Wright and colleagues investigate the role of Pdx1 in cell fate reprogramming and differentiation in organogenesis. In progenitor cells and embryonic cells, induced expression of Pdx1 in α-cells resulted in increased β-cell numbers and trans-differentiation to β-cells postnatally, respectively. Mature α-cells expressing exogenous Pdx1 suppress glucagon expression, but are not reprogrammed into insulin-expressing β-like cells. These results provide insight into single-factor-based reprogramming at different competence states.
Keywords: pancreas, endocrine progenitors, α and β cells, reprogramming, Pdx1
Abstract
Using single transcription factors to reprogram cells could produce important insights into the epigenetic mechanisms that direct normal differentiation, or counter inappropriate plasticity, or even provide new ways of manipulating normal ontogeny in vitro to control lineage diversification and differentiation. We enforced Pdx1 expression from the Neurogenin-3-expressing endocrine commitment point onward and found during the embryonic period a minor increased β-cell allocation with accompanying reduced α-cell numbers. More surprisingly, almost all remaining Pdx1-containing glucagon/Arx-producing cells underwent a fairly rapid conversion at postnatal stages, through glucagon–insulin double positivity, to a state indistinguishable from normal β cells, resulting in complete α-cell absence. This α-to-β conversion was not caused by activating Pdx1 in the later glucagon-expressing state. Our findings reveal that Pdx1 can work single-handedly as a potent context-dependent autonomous reprogramming agent, and suggest a postnatal differentiation evaluation stage involved in normal endocrine maturation.
A major hurdle for cell replacement-based diabetes therapy is the difficulty of supplying vast numbers of functioning insulin-producing β cells. One method could be through the reprogramming of alternative cell types. While this process might be easier with closely lineage-related cells, even substantially different cells may be susceptible (e.g., Zhou et al. 2008).
Recent studies reveal significant plasticity between pancreatic α and β cells under certain induced conditions, implying a potential route to β cells through α cells. In a near-total β-cell destruction and regeneration model in adult mice, a proportion of new β cells were produced from α cells via a bihormonal glucagon+insulin+ (Gcg+Ins+) transitional state (Thorel et al. 2010). The interconversion presumably occurs in response to a combination of the physiological need to replenish β cells and regeneration-induced stress, raising questions as to the local or systemic signals triggered by such lesions. Direct superimposition of a pro-β-lineage condition was reported when Pax4 expression was forced in pancreatic or endocrine progenitors or in embryonic α cells to redirect endocrine differentiation or coax pre-existing α cells into β cells. The converted cells seemed similar to normal β cells and temporarily improved glycemia under induced diabetes, although the effect was superseded by uncontrolled α-cell neogenesis and fatality caused by extreme hyperglycemia (Collombat et al. 2009). These studies on the ability of a single lineage-allocating transcription factor to sustain complete cell fate conversion suggest that similar analyses for other transcription factors could be insightful. Determining which factors induce specific types of lineage reprogramming, as well as the repertoire of cellular competence states amenable to fate switching, could lead to pharmacological intervention to activate such factors in vivo, or to improved differentiation of embryonic stem cells to β cells.
Clues to the fate-instructing capacity of Pdx1 as a β-cell selector are inferred from its enriched expression in embryonic and mature β cells. Ectopic Pdx1 alone can induce incomplete reprogramming of liver or pancreatic acinar cells (e.g., Ferber et al. 2000; Heller et al. 2001). A synergistic effect between Pdx1, Neurog3, and MafA was observed when acinar cells were converted into β-like cells (Zhou et al. 2008), which inefficiently ameliorated hyperglycemia caused by loss of endogenous β cells, perhaps because the reprogrammed cells did not assemble into islet-like clusters. Rather than triggering a redirection into endocrine cells, forced Pdx1 expression in Ptf1a-expressing cells caused late stage acinar-to-ductal hyperplasia (Miyatsuka et al. 2006). While these studies suggest that Pdx1 alone is contextually sufficient to induce partial trans-differentiation or trans-determination, little is known about how different competence states affect the response to this single factor.
Here, we report on the previously unknown sufficiency for Pdx1 as a potent regulator of endocrine lineage allocation and maintenance of the mature state. With Pdx1 expression enforced from the Neurog3+ endocrine progenitor state onward, two periods of dominant lineage redirection occurred: (1) during early organogenesis, a minor reproducible reduction in cells directed to the α fate, and (2) a surprising peri/postnatal redirection of Pdx1-expressing α cells, with rapid reprogramming into Ins+ cells that are indistinguishable from normal β cells. The delayed conversion occurred despite α cells having expressed exogenous Pdx1 from their endocrine commitment point onward, suggesting the possibility of a cryptic chromatin-priming effect. In contrast, exogenous Pdx1 in Gcg+ embryonic or adult α cells suppressed Gcg expression but did not induce α/β fate switching. Our findings reveal differential α-to-β plasticity between endocrine progenitors and hormone-secreting cells in response to Pdx1. We speculate on the epigenetic ramifications of these differential lineage-switching findings.
Results and Discussion
Exogenous Pdx1 expression in endocrine progenitors
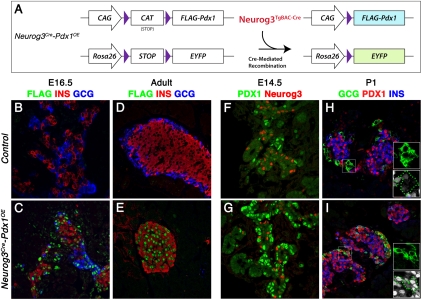
Forced “constitutive” Pdx1 expression was derived from a CAG-CAT-Pdx1 allele (Miyatsuka et al. 2006) via a BAC transgene driving Cre from Neurog3 regulatory elements (Neurog3TgBAC-Cre) (Schonhoff et al. 2004). Neurog3TgBAC-Cre-mediated CAT excision led to Flag-tagged Pdx1 (FlagPdx1) production in Neurog3+ descendants from the ubiquitously active CAG promoter (Fig. 1A). We compared tissues from Neurog3TgBAC-Cre;CAG-CAT-Pdx1;R26REYFP mice (referred to as Neurog3Cre-Pdx1OE hereafter) with those from Neurog3TgBAC-Cre;R26REYFP littermate controls.
Figure 1.
Neurog3Cre-mediated exogenous Pdx1 expression. (A) Schematic presentation of CAG-CAT-Pdx1 and R26REYFP and Cre recombination. Exogenous Flag-tagged Pdx1 (Flag-Pdx1) and EYFP expression is activated after CAT or STOP cassette excision. (B–E) FlagPdx1 was only detected in the Neurog3Cre-Pdx1OE pancreas. No Gcg was detected in adult Neurog3Cre-Pdx1OE. (F,G) Increased Pdx1+ cells were detected in E14.5 Neurog3Cre-Pdx1OE compared with control. (H,I) Ectopic Pdx1 in Gcg+ cells in Neurog3Cre-Pdx1OE but not control (dashed lines in insets; white, Pdx1).
Two key criteria were evaluated for Neurog3-dependent activation of CAG-CAT-Pdx1. First, selective FlagPdx1 production within endocrine cells was confirmed by Flag immunostaining, with signal only in Neurog3Cre-Pdx1OE pancreas, from embryonic day 16.5 (E16.5) to postnatal stages (Fig. 1B–E). Second, FlagPdx1 immunodetection with Pdx1 antibodies labeled cell types that normally do not express Pdx1 at high levels (Pdx1HI). A large increase occurred in the number of Pdx1HI cells in E14.5 Neurog3Cre-Pdx1OE pancreatic epithelium compared with equivalent control tissue (Fig. 1F,G). Ectopic Pdx1 was detected in non-β/non-δ endocrine cells (i.e., in α, PP, and ε cells). We found Pdx1HI Gcg+ α cells in postnatal day 1 (P1) Neurog3Cre-Pdx1OE pancreas, while control α cells were Gcg+Pdx1− (Fig. 1H,I). This evidence demonstrates a spatiotemporally defined system for endocrine–progenitor-selective exogenous Pdx1 expression in the Neurog3Cre-Pdx1OE pancreas.
Progressive reduction of α cells in the Neurog3Cre-Pdx1OE pancreas
Global glucose homeostasis and the overall size and outward appearance of the pancreas in Neurog3Cre-Pdx1OE mice were similar to controls (Supplemental Fig. 2). However, a drastic Gcg+ cell loss (>99%) was found in the adult pancreas, while other hormone-producing cell types were similar in number and location (Fig. 1E; Supplemental Fig. 1). The numbers of α (Gcg+) or β (Ins+) cells were quantified at key stages. At each stage, the combined number of α and β cells (“α + β”) was similar between Neurog3Cre-Pdx1OE and control, despite a significant difference in the α versus β representation (Fig. 2I; Supplemental Fig. 3). This observation strongly suggests a scenario of lineage diversion, wherein one cell population expands at the expense of the other under a constant total number. We identified two phases of lineage conversion that ultimately contributed to a complete α-cell loss by the early adult stage. First, a significant decrease in Gcg+ cell number (control 47% representation reduced to 35%) and accompanying increase in Ins+ cells was detected in the E16.5 Neurog3Cre-Pdx1OE pancreas, shortly after the peak of Neurog3 expression at approximately E15 (Gu et al. 2002). Coexpression of Ins and α-cell-specific factors, such as Arx, suggesting an early movement toward β-cell-directed trans-differentiation, was not detected at this stage (data not shown). The early phase shift in α/β representation suggests that exogenous Pdx1 biases the behavior of a fraction of early endocrine progenitors, increasing flux toward the β lineage, disfavoring the α lineage. The reason for only 25% of Gcg+ cells being affected could be related to nonuniformity in Pdx1 accumulation within Neurog3+ progenitor cells and/or their immediate progeny.
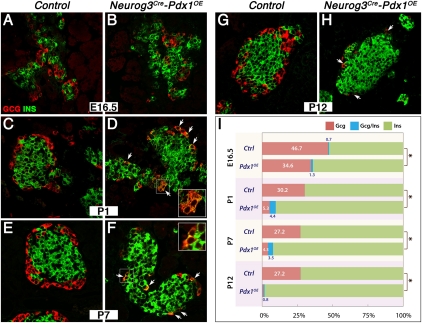
Figure 2.
Progressive decrease of Gcg+ cells concomitant with increase of Ins+ cells. (A–H) Ins and Gcg expression at E16.5, P1, P7, and P12. (Arrows) Gcg+Ins+-coexpressing cells. (I) Quantitative analysis of cells with Gcg, Ins, or Gcg-Ins expression in control and Neurog3Cre-Pdx1OE; (*) P < 0.05.
Second, a major progressive loss of Gcg+ cells concurrent with increased Ins+ numbers was detected at P1–P12 (Fig. 2I). The number of Gcg+ cells in the P1 Neurog3Cre-Pdx1OE pancreas was 32% of controls, suggesting that loss of Gcg expression in some cells had been initiated around birth. By P12, few Gcg+ cells were present, which were usually GcgLO by immunodetection (Fig. 2H). Importantly, numerous mantle-located Gcg+Ins+ cells were detected (Fig. 2D,F), representing intermediate state α cells undergoing conversion. The presence of Gcg+Ins+ cells in Neurog3Cre-Pdx1OE suggests that suppression of Gcg and induction of Ins occurred concurrently. Consistently, we did not detect Synaptophysin+ endocrine cells that were not producing islet hormones (Pdx1HIGcg−Ins− cells) (data not shown). We therefore propose that the large numbers of Pdx1HI α cells produced after endocrine specification (Pdx1OE via Neurog3 promoter activity), but only after a considerable delay until the peri/postnatal period, undergo a remarkable transformation toward Ins-expressing cell types.
Further validation by quantitative RT–PCR (qRT–PCR) of pancreatic tissue (Neurog3-Pdx1OE islets could not be isolated) (see Supplemental Fig. 4),examining a panel of differentiation pathway transcriptional effectors and hormones, confirmed the postnatal alteration in α/β-cell proportions. Expression of Gcg and Pou3f4, encoding an α-cell factor involved in Gcg transactivation (Hussain et al. 1997), was significantly reduced and increased Ins and Nkx6.1 expression was detected with increased Pdx1 expression (Supplemental Fig. 4). The considerable increase in Sst and Ppy RNA in Neurog3Cre-Pdx1OE tissue was not associated with an overt difference in δ- or PP-cell numbers, or coexpression of Ins with Sst or Ppy, but could represent increased expression per cell (Supplemental Fig. 1).
It was important to address whether the loss of Gcg+ α cells in Neurog3Cre-Pdx1OE might be caused by the death of Pdx1HI α cells or overproliferation of other endocrine cells outcompeting or stifling Pdx1HI α cells. Any dying cells should have been relatively easily detected in the mantle location over the P1–P14 postnatal reprogramming period. No difference was observed in general apoptosis or cell proliferation (TUNEL- or BrdU/Ki67-labeling assay) between Neurog3Cre-Pdx1OE and control (Supplemental Fig. 5), supporting the hypothesis that Pdx1HI α cells become actively reprogrammed via a Gcg+Ins+ transitional state into β cells. The comparable α + β population size between genotypes at all stages also supports our interpretation of a Pdx1-induced delayed reprogramming at the perinatal hormone-expressing stage, with no net loss of total endocrine cells.
Postnatal completion of α-to-β reprogramming induced by Pdx1
Ideally, pulse-chase lineage tracing of α-cell progenitors, with specific labeling concurrent with or just following the Neurog3+ state, should unequivocally demonstrate the proposed α-to-β-cell conversion. Unfortunately, we could not perform Cre-based lineage tracing on top of our Cre-based Pdx1 activation method, and a suitable genetic tool for this test (i.e., an ArxCreER strain) is not available. We therefore directly examined the production of Arx, a crucial α-cell transcription factor (Collombat et al. 2003), as an alternative approach to evaluate the α-cell reprogramming. In P1 Neurog3Cre-Pdx1OE, all Gcg+ cells, which expressed Pdx1 ectopically (Fig. 1I), were Arx+, as in controls (Fig. 3B). The total number of Arx+ cells and the Arx mRNA level were comparable with controls at P1 (Fig. 3B; Supplemental Fig. 4). The presence of both Arx+Gcg+ and Arx+Ins+ cells is most consistent with the idea that normal numbers of α cells were initially produced and that the reprogramming was far from complete at birth. Arx was also present in the Gcg+Ins+ cells over the postnatal period. In adult Neurog3Cre-Pdx1OE tissue, we reproducibly detected Arx in scarce, mantle-located Ins+ monohormonal cells. These Arx+Ins+ cells were detected in few islets, although sometimes several per islet, and were absent in controls (Fig. 3D). The Ins signal in these cells was similar to the other Ins+ cells. Intriguingly, the decrease in total Arx+ cell number and mRNA level became apparent only at P7–P12, while the Gcg+ cell number and the mRNA level were already reduced by P1 (Fig. 2I; Supplemental Figs. 4, 6). The conversion of hormone expression (Gcg to Ins) seems to precede down-regulation of the α-cell progenitor factor Arx, with most Pdx1HI α cells acquiring a more completely reprogrammed state by P7–P12. Despite their small number, the presence of adult Arx+Ins+ cells with apparently normal Ins immunodetectability per cell agrees with the idea that Pdx1 is dominant over Arx in determining the overt hormone-expressing state, even when in direct competition with the α-cell determinant.
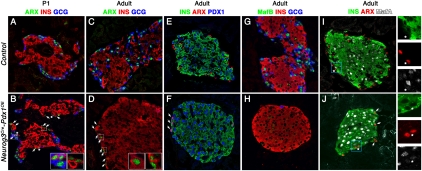
Figure 3.
Loss of α-cell character in the Neurog3Cre-Pdx1OE pancreas. (A–F) Arx was expressed in Gcg+ cells in control in P1 (A) and adult pancreas (C). In B, Arx was found in Gcg+, Gcg+Ins+ (asterisks), and some Ins+ cells in P1 Neurog3Cre-Pdx1OE (arrows, respectively). Only a few islet mantle Ins+ cells expressed Arx in adult Neurog3Cre-Pdx1OE (arrows in D and inset). They also expressed Pdx1HI (arrows in F). (G,H) MafB was detected in Gcg+ cells in the control adult pancreas but not Neurog3Cre-Pdx1OE. (I,J) Most Ins+ cells, except for a few Arx+Ins+ cells (arrows; also asterisks and separated channels), expressed MafA in Neurog3Cre-Pdx1OE adult at levels comparable with control.
Consistent with the massive postnatal reprogramming of Neurog3Cre-Pdx1OE α cells, a decrease in MafB+ cells in P12 and adult pancreas (Fig. 3H; Supplemental Fig. 6) confirmed the lack of normally differentiated mature α cells and the absence of MafB in Ins+ cells (whether reprogrammed or not). Accordingly, a large decrease in MafB mRNA was found in P12 Neurog3Cre-Pdx1OE (Supplemental Fig. 4). Besides its endocrine progenitor function, Pax6 was implicated in the control of the transcription of several α-cell-associated genes, such as Gcg and MafB (Gosmain et al. 2010). Interestingly, Pax4 could repress Pax6-mediated transactivation in vitro (Ritz-Laser et al. 2002). We speculate that the Pax6 reduction at P12 (Supplemental Fig. 4) results indirectly from the ectopic Pax4 in Gcg+ cells, described below.
Next, we assessed whether a complete β-cell differentiation program was deployed in the reprogrammed α cells. MafA, a marker associated with “final maturation” of β cells (Artner et al. 2010), at a level comparable with that in control Ins+ cells, was found in all Ins+ cells in Neurog3Cre-Pdx1OE tissue, irrespective of core or peripheral location. The rare adult Arx+Ins+ cells were always MafA− (Fig. 3J), suggesting that these former α cells resist complete reprogramming. Equivalent results were found for two more β-cell maturity markers: Nkx6.1 and Glut2 (Supplemental Fig. 7; Guillam et al. 1997; Sander et al. 2000). All of our evidence is therefore consistent with the idea that the vast majority of Pdx1HI α cells present at the perinatal stage undergo reprogramming toward authentic β cells, exhibiting both a normal mature state and complete loss of preceding α-cell identity. Pdx1 is dominant over Arx with respect to driving cells toward an Ins+Gcg− state, and Arx repression is prerequisite for thorough reprogramming.
The qRT–PCR analysis on postnatal Neurog3Cre-Pdx1OE pancreas showed significant up-regulation of genes encoding β-cell factors (MafA, Nkx6.1, and Pax4) (Supplemental Fig. 4), likely caused by the increased number of converted β cells (Fig. 2I) as well as the additional exogenous Pdx1 expression within all β cells. We note a likely normal function of these β cells, because the mice had normal to slightly decreased glycemia (the latter reflecting the lack of counterregulatory Gcg), and glucose tolerance analogous to controls (Supplemental Fig. 2). Pax4, previously reported to function as an α-cell reprogrammer (Collombat et al. 2009), was ectopically expressed in a proportion of Pdx1HI α cells (Supplemental Fig. 6). While Nkx6.1 expression was directly correlated with increased Pdx1 (e.g., at P1), Pax4 showed a late response (increased by P12 but not P1). This Pax4 pattern complements the graded decrease of Arx (Supplemental Fig. 4), agreeing with their proposed antagonistic relationship (Collombat et al. 2003). We propose that the initial hormone switching, which begins at P1 and precedes Pax4 elevation, could be Pax4-independent. It will be important to determine whether Pdx1 initiates a postnatal induction of Pax4, and whether their mutual reinforcement of expression works in a synergistic feed-forward loop to engage full reprogramming. Notably, Pdx1 overexpression triggered postnatal α-to-β reprogramming via an intermediate cell type (Ins+Gcg+) and did not lead to oversized islets, profound α-cell neogenesis, and failure to maintain euglycemia (Supplemental Figs. 2, 3), phenotypes therefore distinct from those caused by Pax4 manipulation (Collombat et al. 2009). On the other hand, exogenous Pax4 was sufficient to coax Gcg+ embryonic α cells into β cells while Pdx1 was not. Further study could address the detailed gene regulatory network responses to Pax4 and Pdx1.
Collectively, our data show that the majority of Pdx1HI α cells were successfully reprogrammed toward a mature β-cell profile. While Gcg and Pou3f4 expression was rapidly reduced, we believe that it is the repression of Arx in α cells that allows full-scale transformation toward an authentic β-cell program. Although further analyses will be required to assess the early-acting effects of Pdx1HI that are imposed at the early Neurog3+ state and/or continued during islet α-cell differentiation, we hypothesize that an early progenitor period priming is crucial for the later (peri/postnatal) override of the Arx-induced α-cell program. The peri/postnatal fate conversion could reflect a final fate assessment and switching of cellular maturation (with dominant establishment of Pdx1-centered regulatory networks) in response to the early, but initially cryptic, establishment of a Pdx1HI condition in developing α cells. With this reasoning, we asked whether the effect noted when expressing Pdx1HI from the endocrine progenitor state onward was different if Pdx1HI expression was initiated within cells slightly further along the differentiation pathway, having already started Gcg expression.
Glucagon suppression but incomplete reprogramming of embryonic and mature α cells
Activating the Pdx1HI condition in embryonic or adult α cells produced evidence that embryonic α cells have already acquired resistance to reprogramming. Because exogenous Pdx1 selectively affected the α lineage, we first tested the effect of Pdx1 overexpression in embryonic Gcg+ cells. GcgTgCre mice (Herrera 2000) were used to generate GcgTgCre;CAG-CAT-Pdx1;R26REYFP mice (GcgCre-Pdx1OE hereafter), driving exogenous Pdx1 expression in Gcg+ cells, with GcgTgCre;R26REYFP littermate controls. Similar lineage-labeled YFP+ cell numbers were detected within the islet mantle of GcgCre-Pdx1OE adult pancreas and controls (aged 3–3.5 mo). In GcgCre-Pdx1OE, but not control, exogenous Pdx1HI was detected in a majority of lineage-labeled α cells (∼56% YFP+Pdx1HI cells among all YFP+ cells, compared with 0% in control) (Fig. 4I), possibly reflecting nonparallel recombination of R26REYFP and CAG-CAT-Pdx1. While Gcg was not detected in most (>94%) YFP+Pdx1HI cells, only a low number (<14%) showed any Ins signal, thereby leaving ∼90% of Pdx1HI α cells expressing neither hormone (YFP+Pdx1HIGcg−Ins− cells) (Fig. 4K). Therefore, despite these Gcg+ cells having expressed Pdx1HI for roughly the same period as those in Neurog3Cre-Pdx1OE (late embryogenesis through adult), the α-to-β conversion in GcgCre-Pdx1OE was dramatically less efficient than that in Neurog3Cre-Pdx1OE (Fig. 2I). The YFP+Pdx1HI α cells showed diminished MafB but maintained Arx (Supplemental Fig. 8), suggesting a substantial barrier against full-scale suppression of the intrinsic α-cell identity. Failure to repress Arx in the great majority of α cells (as in the few residual Arx+Ins+ cells in Neurog3Cre-Pdx1OE adults) (Fig. 3D) could be a limiting factor for successful conversion. Our findings indicate qualitatively discrete competencies between Neurog3+ endocrine progenitor cells and nascent Gcg+ α cells with respect to their plasticity toward Pdx1-enforced reprogramming. The features of lineage-labeled α cells in GcgCre-Pdx1OE by molecular marker analysis overlap those of Neurog3Cre-Pdx1OE α cells, except that the latter move forward and convert to a β-cell program. No difference in Sst and Ppy expression between GcgCre-Pdx1OE and control animals was detected (Supplemental Fig. 9).
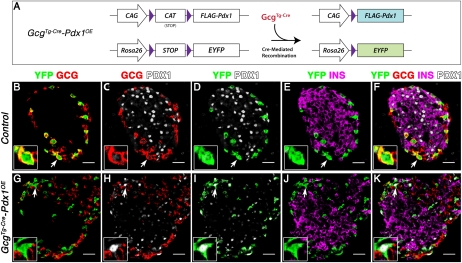
Figure 4.
Exogenous Pdx1 in Gcg+ cells does not lead to α-cell reprogramming. (A) Schematic presentation of CAG-CAT-Pdx1 and R26REYFP alleles before and after recombination. (B–K) Arrows denote representative cells in insets shown with separated channels. Lineage-labeled YFP+ cells were restricted to mantle Gcg+ cells (B), and ectopic Pdx1 was only detected in GcgCre-Pdx1OE but not in the control among YFP+ lineage-labeled cells (D,I). (K) Gcg expression was repressed in most YFP+ cells in GcgCre-Pdx1OE, but Ins expression was not detected.
We found an even greater resistance to reprogramming of adult α cells when we used a doxycycline-inducible genetic recombination system (Thorel et al. 2010) to delay the time of Pdx1 activation within Gcg+ cells until 2–3.5 mo of age (see Supplemental Fig. 10 for details). Most of the lineage-labeled adult α cells (95% of YFP+Pdx1HI cells) had lost Gcg expression, and <1% showed evidence of Ins expression (i.e., YFP+Pdx1HIGcg−Ins−) (Supplemental Fig. 10). These data clearly show that exogenous Pdx1 is insufficient to reprogram mature α cells, even over a longer time frame (4–6 wk) than the observed postnatal conversion (∼2 wk) effected by expressing Pdx1 from the Neurog3+ stage onward.
Overall, our findings demonstrate (1) a context-dependent differential competence of Pdx1 in directing endocrine fate allocation and differentiation and (2) a delayed peri/postnatal response to the early Neurog3-based imposition of a cell-autonomous reprogramming stimulus. With respect to affecting the embryonic α- versus β-lineage-commitment process, exogenous Pdx1 expression within the Neurog3+ cell population might not occur early enough in all progenitors. With this reasoning, the cells that move forward initially as “unaffected” α cells (i.e., Arx+Gcg+Pdx1HIIns−) then experience an unknown peri/postnatal trigger to initiate a rapid yet progressive α-to-β conversion. Delaying the activation of exogenous Pdx1 until the Gcg+ state, either embryonic or adult, failed to induce any reprogramming, effectively producing an “unprogrammed” state with loss of Gcg/MafB but not suppression of Arx or gain of Ins. The differential response strongly suggests a requirement for Pdx1-mediated early competence priming in order to exert future cell conversion, and further, that reprogramming might be separable into modes of either unprogramming or a more profound switching of cell fate.
The unexpectedly delayed postnatal α-to-β conversion leads to speculation on the mechanism of this temporally cryptic priming, which could be referred to as a “chromotypic effect”—being encoded at the chromatin level, without necessarily causing an immediate explicit phenotypic alteration. Learning how Pdx1 pioneers chromatin priming or epigenetic landscaping could be important with respect to cell reprogramming in vitro and in vivo. It is pertinent to note that Pdx1 has been implicated in epigenetic modification via interaction with a pancreatic islet-enriched histone methyltransferase, Set7/9, a Pdx1-responsive factor proposed to enhance chromatin accessibility and transcription of β-cell genes (Deering et al. 2009; Ogihara et al. 2009). A switch of Set7/9 subcellular localization from cytoplasmic/nuclear (α cell) to exclusively nuclear (β cell) was observed in adult Pdx1HI α cells in our various genetic conditions (Supplemental Fig. 11). This finding suggests that exogenous Pdx1 begins to initiate an epigenetic reconfiguration from α to β, potentially influencing the recruitment of Set7/9 to certain β-cell-specific loci. We also note that, despite Pdx1-driven Set7/9 nuclear entry, the Gcg-mediated Pdx1OE α cells (embryonic and adult) remain refractory to reprogramming (Fig. 4; Supplemental Fig. 10). Identifying ways to switch certain forms of epigenetic coding associated with this resistance, perhaps determined via flow cytometry-based cell capture, could improve reprogramming efficiency.
The peri/postnatal conversion of Neurog3Cre-Pdx1OE α cells raises the possibility that part of the final process of normal islet cell development involves cells checking their “internal transcriptional status,” with considerable pathway shifting still possible if the epigenetic state is not completely fixed toward specific fates. The nature of any associated peri/postnatal developmental cue is unknown, and could be organ-local, systemic, or presumably even metabolic. How these ideas relate to MafA/MafB resolution (the change from common production of both factors in immature β cells toward the mature pattern of MafA+ cells or MafB+ α cells), which also occurs perinatally (Artner et al. 2010), is a potentially fruitful area for future study. Determining the nature of regulatory checkpoints in the final maturation of endocrine cells could be germane to β-cell differentiation in vitro. The genetic models here could also provide a novel platform for identifying and manipulating cellular maturation and understanding how plasticity could be induced or restricted. Such principles might be applicable to multiple cell types in the pancreas and, more generally, to other organ systems.
Materials and methods
Mice
Information on mouse strains is in the Supplemental Material. All animals and embryos used were PCR-genotyped. Animal handling was under protocols approved by Vanderbilt University Medical Center Institutional Animal Care and Use Committee, or according to the Direction Générale de la Santé of the Canton de Genève.
Immunohistochemistry and morphometric analysis
Tissues were prepared as described (Fujitani et al. 2006). Information on antibodies and morphometric methods are in the Supplemental Material. Statistical analysis was performed using single-factor ANOVA tests and significance determined by P < 0.05.
qRT–PCR
RNA isolation (Trizol, Invitrogen), DNase treatment (Ambion), cDNA synthesis, and qPCR (SYBR green, Bio-Rad) were performed. Three samples per genotype per stage were collected, and qPCR was performed at least twice on each sample to determine ΔCT. Results were shown as ΔCT ± SEM and subjected to Student's t-test to determine significance (P < 0.05).
Acknowledgments
We thank A. Leiter for Neurog3TgBAC-Cre; T. Miyatsuka and H. Kaneto for CAG-CAT-Pdx1 mice; and P. Collombat, G. Gu, and B. Sosa-Pineda for Arx, Neurog3, and Pax4 antibodies. We also thank R. Stein, G. Gu, M. Gannon, and members of Wright laboratory for discussions. The work was supported by JDRF to Y.P.Y. and NIH U19 DK 042502 and U01 DK 089570 to C.V.E.W.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.16875711.
References
- Artner I, Hang Y, Mazur M, Yamamoto T, Guo M, Lindner J, Magnuson MA, Stein R 2010. MafA and MafB regulate genes critical to β-cells in a unique temporal manner. Diabetes 59: 2530–2539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collombat P, Mansouri A, Hecksher-Sorensen J, Serup P, Krull J, Gradwohl G, Gruss P 2003. Opposing actions of Arx and Pax4 in endocrine pancreas development. Genes Dev 17: 2591–2603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collombat P, Xu X, Ravassard P, Sosa-Pineda B, Dussaud S, Billestrup N, Madsen OD, Serup P, Heimberg H, Mansouri A 2009. The ectopic expression of Pax4 in the mouse pancreas converts progenitor cells into α and subsequently β cells. Cell 138: 449–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deering TG, Ogihara T, Trace AP, Maier B, Mirmira RG 2009. Methyltransferase Set7/9 maintains transcription and euchromatin structure at islet-enriched genes. Diabetes 58: 185–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferber S, Halkin A, Cohen H, Ber I, Einav Y, Goldberg I, Barshack I, Seijffers R, Kopolovic J, Kaiser N, et al. 2000. Pancreatic and duodenal homeobox gene 1 induces expression of insulin genes in liver and ameliorates streptozotocin-induced hyperglycemia. Nat Med 6: 568–572 [DOI] [PubMed] [Google Scholar]
- Fujitani Y, Fujitani S, Boyer DF, Gannon M, Kawaguchi Y, Ray M, Shiota M, Stein RW, Magnuson MA, Wright CV 2006. Targeted deletion of a cis-regulatory region reveals differential gene dosage requirements for Pdx1 in foregut organ differentiation and pancreas formation. Genes Dev 20: 253–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosmain Y, Marthinet E, Cheyssac C, Guerardel A, Mamin A, Katz LS, Bouzakri K, Philippe J 2010. Pax6 controls the expression of critical genes involved in pancreatic α-cell differentiation and function. J Biol Chem 285: 33381–33393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu G, Dubauskaite J, Melton DA 2002. Direct evidence for the pancreatic lineage: NGN3+ cells are islet progenitors and are distinct from duct progenitors. Development 129: 2447–2457 [DOI] [PubMed] [Google Scholar]
- Guillam MT, Hummler E, Schaerer E, Yeh JI, Birnbaum MJ, Beermann F, Schmidt A, Deriaz N, Thorens B 1997. Early diabetes and abnormal postnatal pancreatic islet development in mice lacking Glut-2. Nat Genet 17: 327–330 [DOI] [PubMed] [Google Scholar]
- Heller RS, Stoffers DA, Bock T, Svenstrup K, Jensen J, Horn T, Miller CP, Habener JF, Madsen OD, Serup P 2001. Improved glucose tolerance and acinar dysmorphogenesis by targeted expression of transcription factor PDX-1 to the exocrine pancreas. Diabetes 50: 1553–1561 [DOI] [PubMed] [Google Scholar]
- Herrera PL 2000. Adult insulin- and glucagon-producing cells differentiate from two independent cell lineages. Development 127: 2317–2322 [DOI] [PubMed] [Google Scholar]
- Hussain MA, Lee J, Miller CP, Habener JF 1997. POU domain transcription factor brain 4 confers pancreatic α-cell-specific expression of the proglucagon gene through interaction with a novel proximal promoter G1 element. Mol Cell Biol 17: 7186–7194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyatsuka T, Kaneto H, Shiraiwa T, Matsuoka TA, Yamamoto K, Kato K, Nakamura Y, Akira S, Takeda K, Kajimoto Y, et al. 2006. Persistent expression of PDX-1 in the pancreas causes acinar-to-ductal metaplasia through Stat3 activation. Genes Dev 20: 1435–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogihara T, Vanderford NL, Maier B, Stein RW, Mirmira RG 2009. Expression and function of Set7/9 in pancreatic islets. Islets 1: 269–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritz-Laser B, Estreicher A, Gauthier BR, Mamin A, Edlund H, Philippe J 2002. The pancreatic β-cell-specific transcription factor Pax4 inhibits glucagon gene expression through Pax6. Diabetologia 45: 97–107 [DOI] [PubMed] [Google Scholar]
- Sander M, Sussel L, Conners J, Scheel D, Kalamaras J, Dela Cruz F, Schwitzgebel V, Hayes-Jordan A, German M 2000. Homeobox gene Nkx6.1 lies downstream of Nkx2.2 in the major pathway of β-cell formation in the pancreas. Development 127: 5533–5540 [DOI] [PubMed] [Google Scholar]
- Schonhoff SE, Giel-Moloney M, Leiter AB 2004. Neurogenin 3-expressing progenitor cells in the gastrointestinal tract differentiate into both endocrine and non-endocrine cell types. Dev Biol 270: 443–454 [DOI] [PubMed] [Google Scholar]
- Thorel F, Nepote V, Avril I, Kohno K, Desgraz R, Chera S, Herrera PL 2010. Conversion of adult pancreatic α-cells to α-cells after extreme β-cell loss. Nature 464: 1149–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Brown J, Kanarek A, Rajagopal J, Melton DA 2008. In vivo reprogramming of adult pancreatic exocrine cells to β-cells. Nature 455: 627–632 [DOI] [PMC free article] [PubMed] [Google Scholar]