Abstract
To analyze the molecular epidemiology of Mycobacterium tuberculosis strains at a hospital in Buenos Aires, Argentina, and mutations related to multidrug-resistant and extensively drug-resistant tuberculosis, we conducted a prospective case–control study. Our findings reinforce the value of incorporating already standardized molecular methods for rapidly detecting resistance.
Keywords: Tuberculosis, tuberculosis and other mycobacteria, molecular epidemiology, drug resistance, Argentina, Mycobacterium tuberculosis, dispatch
During the 1990s, an outbreak of multidrug-resistant (MDR) tuberculosis (TB) in HIV-positive patients occurred at the Muñiz Hospital in Buenos Aires, Argentina (1). Molecular analysis showed that a member of Haarlem2 family of Mycobacterium tuberculosis was responsible (1).
We conducted a prospective case-control study during June 1, 2006–April 30, 2007, at this 300-bed public hospital, which reports ≈40% of new TB cases in the city (2). Our primary aims were to analyze the molecular epidemiology of M. tuberculosis strains circulating at the hospital and the mutations related to MDR TB and extensively drug-resistant TB.
The Study
The strains were isolated and tested for antimicrobial drug susceptibility according to the proportion method (3) at the Muñiz Hospital Mycobacteria Laboratory. A proportion were tested for reserve drugs at the Health Protection Agency National Mycobacteria Reference Unit Laboratory, London, UK. DNA was extracted from cultures at this UK laboratory; spoligotyping was performed according to the manufacturer’s instructions (Isogen Life Science, IJsselstein, the Netherlands); and data were analyzed with SPOTCLUST (http://cgi2.cs.rpi.edu/~bennek/SPOTCLUST.html).
We performed 15-locus variable number tandem repeat (VNTR) using a CEQ 8000 Genetic Analysis System (Beckman Coulter, Inc., Fullerton, CA, USA) (4). Cluster analysis was performed by using Bionumerics software (Applied Maths, St-Martens-Latem, Belgium). Strains lacking a unique pattern were subjected to further analysis with an expanded set of VNTR loci (5).
In-house macroarrays were performed on MDR TB strains (6) to identify mutations in the katG and the inhA genes. Two regions of the rpoB gene of MDR TB strains were sequenced with the CEQ 8000 Genetic Analysis System. Statistical analyses were conducted by using χ2 and Fisher exact tests.
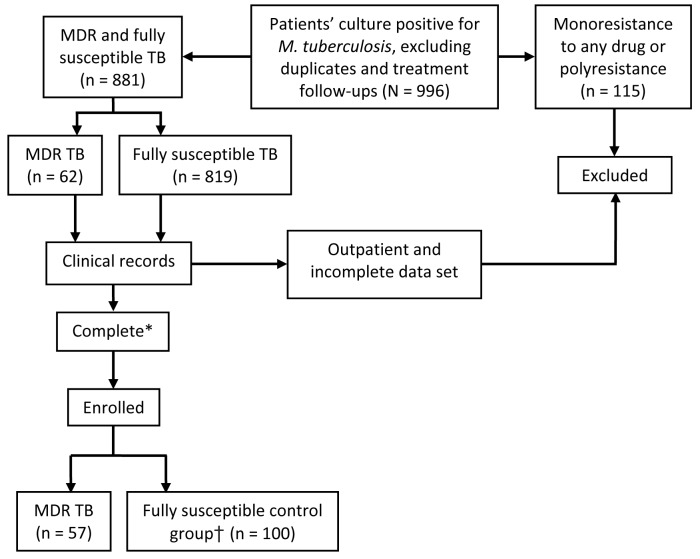
After we excluded duplicates, treatment follow-ups, and strains with susceptibility patterns other than MDR TB or susceptibility to all drugs tested, 881 strains were susceptible to all drugs tested. Patients with a minimum dataset (name, sex, date of birth or age, TB presentation [i.e., pulmonary or nonpulmonary], and at least 1 sign or symptom describing TB illness and treatment received) were enrolled: 57 of 62 hospitalized patients with MDR TB cultures (Table) and 100 fully susceptible unmatched inpatient controls, for a total of 157 patients. This convenience sample (Figure 1) included only admitted patients because of the difficulty of obtaining clinical information about outpatients.
Figure 1.
Multistage cluster sampling method for a study of the molecular epidemiology of Mycobacterium tuberculosis, Buenos Aires, Argentina, June 1, 2006–April 30, 2007. *Complete, having all but one of the following data: name, gender, date of birth or age, tuberculosis (TB) presentation, >1 sign or symptom describing the presentation and treatment received. †Unmatched controls. MDR, multidrug-resistant.
Table. Demographic information for 157 patients in a study of the molecular epidemiology of TB, Buenos Aires, Argentina, June 1, 2006–April 30, 2007*.
| Demographic characteristic |
Patients with MDR TB, n = 57, no. (%)† |
Patients with non–MDR TB, n = 100, no. (%)‡ |
p value, OR (95% CI)§ |
| Sex | |||
| M | 35 (61) | 70 (70) | 0.271, 1.4667 (0.7404–2.9055) |
| F |
22 (39) |
30 (30) |
|
| Location | |||
| Buenos Aires area | 46 (81) | 95 (95) | 0.004, 4.5435 (1.491–13.845) |
| Other |
11 (19) |
5 (5) |
|
| Country of birth | |||
| Argentina | 43 (75) | 66 (66) | 0.2176, 0.632 (0.3041–1.3133) |
| Bolivia | 6 (11) | 20 (20) | |
| Peru | 7 (12) | 8 (8) | |
| Paraguay | 0 | 3 (3) | |
| Uruguay | 1 (2) | 1 (1) | |
| Chile | 0 | 1 (1) | |
| Missing data |
0 |
1 (1) |
|
| Education | |||
| Illiterate or some primary | 16 (28) | 32 (32) | 0.2059, 0.5185 (0.1860–1.4456) |
| Some secondary or tertiary | 7 (12) | 27 (27) | |
| Missing data |
34 (59) |
41 (41) |
|
| Occupation | |||
| Unemployed | 7 (12) | ||
| Construction and manual worker | 20 (35) | ||
| Factory worker | 4 (7) | 14 (14) | |
| Health care worker | 1 (2) | 1 (1) | |
| Education, i.e., student and teacher | 2 (4) | 4 (4) | |
| Housewife | 6 (11) | 5 (5) | |
| Missing data |
17 (30) |
23 (23) |
|
| HIV infection | |||
| Positive | 25 (44) | 27 (27) | 0.04, 0.4737 (0.2308–0.9722) |
| Negative | 25 (44) | 57 (57) | |
| Missing data |
7 (12) |
16 (16) |
|
| Nature of TB contact | |||
| Close (i.e., household, family, co-worker) | 10 (18) | 32 (32) | |
| Institution (i.e., hospital, prison) | 2 (4) | 3 (3) | |
| Casual (e.g., acquaintance) | 5 (9) | 3 (3) | |
| Missing data |
40 (70) |
62 (62) |
|
| TB presentation | |||
| Pulmonary | 36 (63) | 61 (61) | 0.7184, 1.1553 (0.5323–2.5073) |
| Nonpulmonary | 15 (26) | 22 (22) | |
| Missing data |
6 (11) |
17 (17) |
|
| *TB, tuberculosis; MDR, multidrug resistant; OR, odds ratio; CI, confidence interval; IQR, interquartile range. †Median age, y (IQR) for patients with MDR TB: 34 (27–40). ‡Median age, y (IQR) for patients with non-MDR TB: 28.5 (23.0–37.0). §χ2 test. | |||
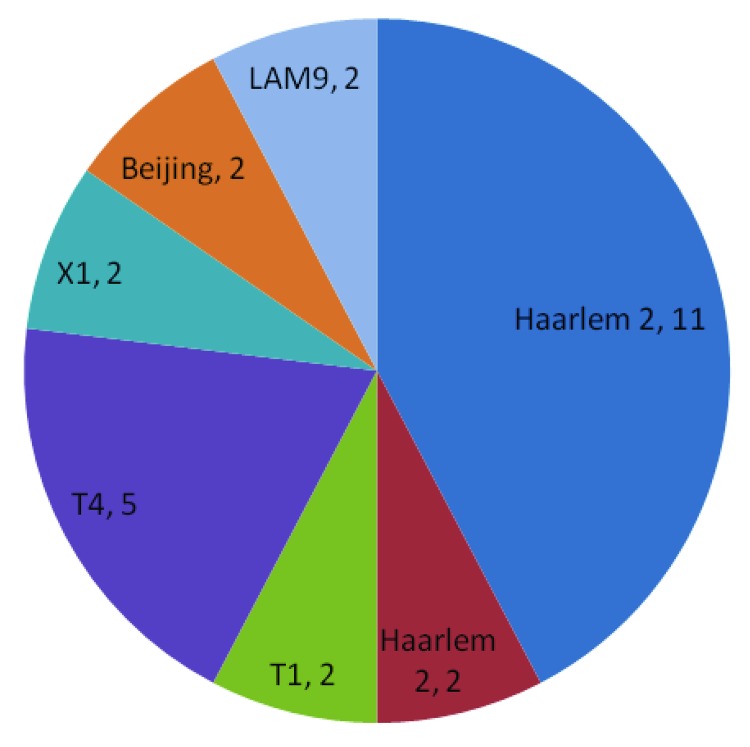
The most common spoligofamilies were T1 (18%), Haarlem 1 (10%), 2 (16%), and 3 (11%), LAM 3 (10%), and LAM 9 (13%). Initial 15-loci VNTR analysis showed 73 strains with unique patterns. Further analysis using 7-loci VNTR was performed on those that did not have a unique pattern. Twenty-six isolates remained clustered (Figure 2).
Figure 2.
Final cluster results in a study of the molecular epidemiology of Mycobacterium tuberculosis, by spoligofamily, Buenos Aires, Argentina, June 1, 2006–April 30, 2007.
Of the 57 MDR TB strains, 43 had a mutation in the katG315 locus, and 6 had a mutation in the inhA region. No strain had mutations in both genes. In 8 strains, resistance to isoniazid was not mediated by mutations in any of them.
Mutations in the rpoB region were detected by sequencing. The most frequent mutation was the S531L. Mutations at >1 site were rare. In 3 cases, we found 2 point mutations in the same codon. Only 1 MDR TB strain had no mutation in the rpoB segment sequenced; it also had a katG and inhA wild type. Complete susceptibility profile for all 57 MDR TB strains is available in Table A1.
In addition, we detected 5 extensively drug-resistant TB strains. None were clustered by 22-loci VNTR typing.
Conclusions
Spoligotyping identified predominance of the Haarlem family among the MDR TB cases (family responsible for the 1990s [1] outbreak) as well as the LAM and T families. A similar strain family distribution was reported for the French Departments of the Americas (7) and Turkey (8). The Beijing family was seldom encountered in these areas, which is in line with recent observations in 7 countries in South America, including Argentina (9).
The MDR TB Haarlem2 strain appears to be more successful than other circulating MDR TB strains and than its susceptible counterpart (of 25 Haarlem2 strains, 20 were MDR TB). This phenomenon could be associated with a bias in the sample resulting from the specialized nature of the hospital or it could be that the MDR TB version has become predominant in the population because of the low fitness cost of its 2 mutations, katG315 and S531L (10,11))). In addition, the presence of clusters suggests that even though new technologies have reduced the time taken to diagnose drug resistance, more rapid initial diagnosis of MDR TB to reduce transmission still is needed (12).
All except 1 of the rpoB mutations in the MDR TB strains were at nt positions 1303–1375. This finding reinforces the value of incorporating already standardized molecular methods for rapidly detecting resistance. Cost is the main reason they are currently not available, but the macroarrays used in this project are inexpensive and have the additional advantage of analyzing katG and inhA mutations independently.
Acknowledgments
We thank Juan Metrebian and Edward Hogg for their constant support and encouragement and Preya Vekaria for her advice and help.
Biography
Dr Gonzalo is a clinician and microbiologist working in the private and public health sector in Argentina on HIV, tuberculosis, and clinical microbiology. Her research interests include tuberculosis, mycobacteria, molecular epidemiology, and HIV.
Table A1. Drug susceptibility profiles for 57 patients with MDR TB, Buenos Aires, Argentina, June 1, 2006–April 30, 2007*.
| Patient no. | INH | Rif | P | E | Str | PAS | Cs | Pto | Eto | Cm | Am | Km | Cip | Ofx | Lfx | Mfx |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | R | R | S | S | R | S | S | – | – | – | – | S | – | – | – | – |
| 2 | R | R | R | – | R | – | – | – | S | – | – | – | – | – | – | – |
| 3 | R | R | S | S | R | S | – | – | – | – | – | S | – | – | – | – |
| 4 | R | R | R | R | R | S | S | – | – | – | – | R | – | R | – | – |
| 5 | R | R | R | R | R | S | S | – | – | – | – | – | – | – | – | – |
| 6 | R | R | R | R | R | R | S | – | – | S | – | R | – | S | – | S |
| 7 | R | R | – | R | R | – | – | – | – | – | – | – | – | – | – | – |
| 8 | R | R | R | S | R | S | S | – | – | – | – | R | – | – | – | – |
| 9 | R | R | – | S | R | S | S | – | – | – | – | – | – | – | – | – |
| 10 | R | R | R | R | R | – | – | – | – | – | R | – | – | – | – | – |
| 11 | R | R | R | R | R | S | S | – | – | – | – | – | – | – | – | – |
| 12 | R | R | R | R | R | S | S | – | – | – | – | – | – | – | – | – |
| 13 | R | R | R | R | R | S | S | – | – | – | – | – | – | – | – | – |
| 14 | R | R | S | R | R | S | S | – | – | – | – | – | – | – | – | – |
| 15 | R | R | R | R | R | S | S | S | S | R | R | R | – | S | – | S |
| 16 | R | R | R | R | R | – | – | S | S | R | R | R | – | S | – | S |
| 17 | R | R | R | R | R | S | S | – | – | – | – | R | – | – | – | – |
| 18 | R | R | R | R | R | S | S | S | R | R | R | R | – | S | – | S |
| 19 | R | R | R | R | R | S | S | S | S | R | R | R | S | S | – | S |
| 20 | R | R | R | S | R | S | – | S | S | R | R | R | – | S | – | S |
| 21 | R | R | R | R | R | – | – | S | S | R | R | R | S | S | – | S |
| 22 | R | R | R | R | S | S | – | – | – | – | – | R | – | – | – | – |
| 23 | R | R | R | R | R | – | – | – | – | R | R | R | – | R | – | R |
| 24 | R | R | R | R | R | S | R | R | R | S | S | R | – | S | – | S |
| 25 | R | R | S | R | R | S | S | – | – | – | – | S | – | – | – | – |
| 26 | R | R | R | R | R | S | S | – | – | – | – | – | – | – | – | – |
| 27 | R | R | R | R | R | R | S | – | – | – | – | R | – | R | – | – |
| 28 | R | R | R | R | R | R | S | – | – | – | – | R | – | S | – | – |
| 29 | R | R | R | – | S | S | S | – | – | – | – | – | – | – | – | – |
| 30 | R | R | R | S | S | S | S | – | – | – | – | S | – | – | – | – |
| 31 | R | R | S | S | R | S | S | – | – | – | – | S | – | – | – | – |
| 32 | R | R | S | S | R | S | S | – | – | – | – | S | – | – | – | – |
| 33 | R | R | R | R | S | S | – | – | – | – | – | S | – | – | – | – |
| 34 | R | R | – | S | R | S | – | – | – | – | – | – | – | – | – | – |
| 35 | R | R | R | R | S | S | S | – | – | – | – | S | – | S | – | – |
| 36 | R | R | S | R | S | S | S | – | – | – | – | – | – | – | – | – |
| 37 | R | R | S | S | S | S | – | – | – | – | – | – | – | – | – | – |
| 38 | R | R | R | S | S | S | S | – | – | – | – | S | – | – | – | – |
| 39 | R | R | R | R | R | S | S | – | – | – | R | R | – | R | – | – |
| 40 | R | R | S | R | R | S | S | – | – | S | – | S | – | S | – | – |
| 41 | R | R | R | R | S | S | S | – | – | – | – | S | – | – | – | – |
| 42 | R | R | – | R | R | S | S | – | – | – | – | R | – | – | – | – |
| 43 | R | R | – | S | S | S | – | S | R | S | S | S | – | S | – | S |
| 44 | R | R | R | R | R | R | S | – | – | – | – | – | – | S | – | – |
| 45 | R | R | R | R | R | S | S | – | – | – | – | R | – | – | – | – |
| 46 | R | R | R | R | R | S | S | – | – | – | – | R | – | – | – | – |
| 47 | R | R | S | S | R | S | – | – | – | – | – | S | – | – | – | – |
| 48 | R | R | R | S | S | S | – | – | – | – | – | – | – | – | – | – |
| 49 | R | R | S | S | R | S | S | – | – | – | – | S | – | – | – | – |
| 50 | R | R | R | R | R | R | R | – | – | S | – | R | R | R | – | S |
| 51 | R | R | S | R | S | S | – | – | – | – | – | – | – | – | – | – |
| 52 | R | R | S | S | S | S | S | – | – | – | – | – | – | – | – | – |
| 53 | R | R | R | S | S | S | – | R | R | – | – | S | – | S | – | – |
| 54 | R | R | R | R | R | S | S | – | – | – | – | R | – | – | – | – |
| 55 | R | R | – | S | S | – | – | – | – | – | – | – | – | – | – | – |
| 56 | R | R | – | S | S | S | S | – | – | – | – | S | – | – | – | – |
| 57 | R | R | S | R | R | S | S | – | – | – | – | S | – | – | – | – |
*MDR, multidrug resistant; TB, tuberculosis; INH, isoniazid; Rif, rifampicin; P, pyrazinamide; E, ethambutol; Str, streptomycin; PAS, p-aminosalicylic acid; Cs, cycloserine; Pto, proteonamide; Eto, ethionamide; Cm, capreomycin; Am, amikacin; Km, kanamycin; Cip, ciprofloxacin; Ofx, ofloxacin; Lfx, levofloxacin; Mfx, moxifloxacin; R, resistant; S, susceptible; –, drugs not tested for those strains.
Footnotes
Suggested citation for this article: Gonzalo X, Ambroggi M, Cordova E, Brown T, Poggi S, Drobniewski F. Molecular epidemiology of Mycobacterium tuberculosis, Buenos Aires, Argentina. Emerg Infect Dis [serial on the Internet]. 2011 Mar [date cited]. http://dx.doi.org/10.3201/eid1703100394
References
- 1.Ritacco V, Di Lonardo M, Reniero A, Ambroggi M, Barrera L, Dambrosi A, et al. Nosocomial spread of human immunodeficiency virus–related multidrug-resistant tuberculosis in Buenos Aires. J Infect Dis. 1997;176:637–42. 10.1086/514084 [DOI] [PubMed] [Google Scholar]
- 2.Comes Y, Forlenza R. Analisis de situación de salud de la población de la ciudad de Buenos Aires según la distribución geográfica de comunas.2006. Departamento de Epidemiología, Dirección General Adj. de Aps, Comité de Análisis de Situación de Salud, Ministerio de Salud, Gobierno de la Ciudad de Buenos Aires [cited 2010 Nov 15]. http://estatico.buenosaires.gov.ar/areas/salud/epidemiologia/archivos/asis/asis%202006%20n3.pdf
- 3.Canetti G, Froman S, Grosset J, Hauduroy P, Langerova M, Mahler HT, et al. Mycobacteria: laboratory methods for testing drug sensitivity and resistance. Bull World Health Organ. 1963;29:565–78. [PMC free article] [PubMed] [Google Scholar]
- 4.Gibson A, Brown T, Baker L, Drobniewski F. Can 15-locus mycobacterial interspersed repetitive unit–variable-number tandem repeat analysis provide insight into the evolution of Mycobacterium tuberculosis? Appl Environ Microbiol. 2005;71:8207–13. 10.1128/AEM.71.12.8207-8213.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iwamoto T, Yoshida S, Suzuki K, Tomita M, Fujiyama R, Tanaka N, et al. Hypervariable loci that enhance the discriminatory ability of newly proposed 15-loci and 24-loci variable-number tandem repeat typing method on Mycobacterium tuberculosis strains predominated by the Beijing family. FEMS Microbiol Lett. 2007;270:67–74. 10.1111/j.1574-6968.2007.00658.x [DOI] [PubMed] [Google Scholar]
- 6.Brown TJ, Herrera-Leon L, Anthony RM, Drobniewski FA. The use of macroarrays for the identification of MDR Mycobacterium tuberculosis. J Microbiol Methods. 2006;65:294–300. 10.1016/j.mimet.2005.08.002 [DOI] [PubMed] [Google Scholar]
- 7.Brudey K, Filliol I, Ferdinand S, Guernier V, Duval P, Maubert B, et al. Long-term population-based genotyping study of Mycobacterium tuberculosis complex isolates in the French Departments of the Americas. J Clin Microbiol. 2006;44:183–91. 10.1128/JCM.44.1.183-191.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Durmaz R, Zozio T, Gunal S, Yaman A, Cavusoglu C, Guney C, et al. Genetic diversity and major spoligotype families of drug-resistant Mycobacterium tuberculosis clinical isolates from different regions in Turkey. Infect Genet Evol. 2007;7:513–9. 10.1016/j.meegid.2007.03.003 [DOI] [PubMed] [Google Scholar]
- 9.Ritacco V, López B, Cafrune P, et al. Mycobacterium tuberculosis strains of the Beijing genotype are rarely observed in tuberculosis patients in South America. Mem Inst Oswaldo Cruz. 2008;103:489–92. 10.1590/S0074-02762008000500014 [DOI] [PubMed] [Google Scholar]
- 10.Palmero D, Cusmano L, Bucci Z, Romano M, Ruano S, Waisman J. Infectiousness and virulence of multidrug-resistant and drug susceptible tuberculosis in adult contacts. Medicina (B Aires). 2002;62:221–5. [PubMed] [Google Scholar]
- 11.Borrell S, Gagneux S. Infectiousness, reproductive fitness and evolution of drug-resistant Mycobacterium tuberculosis. Int J Tuberc Lung Dis. 2009;13:1456–66. [PubMed] [Google Scholar]
- 12.Waisman JL, Palmero DJ, Güemes-Gurtubay JL, Videla JJ, Moretti B, Cantero M, et al. Evaluación de las medidas de control adoptadas frente a la epidemia de tuberculosis multirresistente asociada al SIDA en un hospital hispanoamericano. Enferm Infecc Microbiol Clin. 2006;24:71–6. 10.1157/13085010 [DOI] [PubMed] [Google Scholar]