These fungi disperse slowly, leading to changes in structure at different geographic locations.
Keywords: Chromoblastomycosis, Fonsecaea, distribution, amplified fragment-length polymorphism, AFLP, fungi, research
Abstract
To assess population diversities among 81 strains of fungi in the genus Fonsecaea that had been identified down to species level, we applied amplified fragment-length polymorphism (AFLP) technology and sequenced the internal transcribed spacer regions and the partial cell division cycle, β-tubulin, and actin genes. Many species of the genus Fonsecaea cause human chromoblastomycosis. Strains originated from a global sampling of clinical and environmental sources in the Western Hemisphere, Asia, Africa, and Europe. According to AFLP fingerprinting, Fonsecaea isolates clustered in 5 groups corresponding with F. pedrosoi, F. monophora, and F. nubica: the latter 2 species each comprised 2 groups, and F. pedrosoi appeared to be of monophyletic origin. F. pedrosoi was found nearly exclusively in Central and South America. F. monophora and F. nubica were distributed worldwide, but both showed substantial geographic structuring. Clinical cases outside areas where Fonsecaea is endemic were probably distributed by human migration.
The genus Fonsecaea comprises etiologic fungal agents of human chromoblastomycosis (1–3), a chronic cutaneous and subcutaneous infection characterized by slowly expanding nodules that eventually lead to emerging, cauliflower-like, mutilating and disfiguring eruptions. Infection proceeds with muriform cells in tissue provoking a granulomatous immune response. In areas where it is endemic, disease incidence is high. Yegres et al. (4) and Yëgues-Rodriguez et al. (5) noted a frequency of 16 cases/1,000 population under arid climatic conditions in rural communities of Venezuela; chromoblastomycosis in that region is caused mainly by Cladophialophora carrionii. In contrast, Fonsecaea spp. are prevalent in humid tropical climates. Esterre et al. (6) reported 1,343 cases of chromoblastomycosis from Madagascar, 61.8% of which were caused by Fonsecaea spp. Kombila et al. (7) reported 64 cases in Gabon (equatorial Africa), all caused by Fonsecaea spp., and Silva et al. (8) cited 325 cases in the Amazon region of Brazil, 98% of which had Fonsecaea spp. as the etiologic agent. In Sri Lanka, 94% of 71 chromoblastomycosis cases were caused by Fonsecaea spp (9).
Fonsecaea contains anamorphic ascomycetes belonging to the family Herpotrichiellaceae (order Chaetothyriales), which includes black yeasts and relatives (10–12). The genus comprises 3 sibling species: F. pedrosoi, F. monophora, and F. nubica, each of which has pathogenic potential (10,13,14). Infection process and routes of dispersal are insufficiently clarified. Humans presumably acquire the infection after being pricked by contaminated thorns or wood splinters, but some agents are substantially more clinically prevalent than their predominantly (hitherto unnamed) environmental counterparts (15), which indicates that infection is not a random process. In many published case reports, etiologic agents were referred to as Phialophora pedrosoi or identified with the obsolete name F. compacta, now known to be a mutant F. pedrosoi (9,13,16). Strains are no longer accessible for molecular verification. Hence, no data are available on the epidemiology of the species as defined by sequence data.
Phylogenetically, Fonsecaea spp. agents of chromoblastomycosis are flanked by nonpathogenic species (10) growing on plant debris. Discovery of natural habitat and source of infection by entities emerging on the human host is essential for understanding the evolution of pathogenicity. We present an amplified fragment-length polymorphism (AFLP) DNA fingerprinting study of a worldwide collection of clinical isolates that were identified as Fonsecaea spp. by state-of-the-art sequencing methods, supplemented with environmental isolates of the same species. The AFLP technique is a powerful method for discrimination between fungal species and for providing high-resolution fingerprinting data within species (17–19).
Materials and Methods
Fungal Strains and Culture Conditions
We studied 81 isolates representing the 3 currently recognized Fonsecaea spp. Geographic origins and hosts of the strains are listed in the Table A1; the set include reference strains from the Centraalbureau voor Schimmelcultures (CBS-KNAW Fungal Biodiversity Centre, Utrecht, the Netherlands) and fresh isolates from patients and from the environment. Stock cultures were maintained on slants of 2% malt extract agar and oatmeal agar at 24°C.
DNA Extraction and Identification
Approximately 1 cm2 of 14- to 21-day-old cultures were transferred to 2 mL Eppendorf tubes containing 400 µL TEx buffer (Sigma-Aldrich, Zwijndrecht, the Netherlands), pH 9.0 (100 mmol Tris, 40 mmol Na-EDTA) and glass beads (Sigma G9143, Sigma-Aldrich). The fungal material was homogenized with a MoBio vortex (Bohemia, New York, USA) for 1 min. Subsequently, 120 µL of a 10% sodium dodecyl sulfate solution and 10 µL proteinase K (10 mg/mL, Sigma-Aldrich) were added and incubated for 30 min at 55°C; the mixture was vortexed for 3 min. After addition of 120 µL of 5M NaCl and 1/10 vol 10% cetyltrimethylammonium bromide solution (Sigma-Aldrich), the material was incubated for 60 min at 55°C. Then the mixture was vortexed for 3 min. Subsequently, 700 µL SEVAG (24:1, chloroform: isoamyl alcohol) was added, mixed carefully, and centrifuged for 5 min at 4°C at 20,400 × g. The supernatant was transferred to a new Eppendorf tube with 225 µL 5M NH4 acetate (Sigma-Aldrich), mixed carefully by inverting, incubated for 30 min on ice water, and centrifuged again for 5 min at 4°C at 20,400 × g. The supernatant was then transferred to another Eppendorf tube with 0.55 vol isopropanol and centrifuged for 5 min at 20,400 × g. Finally, the pellet was washed with 1 mL ice cold 70% ethanol. After drying at room temperature, it was resuspended in 48.5 µL TE buffer (Sigma-Aldrich) (Tris 0.12% wt/vol, Na-EDTA 0.04% wt/vol) and 1.5 µL of RNase (Sigma-Aldrich) and incubated in 37°C for 20–30 min. Quality of genomic DNA was verified on agarose gel. Species were identified on the basis of internal transcribed spacer (ITS), partial cell division cycle (CDC42), β-tubulin (BT2), and ACT sequences (10–14).
AFLP Fingerprinting
We followed a protocol provided by the manufacturer (Applied Biosystems, Nieuwerkerk aan de IJssel, the Netherlands), with some minor modifications (20–23). Analyses were performed with 100–200 ng DNA.
Restriction and Ligation of Adaptors
Two μL of DNA (100 ng/μL) was added to 9 μL restriction and ligation mixture (1.1 μL T4 DNA ligase buffer [Applied Biosystems]), 1.1 μL M NaCl, 2 U MseI endonuclease, 10 U EcoRI endonuclease (New England Biolabs, Ipswich, UK), 30 U T4 DNA ligase, 1 μL MseI-adaptor, 1 μL EcoRI-adaptor, and 3 μL dH20 and incubated at 37°C for 2.5 h. Subsequently, each restriction/ligation reaction was diluted ≈3× by adding 25 μL demineralized water.
Preselective and Selective PCR
In preselective PCR, 2 μL of diluted restriction/ligation product was added to 7.5 μL of AFLP core mix (Applied Biosystems), 0.25 μL of the EcoRI core sequence (5′-GAC TGC GTA CCA ATTC-3′), and 0.25 μL of the MseI core sequence (5′-GAT GAG TCC TGA GTAA-3′). The mixture was amplified in an iCycler (Bio-Rad, Hercules, CA, USA) under the following conditions: 2 min at 72°C, followed by 20 cycles of 20 s at 94°C, 30 s at 56°C, and 2 min at 72°C. Each preselective PCR was diluted 2× by adding 10 μL of dH2O. In selective PCR, 1.5 μL of diluted preselective PCR products was mixed with 8.5 selective PCR mix containing 0.5 μL EcoRI-AC (labeled with FAM [6-carboxy fluorescein]), 0.5 μL MseI-A, and 7.5 μL AFLP core mix (Applied Biosystems). The selective PCR conditions were cycling for 2 min at 94°C, followed by 10 cycles of 20 s at 94°C and 30 s at 66°C (decreasing 1°C with each subsequent cycle), and a final extension of 2 min at 72°C. This sequence was followed by 25 cycles of 20 s at 94°C, 30 s at 56°C, and 2 min at 72°C, and a final incubation of 30 min at 60°C.
AFLP Analysis
FAM-labeled products were prepared for analysis in an ABI PRISM 377 Genetic Analyzer (Applied Biosystems) as follows: the selective PCR products were cleaned with Sephadex G-50, and selective PCR products were mixed with LIZ 500 in the new plate by several times pipetting (first by preparing master mix [8.7 µL demineralized water plus 0.3 µL Liz 500], then mixing this with 1.0 µL of selective PCR product by pipetting). The total volume was adjusted to 10 µL with dH2O. Denaturation was done at 95°C for 5 min, and then the reaction was snap-cooled on ice water. The LIZ 500 internal size standard in each sample was used for normalization of the fingerprint pattern according to the instruction manual. The densitometric curves were analyzed with BioNumerics software package (version 4.61, Applied Maths, Kortrijk, Belgium), by using the cosine similarity coefficient and the unweighted pair group method with arithmetic means cluster analysis. Statistical reliability of the cluster was investigated by using a cophenetic value, which calculates the correlation between the calculated and the dendrogram-derived similarity. Subdivisions in clusters were checked visually if they were supported by the banding patterns.
Results
Profiles of 81 strains were generated with the EcoRI-AC + MseI-A PCR adaptors. Fingerprints contained ≈60–70 bands in a 50–500-bp range. Another selective PCR with EcoRI core sequence+C and MseI core sequence+A primer combination used elsewhere in related fungi (24) resulted in nonscorable fingerprints because of amplification of too many or only faint bands. Dendrograms derived from the AFLP banding patterns of Fonsecaea spp. were generated by using the unweighted pair group method with arithmetic means cluster analysis (Figure A1). At >62.50% similarity, 3 main clusters were found that matched with existing species on the basis of multilocus sequence analysis (ITS, CDC42, BT2, and ACT1), i.e., F. pedrosoi, F. monophora, and F. nubica. At an automatic cutoff value option set at <62.5% similarity, the F. monophora and F. nubica clusters were subdivided in 2 evident groups each, leading to a total of 5 clusters (1–5) interpreted as populations. Clusters 1 and 2 matched with F. nubica, clusters 3 and 4 with F. monophora, and cluster 5 with F. pedrosoi. Individual bands varied within the profiles, but further subclustering was limited, e.g., in a slightly deviating derived subclade in population 5. The groups defined above by AFLP analysis are interpreted as populations (1–5) in the text below. In population 5, some strains were nearly 100% identical, e.g., CBS 122341, 122343, 122345, and 122349, all originating from patients with chromoblastomycosis in Mexico City, Mexico (Figure A1; Table A1).
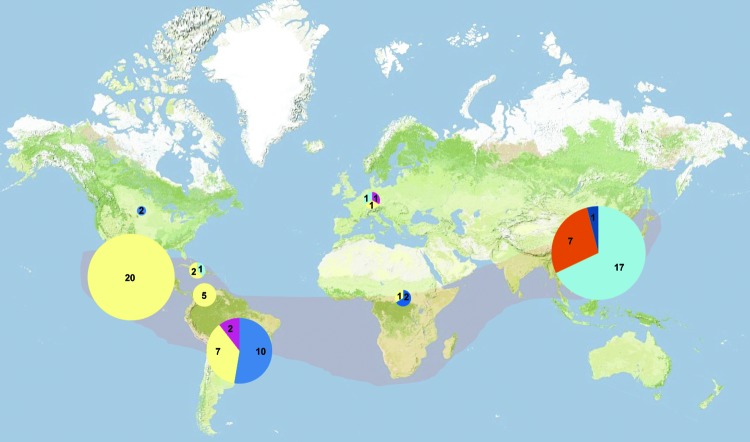
We determined the geographic distributions of the 5 main populations of Fonsecaea strains (Figure). Areas endemic for Fonsecaea, judging from the literature, are in tropical and subtropical climate zones. Population 1 comprised a cluster of F. nubica strains originating from humans with chromoblastomycosis in Guangdong, People’s Republic of China. Population 2 of the same species comprised 4 strains, 2 of which originated from humans with chromoblastomycosis in South America, 1 from France, and 1 with unknown origin. The profiles were too different to trace to any clonal identity. Population 3 (F. monophora) comprised 15 strains, most of which were isolated from humans with chromoblastomycosis in South America; 1 originated from the United States, and 1 originated from Haikou in southern China. Two strains were isolated from decaying plants in Brazil, and the second US strain was derived from a human with a brain infection. Two other strains from human brain infections in Brazil and in Africa had unique profiles that could not be unambiguously linked to any other isolate. Another African strain, from a patient with chromoblastomycosis who lived in Spain and had acquired the infection 36 years earlier in Guinea (25), also had a unique profile. Population 4 of F. monophora comprised 16 strains from Guangdong in southern China, and 1 came from Shandong, ≈1,850 km distant. All had derived from humans with chromoblastomycosis. A single sample originated from a patient with a brain infection who lived in the United Kingdom (26); whether the patient had visited southern China could not be established. In population 5 (F. pedrosoi), most strains originated from chromoblastomycosis patients in Central and South America. Some geographic clustering was visible, i.e., the derived group of strains from South America (uppermost clade of population 5 in the Figure A1) was segregated from those from Central America. Several of the strains from South American originated from soil and were isolated through mouse passage. One strain from an ear of a gazelle in Libya and 1 from a human with chromoblastomycosis in the Netherlands could not directly be linked to any other strain.
Discussion
AFLP typing is comparable to use of other DNA markers, such as random amplified polymorphic DNA, restriction fragment-length polymorphism, or microsatellites, in terms of time and cost efficiency, reproducibility, and resolution (27). The technique has emerged as a major epidemiologic tool with broad application in ecology, population genetics, pathotyping, DNA fingerprinting, and quantitative trait loci mapping (28). AFLP fingerprinting is useful for the molecular characterization of microorganisms with relatively large genomes, including various fungal species (18,19,21–23,29,30). In a preliminary experiment that used different primer combinations, the combination EcoRI-AC + MseI-A adaptors gave excellent results, yielding readable profiles with well-separated bands.
The degree of variation in Fonsecaea appeared to differ between species. The major 5 clusters were separated at <62.5% similarity, with significant differences in the presence of major fragments, several of which were unique to individual isolates or subpopulations. Populations 1 and 2, 3 and 4, and 5 corresponded with species borderlines established recently by Najafzadeh et al. (10,14) on the basis of multilocus sequencing with ITS, CDC42, BT2, and ACT1. Population 5 (F. pedrosoi) varied least at >71.7% similarity, with limited reproducible substructure being discernable. Nearly all isolates of this species originated from South and Central America (Venezuela, Brazil, Mexico, Argentina, Puerto Rico, and Uruguay). One isolate from a human with chromoblastomycosis in the Netherlands was likely to have been imported (13). One isolate from a gazelle ear in Libya, northern Africa, was the only geographic exception that could not be explained. Clusters of strains that could be grouped as being visually identical and with similarities >71.7% (Figure A1; Table A1) were mostly collected at close geographic distance from each other. This finding suggests that vectors of dispersal for Fonsecaea spp. are slow, leading to detectable regional diversification. The relatively low degree of variation of F. pedrosoi and confinement to Central and South America indicate a founder effect, the species being the most recently emerged taxon in Fonsecaea. F. monophora and F. nubica were distributed worldwide but were geographically diverse in that population 4 of F. monophora was nearly confined to China, with highly similar profiles (Figure A1). One strain of this population 4, CBS 117238, originated from a brain infection in a human in the United Kingdom; whether this patient had emigrated from China could not be determined from the original publication (25). F. monophora population 3 was found mainly in the Western Hemisphere, particularly in Brazil. Judging from the near identity of profiles of strains isolated in 1937 (CBS 271.37) and in 1999 (CBS 102245) (Figure A1), we can conclude that clones are maintained locally over decades. The 2 US strains presumably derived from immigrants from South America or Central America. Population 3 was also found in Africa and in Haikou in China, 600 km from Guangdong, where population 4 of F. monophora is prevalent. Strains of F. nubica show a similar bipartition over Asia and the Western Hemisphere, with a prevalently Chinese (population 1) and a prevalently Brazilian (population 2) population, and a presumed infected immigrant in France. Kawasaki et al. (31,32) provided similar data on the basis of restriction fragment-length polymorphism of mitochondrial DNA, showing that Fonsecaea spp. from Japan and China differed consistently from isolates from Central and South America.
Nearly all Fonsecaea spp. isolates available in culture collections originate from mammals, mostly humans with chromoblastomycosis, and were rarely recovered from the environment of symptomatic patients despite several attempts (33). Occasionally, F. pedrosoi was isolated from mice that were euthanized for isolation of black yeasts after they had been inoculated with environmental samples (34). This information suggests that Fonsecaea spp., particularly F. pedrosoi, have a competitive advantage by using this enrichment source. Mouse passage proved to be more efficient for environmental isolation of etiologic agents of chromoblastomycosis than general methods such as oil flotation (35). The latter technique mostly isolates other environmental Fonsecaea spp. that are not known to be pathogenic to humans (33).
In humans with chromoblastomycosis, the male:female ratio of patients is 63:2. This male preponderance of 97% cannot be explained by different exposition rates. Distinct male preponderance is also noted in the neurotropic relative, Cladophialophora bantiana (G.S. de Hoog, unpub. data). Population 3 of F. monophora has a wider clinical spectrum than the remaining groups, comprising, in addition to chromoblastomycosis, several isolates from human brain infection. This population also comprised some isolates from soil and plant debris acquired without use of mammal baits. Coexistence of closely interrelated entities differing in pathogenicity and virulence seems likely in Fonsecaea spp., as was also suggested for black yeasts (A.H.G. Gerrits van den Ende et al., unpub. data).
Our data demonstrate that AFLP fingerprinting is a tool that produces highly reproducible results for molecular epidemiology. The use of AFLP showed that local Fonsecaea agents of chromoblastomycosis seem able to be maintained over 70 years, and therefore epidemiologic profiles take the structure of expanding clones. By locality, patients are infected by only a limited number of genotypes. The fungi disperse slowly, leading to appreciable geographic structuring, which ultimately may lead to allopatric speciation (diversification resulting from geographic barriers). Few environmental strains have been recovered during repeated isolation experiments, whereas Fonsecaea spp. accumulates substantially in the human host. The mechanisms behind their pathology remain unexplained.
Acknowledgments
We are grateful to Liyan Xi and Flavio Queiroz-Telles for providing Fonsecaea spp. strains.
M.J.N. was supported by the Ministry of Health and Medical Education of Iran and Mashhad University of Medical Sciences, Mashhad, Iran. J.S. was partly supported by International Program of Project 985, Sun Yat-Sen University, China. V.A.V. was supported by Coordenação de Aperfeiçoamento Pessoal de Nivel Superior/Brazil.
Biography
Dr Najafzadeh is a PhD student of medical mycology at CBS-KNAW Fungal Biodiversity Centre, Utrecht, the Netherlands. His research interest is molecular epidemiology and phylogenic study of medically important black fungi.
Table A1. Fonsecaea spp. isolates used for amplified fragment-length polymorphism analyses.
| Taxonomic name | CBS number | Other reference(s) | Origin | Host/sex | Location | Population |
|---|---|---|---|---|---|---|
| F. nubica | CBS 121733 | dH 18411, SUMS 0011 | Chromoblastomycosis | Human/M | China, Guangdong | 1 |
| CBS 125199 | dH 20427 | Chromoblastomycosis | Human/F | China, Guangdong | 1 | |
| CBS 125186 | dH 20429 | Chromoblastomycosis | Human/M | China, Guangdong | 1 | |
| CBS 125200 | dH 20425 | Chromoblastomycosis | Human/M | China, Guangdong | 1 | |
| CBS 121720 | dH 18398, SUMS 0251 | Chromoblastomycosis | Human/M | China, Guangdong | 1 | |
| CBS 125198 | dH 20418 | Chromoblastomycosis | Human/M | China, Guangdong | 1 | |
| CBS 121734 | dH 18412, SUMS 0255 | Chromoblastomycosis | Human/M | China, Guangdong | 1 | |
| CBS 271.33 | dH 15659, ATCC 18658, IMI 134458 | Chromoblastomycosis | Human/M | South America | 2 | |
| CBS 557.76 | ATCC 28174 | Unknown | Unknown | Unknown | 2 | |
| CBS 270.37 | dH 15657 | Unknown | Unknown | France | 2 | |
|
|
CBS 277.29 |
dH 15668 |
Chromoblastomycosis |
Human/M |
Brazil |
2 |
| F. monophora | CBS 102243 | dH 11607 | Chromoblastomycosis | Human/M | Brazil, Parana, Ibituva | 3 |
| CBS 117236 | dH 15330, UTHSC 04-2904 | Brain | Human/M | United States | 3 | |
| CBS 102246 | dH 11611 | Chromoblastomycosis | Human/M | Brazil, Parana, Campo Largo | 3 | |
| CBS 102242 | dH 11606 | Chromoblastomycosis | Human/M | Brazil, Santa Catarina, Curitibanos | 3 | |
| CSB 102225 | dH 11585 | Decaying wood | Plant | Brazil, Parana, Colombo | 3 | |
| CSB 269.37 | dH 12659 | Chromoblastomycosis | Human | South America | 3 | |
| CSB 102238 | dH 11602 | Soil | Soil | Brazil, Parana, Tibagi River | 3 | |
| CBS 117237 | dH 15331, UTHSC 04-2631 | Chromoblastomycosis | Human/M | United States | 3 | |
| CBS 102229 | dH 11590 | Decaying vegetable cover | Plant | Brazil, Parana, Piraquara | 3 | |
| CBS 397.48 | dH 15828, ATCC 9475 | Chromoblastomycosis | Human/M | South America | 3 | |
| CBS 115830 | dH 12978 | Brain | Human/M | Brazil | 3 | |
| CBS125189 | dH 20421 | Chromoblastomycosis | Human/M | China, Haikou | 3 | |
| CBS 100430 | ATCC 32280 | Brain | Human/M | Africa | 3 | |
| CBS 123849 | dH 20215 | Chromoblastomycosis | Human/F | Africa, Guinea | 3 | |
| CBS 102248 | dH 11613 | Chromoblastomycosis | Human/M | Brazil, Parana, Piraquara | 3 | |
| CBS 121725 | dH 18403, SUMS 0250 | Chromoblastomycosis | Human/M | China, Guangdong | 4 | |
| CBS 121728 | dH 18406, SUMS 0158 | Chromoblastomycosis | Human/M | China, Guangdong | 4 | |
| CBS 121726 | dH 18404, SUMS 0192 | Chromoblastomycosis | Human/M | China, Guangdong | 4 | |
| CBS 121727 | dH 18405, SUMS 0190 | Chromoblastomycosis | Human/M | China, Guangdong | 4 | |
| CBS 121721 | dH 18399, SUMS 0246 | Chromoblastomycosis | Human/M | China, Guangdong | 4 | |
| CBS 125193 | dH 20426 | Chromoblastomycosis | Human/M | China, Guangdong | 4 | |
| CBS 125195 | dH 20417 | Chromoblastomycosis | Human/M | China, Guangdong | 4 | |
| CBS 125196 | dH 20419 | Chromoblastomycosis | Human/M | China, Guangdong | 4 | |
| CBS 125197 | dH 20420 | Chromoblastomycosis | Human/M | China, Guangdong | 4 | |
| CBS 121732 | dH 18410, SUMS 0012 | Chromoblastomycosis | Human/M | China, Guangdong | 4 | |
| CBS 125190 | dH 20422 | Chromoblastomycosis | Human/M | China, Guangdong | 4 | |
| CBS 125192 | dH 20424 | Chromoblastomycosis | Human/M | China, Guangdong | 4 | |
| CBS 117238 | dH 13130, UTHSC R-3486 | Brain | Human | United Kingdom, England | 4 | |
| CBS 121731 | dH 18409, SUMS 0013 | Chromoblastomycosis | Human/M | China, Guangdong | 4 | |
| CBS 121730 | dH 18408, SUMS 0014 | Chromoblastomycosis | Human/M | China, Guangdong | 4 | |
| CBS 121722 | dH 18400, SUMS 0247 | Chromoblastomycosis | Human/M | China, Guangdong | 4 | |
| CBS 122742 | dH 19251, SUMS 0147 | Chromoblastomycosis | Human | China, Shandong | 4 | |
|
|
CBS 121724 |
dH 18402, SUMS 0200 |
Chromoblastomycosis |
Human/M |
China, Guangdong |
4 |
| F. pedrosoi | CBS 273.66 | dH 15663 | Mouse passage | Soil | Venezuela | 5 |
| CBS 271.37 | dH 15659, ATCC 18658, IMI 134458 | Chromoblastomycosis | Human/M | South America | 5 | |
| CBS 671.66 | dH 16159 | Mouse passage | Soil | Venezuela | 5 | |
| CBS 274.66 | dH 15665 | Mouse passage | Soil | Venezuela | 5 | |
| CBS 102245 | dH 11610 | Chromoblastomycosis | Human/M | Brazil, Parana, Ampere | 5 | |
| CBS 659.76 | dH 16142, ATCC 28303 | Chromoblastomycosis | Human/M | Argentina | 5 | |
| CBS 102247 | dH 11612 | Chromoblastomycosis | Human/M | Brazil, Parana | 5 | |
| CBS 122740 | dH 18430, Bonifaz 002200 | Chromoblastomycosis | Human/M | Mexico, Mexico City | 5 | |
| CBS 122737 | dH 18896 | Chromoblastomycosis | Human/M | Mexico, Mexico City | 5a | |
| CBS 122735 | dH 18898 | Chromoblastomycosis | Human/M | Mexico, Mexico City | 5a | |
| CBS 122736 | dH 18897 | Chromoblastomycosis | Human/M | Mexico, Mexico City | 5a | |
| CBS 122739 | dH 18894 | Chromoblastomycosis | Human/M | Mexico, Mexico City | 5a | |
| CBS 122732 | dH 18901 | Chromoblastomycosis | Human/M | Mexico, Mexico City | 5 | |
| CSB 122733 | dH 18900 | Chromoblastomycosis | Human/M | Mexico, Mexico City | 5 | |
| CBS 122849 | dH 18902 | Chromoblastomycosis | Human/M | Mexico, Mexico City | 5b | |
| CBS 122738 | dH 18895 | Chromoblastomycosis | Human/M | Mexico, Mexico City | 5b | |
| CBS 122731 | dH 18903 | Chromoblastomycosis | Human/M | Mexico, Mexico City | 5b | |
| CBS 122734 | dH 18899 | Chromoblastomycosis | Human/M | Mexico, Mexico City | 5b | |
| CBS 102244 | dH 11608 | Chromoblastomycosis | Human/M | Brazil, Parana, Ipora | 5 | |
| CBS 122729 | dH 18905 | Chromoblastomycosis | Human/M | Mexico, Mexico City | 5 | |
| CBS 122730 | dH 18904 | Chromoblastomycosis | Human/M | Mexico, Mexico City | 5 | |
| CBS 285.47 | dH 15680, ATCC 10222 | Chromoblastomycosis | Human/M | Puerto Rico | 5 | |
| CBS 342.34 | dH 15773 | Chromoblastomycosis | Human/M | Puerto Rico | 5 | |
| CBS 670.66 | dH 16157 | Mouse passage | Soil | Venezuela | 5 | |
| CBS 122741 | dH 18431, Bonifaz 02300 | Chromoblastomycosis | Human/M | Mexico, Mexico City | 5 | |
| CBS 122729 | dH 18905 | Chromoblastomycosis | Human | Mexico, Mexico City | 5 | |
| CBS 212.77 | dH 15549 | Chromoblastomycosis | Human/M | Netherlands, Amsterdam | 5 | |
| CBS 117910 | dH 14477 | Chromoblastomycosis | Human/M | Venezuela, Coro, Falcón State | 5 | |
| CBS 272.37 | dH 15661 | Chromoblastomycosis | Human | Brazil | 5 | |
| CBS 122345 | dH 18914, Bonifaz 121-06 | Chromoblastomycosis | Human/M | Mexico, Mexico City | 5c | |
| CBS 122343 | dH 18916, Bonifaz 122-07 | Chromoblastomycosis | Human/M | Mexico, Mexico City | 5c | |
| CBS 122341 | dH 18918, Bonifaz 345-07 | Chromoblastomycosis | Human/M | Mexico, Mexico City | 5c | |
| CBS 122349 | dH 18910, Bonifaz 234-04 | Chromoblastomycosis | Human/M | Mexico, Mexico City | 5c | |
| CBS 122347 | dH 18912, Bonifaz 0257-05 | Chromoblastomycosis | Human/M | Mexico, Mexico City | 5c | |
| CBS 122346 | dH 18913, Bonifaz 333-05 | Chromoblastomycosis | Human/M | Mexico, Mexico City | 5c | |
| CBS 253.49 | dH 15620 | Chromoblastomycosis | Human | Uruguay, Montevideo | 5 | |
| CBS 201.31 | dH 15523 | Gazelle, ear | Animal | Libya, Cyrenaica, Derna | 5 |
*ATCC, American Type Culture Collection, Manassas, VA, USA; CBS, Centraalbureau voor Schimmelcultures, Utrecht, the Netherlands; dH, G.S. de Hoog working collection; SUMS, Sun Yat-Sen University Medical Science, Guangzhou, People’s Republic of China; IMI, International Mycological Institute, London, UK; UTHSC, Fungus Testing Laboratory, Department of Pathology, University of Texas Health Science Center at San Antonio, San Antonio, TX, USA.
Figure.
Geographic distribution of Fonsecaea spp. samples analyzed by using amplified fragment-length polymorphism. Light pink shading indicates zone of clinical Fonsecaea spp. endemicity, according to published case reports. Sizes of pies and numbers reported within the pies denote the number of strains examined; colors represent Fonsecaea spp. populations: orange, F. nubica population 1; fuchsia, F. nubica population 2; dark blue, F. monophora population 3; light blue, F. monophora population 4; yellow, F. pedrosoi population 5.
Figure A1.
Clustering of amplified fragment-length polymorphism banding pattern of isolates of Fonsecaea spp. analyzed by using unweighted pair group method with arithmetic means. Red bars indicate clonal dispersal. Clusters 1 and 2 are F. nubica, clusters 3 and 4 are F. monophora, and cluster 5 is F. pedrosoi.
Footnotes
Suggested citation for this article: Najafzadeh MJ, Sun J, Vicente VA, Klaassen CHW, Bonifaz A, Gerrits van den Ende AHG, et al. Molecular epidemiology of Fonsecaea species. Emerg Infect Dis [serial on the Internet]. 2011 Mar [date cited]. http://dx.doi.org/10.3201/1703.100555
References
- 1.Queiroz-Telles F, Esterre P, Perez-Blanco M, Vitale RG, Salgado CG, Bonifaz A. Chromoblastomycosis: an overview of clinical manifestations, diagnosis and treatment. Med Mycol. 2009;47:3–15. 10.1080/13693780802538001 [DOI] [PubMed] [Google Scholar]
- 2.López Martínez R, Méndez Tovar LJ. Chromoblastomycosis. Clin Dermatol. 2007;25:188–94. 10.1016/j.clindermatol.2006.05.007 [DOI] [PubMed] [Google Scholar]
- 3.Bonifaz A, Carrasco-Gerard E, Saul A. Chromoblastomycosis: clinical and mycologic experience of 51 cases. Mycoses. 2001;44:1–7. 10.1046/j.1439-0507.2001.00613.x [DOI] [PubMed] [Google Scholar]
- 4.Yegres FR-YN, Medina-Ruiz E, González-Vivas R. Cromomicosis por Cladosporium carrionii en criadores de caprinos del estado Falcon. Investigaciόn. Clinica. 1985;26:235–46. [Google Scholar]
- 5.Yegüez-Rodriguez J R-YN, Yegres F, Rodríguez Larralde A. Cromomicosis: susceptibilidad genética en grupos familiares de la zona endémica en Venezuela. Acta Cientίfica Venezuelana 1992;43:98–102.
- 6.Esterre P, Andriantsimahavandy A, Ramarcel ER, Pecarrere JL. Forty years of chromoblastomycosis in Madagascar: a review. Am J Trop Med Hyg. 1996;55:45–7. [DOI] [PubMed] [Google Scholar]
- 7.Kombila M, Gomez de Diaz M, Richard-Lenoble D, Renders A, Walter P, Billiault X, et al. Chromoblastomycosis in Gabon. Study of 64 cases [in French]. Sante. 1995;5:235–44. [PubMed] [Google Scholar]
- 8.Silva JP, de Souza W, Rozental S. Chromoblastomycosis: a retrospective study of 325 cases on Amazonic Region (Brazil). Mycopathologia. 1998–1999;143:171–5. 10.1023/A:1006957415346 [DOI] [PubMed] [Google Scholar]
- 9.Attapattu MC. Chromoblastomycosis—a clinical and mycological study of 71 cases from Sri Lanka. Mycopathologia. 1997;137:145–51. 10.1023/A:1006819530825 [DOI] [PubMed] [Google Scholar]
- 10.Najafzadeh MJ, Gueidan C, Badali H, van den Ende AH, Xi L, de Hoog GS. Genetic diversity and species delimitation in the opportunistic genus Fonsecaea. Med Mycol. 2009;47:17–25. 10.1080/13693780802527178 [DOI] [PubMed] [Google Scholar]
- 11.Xi L, Sun J, Lu C, Liu H, Xie Z, Fukushima K, et al. Molecular diversity of Fonsecaea (Chaetothyriales) causing chromoblastomycosis in southern China. Med Mycol. 2009;47:27–33. 10.1080/13693780802468209 [DOI] [PubMed] [Google Scholar]
- 12.Badali H, Gueidan C, Najafzadeh MJ, Bonifaz A, van den Ende AHG, de Hoog GS. Biodiversity of the genus Cladophialophora. Stud Mycol. 2008;61:175–91. 10.3114/sim.2008.61.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Hoog GS, Attili-Angelis D, Vicente VA, Gerrits van den Ende AHG, Queiroz-Telles F. Molecular ecology and pathogenic potential of Fonsecaea species. Med Mycol. 2004;42:405–16. 10.1080/13693780410001661464 [DOI] [PubMed] [Google Scholar]
- 14.Najafzadeh MJ, Sun J, Vicente V, Xi L, van den Ende AH, de Hoog GS. Fonsecaea nubica sp. nov, a new agent of human chromoblastomycosis revealed using molecular data. Med Mycol. 2010;48:800–6. 10.3109/13693780903503081 [DOI] [PubMed] [Google Scholar]
- 15.de Hoog GS, Nishikaku AS, Fernandez-Zeppenfeldt G, Padin-Gonzalez C, Burger E, Badali H, et al. Molecular analysis and pathogenicity of the Cladophialophora carrionii complex, with the description of a novel species. Stud Mycol. 2007;58:219–34. 10.3114/sim.2007.58.08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nishimoto K. Chromomycosis in Japan. Ann Soc Belg Med Trop. 1981;61:405–12. [PubMed] [Google Scholar]
- 17.Klaassen CHW, Osherov N. Aspergillus strain typing in the genomics era. Stud Mycol. 2007;59:47–51. 10.3114/sim.2007.59.06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Borst A, Theelen B, Reinders E, Boekhout T, Fluit AC, Savelkoul PH. Use of amplified fragment length polymorphism analysis to identify medically important Candida spp., including C. dubliniensis. J Clin Microbiol. 2003;41:1357–62. 10.1128/JCM.41.4.1357-1362.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Warris A, Klaassen CH, Meis JF, De Ruiter MT, De Valk HA, Abrahamsen TG, et al. Molecular epidemiology of Aspergillus fumigatus isolates recovered from water, air, and patients shows two clusters of genetically distinct strains. J Clin Microbiol. 2003;41:4101–6. 10.1128/JCM.41.9.4101-4106.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boekhout T, Theelen B, Diaz M, Fell JW, Hop WC, Abeln EC, et al. Hybrid genotypes in the pathogenic yeast Cryptococcus neoformans. Microbiology. 2001;147:891–907. [DOI] [PubMed] [Google Scholar]
- 21.Theelen B, Silvestri M, Gueho E, van Belkum A, Boekhout T. Identification and typing of Malassezia yeasts using amplified fragment length polymorphism (AFLP), random amplified polymorphic DNA (RAPD) and denaturing gradient gel electrophoresis (DGGE). FEM Yeast Res. 2001;1:79–86. 10.1111/j.1567-1364.2001.tb00018.x [DOI] [PubMed] [Google Scholar]
- 22.Ball LM, Bes MA, Theelen B, Boekhout T, Egeler RM, Kuijper EJ. Significance of amplified fragment length polymorphism in identification and epidemiological examination of Candida species colonization in children undergoing allogeneic stem cell transplantation. J Clin Microbiol. 2004;42:1673–9. 10.1128/JCM.42.4.1673-1679.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gupta AK, Boekhout T, Theelen B, Summerbell R, Batra R. Identification and typing of Malassezia species by amplified fragment length polymorphism and sequence analyses of the internal transcribed spacer and large-subunit regions of ribosomal DNA. J Clin Microbiol. 2004;42:4253–60. 10.1128/JCM.42.9.4253-4260.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sudhadham M, de Hoog GS, Menken SBJ, Gerrits van den Ende AHG, Sihanonth P. Elucidation of distribution patterns and possible infection routes of the neutropic black yeast Exophiala dermatitidis using AFLP. Fungal Biol. 2011. In press. [DOI] [PubMed] [Google Scholar]
- 25.Najafzadeh MJ, Rezusta A, Cameo MI, Zubiri ML, Yus MC, Badali H, et al. Successful treatment of chromoblastomycosis of 36 years duration caused by Fonsecaea monophora. Med Mycol. 2010;48:390–3. 10.3109/13693780903008813 [DOI] [PubMed] [Google Scholar]
- 26.Surash S, Tyagi A, de Hoog GS, Zeng JS, Barton RC, Hobson RP. Cerebral phaeohyphomycosis caused by Fonsecaea monophora. Med Mycol. 2005;43:465–72. 10.1080/13693780500220373 [DOI] [PubMed] [Google Scholar]
- 27.Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, Hornes M, et al. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 1995;23:4407–14. 10.1093/nar/23.21.4407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mueller UG, Wolfenbarger LL. AFLP genotyping and fingerprinting. Trends Ecol Evol. 1999;14:389–94. 10.1016/S0169-5347(99)01659-6 [DOI] [PubMed] [Google Scholar]
- 29.Neyra E, Fonteyne PA, Swinne D, Fauche F, Bustamante B, Nolard N. Epidemiology of human sporotrichosis investigated by amplified fragment length polymorphism. J Clin Microbiol. 2005;43:1348–52. 10.1128/JCM.43.3.1348-1352.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Savelkoul PH, Aarts HJ, de Haas J, Dijkshoorn L, Duim B, Otsen M, et al. Amplified-fragment length polymorphism analysis: the state of an art. J Clin Microbiol. 1999;37:3083–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kawasaki M, Aoki M, Ishizaki H, Miyaji M, Nishimura K, Nishimoto K, et al. Molecular epidemiology of Fonsecaea pedrosoi using mitochondrial DNA analysis. Med Mycol. 1999;37:435–40. [PubMed] [Google Scholar]
- 32.Kawasaki M. Typing and molecular epidemiology of some black fungi based on analysis of the restriction fragment length polymorphism in the mitochondrial DNA. Jpn J Med Mycol. 1996;37:129–33. 10.3314/jjmm.37.129 [DOI] [Google Scholar]
- 33.Vicente VA, Attili-Angelis D, Pie MR, Queiroz-Telles F, Cruz LM, Najafzadeh MJ, et al. Environmental isolation of black yeast-like fungi involved in human infection. Stud Mycol. 2008;61:137–44. 10.3114/sim.2008.61.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gezuele E, Mackinnon JE, Conti-Diaz IA. The frequent isolation of Phialophora verrucosa and Phialophora pedrosoi from natural sources. Sabouraudia. 1972;10:266–73. [PubMed] [Google Scholar]
- 35.Satow MM, Attili-Angelis D, de Hoog GS, Angelis DF, Vicente VA. Selective factors involved in oil flotation isolation of black yeasts from the environment. Stud Mycol. 2008;61:157–63. 10.3114/sim.2008.61.16 [DOI] [PMC free article] [PubMed] [Google Scholar]