X-chromosome inactivation in placental mammals is mediated by Xist RNA. Using an inducible expression system, Wutz and colleagues find that Tsix RNA, the antisense repressor of Xist, has a defined window of activity during extraembryonic development, during which it can inactivate Xist and reactivate the inactive X chromosome. Interestingly, the work also highlights that different lineages appear to have different degrees of X-inactivation maintenance.
Keywords: noncoding RNA, Tsix, X-chromosome inactivation, development, Xi reactivation, reprogramming
Abstract
The noncoding Tsix RNA is an antisense repressor of Xist and regulates X inactivation in mice. Tsix is essential for preventing the inactivation of the maternally inherited X chromosome in extraembryonic lineages where imprinted X-chromosome inactivation (XCI) occurs. Here we establish an inducible Tsix expression system for investigating Tsix function in development. We show that Tsix has a clear functional window in extraembryonic development. Within this window, Tsix can repress Xist, which is accompanied by DNA methylation of the Xist promoter. As a consequence of Xist repression, reactivation of the inactive X chromosome (Xi) is widely observed. In the parietal endoderm, Tsix represses Xist and causes reactivation of an Xi-linked GFP transgene throughout development, whereas Tsix progressively loses its Xist-repressing function from embryonic day 9.5 (E9.5) onward in trophoblast giant cells and spongiotrophoblast, suggesting that Tsix function depends on a lineage-specific environment. Our data also demonstrate that the maintenance of imprinted XCI requires Xist expression in specific extraembryonic tissues throughout development. This finding shows that reversible XCI is not exclusive to pluripotent cells, and that in some lineages cell differentiation is not accompanied by a stabilization of the Xi.
Numerous cases of sense–antisense transcript pairs have been discovered in mammalian genomes (Katayama et al. 2005). Expression patterns of pairs of sense and antisense transcripts suggest that antisense transcription can activate or repress gene expression (Okada et al. 2008). Antisense transcription has been implicated in a wide range of mechanisms, including development, genomic imprinting, hypermutation of immunoglobulin and T-cell receptor genes, and human diseases such as α-Thalassaemia, Alzheimer's disease, and cancer (Faghihi and Wahlestedt 2009). One of the well-characterized antisense RNAs is Tsix, which represses Xist and regulates X inactivation in mice.
X-chromosome inactivation (XCI) equalizes the dosage of X-linked genes between females and males in placental mammals. In mice, random XCI of either the paternal or maternal X chromosome is observed in all female somatic tissues, whereas the paternal X chromosome is predetermined as the inactive X chromosome (Xi) in preimplantation embryos and extraembryonic tissues (imprinted X inactivation) (Heard and Disteche 2006). Specifically, imprinted XCI is observed in cells of the trophoblast lineage and in primitive endoderm-derived cells of the visceral and parietal yolk sac, whereas placental blood vessels derived from extraembryonic mesoderm exhibit random XCI (Hemberger 2002). At the initiation of XCI, Xist expression is activated and Xist RNA accumulates on the future Xi. Localization of Xist to the Xi requires the nuclear scaffold protein hnRNPU/SP120/SAF-A (Hasegawa et al. 2010). Xist recruits chromatin-modifying complexes of the Polycomb group; Polycomb-repressive complex 2 (PRC2) establishes trimethylation of histone H3 Lys 27 (H3K27me3) (Silva et al. 2003), and PRC1 establishes monoubiquitination of histone H2A (ubH2A) on the Xi (Plath et al. 2004). During cell differentiation, silencing of the Xi becomes further stabilized. Whereas Xist is required for the initiation of X inactivation, it is dispensable for maintenance of the Xi in differentiated cells (Brown and Willard 1994; Csankovszki et al. 1999; Wutz and Jaenisch 2000). Gene silencing on the Xi is highly stable in differentiated somatic cells. In contrast, Xi reactivation is observed in cells of the inner cell mass (ICM) of the blastocyst that develop into the epiblast lineage (Huynh and Lee 2003; Mak et al. 2004; Okamoto et al. 2004) and in migrating primordial germ cells (PGCs) (de Napoles et al. 2007; Sugimoto and Abe 2007; Chuva de Sousa Lopes et al. 2008). Furthermore, the reprogramming of induced pluripotent stem (iPS) cells is also accompanied by Xi reactivation (Maherali et al. 2007). Xi reactivation is presently viewed as an important marker for the naive state of pluripotency and has been used to investigate changes in the epigenetic context during reprogramming (Silva et al. 2008; Stadtfeld et al. 2008; Hanna et al. 2010; Lengner et al. 2010).
Tsix is a paradigm for antisense regulation and for understanding the regulation of XCI. Tsix is an antisense transcript through the Xist locus covering the entire Xist transcription unit (Lee et al. 1999). Disruption of Tsix results in ectopic expression of Xist in embryonic stem (ES) cells and mice, suggesting that Tsix acts as a negative regulator of Xist (Lee and Lu 1999; Lee 2000; Sado et al. 2001). Tsix expression is observed from both X chromosomes before the onset of X inactivation in undifferentiated female mouse ES cells. At the onset of X inactivation, Tsix expression becomes monoallelic and is associated with Xist repression on the active X chromosome (Xa) (Lee et al. 1999). At later stages of differentiation, Tsix expression is lost and Xist repression on the Xa is maintained by other mechanisms, including DNA methylation (Beard et al. 1995; Barr et al. 2007). These observations suggest that Tsix regulates the initiation of Xist silencing, but not its maintenance.
We previously showed that transcription overlapping the Xist promoter region is necessary for Tsix function (Ohhata et al. 2008). Furthermore, several studies have shown that the endogenous Tsix promoter can be substituted for a constitutive or a tetracycline-responsive (Tet) promoter (Luikenhuis et al. 2001; Stavropoulos et al. 2001). Recruitment of DNA methylation and histone modifications such as H3K27me3 around the Xist promoter by Tsix have been reported in differentiated ES cells (Navarro et al. 2006; Nesterova et al. 2008), embryos (Sado et al. 2005), and visceral endoderm (Ohhata et al. 2008). Interactions of Tsix RNA with the DNA methyltransferase Dnmt3a (Sun et al. 2006) and the PRC2 protein Ezh2 (Zhao et al. 2008) have been reported with implications for the mechanism of Tsix-mediated recruitment of these modifications on the Xist promoter. However, DNA methylation is not required for initial repression of Xist (Sado et al. 2004). Moreover, Xist repression is not affected in ES cells with a disruption of the Eed gene that lack PRC2 function (Schoeftner et al. 2006). Importantly, random X inactivation is initiated normally in Eed-deficient female embryos (Kalantry and Magnuson 2006). This suggests that the regulation of Xist by Tsix is likely complex, and neither PRC2 nor Dnmt3a are absolutely required for Xist repression by Tsix.
Here, we establish an inducible Tsix expression system and use it to investigate Tsix function in development. Focusing on its role in imprinted X inactivation, we show that Tsix function is restricted in trophoblast development, whereas Tsix repressed Xist during the entire gestation period in parietal endoderm. On the molecular level, DNA methylation but not histone modifications such as dimethylation of histone H3 Lys 9 (H3K9me2) and H3K27me3 accompanied Xist repression by Tsix. Notably, Xist repression by Tsix induction led to reactivation of the Xi in the extraembryonic lineages. In parietal endoderm, maintenance of imprinted X inactivation required Xist expression throughout development.
Results
Generation of an inducible Tsix allele in mice
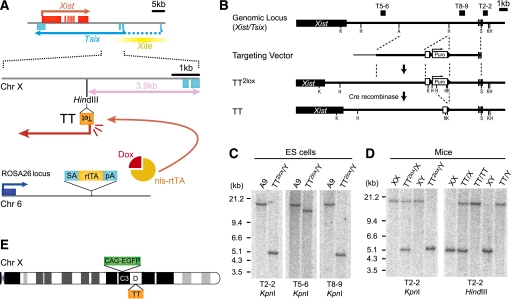
In order to investigate Tsix function in development, we focused on the extraembryonic lineages where Tsix function is required for repression of Xist. Deletion of Tsix causes Xist expression from the maternally inherited X chromosome in extraembryonic lineages of male and female embryos, whereas in the embryonic lineages other mechanisms repress Xist in parallel with Tsix (Ohhata et al. 2006). We generated an inducible Tsix allele in mice taking advantage of the tetracycline-regulated expression system (Gossen et al. 1995). We introduced a Tet promoter 3.9 kb downstream from the Tsix major promoter in ES cells to generate the tetracycline-inducible Tsix (TT) allele (Fig. 1A). Insertion of a Tet promoter at this position has been previously shown not to affect the endogenous regulation of Tsix, and induction of expression from this site recaptures Tsix function in ES cells (Luikenhuis et al. 2001). Targeting was confirmed by Southern analysis, and mice were generated by ES cell injection into C57Bl6 eight-cell embryos (TT2lox, Fig. 1B–D). Mice with >90% contribution from the ES cells were obtained and transmitted the TT2lox allele through the germline. Subsequently, mice were crossed to β-actin Cre-expressing mice to remove the selection cassette (TT) (Fig. 1D). Further crosses to eliminate the β-actin Cre transgene and introduce a ROSA26 targeted tetracycline-inducible transactivator (ROSA26 nls-rtTA) (Savarese et al. 2006) and an X-linked CAG-EGFP transgene (Nakanishi et al. 2002) to visualize the expression status of X-linked genes resulted in the TT/GFP mouse line (Fig. 1E). The TT allele was transmitted through the male and female germline in the expected Mendelian ratio, and both homozygous XTT/GFPXTT/GFP females and XTT/GFPY males that were also homozygous for the ROSA26 nls-rtTA allele appeared normal and were fertile, suggesting that the TT allele did not disrupt regulation of Tsix before induction (Supplemental Fig. S1A,B).
Figure 1.
Generation of Tsix-inducible mice. (A) The genomic structure of the TT (Tet Tsix) allele is shown below the Xist/Tsix locus. (Tet) Tet operator with CMV minimal promoter. The transactivator nls-rtTA is expressed from the ROSA26 locus, allowing induction of Tsix via Dox administration from the TT allele. (B) Targeting and genotyping strategy. (A) ApaI; (H) HindIII; (K) KpnI; (S) SmaI. (C,D) Southern analysis confirming homologous recombination in A9 ES cells (C), germline transmission, and removal of Puro selection cassette in mice (D). Probes and enzymes are indicated below. (E) Schematic representation of the X chromosome in TT/GFP mice. Xist/Tsix and the X-linked CAG-EGFP transgene map to the D and C3 bands, respectively.
Tsix induction blocks imprinted X inactivation
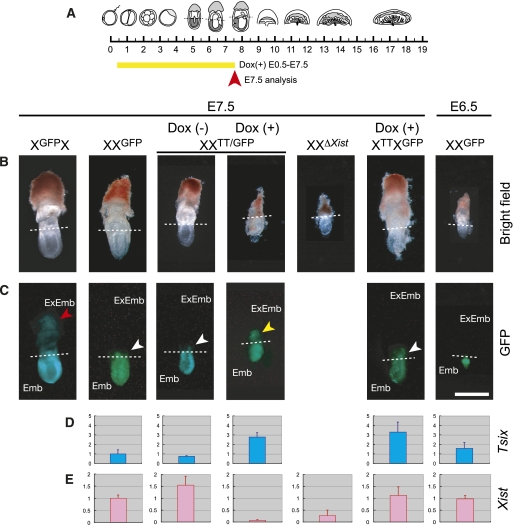
To investigate whether Tsix transcription from the Tet promoter can be induced, we administered doxycycline (Dox) from embryonic day 0.5 (E0.5) to E7.5 (Fig. 2A). As expected, Dox administration through the drinking water of pregnant females led to around twice the level of Tsix compared with endogenous Tsix expression in the ectoplacental cone of XXTT/GFP embryos (maternal allele first, paternal allele second in all genotypes) where imprinted XCI takes place (Fig. 2D). Induction of Tsix expression from the TT allele on the paternally inherited X chromosome repressed Xist to a similar level as in the ectoplacental cone of XXΔXist embryos that have a paternal deletion of Xist (Fig. 2E). Furthermore, Tsix induction resulted in a severe developmental defect in female XXTT/GFP embryos, comparable with the phenotype of the Xist deletion in XXΔXist embryos (Fig. 2B). Importantly, both maternal and paternal transmission of the TT allele did not lead to developmental defects or a significant reduction in the size of E7.5 embryos, when Tsix expression was not induced (Supplemental Fig. S1C). These observations indicate that imprinted XCI was disrupted in XXTT/GFP embryos after Tsix induction.
Figure 2.
Tsix induction from E0.5 represses Xist and blocks XCI in E7.5 ectoplacental cone. (A) Schematic representation of development indicating the timing of Dox administration and analysis (embryos until E8.0 and placenta from E9.5 are shown). (B,C) Bright-field (B) and GFP (C) images of embryos. The dashed line divides extraembryonic (top, ExEmb) and embryonic (bottom, Emb) tissues. GFP on the paternal X chromosome is silenced in the extraembryonic portion because of imprinted XCI. However, some GFP expression in the extraembryonic region (white arrow) is due to allantois and exocoelum derived from mesoderm (random XCI). In contrast, GFP is widely observed in the extraembryonic tissues from the maternally inherited X (red arrow) and from the paternally inherited Xi in XXTT/GFP embryos after Tsix induction by Dox from E0.5 to E7.5 (yellow arrow). The embryonic phenotype of a deletion of Xist on the paternally inherited X chromosome is shown for comparison (XXΔXist). Bar, 0.5 mm. (D,E) Quantitative RT–PCR (qRT–PCR) of Tsix (D) and Xist (E) in the ectoplacental cone, normalized with Gapdh (n = 2∼5). The expression of Tsix and Xist in E7.5 XXGFP embryos was set to 1. Error bars represent standard deviation.
We used the X-linked GFP transgene to investigate XCI after Tsix induction (Fig. 2C). The paternal X chromosome is normally inactivated due to imprinted XCI in the ectoplacental cone, which leads to silencing of the GFP transgene in XXGFP embryos (Fig. 2C). We observed that GFP was not inactivated in E7.5 ectoplacental cone of XXTT/GFP embryos after Tsix induction (Fig. 2C). Since Tsix induction caused a developmental delay, we also investigated imprinted XCI in control XXGFP embryos at E6.5, which showed similar developmental morphology to XXTT/GFP embryos after induction (Fig. 2B). In E6.5 control embryos, X-linked GFP expression was already silenced and Xist was highly expressed (Fig. 2C,E). These observations confirm that the GFP expression observed in the ectoplacental cone of XXTT/GFP embryos after Tsix induction is due to the failure of imprinted XCI but is not due to a developmental delay. Furthermore, induction of Tsix from the maternal X chromosome of XTTXGFP embryos showed high Tsix expression, which did not lead to Xist repression in the ectoplacental cone (Fig. 2D,E). GFP was not activated from the paternally inherited Xi, nor was a developmental phenotype observed (Fig. 2B,C). We conclude that Tsix expression could be induced from the TT allele in mouse embryos and repressed Xist in cis. Tsix induction on the paternally inherited Xi in turn led to a failure of imprinted XCI in the extraembryonic lineages and caused a developmental defect similar to a deletion of Xist. Importantly, induction of Tsix from a maternally inherited TT allele did not affect imprinted X inactivation, showing that Tsix did not cause effects in trans.
Effect of Tsix induction at different time points in placental development
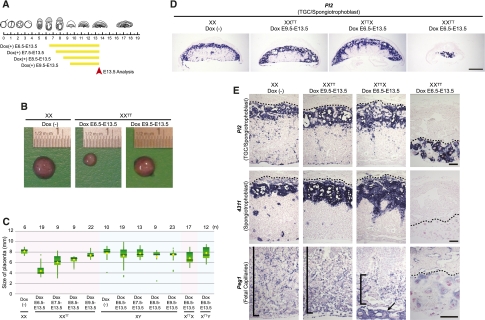
To investigate whether Tsix functions at later time points in development, we administrated Dox starting from E6.5, E7.5, E8.5, and E9.5, and analyzed the placenta at E13.5 (Fig. 3A). Tsix induction from the paternally inherited X chromosome in XXTT embryos from E6.5 resulted in a clear reduction of placental size at E13.5 (Fig. 3B). Placental size was also reduced when Dox was administrated from E7.5 (Fig. 3C). Dox administration from E8.5 had a weaker effect and, finally, no size difference between control and XXTT placenta was observed when Dox was administered from E9.5 (Fig. 3B,C). These observations indicated that the phenotypic effect of Tsix induction became less severe at later time points in placental development.
Figure 3.
Effect of Tsix induction on placental development. (A) Scheme of Dox administration and analysis. (B) The representative images of placentas and Dox administration as indicated. (C) Box plots of placental size for each genotype, with Dox administration indicated. The green box spans the middle of 50% of the data. The top and bottom edges indicate 75th and 25th percentile. Median and average are shown as a horizontal bar and a diamond, respectively. Vertical lines indicate the minimum or maximum data values unless outliers (dots) are present, in which case the vertical lines extend to a maximum of 1.5 times the interquartile range. The number of the placentas (n) is above. (D,E) In situ hybridization of E13.5 placental sections as indicated. Control placenta [XX Dox (−)] is shown as reference. No obvious phenotype is observed in XXTT placenta with Dox administration from E9.5. Fetal capillaries (Peg1) are affected in XTTX placenta after Dox from E6.5 (black bar demarks location of fetal capillaries; arrow indicates Peg1-positive chorionic plate blood vessels). Fetal capillaries and spongiotrophoblast are severely affected and giant cells are abnormally enlarged in XXTT placenta after Dox administration from E6.5. Bars: D, 1 mm; E, 0.2 mm. The dotted line demarks the border between deciduas and trophoblast giant cells.
In placenta, imprinted XCI is observed in trophoblast giant cells, spongiotrophoblast, and syncytiotrophoblast, whereas random XCI is observed in the fetal capillaries that invade the labyrinth layer via the allantois and later umbilical cord (Hemberger 2002). To characterize the placental phenotype of Tsix induction in greater detail, we used in situ hybridization with the probes specific for spongiotrophoblast (4311, also known as Tpbpa) and fetal capillaries (Peg1, also known as Mest), and a probe that marks both trophoblast giant cells and spongiotrophoblast (Pl2, also known as Prl3b1). Induction of Tsix in XXTT embryos from E9.5 did not change overall placental morphology nor the expression pattern of these markers at E13.5, consistent with our observation that induction from this late time point did not lead to a reduction in placental size (Fig. 3D,E). Induction of Tsix at E6.5 from the maternally inherited X chromosome in XTTX placentas led to a reduced density of fetal capillaries in the placental labyrinth as visualized by Peg1 staining, which is the cell type in which random XCI takes place (Fig. 3C–E; Supplemental Fig. S2). Induction of Tsix at E6.5 from the paternally inherited X chromosome in XXTT embryos resulted in a severe defect in all trophoblast cell types and fetal capillaries, such that in the most extreme cases, the placental labyrinth and spongiotrophoblast layers were completely absent and trophoblast giant cells were overabundant and massively enlarged (Fig. 3D,E; Supplemental Fig. S2). These observations suggest that Tsix induction caused a combined disruption of both random and imprinted XCI.
Xist repression by Tsix is accompanied by DNA methylation
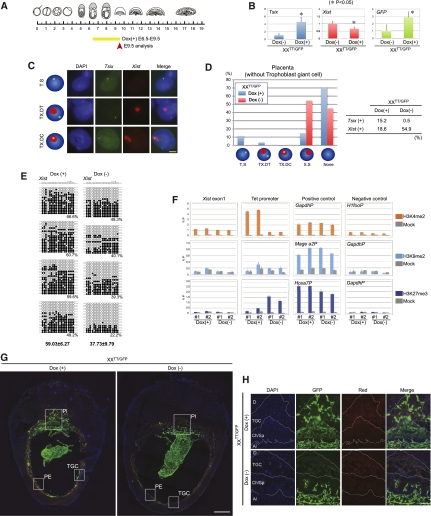
The severe developmental defect after Tsix induction from E6.5 from the paternally inherited Xi prompted us to investigate the repression of Xist after Tsix induction. Since the structure of the placenta was severely disrupted at E13.5 (Fig. 3D), we analyzed E9.5 placenta, 3 d after Dox administration from E6.5 (Fig. 4A), when placental size was not yet affected. After Dox administration, Tsix expression was induced around fivefold and led to a reduction in Xist (Fig. 4B). The remaining Xist expression was presumably from the allantois, where random XCI takes place. To further analyze the function of Tsix, we performed strand-specific Tsix/Xist double RNA-FISH using ssDNA probes on placental sections. We observed clear Tsix signals only after Dox treatment, indicating that our RNA-FISH protocol detected only highly induced Tsix RNA from the TT allele, but not the lower expression of endogenous Tsix. With Dox administration, three different types of Tsix-positive cells were observed: first, “pinpoint Tsix without Xist,” suggesting that Tsix is functional; second, “pinpoint Tsix and accumulated Xist from a different chromosome,” presumably arising from extraembryonic mesoderm cells in which the pinpoint Tsix would be expressed from the TT allele on the paternal X chromosome and Xist on the maternally inherited X chromosome; and third, “pinpoint Tsix within accumulated Xist,” indicating that Tsix did not repress Xist expression (Fig. 4C). In control XXTT/GFP placentas, Xist clusters were detected in 55% of the cells under our conditions. After Dox administration from E6.5, the number of cells showing Xist clusters was decreased to 19% and the number of cells showing a pinpoint Tsix signal without an Xist cluster was significantly increased in XXTT/GFP placenta compared with uninduced samples (Fig. 4D; Supplemental Fig. S3C). Detection of Tsix on sections was inefficient, and we were able to observe clear signals in only 15% of the cells after induction (Fig. 4D). However, given the significant reduction in Xist clusters, these results demonstrate that Tsix induction from E6.5 repressed Xist in placenta.
Figure 4.
Tsix induction from E6.5 represses Xist and causes Xi reactivation in the placenta. (A) Scheme of Dox administration and analysis. (B) qRT–PCR of Tsix, Xist, and GFP expression in E9.5 placenta after Tsix induction at E6.5, normalized by Gapdh (n = 3). Error bars represent standard deviations (asterisk represents significant differences). (C) Strand-specific Tsix (green) and Xist (red) RNA-FISH. Three types of Tsix-positive cells are observed in E9.5 placenta after Dox administration from E6.5: pinpoint Tsix without Xist (Tsix single, T.S), pinpoint Tsix and accumulated Xist from different chromosomes (Tsix and Xist double in trans, TX.DT), and Tsix within accumulated Xist (Tsix and Xist double in cis, TX.DC). Bar, 5 μm. (D) Summary of Tsix/Xist RNA-FISH on E9.5 placenta without trophoblast giant cells. No Tsix signal with Xist accumulation (Xist single, X.S) and neither Tsix nor Xist signal (none) are counted. [Dox(+), n = 797; Dox(−), n = 813). The distribution of these cells on placenta section is shown in Supplemental Figure S3. (E) DNA methylation analysis by bisulfite sequencing of the Xist promoter in E9.5 XXTT/GFP placenta. Around 20 sequences ([dark circle] methylated C; [open circle] unmethylated C) are shown, and the percentage methylation is given below. (F) Analysis of chromatin modifications by directed ChIP in E9.5 XXTT/GFP placenta as indicated. Error bars represent standard deviation. (G) Immunofluorescence image showing GFP expression in sections of whole deciduas. (Pl) Placenta; (PE) parietal endoderm; (TGC) trophoblast giant cells. The squares indicate regions magnified in H and Supplemental Figure S5, B and C. Bar, 0.5 mm. (H) Magnified region showing GFP expression and autofluorescence of the tissue (red) in placenta. (D) Deciduas; (TGC) trophoblast giant cells; (Ch) chorion; (Sp) spongiotrophoblast; (Al) allantois. Borders between different layers were identified from the DAPI image using the large nuclei of trophoblast giant cells as a reference. Bar, 100 μm.
We also investigated the function of Tsix in parietal endoderm and trophoblast giant cells, which also show imprinted XCI in mice. RNA-FISH on sections of E9.5 embryos showed that in both parietal endoderm and trophoblast giant cells, the number of Xist-positive cells was decreased after Tsix induction from E6.5, indicating that Tsix repressed Xist (Supplemental Fig. S3D,E). However, in trophoblast giant cells, the number of cells with a pinpoint Tsix within an Xist cluster was increased, showing that Tsix is unable to repress Xist in some trophoblast giant cells already at E9.5 (Supplemental Fig. S3E).
To investigate the molecular basis of Xist repression by Tsix, we analyzed DNA methylation on the Xist promoter in placenta. We found that DNA methylation was significantly increased after Tsix induction (59.03% ± 6.27%) compared with uninduced controls (37.73% ± 9.79%, P < 0.05, n = 4) (Fig. 4E; Supplemental Fig. S4). We also analyzed histone modifications on the Xist promoter after Tsix induction. Previously, an increase of H3K9me2 and H3K27me3 and a decrease of H3K4me2 was reported at Xist exon1, depending on Tsix transcription in the visceral endoderm at E13.5 (Ohhata et al. 2008). Although the specificity of the antibodies for the chromatin immunoprecipitation (ChIP) experiment was confirmed with positive and negative control loci, we did not detect either an increase in H3K9me2 and H3K27me3 or a decrease in H3K4me2 at Xist exon1 after Tsix induction in the placenta (Fig. 4F). We conclude that Tsix-induced repression of Xist is accompanied by DNA methylation but without a detectable change in histone modifications in E9.5 placenta.
Xist repression by Tsix from E6.5 leads to reactivation of the Xi
We next investigated whether repression of Xist by Tsix induction at E6.5 would cause reactivation of the paternally inherited Xi in the extraembryonic lineages. We detected increased expression of the X-linked GFP transgene from the paternal X chromosome in XXTT/GFP placenta after Dox administration in E9.5 placenta (Fig. 4B). To investigate the GFP expression in individual cells of different lineages in the placenta, immunofluorescence staining was performed. After Dox administration, clear GFP expression was observed in chorion, spongiotrophoblast, trophoblast giant cells, and parietal endoderm (Fig. 4G,H; Supplemental Fig. S5). In control embryos without induction, no or very few cells showed GFP staining, which were probably derived from the allantois. Since the X-linked GFP transgene was already inactivated in the ectoplacental cone of E6.5 control XXGFP embryos (Fig. 2C), our observations suggest that Tsix induction at E6.5 had led to reactivation of transgene expression on the paternally inherited Xi in these various extraembryonic cell types.
The function of Tsix for Xist repression is lost at E9.5 in spongiotrophoblast
Since Tsix induction from the paternally inherited Xi at E9.5 did not result in a defect in XXTT placentas at E13.5 (Fig. 3), we analyzed the expression of Tsix and Xist (Fig. 5A). After Dox administration from E9.5, Tsix levels at E13.5 were sixfold to sevenfold higher than in uninduced control samples, confirming that Tsix could be induced (Fig. 5B). However, Xist expression was not reduced after Tsix induction from E9.5, suggesting that Tsix might have lost its repressor function (Fig. 5B). To confirm this result, we performed Tsix/Xist RNA-FISH. In the labyrinth layer, which consists of syncytiotrophoblast (imprinted XCI) and fetal capillaries (random XCI), the number of Xist-positive cells was decreased by ∼25% and the number of cells with a pinpoint Tsix without Xist was increased compared with uninduced controls (Fig. 5C,D; Supplemental Fig. S6). This suggests that Tsix is still functional beyond E9.5 in a subset of cells in the labyrinth layer even though the number of Tsix-expressing cells was decreased. We observed a small number of cells with a pinpoint Tsix and a nonoverlapping Xist cluster on a different chromosome, which we attribute to random XCI in the cells of the fetal capillaries (Fig. 5C).
Figure 5.
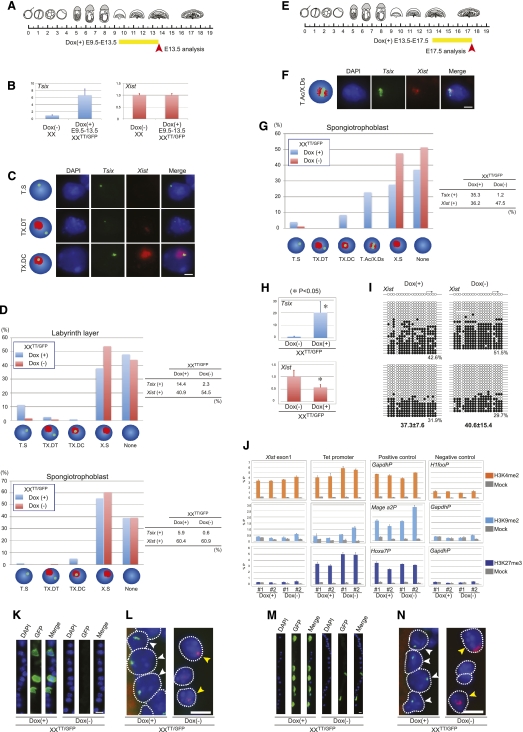
Tsix loses its function from E9.5 in spongiotrophoblast. (A) Scheme of Dox administration and analysis in B–D, K, and L. (B) qRT–PCR of Tsix and Xist using placenta, normalized by Gapdh (n = 3) as in Figure 4B. (C) Strand-specific Tsix (green) and Xist (red) RNA-FISH on placental sections as in Figure 4C. Bar, 5 μm. (D) Summary of Tsix/Xist RNA-FISH on E13.5 labyrinth layer and spongiotrophoblast as in Figure 4D. Labyrinth layer: Dox(+), n = 514; Dox(−), n = 972. Spongiotrophoblast: Dox(+), n = 657; Dox(−), n = 711. The distribution of cells on placental sections is shown in Supplemental Figure S6. (E) Scheme of Dox administration and analysis in F–J, M, and N. (F) A new type of Tsix-positive cell showing accumulated Tsix (green) overlapping dispersed Xist (red) (T.ac/X.ds) is observed in placenta. Bar, 5 μm. (G) Summary of Tsix/Xist RNA-FISH on E17.5 spongiotrophoblast as in Figure 4D. Dox(+), n = 224; Dox(−), n = 162. The distribution of cells on placental sections is shown in Supplemental Figure S7. (H) qRT–PCR of Tsix and Xist expression in E17.5 spongiotrophoblast, normalized by Gapdh (n = 2) as in Figure 4B. (I) DNA methylation of the Xist promoter in E17.5 spongiotrophoblast as in Figure 4E. (J) Analysis of chromatin modifications in E17.5 spongiotrophoblast as in Figure 4F. All error bars show the standard deviation. (K,M) Immunofluorescence showing GFP expression in the parietal endoderm at E13.5 (K) and E17.5 (M). Bars, 10 μm. (L,N) RNA FISH analysis of Tsix (green) and Xist (red) expression in parietal endoderm at E13.5 (L) and E17.5 (N). White arrows identify cells with Tsix expression, and yellow arrows identify Xist-expressing cells. Bars, 10 μm.
In the spongiotrophoblast, we observed a significant increase in cells with a pinpoint Tsix within an Xist cluster in XXTT/GFP placenta at E13.5 after Tsix induction from E9.5 (Fig. 5C,D; Supplemental Fig. S6). Overlapping Tsix and Xist expression suggests that in the spongiotrophoblast layer, Tsix had lost its function for repressing Xist. This interpretation is further consistent with the observation that the total number of Xist-expressing cells was unchanged after Tsix induction.
We next examined a late gestational stage of placental development, inducing Tsix from E13.5, followed by analysis at E17.5 (Fig. 5E). In the labyrinth layer, the number of cells with a pinpoint Tsix without Xist signal could be observed even though the number of Tsix-expressing cells was small (Supplemental Fig. S7). However, the number of Xist-expressing cells was not decreased, indicating that Tsix induction at E13.5 did not lead to Xist repression in the majority of cells of the labyrinth layer (Supplementa Fig. S7). In the spongiotrophoblast layer, Tsix was detected in a large number of cells after Dox administration at E13.5. A new type of Tsix-positive cell was observed with an accumulation of Tsix and a dispersed Xist distribution (Fig. 5F,G; Supplemental Fig. S7). In fact, the majority of Tsix signals overlapped with Xist, suggesting that Tsix was not functional in these spongiotrophoblast cells (Fig. 5G). However, RT–PCR detected a significant decrease in the amount Xist RNA after Tsix induction (Fig. 5H). To distinguish whether the reduction of Xist RNA occurred due to transcriptional repression or post-transcriptionally, we analyzed DNA methylation on the Xist promoter and histone modification on the Xist exon1. We were not able to detect an increase in DNA methylation (Fig. 5I) or changes in histone modifications in E17.5 spongiotrophoblast after Tsix induction from E13.5 (Fig. 5J). This observation suggests that accumulation of Tsix led to a reduction in steady-state Xist RNA levels without causing transcriptional silencing. We conclude that Tsix induction from E9.5 or later did not cause repression of Xist in the spongiotrophoblast lineage, suggesting that Tsix function was dependent on a developmental window.
Xist is required for maintenance of X inactivation throughout parietal endoderm development
We next assessed the function of Tsix in parietal endoderm after E9.5 by studying X-linked GFP expression. In control XXTT/GFP embryos without induction, the paternally inherited Xi was silent, as no or very few GFP-positive cells were observed. The number of cells expressing the X-linked GFP was drastically increased in parietal endoderm after Tsix induction from the paternally inherited Xi at E9.5 [Dox(+), 21.9%, n = 178; Dox(−); 3.6%, n = 138] (Fig. 5K). This suggested that Xist was repressed by Tsix at E9.5, resulting in reversal of imprinted XCI. Consistent with this, Xist/Tsix double RNA-FISH showed that the number of cells with a pinpoint Tsix without Xist was significantly increased in samples where Tsix had been induced compared with controls [Dox(+), 47.9%, n = 399; Dox(−), 0%, n = 136] (Fig. 5L).
Similarly, when Dox was administered to XXTT/GFP embryos from E13.5, the number of GFP-positive cells was increased at E17.5 in parietal endoderm [Dox(+), 22.6%, n = 266; Dox(−), 4.1%, n = 245] (Fig. 5M) and the number of cells with a pinpoint Tsix without Xist signal was increased [Dox(+), 57.0%, n = 107; Dox(−), 0%, n = 47] (Fig. 5N). Taken together, our data demonstrate that Tsix repressed Xist at late stages of parietal endoderm development, and loss of Xist in turn led to a defect in the maintenance of imprinted XCI.
Tsix-mediated Xist repression is reversible in preimplantation embryos
In order to address whether Tsix would repress Xist already in preimplantation embryos, we performed Tsix/Xist double RNA-FISH on XXTT/GFP blastocysts treated with Dox at several time points (Fig. 6A). For this, we isolated two-cell embryos (E1.5) and cultured them to the blastocyst stage. Female embryos were identified based on a GFP transgene on the paternal X chromosome (Supplemental Fig. S8). In control female XXGFP blastocysts, Xist clusters were observed in most of the blastomeres when Dox was present or not (Fig. 6B,C). However, the number of Xist-positive cells was significantly reduced in XXTT/GFP blastocysts cultured with Dox (Fig. 6B,C). Thus, Tsix induction from the paternally inherited Xi can repress Xist in preimplantation embryos.
Figure 6.
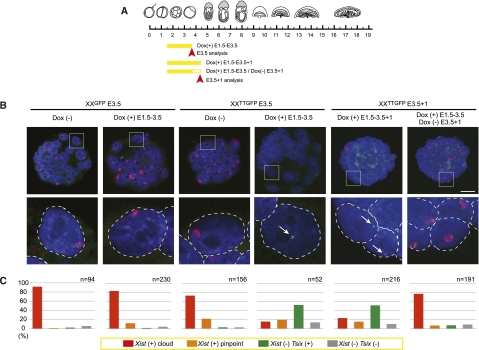
Repression of Xist by Tsix in preimplantation embryos is reversible. (A) Scheme of Dox administration and analysis. (B) Tsix (green, white arrow) and Xist (red) RNA-FISH on blastocyst with or without Dox administration. Enlarged regions below are indicated by boxes. Bar, 25 μm. (C) Bar graph showing the percentage of cells with Xist clusters with or without Tsix, pinpoint Xist with or without Tsix, without Xist and pinpoint Tsix, and no Xist or Tsix in blastocysts. The number of the blastomeres is shown above the graph.
To investigate further whether Xist repression by Tsix could be reversed, we cultured the XXTT/GFP blastocysts for one more day without Dox treatment (Fig. 6A). Clear reactivation and accumulation of Xist was observed 1 d after Dox removal (Fig. 6B,C). In contrast, Xist remained repressed in the continuous presence of Dox (Fig. 6B,C). This observation demonstrates that Xist repression was not stabilized and required continued Tsix induction in blastocysts. We further tested whether re-expression of Xist would re-establish imprinted XCI. For this, we transferred blastocysts cultured in the presence of Dox into recipient females and analyzed embryonic development at E8.5 (Supplemental Fig. S9). Embryos developed normally and showed repression of the X-linked GFP transgene in the ectoplacental cone. This result indicates that blocking of Xist expression throughout preimplantation development does not cause an irreversible defect, and restoration of imprinted XCI is possible by Xist expression from E3.5 to sustain normal development.
Discussion
Defining the functional window of Tsix in imprinted X inactivation
Here, we report an inducible Tsix expression system in mice and use it to define the functional window of Tsix in imprinted XCI (Fig. 7A). Tsix function for repressing Xist was already observed in preimplantation blastocyst stage embryos. Xist repression is reversible and depends on continued Tsix expression at this stage. This observation is consistent with the idea that Tsix-mediated repression of Xist has an initiation and a maintenance mechanism. In differentiated female cells, Xist repression is thought to be maintained without Tsix (Lee et al. 1999). Indeed, we did not observe endogenous Tsix expression by RNA-FISH in E9.5, E13.5, and E17.5 placenta. Since at these stages Xist remained repressed without detectable Tsix expression, we suggest that a shift from reversible to irreversible Xist repression might occur in placental development. Tsix is critical for the initiation but not maintenance of Xist repression, similar to the role of Xist for chromosome-wide silencing in X inactivation (Brown and Willard 1994; Csankovszki et al. 1999; Wutz and Jaenisch 2000).
Figure 7.
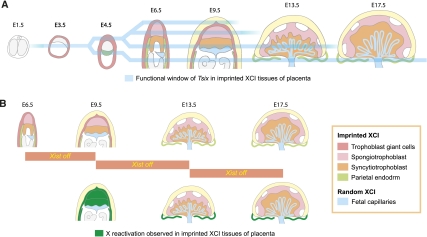
Summary: functional window of Tsix and X reactivation in imprinted XCI. (A) A summary of the context for Tsix function in imprinted XCI. Tsix represses Xist in a developmental window beginning in preimplantation embryos. Tsix starts to lose its function in some trophoblast giant cells at E9.5, and in spongiotrophoblast from E9.5 to E13.5. In parietal endoderm, Tsix maintains its function during the entire developmental period analyzed. (B) A summary of the reactivation of X-linked GFP after Tsix induction observed in this study. Xi reactivation is widely observed in extraembryonic tissues at E9.5, when Xist is repressed by Tsix induction from E6.5. In parietal endoderm, Xi reactivation is also observed when Tsix is induced from E9.5 or E13.5.
In the post-implantation embryo, Tsix functioned at E6.5 in all of the extraembryonic lineages we investigated. Repression of Xist on the paternally inherited Xi led to reactivation of an X-linked GFP transgene, suggesting that Xist is required for maintenance of X inactivation in a wide range of cell types at this stage. Importantly, our data demonstrate that Tsix induction at E6.5 can repress the fully activated Xist promoter. This observation indicates that Tsix uses an active repression mechanism, as opposed to merely preventing Xist activation. At later stages of development, Tsix lost its function for repressing Xist in a lineage-specific manner. At E9.5, we observed Tsix coexpression with Xist from the same chromosome in spongiotrophoblast, indicating that Tsix did not repress Xist. However, in parietal endoderm and potentially also in syncytiotrophoblast, Tsix continued to repress Xist. Lineage-specific function of Tsix suggests that Tsix-mediated Xist repression requires cell type-specific factors or pathways.
Surprisingly, induction of Tsix at E13.5 resulted in accumulation of Tsix RNA in a small cluster in some spongiotrophoblast cells. In these cells, Xist was observed in a dispersed pattern. It is unclear how Tsix accumulation is mediated. A potential explanation could be the presence of multiple copies of the Tsix locus due to endoreduplication of the genome in trophoblast cells, whereby Tsix RNA would be expressed from many sites in close vicinity, resulting in a cluster-like appearance. At this stage, Tsix did not repress Xist, and we suggest that strong Tsix expression from the tetracycline-inducible promoter from the same chromosome could induce interactions between Tsix and Xist affecting either the stability of Tsix or mediating its retention in the vicinity of the chromosome. However, further investigation is required to clarify whether this observation is relevant for the mechanism of endogenous Tsix.
DNA methylation accompanies Xist repression by Tsix in placenta
We observe that an increase in DNA methylation on the Xist promoter accompanies the repression of Xist by Tsix in the placenta. A recent study clearly showed that extraembryonic and, specifically, trophoblast cell types can form in the absence of all DNA methylation (Sakaue et al. 2010). However, in trophoblast stem cells with mutations in all three DNA methyltransferase genes (Dnmt1, Dnmt3a, and Dnmt3b), Xist expression is found up-regulated to some extent (Supplemental Fig. S10). This suggests that DNA methylation contributes to Xist repression in the extraembryonic lineages but might not be essential.
A previous study showed that H3K9me2 and H3K27me3 were increased at Xist exon1 in visceral endoderm, depending on Tsix transcription (Ohhata et al. 2008). However, Xist is repressed in Eed-deficient and G9a-deficient embryos (Wang et al. 2001; Ohhata et al. 2004). In this study, we also observe that Xist repression by Tsix did not result in a measurable increase of H3K27me3 and H3K9me2 on Xist, indicating that these chromatin marks are less important for Tsix-mediated Xist repression in placenta. Therefore, it is likely that additional and potentially lineage-specific mechanisms contribute to Xist repression by Tsix in the extraembryonic lineages.
Xist is required for the maintenance of imprinted X inactivation
One interesting observation we made in our study is the finding that X inactivation is reversible and Xist-dependent in a wide range of placental cell types (Fig. 7B). Reactivation of a GFP transgene on the Xi was widely observed in trophoblast lineages after Tsix induction. Moreover, in the parietal endoderm, X inactivation appears to remain Xist-dependent until E13.5, at which placental development is thought to be complete. Albeit our results using an X-linked GFP transgene do not necessarily reflect the sate of all genes on the Xi, they demonstrate a clear difference in the maintenance of X inactivation between extraembryonic and embryonic cells. X inactivation is remarkably stable in differentiated somatic cells, and reactivation of genes on the Xi has not been achieved efficiently in cells from E13.5 embryos (Csankovszki et al. 1999). Furthermore, DNA methylation is required for maintenance of X inactivation in embryonic lineages but not extraembryonic lineages (Sado et al. 2000). These observations demonstrate a clear difference between the mechanisms for maintaining X inactivation in embryonic and extraembryonic lineages.
Xi reactivation is also seen as a marker for the naive state of iPS cells during reprogramming (Silva et al. 2008; Stadtfeld et al. 2008; Hanna et al. 2010; Lengner et al. 2010). Our observation of readily reversible X inactivation in extraembryonic cell types suggests that the underlying chromatin structure might not be unique to naive pluripotent stem cells. It also raises the question of whether extraembryonic cells are more amenable for reprogramming to iPS cells. This observation could be useful for determining the extent to which the chromatin composition or epigenetic milieu of cells affects reprogramming. Investigating the maintenance of XCI in embryogenesis will ultimately contribute to a better understanding of epigenetic regulation in development and reprogramming.
Materials and methods
Vector construction and generation of mouse strains
A 9.2-kb SmaI–ApaI fragment containing a part of Tsix exon2 and exon3 was isolated from an 11.9-kb SpeI–ApaI genomic fragment (Sado et al. 2001). A loxP-flanked puromycine resistance cassette (loxP–puro–loxP) and a Tet promoter were integrated into the HindIII site. The targeting vector was linearized and electroporated into A9 129/B6 F1 hybrid ES cells, which were established from blastocysts isolated from C57Bl6 females mated to 129 males. The conditions of ES cell cultures and electroporation were described previously (Wutz and Jaenisch 2000). The selection cassette was removed by crossing chimeric mice with β-actin Cre transgenic mice as described previously (Savarese et al. 2006). Mice harboring a disruption of the Xist gene (ΔXist) were generated from a conditional Xist2lox allele (Csankovszki et al. 1999) to β-actin Cre transgenic mice. The Cre transgene was removed through subsequent crosses whereby the ΔXist was transmitted through the maternal germline. Dox was administered via drinking water containing 1 g of Dox (Sigma) and 100 g of sucrose (Fluka) per liter. All animal experiments were performed following institutional guidelines.
Quantitative RT–PCR
Total RNA was extracted from dissected ectoplacental cones (E7.5), placentas (E13.5), and spongiotrophoblast (E17.5) using RNeasy (Qiagen), and cDNA was prepared using SuperScript II and random primers (Invitrogen). Reactions were performed in duplicate using SYBR Green PCR Master Mix (ABI) with a StepOnePlus Real-Time PCR system (ABI) and primers listed in Supplemental Table S1. The standard curve method was used for quantification, and expression levels were normalized to Gapdh.
RNA-FISH
Freshly dissected whole deciduas (E9.5) or placentas (E13.5 and E17.5) were embedded in OCT compounds (Tissue Tek). Sections (7 μm) were cut on a cryostat (Leica) and mounted on Super Frost Plus slides (VWR). For preimplantation embryos, two-cell stage embryos were flushed from oviducts and cultured in KSOM medium (Millipore) with 1 μg/mL Dox or without. After sexing using a fluorescent stereomicroscope, the zona pellucida was removed using Acid Tyrode's solution (Sigma). Subsequently, embryos were attached on slides coated with Denhardt's solution (Okamoto et al. 2004). Samples were permeabilized in CSK buffer with 0.5% Triton X-100, 1 mM EGTA, and 2 mM Vanadyl Ribonucleotide complex (NEB) for 5 min at 4°C; fixed by 4% PFA in PBS for 10 min at 4°C; and dehydrated through a 70%, 80%, 95%, and 100% ethanol series. RNA-FISH was performed as described (Ohhata et al. 2008). Details of strand-specific probes are in the Supplemental Material. Different layers of the placenta were determined on sequential sections stained with hematoxylin and eosin (H&E).
In situ hybridization
Placentas were fixed overnight in 4% PFA in PBS, processed for routine paraffin histology, embedded, and cut into 7-μm sections using a Leica paraffin microtome. Digoxigenin-labeled antisense riboprobes were generated from linearized plasmids containing Pl2, Tpbpa (4311), and Peg1 cDNA sequences using a commercial system (Roche). Probes were hybridized to sections at 52°C following a standard protocol. Signals were detected using an anti-DIG alkaline phosphatase-conjugated antibody (Roche), followed by color reaction using NBT and BCIP (Promega), and imaged after counterstaining with nuclear fast red (Sigma).
Immunofluorescence
Freshly dissected whole deciduas (E9.5) or placentas (E13.5 and E17.5) were fixed in 4% PFA in PBS overnight at 4°C. For cryoprotection, fixed samples were incubated at 4°C in PBS containing 15% sucrose for 3 h, then overnight in 30% sucrose, equilibrated in OCT for 1 h at 4°C, and embedded in OCT. Sections (7 μm) were cut on a cryostat (Leica) and mounted on Super Frost Plus slides (VWR). For GFP immunofluorescence, samples were permeabilized by PBST (PBS with 0.1% Tween) with 0.5% BSA for 5 min, blocked with 5% BSA in PBST for 1 h, and incubated overnight with antiGFP antibody (ab290, Abcam) diluted 1:500 in blocking buffer at room temperature. After three washes in PBST for 10 min, slides were incubated for 1 h with donkey anti-rabbit Alexa 488 antiserum (A21206, Molecular Probes) diluted 1:1000 in blocking buffer at room temperature, washed three times in PBST for 10 min, and mounted using Vectershield with DAPI (Vector Laboratories).
Bisulfite sequencing
Genomic DNA prepared from dissected placentas (E9.5) and spongiotrophoblast (E17.5) were digested with EcoRI and treated with bisulfite according to manufacturer's procedures (Imprint DNA modification kit, Sigma). Two rounds of PCR were performed for amplification of the Xist promoter region using primer sets Pr1/Pr2 and Pr2/Pr3 (McDonald et al. 1998). PCR conditions were as follows: eight cycles of 30 sec at 95°C, 1 min at 60°C (progressively decreased by 0.5°C every cycle), and 1 min at 72°C; followed by 25 cycles of 30 sec at 95°C, 1 min at 56°C, and 1 min at 72°C; and a final extension for 5 min at 72°C. Amplified products were cloned using the Topo TA cloning kit (Invitrogen). Individual clones were amplified using TempliPhi (GE Hearthcare), and 24 clones were sequenced for each sample.
Quantitative ChIP assay
ChIP was performed as described previously with minor modifications at the step of sample preparation (Ohhata et al. 2008). Dissected placentas (E9.5) and spongiotrophoblast (E17.5) were fixed in 1% formaldehyde in PBS for 10 min, and homogenized using a dounce homogenizer in lysis buffer without SDS (50 mM Tris-HCl at pH 8.0, 10 mM EDTA at pH 8.0 with protease inhibitor complete [Roche]). Homogenate was filtered using a 40-μm cell strainer (BD Biosciences). The following antibodies were used: rabbit monoclonal H3K4me2 (#9725, Cell Signaling), rabbit monoclonal H3K9me2 (#4658, Cell Signaling), rabbit monoclonal H3K27me3 (#9733, Cell Signaling), and mock (normal rabbit IgG; I5006, Sigma). Quantitative PCR was performed as described using primers listed in Supplemental Table S1.
Acknowledgments
We thank M. Okabe for EGFP transgenic mice; T. Sado for the genomic Tsix fragment and critical discussions; T. Nagano and S. Tomizawa for technical advice; C.H. Theussl for generating chimeric mice; M.H. Idarraga-Amado and M. Saltik for genotyping; M. Okano, D. Barlow, and her laboratory members for critical discussions; and P. Humphreys, M. McLeish, S. Jameson, C. Dumeau, and B. Mansfield for technical support. This work was supported by the Lise Meitner program of the Austrian Science Fund to T.O. (M1022-B09) and a Wellcome Trust Senior Research Fellowship to A.W. (grant reference 087530/Z/08/A).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.16997911.
References
- Barr H, Hermann A, Berger J, Tsai HH, Adie K, Prokhortchouk A, Hendrich B, Bird A 2007. Mbd2 contributes to DNA methylation-directed repression of the Xist gene. Mol Cell Biol 27: 3750–3757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beard C, Li E, Jaenisch R 1995. Loss of methylation activates Xist in somatic but not in embryonic cells. Genes Dev 9: 2325–2334 [DOI] [PubMed] [Google Scholar]
- Brown CJ, Willard HF 1994. The human X-inactivation centre is not required for maintenance of X-chromosome inactivation. Nature 368: 154–156 [DOI] [PubMed] [Google Scholar]
- Chuva de Sousa Lopes SM, Hayashi K, Shovlin TC, Mifsud W, Surani MA, McLaren A 2008. X chromosome activity in mouse XX primordial germ cells. PLoS Genet 4: e30 doi: 10.1371/journal.pgen.0040030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csankovszki G, Panning B, Bates B, Pehrson JR, Jaenisch R 1999. Conditional deletion of Xist disrupts histone macroH2A localization but not maintenance of X inactivation. Nat Genet 22: 323–324 [DOI] [PubMed] [Google Scholar]
- de Napoles M, Nesterova T, Brockdorff N 2007. Early loss of Xist RNA expression and inactive X chromosome associated chromatin modification in developing primordial germ cells. PLoS ONE 2: e860 doi: 10.1371/journal.pone.0000860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faghihi MA, Wahlestedt C 2009. Regulatory roles of natural antisense transcripts. Nat Rev Mol Cell Biol 10: 637–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossen M, Freundlieb S, Bender G, Muller G, Hillen W, Bujard H 1995. Transcriptional activation by tetracyclines in mammalian cells. Science 268: 1766–1769 [DOI] [PubMed] [Google Scholar]
- Hanna J, Cheng AW, Saha K, Kim J, Lengner CJ, Soldner F, Cassady JP, Muffat J, Carey BW, Jaenisch R 2010. Human embryonic stem cells with biological and epigenetic characteristics similar to those of mouse ESCs. Proc Natl Acad Sci 107: 9222–9227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa Y, Brockdorff N, Kawano S, Tsutui K, Nakagawa S 2010. The matrix protein hnRNP U is required for chromosomal localization of Xist RNA. Dev Cell 19: 469–476 [DOI] [PubMed] [Google Scholar]
- Heard E, Disteche CM 2006. Dosage compensation in mammals: fine-tuning the expression of the X chromosome. Genes Dev 20: 1848–1867 [DOI] [PubMed] [Google Scholar]
- Hemberger M 2002. The role of the X chromosome in mammalian extra embryonic development. Cytogenet Genome Res 99: 210–217 [DOI] [PubMed] [Google Scholar]
- Huynh KD, Lee JT 2003. Inheritance of a pre-inactivated paternal X chromosome in early mouse embryos. Nature 426: 857–862 [DOI] [PubMed] [Google Scholar]
- Kalantry S, Magnuson T 2006. The Polycomb group protein EED is dispensable for the initiation of random X-chromosome inactivation. PLoS Genet 2: e66 doi: 10.1371/journal.pgen.0020066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, et al. 2005. Antisense transcription in the mammalian transcriptome. Science 309: 1564–1566 [DOI] [PubMed] [Google Scholar]
- Lee JT 2000. Disruption of imprinted X inactivation by parent-of-origin effects at Tsix. Cell 103: 17–27 [DOI] [PubMed] [Google Scholar]
- Lee JT, Lu N 1999. Targeted mutagenesis of Tsix leads to nonrandom X inactivation. Cell 99: 47–57 [DOI] [PubMed] [Google Scholar]
- Lee JT, Davidow LS, Warshawsky D 1999. Tsix, a gene antisense to Xist at the X-inactivation centre. Nat Genet 21: 400–404 [DOI] [PubMed] [Google Scholar]
- Lengner CJ, Gimelbrant AA, Erwin JA, Cheng AW, Guenther MG, Welstead GG, Alagappan R, Frampton GM, Xu P, Muffat J, et al. 2010. Derivation of pre-X inactivation human embryonic stem cells under physiological oxygen concentrations. Cell 141: 872–883 [DOI] [PubMed] [Google Scholar]
- Luikenhuis S, Wutz A, Jaenisch R 2001. Antisense transcription through the Xist locus mediates Tsix function in embryonic stem cells. Mol Cell Biol 21: 8512–8520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maherali N, Sridharan R, Xie W, Utikal J, Eminli S, Arnold K, Stadtfeld M, Yachechko R, Tchieu J, Jaenisch R, et al. 2007. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell 1: 55–70 [DOI] [PubMed] [Google Scholar]
- Mak W, Nesterova TB, de Napoles M, Appanah R, Yamanaka S, Otte AP, Brockdorff N 2004. Reactivation of the paternal X chromosome in early mouse embryos. Science 303: 666–669 [DOI] [PubMed] [Google Scholar]
- McDonald LE, Paterson CA, Kay GF 1998. Bisulfite genomic sequencing-derived methylation profile of the xist gene throughout early mouse development. Genomics 54: 379–386 [DOI] [PubMed] [Google Scholar]
- Nakanishi T, Kuroiwa A, Yamada S, Isotani A, Yamashita A, Tairaka A, Hayashi T, Takagi T, Ikawa M, Matsuda Y, et al. 2002. FISH analysis of 142 EGFP transgene integration sites into the mouse genome. Genomics 80: 564–574 [DOI] [PubMed] [Google Scholar]
- Navarro P, Page DR, Avner P, Rougeulle C 2006. Tsix-mediated epigenetic switch of a CTCF-flanked region of the Xist promoter determines the Xist transcription program. Genes Dev 20: 2787–2792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesterova TB, Popova BC, Cobb BS, Norton S, Senner CE, Tang YA, Spruce T, Rodriguez TA, Sado T, Merkenschlager M, et al. 2008. Dicer regulates Xist promoter methylation in ES cells indirectly through transcriptional control of Dnmt3a. Epigenetics Chromatin 1: 2 doi: 10.1186/1756-8935-1-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohhata T, Tachibana M, Tada M, Tada T, Sasaki H, Shinkai Y, Sado T 2004. X-inactivation is stably maintained in mouse embryos deficient for histone methyl transferase G9a. Genesis 40: 151–156 [DOI] [PubMed] [Google Scholar]
- Ohhata T, Hoki Y, Sasaki H, Sado T 2006. Tsix-deficient X chromosome does not undergo inactivation in the embryonic lineage in males: implications for Tsix-independent silencing of Xist. Cytogenet Genome Res 113: 345–349 [DOI] [PubMed] [Google Scholar]
- Ohhata T, Hoki Y, Sasaki H, Sado T 2008. Crucial role of antisense transcription across the Xist promoter in Tsix-mediated Xist chromatin modification. Development 135: 227–235 [DOI] [PubMed] [Google Scholar]
- Okada Y, Tashiro C, Numata K, Watanabe K, Nakaoka H, Yamamoto N, Okubo K, Ikeda R, Saito R, Kanai A, et al. 2008. Comparative expression analysis uncovers novel features of endogenous antisense transcription. Hum Mol Genet 17: 1631–1640 [DOI] [PubMed] [Google Scholar]
- Okamoto I, Otte AP, Allis CD, Reinberg D, Heard E 2004. Epigenetic dynamics of imprinted X inactivation during early mouse development. Science 303: 644–649 [DOI] [PubMed] [Google Scholar]
- Plath K, Talbot D, Hamer KM, Otte AP, Yang TP, Jaenisch R, Panning B 2004. Developmentally regulated alterations in Polycomb repressive complex 1 proteins on the inactive X chromosome. J Cell Biol 167: 1025–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sado T, Fenner MH, Tan SS, Tam P, Shioda T, Li E 2000. X inactivation in the mouse embryo deficient for Dnmt1: distinct effect of hypomethylation on imprinted and random X inactivation. Dev Biol 225: 294–303 [DOI] [PubMed] [Google Scholar]
- Sado T, Wang Z, Sasaki H, Li E 2001. Regulation of imprinted X-chromosome inactivation in mice by Tsix. Development 128: 1275–1286 [DOI] [PubMed] [Google Scholar]
- Sado T, Okano M, Li E, Sasaki H 2004. De novo DNA methylation is dispensable for the initiation and propagation of X chromosome inactivation. Development 131: 975–982 [DOI] [PubMed] [Google Scholar]
- Sado T, Hoki Y, Sasaki H 2005. Tsix silences Xist through modification of chromatin structure. Dev Cell 9: 159–165 [DOI] [PubMed] [Google Scholar]
- Sakaue M, Ohta H, Kumaki Y, Oda M, Sakaide Y, Matsuoka C, Yamagiwa A, Niwa H, Wakayama T, Okano M 2010. DNA methylation is dispensable for the growth and survival of the extraembryonic lineages. Curr Biol 20: 1452–1457 [DOI] [PubMed] [Google Scholar]
- Savarese F, Flahndorfer K, Jaenisch R, Busslinger M, Wutz A 2006. Hematopoietic precursor cells transiently reestablish permissiveness for X inactivation. Mol Cell Biol 26: 7167–7177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeftner S, Sengupta AK, Kubicek S, Mechtler K, Spahn L, Koseki H, Jenuwein T, Wutz A 2006. Recruitment of PRC1 function at the initiation of X inactivation independent of PRC2 and silencing. EMBO J 25: 3110–3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J, Mak W, Zvetkova I, Appanah R, Nesterova TB, Webster Z, Peters AH, Jenuwein T, Otte AP, Brockdorff N 2003. Establishment of histone h3 methylation on the inactive x chromosome requires transient recruitment of eed-enx1 polycomb group complexes. Dev Cell 4: 481–495 [DOI] [PubMed] [Google Scholar]
- Silva J, Barrandon O, Nichols J, Kawaguchi J, Theunissen TW, Smith A 2008. Promotion of reprogramming to ground state pluripotency by signal inhibition. PLoS Biol 6: e253 doi: 10.1371/journal.pgen.0060253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Maherali N, Breault DT, Hochedlinger K 2008. Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell Stem Cell 2: 230–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavropoulos N, Lu N, Lee JT 2001. A functional role for Tsix transcription in blocking Xist RNA accumulation but not in X-chromosome choice. Proc Natl Acad Sci 98: 10232–10237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto M, Abe K 2007. X chromosome reactivation initiates in nascent primordial germ cells in mice. PLoS Genet 3: e116 doi: 10.1371/journal.pgen.0030116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun BK, Deaton AM, Lee JT 2006. A transient heterochromatic state in Xist preempts X inactivation choice without RNA stabilization. Mol Cell 21: 617–628 [DOI] [PubMed] [Google Scholar]
- Wang J, Mager J, Chen Y, Schneider E, Cross JC, Nagy A, Magnuson T 2001. Imprinted X inactivation maintained by a mouse Polycomb group gene. Nat Genet 28: 371–375 [DOI] [PubMed] [Google Scholar]
- Wutz A, Jaenisch R 2000. A shift from reversible to irreversible X inactivation is triggered during ES cell differentiation. Mol Cell 5: 695–705 [DOI] [PubMed] [Google Scholar]
- Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT 2008. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 322: 750–756 [DOI] [PMC free article] [PubMed] [Google Scholar]