The effects of systemic MdmX inhibition in normal tissues and in tumors was determined. MdmX inhibition was modeled by restoring p53 function in MdmX-deficient mice in a p53ERTAM knock-in background. The effects of transient p53 restoration in the absence of MdmX are nonlethal and reversible, unlike transient p53 restoration in the absence of Mdm2, which is lethal. Thus, MdmX inhibition can be considered as a therapeutic strategy for restoring p53 function in tumors that retain wild-type p53.
Keywords: Mdm2, MdmX, cancer therapy, p53
Abstract
MdmX, also known as Mdm4, is a critical negative regulator of p53, and its overexpression serves to block p53 tumor suppressor function in many cancers. Consequently, inhibiting MdmX has emerged as an attractive approach to restoring p53 function in those cancers that retain functional p53. However, the consequences of acute systemic MdmX inhibition in normal adult tissues remain unknown. To determine directly the effects of systemic MdmX inhibition in normal tissues and in tumors, we crossed mdmX−/− mice into the p53ERTAM knockin background. In place of wild-type p53, p53ERTAM knockin mice express a variant of p53, p53ERTAM, that is completely dependent on 4-hydroxy-tamoxifen for its activity. MdmX inhibition was then modeled by restoring p53 function in these MdmX-deficient mice. We show that MdmX is continuously required to buffer p53 activity in adult normal tissues and their stem cells. Importantly, the effects of transient p53 restoration in the absence of MdmX are nonlethal and reversible, unlike transient p53 restoration in the absence of Mdm2, which is ineluctably lethal. We also show that the therapeutic impact of restoring p53 in a tumor model is enhanced in the absence of MdmX, affording a significant extension of life span over p53 restoration in the presence of MdmX. Hence, systemic inhibition of MdmX is both a feasible therapeutic strategy for restoring p53 function in tumors that retain wild-type p53 and likely to be significantly safer than inhibition of Mdm2.
The p53 transcription factor coordinates responses in somatic cells to a variety of stresses, including DNA damage, hypoxia, and oncogene activation, by triggering replicative arrest or death of the damaged cell. The pivotal role that p53 plays in tumor suppression is evidenced by the fact that the p53 pathway is functionally inactivated in the majority of human cancers and that p53-deficient mice develop cancers at high frequencies (Kruse and Gu 2009; Vousden and Prives 2009). Intriguingly, many cancers retain expression of wild-type p53, inactivating instead upstream or downstream p53 pathway components, and intense research has focused on finding ways to reactivate p53 function selectively in such p53-competent tumors. Hitherto, most pharmacological strategies have sought to inhibit Mdm2, a critical negative regulator of p53 (Issaeva et al. 2004; Vassilev et al. 2004; Shangary et al. 2008; Sun et al. 2008; Vazquez et al. 2008; Canner et al. 2009; Hedstrom et al. 2009). While such studies confirm that inhibition of Mdm2 can activate latent p53 in tumor cells, it also triggers pathological p53 function in normal tissues (Boesten et al. 2006; Francoz et al. 2006; Grier et al. 2006; Ringshausen et al. 2006; Xiong et al. 2006; Maetens et al. 2007; Valentin-Vega et al. 2008), narrowing the therapeutic window available for Mdm2 inhibition in cancer therapy.
MdmX, also known as Mdm4, has recently emerged as a discrete critical negative regulator of p53 (Marine and Jochemsen 2005; Marine et al. 2007; Wade and Wahl 2009; Mancini et al. 2010). While MdmX is structurally related to Mdm2, it does not target p53 for degradation via ubiquitin ligation, but instead acts as a direct transcriptional squelcher of p53 (Marine and Jochemsen 2005; Marine et al. 2007; Kruse and Gu 2009; Wade and Wahl 2009). MdmX also modulates Mdm2 stability and function, thereby indirectly regulating p53 (Jackson et al. 2001; Uldrijan et al. 2007; Linke et al. 2008; Okamoto et al. 2009). Like Mdm2, MdmX is overexpressed in many cancers, especially those of breast, colon, and lung, as well as in glioma, lymphoma, and retinoblastoma (Danovi et al. 2004; Laurie et al. 2006), where it is thought to promote tumorigenesis by suppressing p53 function. However, knockout (Parant et al. 2001; Migliorini et al. 2002) and conditional deletion (Francoz et al. 2006; Grier et al. 2006; Xiong et al. 2006) studies have shown that MdmX, like Mdm2, is required as a buffer against precocious and lethal p53 activity at various bottlenecks in embryonic development. Ascertaining whether acute MdmX inhibition also shares the same disquieting toxicity as does Mdm2 inhibition in p53-competent adult tissues has been confounded by this embryonic lethality in p53-competent mdmX knockout mice, as has evaluation of the utility of MdmX inhibition in cancer therapy.
To model directly the side effects of acute systemic MdmX inhibition in adult p53-competent animals, together with the therapeutic potential of MdmX inhibition in established tumors, we crossed mdmX+/− mice with p53ERTAM knockin (p53KI) animals. p53KI mice express p53ERTAM, a 4-hydroxy-tamoxifen (4-OHT)-dependent variant of endogenous p53, in place of wild-type p53 protein. p53KI mice are effectively p53-deficient in the absence of 4-OHT ligand and are p53 wild type in its presence (Christophorou et al. 2005), allowing for acute and reversible systemic functional restoration of wild-type p53 function in adult mdmX−/−;p53KI mice. We show that MdmX, like Mdm2, is indeed continuously required to suppress untoward p53 function in adult tissues and their stem cells. However, unlike even fleeting p53 restoration in mdm2−/− mice, which is rapidly and irrevocably fatal, transient p53 function is surprisingly well tolerated by mdmX−/− animals. We show that the milder impact of p53 restoration in mdmX−/− mice compared with restoration in mdm2−/− mice is likely a consequence of the fact that Mdm2 retains partial functionality in the absence of MdmX. Furthermore, we show that absence of MdmX potentiates the therapeutic benefit of p53 restoration in a lymphoma model. These data intimate that inhibition of MdmX would be an effective therapeutic strategy for restoring p53 function in cancer that is better tolerated than is inhibition of Mdm2.
Results
p53 is spontaneously active in MdmX-deficient adult tissues
To determine the extent to which MdmX continually restrains spontaneous p53 activity in normal adult mouse tissues, we crossed p53KI/KI mice with mdmX+/− mice. No pups harboring two copies of the p53KI allele were born in an mdmX−/− background, indicating that, as reported with mdm2−/−;p53KI/KI mice, even in the absence of 4-OHT there is some residual p53 activity in mdmX−/− mice from two copies of p53ERTAM that is sufficient to induce embryonic lethality. However, mdmX−/− mice harboring only a single p53ERTAM allele (p53KI/−) were born at expected Mendelian ratios (as were mdmX+/+;p53KI/− and mdmX+/−;p53KI/− animals), indicating no biologically significant leakiness from a single copy of p53ERTAM; consequently, mdmX−/−;p53KI/− animals were used in all subsequent experiments.
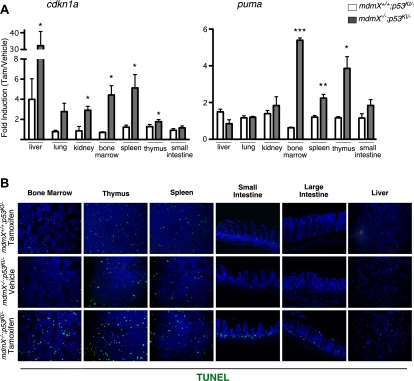
To ascertain the extent of spontaneous p53 activity in adult tissues in the absence of MdmX, p53 was restored in mdmX−/−;p53KI/− mice by systemic administration of tamoxifen and selected organs were harvested for analysis 6 h later. For direct evidence of p53 activity in the absence of MdmX, we assayed for the expression of known p53 target genes cdkn1a (encoding the Cdk inhibitor p21cip1) and puma (encoding the proapoptotic BH3 protein PUMA). We noted rapid and dramatic induction of these two p53 target genes in tissues of tamoxifen-treated mdmX−/−;p53KI/−, mice confirming that p53 is indeed spontaneously active in adult tissues of MdmX-deficient mice (Fig. 1A). However, quite unlike the situation following p53 functional restoration in the absence of Mdm2 (Ringshausen et al. 2006), where both cdkn1a and puma are profoundly induced in all tissues irrespective of whether or not that specific tissue then undergoes apoptosis, restoration of p53 function in mdmX−/−;p53KI/− tissues elicited more specific p53 target gene induction. While, with the exception of small intestine, p53 restoration potently induced cdkn1a in all tested tissues, the proapoptotic gene puma was induced only in classically radiosensitive tissues (bone marrow, spleen, thymus, and intestinal epithelium) (Fig. 1A).
Figure 1.
p53 is spontaneously active in tissues of p53KI/−;mdmX−/− mice. (A) RNA was isolated from organs of mice treated with either vehicle (oil) or tamoxifen for 6 h, and cdkn1a and puma gene expression was quantified by TaqMan analysis. Fold induction (expression in tamoxifen-treated tissues over vehicle-treated tissues) is plotted. Fold inductions that were statistically significant are marked with an asterisk. Error bars show the SEM of triplicates. (B) A single bolus of either vehicle or tamoxifen was administered to mice of the indicated genotypes, and organs were collected 6 h later. Apoptosis was detected in tissue sections by TUNEL staining. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001.
This unexpected selectivity of p53 target gene induction in different mdmX−/− tissues was broadly mirrored by the biological outcomes of p53 restoration. Radiosensitive tissues (bone marrow, thymus, spleen, and intestinal epithelium) of tamoxifen-treated mdmX−/−;p53KI/− mice exhibited a marked increase in apoptosis compared with controls (mdmX+/+;p53KI/− and mdmX−/−;p53−/− mice treated with tamoxifen, or mdmX−/−;p53KI/− mice treated with oil) (Fig. 1B; data not shown). In contrast, no detectable apoptosis was observed in classically radio-resistant tissues like liver, kidney, lung, and heart (Fig. 1B; data not shown). Of note, despite the crucial role played by MdmX inhibition of p53 during development of the cerebellum, especially with regard to suppression of p53-dependent apoptosis in post-mitotic cerebellar neurons in embryos (Francoz et al. 2006), we saw no apoptosis or any other evident impact of p53 restoration on histology or cell viability in the fully formed adult cerebellum of mdmX−/− mice, even though significant p53 activity was evident from the induction of cdkn1a (Supplemental Fig. S1). p53-induced apoptosis was evident only in the continuously proliferating subventricular zones of mdmX−/−;p53KI/− mouse brains (Supplemental Fig. S1).
mdmX−/− mice survive transient p53 restoration
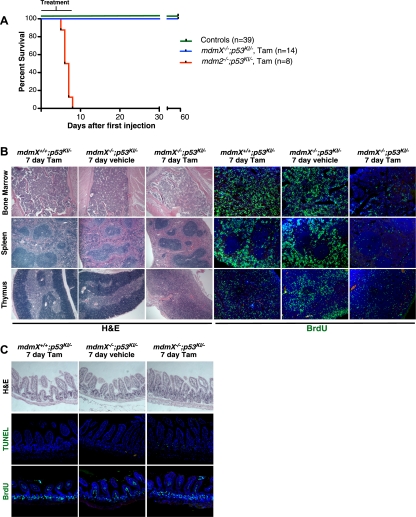
Substantial evidence indicates that both Mdm2 and MdmX are crucial for reining in the toxic effects of untoward p53 activity. Indeed, we previously showed that even very transient restoration of p53 (i.e., a single tamoxifen injection) is lethal to adult Mdm2-deficient mice within 6 d (Ringshausen et al. 2006). In stark contrast, even 7 d of sustained (i.e., daily tamoxifen injections) p53 restoration was not lethal to MdmX-deficient mice (Fig. 2A), even though histological analysis indicated significant degenerative tissue-specific pathologies, including striking suppression of proliferation and decreased cellularity in the bone marrow and red pulp of the spleen together with loss of structural integrity in the thymus (Fig. 2B). Unexpectedly, given that intestines of tamoxifen-treated mdm2−/−;p53KI/− mice rapidly succumb to severe and irrevocable damage (Supplemental Fig. S2; Ringshausen et al. 2006), intestinal epithelia of tamoxifen-treated mdmX−/−;p53KI/− mice maintained normal architecture and barrier function throughout the 1 wk of p53 restoration, despite exhibiting significant ongoing levels of p53-dependent apoptosis (which, incidentally, is evidence that restored p53 function is indeed maintained throughout the 7 d of tamoxifen treatment) (Fig. 2C). One likely reason for the sustained structural integrity of mdmX−/− intestinal epithelia is that proliferation in mdmX−/− intestines continues throughout p53 restoration (Fig. 2C), a surprising observation that is nonetheless consistent with the failure of p53 to induce p21cip1 in that tissue (Fig. 1A).
Figure 2.
Transient p53 restoration in the absence of MdmX is well tolerated. (A) Kaplan-Meier curve showing survival of mice of the indicated genotypes given daily injections of vehicle or tamoxifen for 1 wk. Controls are p53KI/− mice treated with vehicle (n = 8), p53KI/− treated with tamoxifen (n = 8), mdm2−/−;p53KI/− mice treated with vehicle (n = 6), mdmX−/−;p53KI/− mice treated with vehicle (n = 13), and mdmX−/−;p53−/− mice treated with tamoxifen (n = 4). (B) Mice of the indicated genotypes were treated daily with either vehicle or tamoxifen for 7 d. The left panels show H&E staining of bone marrow, spleen, and thymus. The right panels show BrdU staining of the same organs (BrdU was administered 2 h prior to sacrifice). (C) H&E, TUNEL, and BrdU staining of sections of small intestine from mice treated as in B.
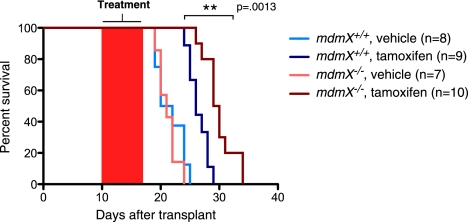
To ascertain whether mdmX−/− mice can survive permanent p53 restoration, mdmX−/−;p53KI/− mice were repeatedly treated daily with tamoxifen and their health and viability were monitored. Long-term sustained p53 restoration was eventually lethal to mdmX−/− mice (median survival of 29 d) (Fig. 3A), albeit greatly delayed relative to the rapid demise (6 d) that succeeds p53 restoration in mdm2−/− mice. At the time of death, bone marrow, spleen, and thymus of tamoxifen-treated mdmX−/−;p53KI/− mice exhibited severe attrition, with decreased cellularity and loss of architectural integrity, reminiscent of that accompanying the far more rapid deaths of tamoxifen-treated mdm2−/−;p53KI/− mice (Fig. 3B). Once again, however, the dramatic loss of intestinal epithelial integrity that p53 restoration elicits in mdm2−/− animals (Supplemental Fig. S2; Ringshausen et al. 2006) was absent from mdmX−/− animals: Overall integrity of both the small and large intestines of mdmX−/−;p53KI/− mice, as well as villus/crypt structure and ratio, were maintained even after long-term (26 d) sustained p53 restoration (Fig. 3C). Detailed histological analysis of such intestines indicated an abnormal expansion of Paneth cells in the crypts and the concurrence of both sustained intestinal crypt proliferation (again, consistent with the absence of p21cip1 induction in intestines) and apoptosis (Fig. 3C).
Figure 3.
Permanent p53 restoration is lethal in the absence of MdmX. (A) Kaplan-Meier curve showing survival of mice of the indicated genotypes given continuous daily injections of vehicle or tamoxifen. Controls are mdmX+/+;p53KI/− mice treated with tamoxifen (n = 13), mdmX−/−;p53KI/− mice treated with vehicle (n = 9), and mdmX−/−;p53−/− mice treated with tamoxifen (n = 6). (B) H&E staining of organs harvested from mdmX+/+;p53KI/− and mdmX−/−;p53KI/− mice treated continuously with daily tamoxifen for 26 d. (C) H&E, TUNEL, and Ki67 staining of small intestine harvested from mdmX+/+;p53KI/− and mdmX−/−;p53KI/− mice treated continuously with daily tamoxifen for 26 d.
Given this long-term maintenance of intestinal integrity in tamoxifen-treated mdmX−/−;p53KI/− mice, the most plausible cause of their eventual demise is bone marrow failure: Indeed, 26-d tamoxifen-treated mdmX−/− mice were severely anemic (reduced red blood cell count, reduced hemoglobin and hematocrit) and exhibited overall reduction of blood counts (Supplemental Fig. S3).
The pathologies induced in MdmX-deficient tissues by transient p53 functional restoration are fully reversible
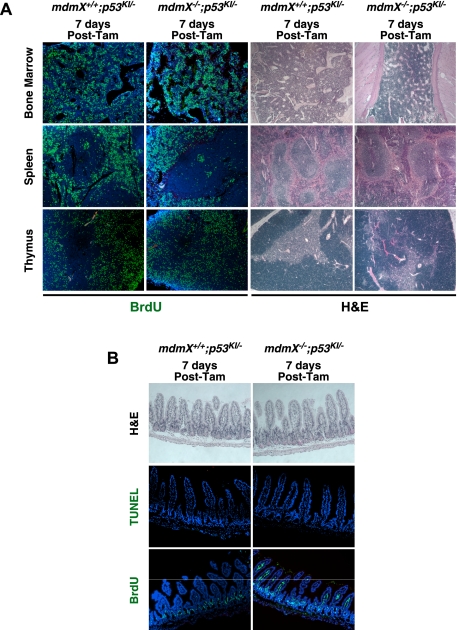
To ascertain whether transient p53 restoration in mdmX−/− mice elicits irreversible damage in tissues, p53 was continuously restored in mdmX−/−;p53KI/− mice for 7 d (daily injections), and tamoxifen was then withdrawn to assess the capacity of tissues to regenerate. All affected tissues (bone marrow, spleen, thymus, and intestine) quickly recovered, appearing largely normal after only 1 wk: Proliferation resumed in lymphoid organs (Fig. 4A), and apoptosis declined in intestinal epithelia (Fig. 4B). Thereafter, such mice lived long term and exhibited no adverse consequences of transient systemic p53 activation. Hence, the extended survival of transiently tamoxifen-treated mdmX−/−;p53KI/− (relative to mdm2−/− mice) is accompanied by a greatly enhanced capacity to recover from the depredations of p53 restoration.
Figure 4.
The pathological effects of transient p53 restoration in the absence of MdmX are reversible. (A) Mice of the indicated genotypes were treated daily with tamoxifen for 7 d and sacrificed 7 d after. The left panels show BrdU staining of sections of bone marrow, spleen, and thymus (BrdU was administered 2 h prior to sacrifice). The right panels show H&E staining of the same organs. (B) H&E, TUNEL, and BrdU staining of small intestine sections from mice treated as in A.
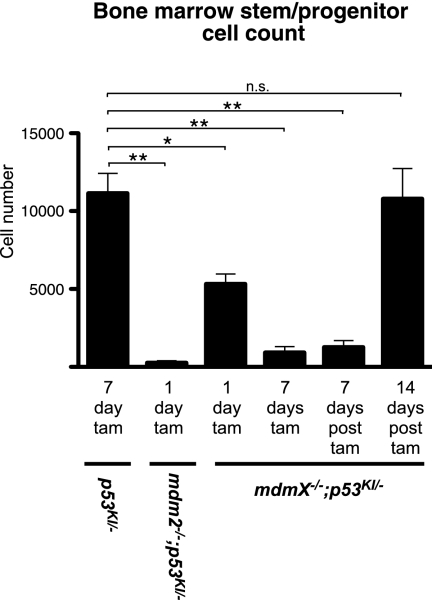
One possible explanation for both the maintenance of intestinal integrity and the expeditious recovery from p53 restoration in tamoxifen-treated mdmX−/−;p53KI/− compared with tamoxifen-treated mdm2−/−;p53KI/− animals is that the stem cell populations required to regenerate damaged tissues are less debilitated by the absence of MdmX than of Mdm2. To test this directly, we turned to the hematopoietic system in which the stem cell population is relatively well-characterized. Bone marrow cells were isolated from tamoxifen-treated p53KI/−, mdmX−/−;p53KI/−, and mdm2−/−;p53KI/− mice and immunostained, and the hematopoietic stem/progenitor cell compartment was quantitated by flow cytometry. While p53 restoration caused a slight reduction in whole bone marrow counts in mdmX−/−;p53KI/− mice, p53 restoration in mdm2−/−;p53KI/− mice triggered dramatic loss of total bone marrow cellularity within only 1 d: By 5 d, the bone marrow had been essentially annihilated—only a few hundred cells (1655.3 ± 558.2) were recoverable, making it impossible to conduct meaningful analysis of stem cells at this or later time points in these mice (Supplemental Fig. S4). Control mdmX−/−;p53KI/− mice treated with vehicle maintained similar cell counts to tamoxifen-treated mdmX+/+;p53KI/− mice (data not shown). Direct quantitation of stem/progenitor cells (Lin−, Sca1+, CD150+, CD48−) (Kiel et al. 2005) indicated that they are effectively ablated in mdm2−/−;p53KI/− mice after only 1 d of p53 restoration (Fig. 5). In comparison, although they are significantly decreased in number compared with wild-type controls, more stem/progenitor cells can be recovered from tamoxifen-treated mdmX−/−;p53KI/− mice even after 7 d of sustained p53 restoration than from 1-d-treated mdm2−/−;p53KI/− animals (Fig. 5). Furthermore, although the numbers of stem/progenitor cells remained low in mdmX−/−;p53KI/− bone marrow 1 wk after the end of tamoxifen treatment, their numbers were fully recovered when measured 1 wk later. Hence, hematopoietic stem/progenitor cells recover well from short-term p53 activity in the absence of MdmX but not of Mdm2. In conclusion, while both Mdm2 and MdmX are required for the long-term maintenance of hematopoietic stem/progenitor cells in the presence of functional p53, it is clear that Mdm2 is the most exigent restraint to p53 toxicity.
Figure 5.
Stem/progenitor cells can recover from p53 restoration in the absence of MdmX. Hematopoietic stem/progenitor cells (Lin−, Sca1+, CD150+, and CD48−) were quantified in mice of the indicated genotypes, treated with tamoxifen as indicated. The last two columns show counts from mdmX−/−;p53KI/− mice 7 d and 14 d after a 7-d tamoxifen treatment, respectively. (*) P < 0.05; (**) P < 0.01; (n.s.) not significant.
Mdm2 restrains p53 activity even when MdmX is absent, blunting the pathological impact of MdmX deficiency in p53-competent tissues
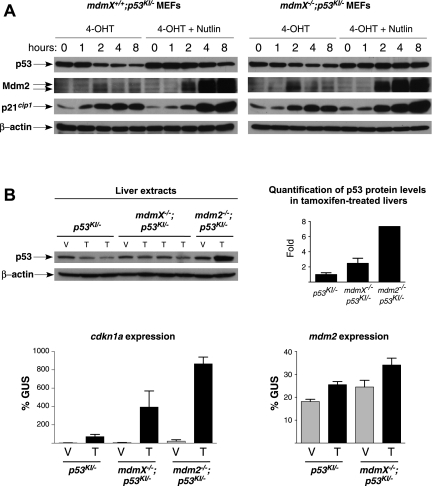
Why should p53 be less toxic in the absence of MdmX than in the absence of Mdm2? It is well established that p53 and Mdm2 form a negative feedback loop whereby active p53 induces expression of Mdm2 that in turn down-regulates p53 (for review, see Kruse and Gu 2009). We hypothesized that, even in the absence of functional MdmX, Mdm2 can still restrain excessive p53 activity, thus mitigating p53 toxicity. To test this, we took advantage of the fact that our acutely switchable p53KI/KI model allows us to follow engagement of the p53–Mdm2 negative feedback loop in real time. We previously showed that upon acute p53 restoration, p53 levels fall progressively—a down-regulation that is compromised in the absence of Mdm2 (Ringshausen et al. 2006). To determine whether p53 is also down-regulated by endogenous Mdm2 in the absence of MdmX, we isolated mdmX+/+;p53KI/− and mdmX−/−;p53KI/− mouse embryonic fibroblasts (MEFs) and treated them with 4-OHT in the presence or absence of the Mdm2 inhibitor Nutlin-3a (Vassilev et al. 2004). Upon acute restoration of p53 function in mdmX+/+;p53KI/− MEFs following 4-OHT addition, overt p53 activity was rapidly engaged by the known p53-activating stresses of in vitro culture, as evidenced by the induction of p21cip1. Concurrently, p53ERTAM levels were down-regulated and Mdm2 levels rose, again confirming that p53, once restored, is activated. The decline in p53 level was partially abrogated when the mdmX+/+;p53KI/− MEFs were cotreated with Nutlin-3a, indicating that Mdm2 contributes to the reduction in p53 levels and the regulation of p53 activity in 4-OHT-treated mdmX+/+ cells (Fig. 6A). Likewise, p53 levels dropped in mdmX−/−;p53KI/− MEFs after addition of 4-OHT, together with a concomitant increase in p53 activity and induction of Mdm2. This p53 down-regulation was also inhibited by Nutlin-3a, resulting in yet higher p53 activity (Fig. 6A), which demonstrates that endogenous Mdm2 still contributes significantly to the down-regulation of p53 levels and activity even in the absence of MdmX. Interestingly, p53 protein levels were higher in mdmX−/−;p53KI/− MEFs compared with wild-type controls, suggesting a role for MdmX in the regulation of p53 protein levels and consistent with previous studies that ascribe such a role to MdmX, principally through modulation of Mdm2 function and stability (Jackson et al. 2001; Gu et al. 2002; Okamoto et al. 2009).
Figure 6.
Mdm2 is functional and contributes to p53 down-regulation in the absence of MdmX. (A) mdmX+/+;p53KI/− and mdmX−/−;p53KI/− MEFs were treated with either 4-OHT or 4-OHT plus 10 μM Nutlin-3a for the indicated times. Immunoblotting was used to estimate levels of p53, Mdm2, and p21cip1 at each time point. (B) Mice of the indicated genotypes were treated with either vehicle (V) or tamoxifen (T) for 1 d. The levels of p53 in the livers of these mice are presented (immunoblot; each lane is a separate mouse), together with a quantification of the p53 levels in tamoxifen-treated mice (from the blot shown). The relative induction of p53 target genes cdkn1a and mdm2 (by TaqMan; gus is the control gene) in these mice is also shown. Error bars show the SEM from triplicates.
To test whether endogenous Mdm2 also restrains p53 level and activity in mdmX−/− tissues in vivo, we treated p53KI/−, mdmX−/−;p53KI/−, and mdm2−/−;p53KI/− mice for 1 d with either vehicle or tamoxifen to restore p53 and then monitored p53 levels over time in the liver. As predicted, p53 levels dropped in the livers of p53KI/− mice after p53 restoration, with little increase in p53 activity (measured by induction of cdkn1a mRNA) (Fig. 6B). p53 levels also dropped after restoration in livers of tamoxifen-treated mdmX−/−;p53KI/− mice, albeit not as efficiently, consistent again with a previously reported contribution of MdmX to the regulation of p53 protein levels (Fig. 6B). Nonetheless, despite this fall in overall p53 levels, p53 activity rose in the absence of MdmX, as evidenced by a clear increase in cdkn1a (Fig. 6B). Of note, Mdm2 was also induced in mdmX−/−;p53KI/− livers after tamoxifen administration, where it presumably contributes to p53 down-regulation (Fig. 6B). In contrast, after p53 restoration in mdm2−/−;p53KI/− livers, p53 levels fail to drop and, indeed, increase, resulting in even higher p53 activity (evidenced by robust induction of cdkn1a) (Fig. 6B). Similar dynamics were observed in the spleen and thymus (Supplemental Fig. S5). Quantification of p53 protein levels in tamoxifen-treated p53KI/−, mdmX−/−;p53KI/−, and mdm2−/−;p53KI/− tissues showed that MdmX plays at least some role in the regulation of p53 protein level also in vivo. Nonetheless, it remains clear that Mdm2 is the principal regulator of p53 protein level in vivo, since Mdm2-null tissues exhibit the highest levels of p53 protein after restoration (Fig. 6B; Supplemental Fig. S4). Overall, the stark differences in down-regulation of p53ERTAM after its functional restoration in organs of mdmX−/− versus mdm2−/− mice affirms that, even in the absence of MdmX, endogenous Mdm2 retains some capacity to mitigate p53 levels and activity in vivo. Taken together, the persistence of the p53–Mdm2 negative feedback loop in mdmX−/− cells and tissues offers the most plausible explanation for the unexpectedly mild and recoverable impact of p53 in the absence of MdmX.
The therapeutic impact of restoring p53 in tumors is augmented by inactivity of MdmX
Several studies have indicated that restoring p53 function in tumors may be therapeutically beneficial (Martins et al. 2006; Ventura et al. 2007; Xue et al. 2007; Junttila et al. 2010). Since our data establish that transient p53 restoration in MdmX-deficient mice has only limited and delayed toxicity, we asked whether the absence of MdmX offers a useful therapeutic window by which to enhance the therapeutic benefit of restoring p53 in tumors in vivo. We crossed mdmX−/−;p53KI/− mice to the lymphoma-prone Eμ-Myc transgenic strain (Adams et al. 1985). Tumors from mdmX+/+;p53KI/−;EμMyc and mdmX−/−;p53KI/−;EμMyc mice were then isolated and transplanted into multiple wild-type recipients. Ten days after transplantation, recipient mice were treated for 1 wk with either vehicle or tamoxifen. Consistent with our previous study (Martins et al. 2006), tamoxifen treatment significantly extended the life span of mice harboring mdmX+/+;p53KI/−;EμMyc tumors compared with sham-treated controls (Fig. 7). Vehicle-treated mice bearing mdmX−/−;p53KI/−;EμMyc tumors showed no increase in overall survival relative to vehicle-treated MdmX-proficient controls, demonstrating that absence of MdmX alone does not affect tumor growth per se. However, p53 restoration in the absence of MdmX significantly increased overall survival relative to tamoxifen-treated mdmX+/+;p53KI/−;EμMyc controls (Fig. 7). Hence, p53 restoration in the absence of MdmX confers a significant therapeutic benefit over p53 restoration in its presence.
Figure 7.
Restoration of p53 in the absence of MdmX extends overall survival in lymphoma-bearing mice. mdmX+/+;p53KI/−;EμMyc and mdmX−/−;p53KI/−;EμMyc tumors (the graph legend indicates the mdmX genotype only) were transplanted into wild-type recipients. Ten days after transplant, recipient mice were treated with either vehicle or tamoxifen for 1 wk. Their survival is plotted as a Kaplan-Meier curve.
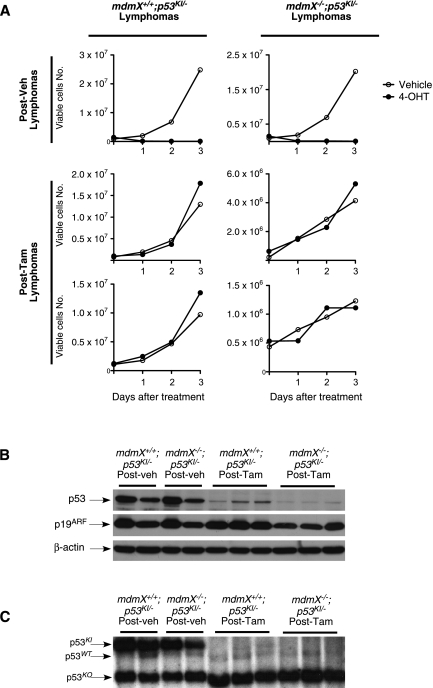
p53 restoration therapy in Eμ-Myc tumors is eventually compromised by the emergence of resistant tumor clones harboring additional mutations in the p53 pathway (Martins et al. 2006). To investigate whether this remains a significant limitation, even when p53 is restored in the absence of MdmX, we harvested tumors from treated host mice and cultured them in vitro in the presence or absence of 4-OHT. Tumor cells derived from vehicle-treated mice were rapidly killed when exposed to 4-OHT, regardless of MdmX genotype, indicating that the absence of MdmX alone does not promote outgrowth of resistant clones (Fig. 8A). In contrast, mdmX+/+ and mdmX−/− tumor cells isolated from relapsed tamoxifen-treated mice survived and grew in the presence of 4-OHT, confirming that p53 restoration does indeed still select for resistant clones irrespective of MdmX status (Fig. 8A).
Figure 8.
p53 restoration in the absence of MdmX does not prevent the emergence of resistant clones that deleted p53 in EμMyc tumors. (A) mdmX+/+;p53KI/−;EμMyc and mdmX−/−;p53KI/−;EμMyc transplanted tumors isolated from recipient mice previously treated with either vehicle (designated as Post-veh) or tamoxifen (designated as Post-Tam) in vivo were cultured in the presence or absence of 4-OHT. In vitro growth of cells derived from representative tumors is presented. (B) Immunoblot analysis of p53 and p19ARF levels in transplanted tumors isolated from recipient mice treated with vehicle (designated as Post-veh) or tamoxifen (designated as Post-Tam). Each lane represents an individual tumor. (C) Southern blot analysis of DNA isolated from the same tumors as in B. The p53 probe detects a 5.9-kb band from the p53KI locus, a 5.0-kb band from the wild-type p53 locus (from the host tissue), and a 3.0-kb band from the p53 knockout locus.
Resistance to p53 restoration arises in Eμ-Myc tumors by either loss of p53 itself or loss of p19ARF, the essential upstream conduit for oncogenic activation of p53 (Zindy et al. 2003; Martins et al. 2006). To establish the mechanism of acquired resistance to p53 restoration in MdmX-deficient tumors, we determined the status of p53 and p19ARF expression in transplanted mdmX+/+;p53KI/−;EμMyc and mdmX−/−;p53KI/−;EμMyc tumors. Tumors from vehicle-treated recipients all exhibited high p19ARF and readily detectable levels of p53, regardless of MdmX genotype. In contrast, mdmX+/+;p53KI/−;EμMyc tumors from tamoxifen-treated recipients had high p19ARF levels but substantially decreased p53 (Fig. 8B). mdmX−/−;p53KI/−;EμMyc tumors from tamoxifen-treated recipients also exhibited high p19ARF levels but had yet lower levels of p53 protein than even their mdmX+/+ counterparts (Fig. 8B). Southern blot analysis revealed that the loss of p53 expression arises through wholesale deletion of the p53KI locus (Fig. 8C). Hence, even though p53 restoration therapy in the absence of MdmX confers a significant extension of survival in lymphoma-bearing mice, such therapeutic benefit is still limited by the emergence of resistant clones in which the restored p53 pathway has been incapacitated.
Discussion
MdmX is a critical negative regulator of p53 (Marine and Jochemsen 2005; Marine et al. 2007; Wade and Wahl 2009; Mancini et al. 2010), necessary to restrain the lethal consequences of unbridled p53 activity at various points in normal development (Parant et al. 2001; Migliorini et al. 2002; Francoz et al. 2006; Grier et al. 2006; Xiong et al. 2006). However, the embryonic lethality of p53-competent MdmX-null mice has confounded genetic analysis of any role that MdmX might play in governing p53 activity in adult tissues. Here, we used a switchable p53 mouse model, in which endogenous p53 function can be systemically and reversibly enabled or disabled at will (Christophorou et al. 2005), to establish the acute and long-term consequences for adult tissues of p53 function in the absence of MdmX. Our data show that p53 is spontaneously active in unperturbed mdmX−/− adult mouse tissues, in the absence of any overt or detectable upstream p53-activating signal, inducing p53 target genes in all tested organs and widespread apoptosis in radiosensitive tissues. Moreover, sustained systemic p53 function in such mdmX−/− mice is ultimately lethal. This unequivocally establishes MdmX, like Mdm2 (Ringshausen et al. 2006), as critically and continuously required to rein in precocious p53 activity in normal adult tissues.
Nonetheless, while both Mdm2 and MdmX serve as constitutive buffers against untoward p53 activity in adult tissues in vivo, their roles are quantitatively and qualitatively quite distinct. Both the anti-proliferative gene cdkn1a and the proapoptotic gene puma are indiscriminately induced in all mdm2−/− tissues following acute p53 restoration, irrespective of whether the cells within that tissue then undergo apoptosis or viable cell cycle arrest (Ringshausen et al. 2006). In contrast, p53 target gene induction is more selective in tamoxifen-treated mdmX−/−;p53KI/− tissues: cdkn1a is still induced in most mdmX−/− tissues (with the notable exception of intestinal epithelium), but, in the main, puma up-regulation is restricted to those (classically radiosensitive) mdmX−/− tissues that undergo apoptosis. Given the evidence suggesting that cdkn1a and puma are induced by different threshold levels of p53 activity (Vousden and Prives 2009; Morachis et al. 2010), the most plausible explanation for such differential target gene induction is the markedly lower overall p53 activities elicited by p53 restoration in mdmX−/−;p53KI/− tissues compared with those in their mdm2−/− counterparts. Presumably, it is this higher level of p53 activity in mdm2-deficient tissues that is responsible for the far more precipitous and deleterious effects of p53 restoration in mdm2−/− tissues. Of note, our observations are not consistent with a simple binary model whereby loss of Mdm2 directs p53 to induce apoptosis while loss of MdmX directs p53 to engage growth arrest (Barboza et al. 2008). For example, both the proapoptotic gene puma and overt apoptosis are potently induced in classically radiosensitive adult tissues of p53KI/-;mdmX−/− mice after acute p53 restoration. Conversely, while puma is potently induced by p53 restoration even in radio-resistant mdm2−/− tissues, no apoptosis occurs even if those tissues are forced to proliferate (Ringshausen et al. 2006). Hence, the proclivity of any adult tissue to undergo p53-induced apoptosis appears to be an intrinsic attribute of that tissue and not the purview of either Mdm2 or MdmX.
There is still debate as to the extent to which MdmX plays a role in controlling p53 protein stability. Some studies suggest that MdmX principally controls p53 activity but not its level, the latter being reserved for the E3-ubiquitin ligase activity of Mdm2 (Boesten et al. 2006; Francoz et al. 2006; Barboza et al. 2008). However, other studies implicate MdmX in the control of p53 protein levels, most likely through regulation of Mdm2 protein stability (Sharp et al. 1999; Stad et al. 2000; Jackson et al. 2001; Gu et al. 2002; Okamoto et al. 2009). In this study, we found that the levels of p53 protein are consistently higher in tamoxifen-treated MdmX-null tissues compared with mdmX wild-type controls. It is quite possible that such elevated p53 levels contribute to the higher overall p53 activity seen in MdmX-null tissues. The role of MdmX as a negative regulator of p53 activity being well established, our data also further support the notion that MdmX plays at least some role (most likely indirect) in the regulation of p53 levels in vivo. Consistent with many existing studies, however, our new data nonetheless confirm that Mdm2 is the principal regulator of p53 levels in vivo, as evidenced by our observations that Mdm2-null tissues exhibit the highest levels of p53 protein after restoration.
Of late, restoration of p53 function has been vigorously pursued as a therapeutic approach to treat cancers and, moreover, has recently been shown to be therapeutically beneficial in several preclinical mouse cancer models (Martins et al. 2006; Ventura et al. 2007; Xue et al. 2007). Accordingly, numerous efforts have focused on blocking endogenous inhibitors of p53, in the main Mdm2, as a strategy for reactivating p53 in tumors (Issaeva et al. 2004; Vassilev et al. 2004; Shangary et al. 2008; Sun et al. 2008; Canner et al. 2009; Hedstrom et al. 2009). Unfortunately, efficient inhibition of Mdm2 triggers p53 not only in tumor cells but also in all normal cells, raising the specter that injudicious pharmacological inhibition of Mdm2 inhibition would elicit rapid, irreversible, and eventually lethal side effects (Ringshausen et al. 2006). In contrast, our data indicate that even complete inhibition of MdmX, if relatively transient, is a far less intrinsically toxic and hazardous therapeutic strategy for reactivating p53: Although transient p53 activity in the absence of MdmX does some damage to lymphoid organs and bone marrow, it has unexpectedly mild effects on intestinal epithelium—arguably the most critical tissue limiting therapeutic toxicity, since its failure is rapidly and irreversibly fatal due to fluid loss and bacterial incursion. Our data indicate that, provided that intestinal integrity is preserved, all other sequelae arising from transient p53 activity in the absence of MdmX are reversible, with all mice recovering and thereafter living long term. Furthermore, we show that absence of MdmX does indeed augment the therapeutic impact of restoring p53 function in treating lymphoma. This, together with the muted toxicity of p53 in the absence of MdmX compared with the absence of Mdm2 and the complete reversibility of its pathological side effects, supports the notion of MdmX inhibition as a feasible strategy for restoring p53 function in tumors that retain expression of wild-type p53.
Increasing evidence now implicates p53 in the regulation of adult stem cell homeostasis. Most notably, chemical inhibition or genetic ablation of p53 elicits defects in stem cell self-renewal and increased proliferation rates that eventually lead to an expansion of both hematopoietic and neural stem cells (TeKippe et al. 2003; Meletis et al. 2006; Liu et al. 2009; Leonova et al. 2010). Our data now add to the emerging view that preternatural p53 activity causes attrition of hematopoietic stem cells (Liu et al. 2010; Abbas et al. 2010). However, consistent with the very different toxicities associated with p53 activity in the absence of MdmX versus Mdm2, the severity and rapidity of stem cell attrition were very different in each instance. Even fleeting p53 restoration in mdm2-deficient mice rapidly and irreversibly wiped out hematopoietic stem/progenitor cells, together with any regenerative capacity in the bone marrow once p53 function was subsequently deactivated. In contrast, absence of MdmX provoked a much milder and protracted erosion of stem/progenitor cells, which were able to recover once p53 was removed, thus allowing for complete bone marrow regeneration and recovery of animals even after extended periods of p53 restoration. This difference has important practical implications. Although our data indicate a previously unappreciated role for both Mdm2 and MdmX in buffering p53 activity in adult hematopoietic stem cells, they also indicate that overly aggressive pharmacological inhibition of Mdm2 (even if transient) would rapidly and irreversibly deplete stem cell pools, whereas efficient MdmX inhibition would not. That this might also hold true for stem cells in other tissues is suggested by our observation that intestinal epithelia are relatively unaffected by p53 restoration in the absence of MdmX compared with restoration in the absence of Mdm2, indicating that intestinal stem cells continue to regenerate and maintain epithelial integrity in mdmX−/− mice after p53 restoration, but not in their mdm2−/− counterparts.
We previously demonstrated that, despite the high levels of p19ARF present in Eμ-Myc tumors, p53 is nonetheless not maximally active when functionally restored in such lymphoma cells, since its activity can be yet further augmented via an overt p53-activating insult, such as irradiation (Martins et al. 2006). This suggests that factors in addition to Mdm2 are suppressing maximal p53 activity, an observation consistent with our data indicating that MdmX plays an additional and independent role in buffering unbridled p53 activity. We now show that absence of MdmX enhances p53 restoration therapy in a transplant model of lymphoma. In this context, it is intriguing that irradiation-induced activation of p53 in lymphoma tumors has recently been shown to depend greatly on effective down-regulation of MdmX in these tumor cells (Wang et al. 2009), which makes MdmX inhibition an even more attractive strategy for enhancing the therapeutic impact of p53 restoration in tumors.
Ultimately, however, the capacity of p53 restoration therapy to prolong overall survival is limited by the probability that further mutations will arise that again incapacitate the restored p53 pathway—mutations that either inactivate p53 itself or interrupt critical upstream or downstream components of the greater p53 tumor suppressor pathway. One notable example of this is inactivation of p19ARF, the principal effector linking upstream oncogenic signals and p53 activation (Martins et al. 2006; Junttila et al. 2010; Young and Jacks 2010). Our data show that such resistant secondary tumors do indeed arise relatively rapidly following p53 restoration in MdmX-deficient tumors, in our case specifically by deletion of p53, which renders the absence of MdmX moot. However, such wholesale deletion of p53 may be less likely in human tumors, since it would necessarily entail loss of both copies of p53, in which case MdmX inhibition may still prove to be an effective therapeutic strategy in human tumors. Given that p53 pathway-defective cells presumably pre-exist in the Eμ-Myc tumors prior to the initial p53 restoration, and it is to these exapted clones that the mice eventually succumb, how might inactivation of MdmX enhance the therapeutic potential of p53 restoration? One possibility is suggested by our immunoblot analysis, which indicates that a low level of p53 remains in the resistant tumors. Since deletion of the p53 gene is the mechanism of secondary resistance to tamoxifen in such recurring tumors, such low-level p53 presumably indicates the persistence of a minor remnant population of p53-positive cells. Of note, the residual level of p53 in recurring MdmX-deficient tumors is lower than that in their MdmX-competent counterparts, likely indicating that fewer p53-competent tumor cells survive the initial p53 restoration. A better initial “kill” of tumor cells in the absence of MdmX would be consistent with the extended survival of the tamoxifen-treated mice bearing MdmX-deficient tumors. It has also recently emerged that tumors can evade p53-mediated tumor suppression by failing to breach the signaling threshold required to trigger the p53 pathway (Sarkisian et al. 2007; Murphy et al. 2008; Junttila et al. 2010). In such cases, inhibiting MdmX has the potential to lower the p53-activating threshold, thus engaging the p53 pathway in a tumor-selective manner. The recent identification of the first small-molecule inhibitor of MdmX (Reed et al. 2010) may therefore be an important step forward in the genesis of novel therapies that are truly tumor-specific.
Materials and methods
Genetically engineered mice and in vivo studies
p53ERTAMKI (p53KI/KI) mice (Christophorou et al. 2005) were crossed with p53−/− (Jacks et al. 1994), mdmX−/− (Parant et al. 2001), mdm2−/− (Montes de Oca Luna et al. 1995), and Eμ-myc mice (Adams et al. 1985) to generate the appropriate genotypes. All animals were kept under SPF conditions, and all experimental procedures adhered to approved University of California at San Francisco Institutional Animal Care and Use Committee protocols.
To restore p53, mice were injected intraperitoneally with 100 μL of 10 mg/mL tamoxifen (Sigma, T5648) in peanut oil at the indicated frequencies. To restore p53 in vitro, cultured cells were exposed to 100 nM 4-OHT (Sigma, H7904). Nutlin-3a was purchased from Cayman Chemical Company (catalog no. 18585). Where appropriate, mice were injected with a single bolus of 100 μL of 10 mg/mL BrdU (Sigma, B5002) 2 h prior to sacrifice.
Prior to lymphoma transplantation, 4 Gy of γ-radiation was administered to recipient wild-type mice for 4 h using a Mark 1-68 cesium-137 source (1.972 Gy/min) prior to tail vein transplantation of lymphomas. Lymphoma transplantation and in vivo treatment were performed as described (Martins et al. 2006). Lymphoma cell cultures were established and maintained as described (Martins et al. 2006). Statistical analysis of survival was performed with the Kaplan-Meier log-rank test.
TaqMan analysis
Total RNA was isolated with Trizol (Invitrogen, catalog no. 15596026) and DNase-treated (Invitrogen, catalog no. 18065-015) prior to reverse transcription (iScript, Bio-Rad, catalog no. 170-8891). TaqMan analysis was performed as previously described (Christophorou et al. 2005). All data were normalized to gus expression (control gene).
Immunoblotting
Proteins were extracted from organs and cells using standard RIPA buffer. Protein lysates (50–100 μg) were fractionated on SDS polyacrylamide gels and blotted onto PVDF membranes (Immobilon-P, Millipore, IPVH00010). The antibodies used were anti-ER (Santa Cruz Biotechnology, sc-542), anti-p21 (BD Pharmingen, catalog no. 556431), anti-p19ARF (Novus, clone C3), anti-Mdm2 (clone 2A10, Calbiochem OP115, and clone 4B2, Calbiochem OP145), and anti-β-actin (AC-15, Sigma). When indicated, blots were quantified using ImageJ software (NIH).
Immunohistochemistry
All harvested organs were fixed overnight, washed, processed, and embedded in paraffin. BrdU staining and TUNEL were performed using kits (Roche, reference 11299964001, and Millipore, catalog no. S1760, respectively), following the manufacturers' instructions. Anti-Ki-67 antibody was from NeoMarkers (SP6).
Flow cytometry
Leg, hip, and arm bones from individual mice were pooled and crushed to harvest bone marrow cells. Following lysis of erythrocytes with ACK, bone marrow cells were separated from bone residue and dead cells by FICOL gradient centrifugation. Bone marrow cells were then resuspended in Hank's buffered saline (HBSS) containing 2% heat-inactivated FBS (Hyclone), and total viable cells numbers were quantitated using a VI-CELL XR (Beckman-Coulter) imaging hemacytometer. For hematopoietic stem cell analysis, 10 × 106 bone marrow cells were incubated with rat unconjugated lineage antibodies (CD3, CD4, CD5, CD8, B220, Ter119, Mac1, and Gr1), stained with PE-Cy5-conjugated goat anti-rat IgG (Caltag), and blocked with rat IgG (Sigma). Cells were then stained with Sca1-PB, CD150-PE, and CD48-APC (BioLegend). For mature analysis, 1 × 106 cells were stained for B cells (B220-APC, CD19-PE, eBioscience) and myeloid cells (Mac1-PE-Cy7, Gr1-PB, eBioscience). After staining, cells were resuspended in HBSS + 2% FBS and 1 μg/mL propidium iodide for dead cell exclusion and analyzed on an LSRII (Becton Dickinson).
Southern blot analysis
For Southern analysis, 10 μg of genomic DNA per sample was digested with EcoRV, fractionated in a 0.7% agarose gel, transferred to a nylon membrane, and hybridized with 32[P]-labeled probe (Rediprime II RPN 1633, Amersham). The p53 probe was generated by PCR with the following primers: 5′-GGTACCTTATGAGCCACCCGAGG-3′ (forward) and 5′-CGAACCTCAAAGCTGTCCCGTCC-3′ (reverse).
Statistics
An unpaired t-test and Kaplan-Meier log-rank test were used to assess statistical difference. Two-tailed P-values of <0.05 were considered significant.
Acknowledgments
We thank Dr. G. Lozano for the mdm2 and mdmX knockout mice. We also thank all of the members of the Evan laboratory for critical comments and advice, and F. Rostker for help with the animal work. This work was supported by grants NCI CA98018, NCI CA100193, the Ellison Medical Foundation, and the Samuel R. Waxman Cancer Research Foundation (all to G.I.E.). D.G. was supported by NIGMS grant #1 R25 GM56847.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.16722111.
References
- Abbas HA, Maccio DR, Coskun S, Jackson JG, Hazen AL, Sills TM, You MJ, Hirschi KK, Lozano G 2010. Mdm2 is required for survival of hematopoietic stem cells/progenitors via dampening of ROS-induced p53 activity. Cell Stem Cell 7: 606–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams JM, Harris AW, Pinkert CA, Corcoran LM, Alexander WS, Cory S, Palmiter RD, Brinster RL 1985. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature 318: 533–538 [DOI] [PubMed] [Google Scholar]
- Barboza J, Iwakuma T, Terzian T, El-Naggar A, Lozano G 2008. Mdm2 and Mdm4 loss regulates distinct p53 activities. Mol Cancer Res 6: 947–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boesten LS, Zadelaar SM, De Clercq S, Francoz S, van Nieuwkoop A, Biessen EA, Hofmann F, Feil S, Feil R, Jochemsen AG, et al. 2006. Mdm2, but not Mdm4, protects terminally differentiated smooth muscle cells from p53-mediated caspase-3-independent cell death. Cell Death Differ 13: 2089–2098 [DOI] [PubMed] [Google Scholar]
- Canner JA, Sobo M, Ball S, Hutzen B, DeAngelis S, Willis W, Studebaker AW, Ding K, Wang S, Yang D, et al. 2009. MI-63: a novel small-molecule inhibitor targets MDM2 and induces apoptosis in embryonal and alveolar rhabdomyosarcoma cells with wild-type p53. Br J Cancer 101: 774–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christophorou MA, Martin-Zanca D, Soucek L, Lawlor ER, Brown-Swigart L, Verschuren EW, Evan GI 2005. Temporal dissection of p53 function in vitro and in vivo. Nat Genet 37: 718–726 [DOI] [PubMed] [Google Scholar]
- Danovi D, Meulmeester E, Pasini D, Migliorini D, Capra M, Frenk R, de Graaf P, Francoz S, Gasparini P, Gobbi A, et al. 2004. Amplification of Mdmx (or Mdm4) directly contributes to tumor formation by inhibiting p53 tumor suppressor activity. Mol Cell Biol 24: 5835–5843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francoz S, Froment P, Bogaerts S, De Clercq S, Maetens M, Doumont G, Bellefroid E, Marine JC 2006. Mdm4 and Mdm2 cooperate to inhibit p53 activity in proliferating and quiescent cells in vivo. Proc Natl Acad Sci 103: 3232–3237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grier JD, Xiong S, Elizondo-Fraire AC, Parant JM, Lozano G 2006. Tissue-specific differences of p53 inhibition by Mdm2 and Mdm4. Mol Cell Biol 26: 192–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J, Kawai H, Nie L, Kitao H, Wiederschain D, Jochemsen AG, Parant J, Lozano G, Yuan ZM 2002. Mutual dependence of MDM2 and MDMX in their functional inactivation of p53. J Biol Chem 277: 19251–19254 [DOI] [PubMed] [Google Scholar]
- Hedstrom E, Issaeva N, Enge M, Selivanova G 2009. Tumor-specific induction of apoptosis by a p53-reactivating compound. Exp Cell Res 315: 451–461 [DOI] [PubMed] [Google Scholar]
- Issaeva N, Bozko P, Enge M, Protopopova M, Verhoef LG, Masucci M, Pramanik A, Selivanova G 2004. Small molecule RITA binds to p53, blocks p53–HDM-2 interaction and activates p53 function in tumors. Nat Med 10: 1321–1328 [DOI] [PubMed] [Google Scholar]
- Jacks T, Remington L, Williams BO, Schmitt EM, Halachmi S, Bronson RT, Weinberg RA 1994. Tumor spectrum analysis in p53-mutant mice. Curr Biol 4: 1–7 [DOI] [PubMed] [Google Scholar]
- Jackson MW, Lindstrom MS, Berberich SJ 2001. MdmX binding to ARF affects Mdm2 protein stability and p53 transactivation. J Biol Chem 276: 25336–25341 [DOI] [PubMed] [Google Scholar]
- Junttila M, Karnezis A, Garcia D, Madriles F, Kortlevel R, Rostker F, Brown-Swigart L, Pham D, Seo YR, Evan G, et al. 2010. Selective activation of p53-mediated tumour suppression in high-grade tumours. Nature 468: 567–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiel MJ, Yilmaz OH, Iwashita T, Yilmaz OH, Terhorst C, Morrison SJ 2005. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell 121: 1109–1121 [DOI] [PubMed] [Google Scholar]
- Kruse JP, Gu W 2009. Modes of p53 regulation. Cell 137: 609–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurie NA, Donovan SL, Shih CS, Zhang J, Mills N, Fuller C, Teunisse A, Lam S, Ramos Y, Mohan A, et al. 2006. Inactivation of the p53 pathway in retinoblastoma. Nature 444: 61–66 [DOI] [PubMed] [Google Scholar]
- Leonova KI, Shneyder J, Antoch MP, Toshkov IA, Novototskaya LR, Komarov PG, Komarova EA, Gudkov AV 2010. A small molecule inhibitor of p53 stimulates amplification of hematopoietic stem cells but does not promote tumor development in mice. Cell Cycle 9: 1434–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linke K, Mace PD, Smith CA, Vaux DL, Silke J, Day CL 2008. Structure of the MDM2/MDMX RING domain heterodimer reveals dimerization is required for their ubiquitylation in trans. Cell Death Differ 15: 841–848 [DOI] [PubMed] [Google Scholar]
- Liu Y, Elf SE, Miyata Y, Sashida G, Liu Y, Huang G, Di Giandomenico S, Lee JM, Deblasio A, Menendez S, et al. 2009. p53 regulates hematopoietic stem cell quiescence. Cell Stem Cell 4: 37–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Ou L, Clemenson GD Jr, Chao C, Lutske ME, Zambetti GP, Gage FH, Xu Y 2010. Puma is required for p53-induced depletion of adult stem cells. Nat Cell Biol 12: 993–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maetens M, Doumont G, Clercq SD, Francoz S, Froment P, Bellefroid E, Klingmuller U, Lozano G, Marine JC 2007. Distinct roles of Mdm2 and Mdm4 in red cell production. Blood 109: 2630–2633 [DOI] [PubMed] [Google Scholar]
- Mancini F, Di Conza G, Monti O, Macchiarulo A, Pellicciari R, Pontecorvi A, Moretti F 2010. Puzzling over MDM4–p53 network. Int J Biochem Cell Biol 42: 1080–1083 [DOI] [PubMed] [Google Scholar]
- Marine J, Jochemsen A 2005. Mdmx as an essential regulator of p53 activity. Biochem Biophys Res Commun 331: 750–760 [DOI] [PubMed] [Google Scholar]
- Marine JC, Dyer MA, Jochemsen AG 2007. MDMX: from bench to bedside. J Cell Sci 120: 371–378 [DOI] [PubMed] [Google Scholar]
- Martins CP, Brown-Swigart L, Evan GI 2006. Modeling the therapeutic efficacy of p53 restoration in tumors. Cell 127: 1323–1334 [DOI] [PubMed] [Google Scholar]
- Meletis K, Wirta V, Hede SM, Nister M, Lundeberg J, Frisen J 2006. p53 suppresses the self-renewal of adult neural stem cells. Development 133: 363–369 [DOI] [PubMed] [Google Scholar]
- Migliorini D, Lazzerini Denchi E, Danovi D, Jochemsen A, Capillo M, Gobbi A, Helin K, Pelicci PG, Marine JC 2002. Mdm4 (Mdmx) regulates p53-induced growth arrest and neuronal cell death during early embryonic mouse development. Mol Cell Biol 22: 5527–5538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montes de Oca Luna R, Wagner DS, Lozano G 1995. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature 378: 203–206 [DOI] [PubMed] [Google Scholar]
- Morachis JM, Murawsky CM, Emerson BM 2010. Regulation of the p53 transcriptional response by structurally diverse core promoters. Genes Dev 24: 135–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy DJ, Junttila MR, Pouyet L, Karnezis A, Shchors K, Bui DA, Brown-Swigart L, Johnson L, Evan GI 2008. Distinct thresholds govern Myc's biological output in vivo. Cancer Cell 14: 447–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K, Taya Y, Nakagama H 2009. Mdmx enhances p53 ubiquitination by altering the substrate preference of the Mdm2 ubiquitin ligase. FEBS Lett 583: 2710–2714 [DOI] [PubMed] [Google Scholar]
- Parant J, Chavez-Reyes A, Little NA, Yan W, Reinke V, Jochemsen AG, Lozano G 2001. Rescue of embryonic lethality in Mdm4-null mice by loss of Trp53 suggests a nonoverlapping pathway with MDM2 to regulate p53. Nat Genet 29: 92–95 [DOI] [PubMed] [Google Scholar]
- Reed D, Shen Y, Shelat AA, Arnold LA, Ferreira AM, Zhu F, Mills N, Smithson DC, Regni CA, Bashford D, et al. 2010. Identification and characterization of the first small molecule inhibitor of MDMX. J Biol Chem 285: 10786–10796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringshausen I, Oshea C, Finch A, Swigart L, Evan G 2006. Mdm2 is critically and continuously required to suppress lethal p53 activity in vivo. Cancer Cell 10: 501–514 [DOI] [PubMed] [Google Scholar]
- Sarkisian CJ, Keister BA, Stairs DB, Boxer RB, Moody SE, Chodosh LA 2007. Dose-dependent oncogene-induced senescence in vivo and its evasion during mammary tumorigenesis. Nat Cell Biol 9: 493–505 [DOI] [PubMed] [Google Scholar]
- Shangary S, Qin D, McEachern D, Liu M, Miller RS, Qiu S, Nikolovska-Coleska Z, Ding K, Wang G, Chen J, et al. 2008. Temporal activation of p53 by a specific MDM2 inhibitor is selectively toxic to tumors and leads to complete tumor growth inhibition. Proc Natl Acad Sci 105: 3933–3938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp DA, Kratowicz SA, Sank MJ, George DL 1999. Stabilization of the MDM2 oncoprotein by interaction with the structurally related MDMX protein. J Biol Chem 274: 38189–38196 [DOI] [PubMed] [Google Scholar]
- Stad R, Ramos YF, Little N, Grivell S, Attema J, van Der Eb AJ, Jochemsen AG 2000. Hdmx stabilizes Mdm2 and p53. J Biol Chem 275: 28039–28044 [DOI] [PubMed] [Google Scholar]
- Sun SH, Zheng M, Ding K, Wang S, Sun Y 2008. A small molecule that disrupts Mdm2–p53 binding activates p53, induces apoptosis and sensitizes lung cancer cells to chemotherapy. Cancer Biol Ther 7: 845–852 [DOI] [PubMed] [Google Scholar]
- TeKippe M, Harrison DE, Chen J 2003. Expansion of hematopoietic stem cell phenotype and activity in Trp53-null mice. Exp Hematol 31: 521–527 [DOI] [PubMed] [Google Scholar]
- Uldrijan S, Pannekoek WJ, Vousden KH 2007. An essential function of the extreme C-terminus of MDM2 can be provided by MDMX. EMBO J 26: 102–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentin-Vega YA, Okano H, Lozano G 2008. The intestinal epithelium compensates for p53-mediated cell death and guarantees organismal survival. Cell Death Differ 15: 1772–1781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, et al. 2004. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 303: 844–848 [DOI] [PubMed] [Google Scholar]
- Vazquez A, Bond EE, Levine AJ, Bond GL 2008. The genetics of the p53 pathway, apoptosis and cancer therapy. Nat Rev Drug Discov 7: 979–987 [DOI] [PubMed] [Google Scholar]
- Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L, Newman J, Reczek EE, Weissleder R, Jacks T 2007. Restoration of p53 function leads to tumour regression in vivo. Nature 445: 661–665 [DOI] [PubMed] [Google Scholar]
- Vousden KH, Prives C 2009. Blinded by the light: the growing complexity of p53. Cell 137: 413–431 [DOI] [PubMed] [Google Scholar]
- Wade M, Wahl GM 2009. Targeting Mdm2 and Mdmx in cancer therapy: better living through medicinal chemistry? Mol Cancer Res 7: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YV, Leblanc M, Wade M, Jochemsen AG, Wahl GM 2009. Increased radioresistance and accelerated B cell lymphomas in mice with Mdmx mutations that prevent modifications by DNA-damage-activated kinases. Cancer Cell 16: 33–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong S, Van Pelt CS, Elizondo-Fraire AC, Liu G, Lozano G 2006. Synergistic roles of Mdm2 and Mdm4 for p53 inhibition in central nervous system development. Proc Natl Acad Sci 103: 3226–3231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue W, Zender L, Miething C, Dickins RA, Hernando E, Krizhanovsky V, Cordon-Cardo C, Lowe SW 2007. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature 445: 656–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young NP, Jacks T 2010. Tissue-specific p19Arf regulation dictates the response to oncogenic K-ras. Proc Natl Acad Sci 107: 10184–10189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zindy F, Williams RT, Baudino TA, Rehg JE, Skapek SX, Cleveland JL, Roussel MF, Sherr CJ 2003. Arf tumor suppressor promoter monitors latent oncogenic signals in vivo. Proc Natl Acad Sci 100: 15930–15935 [DOI] [PMC free article] [PubMed] [Google Scholar]