Abstract
TGFβ can override the proliferative effects of EGF and other Ras-activating mitogens in normal epithelial cells. However, epithelial cells harboring oncogenic Ras mutations often show a loss of TGFβ antimitogenic responses. Here we report that oncogenic Ras inhibits TGFβ signaling in mammary and lung epithelial cells by negatively regulating the TGFβ mediators Smad2 and Smad3. Oncogenically activated Ras inhibits the TGFβ-induced nuclear accumulation of Smad2 and Smad3 and Smad-dependent transcription. Ras acting via Erk MAP kinases causes phosphorylation of Smad2 and Smad3 at specific sites in the region linking the DNA-binding domain and the transcriptional activation domain. These sites are separate from the TGFβ receptor phosphorylation sites that activate Smad nuclear translocation. Mutation of these MAP kinase sites in Smad3 yields a Ras-resistant form that can rescue the growth inhibitory response to TGFβ in Ras-transformed cells. EGF, which is weaker than oncogenic mutations at activating Ras, induces a less extensive phosphorylation and cytoplasmic retention of Smad2 and Smad3. Our results suggest a mechanism for the counterbalanced regulation of Smad2/Smad3 by TGFβ and Ras signals in normal cells, and for the silencing of antimitogenic TGFβ functions by hyperactive Ras in cancer cells.
Keywords: Growth inhibition, MAP kinase, Ras, Smad, TGFβ
The cytokine TGFβ plays a dual role in tumorigenesis. On one hand, TGFβ inhibits the proliferation of normal epithelial, endothelial, and hematopoietic cells, thus being crucial for the homeostasis of these tissues (Massagué 1990; Roberts and Sporn 1993; Alexandrow and Moses 1995). TGFβ signaling is lost in some cancers by mutational inactivation of TGFβ signal-transduction components. A substantial proportion of colorectal or pancreatic cancers harbor inactivating mutations in the genes encoding the TGFβ type II receptor (TβR-II) or its mediators, Smad2 and Smad4 (Markowitz et al. 1995; Eppert et al. 1996; Hahn et al. 1996; Heldin et al. 1997; Chen et al. 1998; Goggins et al. 1998; Massagué 1998; Grady et al. 1999). In the mouse, expression of a dominant-negative TβR-II transgene in the pancreas, mammary gland, or skin causes abnormal growth of these tissues (Böttinger et al. 1996; Wang et al. 1997; Gorska et al. 1998). Furthermore, Smad3 mutant mice (Zhu et al. 1998) or the combined loss of wild-type Smad4 and APC alleles in compound heterozygotes (Takaku et al. 1998) lead to formation of invasive intestinal tumors.
On the other hand, TGFβ can exacerbate the malignant phenotype at later stages of tumorigenesis (Cui et al. 1996; Barrack 1997; Factor et al. 1997; Reiss and Barcellos-Hoff 1997). TGFβ is abundantly expressed in various tumors of epithelial origin (Derynck et al. 1985; Keski-Oja et al. 1987) in which it can suppress immune surveillance (Letterio and Roberts 1998), foster tumor invasion (Cui et al. 1996), and promote the development of metastases (Welch et al. 1990; Yin et al. 1999). These effects become manifest in tumor cells that retain TGFβ receptors but have lost the capacity to respond to TGFβ with growth arrest. Such a state of altered TGFβ responsiveness is observed in Ras-transformed cells. These cells typically exhibit a limited growth inhibitory response to TGFβ (Schwarz et al. 1988; Houck et al. 1989; Valverius et al. 1989; Longstreet et al. 1992; Filmus and Kerbel 1993) but may respond to TGFβ with invasive activity (Oft et al. 1996) and metastatic behavior (Oft et al. 1998; Yin et al. 1999).
TGFβ exerts growth inhibitory and transcriptional responses through Smad2 and the highly related protein Smad3, which are direct TGFβ receptor substrates, whereas Smad1 is a substrate and mediator of bone morphogenetic protein (BMP) receptors (Heldin et al. 1997; Massagué 1998). Receptor-mediated phosphorylation of these Smads, which occurs at serine residues in the carboxy-terminal SSXS sequence (Macias-Silva et al. 1996; Kretzschmar et al. 1997b), induces their association with the shared partner Smad4 followed by translocation into the nucleus in which these complexes activate transcription of specific genes (Heldin et al. 1997; Massagué 1998). Smad proteins contain a conserved amino-terminal domain that binds DNA (Shi et al. 1998), and a conserved carboxy-terminal domain that binds receptors, partner Smads, and transcription coactivators (Shi et al. 1997; X. Chen et al. 1998). These two domains are separated by a more divergent linker region.
How oncogenic Ras counteracts the growth inhibitory effects of TGFβ has remained unknown. Although oncogenic Ras can prevent the antimitogenic effects of TGFβ, TGFβ potently overcomes the mitogenic effects of Ras-activating factors such as EGF in epithelial cells (Massagué 1990; Roberts and Sporn 1993; Alexandrow and Moses 1995). To investigate the molecular basis for these interactions, we focused on Smad2 and Smad3 as possible targets of inhibition by Ras. Here we show that Ras activation by oncogenic mutations or, to a lesser extent, by EGF receptor signals, inhibits the TGFβ-induced nuclear accumulation of Smad2 and Smad3. These effects are mediated by phosphorylation of specific sites in Smad2 and Smad3, and we demonstrate that these sites are distinct from the TGFβ receptor phosphorylation sites. We present evidence that this mechanism mediates the silencing of TGFβ antimitogenic responses in Ras-transformed cells, whereas in normal cells this mechanism serves to adjust the level of TGFβ/Smad signaling according to the level of Ras activity in the cell. These results reconcile a diverse body of observations on the interaction between the TGFβ and Ras pathways, and provide insights into the subversion of TGFβ signaling by oncogenic Ras mutations in cancer.
Results
Ras inhibition of Smad-dependent TGFβ responses
We investigated Ras as an antagonist of TGFβ signaling using a well-characterized mouse mammary epithelial cell system (Oft et al. 1996). The parental cell line EpH4 is nontumorigenic and responds to TGFβ with growth inhibition, whereas its v-Ha-Ras-transformed derivative, EpRas, responds with a transition into a fibroblastoid phenotype that is not growth inhibited and is highly invasive in vivo (Oft et al. 1996). We investigated whether loss of growth inhibitory responses to TGFβ in EpRas cells might be related to Ras inhibition of Smad transcriptional functions. One of the best understood Smad target promoters is that of the Mix.2 gene from Xenopus laevis (Y. Chen et al. 1996; X. Chen et al. 1997; Liu et al. 1997). On translocation into the nucleus, receptor-activated Smad2 or Smad3 associate with the DNA-binding protein FAST1 to form a transcriptional complex on the activin/TGFβ response element (ARE) of the Mix.2 promoter (Y. Chen et al. 1996; X. Chen et al. 1997; Liu et al. 1997). Strong (up to 14-fold) activation of an ARE reporter construct (A3–Luc) by TGFβ was achieved in EpH4 cells cotransfected with FAST1 (data not shown) or its mammalian homolog FAST2 (Labbé et al. 1998; Zhou et al. 1998; Liu et al. 1999) (Fig. 1A). In contrast, FAST2-transfected EpRas cells showed only a weak (twofold) activation of A3–Luc by TGFβ, suggesting that Smad signaling was impaired (Fig. 1A).
Figure 1.
Oncogenic H-Ras inhibits Smad-dependent TGFβ transcriptional responses. (A) Activation of the A3–Luciferase reporter construct by the indicated concentrations of TGFβ was analyzed in EpH4 and EpRas mammary epithelial cells. (█) EpH4; (▴) EpRas. (B) TGFβ- and FAST2-dependent activation of the A3–Luc reporter in Mv1Lu mink lung epithelial cells transfected with or without H-RasV12. (█) −RasV12 + FAST-2; (▴) +RasV12 + FAST-2; (♦) −RasV12; (●) +RasV12. (C) TGFβ antiproliferative response in Mv1Lu cells transfected with or without H-RasV12. (█) −RasV12; (▴) +RasV12. G1 progression activity was determined with an E2F reporter construct driving luciferase expression. In all cases, data are the average of triplicates ±s.d.
EpRas cells may have accumulated Ras-independent defects in TGFβ signaling over time. Therefore, we verified the ability of oncogenic Ras to inhibit A3–Luc activation by transiently transfecting a H-RasV12 oncogene into Mv1Lu mink lung epithelial cells. This cell line is highly sensitive to growth inhibition and A3–Luc activation by TGFβ. The TGFβ-induced activation of A3–Luc in these cells was strongly inhibited by cotransfection of H-RasV12 (Fig. 1B). Transient transfection of H-RasV12 also inhibited the antiproliferative response to TGFβ, as determined with an E2F-1 reporter construct whose activity is a measure of G1 progression (Lukas et al. 1997; Fig. 1C). Thus, in a stably transfected mammary epithelial cell line and in a transiently transfected lung epithelial cell line, the presence of oncogenic Ras caused not only a loss of antiproliferative responses to TGFβ but also of Smad-dependent transcriptional responses.
Inhibition of Smad2/Smad3 nuclear accumulation by oncogenic Ras
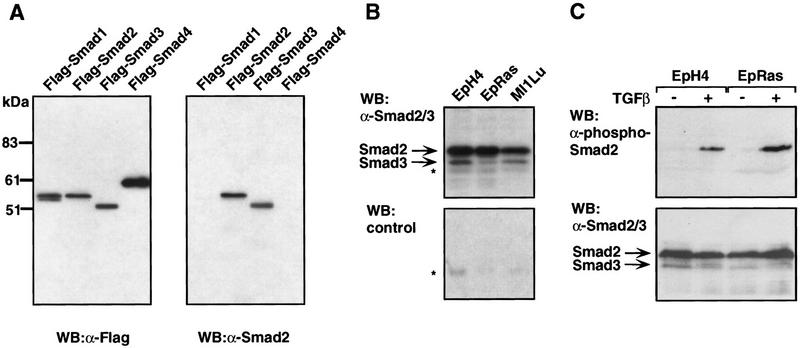
Translocation of Smad2 and Smad3 into the nucleus on receptor-mediated phosphorylation is a central event in TGFβ signal transduction (Heldin et al. 1997; Massagué 1998). Growth factors that signal via Ras, such as EGF, can inhibit the BMP-induced nuclear accumulation of Smad1 by inducing its phosphorylation at four PXSP Erk sites (Kretzschmar et al. 1997a). These four sites are not conserved in Smad2 or Smad3. Nevertheless, we decided to investigate whether oncogenic Ras might inhibit nuclear accumulation of endogenous Smads in response to TGFβ. Rabbit polyclonal antibodies were raised against the recombinant linker region of Smad2 and tested by Western immunoblotting on transfected COS cells expressing different Flag-tagged Smads. These antibodies specifically recognized Smad2, and to a lesser extent Smad3, but not Smad1 or Smad4 (Fig. 2A). This cross-reactivity profile is consistent with the high level of amino acid sequence similarity of the Smad2 and Smad3 linker region and the extensive divergence of this region in the other Smads (Zhang et al. 1996). Immunoblot assays of untransfected EpH4, EpRas, and Mv1Lu cells with these antibodies demonstrated the presence of Smad2 and lower levels of Smad3 in each cell line (Fig. 2B). Using antibodies that specifically recognize the TGFβ receptor-phosphorylated Smad2, we determined that endogenous Smad2 in both EpH4 and EpRas cells is phosphorylated in response to TGFβ (Fig. 2C). Thus, Ras transformation did not interfere with TGFβ receptor-mediated phosphorylation of Smad2 in EpRas cells.
Figure 2.

TGFβ-induced nuclear accumulation of Smad2 and Smad3 in EpH4 and EpRas cells. (A) COS cells transfected with Flag-tagged versions of Smad1, Smad2, Smad3, or Smad4 were subjected to Western immunoblotting with anti-Flag antibodies as a positive control, or antibodies raised against the linker region of Smad2. (B) Untransfected EpH4, EpRas, and Mv1Lu cell extracts were subjected to Western immunoblotting with affinity-purified anti-Smad2/Smad3 (top) or nonimmune serum (bottom). (Asterisks) Nonspecific bands. (C) EpRas and EpH4 cells were stimulated with TGFβ for 30 min. Cell lysates were subjected to Western immunoblotting with either antibodies against receptor-phosphorylated Smad2 (top) or anti-Smad2/Smad3 antibodies (bottom). (D) In a similar experiment, endogenous Smad2 and Smad3 were visualized by anti-Smad2/Smad3 immunofluorescence. (E) EpH4 (█) and EpRas (▴) cells were treated with TGFβ for different time periods. Following immunofluorescence staining, the percentage of cells with Smad2/Smad3 staining predominantly or exclusively in the nucleus was determined.
We determined the immunofluorescence pattern of endogenous proteins in EpH4 and EpRas using anti-Smad2/Smad3 antibodies (Fig. 2D). In the absence of TGFβ, both cell lines showed anti-Smad2/Smad3 staining throughout the cell. On TGFβ addition, >95% of the EpH4 cells showed an accumulation of all the Smad2/Smad3 immunostaining in the nucleus (Fig. 2D). This nuclear accumulation was rapid and lasted for at least 3 hr (Fig. 2E). Compared to EpH4 cells, EpRas cells responded to TGFβ with a limited accumulation of Smad2/Smad3 in the nucleus. Furthermore, this accumulation was slow and only partial, as most cells with predominant nuclear staining still showed an extensive staining in the cytoplasm (Fig. 2D,E). We also examined the pattern of Smad2/Smad3 nuclear accumulation in five human colon carcinoma cell lines, three of which contain oncogenic K-Ras mutations (Table 1). Nuclear accumulation of Smad2/Smad3 in response to an intermediate (10 pm) TGFβ concentration was poor or absent in these three cell lines but extensive in the two cell lines containing wild-type ras alleles.
Table 1.
TGFβ-induced nuclear accumulation of Smad2/3 in human colon cancer cell lines
|
Cell line
|
ras allelea
|
Other alterationsa
|
Smad2/Smad 3, % nuclearb
|
||
|---|---|---|---|---|---|
|
nontransfected
|
vector
|
H-RasV12
|
|||
| HT-29 | wild type | p53−/APC−/src | 93 | 75 | 12 |
| Colo205 | wild type | p53−/APC−/lck/src/myb | 45 | 48 | 4 |
| SW620 | K-rasV12 | p53−/APC−/src/myb | 10 | N.T. | N.T. |
| LoVo | K-rasD12 | p53−/APC−/src | <2 | N.T. | N.T. |
| DLD-1 | K-rasD13 | p53−/APC− | <2 | N.T. | N.T. |
ras status and other known oncogenic alterations according to Sepp-Lorenzino et al. (1995). p53 and APC are inactivated by mutation or not expressed. The oncogenes listed are mutated or overexpressed.
Cells were either not transfected, transfected with GFP vector and empty vector (vector), or GFP vector and H-RasV12 vector (H-RasV12). Cells were incubated with 10 pm TGFβ for 30 min and subjected to anti-Smad2/3 immunofluorescence. The percentage of cells with predominant nuclear staining in the total nontransfected population or in the transfected (GFP positive) population is shown. (N.T.) Not tested.
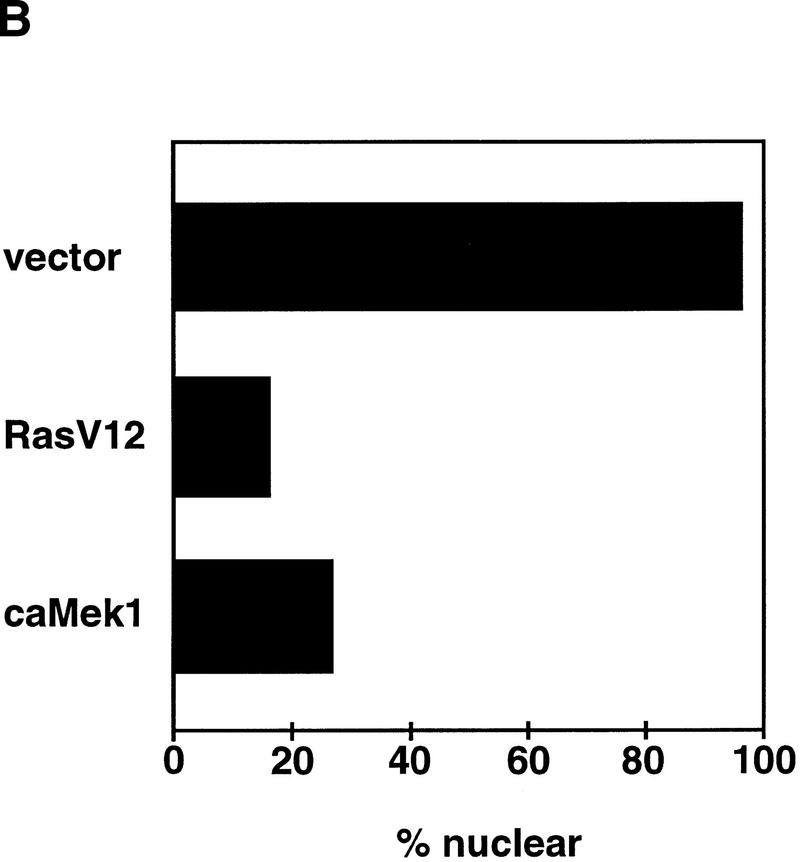
To directly test whether oncogenic Ras can interfere with nuclear accumulation of endogenous Smad2 and Smad3, we transiently transfected Mv1Lu cells with H-RasV12. Cotransfection of the green fluorescence protein (GFP) served to distinguish transfected from nontransfected cells in the dish. Up to 95% of the control vector transfected (GFP positive) cells showed nuclear accumulation of endogenous Smad2/Smad3 in response to TGFβ (Fig. 3A). In contrast, only a small proportion (16%) of the H-RasV12 transfected cells showed nuclear accumulation of Smad2/Smad3 (Fig. 3A). A similar effect was observed in cells expressing a constitutively active form of Mek1 (Fig. 3B), which is an activator of Erk MAP kinases downstream of Ras (Davis 1993). GFP-negative cells, which served as internal controls in these assays, had extensive Smad2/Smad3 staining in the nucleus in the presence of TGFβ (Fig. 3). A similar inhibition of Smad2/Smad3 nuclear accumulation by transfection of a H-RasV12 vector was observed in the two tumor cell lines containing wild-type ras alleles (Table 1).
Figure 3.

Nuclear accumulation of Smads in response to TGFβ is inhibited by oncogenic H-RasV12 or constitutively active Mek1. Mv1Lu cells were transiently transfected with empty vector, H-RasV12 or constitutively active Mek1 (ca Mek1), and then treated with TGFβ for 30 min before fixation. Transfected cells were marked by cotransfection with GFP. Endogenous proteins were visualized by anti-Smad2/Smad3 immunofluorescence by a rhodamine-coupled system. (Arrowheads) GFP-positive cells. (A) Representative micrographs of the control and H-RasV12 transfectants visualizing the GFP and anti-Smad2/Smad3 signals. (B) Percentage of GFP-positive cells with Smad2/Smad3 predominantly in the nucleus under the three experimental conditions.
Ras induces phosphorylation of Smad2 and Smad3 in the linker region
Next, we investigated whether Ras inhibition of Smad nuclear accumulation involves Smad phosphorylation. Activated Ras propagates signals through the activation of protein kinases including the MAP kinases p44Erk1 and p42Erk2 (Davis 1993). The levels of these two proteins were similar in EpH4 and EpRas cells, as determined by Western immunoblotting with anti-Erk antibodies (Fig. 4A, bottom). However, the level of Erk activity was higher in EpRas cells than in EpH4 cells, as determined by measuring the kinase activity of Erk immunoprecipitates (Fig. 4A, top). This result was consistent with the presence of hyperactive Ras in EpRas cells. Interestingly, immunoprecipitation of endogenous Smad2 and Smad3 from 32P-labeled cells showed that the level of phosphorylation of these proteins was higher in EpRas cells than in EpH4 cells (Fig. 4B). Elution of the 32P-labeled EpRas Smad band from these gels followed by digestion with trypsin yielded labeled products of no less than 13 kD, whereas digestion with α-chymotrypsin yielded mostly products of <5 kD (data not shown). On the basis of the predicted proteolytic sites of Smad2 and Smad3 (Zhang et al. 1996), the observed digestion pattern suggests that most of the phosphorylation resides in the linker region of these proteins.
Figure 4.
Elevated Erk activity and Smad2/Smad3 phosphorylation levels in EpRas cells. (A) The kinase activity associated with anti-Erk immunoprecipitates from EpH4 and EpRas cells was determined with myelin basic protein (MBP) as a substrate. The relative levels of Erk1 and Erk2 were determined by anti-Erk Western immunoblotting. (B) The phosphorylation level of Smad2 and Smad3 in EpH4 and EpRas cells was determined by immunoprecipitation from 32P-labeled cells. The relative levels of Smads were determined by anti-Smad2/Smad3 Western immunoblotting of unlabeled cell extracts prepared in parallel.
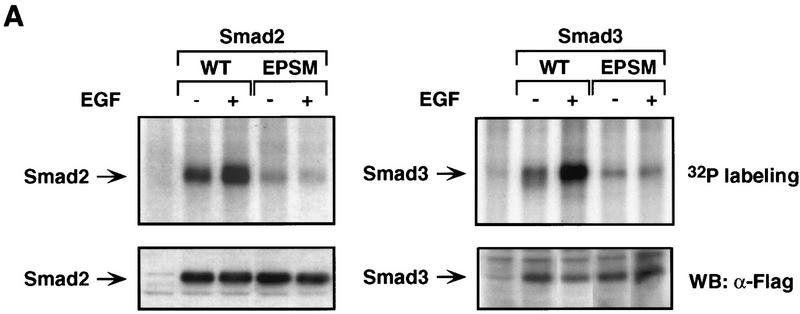
Cell stimulation with EGF has been shown to inhibit the BMP-induced nuclear accumulation of Smad1, and this effect requires the phosphorylation of four Erk kinase sites (PXS/TP) in the linker region of Smad1 (Kretzschmar et al. 1997a). Smad2 has only one Erk site in the linker region (Thr-220) and Smad3 has two (Thr-178, Ser-212) (Fig. 5A). However, the Smad2 linker region has three Ser–Pro sequences (Ser-245, Ser-250, Ser-255) and Smad3 has two (Ser-203, Ser-207). These sequences can serve as phosphorylation sites for proline-directed protein kinases including Erk (Davis 1993). Therefore, we asked whether Ras pathway activation causes phosphorylation of these sites. Smad2 and Smad3 constructs were generated that encoded serine-to-alanine or threonine-to-valine mutations at all these sites (Erk/Pro-directed kinase site mutant constructs, EPSM). Wild-type or EPSM Smad constructs containing a amino-terminal Flag epitope were transiently transfected either alone or together with H-RasV12 or activated Mek1 vectors into Mv1Lu/R1B-L17 cells. Cell labeling with 32P followed by anti-Flag immunoprecipitation showed that Smad2 and Smad3 were phosphorylated under normal culture conditions, and the phosphorylation level of these proteins was increased by transfection of H-RasV12 or activated Mek1 vectors (Fig. 5B,C). Treatment of the Ras-transfected cells with the inhibitor of Mek1 activation PD98059 (Alessi et al. 1995) or the inhibitor of phosphatidylinositol 3-kinase, Wortmannin (Ui et al. 1995), partially inhibited Smad2 and Smad3 phosphorylation, implicating these kinases in the Ras-induced phosphorylation (Fig. 5B,C). The results suggest that the basal activity of the Ras pathway and, to a larger extent, the hyperactivation of this pathway by H-RasV12 or activated Mek1, cause the phosphorylation of Smad2 and Smad3.
Figure 5.
Oncogenic H-RasV12 causes phosphorylation of Smad2 and Smad3 through Mek1 at Ser/Thr–Pro sites in the linker region. (A) Schematic representation of phosphorylation sites in the region that links the DNA binding domain (MH1 domain) and the receptor-interaction/transcriptional domain (MH2 domain) in Smad1, Smad2, and Smad3. (●) Erk consensus sites (PXS/TP); (□) serine/proline motifs, which may serve as phosphorylation sites for proline-directed kinases; (⋄) receptor phosphorylation sites. (B,C) Vectors encoding Flag-tagged wild-type (WT) Smads or mutant Smads lacking all four potential phosphorylation sites in the linker region (EPSM constructs), were used. These constructs were cotransfected with vectors encoding H-RasV12 or constitutively active Mek1 into Mv1Lu/R1B-L17 cells as indicated. The phosphorylation level of transfected Smads was determined by anti-Flag immunoprecipitation from 32P-labeled cells. PD98058 (a Mek1 inhibitor) or wortmannin (a phosphatidyl inositol 3′-kinase inhibitor) were added 1.5 hr prior to cell lysis, as indicated. Smad expression was monitored by anti-Flag immunoblotting of cell lysates. (D) Purified, recombinant Smad proteins (wild-type or EPSM mutants) were tested as substrates for recombinant activated Erk2 in in vitro kinase assays. Identical amounts of recombinant Smad protein was added to all assays. (E) Effects of H-RasV12 cotransfection or EGF addition (18 nm, 30 min) on the phosphorylation of Flag-tagged Smad constructs containing Ser → Ala mutation in the carboxy-terminal SSXS receptor phosphorylation sites. Other assay conditions and controls were as described above.
Erk kinase phosphorylation sites in the linker region
Compared with the transfected Smad2 and Smad3, their corresponding EPSM constructs showed very low levels of phosphorylation, and these levels were not increased by H-RasV12 (Fig. 5B,C). To verify that Erk kinases can directly phosphorylate Smad2 and Smad3 at the identified sites, we performed in vitro kinase assays using recombinant forms of these proteins. Recombinant, activated Erk2 phosphorylated Smad2 and Smad3 to a much greater extent than it phosphorylated Smad2–EPSM or Smad3–EPSM (Fig. 5D). We also tested Smad2 and Smad3 constructs containing hydroxyl residue mutations either in the PXS/TP sites or in the other SP sites. Each of these four mutants showed a partial loss of phosphorylation compared with the EPSM constructs, both in Ras transfected cells and in in vitro Erk2 phosphorylation assays (data not shown).
We also tested Smad2(3SA) and Smad3(3SA) constructs that contain alanine mutations in the three serines of the carboxy-terminal sequence SSXS (Kretzschmar et al. 1997b). These serines serve as TGFβ receptor phosphorylation sites and are required for Smad translocation into the nucleus (Macias-Silva et al. 1996). H-RasV12 transfection of Mv1Lu/R1B-L17 cells induced phosphorylation of both 3SA constructs (Fig. 5E), arguing that the TGFβ-receptor phosphorylation sites are not required for Ras-induced phosphorylation. Collectively, these results suggest that oncogenic Ras, acting through Mek1 and Erk kinases, induces the phosphorylation of Smad2 and Smad3 at a cluster of Ser/Thr-Pro sites in the linker region.
Effects of EGF on Smad2 and Smad3 phosphorylation and nuclear accumulation
As EGF is a physiological activator of the Ras pathway (Vojtek and Der 1998), we investigated its effects on Smad2 and Smad3 phosphorylation and nuclear accumulation. EGF was reported to cause an increase in the phosphorylation levels of an endogenous Smad2 in Mv1Lu cells (deCaesteker et al. 1998). In agreement with this result, we observed that EGF induces phosphorylation of exogenous Smad2 and Smad3 in transfected Mv1Lu/R1B-L17 cells (Fig. 6A). However, Ras activation in response to EGF is thought to be less extensive than Ras activation by oncogenic mutations. The increase in Smad2 or Smad3 phosphorylation induced by EGF in Mv1Lu cells was more limited than that induced by H-RasV12. The EGF-induced phosphorylation requires the same cluster of sites in the linker region, as EGF did not elevate the phosphorylation of Smad2–EPSM or Smad3–EPSM, which lack these sites (Fig. 6A). Like H-RasV12, EGF elevated the phosphorylation of Smad2(3SA) and Smad3(3SA), which lack the TGFβ receptor phosphorylation sites (Fig. 5E).
Figure 6.
EGF induces phosphorylation and inhibits TGFβ-dependent nuclear accumulation of Smad2 and Smad3. (A) Mv1Lu/R1B-L17 cells transfected with vectors encoding the indicated Flag-tagged Smad constructs were labeled with 32P, and incubated with EGF for 30 min. The phosphorylation level of the transfected Smads was determined by anti-Flag immunoprecipitation. Smad expression was controlled by anti-Flag immunoblotting of cell lysates. (B) Mv1Lu cells were incubated with EGF for 60 min before fixation (bottom) and/or TGFβ for 30 min before fixation (right), and endogenous Smad2/3 visualized by immunofluorescence. Note the lack of cytoplasmic staining in cells treated with TGFβ alone, and the cytoplasmic staining remaining in cells treated with TGFβ plus EGF.
Next, we examined the effect of EGF on TGFβ-induced nuclear accumulation of endogenous Smads. EGF added alone had no discernible effect on the staining pattern of endogenous Smad2/Smad3 in Mv1Lu cells. Whether added alone or together with EGF, TGFβ induced an accumulation of Smad2/Smad3 in the nucleus (Fig. 6B). However, EGF clearly limited the extent of this accumulation. When TGFβ was added alone, almost all cells in the population showed an intense nuclear accumulation of Smad2/Smad3 with very little immunostaining left in the cytoplasm. In the presence of EGF, the nuclear accumulation of Smad2/Smad3 was less extensive, and a substantial level of Smad2/Smad3 staining remained in the cytoplasm in most cells (Fig. 6B). EGF was more effective at inhibiting the effect of 10 pm TGFβ (Fig. 6B) than that of 100 pm TGFβ (data not shown), suggesting that the extent of Smad2/Smad3 nuclear accumulation depends on the relative strength of countervailing TGFβ and EGF signals.
Linker phosphorylation inhibits nuclear accumulation
These results showed a correlation between a Ras-induced phosphorylation of a cluster of MAP kinase sites in the linker region of Smad2/Smad3 and an inhibition of TGFβ-induced nuclear accumulation of these Smads. To directly test whether phosphorylation of these sites is required for inhibition of nuclear accumulation, we cotransfected vectors encoding activated Mek1 and either Flag-tagged Smad3 or Smad3–EPSM constructs into Mv1Lu cells. Similar to its effect on the nuclear accumulation of endogenous Smad2/Smad3 (see Fig. 3), the activated Mek1 inhibited the TGFβ-induced nuclear accumulation of Flag–Smad3 (Fig. 7A,B). However, Flag–Smad3–EPSM was resistant to this inhibitory effect, as it accumulated in the nucleus in response to TGFβ even in the presence of activated Mek1 (Fig. 7A,B). Inhibition of Smad3 nuclear accumulation by activated Mek1 therefore requires the linker region MAP kinase phosphorylation sites.
Figure 7.
Mek1 inhibition of Smad3 nuclear accumulation requires phosphorylation in the linker domain. (A) Mv1Lu cells were cotransfected with Flag-tagged Smad3 [wild-type (WT) or mutant (EPSM)] and empty vector or constitutively active Mek1 as indicated. TGFβ was added 30 min prior to fixation. Flag–Smad3 was visualized by anti-Flag immunofluorescence. (B) Nuclear accumulation of Flag–Smad3 in the TGFβ-treated cells of A was quantitated by determining the proportion of cells with Smad2/Smad3 staining exclusively in the nucleus (e.g., as in A, far right; solid area) and cells with predominant but not exclusive nuclear staining (e.g., as in A, second from right; stippled area).
Ras-resistant Smad3 rescues TGFβ antimitogenic and transcriptional responses
Finally, we used the Smad3–EPSM construct to investigate whether the TGFβ responses lost in EpRas cells could be restored by a Ras-resistant Smad3 mutant. TGFβ addition inhibited the activity of the E2F-1–luciferase reporter assay in EpH4 cells but not in EpRas cells (Fig. 8). This result is consistent with the previously described loss of antiproliferative responses in EpRas cells (Oft et al. 1997). Transfection of either wild-type Smad3 or Smad3–EPSM into EpH4 cells did not significantly enhance this antimitogenic effect of TGFβ (Fig. 8A). However, Smad3–EPSM restored the ability of TGFβ to inhibit the E2F-1 reporter in EpRas cells, whereas wild-type Smad3 had only a small effect (Fig. 8A).
Figure 8.

Ras-resistant Smad3 rescues TGFβ antimitogenic and transcriptional responses in EpRas cells. EpH4 and EpRas cells were cotransfected with wild-type Smad3, Ras-resistant (EPSM) Smad3 mutant, or the corresponding empty vector. These vectors were cotransfected with the E2F–luciferase reporter construct (6×E2F–Luc) in A, or the Smad-responsive ARE–luciferase reporter construct (A3–Luc) and FAST2 in B. Cells were incubated with or without TGFβ and luciferase activity was then measured. Data are the average of triplicates ±s.d.
We also tested the effect of Smad3 or Smad3–EPSM transfection on the activation of the A3–Luc reporter (Fig. 8B). A3–Luc activation by TGFβ in EpH4 cells was only slightly increased by transfection of Smad3 or Smad3–EPSM vectors. However, transfection of Smad3–EPSM into EpRas cells strongly enhanced the otherwise limited response of A3–Luc to TGFβ, whereas transfection of the wild-type Smad3 had only a small effect (Fig. 8B). EpRas cells transfected with Smad3–EPSM vector had a high level of basal luciferase activity that may be due to spontaneous accumulation of the overexpressed product in the nucleus. These results further suggest that Smad phosphorylation by Ras-activated MAP kinases is responsible for the decreased responsiveness of Ras-transformed cells to TGFβ.
Discussion
We have investigated the mechanism of inhibition of TGFβ signaling by the Ras pathway in epithelial cells. Our results suggest that Ras activation induces the phosphorylation of MAP kinase sites in the linker region of Smad2 and Smad3, inhibiting the nuclear accumulation of the Smads and their ability to mediate TGFβ antiproliferative responses and other effects. In normal epithelial cells, this mechanism may function to adjust the level of Smad signaling according to the level of Ras activity. However, in the case of Ras hyperactivation by oncogenic mutations, a common event in several human cancers, the same mechanism may cause the loss of growth inhibition by TGFβ.
Repression of TGFβ/Smad signaling by the Ras pathway
Numerous reports have described a correlation between Ras transformation and deficient TGFβ responsiveness, particularly with regards to TGFβ antimitogenic responses. Ras transformation of lung, intestinal, liver, or mammary epithelial cells confers resistance to growth inhibition by TGFβ (Schwarz et al. 1988; Houck et al. 1989; Valverius et al. 1989; Longstreet et al. 1992; Filmus and Kerbel 1993). Microinjection of oncogenic H-Ras protein into TGFβ-arrested mink lung epithelial cells overcomes TGFβ growth inhibition and allows cell-cycle progression into S phase (Howe et al. 1993). Here, we describe a mechanism by which oncogenic Ras can induce such losses of TGFβ responsiveness.
Our results show that oncogenic Ras inhibits the TGFβ signal transduction pathway at the level of Smad. As a readout of Smad transcriptional activity we used the A3–Luc reporter because activation of the enhancer element ARE that drives gene expression in A3–Luc depends on the direct binding of a TGFβ-induced Smad (a TGFβ-induced Smad complex) (Chen et al. 1997; Liu et al. 1997). Oncogenic Ras inhibits the A3–Luc transcriptional response to TGFβ, both in mammary epithelial cells and lung epithelial cells. H-RasV12 decreases the amplitude of the A3–Luc response by as much as 90%. This is accompanied by a similar inhibition of the TGFβ antimitogenic response.
Ras signaling was shown previously to induce the phosphorylation of the BMP mediator Smad1 at four Erk consensus sites in the linker domain (Kretzschmar et al. 1997a). Phosphorylation of these four sites inhibits Smad1 accumulation in the nucleus without interfering with BMP-induced phosphorylation of Smad1. Many other serine residues present in the Smad1 linker region are not required for this effect (Kretzschmar et al. 1997a). A similar mechanism of regulation was not predicted for Smad2 or Smad3. These proteins are divergent from Smad1 in the linker region and have only one or two consensus Erk sites. However, our evidence suggests that clusters of four Ser/Thr–Pro sites including these Erk sites in the linker region of Smad2 and Smad3 are functionally equivalent to the four Erk sites of Smad1.
The following lines of evidence support the conclusion that Ras-induced phosphorylation of these sites prevents accumulation of Smad2 and Smad3 in the nucleus and inhibits TGFβ signaling. First, expression of oncogenic Ras elevates the phosphorylation of endogenous as well as exogenous Smad2 and Smad3. Second, Ras-induced phosphorylation of Smad2 and Smad3 is mimicked by expression of an activated Mek1 construct and is inhibited by addition of a specific Mek1 inhibitor. Third, these Ras- and Mek1-induced phosphorylations are prevented by mutation of the four Ser/Thr-Pro sites in the linker region of Smad2 and Smad3. These mutations also inhibit the phosphorylation of Smad2 and Smad3 by Erk2 in vitro. In contrast, mutation of the carboxy-terminal serines that serve as TGFβ receptor phosphorylation sites does not inhibit Ras-induced phosphorylation of Smad2 or Smad3. Fourth, expression of oncogenic Ras or constitutively active Mek1 inhibit the TGFβ-induced accumulation of Smad2 and Smad3 in the nucleus. Fifth, Mek1 does not inhibit the TGFβ-induced accumulation of a mutant Smad3 lacking the four Ras-dependent phosphorylation sites. Finally, expression of this Ras-resistant Smad3 mutant restores growth inhibitory and transcriptional responses to TGFβ in Ras-transformed cells.
Smad2 and Smad3 integrate antagonistic TGFβ and Ras signals
What is the role of this inhibitory mechanism in epithelial cells that contain a normal Ras pathway? We have approached this question by studying the effect of EGF, a physiological activator of this pathway (Vojtek and Der 1998). It should be noted that TGFβ is a potent inhibitor of the mitogenic response to EGF in Mv1Lu cells and other nontransformed epithelial cells (Like and Massagué 1986; Massagué 1990; Alexandrow and Moses 1995). TGFβ antagonizes these signals by inhibiting G1 cyclin-dependent kinases (Koff et al. 1993; Hannon and Beach 1994; Polyak et al. 1994; Iavarone and Massagué 1997; Reynisdóttir and Massagué 1997). Therefore, EGF may not be expected to cause an extensive inhibition of TGFβ signaling, and our results agree with this. Like oncogenic Ras, EGF elevates the phosphorylation of Smad2 and Smad3, and this requires the four Ser/Thr–Pro sites in the linker region but not the carboxy-terminal receptor phosphorylation sites. However, EGF, which is weaker than oncogenic mutations at activating Ras, only tones down the nuclear accumulation of Smad2 and Smad3. On balance, TGFβ prevails over EGF in the regulation of Smad2/Smad3 nuclear accumulation. Thus, physiological signals and oncogenic mutations that activate Ras induce phosphorylation of the same inhibitory sites in Smad2 and Smad3, but with different intensities and different outcomes. Physiological activators such as EGF may use this mechanism to adjust the ability of Smad2 and Smad3 to convey TGFβ signals, whereas oncogenic Ras mutations may use it to disrupt TGFβ signaling.
Adjusting the ability of Smad2 and Smad3 to convey TGFβ signals may be important in processes that are cooperatively stimulated by TGFβ and Ras signals. Activin-like TGFβ family members cooperate with Ras signals in the induction of mesoderm in Xenopus (Whitman and Melton 1992). During endoderm formation in Drosophila, Ras signals and the TGFβ family member Dpp synergize in the induction of a common target gene, Ubx, by activating separate sites in the Ubx promoter (Szüts et al. 1998). TGFβ and EGF acting together allow the proliferation of fibroblasts under anchorage-deficient conditions (Roberts and Sporn 1990). TGFβ may allow the mitogenic stimulation of anchorage-deprived fibroblasts by inducing formation of extracellular matrix and cell adhesion to this matrix (Ignotz and Massagué 1986, 1987). Integrin-mediated adhesion to an extracellular matrix is required for effective mitogenic stimulation (Assoian and Zhu 1997).
EGF and hepatocyte growth factor (HGF) have been shown to activate the TGFβ reporter plasmid p3TP–Lux (Cárcamo et al. 1995; de Caestecker et al. 1998). On the basis of these and related observations, it has been proposed that EGF and HGF may signal through Smad2 (de Caestecker et al. 1998). Compared with TGFβ, however, these factors induce minimal or no accumulation of Smad2 in the nucleus (de Caestecker et al. 1998; present results). It should be noted that the p3TP–Lux promoter contains three AP-1 sites (Cárcamo et al. 1995), and EGF can activate AP-1 directly through the Ras/MAPK pathway (Hunter and Karin 1992; Davis 1993). Other AP-1 activators such as phorbol esters also activate the 3TP–Lux construct (Cárcamo et al. 1995). Ras signaling slightly elevates the transcriptional activity of a GAL4–Smad2 fusion construct (de Caestecker et al. 1998), but Ras signaling has a general effect on transcription (Abdellatif et al. 1994) and can stimulate general transcriptional coactivators (Xu et al. 1998). Our results do not support the notion that EGF specifically signals through Smad2.
Implications for cancer
One physiological function of TGFβ is to constrain the proliferation of epithelial, endothelial, and hematopoietic cells, thus contributing to the maintenance of homeostasis in these tissues (Massagué 1990; Roberts and Sporn 1993; Alexandrow and Moses 1995). This function of TGFβ is often lost in cancer as a result of mutations that directly inactivate components of the growthinhibitory TGFβ/Smad signaling pathway including TβR-II, Smad2, and Smad4 (Markowitz et al. 1995; Eppert et al. 1996; Hahn et al. 1996; Heldin et al. 1997; T. Chen et al. 1998; Goggins et al. 1998; Massagué 1998; Grady et al. 1999). However, many tumor cells without known mutations in these components are refractory to growth inhibition by TGFβ. Understanding the mechanisms by which certain tumor cells selectively lose growth-inhibitory responses to TGFβ is therefore important for a better understanding of oncogenic processes.
We have investigated this problem in Ras-transformed mammary and lung epithelial cells. Our results suggest that hyperactive Ras can interfere with the TGFβ/Smad signaling pathway by inhibiting nuclear accumulation of Smad2/Smad3. This mechanism may be operative in tumors that harbor a hyperactive Ras pathway. A substantial proportion of colon carcinomas and pancreatic carcinomas contain oncogenic Ras mutations (Fearon and Vogelstein 1990; Kern 1998), whereas breast carcinomas often contain HER2 and EGFR receptor tyrosine kinase amplifications (Clark and Der 1995). The colon carcinoma cell lines of known Ras status that we have screened to date show a correlation between the presence of oncogenic Ras mutations and a deficient nuclear accumulation of Smad2/Smad3. Tumors that contain inactivating mutations in TβRII, Smad2, or Smad4 may suffer a more complete loss of TGFβ signaling than tumors containing Ras mutations. However, Ras mutations and mutations in TGFβ pathway components diminish TGFβ signaling with different outcomes. Ras transformation not only diminishes growth inhibition by TGFβ, but it subverts this pathway into stimulating epithelial-mesenchymal transdifferentiation, invasion, and metastasis (Welch et al. 1990; Caulin et al. 1995; Oft et al. 1996, 1998; Sehgal et al. 1996; Farina et al. 1998; Yin et al. 1999). The inhibitory mechanism described here may allow the emergence of these tumorigenic responses through the residual activity of Smads remaining in the tumor cells or through as yet unknown Smad-independent pathways.
Materials and methods
Cell lines
Parental Mv1Lu cells were obtained from the American Type Culture Collection. Mv1Lu/R1B-L17 is a derivative cell line defective in the TGFβ type I receptor (TβRI) that has a high transfection efficiency and responds to TGFβ on transfection of TβR-I (Wrana et al. 1994). EpH4 and EpRas were a generous gift of E. Reichmann (ISREC, Lausanne, Switzerland). Colon cancer cell lines were provided by N. Rosen (Memorial Sloan-Kettering Cancer Center, New York, NY).
Transfections and metabolic labeling
Most procedures were essentially carried out as described (Kretzschmar et al. 1997b). In brief, Mv1Lu/R1B-L17 mink lung epithelial cells were transiently transfected with Flag-tagged Smad2 or Smad3 constructs and H-RasV12 or caMek1 vectors as indicated. Smad constructs and H-RasV12 were in pCMV5 or pCS2, caMek1 was in pMCL. Mutant Smad constructs were obtained by standard in vitro mutagenesis procedures and were confirmed by sequencing. Three days post-transfection, cells were metabolically labeled for 3 hr with 32P and lysed. Where indicated, cells were treated with EGF (18 nm; R&D systems) for the indicated time, or with the Mek1 inhibitor PD98059 (100 μm; New England Biolabs), or the PI3-kinase inhibitor wortmannin (0.1 μm; Calbiochem) for 1.5 hr prior to cell lysis. After cell lysis, Flag–Smad2 and Flag–Smad3 were precipitated with anti-Flag antibody, resolved by SDS-PAGE, and visualized by autoradiography. Parallel cultures were treated equivalently, lysed, and subjected to Western immunoblotting with anti-Flag antibody and chemiluminescence (ECL, Amersham).
Reporter assays
Luciferase assays were essentially carried out as described (Kretzschmar et al. 1997a). The A3–Luc reporter construct was generated by subcloning the region comprising the three AREs and the core promoter from the previously described A3–CAT construct (Huang et al. 1995) into the pGL2–Basic luciferase vector (Promega). For A3–Luc assays, cells were transiently cotransfected with FAST-2 and A3–Luc vectors, serum-starved for 6 hr, treated with TGFβ at the indicated concentrations, and assayed for luciferase activity 18 hr after factor addition. For E2F–luciferase assays, cells were transiently transfected with empty vector or vector encoding wild-type Smad3 or Smad3–EPSM, as indicated, and the E2F–luciferase reporter construct 6×E2F–Luc (Lukas et al. 1997) as specified. Assays were carried out equivalently except that cells were not serum starved. EpH4 and EpRas cells were grown as described previously (Oft et al. 1996).
Antibodies
Anti-Smad2/Smad3 antibodies were raised in rabbits by immunization with the recombinant linker region of human Smad2 (amino acids 183–273), which is almost identical across species and highly homologous to the corresponding region in Smad3. These antibodies, referred to as anti-Smad2/Smad3, were affinity purified with immobilized Smad2 prior to use. Rabbit polyclonal antibodies raised against a peptide corresponding to amino acids 457–467 of Smad2, which included phosphorylated serine residues at positions 465 and 467 (Upstate Biotechnology, Lake Placid, NY) was used for Western immunoblotting of carboxyl terminus phosphorylated Smad2. Rabbit polyclonal anti-Erk antibodies used for Western immunoblotting (Upstate Biotechnology) and immunoprecipitation (Santa Cruz) recognize both Erk1 and Erk2. Monoclonal anti-Flag antibody (M2) was from Kodak Scientific.
Immunofluorescence assays
Vector, H-RasV12, caMek1, GFP, or Flag–Smad3 constructs were transiently transfected 48 hr prior to factor treatment. Cells were treated with TGFβ and/or EGF as specified and processed for immunofluorescence by a triple sandwich method (Harlow and Lane 1988). Endogenous Smad proteins were visualized with affinity-purified anti-Smad2/Smad3 antibodies, biotin-conjugated anti-rabbit immunoglobulin and either FITC- or rhodamine-conjugated streptavidin (Jackson ImmunoResearch, West Grove, PA). Exogenous Flag-tagged Smad proteins were visualized with the triple sandwich method by use of M2 anti-Flag monoclonal antibodies (Kodak) as primary antibodies. All slides were counterstained with DAPI to visualize cell nuclei (data not shown). In all cases, at least 100 stained cells were scored.
Kinase assays
Smad constructs were subcloned into a pET expression vector (Novagen) encoding an amino-terminal hexahistidine tag. Bacterial expression and purification of recombinant proteins were carried out as described (Kretzschmar et al. 1997b). Equivalent amounts of recombinant Smad3 proteins, as quantified by Coomassie staining and protein concentration assays, were incubated with recombinant, activated Erk2 MAP kinase (New England Biolabs) in the presence of [γ-32P]ATP at 28–30°C for 20 min. Reactions were stopped by addition of SDS loading buffer and proteins resolved by SDS-PAGE.
Phosphopeptide analysis
EpRas cells were metabolically labeled for 3 hr with 32P and lysed. Endogenous Smad2/Smad3 was immunoprecipitated with affinity-purified anti-Smad2/Smad3 and separated by SDS-PAGE. Endogenous Smad2/Smad3 was visualized by autoradiography and then electroeluted from the gel. The eluted protein was concentrated and aliquots incubated with trypsin or α-chymotrypsin (both from Worthington, Lakewood, NJ). Resulting phosphopeptides were resolved by SDS-PAGE and visualized by autoradiography.
Acknowledgments
We thank E. Reichmann for kindly providing the EpH4 and EpRas cell lines, N. Rosen and L. Sepp-Lorenzino for tumor cell lines, E. Lai for the FAST2 vector, R. Derynck for the Smad3 vector, S. Mansour for the caMek1 vector, and A. Fattaey for the E2F-luciferase reporter. We also acknowledge the use of the Molecular Cytology Core Facility at Memorial Sloan-Kettering Cancer Center. This work was supported by Breast SPORE (Specialized Program of Research Excellence) and Cancer Center, National Institutes of Health grants to J.M. and to Memorial Sloan-Kettering Cancer Center. M.K. was a recipient of a postdoctoral fellowship from the Deutsche Forschungsgemeinschaft. J.M. is an Investigator of the Howard Hughes Medical Institute.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL j-massague@ski.mskcc.org; FAX (212) 717-3298.
References
- Abdellatif M, MacLellan WR, Schneider MD. p21Ras as a governor of global gene expression. J Biol Chem. 1994;269:15423–15426. [PubMed] [Google Scholar]
- Alessi DR, Cuenda A, Cohen P, Dudley DT, Saltiel AR. PD 098059 is a specific inhibitor of the activation of mitogen-activated protein kinase kinase in vitro and in vivo. J Biol Chem. 1995;270:27489–27494. doi: 10.1074/jbc.270.46.27489. [DOI] [PubMed] [Google Scholar]
- Alexandrow MG, Moses HL. Transforming growth factor β and cell cycle regulation. Cancer Res. 1995;55:1452–1457. [PubMed] [Google Scholar]
- Assoian RK, Zhu X. Cell anchorage and the cytoskeleton as partners in growth factor dependent cell cycle progression. Curr Opin Cell Biol. 1997;9:93–98. doi: 10.1016/s0955-0674(97)80157-3. [DOI] [PubMed] [Google Scholar]
- Barrack ER. TGF-β in prostate cancer: A growth inhibitor that can enhance tumorigenicity. Prostate. 1997;31:61–70. doi: 10.1002/(sici)1097-0045(19970401)31:1<61::aid-pros10>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- Böttinger EP, Factor VM, Tsang MLS, Weatherbee JA, Kopp JB, Qian SW, Wakefield LM, Roberts AB, Thorgeirsson SS, Sporn MB. The recombinant proregion of transforming growth factor β1 (latency-associated peptide) inhibits active transforming growth factor β1 in transgenic mice. Proc Natl Acad Sci. 1996;93:5877–5882. doi: 10.1073/pnas.93.12.5877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cárcamo J, Zentella A, Massagué J. Disruption of TGF-β signaling by a mutation that prevents transphosphorylation within the receptor complex. Mol Cell Biol. 1995;15:1573–1581. doi: 10.1128/mcb.15.3.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caulin C, Scholl FG, Frontelo P, Gamallo C, Quintanilla M. Chronic exposure of cultured transformed mouse epidermal cells to transforming growth factor-β1 induces an epithelial-mesenchymal transdifferentiation and a spindle tumoral phenotype. Cell Growth Differ. 1995;6:1027–1035. [PubMed] [Google Scholar]
- Chen T, Carter D, Garrigue-Antar L, Reiss M. Transforming growth factor β type I receptor kinase mutant associated with metastatic breast cancer. Cancer Res. 1998;58:4805–4810. [PubMed] [Google Scholar]
- Chen X, Weisberg E, Fridmacher V, Watanabe M, Naco G, Whitman M. Smad4 and FAST-1 in the assembly of activin-responsive factor. Nature. 1997;389:85–89. doi: 10.1038/38008. [DOI] [PubMed] [Google Scholar]
- Chen Y, Rubock MJ, Whitman M. A transcriptional partner of MAD proteins in TGF-β signalling. Nature. 1996;383:691–696. doi: 10.1038/383691a0. [DOI] [PubMed] [Google Scholar]
- Chen YG, Hata A, Lo RS, Wotton D, Shi Y, Pavletich N, Massagué J. Determinants of specificity in TGF-β signal transduction. Genes & Dev. 1998;12:2144–2152. doi: 10.1101/gad.12.14.2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark GJ, Der CJ. Aberrant function of the Ras signal transduction pathway in human breast cancer. Breast Cancer Res Treat. 1995;35:133–144. doi: 10.1007/BF00694753. [DOI] [PubMed] [Google Scholar]
- Cui W, Fowlis DJ, Bryson S, Duffie E, Ireland H, Balmain A, Akhurst RJ. TGFβ1 inhibits the formation of benign skin tumors, but enhances progression to invasive spindle carcinomas in transgenic mice. Cell. 1996;86:531–542. doi: 10.1016/s0092-8674(00)80127-0. [DOI] [PubMed] [Google Scholar]
- Davis RJ. The mitogen-activated protein kinase signal transduction pathway. J Biol Chem. 1993;268:14553–14556. [PubMed] [Google Scholar]
- de Caestecker MP, Parks WT, Frank CJ, Castagnino P, Bottaro DP, Roberts AB, Lechleider RJ. Smad2 transduces common signals from receptor serine-threonine and tyrosine kinases. Genes & Dev. 1998;12:1587–1593. doi: 10.1101/gad.12.11.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derynck R, Jarrett JA, Chen EY, Eaton DH, Bell JR, Assoian RK, Roberts AB, Sporn MB, Goeddel DV. Human transforming growth factor-β complementary DNA sequence and expression in normal and transformed cells. Nature. 1985;316:701–705. doi: 10.1038/316701a0. [DOI] [PubMed] [Google Scholar]
- Eppert K, Scherer SW, Ozcelik H, Pirone R, Hoodless P, Kim H, Tsui L-C, Bapat B, Gallinger S, Andrulis IL, Thomsen GH, Wrana JL, Attisano L. MADR2 maps to 18q21 and encodes a TGFβ-regulated MAD-related protein that is functionally mutated in colorectal carcinoma. Cell. 1996;86:543–552. doi: 10.1016/s0092-8674(00)80128-2. [DOI] [PubMed] [Google Scholar]
- Factor VM, Cao C-Y, Santoni-Rugiu E, Woitach JT, Jensen MR, Thorgeison SS. Constitutive expression of mature transforming growth factor β1 in the liver accelerates hepatocarcinogenesis in transgenic mice. Cancer Res. 1997;57:2089–2095. [PubMed] [Google Scholar]
- Farina AR, Coppa A, Tiberio A, Tacconelli A, Turco A, Colletta G, Gulino A, Mackay AR. Transforming growth factor-β1 enhances the invasiveness of human MDA-MB-231 breast cancer cells by up-regulating urokinase activity. Int J Cancer. 1998;75:721–730. doi: 10.1002/(sici)1097-0215(19980302)75:5<721::aid-ijc10>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- Filmus J, Kerbel RS. Development of resistance mechanisms to growth-inhibitory effects of transforming growth factor-β during tumor progression. Curr Opin Oncol. 1993;5:123–129. [PubMed] [Google Scholar]
- Goggins M, Shekher M, Turnacioglu K, Yeo CJ, Hruban RH, Kern SE. Genetic alterations of the transforming growth factor β receptor genes in pancreatic and biliary adenocarcinomas. Cancer Res. 1998;58:5329–5332. [PubMed] [Google Scholar]
- Gorska AE, Joseph H, Derynck R, Moses HL, Serra R. Dominant-negative interference of the transforming growth factor β type II receptor in mammary gland epithelium results in alveolar hyperplasia and differentiation in virgin mice. Cell Growth Differ. 1998;9:229–238. [PubMed] [Google Scholar]
- Grady WM, Myeroff LL, Swinler SE, Rajput A, Thiagalingam S, Lutterbaugh JD, Neumann A, Chang J, Kim S-J, Kinzler KW, Vogelstein B, Willson JKV, Markowitz S. Mutational inactivation of transforming growth factor β receptor type II in microsatelite stable colon cancers. Cancer Res. 1999;59:320–324. [PubMed] [Google Scholar]
- Hahn SA, Schutte M, Hoque ATMS, Moskaluk CA, da Costa LT, Rozenblum E, Weinstein CL, Fischer A, Yeo CJ, Hruban RH, Kern SE. DPC4, a candidate tumor suppressor gene at human chromosome 18q21.1. Science. 1996;271:350–353. doi: 10.1126/science.271.5247.350. [DOI] [PubMed] [Google Scholar]
- Hannon GJ, Beach D. p15INK4B is a potential effector of TGF-β-induced cell cycle arrest. Nature. 1994;371:257–261. doi: 10.1038/371257a0. [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane D. Antibodies: A laboratory handbook. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- Heldin C-H, Miyazono K, ten Dijke P. TGF-β signalling from cell membrane to nucleus through SMAD proteins. Nature. 1997;390:465–471. doi: 10.1038/37284. [DOI] [PubMed] [Google Scholar]
- Houck KA, Michalopoulos GK, Strom SC. Introduction of a Ha-ras oncogene into rat liver epithelial cells and parenchymal hepatocytes confers resisitance to the growth inhibitory effects of TGF-β. Oncogene. 1989;4:19–25. [PubMed] [Google Scholar]
- Howe PH, Dobrowolski SF, Reddy KB, Stacey DW. Release from G1 growth arrest by transforming growth factor β1 requires ras activity. J Biol Chem. 1993;268:21448–21452. [PubMed] [Google Scholar]
- Huang H-C, Murtaugh LC, Vize PD, Whitman M. Identification of a potential regulator of early transcriptional responses to mesoderm inducers in the frog embryo. EMBO J. 1995;14:5965–5973. doi: 10.1002/j.1460-2075.1995.tb00285.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter T, Karin M. The regulation of transcription by phosphorylation. Cell. 1992;70:375–387. doi: 10.1016/0092-8674(92)90162-6. [DOI] [PubMed] [Google Scholar]
- Iavarone A, Massagué J. Repression of the CDK activator Cdc25A and cell-cycle arrest by cytokine TGF-β in cells lacking the CDK inhibitor p15. Nature. 1997;387:417–422. doi: 10.1038/387417a0. [DOI] [PubMed] [Google Scholar]
- Ignotz RA, Massagué J. Transforming growth factor-β stimulates the expression of fibronectin and collagen and their incorporation into the extracellular matrix. J Biol Chem. 1986;261:4337–4345. [PubMed] [Google Scholar]
- ————— Cell adhesion protein receptors as targets for transforming growth factor-β. Cell. 1987;51:189–197. doi: 10.1016/0092-8674(87)90146-2. [DOI] [PubMed] [Google Scholar]
- Kern SE. Advances from genetic clues in pancreatic cancer. Curr Opin Oncol. 1998;10:74–80. doi: 10.1097/00001622-199801000-00012. [DOI] [PubMed] [Google Scholar]
- Keski-Oja J, Leof EB, Lyons RM, Coffey RJJ, Moses HL. Transforming growth factors and control of neoplastic cell growth. J Cell Biochem. 1987;33:95–107. doi: 10.1002/jcb.240330204. [DOI] [PubMed] [Google Scholar]
- Koff A, Ohtsuki M, Polyak K, Roberts JM, Massagué J. Negative regulation of G1 in mammalian cells: Inhibition of cyclin E-dependent kinase by TGF-β. Science. 1993;260:536–539. doi: 10.1126/science.8475385. [DOI] [PubMed] [Google Scholar]
- Kretzschmar M, Doody J, Massagué J. Opposing BMP and EGF signalling pathways converge on the TGFβ family mediator Smad1. Nature. 1997a;389:618–622. doi: 10.1038/39348. [DOI] [PubMed] [Google Scholar]
- Kretzschmar M, Liu F, Hata A, Doody J, Massagué J. The TGF-β mediator Smad1 is directly phosphorylated and functionally activated by the BMP receptor kinase. Genes & Dev. 1997b;11:984–995. doi: 10.1101/gad.11.8.984. [DOI] [PubMed] [Google Scholar]
- Labbé E, Silvestri C, Hoodless PA, Wrana JL, Attisano L. Smad2 and Smad3 positively and negatively regulate TGFβ-dependent transcription through the forkhead DNA-binding protein FAST2. Mol Cell. 1998;2:109–120. doi: 10.1016/s1097-2765(00)80119-7. [DOI] [PubMed] [Google Scholar]
- Letterio JL, Roberts AB. Regulation of immune responses by TGF-β. Annu Rev Immunol. 1998;16:137–161. doi: 10.1146/annurev.immunol.16.1.137. [DOI] [PubMed] [Google Scholar]
- Like B, Massagué J. The antiproliferative effect of type β transforming growth factor occurs at a level distal from receptors for growth-activating factors. J Biol Chem. 1986;261:13426–13429. [PubMed] [Google Scholar]
- Liu B, Dou C, Prabhu L, Lai E. Fast2 is a mammalian winged helix protein that mediates TGFβ signals. Mol Cell Biol. 1999;19:424–430. doi: 10.1128/mcb.19.1.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Pouponnot C, Massagué J. Dual role of the Smad4/DPC4 tumor suppressor in TGFβ-inducible transcriptional responses. Genes & Dev. 1997;11:3157–3167. doi: 10.1101/gad.11.23.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longstreet M, Miller B, Howe PH. Loss of transforming growth factor β 1 (TGF-β1)-induced growth arrest and p34cdc2 regulation in ras-transfected epithelial cells. Oncogene. 1992;7:1549–1556. [PubMed] [Google Scholar]
- Lukas J, Herzinger T, Hansen K, Moroni MC, Resnitzky D, Helin K, Reed SI, Bartek J. Cyclin E-induced S phase without activation of the pRb/E2F pathway. Genes & Dev. 1997;11:1479–1492. doi: 10.1101/gad.11.11.1479. [DOI] [PubMed] [Google Scholar]
- Macias-Silva M, Abdollah S, Hoodless PA, Pirone R, Attisano L, Wrana JL. MADR2 is a substrate of the TGFβ receptor and phosphorylation is required for nuclear accumulation and signaling. Cell. 1996;87:1215–1224. doi: 10.1016/s0092-8674(00)81817-6. [DOI] [PubMed] [Google Scholar]
- Markowitz S, Wang J, Myeroff L, Parsons R, Sun L, Lutterbaugh J, Fan RS, Zborowska E, Kinzler KW, Vogelstein B, Brattain M, Wilson JKV. Inactivation of the type II TGF-β receptor in colon cancer cells with microsatellite instability. Science. 1995;268:1336–1338. doi: 10.1126/science.7761852. [DOI] [PubMed] [Google Scholar]
- Massagué J. The transforming growth factor-β family. Annu Rev Cell Biol. 1990;6:597–641. doi: 10.1146/annurev.cb.06.110190.003121. [DOI] [PubMed] [Google Scholar]
- ————— TGFβ signal transduction. Annu Rev Biochem. 1998;67:753–791. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- Oft M, Peli J, Rudaz C, Schwarz H, Beug H, Reichmann E. TGF-β1 and Ha-Ras collaborate in modulating the phenotypic plasticity and invasiveness of epithelial tumor cells. Genes & Dev. 1996;10:2462–2477. doi: 10.1101/gad.10.19.2462. [DOI] [PubMed] [Google Scholar]
- Oft M, Heider KH, Burg H. TGF-β signaling is necessary for carcinoma cell invasiveness and metastasis. Curr Biol. 1998;8:1243–1252. doi: 10.1016/s0960-9822(07)00533-7. [DOI] [PubMed] [Google Scholar]
- Polyak K, Lee M-H, Erdjument-Bromage H, Koff A, Roberts JM, Tempst P, Massagué J. Cloning of p27kip1, a cyclin-dependent kinase inhibitor and a potential mediator of extracellular antimitogenic signals. Cell. 1994;78:59–66. doi: 10.1016/0092-8674(94)90572-x. [DOI] [PubMed] [Google Scholar]
- Reiss M, Barcellos-Hoff MH. Transforming growth factor-β in breast cancer: A working hypothesis. Breast Cancer Res Treat. 1997;45:81–95. doi: 10.1023/a:1005865812918. [DOI] [PubMed] [Google Scholar]
- Reynisdóttir I, Massagué J. The subcellular location of p15Ink4b and p27Kip1 coordinate their inhibitory interactions with cdk4 and cdk2. Genes & Dev. 1997;11:492–503. doi: 10.1101/gad.11.4.492. [DOI] [PubMed] [Google Scholar]
- Roberts AB, Sporn MB. The transforming growth factor-betas. In: Sporn MB, Roberts AB, editors. Peptide growth factors and their receptors. Heidelberg, Germany: Springer-Verlag; 1990. pp. 419–472. [Google Scholar]
- ————— Physiological actions and clinical applications of transforming growth factor-β (TGF-β) Growth Factors. 1993;8:1–9. doi: 10.3109/08977199309029129. [DOI] [PubMed] [Google Scholar]
- Schwarz LC, Gingras MC, Goldberg G, Greenberg AH, Wright JA. Loss of growth factor dependence and conversion of transforming growth factor-β1 inhibition to stimulation in metastatic H-ras-transformed murine fibroblasts. Cancer Res. 1988;48:6999–7003. [PubMed] [Google Scholar]
- Sehgal I, Baley PA, Thompson TC. Transforming growth factor β1 stimulates contrasting responses in metastatic versus primary mouse prostate cancer-derived cell lines in vitro. Cancer Res. 1996;56:3359–3365. [PubMed] [Google Scholar]
- Sepp-Lorenzino L, Ma Z, Rands E, Kohl NE, Gibbs JB, Oliff A, Rosen N. A peptidomimetic inhibitor of farnesyl:protein transferase blocks the anchorage-dependent and -independent growth of human tumor cell lines. Cancer Res. 1995;55:5302–5309. [PubMed] [Google Scholar]
- Shi Y, Hata A, Lo RS, Massagué J, Pavletich NP. A structural basis for the mutational inactivation of the Smad4 tumour suppressor. Nature. 1997;388:87–93. doi: 10.1038/40431. [DOI] [PubMed] [Google Scholar]
- Shi Y, Wang Y-F, Jayaraman L, Yang H, Massagué J, Pavletich N. Crystal structure of Smad MH1 domain bound to DNA: Insights on DNA-binding and TGFβ signaling. Cell. 1998;94:585–594. doi: 10.1016/s0092-8674(00)81600-1. [DOI] [PubMed] [Google Scholar]
- Szüts D, Eresh S, Bienz M. Functional intertwining of Dpp and EGFR signaling during Drosophila endoderm induction. Genes & Dev. 1998;12:2022–2035. doi: 10.1101/gad.12.13.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaku K, Oshima M, Miyoshi H, Matsui M, Seldin MF, Taketo MM. Instestinal tumorigenesis in compound mutant mice of both Dpc4 (Smad4) and APC genes. Cell. 1998;92:645–656. doi: 10.1016/s0092-8674(00)81132-0. [DOI] [PubMed] [Google Scholar]
- Ui M, Okada T, Hazeki K, Hazeki O. Wortmannin as a unique probe for an intracellular signalling protein, phosphoinositide 3-kinase. Trends Biochem Sci. 1995;20:303–307. doi: 10.1016/s0968-0004(00)89056-8. [DOI] [PubMed] [Google Scholar]
- Valverius EM, Walker-Jones D, Bates SE, Stampfer MR, Clark R, McCormick F, Dickson RB, Lippman ME. Production and responsiveness to transforming growth factor-β in normal and oncogene-transformed human mammary epithelial cells. Cancer Res. 1989;49:6269–6274. [PubMed] [Google Scholar]
- Vojtek AB, Der CJ. Increasing complexity of the Ras signaling pathway. J Biol Chem. 1998;273:19925–19928. doi: 10.1074/jbc.273.32.19925. [DOI] [PubMed] [Google Scholar]
- Wang XJ, Greenhalgh DA, Bickenbach JR, Jiang A, Bundman DS, Krieg T, Derynck R, Roop DR. Expression of a dominant-negative type II transforming growth factor β (TGF-β) receptor in the epidermis of transgenic mice blocks TGF-β-mediated growth inhibition. Proc Natl Acad Sci. 1997;94:2386–2391. doi: 10.1073/pnas.94.6.2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch DR, Fabra A, Nakajima M. Transforming growth factor β stimulates mammary adenocarcinoma cell invasion and metastatic potential. Proc Natl Acad Sci. 1990;87:7678–7682. doi: 10.1073/pnas.87.19.7678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitman M, Melton DA. Involvement of p21ras in Xenopus mesoderm induction. Nature. 1992;357:252–254. doi: 10.1038/357252a0. [DOI] [PubMed] [Google Scholar]
- Wrana JL, Attisano L, Wieser R, Ventura F, Massagué J. Mechanism of activation of the TGF-β receptor. Nature. 1994;370:341–347. doi: 10.1038/370341a0. [DOI] [PubMed] [Google Scholar]
- Xu L, Lavinsky RM, Dasen JS, Flynn SE, McInerney EM, Mullen T-M, Heinzel T, Szeto D, Korzus E, Kurokawa R, Aggarwal AK, Rose DW, Glass CK, Rosenfeld MG. Signal-specific co-activator domain requirements for Pit-1 activation. Nature. 1998;395:301–306. doi: 10.1038/26270. [DOI] [PubMed] [Google Scholar]
- Yin JJ, Selander K, Chirwing JM, Dallas M, Grubbs BG, Wieser R, Massagué J, Mundy GR, Guise TA. Blockade of transforming growth factor β signaling inhibits parathyroid hormone-related protein secretion by breast cancer cells and the development of bone metastases. J Clin Invest. 1999;103:197–206. doi: 10.1172/JCI3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Feng X-H, Wu R-Y, Derynck R. Receptor-associated Mad homologs synergize as effectors of the TGFβ response. Nature. 1996;383:168–172. doi: 10.1038/383168a0. [DOI] [PubMed] [Google Scholar]
- Zhou S, Zawel L, Lengauer C, Kinzler KW, Vogelstein B. Characterization of human FAST-1, a TGFβ and activin signal transducer. Mol Cell. 1998;2:121–127. doi: 10.1016/s1097-2765(00)80120-3. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Richardson JA, Parada LF, Graff JM. Smad3 mutant mice develop metastatic colorectal carcinoma. Cell. 1998;94:703–714. doi: 10.1016/s0092-8674(00)81730-4. [DOI] [PubMed] [Google Scholar]