbpj with Rbpjl in the PTF1 complex controls the final maturation of pancreatic acinar cells. Gastroenterology 139: 270–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minoguchi S, Taniguchi Y, Kato H, Okazaki T, Strobl LJ, Zimber-Strobl U, Bornkamm GW, Honjo T 1997. RBP-L, a transcription factor related to RBP-Jκ. Mol Cell Biol 17: 2679–2687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R, Pedersen TA, Hagenbeek D, Moulos P, Siersbaek R, Megens E, Denissov S, Borgesen M, Francoijs KJ, Mandrup S, et al. 2008. Genome-wide profiling of PPARγ:RXR and RNA polymerase II occupancy reveals temporal activation of distinct metabolic pathways and changes in RXR dimer composition during adipogenesis. Genes & Dev 22: 2953–2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pare JF, Malenfant D, Courtemanche C, Jacob-Wagner M, Roy S, Allard D, Belanger L 2004. The fetoprotein transcription factor (FTF) gene is essential to embryogenesis and cholesterol homeostasis and is regulated by a DR4 element. J Biol Chem 279: 21206–21216 [DOI] [PubMed] [Google Scholar]
- Rausa FM, Galarneau L, Belanger L, Costa RH 1999. The nuclear receptor fetoprotein transcription factor is coexpressed with its target gene HNF-3β in the developing murine liver, intestine and pancreas. Mech Dev 89: 185–188 [DOI] [PubMed] [Google Scholar]
- Rose SD, Swift GH, Peyton MJ, Hammer RE, MacDonald RJ 2001. The role of PTF1-P48 in pancreatic acinar gene expression. J Biol Chem 276: 44018–44026 [DOI] [PubMed] [Google Scholar]
- Roux E, Strubin M, Hagenbuchle O, Wellauer PK 1989. The cell-specific transcription factor PTF1 contains two different subunits that interact with the DNA. Genes Dev 3: 1613–1624 [DOI] [PubMed] [Google Scholar]
- Sawano A, Miyawaki A 2000. Directed evolution of green fluorescent protein by a new versatile PCR strategy for site-directed and semi-random mutagenesis. Nucleic Acids Res 28: E78 doi: 10.1093/nar/28.16.e78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nest GA, MacDonald RJ, Raman RK, Rutter WJ 1980. Proteins synthesized and secreted during rat pancreatic development. J Cell Biol 86: 784–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitby RJ, Stec J, Blind RD, Dixon S, Leesnitzer LM, Orband-Miller LA, Williams SP, Willson TM, Xu R, Zuercher WJ, et al. 2011. Small molecule agonists of the orphan nuclear receptors steroidogenic factor-1 (SF-1, NR5A1) and liver receptor homologue-1 (LRH-1, NR5A2). J Med Chem 54: 2266–2281 [DOI] [PMC free article] [PubMed] [Google Scholar]