Lei and colleagues explore the role of AGO2 in chromatin organization in Drosophila. AGO2 is found to localize to euchromatin, where it is required for CTCF/CP190-dependent Fab-8 insulator function in a manner independent of its catalytic activity and of Dicer-2. The work points to an RNAi-independent role for AGO2 in defining transcriptional domains through chromosomal looping.
Keywords: RNAi, chromatin, insulator, Drosophila, ChIP-seq, nuclear organization
Abstract
A major role of the RNAi pathway in Schizosaccharomyces pombe is to nucleate heterochromatin, but it remains unclear whether this mechanism is conserved. To address this question in Drosophila, we performed genome-wide localization of Argonaute2 (AGO2) by chromatin immunoprecipitation (ChIP)-seq in two different embryonic cell lines and found that AGO2 localizes to euchromatin but not heterochromatin. This localization pattern is further supported by immunofluorescence staining of polytene chromosomes and cell lines, and these studies also indicate that a substantial fraction of AGO2 resides in the nucleus. Intriguingly, AGO2 colocalizes extensively with CTCF/CP190 chromatin insulators but not with genomic regions corresponding to endogenous siRNA production. Moreover, AGO2, but not its catalytic activity or Dicer-2, is required for CTCF/CP190-dependent Fab-8 insulator function. AGO2 interacts physically with CTCF and CP190, and depletion of either CTCF or CP190 results in genome-wide loss of AGO2 chromatin association. Finally, mutation of CTCF, CP190, or AGO2 leads to reduction of chromosomal looping interactions, thereby altering gene expression. We propose that RNAi-independent recruitment of AGO2 to chromatin by insulator proteins promotes the definition of transcriptional domains throughout the genome.
RNA silencing pathways are evolutionarily conserved mechanisms that control gene expression via sequence-specific interactions mediated by a small RNA bound to an Argonaute (AGO) effector protein. The paradigm for how RNA silencing controls gene expression at the chromatin level comes from studies in fission yeast, in which the RNAi machinery establishes heterochromatin at the centromere and mating type locus to ensure proper chromosome segregation and to promote stability of repetitive regions. At the centromere, RNAs transcribed from pericentromeric repeats are processed by the Dcr1 endonuclease and Ago1 Argonaute protein, which leads to the recruitment of the histone H3K9 methyltransferase and Swi6/HP1 binding (for review, see Grewal and Elgin 2007).
In Drosophila, it remains unclear whether the RNAi pathway is involved directly in heterochromatin formation. The primary endogenous function of the RNAi/siRNA pathway is to silence the expression of transposable elements (TEs) in the soma (for review, see Okamura and Lai 2008). Silencing is achieved by Dcr-2-mediated cleavage of dsRNAs into 21- to 22-nucleotide (nt) siRNA that are loaded into AGO2, which cleaves the target TE mRNA using its Slicer activity. Less well understood is the function of non-TE endo-siRNAs also produced by Dcr-2 activity and loaded into AGO2, which are generated from hairpin transcripts and regions of 3′ overlap of convergent transcripts (3′ cis-NATs). Two studies implicated AGO2 in heterochromatin formation based on mislocalization of HP1 and desilencing of pericentromeric transcriptional reporters in AGO2 mutants (Deshpande et al. 2005; Fagegaltier et al. 2009). However, direct analysis of HP1 recruitment by chromatin immunoprecipitation (ChIP) and HP1-dependent silencing at small RNA-generating loci led to the suggestion that AGO2 and other Argonaute genes may not be required for heterochromatin formation in the soma (Moshkovich and Lei 2010). Nevertheless, AGO2 or other RNA silencing factors appear to play important roles in chromatin and nuclear organization, such as formation of Polycomb group (PcG) repression bodies (Grimaud et al. 2006) and gypsy chromatin insulator bodies (Lei and Corces 2006).
Chromatin insulators are DNA–protein complexes defined functionally as either barriers that prevent the spread of silent chromatin or enhancer blockers that constrain enhancer–promoter communication. Unlike vertebrates, which possess only one known insulator protein, CTCF (for review, see Phillips and Corces 2009), Drosophila employs at least five different insulator complexes. Two well-characterized insulators are the gypsy [also known as Su(Hw)] insulator and the Fab-8 insulator of the Abd-B locus in the bithorax complex (BX-C) (for review, see Bushey et al. 2008). The gypsy and Fab-8 insulators harbor binding sites for the zinc finger DNA-binding proteins Su(Hw) and CTCF, respectively, and both insulator complexes share a common component: CP190. Despite thousands of distinct DNA-binding sites throughout the genome, insulator proteins concentrate at a small number of nuclear foci, termed insulator bodies, which are dependent on CP190 for their integrity. Highly correlated at least with gypsy insulator function, insulator bodies have been proposed to serve as tethering sites for large chromosomal loops or other higher-order chromatin structures.
It has become increasingly apparent that DNA topology is a critical determinant of gene regulation. While enhancers activate their target promoters over long distances, insulators act to restrict these communications (for review, see Wallace and Felsenfeld 2007). Insulators and other cis-regulatory regions in the Abd-B locus engage in numerous interactions, and the precise topology of the locus has been postulated to be a central mechanism of tissue-specific Abd-B regulation (Cleard et al. 2006; Lanzuolo et al. 2007; Kyrchanova et al. 2008; Bantignies et al. 2011). However, the mechanism by which chromosome looping is achieved at this locus has not been elucidated. Vertebrate CTCF has been demonstrated to mediate chromosomal looping at several developmentally regulated loci in concert with cohesin (for review, see Merkenschlager 2010), but it is not known whether Drosophila CTCF, which only shares homology in the zinc finger DNA-binding domain, retains the capacity to promote looping.
In order to address whether AGO2 functions on chromatin, we performed ChIP-seq analysis of AGO2 in two Drosophila cell lines. Instead of repetitive sequence, AGO2 associates primarily with euchromatic sites, the majority of which correspond to chromatin insulators. Intriguingly, AGO2 chromatin association does not correspond to regions of the genome that produce endo-siRNAs. We demonstrate that AGO2, but not its catalytic activity or other RNAi components, is required for CTCF/CP190-dependent Fab-8 insulator function. Additionally, AGO2 interacts physically with CP190, and depletion of either CP190 or CTCF results in a decrease in AGO2 recruitment throughout the genome. Chromosome conformation capture (3C) experiments demonstrate that CTCF/CP190-dependent looping interactions may regulate AGO2 recruitment to chromatin. Therefore, we propose an RNAi-independent role for AGO2 to promote or stabilize insulator-dependent looping interactions to define transcriptional domains throughout the genome.
Results
AGO2 associates with euchromatin and not repetitive sequences
In order to obtain high-resolution information about the genome-wide chromatin association profile of AGO2, we performed ChIP-seq analysis of AGO2 in S2 and S3 Drosophila embryonic cell lines. ChIP was performed using a previously characterized monoclonal antibody, 9D6, capable of isolating AGO2 and associated small RNAs (Miyoshi et al. 2005; Kawamura et al. 2008). Greater than 9 million reads per input or immunoprecipitation sample were obtained, leading to the identification of 3367 AGO2-bound sites between both cell types using a 5% false discovery rate threshold with the MACS algorithm (Zhang et al. 2008). Approximately 86% of AGO2 sites in S2 overlap with those found in S3, suggesting that AGO2 genome-wide localization is mainly consistent between cell types. Comparing the fraction of total reads mapping to repetitive sequences indicates no enrichment of repetitive sequences in the immunoprecipitation versus input (χ2 test, P < 2 × 10−16); therefore, we conclude that AGO2 localizes predominantly to euchromatic regions.
Strikingly, the majority of AGO2 sites overlap with known chromatin insulator sites throughout the genome. As a model region, we inspected the 300-kb BX-C Hox gene cluster and observed association of AGO2 with all known cis-regulatory domain boundaries in both cell lines (Fig. 1). These insulators include the Abd-B locus boundary elements Mcp, Fab-6, Fab-7, and Fab-8. We obtained a similar ChIP-seq profile with lower signal using an independent α-AGO2 polyclonal antibody (Meyer et al. 2006). Moreover, three independent antibodies capable of immunoprecipitating AGO2 (Jiang et al. 2005; Meyer et al. 2006; Czech et al. 2008) show similar enrichment profiles at the Abd-B locus, as determined by ChIP followed by quantitative PCR (qPCR) (Supplemental Fig. S1). For subsequent genome-wide binding site analyses, we use 9D6 data exclusively because of its high signal-to-noise ratio and well-characterized specificity (Fig. 1; Supplemental Fig. S2; data not shown; Kawamura et al. 2008).
Figure 1.
ChIP-seq profiles of AGO2 in S2 and S3 cells at BX-C. AGO2 ChIP-seq profiles of input DNA and immunoprecipitations in S2 and S3 cells compared with tiling array ChIP data for CTCF, CP190, GAF (Negre et al. 2010), Trx-N, Pho, and Pc (Schuettengruber et al. 2009) in indicated cell types or embryos over the BX-C region (top) and Abd-B locus (bottom). Coding sequences, promoters, and cis-regulatory regions are shown. ChIP-seq scales are in reads per million unique mapped reads. Input samples are shown on the same scale relative to respective immunoprecipitation and are therefore directly comparable. ChIP–chip data are expressed as either log2 (IP/input) or MA2C score. The bottom of each scale bar indicates 0.
AGO2 colocalizes with chromatin insulator sites throughout the genome
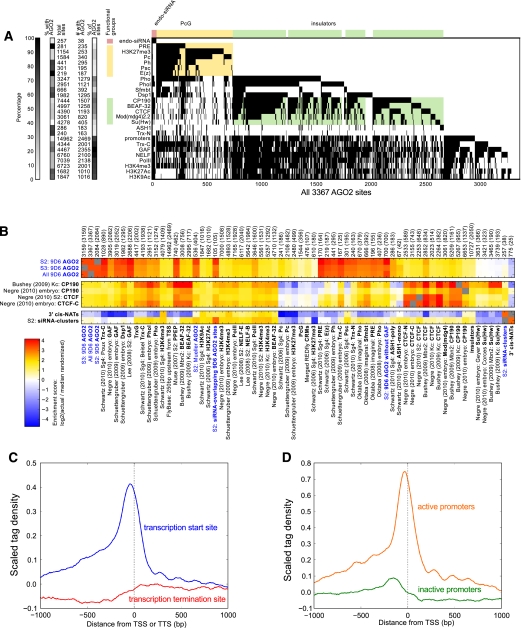
Consistent with binding at BX-C boundary sites, ∼62% of AGO2 sites overlap with known chromatin insulator proteins. Comparison of AGO2 ChIP-seq profiles with previously determined genome-wide ChIP tiling array analyses indicates extensive overlap with the insulator proteins CP190, CTCF, and BEAF-32, and modest similarity to Mod(mdg4)2.2 compared with random expectation (Fig. 2A,B; Supplemental Fig. S3). In contrast, AGO2 sites display no statistically significant overlap with the gypsy insulator protein Su(Hw), indicating specificity of the AGO2 correspondence with CTCF/CP190 insulators.
Figure 2.
Overlap between genome-wide binding sites of AGO2, insulator, TrxG/PcG, transcription-related factors, and promoters. (A) Binary heat map of AGO2-binding sites ordered by supervised hierarchical clustering. Each column represents one of the 3367 AGO2-binding sites across both S2 and S3 cell types, and each row represents overlapping binding sites for a particular factor across all available data sets. A mark in a row indicates that the indicated protein colocalizes with AGO2 at that site. AGO2 sites are classified into functional groups (endo-siRNA, PcG, and insulators). Feature counts for each factor and the number of features that intersect with the set of all AGO2 sites are shown. (Left) Corresponding percentages of overlap for each factor or for AGO2 are represented as grayscale values. (B) Heat map of log2 enrichment scores for pairwise comparisons of binding sites for AGO2, CP190, CTCF, 3′ cis-NATs, and endo-siRNA clusters with additional data sets. Enrichment score was calculated by dividing the actual overlapping feature count by the median overlapping feature count from 1000 random shufflings of features. Empirical P-values reported in the text are the percentile of the actual overlapping feature count in this null distribution. (Left) Color scale corresponding to enrichment value is indicated. Positive values indicate significant enrichment, while negative values indicate significant negative correlation of enrichment. Self–self comparisons are indicated in gray, and pairwise comparisons that are not statistically significant (P > 0.001) are indicated in white. Numbers along the top of each column indicate the total number of features in each data set, and the number of sites that interact with all AGO2 sites are indicated in parentheses. Full heat map with hierarchical clustering is shown in Supplemental Figure S3. (C) Half of AGO2-binding sites correspond to promoters. Profile of S2 AGO2 ChIP-seq tag density subtracted by input density around TSSs (blue) or transcription termination sites (red) from coding genes (FlyBase release 5.23) generated using CEAS. (D) AGO2 associates preferentially with active promoters. Profile of S2 AGO2 ChIP-seq tag density subtracted by input density around TSSs associated (orange) or not associated (green) with H3K4me2 and Pol II 250 bp upstream or 750 bp downstream.
In order to confirm the genome-wide colocalization of AGO2 with insulator proteins and specific association with euchromatin, we stained highly replicated polytene chromosomes of third instar larvae by indirect immunofluorescence. AGO2 staining is mainly observed at euchromatic DAPI interbands, which correspond to decondensed regions of the genome bearing the majority of transcribed genes (Supplemental Fig. S2). In contrast, AGO2 is not visible at the heterochromatic chromocenter, at which the centromere of each chromosome coalesces. In AGO251B-null mutants (Xu et al. 2004), this staining pattern is dramatically reduced, verifying the specificity of the antibody. In wild type, modest genome-wide colocalization is observed between AGO2 and CTCF, while more extensive overlap is seen between AGO2 and CP190, consistent with our ChIP-seq results (Supplemental Fig. S2).
AGO2 associates with active promoters
Like insulator proteins, over half of AGO2-binding sites are located at promoters. Extensive promoter association has been reported for the insulator proteins CP190, CTCF, Mod(mdg4)2.2, and BEAF-32, but not Su(Hw) (Bushey et al. 2009; Jiang et al. 2009; Smith et al. 2009), with a preference for active promoters (Negre et al. 2010). Genome-wide, 61% of AGO2 sites in S2 cells are found within 250 base pairs (bp) upstream of a transcription start site (TSS), with a slight bias upstream of the TSS (Fig. 2C). In contrast, no enrichment of binding is seen proximal to transcription termination sites. We next compared AGO2-binding sites at TSSs associated with RNA Pol II and H3K4me3 in the body of the gene with those lacking these active marks of transcription and found that AGO2 associates preferentially with active promoters (Fig. 2D), corresponding to ∼14% of all active promoters. Consistent with this finding, AGO2 associates with all five active Abd-B promoters in S3 cells but with only the RB and RE (also known as m and γ, respectively) inactive promoters in S2 cells (Fig. 1). Moreover, AGO2 associates with the iab-8 enhancer in S3 but not in S2 cells, suggesting that its chromatin association with certain enhancers and promoters may be dependent on active transcription.
AGO2 chromatin association does not correspond to regions of the genome that produce endo-siRNA
In contrast to its association with insulator proteins, AGO2 genome-wide localization does not coincide with regions of the genome that produce Dicer-dependent endo-siRNA. First, we compared the genome-wide distribution of AGO2-binding sites with a set of 257 clusters of high-density AGO2-bound unique endo-siRNAs in S2 cells (Czech et al. 2008; Ghildiyal et al. 2008; Kawamura et al. 2008; Okamura et al. 2008) and found overlap with <1% of AGO2 sites, which is not statistically significant compared with random expectations (P = 0.35) (Fig. 2A,B). As an additional test, we calculated the densities of unique endo-siRNA-matching AGO2 chromatin-binding sites in comparison with regions of the genome known to produce endo-siRNAs. Only 30% of 3′ cis-natural antisense transcripts (cis-NATs) have been shown to produce Dicer-dependent endo-siRNAs (Okamura et al. 2008; Roy et al. 2010). We used the endo-siRNA densities of all known 3′ cis-NATs to perform a conservative comparison with AGO2 chromatin-associated sites. We then calculated the endo-siRNA densities of sets of AGO2-binding regions and 3′ cis-NATs shuffled throughout the genome in order to randomize their positions. Normalized to relative random expectations, substantially more 3′ cis-NATs produce more than eight endo-siRNAs per kilobase compared with AGO2 sites, which produce much lower levels of endo-siRNA (Supplemental Fig. S4A).
Production of such a low level of endo-siRNA at AGO2 sites may be due to the fact that AGO2-bound sites are associated with active transcription. We hypothesized that actively transcribed regions may produce more endo-siRNA than transcriptionally silent regions. In fact, the top 5% highest endo-siRNA density AGO2 sites cluster with marks of active transcription in our heat map (Supplemental Fig. S3). Therefore, we repeated the endo-siRNA density analysis using Pol II- and H3K27me3-bound regions, which represent transcriptionally active and inactive sites, respectively. Overall, we found that Pol II-bound regions produce moderately higher levels of endo-siRNA than AGO2 sites, while H3K27me3-bound regions produce lower levels of endo-siRNA compared with respective random expectations (Supplemental Fig. S4A,B). Similar results were obtained using a nuclear library of S2 endo-siRNA (Supplemental Fig. S4C,D; Fagegaltier et al. 2009). These results suggest that regions of active transcription tend to produce low levels of endo-siRNA, and the majority of AGO2-binding sites correspond to little or no endo-siRNA production.
AGO2 associates with polycomb response elements (PREs) and overlaps extensively with TrxG and PcG proteins
Approximately 15% of AGO2 sites correspond to regions that can be regulated by both TrxG and PcG proteins. The TrxG and PcG complexes maintain transcriptional activation or repression, respectively, of critical developmental regulators and are recruited by DNA-binding proteins that recognize PREs, which are frequently juxtaposed to chromatin insulators (for review, see Simon and Kingston 2009). This close configuration is particularly evident in the BX-C locus, in which insulators act as barriers to constrain PRE activity directionally. The high resolution afforded by ChIP-seq allows AGO2 detection specifically at all known PREs in the BX-C (bx, bxd, iab-2, Fab-6, Fab-7, and Fab-8) despite their close proximity to insulators in this locus (Figs. 1, 2A). Additionally, AGO2 associates with 84% of PREs across the genome, as previously defined (Fig. 2A, left panel; Oktaba et al. 2008; Schwartz et al. 2010). We note that the probability-based enrichment values calculated for the AGO2 overlap with PREs and associated factors are higher than that with insulator proteins; this result is influenced by the small number of sites bound by PcG proteins genome-wide compared with insulator proteins (Fig. 2B). Finally, mild but statistically significant overlap is also detected between AGO2 and annotated cis-regulatory modules (CRMs) in the REDFly database (Gallo et al. 2011), which is biased toward extensively studied TrxG- and PcG-regulated genes.
AGO2 chromatin localization at PREs resembles that of TrxG proteins more closely than that of PcG proteins. Genome-wide, AGO2 overlaps extensively with the TrxG proteins Trx-N, Trx-C, and Ash1, as well as with the recruiter proteins Pho, Phol, Sfmbt, Dsp1, and GAF, which also associate with non-PRE sites in the genome (Fig. 2A,B). Furthermore, AGO2 colocalizes substantially with the sharply peaking PRE-associated PcG proteins E(z), Ph, and Psc, as well as the broadly spreading Pc and H3K27me3; however, AGO2 itself does not bind chromatin in extended domains (Figs. 1, 2A,B). Furthermore, AGO2 associates with both Fab-7 and Fab-8 PREs in S2 cells, in which Abd-B is silent, as well as in S3 cells, in which Abd-B is expressed (Fig. 1). Likewise, recruiter and TrxG proteins bind at Abd-B and its PREs irrespective of transcriptional expression state (Beisel et al. 2007), whereas PcG recruitment at Abd-B is only apparent in S2 cells (Breiling et al. 2004). This observation suggests that AGO2 does not require PcG proteins in order to associate with PREs.
In order to obtain further insight into the specificity of AGO2 chromatin association, we performed de novo motif analysis of AGO2-binding sites. We analyzed the central 500 bp of 500 random AGO2-binding sites using the MEME algorithm (Bailey and Elkan 1995) and identified a GA-rich consensus binding sequence reminiscent of the binding motif for the TrxG and insulator-associated GAGA factor (GAF) (Supplemental Fig. S5; Farkas et al. 1994; Belozerov et al. 2003; Schweinsberg et al. 2004). Similar results were obtained using all or non-GAF-occupied AGO2-binding sites with the GADEM (Li 2009) and Weeder (Pavesi and Pesole 2006) algorithms (data not shown).
AGO2 opposes Polycomb function
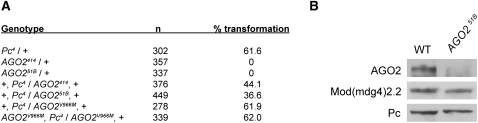
Given the high overlap of AGO2 with TrxG proteins, we tested whether AGO2 affects either TrxG or PcG function. We anticipated that AGO2 may function as a trxG gene, since the genes that encode GAF and Mod(mdg4)2.2 chromatin insulator proteins have been shown to behave as trxG genes (Farkas et al. 1994; Gerasimova and Corces 1998). We examined the classic posterior-to-anterior transformation phenotype of Pc4/+ mutants and determined that 62% of adult males exhibit ectopic sex combs on second and/or third legs (Fig. 3A). The AGO2414/+ mutation results in a mild suppression of the Pc4/+ phenotype, such that a reduced number of double mutant males, 44%, display transformation. Interestingly, the partial loss-of-function AGO2414/+ mutation is not defective for RNAi-dependent silencing in the heterozygous state (Okamura et al. 2004). Furthermore, heterozygous-null AGO251B/+ mutants display stronger suppression of the Pc4/+ phenotype in that only 37% of flies exhibit transformation. Neither AGO2414/+ nor AGO251B/+ mutants, both of which harbor deletions of the first two exons of AGO2 (Okamura et al. 2004; Xu et al. 2004), exhibit developmental delays compared with wild type (data not shown). The AGO2V966M point mutation results in production of wild-type levels of catalytically inactive protein incompetent for RNAi-dependent silencing (Kim et al. 2007) but capable of associating with polytene chromosomes (data not shown). Importantly, the heterozygous AGO2V966M/+ or homozygous AGO2V966M mutations do not affect the Pc4/+ phenotype, indicating that Slicer catalytic activity of AGO2 is not required for the suppression of the Pc4/+ phenotype. This suppression is not due to an indirect effect on Pc gene expression, as Pc protein levels are equivalent in wild type and AGO251B mutants (Fig. 3B). These results indicate that AGO2 behaves as a trxG gene and can counteract PcG function.
Figure 3.
AGO2 behaves as a TrxG protein. (A) Percentage of adult male flies displaying second and/or third legs with at least one ectopic sex comb tooth as an indication of posterior-to-anterior transformation was scored in the indicated genotypes, and number of flies (n) scored is shown. (B) Western blotting of AGO2, Pc, and Mod(mdg4)2.2 in wild-type and AGO251B adult male extracts.
AGO2, but not its catalytic activity, is specifically required for Fab-8 insulator activity
Given the high overlap of AGO2 with insulator sites throughout the genome, particularly of the CP190 class, we wished to determine whether AGO2 is required for activity of the well-characterized CTCF/CP190-dependent insulator Fab-8 of the Abd-B locus. We used a transgenic enhancer-blocking assay in which a genomic fragment containing the Fab-8 insulator and PRE positioned between a mini-white (mini-w+) reporter and w+ enhancer reduces reporter expression, resulting in intermediate levels of pigmentation in the adult eye (Barges et al. 2000). Compared with wild type, AGO2414/+, AGO251B/+, AGO2414, and AGO251B mutants carrying the Fab-8 insulator transgene display increased eye pigmentation corresponding to the strength of AGO2 loss-of-function mutation, indicating a positive role for AGO2 in Fab-8 insulator function (Fig. 4A). Importantly, the AGO2V966M catalytic activity mutant remains fully competent for Fab-8 insulator activity. In comparison, loss-of-function CP1904-1/CP190H31-2 mutants (Pai et al. 2004) that reduce Fab-8 insulator function (Gerasimova et al. 2007) display a more modest increase of mini-w+ expression than AGO251B/+ mutants (Supplemental Fig. S6). No differences compared with wild type are detected in AGO2 mutant flies carrying a transgene containing only the Fab-8 PRE or no cis-regulatory sequence (Fig. 4A), indicating that the effects on the Fab-8 insulator reporter are likely specific to the insulator.
Figure 4.
AGO2, but not its catalytic activity, is required for Fab-8 insulator function. (A) Eye color due to expression of a transgenic construct carrying no regulatory element (top row), Fab-8 insulator and PRE (middle row), or Fab-8 PRE (bottom row) between the mini-white enhancer and its coding sequence in wild-type, AGO2414/+, AGO251B/+ AGO2414, AGO251B, and AGO2V966M flies. (B) Visualization of insulator bodies by indirect immunofluorescence of whole-mount larval imaginal discs using α-CP190 antibodies (red) merged with DAPI staining (blue) in wild type and AGO251B mutants. (C) Polytene chromosome staining of α-CTCF (green), α-CP190 (red), and merged images in wild type and AGO251B mutants. Arrows point to the BX-C locus. (D) Western blotting of CP190, CTCF, and Pep (loading control) in wild-type and AGO251B pupal extracts.
Comprehensive genetic analysis of RNA silencing mutants revealed that AGO2, but not other RNA silencing factors, is required for Fab-8 insulator activity. Using the Fab-8 insulator transgene assay, we found that mutation of Rm62 or Dcr-2 RNAi pathway genes does not affect Fab-8 insulator activity, further suggesting that AGO2 is required for Fab-8 insulator activity in a manner independent of the RNAi pathway (Supplemental Table S1). Moreover, no change in Fab-8 insulator activity is observed when miRNA or piRNA silencing pathways are disrupted.
In order to obtain mechanistic insight into the possible function of AGO2 with respect to Fab-8 insulator activity, we examined the in vivo localization of insulator proteins in AGO2 mutants. Previously, it was shown that positive or negative effects of certain RNA silencing mutants on gypsy insulator activity correlate with the integrity of insulator bodies (Lei and Corces 2006). The AGO251B-null mutation does not appear to reduce Fab-8 function by disrupting the integrity of insulator bodies (Fig. 4B). Furthermore, we examined the localization of CTCF and CP190 on polytene chromosomes of wild type compared with AGO251B mutants, and no overall differences in the ability of CTCF and CP190 to associate with chromatin or, specifically, with the BX-C were observed (Fig. 4C; Supplemental Fig. S2). Finally, Western blotting of wild type and AGO251B mutants indicates no effect on CTCF or CP190 protein levels (Fig. 4D).
AGO2 interacts physically with CTCF and CP190
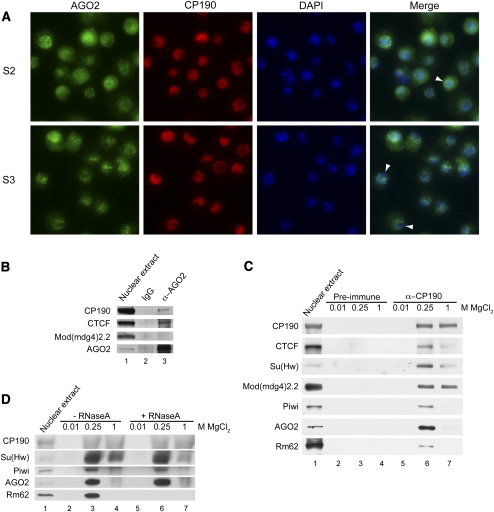
In order to address whether AGO2 influences chromatin insulator activity in a direct manner, we examined its subcellular localization compared with that of CP190. In S2 cells, CP190 localization is mainly diffuse within the nucleus, whereas in S3 cells, CP190 is nuclear but also concentrates into insulator bodies reminiscent of those seen in larval imaginal disc cells (Fig. 5A). In both S2 and S3 cells, AGO2 localizes throughout the cell but concentrates preferentially in the nucleoplasm in the majority of cells. Nuclear signal is reduced upon siRNA knockdown of AGO2 (data not shown). Importantly, AGO2 staining is excluded from the heterochromatic DAPI dot (Fig. 5A) and is mainly nonoverlapping with the heterochromatin protein HP1 (data not shown).
Figure 5.
AGO2 associates physically with CP190 and CTCF. (A) Indirect immunofluorescence of S2 and S3 cells using α-AGO2 (green) and α-CP190 (red) antibodies. DAPI staining (blue) and merged image of α-AGO2 with DAPI are also shown. Arrowheads point to heterochromatic regions that stain intensely with DAPI but are depleted for AGO2. (B) Western blotting of embryonic nuclear extracts immunoprecipitated with α-AGO2 antibodies. Nuclear extract (lane 1) bound to control IgG (lane 2) or α-AGO2 immobilized on ProtA-sepharose (lane 3) at >1.1 M monovalent salt concentration. (C) Western blotting of embryonic nuclear extracts (lane 1) bound to a control preimmune column (lanes 2–4) or α-CP190 column (lanes 5–7) and step-eluted with increasing MgCl2 concentrations as indicated. (D) Western blotting of embryonic nuclear extracts (lane 1) bound to α-CP190 columns either untreated (lanes 2–4) or treated with RNaseA (lanes 5–7) and step-eluted with increasing MgCl2 concentrations as indicated.
We next probed for physical interactions between insulator complexes and RNA silencing components. Immunoprecipitation of AGO2 from embryonic nuclear extracts at high monovalent salt concentrations results in copurification of CTCF and CP190 but not the gypsy insulator protein Mod(mdg4)2.2 (Fig. 5B). In addition, column-based immunoaffinity purification of CP190-associated complexes from nuclear extracts verifies the presence of core gypsy and Fab-8 insulator components CP190, Su(Hw), Mod(mdg4)2.2, and CTCF, and reveals association of the RNA silencing components Rm62, Piwi, and AGO2 (Fig. 5C). Interactions between insulator proteins, Piwi, or AGO2 and CP190 complexes are not affected by RNaseA treatment under conditions that disassociate Rm62 (Fig. 5D), suggesting that RNA does not mediate physical associations between Piwi or AGO2 and CP190. Physical interactions between these RNA silencing components and CP190, either direct or in the context of larger complexes, are consistent with the direct involvement of Piwi and Rm62 in gypsy insulator activity and that of AGO2 in CTCF/CP190 insulator activity.
AGO2 chromatin association requires CP190 and CTCF
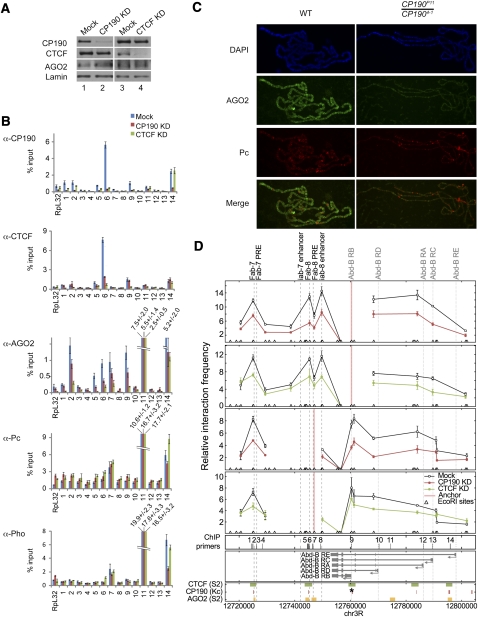
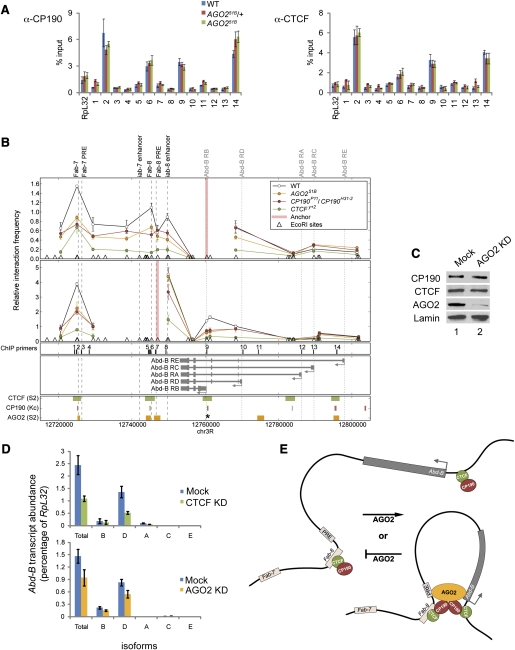
In order to determine whether CP190 and CTCF are required for AGO2 chromatin association, we performed ChIP analysis of the Abd-B locus in S2 cells depleted of CP190 or CTCF. The Abd-B gene, which harbors multiple juxtaposed cis-regulatory elements that control its spatiotemporal expression, serves as an ideal model locus to study the interplay between insulator and TrxG/PcG activities. Transfection of dsRNA corresponding to CP190 or CTCF results in reduction of the respective target protein by ∼90% (Fig. 6A). In control cells, ChIP using α-CP190 or α-CTCF antibodies results in enrichment of the Fab-8 insulator (primer set 6) more than eightfold over the RpL32 reference locus (Fig. 6B; Supplemental Fig. S7). Either CP190 or CTCF knockdown results in at least fourfold reduction of both CP190 and CTCF levels at the Fab-8 insulator, indicating mutual dependence for association with Fab-8.
Figure 6.
CP190 and CTCF are required for AGO2 chromatin association and looping interactions throughout the Abd-B locus. (A) Western blotting of lysates from S2 cells mock-treated (lanes 1,3) or transfected with CP190 (lane 2) or CTCF (lane 4) dsRNA. (B) S2 cells mock-treated (blue) or transfected with CP190 (red) or CTCF (green) dsRNA were subjected to ChIP using α-CP190, α-CTCF, α-AGO2, α-Pho, and α-Pc antibodies. Locations of primer sets are indicated in D. Percent input DNA immunoprecipitated is shown for each primer set, and error bars indicate standard deviation of quadruplicate PCR measurements. IgG-negative control immunoprecipitations for all sites yielded <0.08% input. (C) AGO2 chromatin association is reduced in CP190 mutant polytene chromosomes. Salivary gland polytene chromosome staining with DAPI (blue), α-AGO2 (green), α-Pc (red), and red/green merged images from wild-type (left) and CP190P11/CP1904-1 (right) larvae. (D) 3C looping interactions between cis-regulatory elements of the Abd-B locus are dependent on CP190 and CTCF. Relative interaction frequencies between EcoRI restriction fragments (triangles) and anchor regions (red vertical lines) are shown for mock (open circles), CP190-depleted cells (filled red circles), and CTCF-depleted cells (filled green circles). Samples are normalized by qPCR to an undigested locus. Error shown is standard deviation of quadruplicate TaqMan PCR reactions. Reported peaks for CTCF (red) and CP190 (green) ChIP–chip studies and MACS-determined peaks for AGO2 (orange) are shown (below). The asterisk notes a site (primer set 9) not identified by MACS but with clear enrichment for AGO2 as determined by directed ChIP in B.
Despite binding to more sites than CTCF/CP190 in the Abd-B locus, the majority of AGO2 chromatin association is dependent on both CP190 and CTCF. In control cells, ChIP of AGO2 results in approximately threefold enrichment of Fab-7 (set 2), Fab-8 insulator, and Fab-8 PRE (set 7) compared with RpL32, which shows low AGO2 association. Furthermore, 30-fold enrichment of the Abd-B RE promoter and 14-fold enrichment of an intronic site (set 11) over RpL32 are observed. In CP190- or CTCF-depleted cells, AGO2 recruitment to most sites is decreased ∼1.5-fold to threefold, including sites not bound by either insulator protein, such as the Abd-B intronic site and Fab-8 PRE. Therefore, CP190- and CTCF-dependent chromatin association of AGO2 is unlikely to be achieved exclusively through direct recruitment and may involve a mechanism such as chromatin looping. AGO2 protein levels are not decreased in CP190- or CTCF-depleted cells (Fig. 6A).
In order to rule out the possibility that depletion of CTCF or CP190 causes a general disruption of chromatin, we performed ChIP of the PcG components Pho and Pc. In S2 cells, Abd-B expression is repressed by PcG proteins, and high levels of Pho and Pc are found associated with the locus (Fig. 6B). Knockdown of CTCF or CP190 does not reduce Pho or Pc association at Abd-B, suggesting that loss of AGO2 on chromatin in CTCF and CP190 knockdowns is not due to general chromatin disruption or loss of PcG association. In order to confirm these ChIP results, we examined AGO2 localization on polytene chromosomes of wild type compared with CP190P11/CP1904-1 mutants (Pai et al. 2004) and found that genome-wide AGO2 levels decrease significantly when CP190 insulator function is lost, while Pc chromatin association is unaffected (Fig. 6C). Additionally, no change in GAF recruitment was observed (data not shown).
CP190 and CTCF are required for looping interactions throughout the Abd-B locus
Loss of AGO2 association with noninsulator regions of Abd-B in CP190 and CTCF knockdowns may be the result of changes in looping interactions at this locus. In order to determine whether CP190 or CTCF insulator proteins mediate these or other long-range interactions in this locus, we examined locus-wide interactions by 3C in S2 cells depleted for CP190 or CTCF. We scanned pairwise interactions using available EcoRI restriction sites in an 80-kb region encompassing the Fab-7 insulator to the most distal Abd-B RE promoter (Supplemental Fig. S8). Using an anchor at the Abd-B RB promoter, high interaction frequencies are observed with Fab-7, Fab-8, iab-8 enhancer, and Abd-B RD and RA (also known as C) promoters, but less with proximal or intervening sequences (Fig. 6D). High-frequency interactions are also detected using an anchor at the Fab-8 PRE, which loops to Fab-7 and each of the Abd-B promoters. Knockdown of CP190 or CTCF decreases peaks of high-frequency interactions by approximately twofold, with lesser effects on lower-frequency interactions. These results indicate a requirement for both CP190 and CTCF for looping interactions between insulators, PREs, enhancers, and promoters of the Abd-B locus.
AGO2 associates with chromatin dependent on CTCF and CP190
In order to examine whether AGO2 recruitment to chromatin is downstream from that of CTCF and CP190, we examined chromatin association of these insulator proteins in the absence of AGO2. No changes in CP190 or CTCF recruitment in AGO2 knockdowns were observed; however, a significant amount of residual AGO2 remains on chromatin despite at least 90% depletion of total AGO2 (data not shown). As a more rigorous test, we examined AGO251B-null mutants derived from mothers with AGO251B ovaries by deriving germline clones; these mutants contain no maternal or zygotic protein. ChIP was performed on adult heads of AGO251B/+ or AGO251B mutant siblings derived from the germline clones as well as from wild-type flies. ChIP profiles of CTCF and CP190 in adult head tissue are similar to that observed in S2 and S3 cells but with considerable enrichment at the Fab-7 insulator (Fig. 7A, primer set 2). Importantly, no changes were observed in AGO251B-null mutants compared with heterozygous siblings or with wild type. Pc chromatin association is also unchanged in AGO251B-null mutants (data not shown). These results, in combination with the finding that CP190 and CTCF localization is unchanged in polytene chromosomes of AGO251B-null mutants (Fig. 4C; Supplemental Fig. S2), suggest that AGO2 is not required for CTCF or CP190 recruitment.
Figure 7.
AGO2 is required for looping interactions throughout the Abd-B locus and proper gene expression. (A) CTCF and CP190 chromatin association is unaffected in AGO251B-null mutants. Adult heads of wild type (blue) as well as AGO251B/+ (red) or AGO251B (green) derived from AGO251B germline clones were subjected to ChIP using α-CP190 and α-CTCF. Locations of primer sets are indicated in B. Percent input DNA immunoprecipitated is shown for each primer set, and error bars indicate standard deviation of quadruplicate PCR measurements. (B) 3C looping interactions between cis-regulatory elements of the Abd-B locus are dependent on AGO2. Relative interaction frequencies between EcoRI restriction fragments (triangles) and anchor regions (red vertical lines) are shown for wild type (open circles) and CP190P11/CP190H31-2 (filled red circles), CTCFy+2 (filled green circles), and AGO251B (filled orange circles) mutant larval brains and imaginal discs. (C) Western blotting of lysates from S3 cells mock-treated (lane 1) or transfected with AGO2 siRNA (lane 2). (D) AGO2 and CTCF are required for proper Abd-B expression. RT–PCR to detect a common region or isoforms of Abd-B transcripts relative to RpL32 in oligo(dT) primed cDNA of S3 cells mock-treated (blue), CTCF-depleted (green), and AGO2-depleted (orange). Quantitative SYBR green PCR was performed in quadruplicate using S3 genomic DNA as a standard to normalize for primer efficiencies. (E) Model for AGO2 function with respect to CTCF/CP190 chromatin insulator activity. Looping at the Abd-B locus between the Fab-8 insulator and Abd-B promoter is dependent on CTCF/CP190 insulator interactions. This specialized configuration promotes interactions between Fab-8-associated cis-regulatory elements and the promoters to facilitate proper gene expression. AGO2 is recruited depending on CTCF/CP190 chromatin association and acts to either promote or stabilize looping interactions. Transfer of AGO2 to noninsulator sites may be achieved through CTCF/CP190-dependent looping interactions.
AGO2 is required for looping at the Abd-B locus
We next addressed the possibility that AGO2 is required for insulator-dependent looping interactions at Abd-B. Therefore, we performed 3C in diploid larval brains and imaginal discs of wild type compared with CTCF-null (Gerasimova et al. 2007), CP190-null, and AGO2-null mutants from germline clones. These tissues represent a mixed population with a minority of cells expressing Abd-B. Similar to S2 cells, high levels of interaction are observed between the Abd-B RB promoter anchor and Fab-7, Fab-8, and iab-8 enhancer in wild type (Fig. 7B). These interactions are decreased 1.5-fold to twofold in both CP190P11/CP190H31-2 and AGO251B mutants and further decreased in CTCFy+2-null mutants. In addition, using an anchor at Fab-8, interactions with Fab-7 and the Abd-B RB promoter are decreased approximately twofold in CP190 and AGO2 mutants, with a greater decrease in CTCF mutants compared with wild type, suggesting that AGO2 is required for CTCF/CP190 insulator-dependent looping interactions at Abd-B.
AGO2 is required for proper expression of Abd-B, similar to CTCF
Consistent with the hypothesis that AGO2 is required for CP190/CTCF insulator activity, we found that AGO2 is required for proper expression of Abd-B. Total and isoform-specific expression levels of Abd-B relative to RpL32 were determined in S3 cells depleted efficiently for either CTCF or AGO2 by dsRNA or siRNA knockdown, respectively (Fig. 7C). In both CTCF- and AGO2-depleted cells, an overall reduction of total as well as RD isoform-specific Abd-B levels is observed, as determined by qPCR (Fig. 7D; Supplemental Fig. S9). Reduction of Abd-B transcript levels is detectable in the AGO2 knockdown despite residual AGO2 remaining bound to chromatin, suggesting that Abd-B expression is highly sensitive to AGO2 levels. These results support the hypothesis that AGO2 contributes to CTCF/CP190 chromatin insulator activity at the Abd-B locus.
Discussion
In this study, we provide the first evidence for an Argonaute protein functioning directly on euchromatin to effect changes in gene expression. The genome-wide binding profile of AGO2 displays striking overlap with insulator proteins. Genetic analysis revealed that AGO2, independent of its catalytic activity, promotes Fab-8 insulator activity. Like known insulator proteins, AGO2 also associates with promoters and can oppose PcG function. Genome-wide AGO2 recruitment to chromatin is dependent on CTCF and CP190 binding and may be partially achieved via looping interactions among cis-regulatory regions and promoters. We propose that AGO2 may act to facilitate or stabilize looping that is needed to partition the genome into independent transcriptional domains (Fig. 7E).
AGO2 localizes predominantly to euchromatin and not heterochromatin
Our results suggest that the main function of AGO2 on chromatin resides in euchromatin and not in heterochromatin. Immunofluorescence localization of AGO2 on polytene chromosomes and cell lines indicates exclusion from heterochromatic and HP1-enriched regions. Furthermore, the majority of chromatin-associated AGO2 resides in nonrepetitive euchromatic but not repeat-rich regions, as determined by genome-wide ChIP-seq. We suggest that the role of AGO2 in RNAi-dependent silencing of TEs occurs primarily at the post-transcriptional level and that AGO2 harbors a second RNAi-independent activity to promote chromatin insulator function.
RNAi-independent function for AGO2 at chromatin
Several observations suggest that AGO2 chromatin association is mainly, if not exclusively, independent of the RNAi pathway. First, AGO2 chromatin association does not correspond to regions of the genome that produce high levels of endo-siRNAs, which are dependent on Dcr-2 and AGO2 (Chung et al. 2008; Czech et al. 2008; Ghildiyal et al. 2008; Kawamura et al. 2008; Okamura et al. 2008). Second, AGO2, but not Dcr-2, is required for Fab-8 insulator function. Finally, a catalytically inactive AGO2 protein, which is defective for RNAi, retains the ability to associate with chromatin and is functional with respect to both TrxG function and Fab-8 insulator activity.
An intriguing question raised by these findings is whether or not the functions of AGO2 in RNAi and chromatin insulator activity are completely distinct. We found that CP190 mutants remain competent for silencing using a GMR-wIR hairpin transgene (Lee et al. 2004), suggesting that AGO2 chromatin association is not required for RNAi (data not shown). Nevertheless, it remains possible that chromatin-associated AGO2 is loaded with siRNA. Future work will address how AGO2 subcellular localization and seemingly disparate functions in RNAi and chromatin insulator activities are regulated.
Role of AGO2 in Fab-8 insulator function
We identified a unique positive role for AGO2 but not other RNA silencing factors in Fab-8 insulator function. Importantly, a catalytically inactive mutant form of AGO2 expressed at wild-type levels retains insulator activity, further suggesting that the RNAi pathway is dispensable for Fab-8 insulator function. A significant fraction of AGO2 resides in the nucleus, and physical interaction is observed between AGO2 and CP190. This interaction is insensitive to RNaseA, suggesting that RNA does not mediate the interaction between AGO2 and CP190. It remains possible that AGO2 can interact with siRNA or other RNA while associated with the insulator complex, although we did not obtain any evidence to support this hypothesis.
We show for the first time that chromosomal looping in the Abd-B locus is dependent on CTCF, CP190, and AGO2. Confirming and extending previous studies, we found that the Abd-B RB promoter interacts frequently with Fab-7, Fab-8, and the iab-8 enhancer and, moreover, that the Fab-8 region also contacts Fab-7 as well as multiple Abd-B promoters. Currently, the significance of insulator protein promoter association is unclear, but insulators may be thus situated to control looping interactions between promoters and cis-regulatory elements. Depletion of CP190 or CTCF reduces these high-frequency looping interactions, and loss of this specialized chromatin configuration could result in disassociation of AGO2. Given this possibility, AGO2 may act to detect the insulator-dependent conformation of this locus.
AGO2 is recruited to chromatin insulator sites as well as noninsulator sites in a CTCF/CP190-dependent manner. We speculate that AGO2 chromatin association with insulator sites could result from physical interactions with CP190 complexes, while AGO2 recruitment to other sites may be achieved at least in part by chromatin looping mediated by CP190 and CTCF. In fact, it was recently shown that PcG proteins can be transferred from a PRE to a promoter as a result of intervening insulator–insulator interactions (Comet et al. 2011). Once recruited to chromatin, AGO2 could perform a primarily structural function to promote or stabilize the frequency of CTCF/CP190-dependent looping interactions.
Role of AGO2 in long-range chromosomal interactions
AGO2 appears to promote Fab-8 insulator activity independently of an effect on gypsy insulator body localization. Previous work showed that both the gypsy class and CTCF/CP190 insulators colocalize to insulator bodies, suggesting that these subnuclear structures may be important for both gypsy and Fab-8 activities (Gerasimova et al. 2007). However, since Fab-8 activity is not affected by RNA silencing components that disrupt gypsy insulator body localization, this subnuclear structure appears to be dispensable for Fab-8 function. Recent work indicates that the BX-C harbors multiple redundant cis-regulatory elements that can maintain looping interactions of this locus (Bantignies et al. 2011), suggesting that the configuration of the BX-C may not require a nuclear scaffold such as the gypsy insulator body.
AGO2 mutations suppress the Polycomb phenotype, indicating that AGO2 behaves similarly to trxG genes and opposes PcG function. A previous study proposed that RNA silencing factors promote long-range PRE-dependent chromosomal pairing as well as PcG body formation but did not examine AGO2 (Grimaud et al. 2006). We found that the AGO251B-null mutation has no effect on Fab-X PRE pairing-dependent silencing on sd as assayed in that study (data not shown), and our genetic results suggest that AGO2 is unlikely to promote PRE-dependent interactions or PcG body formation, which are both positively correlated with PcG function. Interestingly, it has recently been shown in the case of AGO2-associated Fab-7 and Mcp boundary elements that long-range interactions are dependent on insulator sequences and not PREs (Li et al. 2011). Future studies will elucidate the complex interplay between PcG and insulator organization as well as the role of AGO2 in the regulation of these structures.
Conclusions
It remains to be seen whether Drosophila AGO2 euchromatin association and function may be conserved in other organisms. In Caenorhabditis elegans, the nuclear NRDE RNAi pathway can block transcriptional elongation of Pol II on a target transcript when treated with exogenous complementary dsRNA (Guang et al. 2010). Interestingly, this negative transcriptional effect is contemporaneous with an increase in H3K9me3. Whether the Argonaute protein NRDE-3/WAGO-12, which lacks Slicer activity, associates with euchromatin to effect this repression is not yet known. Furthermore, the C. elegans Argonaute Csr-1, loaded with 22G endo-siRNAs antisense to mRNAs of holocentric chromosomes, may serve as chromosomal attachment points to promote efficient chromosome segregation (Claycomb et al. 2009; van Wolfswinkel et al. 2009). Recently, it has been shown that Schizosaccharomyces pombe Ago1 participates in surveillance mechanisms to prevent readthrough transcription of mRNA (Gullerova and Proudfoot 2008; Zofall et al. 2009; Halic and Moazed 2010). However, the majority of Ago1 associates with heterochromatic regions (Noma et al. 2004), and it is not clear thus far whether Ago1 directly associates with euchromatin or acts post-transcriptionally. An emerging theme from studies of RNAi in various model systems is that genome integrity and control of gene expression may be achieved by multiple yet overlapping mechanisms.
Materials and methods
Fly strains
Flies were maintained on standard cornmeal medium at room temperature or 25°C. Newly eclosed flies were collected and aged for 24–27 h and examined for eye pigmentation. Larvae for immunostaining of imaginal discs were raised at 25°C. Larvae for immunostaining of polytene chromosomes were raised at 18°C. The Fab-8 insulator + PRE transgene contains a HindIII–EcoRI fragment, and the Fab-8 PRE transgene contains an EcoRI–AflII fragment (Barges et al. 2000). Transgenes were scored as single copy. The AGO251B/+ mutation was tested on five independent Fab-8 insulator + PRE insertion lines, and similar results were observed.
Homozygous AGO251B flies exhibit a high degree of male and female sterility, but these phenotypes appear to be caused by second site mutations unlinked to the AGO2 mutation. Furthermore, AGO251B mutants exhibit a low, variable level of protein, likely maternally deposited. Consequently, homozygous mutant germline clones were produced by recombining the AGO251B mutation with FRT2A and inducing recombination with a ovoD1-marked FRT2A chromosome using a hs-FLP recombinase induced for 1 h in larvae at 5 d and 6 d of age as described previously (Selva and Stronach 2007). These flies were then crossed with AGO251B/+ males to obtain the desired progeny. The progeny were verified by Western blotting, PCR, and ChIP, and the same results were obtained with AGO2321/AGO2454-null mutants (Hain et al. 2010) from AGO2321 germline clones (data not shown).
Indirect immunofluorescence
Preparation and immunostaining of salivary gland polytene chromosomes was performed as described previously (Lei and Corces 2006). Cell staining and whole-mount staining are detailed in the Supplemental Material. Rabbit α-Su(Hw) (Moshkovich and Lei 2010), guinea pig α-CP190 (generated similarly as in Pai et al. 2004), rabbit α-CP190 (Pai et al. 2004), rat α-CTCF (Gerasimova et al. 2007), rabbit α-CTCF (Gerasimova et al. 2007), mouse α-AGO2 (9D6) (Kawamura et al. 2008), and rabbit α-Pc antibodies (a kind gift from D. Moazed and P. O'Farrell, University of California at San Francisco) were used for staining. Samples were mounted with Vectashield (Vector Laboratories), and images were acquired on a Leica DM5000 epifluorescence microscope with Openlab (PerkinElmer) software.
Western blotting
Lysates from whole pupae, the anterior third of third instar larvae, whole flies, or cell lines were prepared as described previously (Lei and Corces 2006). Guinea pig α-CP190, rabbit α-CP190, (generated similarly as in Pai et al. 2004), guinea pig α-Su(Hw) (generated similarly as in Moshkovich and Lei 2010), guinea pig α-Mod(mdg4)2.2 (Moshkovich and Lei 2010), rabbit α-CTCF, rat α-CTCF, mouse α-Pep (Amero et al. 1991), MAD1 mouse α-p68 (Ishizuka et al. 2002), mouse α-AGO2 (4D2) (Okamura et al. 2004), mouse α-AGO2 (9D6), rabbit α-Piwi (Abcam, ab-5207), rabbit α-AGO2 (Abcam, ab-5072), mouse α-Lamin (ADL67.10) (Stuurman et al. 1996), and rabbit α-Pc were used for Western blotting. Specificity of generated and commercial antibodies was verified by blotting mutant fly lysates and/or cells knocked down with the corresponding dsRNA.
Immunoaffinity purification
Immunoaffinity purification with α-CP190 and RNaseA treatment was carried out as described previously (Lei and Corces 2006). Immunoprecipitation with α-AGO2 (9D6) was performed using nuclei isolated from 20 g of 0- to 24-h embryos as described previously (Lei and Corces 2006). Nuclei were lysed by sonication in 5 mL of HBSMT-0.3% + 1 M KCl (50 mM HEPES, 150 mM NaCl, 1 M KCl, 3 mM MgCl2, 0.3% Triton X-100 [v/v] at pH 7) including 1 mM PMSF and Complete protease inhibitor cocktail (Roche), and extracts were prepared as described previously (Lei and Corces 2006). Extract (1.2 mL) was bound overnight at 4°C to 1 mL of α-AGO2 (9D6) tissue culture supernatant or 1.4 μg of mouse IgG (Santa Cruz Biotechnology) prebound to rProtA-sepharose for 1 h at 4°C. Beads were washed three times with HBSMT-0.3% + 1 M KCl, then once with HBSM (50 mM HEPES, 150 mM NaCl, 5 mM KCl, 3 mM MgCl2), and eluted with denaturing sample buffer by boiling for 5 min. Samples were Western blotted as described previously (Pai et al. 2004).
ChIP and ChIP-seq
Preparation of ChIP samples and analysis were performed essentially as described previously (Moshkovich and Lei 2010). S2 and S3 cells were grown at 25°C in Shield and Sangs M3 insect medium (Sigma) supplemented with 0.1% yeast extract, 0.25% bactopeptone, and 10% fetal bovine serum (HyClone). Immunoprecipitations were performed with α-AGO2 (9D6), rabbit α-CP190 (this study), rabbit α-CTCF, rabbit α-Pho (Fritsch et al. 1999), rabbit α-Pc, mouse IgG, and rabbit IgG (Santa Cruz Biotechnology) coupled to rProtein A agarose beads (GE Healthcare). Similar results, but with lower signal, were obtained with rabbit α-AGO2 (Jiang et al. 2005), rabbit α-AGO2 (Meyer et al. 2006), or α-Flag (M2, Sigma) or α-HA (12CA5, Santa Cruz Biotechnology) using chromatin prepared from HA/Flag-AGO2 transgenic flies also expressing wild-type AGO2 (Czech et al. 2008). The primers used are indicated in Supplemental Table S3.
Samples for ChIP-seq from input DNA and AGO2 ChIP were prepared according to the manufacturer's protocol (Illumina). DNA was sequenced on an Illumina Genome Analyzer at the NIDDK Genomics Core. Computational methods are detailed in the Supplemental Material. AGO2 ChIP-seq data are available at Gene Expression Omnibus (GSE22623).
dsRNA and siRNA knockdowns
Amplicons used for dsRNA knockdowns were designed based on recommendations from the Drosophila RNAi Screening Center. Templates were PCR-amplified from genomic DNA using primers containing the T7 promoter sequence. dsRNAs were produced by in vitro transcription of PCR templates using the MEGAscript T7 kit (Ambion) and purified using NucAway spin columns (Ambion). Transfections using 200 ng to 1.25 μg of dsRNA or 100 pmol of siRNA per million cells, or no dsRNA/siRNA for mock treatment, were performed using Cellfectin (Invitrogen), Effectene (Qiagen), or Cell Line Nucleofector kit V (Amaxa Biosystems) transfection reagent using the recommended protocol. Four days to 6 d after transfection, cells were collected and knockdown efficiency was confirmed by Western blotting. The highest knockdown efficiencies were generally obtained using the Amaxa system. No differences were seen with mock treatment, GFP dsRNA, or luciferase dsRNAs. Primers used are indicated in Supplemental Table S3, and 3C methods are detailed in the Supplemental Material.
RT–PCR
Total RNA was isolated from S3 cells using Trizol reagent (Invitrogen) using the recommended protocol. Reverse transcription of 1 μg of total RNA was performed using oligo(dT) as a primer and SuperScript III reverse transcriptase (Invitrogen) using the manufacturer's protocol. Transcript levels were quantified in the linear amplification range by real-time PCR using HotStart-IT SYBR green qPCR Master Mix (USB Corporation) by calibration to a standard curve of genomic DNA to account for differences in primer efficiencies.
Acknowledgments
We thank A. Beyer for α-Pep; F. Fuller-Pace for α-p68; J. Kassis for α-Pho; Q. Liu, A. Mueller, and M. Siomi for α-AGO2; P. O'Farrell and D. Moazed for α-Pc; and C. Berg, J. Birchler, R. Carthew, G. Gavalli, V. Corces, G. Hannon, S. Hou, F. Karch, J. Kassis, E. Lai, G. Shanower, P. Schedl, and P. Zamore for strains. We are indebted to P. Murphy for antibody characterization; M. Emmett for primers; A. Dean for 3C protocols; S. Grewal, F. Karch, and B. Oliver for discussions; and J. Kassis and L. Matzat for critical reading of the manuscript. This work was supported by the Intramural Program of the National Institute of Diabetes and Digestive and Kidney Diseases.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.16651211.
References
- Amero SA, Elgin SC, Beyer AL 1991. A unique zinc finger protein is associated preferentially with active ecdysone-responsive loci in Drosophila. Genes Dev 5: 188–200 [DOI] [PubMed] [Google Scholar]
- Bailey TL, Elkan C 1995. The value of prior knowledge in discovering motifs with MEME. Proc Int Conf Intell Syst Mol Biol 3: 21–29 [PubMed] [Google Scholar]
- Bantignies F, Roure V, Comet I, Leblanc B, Schuettengruber B, Bonnet J, Tixier V, Mas A, Cavalli G 2011. Polycomb-dependent regulatory contacts between distant Hox loci in Drosophila. Cell 144: 214–226 [DOI] [PubMed] [Google Scholar]
- Barges S, Mihaly J, Galloni M, Hagstrom K, Muller M, Shanower G, Schedl P, Gyurkovics H, Karch F 2000. The Fab-8 boundary defines the distal limit of the bithorax complex iab-7 domain and insulates iab-7 from initiation elements and a PRE in the adjacent iab-8 domain. Development 127: 779–790 [DOI] [PubMed] [Google Scholar]
- Beisel C, Buness A, Roustan-Espinosa IM, Koch B, Schmitt S, Haas SA, Hild M, Katsuyama T, Paro R 2007. Comparing active and repressed expression states of genes controlled by the Polycomb/Trithorax group proteins. Proc Natl Acad Sci 104: 16615–16620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belozerov VE, Majumder P, Shen P, Cai HN 2003. A novel boundary element may facilitate independent gene regulation in the Antennapedia complex of Drosophila. EMBO J 22: 3113–3121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiling A, O'Neill LP, D'Eliseo D, Turner BM, Orlando V 2004. Epigenome changes in active and inactive polycomb-group-controlled regions. EMBO Rep 5: 976–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushey AM, Dorman ER, Corces VG 2008. Chromatin insulators: regulatory mechanisms and epigenetic inheritance. Mol Cell 32: 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushey AM, Ramos E, Corces VG 2009. Three subclasses of a Drosophila insulator show distinct and cell type-specific genomic distributions. Genes Dev 23: 1338–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung WJ, Okamura K, Martin R, Lai EC 2008. Endogenous RNA interference provides a somatic defense against Drosophila transposons. Curr Biol 18: 795–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claycomb JM, Batista PJ, Pang KM, Gu W, Vasale JJ, van Wolfswinkel JC, Chaves DA, Shirayama M, Mitani S, Ketting RF, et al. 2009. The Argonaute CSR-1 and its 22G-RNA cofactors are required for holocentric chromosome segregation. Cell 139: 123–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleard F, Moshkin Y, Karch F, Maeda RK 2006. Probing long-distance regulatory interactions in the Drosophila melanogaster bithorax complex using Dam identification. Nat Genet 38: 931–935 [DOI] [PubMed] [Google Scholar]
- Comet I, Schuettengruber B, Sexton T, Cavalli G 2011. A chromatin insulator driving three-dimensional Polycomb response element (PRE) contacts and Polycomb association with the chromatin fiber. Proc Natl Acad Sci 108: 2294–2299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B, Malone CD, Zhou R, Stark A, Schlingeheyde C, Dus M, Perrimon N, Kellis M, Wohlschlegel JA, Sachidanandam R, et al. 2008. An endogenous small interfering RNA pathway in Drosophila. Nature 453: 798–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande G, Calhoun G, Schedl P 2005. Drosophila argonaute-2 is required early in embryogenesis for the assembly of centric/centromeric heterochromatin, nuclear division, nuclear migration, and germ-cell formation. Genes Dev 19: 1680–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagegaltier D, Bouge AL, Berry B, Poisot E, Sismeiro O, Coppee JY, Theodore L, Voinnet O, Antoniewski C 2009. The endogenous siRNA pathway is involved in heterochromatin formation in Drosophila. Proc Natl Acad Sci 106: 21258–21263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas G, Gausz J, Galloni M, Reuter G, Gyurkovics H, Karch F 1994. The Trithorax-like gene encodes the Drosophila GAGA factor. Nature 371: 806–808 [DOI] [PubMed] [Google Scholar]
- Fritsch C, Brown JL, Kassis JA, Muller J 1999. The DNA-binding polycomb group protein pleiohomeotic mediates silencing of a Drosophila homeotic gene. Development 126: 3905–3913 [DOI] [PubMed] [Google Scholar]
- Gallo SM, Gerrard DT, Miner D, Simich M, Des Soye B, Bergman CM, Halfon MS 2011. REDfly v3.0: toward a comprehensive database of transcriptional regulatory elements in Drosophila. Nucleic Acids Res 39: D118–D123 doi: 10.1093/nar/gkq999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerasimova TI, Corces VG 1998. Polycomb and trithorax group proteins mediate the function of a chromatin insulator. Cell 92: 511–521 [DOI] [PubMed] [Google Scholar]
- Gerasimova TI, Lei EP, Bushey AM, Corces VG 2007. Coordinated control of dCTCF and gypsy chromatin insulators in Drosophila. Mol Cell 28: 761–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Seitz H, Horwich MD, Li C, Du T, Lee S, Xu J, Kittler EL, Zapp ML, Weng Z, et al. 2008. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science 320: 1077–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SI, Elgin SC 2007. Transcription and RNA interference in the formation of heterochromatin. Nature 447: 399–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimaud C, Bantignies F, Pal-Bhadra M, Ghana P, Bhadra U, Cavalli G 2006. RNAi components are required for nuclear clustering of Polycomb group response elements. Cell 124: 957–971 [DOI] [PubMed] [Google Scholar]
- Guang S, Bochner AF, Burkhart KB, Burton N, Pavelec DM, Kennedy S 2010. Small regulatory RNAs inhibit RNA polymerase II during the elongation phase of transcription. Nature 465: 1097–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gullerova M, Proudfoot NJ 2008. Cohesin complex promotes transcriptional termination between convergent genes in S. pombe. Cell 132: 983–995 [DOI] [PubMed] [Google Scholar]
- Hain D, Bettencourt BR, Okamura K, Csorba T, Meyer W, Jin Z, Biggerstaff J, Siomi H, Hutvagner G, Lai EC, et al. 2010. Natural variation of the amino-terminal glutamine-rich domain in Drosophila argonaute2 is not associated with developmental defects. PLoS ONE 5: e15264 doi: 10.1371/journal.pone.0015264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halic M, Moazed D 2010. Dicer-independent primal RNAs trigger RNAi and heterochromatin formation. Cell 140: 504–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizuka A, Siomi MC, Siomi H 2002. A Drosophila fragile X protein interacts with components of RNAi and ribosomal proteins. Genes Dev 16: 2497–2508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F, Ye X, Liu X, Fincher L, McKearin D, Liu Q 2005. Dicer-1 and R3D1-L catalyze microRNA maturation in Drosophila. Genes Dev 19: 1674–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang N, Emberly E, Cuvier O, Hart CM 2009. Genome-wide mapping of boundary element-associated factor (BEAF) binding sites in Drosophila melanogaster links BEAF to transcription. Mol Cell Biol 29: 3556–3568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura Y, Saito K, Kin T, Ono Y, Asai K, Sunohara T, Okada TN, Siomi MC, Siomi H 2008. Drosophila endogenous small RNAs bind to Argonaute 2 in somatic cells. Nature 453: 793–797 [DOI] [PubMed] [Google Scholar]
- Kim K, Lee YS, Carthew RW 2007. Conversion of pre-RISC to holo-RISC by Ago2 during assembly of RNAi complexes. RNA 13: 22–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrchanova O, Toshchakov S, Podstreshnaya Y, Parshikov A, Georgiev P 2008. Functional interaction between the Fab-7 and Fab-8 boundaries and the upstream promoter region in the Drosophila Abd-B gene. Mol Cell Biol 28: 4188–4195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzuolo C, Roure V, Dekker J, Bantignies F, Orlando V 2007. Polycomb response elements mediate the formation of chromosome higher-order structures in the bithorax complex. Nat Cell Biol 9: 1167–1174 [DOI] [PubMed] [Google Scholar]
- Lee YS, Nakahara K, Pham JW, Kim K, He Z, Sontheimer EJ, Carthew RW 2004. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell 117: 69–81 [DOI] [PubMed] [Google Scholar]
- Lei EP, Corces VG 2006. RNA interference machinery influences the nuclear organization of a chromatin insulator. Nat Genet 38: 936–941 [DOI] [PubMed] [Google Scholar]
- Li L 2009. GADEM: a genetic algorithm guided formation of spaced dyads coupled with an EM algorithm for motif discovery. J Comput Biol 16: 317–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HB, Muller M, Bahechar IA, Kyrchanova O, Ohno K, Georgiev P, Pirrotta V 2011. Insulators, not Polycomb response elements, are required for long-range interactions between Polycomb targets in Drosophila melanogaster. Mol Cell Biol 31: 616–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkenschlager M 2010. Cohesin: a global player in chromosome biology with local ties to gene regulation. Curr Opin Genet Dev 20: 555–561 [DOI] [PubMed] [Google Scholar]
- Meyer WJ, Schreiber S, Guo Y, Volkmann T, Welte MA, Muller HA 2006. Overlapping functions of argonaute proteins in patterning and morphogenesis of Drosophila embryos. PLoS Genet 2: e134 doi: 10.1371/journal.pgen.0020134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi K, Tsukumo H, Nagami T, Siomi H, Siomi MC 2005. Slicer function of Drosophila Argonautes and its involvement in RISC formation. Genes Dev 19: 2837–2848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moshkovich N, Lei EP 2010. HP1 recruitment in the absence of argonaute proteins in Drosophila. PLoS Genet 6: e1000880 doi: 10.1371/journal.pgen.1000880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negre N, Brown CD, Shah PK, Kheradpour P, Morrison CA, Henikoff JG, Feng X, Ahmad K, Russell S, White RA, et al. 2010. A comprehensive map of insulator elements for the Drosophila genome. PLoS Genet 6: e1000814 doi: 10.1371/journal.pgen.1000814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noma K, Sugiyama T, Cam H, Verdel A, Zofall M, Jia S, Moazed D, Grewal SI 2004. RITS acts in cis to promote RNA interference-mediated transcriptional and post-transcriptional silencing. Nat Genet 36: 1174–1180 [DOI] [PubMed] [Google Scholar]
- Okamura K, Lai EC 2008. Endogenous small interfering RNAs in animals. Nat Rev Mol Cell Biol 9: 673–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Ishizuka A, Siomi H, Siomi MC 2004. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev 18: 1655–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Balla S, Martin R, Liu N, Lai EC 2008. Two distinct mechanisms generate endogenous siRNAs from bidirectional transcription in Drosophila melanogaster. Nat Struct Mol Biol 15: 998. [DOI] [PubMed] [Google Scholar]
- Oktaba K, Gutierrez L, Gagneur J, Girardot C, Sengupta AK, Furlong EE, Muller J 2008. Dynamic regulation by polycomb group protein complexes controls pattern formation and the cell cycle in Drosophila. Dev Cell 15: 877–889 [DOI] [PubMed] [Google Scholar]
- Pai CY, Lei EP, Ghosh D, Corces VG 2004. The centrosomal protein CP190 is a component of the gypsy chromatin insulator. Mol Cell 16: 737–748 [DOI] [PubMed] [Google Scholar]
- Pavesi G, Pesole G 2006. Using Weeder for the discovery of conserved transcription factor binding sites. Curr Protoc Bioinformatics 15: 2.11.1–2.11.19 doi: 10.1002/0471250953.bi0211s15 [DOI] [PubMed] [Google Scholar]
- Phillips JE, Corces VG 2009. CTCF: master weaver of the genome. Cell 137: 1194–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, Ernst J, Kharchenko PV, Kheradpour P, Negre N, Eaton ML, Landolin JM, Bristow CA, Ma L, Lin MF, et al. 2010. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science 330: 1787–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuettengruber B, Ganapathi M, Leblanc B, Portoso M, Jaschek R, Tolhuis B, van Lohuizen M, Tanay A, Cavalli G 2009. Functional anatomy of polycomb and trithorax chromatin landscapes in Drosophila embryos. PLoS Biol 7: e13 doi: 10.1371/journal.pbio.1000013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz YB, Kahn TG, Stenberg P, Ohno K, Bourgon R, Pirrotta V 2010. Alternative epigenetic chromatin states of polycomb target genes. PLoS Genet 6: e1000805 doi: 10.1371/journals.pgen.100805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweinsberg S, Hagstrom K, Gohl D, Schedl P, Kumar RP, Mishra R, Karch F 2004. The enhancer-blocking activity of the Fab-7 boundary from the Drosophila bithorax complex requires GAGA-factor-binding sites. Genetics 168: 1371–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selva EM, Stronach BE 2007. Germline clone analysis for maternally acting Drosophila hedgehog components. Methods Mol Biol 397: 129–144 [DOI] [PubMed] [Google Scholar]
- Simon JA, Kingston RE 2009. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol 10: 697–708 [DOI] [PubMed] [Google Scholar]
- Smith ST, Wickramasinghe P, Olson A, Loukinov D, Lin L, Deng J, Xiong Y, Rux J, Sachidanandam R, Sun H, et al. 2009. Genome wide ChIP–chip analyses reveal important roles for CTCF in Drosophila genome organization. Dev Biol 328: 518–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuurman N, Sasse B, Fisher PA 1996. Intermediate filament protein polymerization: molecular analysis of Drosophila nuclear lamin head-to-tail binding. J Struct Biol 117: 1–15 [DOI] [PubMed] [Google Scholar]
- van Wolfswinkel JC, Claycomb JM, Batista PJ, Mello CC, Berezikov E, Ketting RF 2009. CDE-1 affects chromosome segregation through uridylation of CSR-1-bound siRNAs. Cell 139: 135–148 [DOI] [PubMed] [Google Scholar]
- Wallace JA, Felsenfeld G 2007. We gather together: insulators and genome organization. Curr Opin Genet Dev 17: 400–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Bogert BA, Li W, Su K, Lee A, Gao FB 2004. The fragile X-related gene affects the crawling behavior of Drosophila larvae by regulating the mRNA level of the DEG/ENaC protein pickpocket1. Curr Biol 14: 1025–1034 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nussbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based analysis of ChIP-seq (MACS). Genome Biol 9: R137 doi: 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zofall M, Fischer T, Zhang K, Zhou M, Cui B, Veenstra TD, Grewal SI 2009. Histone H2A.Z cooperates with RNAi and heterochromatin factors to suppress antisense RNAs. Nature 461: 419–422 [DOI] [PMC free article] [PubMed] [Google Scholar]