Abstract
We show that during Caenorhabditis elegans male spicule development, the specification of a glial versus neuronal cell fate in a canonical neurogenic sublineage is dependent on Wnt signaling. Inactivation of a Wnt signaling pathway mediated by the Wnt receptor LIN-17 transforms the SPD sheath cell into its sister, the SPD neuron. We discovered a new mutant, son-1, that displays this same cell fate transformation. The son-1 mutation enhances the phenotypes of reduction-of-function lin-17 mutants in several developmental processes, including vulva development, somatic gonad development, and male tail patterning. son-1 encodes an HMG1/2-like DNA-binding protein and is localized in all cell nuclei through development as revealed by a GFP reporter construct. Disruption of son-1 function by RNA-mediated interference results in the same spicule defect as caused by overexpression of POP-1, a TCF/LEF class HMG protein known to act downstream of the Wnt signaling pathway. Our results provide in vivo evidence for the functional involvement of an HMG1/2-like protein, SON-1, in Wnt signaling. The sequence nonspecific HMG protein SON-1 and the sequence specific HMG protein POP-1 might both act in the Wnt responding cells to regulate gene transcription in opposite directions.
Keywords: Wnt, HMG-1, TCF/LEF, cell fate specification, C. elegans
The Wnt signaling pathway is one of the major signaling pathways controlling animal development. It controls cell proliferation, cell and tissue polarity, and cell fate specification in many developmental processes of many organisms. In recent years, several components of the Wnt signaling pathway have been identified and the mechanisms by which these factors act have begun to be elucidated (for review, see Cadigan and Nusse 1997; Moon et al. 1997). Wnt family genes, which encode secreted glycoproteins, act as intercellular signals. The Frizzled (Fz) family seven transmembrane proteins are putative Wnt receptors that transduce Wnt signals to downstream targets. Activation of the Wnt signaling pathway leads to accumulation of β-catenin in the cytoplasm. β-Catenin interacts with the TCF/LEF family HMG box-containing DNA-binding protein and the complex accumulates in the nucleus, whereby it regulates gene expression of Wnt pathway targets. Although this simplified model appears to be conserved among diverse organisms, the discoveries of new components and the multiplicity of Wnt pathway components have added complexity to this model.
In Caenorhabditis elegans, there are several Wnt signaling pathways that are involved in several aspects of development such as cell fate specification during embryogenesis, vulval formation, somatic gonad development, and tail patterning. Some components are shared by several or all of these processes. During embryogenesis, Wnt signaling pathway genes, mom-2 (Wnt), mom-5 (Fz), wrm-1 (β-catenin), and pop-1 (TCF/LEF), mediate interactions between the P2 and EMS cells (Lin et al. 1995, 1998; Rocheleau et al. 1997; Thorpe et al. 1997). Signaling from the P2 cell promotes asymmetric division of the EMS cell to produce an anterior MS fate and a posterior E fate. Inactivation of the Wnt signaling pathway results in an E-to-MS fate transformation. The Wnt pathway also plays a role in vulval development. Mutants of several Wnt pathway components show various vulva lineage defects, for example, lin-17 (Fz), bar-1 (β-catenin), and mom-1 and mom-3 mutants (Sternberg and Horvitz 1988; Sawa et al. 1996; Thorpe et al. 1997; Eisenmann et al. 1998). In somatic gonad development, lin-17 has been also shown to control several asymmetric cell divisions (Sternberg and Horvitz 1988). In the tail region, a Wnt signal LIN-44 and its putative receptor LIN-17 control the polarity of several asymmetric cell divisions, including B, T, F, and U (Herman and Horvitz 1994; Chamberlin and Sternberg 1995; Herman et al. 1995; Sawa et al. 1996). lin-44, lin-17, and bar-1 also are involved in the specification of P12 fate in the tail region (Eisenmann et al. 1998; Jiang and Sternberg 1998). A common feature of most of these Wnt signaling pathways in C. elegans appears to be that Wnt signaling is required for posterior cell fates, and inactivation of signaling results in posterior to anterior fates transformation.
The TCF/LEF class HMG box proteins have been shown to regulate Wnt target gene expression on pathway activation (van de Wetering et al. 1997). The C. elegans TCF/LEF class protein is encoded by the pop-1 gene (Lin et al. 1995). pop-1 negatively regulates Wnt signaling, as loss-of-function mutation in pop-1 confers a phenotype opposite that of inactivation of Wnt signaling. Moreover, the expression level of POP-1 protein appears to be up-regulated on inactivation of Wnt signaling (Rocheleau et al. 1997; Lin et al. 1998). Therefore, it was proposed that activation of the Wnt signaling pathway relieves the inhibitory effects of POP-1 on gene transcriptional regulation. Yet it is not clear how Wnt target gene expression is regulated and what other unidentified factors are involved in this process.
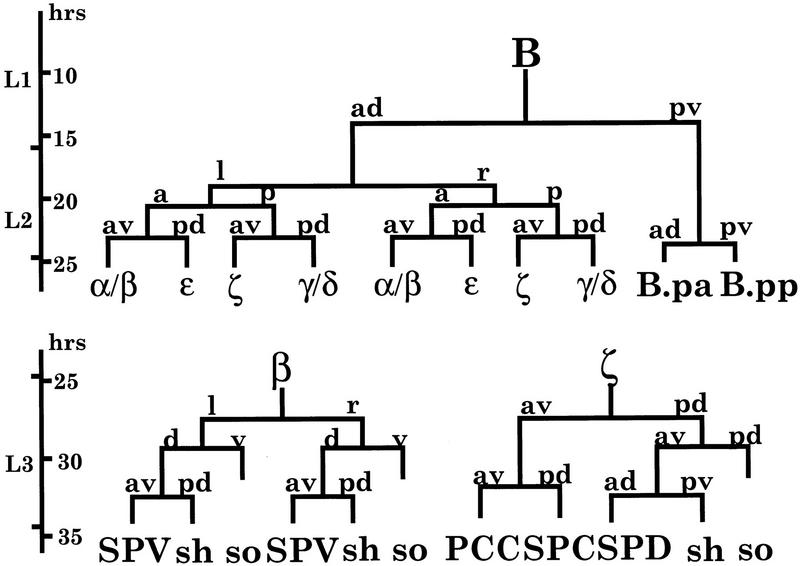
We have been studying cell–cell interactions and cell fate specification during C. elegans male spicule development. Previous studies have shown that cell fate specification in spicule development is mediated by multiple signaling pathways. The C. elegans male spicule is generated by a single male specific B blast cell (Fig. 1). The first division of B cell is asymmetric and is controlled by the Wnt signaling pathway genes, lin-44 and lin-17 (Sternberg and Horvitz 1988; Herman and Horvitz 1994; Chamberlin and Sternberg 1995). At the early third larval stage (L3; B lineage 10-cell stage), the specification of B cell progeny involves a RAS signaling pathway and a C. elegans Notch homolog lin-12 (Chamberlin and Sternberg 1994). During the L3 stage, those cells continue to divide to generate spicule neurons, non-neuronal sheath and socket cells, and connective tissues (Fig. 1). For example, ζ sublineage generates the neuron SPD and its associated sheath (SPDsh) and socket (SPDso) cells.
Figure 1.
The B lineage. The B cell division pattern from 1- to 10-cell stage and the neurogenic sublineages β and ζ are shown (adapted from Sulston et al. 1980; Chamberlin and Sternberg 1993). (sh) Sheath cell; (so) socket cell. The SPV neuron is derived from β sublineage and the SPD neuron from ζ sublineage. The SPD neuron and SPD sheath cell are sister cells derived from ζ.pa.
Using a molecular marker that is specifically expressed in ζ-derived SPD neurons, we studied neuronal versus non-neuronal cell fate specification during male spicule development. Although the cell division pattern of the ζ sublineage resembles the Drosophila sensory organ precursor (SOP) lineage in which the Notch pathway plays a crucial role, we found that lin-12 is not involved in the specification of the SPD neuron fate. Rather, the lin-17-mediated Wnt signaling pathway plays an important role in this neuronal versus non-neuronal fate decision in ζ lineage. Loss-of-function mutants of lin-17 display a SPDsh to neuron fate transformation. To identify new genes involved in this cell fate decision, we conducted a genetic screen and discovered a mutant, son-1(sy549), which shows the same cell fate transformation. Moreover the son-1 (sheath-to-neuron fate transformation) mutation enhances the phenotypes of a lin-17 hypomorph, n698, to lin-17 null-like, with respect to vulval development, somatic gonad development, and male tail patterning. We cloned the son-1 locus and found it encodes an HMG1/2-like DNA-binding protein, which is distinct from the TCF/LEF class HMG proteins. Disruption of son-1 function through RNA-mediated interference (RNAi) leads to defects in several Wnt pathway-mediated developmental processes. Overexpression of POP-1 causes the same spicule defect as loss-of-function son-1. Our results provide in vivo evidence that the HMG1/2-like protein encoded by son-1 plays a specific role in Wnt signaling. SON-1 and POP-1 may both act in the Wnt-responding cells, but in opposite directions, to regulate gene transcription. Taken together, these data provide evidence for further complexity in Wnt responses during development.
Results
lin-17 mutants cause a SPDsh to neuron fate transformation
To study terminal cell fate specification in C. elegans male spicule development, we used a molecular marker, gpa-1::lacZ. In transgenic animals bearing a lacZ reporter gene driven by the gpa-1 promoter, expression is found in male spicules as well as phasmid neurons in the tail region and several cells in the head (J. Mendel and P.W. Sternberg, unpubl.). In wild-type animals, gpa-1::lacZ marker is expressed in one cell per spicule. Cell ablation experiments identified this cell as the spicule neuron SPD (Fig. 2A).
Figure 2.
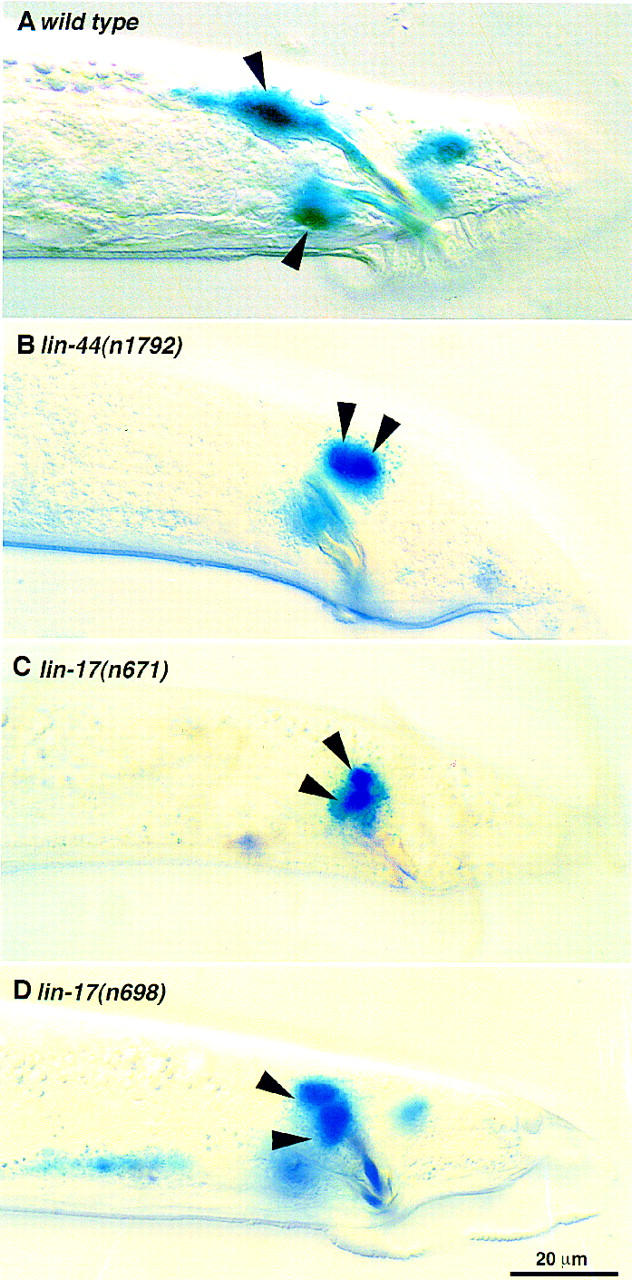
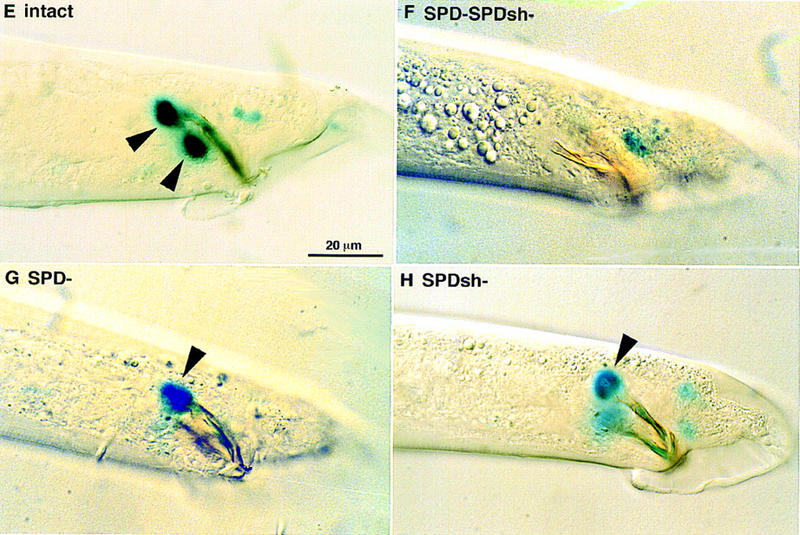
(A) In wild-type males, the gpa-1::lacZ marker is expressed in one cell per spicule. Cell ablation experiment identifies this cell being SPD neuron. Both spicules are shown on this focal plane. (B–D) gpa-1::lacZ is expressed in two cells per spicule in lin-44 and lin-17 mutant males. (B) lin-44(n1792); (C) lin-17(n671); (D) lin-17(n698), early B lineage defect is rare in this weak allele and ∼80% males show a two cells per spicule marker expression pattern. Lateral view, only one spicule is in focus. (E–H) The gpa-1::lacZ marker is expressed in two cells per spicule in son-1(sy549) mutants. Cell ablation experiment indicated that son-1(sy549) confers a SPD sheath to neuron fate transformation. (E) sy549 intact animal, lateral view showing the left spicule. (F) When the precursor of the SPD neuron and the SPD sheath is ablated, no expression is observed in the spicule region. (G) When the SPD neuron is ablated, the SPD marker is expressed in one cell per spicule. (H) When the SPD sheath is ablated, the SPD marker is expressed in one cell per spicule. Scale bar, 20 μm.
Using this SPD neuron specific marker, we examined spicule terminal cell fates in several classes of known mutants. The Notch pathway controls neuronal versus non-neuronal cell fate specification in both the central and peripheral nervous systems during Drosophila embryonic development (Hartenstein and Posakony 1990; Parks and Muskavitch 1993). In the development of Drosophila SOP lineage, the Notch pathway mediates interactions between two daughter cells to ensure one adopt neuronal fate, the other one non-neuronal fate. As the cell division pattern of the spicule neurogenic lineage resembles that of Drosophila SOP lineage (Fig. 1), we examined the possible involvement of the C. elegans Notch homolog lin-12 in the specification of SPD neuron versus SPDsh fate. Neither gain-of-function nor loss-of-function lin-12 mutants displayed abnormal marker expression pattern (n = 50). Activation of the lin-12 pathway at a critical developmental time by overexpressing the cytoplasmic ankyrin repeat region of GLP-1, which has been shown to act as an activated lin-12 or glp-1 in many developmental processes (Roehl and Kimble 1993), did not affect marker expression (n = 50).
The Wnt pathway genes, lin-44 and lin-17, are involved in the first asymmetric division of the B cell. Mutant males bearing strong lin-44 and lin-17 alleles have early B lineage defects, resulting in severely crumpled spicules. From a few escapers in which the first division of B cell is not disrupted, we noticed extra expression of the gpa-1::lacZ marker in the spicule region (Fig. 2B,C). Using a weak lin-17 allele, n698, wherein the first B cell division is usually not disrupted, we found that 80% of the animals show two cells per spicule expressing the SPD marker (Fig. 2D; Table 1).
Table 1.
Both son-1(sy549) and lin-17(n698) confer a SPD sheath to neuron fate transformation
|
Cells ablated
|
lin-17(n698)
|
son-1(sy549)
|
||
|---|---|---|---|---|
|
no.a
|
marker expressionb
|
no.a
|
marker expressionb
|
|
| Intact | 15/17 | 2 | 39/52 | 2 |
| β | 5/6 | 2 | 4/6 | 2 |
| ζ | 15/16 | 0 | 12/15 | 0 |
| ζ a | 13/15 | 2 | 5/5 | 2 |
| ζ p | 5/5 | 0 | 6/9 | 0 |
| ζ pp | 4/7 | 2 | 1/2 | 2 |
| ζ pa | ||||
| (SPD+SPDsh) | 11/11 | 0 | 12/14 | 0 |
| ζ paa (SPD) | 10/13 | 1 | 13/16 | 1 |
| ζ pap (SPDsh) | 13/21 | 1 | 6/7 | 1 |
Cells were ablated at L3 stage and animals recovered to allow reaching adulthood. Adult males were then examined for gpa-1::lacZ marker expression. There are two ζ cells per male; both were ablated.
Numbers of animals operated on and scored that showed the corresponding phenotypes. The rest of the animals showed 0, 1, or 2 cells per spicule expressing the marker due to incomplete penetrance or ablation error or damage.
Numbers indicate number of cells per spicule expressing the gpa-1::lacZ marker.
To determine which cell is responsible for the ectopic expression, we ablated specific cells in the B lineage and examined marker expression in operated animals. Because the ε lineage develops abnormally in lin-17 mutants (Chamberlin and Sternberg 1995; this paper), we first ablated the ε cells at the 10-cell stage and examined the marker expression pattern in operated animals. Ablation of ε does not eliminate the extra gpa-1::lacZ marker-expressing cell (n = 9). Further ablation experiments indicate that the extra expression results from SPDsh, the sister cell of the SPD neuron (Table 1). Ablation of the precursor cell of SPD and SPDsh eliminated marker expression in the spicule region; ablation of either cell resulted in one cell per spicule expressing the marker. We also used other cell type-specific markers to examine the terminal cell fates in lin-17(n698) mutants. osm-6::GFP is normally expressed in SPD and SPV neurons of wild-type spicules and in male sensory ray neurons (Collet et al. 1998). In the lin-17(n698) mutant, osm-6::GFP is expressed in an extra cell associated with the spicules (four of six animals examined), consistent with the extra expression observed with the gpa-1::lacZ marker. Expression of a socket cell marker, egl-17::GFP (Burdine et al. 1998; L.I. Jiang and P.W. Sternberg, in prep.), was not affected by the lin-17(n698) mutation (n = 10), suggesting that the socket cells are correctly specified in lin-17(n698) mutants. Therefore, lin-17 mutants confer a sheath to neuron fate transformation.
son-1 mutant confers the same sheath to neuron fate transformation
To discover additional loci involved in the neurogenic sublineage cell fate specification, we performed an F1 clonal screen looking for mutations that alter gpa-1::lacZ expression specifically in the spicule region. From the screen, we recovered one mutant, sy549, in which two cells per spicule express the gpa-1::lacZ marker (Fig. 2E). Cell ablation experiments revealed that the extra cell that expresses the gpa-1::lacZ marker is the SPDsh cell, as in lin-17 mutants (Fig. 2E–G; Table 1).
sy549 mutant males have wild-type spicules and wild-type B-cell division pattern. Expression patterns of other cell-type-specific markers examined are consistent with the conclusion that sy549 confers a SPDsh to SPD neuron fate transformation (osm-6::GFP: 7 of 11 animals showed extra marker expression in the spicule region; egl-17::GFP: 10 of 10 wild-type expression). Thus, we named this mutant son-1.
son-1(sy549) displays several phenotypes resembling lin-17 mutants
In addition to the male spicule phenotype, son-1(sy549) mutants also display a protruding vulva phenotype with high penetrance. Hermaphrodites retain eggs in uterus, some animals have a disorganized gonad and explode at the vulva, and the brood size is low [∼50 in a syIs20 him-5(e1490) background]. Abnormal vulval lineages and vulval morphology are observed in ∼10% of animals (n > 50). The abnormal vulval lineage occurs in 2° cells; it can occur at either P5.p or P7.p. In most cases, the correct number of cells were generated, but the vulval morphology was abnormal, and an extra vulva invagination was observed in several animals. This phenotype is similar to the vulval defect observed in lin-17 mutants (Ferguson and Horvitz 1985; Ferguson et al. 1987). Occasionally, son-1(sy549) hermaphrodites have gonads with a single arm instead of two, as observed in lin-17 mutants (Sternberg and Horvitz 1988). In addition, like lin-17 mutant males, son-1(sy549) mutant males have ray defects and cannot mate.
son-1(sy549) enhances lin-17(n698) phenotypes to lin-17 null like
To examine the interactions between son-1 and lin-17, we constructed strains defective in both son-1 and lin-17. We first examined the gpa-1::lacZ expression pattern in the double-mutant animals. The SPD neuron marker is expressed in no more than two cells per spicule in the double mutants, confirming the conclusion from the ablation experiment that both confer the same sheath-to-neuron fate transformation.
lin-17 mutants are defective in vulva formation, somatic gonad development, and phasmid and spicule development in the tail. n698 is a reduction-of-function allele of lin-17 (Sawa et al. 1996) and displays weaker phenotypes than a null allele, n671. About 50% of n698 animals have abnormal vulval invaginations, but the extra posterior invagination phenotype is rarely seen (Table 2). In contrast, all n698; sy549 double-mutant animals examined have abnormal vulval invaginations, and 58% of animals show an extra posterior invagination, which is typically seen only in strong lin-17 alleles (Table 2). Approximately 73% of the n698 animals have a T-lineage defect, such that their phasmids fail to take up DiO (Sawa et al. 1996). son-1(sy549) mutants are wild type for DiO staining, whereas 98% of n698; sy549 double-mutant animals fail to uptake DiO (Table 2). Double-mutant hermaphrodites often have a single gonad arm instead of the normal two. Double-mutant males show ectopic meiosis, indicative of no distal tip cells (Sternberg and Horvitz 1988). Double-mutant males also show more severely crumpled spicules and have no rays. Taken together, lin-17(n698); son-1(sy549) double-mutant animals behave like strong lin-17 mutant animals with respect to vulva development, somatic gonad development, and male tail patterning. However, neither lin-17(n698) mutants nor the double mutants exhibit a P12 fate specification defect (Table 2).
Table 2.
son-1(sy549) enhances lin-17(n698) phenotypes to lin-17 null-like mutations
|
Genotype
|
Phenotypes
|
||||
|---|---|---|---|---|---|
| lin-17 | son-1 | bivulvaa | hermaphrodite gonadb | P12.p → P11.pc | abnormal phasmidsd |
| + | sy549 | 2% (n = 108) | 1% (n = 108) | 0 (n = 100) | 4% (n = 53) |
| n698 | + | 2% (n = 46) | 8% (n = 46) | 0 (n = 100) | 73% (n = 346) |
| n698 | sy549 | 58% (n = 98) | 28% (n = 101) | 0 (n = 100) | 98% (n = 57) |
| n671 | + | 64% (n = 66) | 23% (n = 66) | 29%e (n = 41) | 95%f (n = 400) |
| n671 | sy549 | 64% (n = 76) | 20% (n = 69) | 22% (n = 92) | N.D. |
Bivulva phenotype is defined as two vulval invaginations at mid- to late L4 stage.
Hermaphrodite gonad defect is defined as abnormal gonad morphology, single gonadal arm, and ectopic meiosis at L4 stage.
P11/P12 phenotype is scored by the nuclear morphologies and positions of P11.p and P12.pa cells at L4 stage.
L3 and L4 stage worms were tested by DiO filling of amphids and phasmids, as described by Herman and Horvitz (1994). Animals that showed positive for amphid filling and negative for phasmid filling were scored as phasmid defective. (N.D.) Not determined.
Data from Jiang and Sternberg (1998).
Data from Sawa et al. (1996).
Next, we constructed a double-mutant strain carrying son-1(sy549) and a lin-17 null allele, n671. We examined two relatively low penetrance phenotypes, the ectopic posterior vulva invagination phenotype and P12 fate specification. We did not observe any phenotypic enhancement in the double-mutant animals (Table 2). Therefore, son-1 and lin-17 likely act in the same pathway.
son-1 encodes an HMG1/2-like protein
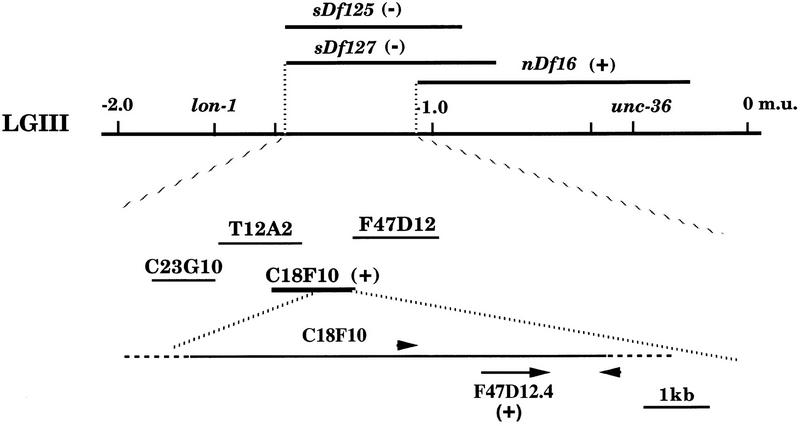
son-1(sy549) is recessive, and on the basis of the spicule phenotype, we mapped it to chromosome III, between lon-1 and unc-36 (Fig. 3). Deficiency mapping put son-1(sy549) between the left end point of sDf127 and the left end point of nDf16. Hemizygous males (sy549/Df) show a similar defect in gpa-1::lacZ marker expression as sy549 homozygotes. Hemizygous hermaphrodites appear to be sterile, possibly as a result of disorganized somatic gonad and bursting at the vulva. Therefore, sy549 is likely to be a reduction-of-function mutant.
Figure 3.
Mapping and cloning of son-1. The son-1 mutation is mapped on the basis of the SPD marker expression pattern in the spicules. It is mapped to linkage group III (LGIII). The son-1 locus is deleted by deficiencies sDf125 and sDf127, but not by nDf16. The cosmid C18F10 rescued son-1 mutation, the adjacent cosmid F47D12 failed to rescue. Primers 1-kb upstream and 1-kb downstream of the F47D12.4 gene amplified a 3-kb fragment from C18F10 but not from F47D12. This 3-kb fragment rescued son-1 phenotypes. (+) Rescuing activity; (−) nonrescue.
To rescue the son-1(sy549) mutation, we tested all of the cosmids (20) that map in this interval of chromosome III. We obtained germ-line transformation rescue of son-1(sy549) by injecting a single cosmid C18F10 (Fig. 3). A 3-kb PCR fragment amplified from this cosmid was able to fully rescue all of the mutant phenotypes (Fig. 3). A single gene, F47D12.4 is, predicted within this 3-kb genomic region. We performed RT–PCR with primers specific to the predicted 5′ start and 3′ stop codon regions of this gene and obtained cDNA clones of ∼600 bp. Sequences of five clones revealed that three clones are the same as predicted by the GeneFinder program; the other two clones appear to be generated through alternative splicing at the end of the first exon, which adds two additional amino acids without changing the rest of the protein coding.
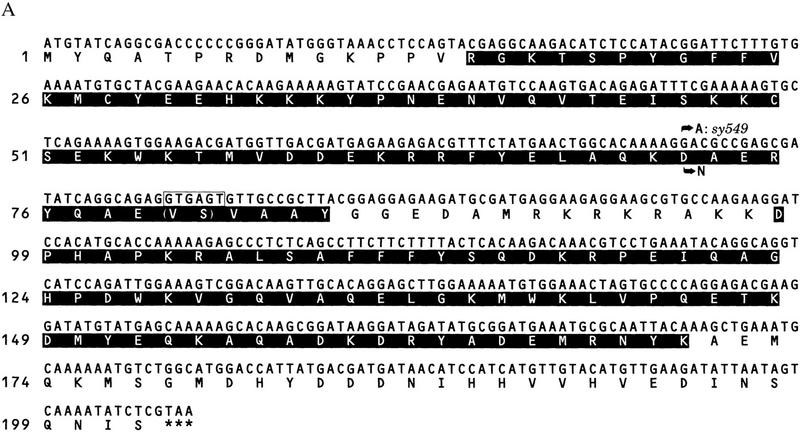
This gene encodes an HMG1-like protein (Fig. 4A). It contains two HMG boxes, box A and box B, and is most homologous to the HMG1/2-like proteins of Drosophila, mouse, and human, exhibiting ∼55% identity within the HMG boxes. It also displays ∼25% identity to TCF/POP/SOX family proteins within the HMG box (Fig. 4B).
Figure 4.
Sequence of the son-1 gene. (A) son-1 cDNA sequence and deduced amino acid sequence. The boxed sequence is derived from alternative splicing at the end of exon 1, which leads to additional two amino acids as indicated in parentheses. This alternative splicing does not change the overall protein structure. (Shaded regions) HMG boxes; (***) stop codon; (arrows) molecular lesion of the sy549 allele. (B) Comparison of the HMG box A and box B sequences between SON-1 and HMG1-like proteins of Drosophila melanogaster, Mus musculus, and Homo sapiens. Consensus resides between SON-1 and other HMG1-like proteins are shaded. (*) Unusual valine residue found in the HMG box A of SON-1 protein.
We sequenced the sy549 allele and identified a missense mutation of g → a at nucleotide position 228, which changes amino acid 72 from aspartic acid (D) to asparagine (N) (Fig. 4A). This amino acid change occurs at a conserved residue in the middle of the third helix of HMG box A.
son-1::GFP is expressed in all somatic cell nuclei through development
We examined the expression pattern of son-1 by generating both transcriptional and translational fusions of son-1 to a GFP reporter gene. The constructs contain either 1- or 3- to 4-kb upstream genomic sequence of son-1. We found that son-1 is expressed in every somatic cell through development (Fig. 5). The transcriptional fusion construct contains only part of HMG box A and does not show nuclear localization of GFP. The translational fusion construct is predicted to encode a fusion protein of full-length SON-1, lacking the last 6 amino acids, followed by GFP. This construct is able to rescue son-1(sy549) phenotypes and displays nuclear localized GFP expression. This expression pattern is consistent with the predicted role for HMG-box containing proteins as DNA-binding proteins.
Figure 5.
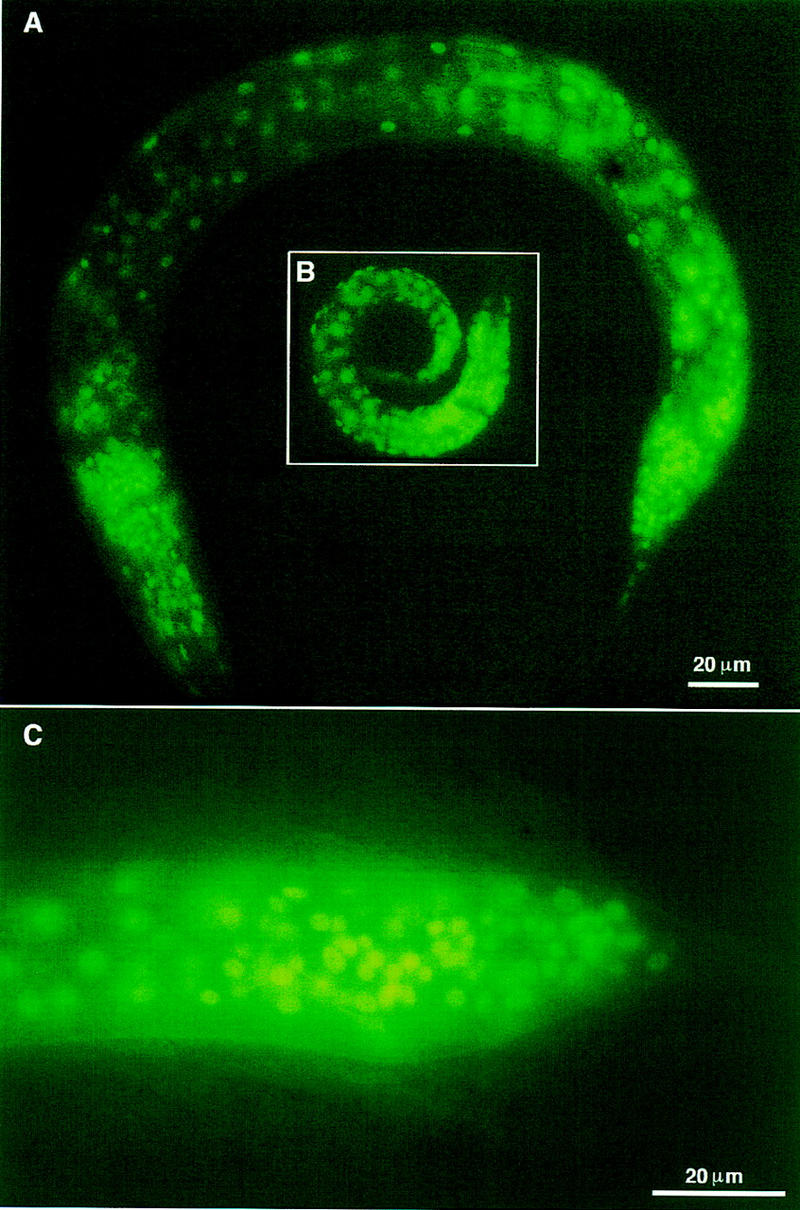
A GFP reporter construct reveals that son-1 is expressed in all somatic cell nuclei through development. (A) An L3 male; (B) an L1 hermaphrodite; (C) male tail at L4 molt. Scale bar, 20 μm.
son-1(sy549) mutants are not generally defective in gene transcription
The HMG1/2-like proteins have been traditionally considered as architectural components of chromatin that have general roles in regulating chromosomal functions (Bustin and Reeves 1996). Although several recent studies have started to suggest specific roles for the HMG1/2-like proteins in regulating gene transcription (Lehming et al. 1994; Zwilling et al. 1995; Zappavigna et al. 1996; Boonyaratanakornkit et al. 1998; Jayaraman et al. 1998), the in vivo functions of this class of HMG proteins during development have not been studied. In spite of the broad expression pattern revealed by the son-1::GFP reporter gene, we do not think that son-1(sy549) generally disrupts transcription. First, son-1(sy549) mutants display specific phenotypes in several developmental processes that resemble lin-17 mutants, and son-1(sy549) genetically interacts with lin-17.
Second, son-1(sy549) mutant males display ectopic transgene expression specifically in the spicule region. If it were a general regulator of transcription, this phenotype would suggest that loss-of-function son-1 might also up-regulate expression of the gpa-1::lacZ reporter elsewhere. Yet, we did not observe ectopic or stronger expression of the gpa-1::lacZ marker in the phasmid neurons or in the head region. Furthermore, other transgenic lines examined did not show any up-regulation or down-regulation of reporter gene expression by son-1(sy549).
Third, son-1(sy549) enhances the phenotypes of a weak allele of lin-17 to null-like phenotypes. This suggests that loss-of-function son-1 may down-regulate gene expression. To further test this possibility, we used a sensitized genetic assay that is able to detect a twofold difference in gene expression (Meneely and Wood 1987). We used a dosage-sensitive allele of the let-2 gene that is involved in developmental processes unrelated to those mediated by son-1. The let-2 gene encodes a type IV α2 collagen, and its different alleles cause animal lethality with varying severity (Sibley et al. 1994). The allele mn114 causes a strong dosage-dependent phenotype: Animals homozygous for mn114 can survive to adulthood but are sterile, whereas hemizygous animals die at larval stages (Meneely and Wood 1987). If son-1 is a general regulator of transcription, it should enhance the phenotype of let-2(mn114). Animals homozygous for both let-2(mn114) and son-1(sy549) can still survive to adulthood as do let-2(mn114) homozygotes. Taken together, our results provide the first evidence that the HMG1/2-like proteins have specific developmental functions in vivo.
Disruption of son-1 function by RNAi results in defects in several lin-17-mediated developmental processes
To obtain possible null phenotypes of son-1, we disrupted son-1 function by RNA-mediated interference. dsRNA were synthesized with a full-length cDNA clone as template and were injected into adult hermaphrodites. We found an embryonic lethal phenotype as well as postembryonic abnormalities. The embryonic lethality is ∼11% penetrant. The reason for the embryonic lethality is not apparent to us. The dead eggs did not show lack of endoderm fates as seen in mom mutants (Rocheleau et al. 1997). Viable hermaphrodites and males grow to adulthood and show a sterile phenotype at ∼30% penetrance. Hermaphrodites often have a single gonad arm, resembling the lin-17 phenotype (Fig. 6A; Sternberg and Horvitz 1988). In males, ectopic meiosis is seen in germ line, indicative of loss of distal tip cells (Fig. 6B; Sternberg and Horvitz 1988). This defect is also seen in lin-17 mutant males a result of Z1 and Z4 lineage defect in which more linker cells are generated at the expense of distal tip cells (Sternberg and Horvitz 1988). Disruption of son-1 function by RNAi also resulted in a vulval lineage defect and bursting at the vulva region at low penetrance. Abnormal gpa-1::lacZ marker expression at the spicule region mimicking sy549 phenotype was also observed (Fig. 6C). Therefore, the phenotypes produced by son-1 RNAi are consistent with those seen in son-1(sy549) mutants. The sy549 allele likely reduces but does not fully eliminate SON-1 functions, consistent with the deficiency result. The strong resemblance of son-1 and lin-17 mutant phenotypes further suggests that son-1 is specifically involved in the lin-17-mediated Wnt signaling.
Figure 6.
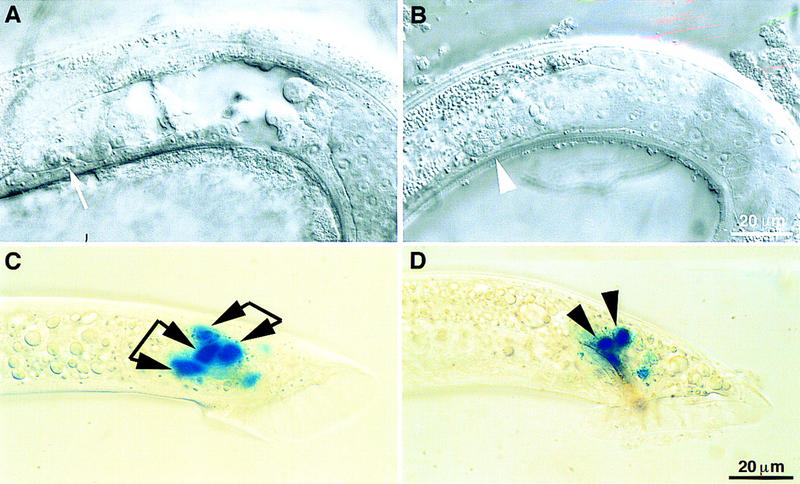
Phenotypes of disruption of son-1 function by RNAi and overexpression of POP-1. (A–C) Phenotypes observed from son-1 RNAi experiment. (A) Hermaphrodite gonad showing abnormally short gonad arm; (arrow) meiosis at the distal end of the gonad arm. (B) Male gonad (white arrowhead) ectopic germ-line meiosis. (C) gpa-1::lacZ is expressed in two cells per spicule. All four blue cells can be seen on this focal plane, two from the left spicule and two from the right one. Cells from the same side are indicated by linkage of two arrowheads. (D) When POP-1 is overexpressed in wild-type males, gpa-1::lacZ expression is observed in two cells per spicule (black arrowheads). Lateral view, showing only the left spicule. Scale bar, 20 μm.
Overexpression of POP-1 results in the same spicule defect as son-1 RNAi
The TCF/LEF class of HMG box-containing proteins, different from the HMG1/2 class of proteins, has been implicated as a component of Wnt signaling pathway to regulate Wnt target gene expression (van de Wetering et al. 1997). The pop-1 gene encodes a C. elegans homolog of the TCF/LEF class HMG protein and has been implicated in Wnt signaling during embryogenesis (Lin et al. 1995, 1998; Rocheleau et al. 1997; Thorpe et al. 1997). As pop-1 is thought to negatively regulate Wnt signaling, we wondered if overexpression of POP-1 would result in similar defects such as loss-of-function of son-1. When POP-1 is overexpressed at early to mid L3 stage in wild-type males, they show two cells per spicule expressing the SPD marker, resembling the son-1(sy549) mutant phenotype (Fig. 6D). Thus, the two HMG box-containing proteins, POP-1 and SON-1, appear to have opposite roles in regulating Wnt signaling. POP-1 negatively regulates Wnt signaling, whereas SON-1 positively regulates Wnt signaling.
Although pop-1(zu189) has a maternal effect embryonic lethal phenotype, this mutant does not display postembryonic defects. Homozygous animals produced by a heterozygous mother are viable and develop normally. Using the spicule SPD neuron-specific marker, we found that pop-1 mutant males have a wild-type marker expression pattern in the spicules. Mutant animals defective in both pop-1 and son-1 genes display additive phenotypes. They still exhibit maternal effect embryonic lethality, whereas all of the postembryonic phenotypes resemble son-1(sy549) mutants.
son-1 does not act upstream of Wnt
To determine how son-1 is involved in Wnt signaling, we first tested whether son-1 acts in the Wnt signaling cells or in the responding cells. The Wnt signal LIN-44 is expressed in the tail region and plays a role in tail patterning (Herman and Horvitz 1994; Herman et al. 1995). We think that LIN-44 is the signal that acts through the LIN-17 receptor during male spicule development. First, during the first division of B lineage, LIN-44 acts through LIN-17 to mediate the asymmetric cell division (Chamberlin and Sternberg 1995). Second, lin-44 mutants exhibit a similar SPD neuron marker expression pattern in the spicule region as lin-17 and son-1 mutants. We reasoned that if son-1 acts in the signaling cells to regulate lin-44 expression, we should be able to rescue son-1 mutant phenotypes by overexpression of the Wnt signal LIN-44. To test this hypothesis, we used a construct containing full-length lin-44 cDNA driven by a heat-shock promoter. This construct has been shown to rescue lin-44 phenotypes (Herman et al. 1995). When LIN-44 overexpression is induced during the L2/L3 stage in son-1(sy549) mutant background, it does not affect the spicule phenotype of son-1(sy549) mutants. This result indicates that son-1 does not act upstream of the Wnt signal to regulate its expression. Because son-1 is expressed in cell nuclei, it is likely that son-1 acts in the Wnt-responding cells to regulate Wnt pathway gene expression. However, we cannot rule out the possibility that son-1 is involved in the post-translational regulation of the Wnt signal.
We then tested whether overexpression of SON-1 is able to suppress lin-17 mutant phenotypes. We engineered a hs-son-1 construct that contains a genomic clone encoding full-length son-1 driven by a heat-shock promoter. In a son-1(sy549) background, overexpression of SON-1 by a single pulse of heat shock at the L3 stage within 4 hr prior to the cell division is able to rescue the spicule sheath to neuron fate transformation phenotype of the mutant. In a lin-17(n698) mutant background, we induced SON-1 expression during embryogenesis to the L1/L2 stage and at the L2/L3 stage and examined the phasmid and spicule phenotypes, respectively. We did not observe any suppression of lin-17 phenotypes by SON-1 overexpression. son-1 might act at the level of Wnt receptor to regulate lin-17 function. Another possibility is that son-1 activity is not rate limiting for Wnt pathway function. Consistent with this hypothesis, simply overexpressing SON-1 in a wild-type background by heat shock or injection of the wild-type gene at high dosage does not result in any gain-of-function phenotype.
Structure/function analysis of SON-1 protein
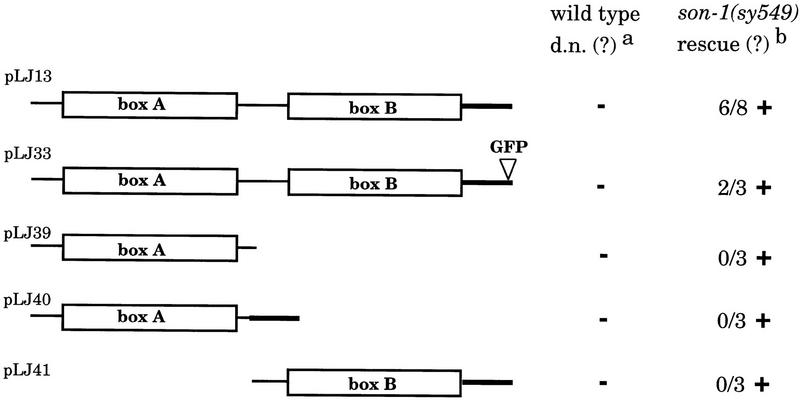
Both HMG boxes A and B of the HMG1/2-like proteins have been shown to be able to bind bent DNA structures and to interact with certain transcription factors independently in vitro (Bianchi et al. 1992; Zappavigna et al. 1996). To examine the in vivo functions of SON-1 HMG boxes A and B, we engineered deletion constructs lacking either individual box and generated transgenic animals bearing these constructs (Fig. 7). In a wild-type background, we did not observe any dominant-negative or gain-of-function effect caused by these constructs. When these transgenes are crossed into a son-1(sy549) mutant background, only the construct containing both HMG boxes is able to rescue the mutant phenotypes. None of the constructs containing a single HMG box were able to rescue the mutant phenotypes (Fig. 7). This result suggests that both HMG boxes A and B are required for the function of the SON-1 protein in vivo.
Figure 7.
Structure/function analysis of HMG boxes. HMG boxes are indicated as open box and labeled. Bold lines indicate the acidic tail. All constructs contain additional 1-kb upstream and 1-kb downstream genomic sequences. (a) Constructs were tested in wild-type background for any dominant-negative or gain-of-function effect. (b) Constructs were tested in son-1(sy549) mutant background for any rescuing activity. The number of transgenic lines tested in which >75% of the animals showed wild-type SPD marker expression is indicated; these were considered positive for rescuing activity. (+) Positive for tested activity; (−) negative for tested activity.
Discussion
Wnt signaling in spicule development
During C. elegans male spicule development, the Wnt pathway genes lin-44 and lin-17 control the first cell division of the spicule lineage, namely, the B lineage (Sternberg and Horvitz 1988; Herman and Horvitz 1994; Chamberlin and Sternberg 1995; Sawa et al. 1996). The first division of the B cell, B.a versus B.p, is asymmetric. Inactivation of the Wnt pathway results in reversed polarity or posterior B.p to anterior B.a fate transformation. Additional functions for lin-17 have also been implicated in several asymmetric cell divisions in both B.a and B.p lineages after the 10-cell stage (Chamberlin and Sternberg 1995). Using a terminal cell fate marker, we discovered that the Wnt pathway genes lin-17 and likely lin-44 are involved in the fate specification of SPD neuron (ζ.xxa) versus SPDsh (ζ.xxp) during the last round cell division of the B lineage. Loss-of-function mutation in lin-17 results in a posterior sheath to anterior neuron fate transformation. Therefore, the lin-17-mediated Wnt signaling pathway is used at the first and the last cell division of B cell development to mediate cell fate specification. Like most of the other Wnt-mediated developmental processes, the Wnt pathway acts to promote the posterior fate in the C. elegans male spicule lineage. We envision that the source of Wnt signal LIN-44, the hyp8-11 cells at the tip of the tail (Herman et al. 1995), sets up the anterior and posterior polarity for general tail patterning.
In C. elegans, several Wnt pathways mediate several distinct developmental processes. Some components are unique, others are shared in several processes. We examined the possible involvement of other Wnt pathway components in this last cell division of male spicule development. In lin-44 mutant males, the abnormal SPD neuron marker expression in the spicule region is observed, resembling that of the lin-17 mutants. However, all of the lin-44 alleles available are null or strong alleles, wherein the early B lineage defect is highly penetrant, making it difficult to rigorously examine its effect on the later stage cell fate specification. mom-1 and mom-3 homozygous males display a severely crumpled spicules phenotype at high penetrance, indicating an early B lineage defect. SPD neuron marker expression appeared to be normal in bar-1(sy324) and bar-1(mu349) mutant males. pop-1 mutant males have wild-type spicules and normal marker expression pattern. However, overexpression of POP-1 at the L3 stage results in extra cell expressing gpa-1::lacZ in the spicules, similar to loss-of-function son-1 mutants.
During the development of C. elegans male spicules, the neurogenic sublineages β and ζ resemble the Drosophila SOP lineage in that a progenitor cell divides twice and generates neuron, sheath, and socket cells. The mechanism used for ζ lineage terminal cell fate specification appears to be different from that for the SOP lineage. In the development of Drosophila SOP lineage, sister cell interaction mediated by the Notch/Delta pathway plays an important role for the specification of neuronal versus non-neuronal cell fate (for review, see Greenwald 1998). However, we did not observe any effect of lin-12 on the specification of SPD versus SPDsh fate in C. elegans male spicule development. Rather, the lin-17-mediated Wnt signaling pathway controls this fate decision. Thus the two different signaling pathways, Notch and Wnt, are used in different organisms to achieve a similar cell fate patterning in a conserved neurogenic lineage.
However, the Wnt pathway is not the sole player for spicule development. The β lineage closely resembles ζ lineage in generating the neuron and non-neuronal structural cells. Yet the Wnt pathway does not seem to be involved to the neuron/sheath fate specification derived from β lineage as revealed by other molecular markers (data not shown). In contrast to the ζ lineage, cell–cell interactions within the β lineage do play an important role during the development of the β lineage, because β lineage can be easily perturbed by cell ablations or mutants of the lin-3/let-23 EGF/EGFR pathway components (Chamberlin and Sternberg 1993, 1994). Therefore, multiple signaling pathways must be utilized during male spicule development.
An HMG1/2-like protein is involved in Wnt signaling
We identified a new gene, son-1, involved in the SPD neuron versus SPDsh fate specification in a similar fashion as lin-17 during C. elegans male spicule development. son-1 encodes an HMG1/2-like protein. A GFP reporter gene revealed that son-1 is broadly expressed through development and its expression is localized in cell nuclei, consistent with its being an HMG-box containing DNA-binding protein.
Although the HMG1/2 class proteins are traditionally considered as architectural components of chromatin, the specific phenotypes of son-1(sy549) suggest that this gene may be specifically involved in Wnt signaling. First, son-1(sy549) causes a similar SPDsh to neuron fate transformation as does a lin-17 hypomorph. The vulva and gonad defects observed in son-1(sy549) mutants, although at low penetrance, do resemble what are seen in lin-17 mutants. Second, we observed specific postembryonic phenotypes that resemble lin-17 mutant phenotypes from son-1 RNAi experiments. Third, son-1(sy549) enhances lin-17(n698) phenotypes to lin-17 null-like but does not enhance phenotypes of other unrelated mutants such as let-2(mn114). Fourth, overexpression of POP-1 gave similar spicule phenotype as disruption of son-1 function. Taken together, son-1 is involved in several Wnt-mediated developmental processes, including vulva formation, somatic gonad development, and male spicule development. Our results provide the first evidence for the specific functions of the HMG1/2 class proteins in vivo.
The HMG1/2-like protein encoded by son-1 has several interesting features that are different from what is known about the HMG1/2 class proteins. The HMG1/2 class of proteins is classified as sequence nonspecific HMG proteins in comparison to the sequence-specific HMG proteins, such as TCF/LEF (Bianchi et al. 1992). The canonical HMG box found in sequence nonspecific HMG proteins and the generalized HMG box found in sequence-specific HMG proteins share substantial sequence homology and identical protein structure. The sequence specificity is thought to reside in the first 12 amino-terminal residues and the last 25 carboxy-terminal residues of the HMG box. In particular, the amino acids at positions 7 and 12 have been suggested to be crucial for sequence-specific binding (Read et al. 1994). The residues at positions 7 and 12 are valine (V) or isoleucine (I) and asparagine (N) in all sequence-specific HMG boxes. Instead, all presently known sequence nonspecific HMG boxes have proline (P) and serine (S) at these two positions. The box B of SON-1 protein appears to be consistent with this finding (positions 102 and 107 of the protein, Fig. 4B). Interestingly, the box A of SON-1 protein has a valine (V) residue at position 7 (position 14 of the protein, Fig. 4B). This raises the possibility that the C. elegans HMG1/2-like protein SON-1 has more specific functions than the HMG1/2-like proteins in other organisms. Furthermore, our data suggest that both HMG boxes are required in vivo for SON-1 functions, in contrast to the in vitro observations that each individual HMG box is able to recognize bent DNA structure and to interact with certain transcription factors in a similar fashion as is the whole protein (Bianchi et al. 1992; Zappavigna et al. 1996). These features of SON-1 may be indicative of its specific functions.
Two classes of HMG proteins, SON-1 and POP-1, play opposite roles in Wnt signaling
The TCF/LEF class sequence-specific HMG proteins have been implicated in Wnt signaling. They can either positively or negatively regulate Wnt target gene expression depending on their transcription cofactors (van de Wetering et al. 1997; Waltzer and Bienz 1998). The C. elegans TCF/LEF homolog, pop-1, appears to negatively regulate Wnt signaling as loss-of-function in pop-1 causes an opposite phenotype as inactivation of the Wnt signaling during C. elegans embryogenesis. In the specification of SPD neuron versus SPDsh fate during male spicule development, overexpression of POP-1 results in the same defect as loss-of-function mutation in lin-17 and son-1. Therefore, a POP-1-like activity also negatively regulates Wnt signaling during C. elegans postembryonic development. Both the sequence-specific TCF/LEF and sequence-nonspecific HMG1/2 class of proteins are involved in lin-17-mediated Wnt signaling and they play opposite roles in regulating Wnt pathway activity.
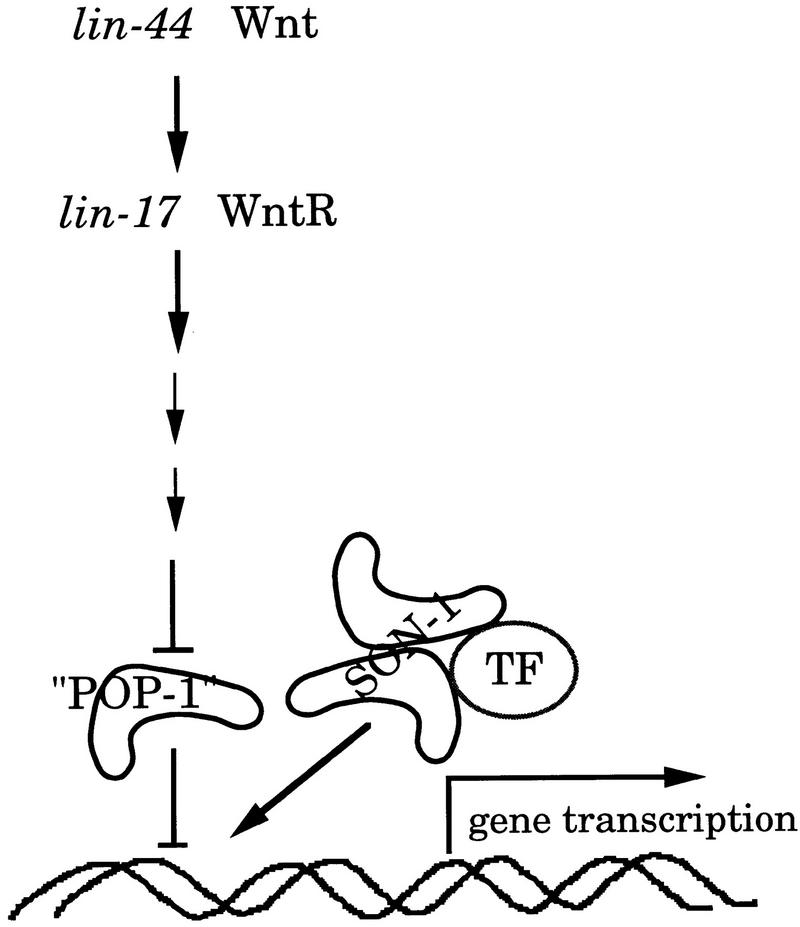
pop-1 has been shown to act downstream of the Wnt receptor and its expression is negatively regulated by Wnt signaling (Rocheleau et al. 1997; Thorpe et al. 1997; Lin et al. 1998). Inactivation of the Wnt pathway up-regulates POP-1 protein level. Here we show that son-1 also acts in the Wnt responding cells. Therefore, both the POP-1-like protein and SON-1 act in the same cell nucleus to regulate gene transcription. The POP-like activity inhibits gene transcription, and SON-1 facilitates gene transcription (Fig. 8). As SON-1 is an HMG-1/2-like protein, we think that SON-1 could activate gene transcription in two ways. First, SON-1 could alter chromatin conformation in favor of gene expression. Second, SON-1 could physically interact with other transcription factors and help stimulate specific gene expression. Proteins of the HMG1/2 class have been shown to interact with POU domain proteins or HOX proteins in vitro and enhance their DNA-binding and transcriptional activities (Zwilling et al. 1995; Zappavigna et al. 1996). Our data suggest that son-1 function may not be rate limiting for Wnt signal transduction. Moreover, son-1::GFP expression does not seem to be altered in a lin-17 mutant background. Therefore, it is possible that SON-1 acts to activate gene transcription after the inhibitory effect of POP-1 is relieved by activation of the Wnt pathway.
Figure 8.
Model for SON-1 function. SON-1 and a POP-1-like protein antagonistically regulate gene expression in Wnt-responding cells. The POP-1-like activity inhibits gene transcription, whereas SON-1 facilitates gene transcription. SON-1 might facilitate gene transcription after the inhibitory effect of the POP-1-like activity is relieved on activation of the Wnt pathway. SON-1 could change the chromatin conformation in favor of gene expression. Alternatively, SON-1 could interact with other transcription factors to help stimulate gene expression. (POP-1) A POP-1-like protein; (TF) transcription factor.
Materials and methods
Strains and alleles
Routine culturing, maintenance and genetic manipulations of C. elegans strains were performed according to standard procedures (Brenner 1974). N2 (Bristol, UK) strain was used as wild type (Brenner 1974). The following strains were used and are referenced in Riddle et al. (1997) or as indicated: LG I–lin-17(n671), lin-17(n698) (Ferguson and Horvitz 1985), pop-1(zu189) (Lin et al. 1995), dpy-5(e61), hT1, hT2; LG II–dpy-10(e128), rol-1(e91) mom-3(or78) (Thorpe et al. 1997), mnC1[dpy-10(e128) unc-52(e444)]; LG III–dpy-17(e164), lon-1(e185), unc-36(e251), unc-32(e189), dpy-18(e364), sDf125, sDf127, sDf135, nDf16, nDf20, nDf21, pha-1(e2123ts) (Granato et al. 1994), hT2, qC1, dpy-19(e1259) lin-12(n137)/unc-32(e189) lin-12(n676n909); LG IV–dpy-20(e1282), dpy-13(e184); LG V–syIs20[gpa-1::lacZ+dpy-20(+)], him-5(e1490), dpy-11(e224); LG X–bar-1(sy324) (A. Golden and P.W. Sternberg, unpubl.), bar-1(mu349) (kindly provided by J. Whangbo and C. Kenyon, University of California, San Franscisco), lon-2(e678), mom-1(or10) unc-6(n102) (Thorpe et al. 1997).
Assay for β-galactosidase activity was performed as described previously (Fire 1992).
Anatomical and lineage analysis/cell ablation/photographs
Animal anatomy and cell division patterns were examined under Nomarski optics at 20°C as described by Sulston and Horvitz (1977). For laser ablation, L3 males were mounted on 5% agar pad and anesthetized with 1%–3% sodium azide. Laser killing cells was performed as described (Sulston and White 1980; Avery and Horvitz 1987, 1989). Worms were then recovered from the agar pad and allowed to grow to adulthood, at which time we examined the gpa-1::lacZ marker expression. Photographs of Nomarski images were taken on Kodak Tech Pan film. Photographs of β-galactosidase expression images were taken on Kodak Royal Gold ASA 25 films. GFP expression was examined with a Chroma High Q GFP LP filter set (450 nm excitation/505 nm emission) and photos were taken on Fuji Provia ASA 400 film.
Isolation of the son-1(sy549) allele
By use of the SPD neuron marker, gpa-1::lacZ, an F1 clonal screen was designed to look for mutants that specifically disrupt the terminal cell fate specification during male spicule development. Standard EMS mutagenesis was used (Brenner 1974). The sy549 allele was recovered from a screen of ∼6300 haploid genomes.
Cloning of son-1
son-1(sy549) was mapped to chromosome III by two-factor mapping. Three-factor mapping put son-1(sy549) between lon-1 and unc-36. Markers used for three-factor mapping were dpy-17(e164) unc-32(e189), dpy-17(e164) unc-36(e251), and lon-1(e185) unc-32(e189). Deficiency mapping indicated that both sDf125 and sDf127 delete son-1(sy549), but nDf16, nDf20, and nDf21 do not. Therefore, son-1 mutation lies between the left end point of sDf127 and the left end point of nDf16. Cosmid pools or a single cosmid in that region were injected into son-1(sy549) pha-1(ts); syIs20 him-5 strain at a concentration of 25 ng/μl each to test for rescue. Those cosmids include C23G10, T12A2, C18F10, F47D12, C56G2, C16A3, C05D11, T26A5, F23F12, K03F8, C09E7, F27B3, F20H11, C13B9, M01G4, F37A4, K07E12, R05H11, F28F5, and R13F6. pha-1 was used as a germ-line transformation marker (Granato et al. 1994). Rescuing activity was scored primarily on the basis of the spicule phenotype and other phenotypes were examined as well. A single cosmid C18F10 rescued son-1(sy549) phenotypes in all respects. Using primers LIJ1(5′-CCCAGTTCTTACTGCTTCGCATC-3′) and LIJ2 (5′-CATGAAAACTCCACTGCAACGG-3′), we amplified a 3-kb fragment by PCR from the 3′ end of C18F10 which contains a single gene F47D12.4. Three independent PCR clones were injected into son-1(sy549) mutants and all of them were able to rescue the mutant phenotypes.
son-1 cDNA was obtained by RT–PCR with wild-type N2 total RNA as template and using primers complementary to son-1 start and stop codon regions on the basis of GeneFinder prediction. The ∼600-bp products from five independent RT–PCR clones were cloned into pBluescript and sequenced. Three clones are consistent with GeneFinder prediction, the other two have an alternative splice site at the end of the first exon and result in an additional 2 amino acids without changing the rest of the protein coding. The 5′ end of the gene was confirmed by RT–PCR by use of SL1 primer and an internal gene-specific primer (LIJ6: 5′-ACAGGGCTCTTTTTGGTGC-3′). A cDNA clone yk74h7 has been reported recently on the web database by Dr. Y. Kohara (National Institute of Genetics, Mishima, Japan). It may predict an extra exon of 32 amino acids at the 5′ end of son-1.
The 1-kb genomic region of the F47D12.4 gene was PCR amplified from small numbers of sy549 mutants (Aroian et al. 1994) and sequenced on an automated sequencing machine (Applied Biosystems). Four independent PCR clones showed the same nucleotide change of g → a at nucleotide position 228 (two plus strands, two minus strands). This change results in a substitution of asparagine for aspartic acid 72.
Molecular biology
son-1-GFP
The transcriptional fusion GFP constructs contain either 1- or 4.5-kb upstream genomic sequence and the first 64 amino acids of son-1 protein (at the EcoRV site). The upstream sequence was PCR amplified from cosmid C18F10, the coding region was derived from the 3-kb rescuing fragment. The translational fusion GFP constructs (pLJ33/pJ35) contain PCR fragment of either 1- or 3.2-kb upstream genomic sequence and full length, but the last 6 amino acids of SON-1 protein cloned into GFP vectors from the Fire laboratory (Carnegie Institute of Washington).
hs-son-1
A 1.4-kb EcoRI fragment of son-1 genomic sequence was cloned into vectors containing heat shock promoter hsp16-2 or hsp16-48 to generate pLJ25 and pLJ26, respectively.
son-1 deletion constructs
To create son-1 deletion constructs, we started from the 3-kb rescuing fragment. The 3-kb fragment was cloned into pCRII vector (Invitrogen; pLJ13). For the box A deletion construct (pLJ41), a SmaI–EcoRV fragment was removed and religated, which deleted amino acids 7–76. For the box B deletion construct (pLJ40), sequences between BamHI and SpeI was deleted, thereby removing amino acids 99–162. For the box A construct only (pLJ39), a stop codon was generated after the first 98 amino acids. All constructs still contain 1-kb upstream and 1-kb downstream genomic sequence of son-1.
Generation of transgenic animals
Transgenic lines carrying an extrachromosomal array of testing plasmids were generated by standard germ-line injection and a DNA transformation method (Mello and Fire 1995). GFP constructs were injected into pha-1(ts); him-5(e1490) strain at a concentration of 20 ng/μl together with pha-1 rescuing plasmid pBX-1 at 100 ng/μl. Heat shock constructs and son-1 deletion constructs were injected into pha-1(ts); syIs20 him-5 strain. Injection concentrations for heat-shock constructs containing son-1 genomic DNA, lin-44 cDNA, and pop-1 cDNA are 50, 100, and 20 or 100 ng/μl, respectively. son-1 deletion constructs were injected at 50 and 100 ng/μl. To test the ability to rescue son-1 phenotypes, transgenic lines were further crossed into son-1(sy549) pha-1(ts); syIs20 him-5 strain.
RNA interference experiments
Double-stranded RNA was synthesized as described (Fire et al. 1998) by use of full-length son-1 cDNA as template. Approximately 40–50 ng/μl dsRNA was injected into syIs20 him-5 hermaphrodites. Eggs laid between 3 and 51 hr after injection were collected at 12-hr intervals. Phenotypes of F1s were analyzed in detail. Unrelated dsRNA and DEPC-H2O were used as controls for the experiment.
Heat shock experiments
Worms were staged under Nomarski optics and heat shocked in a 33°C water bath for 30, 45, or 60 min.
Acknowledgments
We thank Jane Mendel for the syIs20 him-5 strain, Mike Herman for the lin-17(n698) allele and the hs-lin44 constructs, Helen Stewart for the sDf strains and sDf break-point mapping data, Rueyling Lin for the hs-pop-1 construct, Jennifer Whangbo for the bar-1(mu349) allele, the CGC for strains, and the Sanger center for cosmids and sequencing information. We thank members of our laboratory for discussion and comments on the manuscript. This work was supported by a U.S. Public Health Service grant HD23690 to P.W.S. and by the Howard Hughes Medical Institute, with which P.W.S. is an investigator.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL pws@cco.caltech.edu; FAX (626) 568-8012.
References
- Aroian RV, Lesa GM, Sternberg PW. Mutations in the Caenorhabditis elegans let-23 EGFR-like gene define elements important for cell-type specificity and function. EMBO J. 1994;13:360–366. doi: 10.1002/j.1460-2075.1994.tb06269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avery L, Horvitz HR. A cell that dies during wild-type C. elegans development can function as a neuron in a ced-3 mutant. Cell. 1987;51:1071–1078. doi: 10.1016/0092-8674(87)90593-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avery L, Horvitz HR. Pharyngeal pumping continues after laser killing of the pharyngeal nervous system of C. elegans. Neuron. 1989;3:473–485. doi: 10.1016/0896-6273(89)90206-7. [DOI] [PubMed] [Google Scholar]
- Bianchi ME, Falciola L, Ferrari S, Lilley DMJ. The DNA binding site of HMG1 protein is composed of two similar segments (HMG boxes), both of which have counterparts in other eukaryotic regulatory proteins. EMBO J. 1992;11:1055–1063. doi: 10.1002/j.1460-2075.1992.tb05144.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonyaratanakornkit V, Melvin V, Prendergast P, Altmann M, Ronfani L, Bianchi ME, Taraseviciene L, Nordeen SK, Allegretto EA, Edwards DP. High-mobility group chromatin proteins 1 and 2 functionally interact with steroid hormone receptors to enhance their DNA binding in vitro and transcriptional activity in mammalian cells. Mol Cell Biol. 1998;18:4471–4487. doi: 10.1128/mcb.18.8.4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdine RD, Branda CS, Stern MJ. egl-17(fgf) expression coordinates the attraction of the migrating sex myoblasts with vulval induction C. elegans. Development. 1998;125:1083–1093. doi: 10.1242/dev.125.6.1083. [DOI] [PubMed] [Google Scholar]
- Bustin M, Reeves R. High mobility group chromosomal proteins: Architectural components that facilitate chromatin function. Prog Nucleic Acid Res Mol Biol. 1996;54:35–100. doi: 10.1016/s0079-6603(08)60360-8. [DOI] [PubMed] [Google Scholar]
- Cadigan KM, Nusse R. Wnt signaling: A common theme in animal development. Genes & Dev. 1997;11:3286–3305. doi: 10.1101/gad.11.24.3286. [DOI] [PubMed] [Google Scholar]
- Chamberlin HM, Sternberg PW. Multiple cell interactions are required for fate specification during male spicule development in Caenorhaebditis elegans. Development. 1993;118:297–324. doi: 10.1242/dev.118.2.297. [DOI] [PubMed] [Google Scholar]
- ————— The lin-3/let-23 pathway mediates inductive signaling during male spicule development in Caenorhabditis elegans. Development. 1994;120:2713–2721. doi: 10.1242/dev.120.10.2713. [DOI] [PubMed] [Google Scholar]
- ————— Mutations in the Caenorhabditis elegans gene vab-3 reveal distinct roles in fate specification and unequal cytokinesis in an asymmetric cell division. Dev Biol. 1995;170:679–689. doi: 10.1006/dbio.1995.1246. [DOI] [PubMed] [Google Scholar]
- Collet J, Spike CA, Lundquist EA, Shaw JE, Herman RK. Analysis of osm-6, a gene that affects sensory cilium structure and sensory neuron function in Caenorhabditis elegans. Genetics. 1998;148:187–200. doi: 10.1093/genetics/148.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenmann DM, Maloof JN, Simske JS, Kenyon C, Kim SK. The β-catenin homolog BAR-1 and LET-60 RAS coordinately regulate the hox gene lin-39 during Caenorhabditis elegans vulval development. Development. 1998;125:3667–3680. doi: 10.1242/dev.125.18.3667. [DOI] [PubMed] [Google Scholar]
- Ferguson EL, Horvitz HR. Identification and characterization of 22 genes that affect the vulval cell lineages of the nematode Caenorhabditis elegans. Genetics. 1985;110:17–72. doi: 10.1093/genetics/110.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson EL, Sternberg PW, Horvitz HR. A genetic pathway for the specification of the vulval cell lineages of Caenorhabditis elegans. Nature. 1987;326:259–267. doi: 10.1038/326259a0. [DOI] [PubMed] [Google Scholar]
- Fire A. Histochemical techniques for locating Escherichia coli beta-galactosidase activity in transgenic organisms. Genet Anal. 1992;9:151–158. doi: 10.1016/1050-3862(92)90042-4. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Greenwald I. Development of the vulva. In: Riddle DL, Blumenthal T, Meyer BJ, Priess JR, editors. Caenorhabditis elegans II. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 519–541. [PubMed] [Google Scholar]
- Granato M, Schnabel H, Schnabel R. pha-1, a selectable marker for gene-transfer in C. elegans. Nucleic Acid Res. 1994;22:1762–1732. doi: 10.1093/nar/22.9.1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartenstein V, Posakony JW. A dual function of the Notch gene in Drosophila sensillum development. Dev Biol. 1990;142:13–30. doi: 10.1016/0012-1606(90)90147-b. [DOI] [PubMed] [Google Scholar]
- Herman MA, Horvitz HR. The Caenorhabditis elegans gene lin-44 controls the polarity of asymmetric cell divisions. Development. 1994;120:1035–1047. doi: 10.1242/dev.120.5.1035. [DOI] [PubMed] [Google Scholar]
- Herman MA, Vassilieva LL, Horvitz HR, Shaw JE, Herman RK. The C. elegans gene lin-44, which controls the polarity of certain asymmetric cell divisions, encodes a Wnt protein and acts cell nonautonomously. Cell. 1995;83:101–110. doi: 10.1016/0092-8674(95)90238-4. [DOI] [PubMed] [Google Scholar]
- Hill RJ, Sternberg PW. The gene lin-3 encodes an inductive signal for vulval development in C. elegans. Nature. 1992;358:470–476. doi: 10.1038/358470a0. [DOI] [PubMed] [Google Scholar]
- Jayaraman L, Moorthy NC, Murthy KG, Manley JL, Bustin M, Prives C. High mobility group protein-1 (HMG-1) is a unique activator of p53. Genes & Dev. 1998;12:462–472. doi: 10.1101/gad.12.4.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang LI, Sternberg PW. Interactions of EGF, Wnt and HOM-C genes specify the P12 neuroectoblast fate in C. elegans. Development. 1998;125:2337–2347. doi: 10.1242/dev.125.12.2337. [DOI] [PubMed] [Google Scholar]
- Lehming N, Thanos D, Brickman JM, Ma J, Maniatis T, Ptashne M. An HMG-like protein that can switch a transcriptional activator to a repressor. Nature. 1994;371:175–179. doi: 10.1038/371175a0. [DOI] [PubMed] [Google Scholar]
- Lin RL, Thompson S, Priess JR. pop-1 encodes an HMG box protein required for the specification of a mesoderm precursor in early C. elegans embryos. Cell. 1995;83:599–609. doi: 10.1016/0092-8674(95)90100-0. [DOI] [PubMed] [Google Scholar]
- Lin RL, Hill RJ, Priess JR. POP-1 and anterior-posterior fate decisions in C. elegans embryo. Cell. 1998;92:229–239. doi: 10.1016/s0092-8674(00)80917-4. [DOI] [PubMed] [Google Scholar]
- Mello CC, Fire A. Methods in Cell Biology: C. elegans. New York, NY: Academic Press; 1995. [Google Scholar]
- Meneely PM, Wood WB. Genetic analysis of X-chromosome dosage compensation in Caenorhabditis-elegans. Genetics. 1987;117:25–41. doi: 10.1093/genetics/117.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon RT, Brown JD, Torres M. Wnts modulate cell fate and behavior during vertebrate development. Trends Genet. 1997;13:157–162. doi: 10.1016/s0168-9525(97)01093-7. [DOI] [PubMed] [Google Scholar]
- Parks AL, Muskavitch MA. Delta function is required for bristle organ determination and morphogenesis in Drosophila. Dev Biol. 1993;157:484–496. doi: 10.1006/dbio.1993.1151. [DOI] [PubMed] [Google Scholar]
- Read CM, Cary PD, Preston NS, Lenicek-Allen M, Crane-Robinson C. The DNA-sequence specificity of HMG boxes lies in the minor wing of the structure. EMBO J. 1994;13:5639–5646. doi: 10.1002/j.1460-2075.1994.tb06902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle DL, Blumenthal T, Meyer BJ, Priess JR, editors. C. elegans II. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- Rocheleau CE, Downs WD, Lin R, Wittmann C, Bei Y, Cha Y-H, Ali M, Priess JR, Mello CC. Wnt signaling and an APC-related gene specify endoderm in early C. elegans embryos. Cell. 1997;90:707–716. doi: 10.1016/s0092-8674(00)80531-0. [DOI] [PubMed] [Google Scholar]
- Roehl H, Kimble J. Control of cell fate in C. elegans by a GLP-1 peptide consisting primarily of ankyrin repeats. Nature. 1993;364:632–635. doi: 10.1038/364632a0. [DOI] [PubMed] [Google Scholar]
- Sawa H, Lobel L, Horvitz HR. The Caenorhabditis elegans gene lin-17, which is required for certain asymmetric cell divisions, encodes a putative protein similar to seven-transmembrane protein similar to the Drosophila Frizzle protein. Genes & Dev. 1996;10:2189–2197. doi: 10.1101/gad.10.17.2189. [DOI] [PubMed] [Google Scholar]
- Sibley MH, Graham PL, Vonmende N, Kramer JM. Mutations in the alpha-2(IV) basement-membrane collagen gene of Caenorhabditis elegans produce phenotypes of differing severities. EMBO J. 1994;13:3278–3285. doi: 10.1002/j.1460-2075.1994.tb06629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg PW, Horvitz HR. lin-17 mutations of Caenorhabditis elegans disrupt certain asymmetric cell divisions. Dev Biol. 1988;130:67–73. doi: 10.1016/0012-1606(88)90414-9. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Horvitz HR. Postembryonic cell lineages of the nematode Caenorhabditis elegans. Dev Biol. 1977;56:110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- Sulston JE, White JG. Regulation and cell autonomy during postembryonic development of Caenorhabditis elegans. Dev Biol. 1980;78:577–597. doi: 10.1016/0012-1606(80)90353-x. [DOI] [PubMed] [Google Scholar]
- Thorpe C, Schlesinger A, Carter JC, Bowerman B. Wnt signaling polarizes an early C. elegans blastomere to distinguish endoderm from mesoderm. Cell. 1997;90:695–705. doi: 10.1016/s0092-8674(00)80530-9. [DOI] [PubMed] [Google Scholar]
- van de Wetering M, Cavallo R, Dooijes D, van Beest M, van Es J, Loureiro J, Ypma A, Hursh D, Jones T, Bejsovec A, Peifer M, Mortin M, Clevers H. Armadillo coactivates transcription driven by the product of the Drosophila segment polarity gene dTCF. Cell. 1997;88:789–799. doi: 10.1016/s0092-8674(00)81925-x. [DOI] [PubMed] [Google Scholar]
- Waltzer L, Bienz M. Drosophila CBP represses the transcription factor TCF to antagonize Wingless signaling. Nature. 1998;395:521–525. doi: 10.1038/26785. [DOI] [PubMed] [Google Scholar]
- Zappavigna V, Falciola L, Citterich MH, Mavilio F, Bianchi ME. HMG1 interacts with HOX proteins and enhances their DNA binding and transcriptional activation. EMBO J. 1996;15:4981–4991. [PMC free article] [PubMed] [Google Scholar]
- Zwilling S, König H, Wirth T. High mobility group protein 2 functionally interacts with the POU domains of octamer transcription factors. EMBO J. 1995;14:1198–1208. doi: 10.1002/j.1460-2075.1995.tb07103.x. [DOI] [PMC free article] [PubMed] [Google Scholar]