Abstract
In budding yeast, the microtubule-organizing center is called the spindle pole body (SPB) and shares structural components with the centriole, the central core of the animal centrosome. During meiotic interphase I, the SPB is duplicated when DNA replication takes place. Duplicated SPBs are linked and then separate to form a bipolar spindle required for homolog separation in meiosis I. During interphase II, SPBs are duplicated again, in the absence of DNA replication, to form four SPBs that establish two spindles for sister-chromatid separation in meiosis II. Here, we report that the Aurora kinase Ipl1, which is necessary for sister-chromatid cohesion, is also required for maintenance of a tight association between duplicated SPBs during meiosis, which we term SPB cohesion. Premature loss of cohesion leads to SPB overduplication and the formation of multipolar spindles. By contrast, the Polo-like kinase Cdc5 is necessary for SPB duplication and interacts antagonistically with Ipl1 at the meiotic SPB to ensure proper SPB separation. Our data suggest that Ipl1 coordinates SPB dynamics with the two chromosome segregation cycles during yeast meiosis.
Key words: Spindle pole body (SPB), Centrosome, SPB duplication, Aurora kinase, Ipl1, Polo-like kinase, Meiosis
Introduction
Structurally and conceptually, the budding yeast spindle pole body (SPB) is best known from vegetative cells (Adams and Kilmartin, 2000; Jaspersen and Winey, 2004). It is embedded in the nuclear envelope as a layered structure (Moens and Rapport, 1971; Byers and Goetsch, 1974). Duplication of the SPB is initiated with the deposition of the satellite at the distal end of the half bridge, a specialized membrane structure attached to the SPB central plaque (Byers and Goetsch, 1974). One of the major components of the satellite is the SPB core component, Spc42 (Donaldson and Kilmartin, 1996; Bullitt et al., 1997). Duplicated SPBs form a side-by-side configuration and are tethered together by the bridge, which is severed or disassembled upon the formation of a bipolar spindle, promoted by the actions of the mitotic cyclin Cdk and the Polo-like kinase Cdc5 (Kilmartin, 2003; Jaspersen et al., 2004; Crasta et al., 2008). Two known structural components of the half bridge are Sfi1 and Cdc31, homologs of which are essential components in the vertebrate centriole (Li et al., 2006). The meiotic SPB resembles its mitotic counterpart (Moens and Rapport, 1971; Straight et al., 2000) and is perhaps regulated in a similar fashion, but how it is reduplicated at interphase II to coordinate meiotic chromosome segregation remains unknown.
In budding yeast, the Aurora kinase Ipl1 is required for the protection of sister-chromatid cohesion during meiosis (Monje-Casas et al., 2007; Yu and Koshland, 2007). Ipl1 is the founding member of the Aurora kinase family and is closely related to the Aurora B kinase found in higher eukaryotes (Chan and Botstein, 1993). Here, we report that Ipl1 is also required for maintenance of a tight association between duplicated SPBs and prevents SPB overduplication at interphase II, revealing that Ipl1 plays a role similar to that of the Aurora A kinase, being important for centrosome propagation. As in animal cells, where centriole duplication depends on Plk1 (Tsou et al., 2009), licensing of SPB duplication requires Cdc5 in budding yeast meiosis.
Results and Discussion
Ipl1 is required for SPB cohesion during meiosis
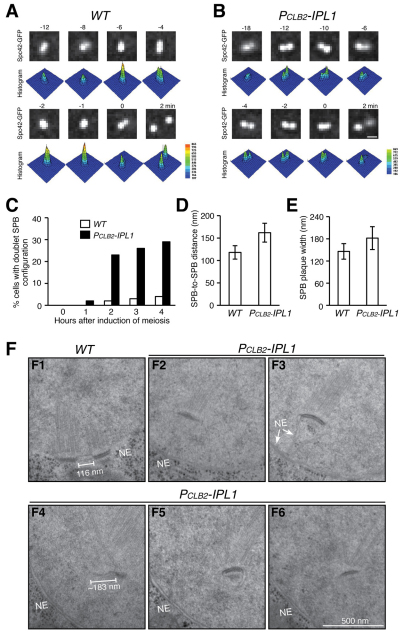
To investigate the dynamic separation of duplicated SPBs in meiosis I (MI), we developed a live-cell culture method of inducing yeast meiosis on a glass slide and observed Spc42–GFP-marked SPBs by fluorescence microscopy (Fig. 1A). In wild-type cells, SPBs were tightly associated after duplication, primarily forming a single Spc42–GFP focus (Fig. 1A). Immediately before their separation, SPBs were resolved by light microscopy as two distinguishable foci (Fig. 1A, t=0), which we refer to as a doublet SPB configuration. For live-cell microscopy, we defined time zero as the point immediately before SPB separation in MI (~4 hours after induction of meiosis), so that we could objectively examine and compare SPB separation and meiotic progression. In contrast to those from the wild type, SPBs in Ipl1-depleted (PCLB2-IPL1) cells were mostly associated at a greater distance and formed the doublet configuration well before their complete separation (Fig. 1B,C), suggesting that the sister SPBs are less cohesive in the absence of Ipl1. To examine the meiotic SPB at a higher resolution, we examined serially sectioned cells by electron microscopy (Fig. 1D–F). Consistent with our light-microscopy observations, duplicated SPBs were linked by the bridge and were tightly associated for an extended period of time in MI (Fig. 1), a situation we termed SPB cohesion. By contrast, sister SPBs were positioned significantly further apart from each other with a collapsed bridge before their complete separation in Ipl1-depleted cells (Fig. 1D, 118 nm in the wild type, 162 nm in the mutant). In addition, the SPB-associated nuclear envelope often became invaginated (Fig. 1F,F2–F6). Even in these cells, the layered structure of SPBs resembled those of the wild type (Fig. 1F). The average widths of SPB layers from wild-type and Ipl1-depleted cells did not differ significantly during MI, but mutant cells showed a greater variation in SPB size (Fig. 1E). Together, these results show that the layered SPB structure appears normal in Ipl1-depleted meiotic cells, but sister SPBs are positioned further apart, suggesting that Ipl1 is required for the maintenance of bridge integrity and therefore SPB cohesion.
Fig. 1.
Requirement for Ipl1 for SPB cohesion during yeast meiosis. (A,B) Fluorescence live-cell microscopy images showing the morphology of duplicated sister SPBs in wild-type (WT, HY1423C) and PCLB2-IPL1 (HY1423) cells in MI. SPBs are marked by Spc42–GFP. Time zero was defined as the point immediately before sister-SPB separation in MI. The time in minutes is shown above each frame. Projected images are shown. Three-dimensional histograms show Spc42–GFP intensity. Pixel intensity counts are shown to the right. Scale bar: 0.5 μm. (C) Quantification of SPB doublet formation. Cells were induced to undergo synchronous meiosis, fixed at the indicated times and visualized under a fluorescence microscope. At least 100 cells were counted for each time point. (D,E) SPB-to-SPB distance and SPB plaque width in MI. Cells were induced to undergo synchronous meiosis, subjected to high-pressure freezing and freeze substitution, serially sectioned and visualized with electron microscopy. MI cells with side-by-side SPBs were identified; SPB-to-SPB distance and plaque width were determined from single sections. WT, n=9; mutant, n=7. Error bars show s.d. (F) Representative images showing sister-SPB configuration from WT (F1) and mutant (F2–F6) cells. Five serial sections are shown for the mutant. Note that the SPB-associated nuclear envelope became invaginated in the mutant. NE, nuclear envelope.
Ipl1 is required for accurate SPB duplication at interphase II
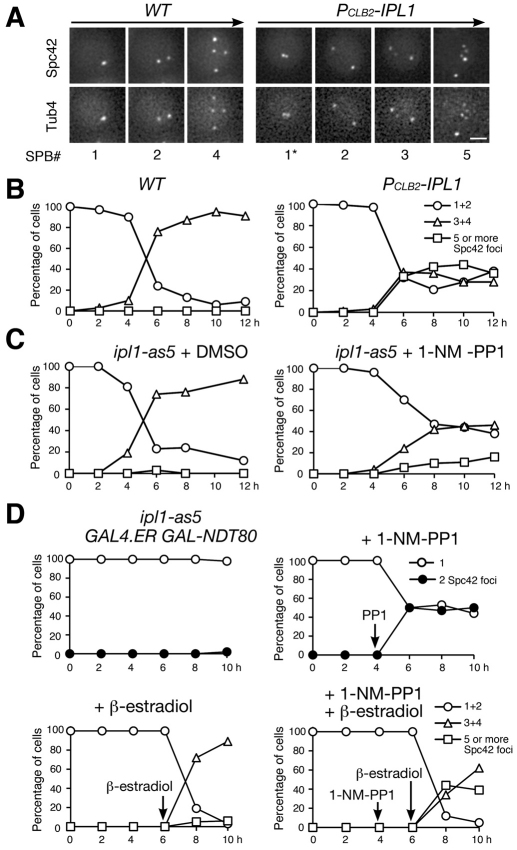
After SPBs separate in MI, they undergo a second round of duplication at interphase II. In wild-type cells, four SPBs were formed after completion of meiosis II (MII) (Fig. 2A,B). By contrast, more than 30% of Ipl1-depleted meiotic cells had formed five or more SPBs 6 hours after induction, a time that corresponded to MII (Fig. 2A,B). These supernumerary SPBs contained the meiotic plaque component Mpc54 (supplementary material Fig. S1), which is characteristic of MII SPBs (Knop and Strasser, 2000). In addition, inactivation of the Ipl1 kinase activity in the ipl1-as5 allele (Pinsky et al., 2006) during meiosis also led to the formation of extra Spc42 foci (Fig. 2C). Together, these results suggest that sister SPBs prematurely lose cohesion and subsequently overduplicate in the absence of Ipl1 activity during meiosis.
Fig. 2.
Requirement for Ipl1 for SPB reduplication during yeast meiosis. (A) Fluorescence live-cell microscopy showing SPB segregation in WT (HY1675) and PCLB2-IPL1 (HY1886) cells during yeast meiosis. SPBs are marked by Spc42–GFP and Tub4–mApple. *Duplicated sister-SPBs formed a doublet configuration in PCLB2-IPL1 cells before separation in MI. Scale bar: 2 μm. (B) The number of Spc42 foci in fixed samples from WT (left) and PCLB2-IPL1 (right) cells. One and two SPB cells were grouped as MI; three and four were grouped as meiosis II (MII); five or more represents cells with overduplicated SPBs. Averages from two independent experiments are shown. (C) Spc42–GFP focus formation in the ipl1-as5 mutant (HY2486) during meiosis. Addition of 1-NM-PP1 inhibits the Ipl1 kinase activity. DMSO treatment (left) serves as a control. Figure symbols are as in B. (D) The execution point of Ipl1 on SPB cohesion and duplication. Yeast cells (HY2627) were induced to undergo synchronous meiosis, subjected to four different treatments and fixed at the indicated times for fluorescence microscopy. Addition of β-estradiol induced the production of Ndt80. Arrows indicate the time of addition.
To determine when Ipl1 is required for SPB duplication and whether SPBs become fragmented in Ipl1-depleted cells, we arrested the cells at prophase I by eliminating the production of Ndt80 by means of the GAL-NDT80 allele (Carlile and Amon, 2008). We then observed SPB separation with and without the Ipl1 kinase activity (Fig. 2D). Sister SPBs failed to separate in cells arrested at prophase I, but inactivation of Ipl1 led to SPB separation in ~50% of these cells, supporting the idea that Ipl1 is required for maintaining SPB cohesion. Importantly, these separated SPBs did not commit to duplication unless Ndt80 was reintroduced and cells proceeded through meiosis (Fig. 2D). These data suggest that supernumerary SPB formation in Ipl1-depleted cells is less likely to be due to SPB fragmentation. We therefore conclude that the Aurora kinase Ipl1 is required for SPB cohesion and prevents SPB overduplication at interphase II.
Ipl1 prevents multipolar spindle formation
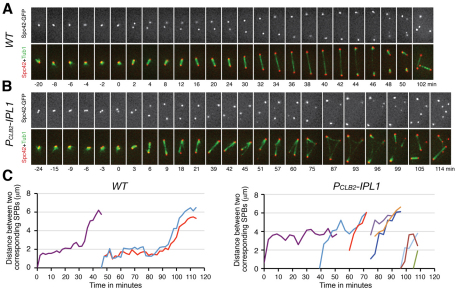
To determine whether overduplicated SPBs in ipl1 mutant cells function to nucleate microtubules, we observed SPB dynamics and spindle formation (Fig. 3A,B). In wild-type cells, sister SPBs remained tightly associated after duplication (Fig. 3A). SPBs were separated to form a 2- to 3-μm-long spindle during metaphase I (Fig. 3A,C). Approximately 30 minutes after SPB separation, the spindle elongated to reach a length of ~6 μm at anaphase I. Then the pole-to-pole distance decreased slightly during an ~8-minute window, before SPBs separated again in MII (Fig. 3C). On average, the interval from the beginning of MI SPB separation to that of MII was 40 (± 3) minutes (n=14). Reduplicated SPBs were kept together before the formation of the MII spindle (Fig. 3A, from t=40 minutes to t=46 minutes), as sister SPBs were in MI. In Ipl1-depleted meiotic cells, well before MI separation, sister SPBs formed the doublet configuration (Fig. 3B, t=−24 minutes). Separated SPBs formed a bipolar spindle. Notably, mutant cells lacked a clear spindle-elongation phase (anaphase I); instead, 30 (±8) minutes (n=12) after MI SPB separation, these cells started to form a third Spc42–GFP focus, apparently originating from an existing SPB; then additional Spc42–GFP foci formed (Fig. 3B,C). The newly formed SPBs were initially present at a very low Spc42–GFP fluorescence intensity (Fig. 3B, t=39 min); they grew to an intensity similar to that of the old MI SPBs in ~20–30 minutes (Fig. 3B). All Spc42–GFP foci formed in Ipl1-depleted cells were able to nucleate microtubules, often resulting in multipolar spindles (Fig. 3B), demonstrating that these SPBs are functional in microtubule organization. The SPB morphology appears normal in Ipl1-depleted cells (Fig. 1; supplementary material Fig. S2), suggesting that these SPBs are fully formed. Collectively, our data support the idea that Ipl1 is required for faithful duplication of existing SPBs and prevention of multipolar spindle formation.
Fig. 3.
Microtubule spindle formation during meiosis. (A,B) Fluorescence live-cell microscopy showing SPBs and microtubule spindle dynamics in WT (HY1737) and PCLB2-IPL1 (HY1738) cells during yeast meiosis. SPBs are marked by Spc42–GFP and microtubules by Tub1–mApple. Time zero was defined as the point of SPB separation in MI. The time in minutes is shown below each frame. The time-lapse was 2 minutes for the wild-type cell and 3 minutes for the PCLB2-IPL1 cell. Movies are provided as supplemental data (supplementary material Movies 1 and 2). Note that sister SPBs form a doublet configuration before MI separation in PCLB2-IPL1 cells. Red, Spc42; green, Tub1. Scale bar: 2 μm. (C) Spindle length as determined by pole-to-pole distance in WT and PCLB2-IPL1 cells, as shown in A and B. The distance in three dimensions was measured. MI spindle, purple; MII spindle, other colors.
Cdc5 licenses SPB duplication
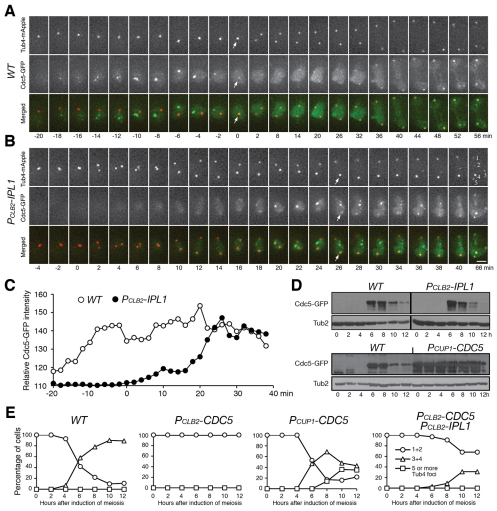
We hypothesized that the completion of SPB separation depends on the formation of a bipolar spindle between the SPBs. The Polo kinase, Cdc5 in yeast, is implicated in SPB separation in vegetative yeast cells and centriole separation in animal cells (Crasta et al., 2008; Tsou et al., 2009). Cdc5, C-terminally tagged with GFP, appeared in the nucleus ~20 minutes before MI SPB separation in wild-type cells, forming foci that were not associated with SPBs (Fig. 4A, from t=−18 to t=−4 minutes, arrows). Cdc5 was concentrated on the SPB during MI SPB separation (Fig. 4A, t=0 minutes, arrows). By contrast, in Ipl1-depleted cells, the complete separation of sister SPBs in MI started at the appearance of Cdc5–GFP, as detected by fluorescence microscopy (Fig. 4B,C), indicating that, upon Cdc5 production, cells immediately initiated SPB separation and bipolar spindle formation. In the mutant cell shown, five Tub4 foci formed 66 minutes after SPB separation and all were enriched with Cdc5–GFP (Fig. 4B). Because the production of Cdc5 appeared at the usual time in Ipl1-depleted cells (Fig. 4D), our data suggest that SPB cohesion prevents premature SPB separation and that Cdc5 is a regulator of this process.
Fig. 4.
Promotion of SPB duplication by Cdc5 at interphase II. (A,B) Live-cell fluorescence microscopy showing Cdc5 localizationinWT(HY1993)and PCLB2-IPL1 (HY2037) cells during meiosis. Cdc5 was tagged with GFP. SPBs are marked by Tub4–mApple. Projected images are shown. Arrows point towards SPB and Cdc5 foci. Red, Tub4; green, Cdc5. Scale bar: 2 μm. (C) Relative intensity of Cdc5–GFP in live meiotic cells. The Cdc5–GFP intensity of an area made up of 400 pixels is plotted against time. This area is centered on the SPBs. The background intensity of Cdc5 is about 110. (D) Western blot showing Cdc5–GFP production during yeast meiosis from WT (HY1993), PCLB2-IPL1 (HY2037) and PCUP1-CDC5 (HY2076) cells. The level of Tub2 serves as a loading control. (E) The number of Tub4–GFP foci in fixed samples from WT (HY1881C), PCLB2-CDC5 (HY2161), PCLB2-CDC5 PCLB2-IPL1 (HY2458) and PCUP1-CDC5 (HY2169) cells. Averages from two independent experiments are shown.
To determine whether Cdc5 is required for SPB duplication at interphase II, we depleted Cdc5 by means of the PCLB2-CDC5 allele (Clyne et al., 2003; Lee and Amon, 2003). In Cdc5-depleted cells, SPBs separated in MI but failed to duplicate at interphase II (Fig. 4E). By contrast, overproduction of Cdc5 by the PCUP1-CDC5 allele dramatically increased the number of meiotic cells with supernumerary SPBs (Fig. 4D,E). In cells depleted of both Ipl1 and Cdc5, ~40% MI SPBs were duplicated to form three or four but not five SPBs (Fig. 4E), suggesting that other factors, in addition to Cdc5, can also promote SPB duplication. Together, these data support the idea that Ipl1 protects SPB cohesion and prevents SPB overduplication, whereas Cdc5 promotes SPB duplication at interphase II.
We propose that SPB cohesion restricts SPB duplication at interphase II, analogous to the notion that engaged centrioles are not licensed for duplication in vertebrates (Tsou and Stearns, 2006). Precocious separation of sister SPBs, presumably due to the collapse of the bridge, licenses them to undergo additional rounds of duplication at interphase II when SPBs are competent for duplication. Meiotic SPB duplication requires the antagonistic interaction between two important cell-cycle regulators, Ipl1 and Cdc5, which have been implicated in numerous activities involving chromosomes and the spindle microtubules through phosphorylation of a variety of cellular substrates. In yeasts, Ipl1 is the only Aurora kinase that functions both on the spindle and at the spindle poles, whereas in metazoans Aurora kinase functions are differentiated, with Aurora A the kinase predominately located at the poles (Barr and Gergely, 2007; Lukasiewicz and Lingle, 2009). In this regard, the kinase activity of Ipl1 in protecting SPB cohesion and preventing SPB overduplication might resemble that of Aurora A kinase in higher eukaryotes. Meanwhile, the Polo-like kinase Cdc5 is required for licensing SPB duplication, presumably by promoting the dissolution of SPB cohesion, as it does with the vertebrate centriole (Tsou et al., 2009). Conceivably, Ipl1 and Cdc5 regulate bridge integrity and SPB cohesion by controlling the phosphorylation status of their substrates at the SPB during budding yeast meiosis.
Materials and Methods
Yeast strains and culture methods
Yeast strains used in the study are diploid SK1 derivatives (supplementary material Table S1). The following mutant alleles have been previously described: PCLB2-IPL1 (Yu and Koshland, 2007), PCLB2-CDC5 (Lee and Amon, 2003), ipl1-as5 (Pinsky et al., 2006), and GAL4.ER and GAL-NDT80 (Carlile and Amon, 2008). We used a PCR-based gene-replacement method to construct PCUP1-CDC5 by replacing the endogenous CDC5 promoter with the CUP1 promoter (Jin et al., 2009). A similar PCR-based method was used to construct SPC42-GFP, CDC5–GFP, MPC54-GFP, TUB1-mApple, and TUB4-mApple. PCR primer information is available upon request.
Yeast cells were grown at 30°C with standard culture methods. Before yeast cells were induced to enter meiosis in 2% potassium acetate, they were grown in the YPA medium with vigorous shaking for about 12 hour, to an optical density (λ=600 nm) of 1.5. To induce PCUP1-CDC5 expression during meiosis, we added 60 μM (final concentration) of CuSO4 to the sporulation medium after induction of meiosis. Inactivation of ipl1-as5 (Pinsky et al., 2006) was induced by addition of 100 μM (final concentration) of 1-NM-PP1 to the sporulation medium after induction of meiosis. To induce GAL-NDT80 expression, we added 100 μM β-estradiol (final concentration) 6 hours after induction of meiosis (Fig. 2D). For monitoring of SPB formation, aliquots of cells were withdrawn at the indicated times, fixed with 1% formaldehyde for 1 hour at room temperature, washed twice with 1× PBS and visualized under a fluorescence microscope.
For live-cell microscopy, we used a concave glass slide as a culture chamber, which was filled with 2% agarose dissolved in 2% potassium acetate. The agarose pad was solidified for 5 minutes at room temperature before use. About 1.5 μl yeast culture was laid on top of the agarose pad then sealed with a glass coverslip. The slide was temperature balanced for 15 minutes at 30°C before microscopy.
Fluorescence microscopy
Live-cell microscopy was carried out on a DeltaVision imaging system (Applied Precision). All live-cell images were acquired at 30°C with a 60× (NA=1.41) objective lens. Seven or eight z-stacks were collected at each time point. Each optical section was 1 μm thick. The exposure time for each optical section was set between 60 and 100 ms. Fixed cells were visualized under an epifluorescence microscope (AxioImager, Zeiss) with a 100× objective lens (NA=1.40).
Electron microscopy
Yeast aliquots (5 ml) were collected 4 and 6 hours after induction of meiosis. Blocks of cells were prepared by a high-pressure freezing and freeze-substitution method, as described previously (Winey et al., 2005). Serial sections of embedded cells were obtained and visualized under a transmission electron microscope (Philips, CM10).
Data analysis and image display
Raw data collected from the DeltaVision imaging system were deconvolved with SoftWorx (Applied Precision). The three-dimensional pole-to-pole length of the SPB (Fig. 3) was determined with the measurement tool provided by SoftWorx. Optical sections were projected into two dimensions for display. Projected images were used to generate the histograms shown in Fig. 1A and Fig. 4B. An area composed of 400 pixels centered on the SPBs is shown, and the pixel size is 0.1070 μm.
Western blot
Western blotting was performed as previously described (Jin et al., 2009). Cdc5-GFP was detected by an anti-GFP antibody (Cat#632569, Clontech). The level of Tub2 (β-tubulin) served as a loading control.
Acknowledgments
We thank A. Amon, S. Biggins and M. Davidson for providing yeast strains and reagents. M. Avey and C. Clarissa provided technical assistance. A. B. Thistle assisted with text editing. This work was supported in part by NIH R01GM51312 to M.W. and by the Florida Biomedical Research Program (08BN-08) and the National Science Foundation (MCB-0718384) to H.Y. Deposited in PMC for release after 12 months.
Footnotes
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.086652/-/DC1
References
- Adams I. R., Kilmartin J. V. (2000). Spindle pole body duplication: a model for centrosome duplication? Trends Cell Biol. 10, 329-335 [DOI] [PubMed] [Google Scholar]
- Barr A. R., Gergely F. (2007). Aurora-A: the maker and breaker of spindle poles. J. Cell Sci. 120, 2987-2996 [DOI] [PubMed] [Google Scholar]
- Bullitt E., Rout M. P., Kilmartin J. V., Akey C. W. (1997). The yeast spindle pole body is assembled around a central crystal of Spc42p. aCell 89, 1077-1086 [DOI] [PubMed] [Google Scholar]
- Byers B., Goetsch L. (1974). Duplication of spindle plaques and integration of the yeast cell cycle. Cold Spring Harb. Symp. Quant. Biol. 38, 123-131 [DOI] [PubMed] [Google Scholar]
- Carlile T. M., Amon A. (2008). Meiosis I is established through division-specific translational control of a cyclin. Cell 133, 280-291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C. S., Botstein D. (1993). Isolation and characterization of chromosome-gain and increase-in-ploidy mutants in yeast. Genetics 135, 677-691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clyne R. K., Katis V. L., Jessop L., Benjamin K. R., Herskowitz I., Lichten M., Nasmyth K. (2003). Polo-like kinase Cdc5 promotes chiasmata formation and cosegregation of sister centromeres at meiosis I. Nat. Cell Biol. 5, 480-485 [DOI] [PubMed] [Google Scholar]
- Crasta K., Lim H. H., Giddings T. H., Jr, Winey M., Surana U. (2008). Inactivation of Cdh1 by synergistic action of Cdk1 and polo kinase is necessary for proper assembly of the mitotic spindle. Nat. Cell Biol. 10, 665-675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson A. D., Kilmartin J. V. (1996). Spc42p: a phosphorylated component of the S. cerevisiae spindle pole body (SPD) with an essential function during SPB duplication. J. Cell Biol. 132, 887-901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen S. L., Winey M. (2004). The budding yeast spindle pole body: structure, duplication, and function. Annu. Rev. Cell Dev. Biol. 20, 1-28 [DOI] [PubMed] [Google Scholar]
- Jaspersen S. L., Huneycutt B. J., Giddings T. H., Jr, Resing K. A., Ahn N. G., Winey M. (2004). Cdc28/Cdk1 regulates spindle pole body duplication through phosphorylation of Spc42 and Mps1. Dev. Cell 7, 263-274 [DOI] [PubMed] [Google Scholar]
- Jin H., Guacci V., Yu H.-G. (2009). Pds5 is required for homologue pairing and inhibits synapsis of sister chromatids during yeast meiosis. J. Cell Biol. 186, 713-725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilmartin J. V. (2003). Sfi1p has conserved centrin-binding sites and an essential function in budding yeast spindle pole body duplication. J. Cell Biol. 162, 1211-1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M., Strasser K. (2000). Role of the spindle pole body of yeast in mediating assembly of the prospore membrane during meiosis. EMBO J. 19, 3657-3667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B. H., Amon A. (2003). Role of Polo-like kinase CDC5 in programming meiosis I chromosome segregation. Science 300, 482-486 [DOI] [PubMed] [Google Scholar]
- Li S., Sandercock A. M., Conduit P., Robinson C. V., Williams R. L., Kilmartin J. V. (2006). Structural role of Sfi1p-centrin filaments in budding yeast spindle pole body duplication. J. Cell Biol. 173, 867-877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukasiewicz K. B., Lingle W. L. (2009). Aurora A, centrosome structure, and the centrosome cycle. Environ. Mol. Mutagen. 50, 602-619 [DOI] [PubMed] [Google Scholar]
- Moens P. B., Rapport E. (1971). Spindles, spindle plaques, and meiosis in the yeast Saccharomyces cerevisiae (Hansen). J. Cell Biol. 50, 344-361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monje-Casas F., Prabhu V. R., Lee B. H., Boselli M., Amon A. (2007). Kinetochore orientation during meiosis is controlled by Aurora B and the monopolin complex. Cell 128, 477-490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinsky B. A., Kung C., Shokat K. M., Biggins S. (2006). The Ipl1-Aurora protein kinase activates the spindle checkpoint by creating unattached kinetochores. Nat. Cell Biol. 8, 78-83 [DOI] [PubMed] [Google Scholar]
- Straight P. D., Giddings T. H., Jr, Winey M. (2000). Mps1p regulates meiotic spindle pole body duplication in addition to having novel roles during sporulation. Mol. Biol. Cell 11, 3525-3537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsou M. F., Stearns T. (2006). Mechanism limiting centrosome duplication to once per cell cycle. Nature 442, 947-951 [DOI] [PubMed] [Google Scholar]
- Tsou M. F., Wang W. J., George K. A., Uryu K., Stearns T., Jallepalli P. V. (2009). Polo kinase and separase regulate the mitotic licensing of centriole duplication in human cells. Dev. Cell 17, 344-354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M., Morgan G. P., Straight P. D., Giddings T. H., Jr, Mastronarde D. N. (2005). Three-dimensional ultrastructure of Saccharomyces cerevisiae meiotic spindles. Mol. Biol. Cell 16, 1178-1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H.-G., Koshland D. E. (2007). The Aurora kinase Ipl1 maintains the centromeric localization of PP2A to protect cohesin during meiosis. J. Cell Biol. 176, 911-918 [DOI] [PMC free article] [PubMed] [Google Scholar]