Abstract
Bacteriophage HK022 Nun protein blocks transcription elongation by Escherichia coli RNA polymerase in vitro without dissociating the transcription complex. Nun is active on complexes located at any template site tested. Ultimately, only the 3′-OH terminal nucleotide of the nascent transcript in an arrested complex can turn over; it is removed by pyrophosphate and restored with NTPs. This suggests that Nun inhibits the translocation of RNA polymerase without abolishing its catalytic activities. Unlike spontaneously arrested complexes, Nun-arrested complexes cannot be reactivated by transcription factor GreB. The various complexes show distinct patterns of nucleotide incorporation and pyrophosphorolysis before or after treatment with Nun, suggesting that the configuration of RNAP, transcript, and template DNA is different in each complex.
Keywords: Phage HK022, Nun protein, RNA polymerase, transcription arrest, translocation
The product of the nun gene of prophage HK022 inhibits superinfection by the related coliphage λ (Robert et al. 1987). The 13-kD Nun protein, like the λ N antitermination protein, binds to the cis-acting λ nut RNA elements through an arginine-rich binding motif (Chattopadhyay et al. 1995). In contrast to λ N, which suppresses transcription termination promoter-distal to nut, Nun provokes termination downstream of these elements in both the λ pL and pR operons (Robert et al. 1987; Robledo et al. 1990; Sloan and Weisberg 1993). Nun-induced transcription termination is inhibited by mutations in nut or in the trans-acting Escherichia coli proteins NusA, NusB, NusE, and NusG (Robert et al. 1987; Robledo et al. 1990, 1991; Baron and Weisberg 1992; E. Burova, S.C. Hung, and M.E. Gottesman, unpubl.).
Transcription arrest by Nun has been duplicated in a defined cell-free system. Purified Nun protein is effective, although the reaction is strongly stimulated by the four Nus proteins (Hung and Gottesman 1995). The binding of Nun protein to the stem–loop boxB sequence of λ nut RNA and the effects of boxB mutations and deletions on Nun binding and transcription arrest have been demonstrated in vitro (Chattopadhyay et al. 1995; Hung and Gottesman 1995). A mutation in NusG that inhibits Nun action in vivo abolishes Nus-mediated stimulation of Nun action in vitro (E. Burova, S.C. Hung, and M.E. Gottesman, unpubl.). Thus, the biological properties of Nun have been reproduced in vitro, with the exception that the arrested complex does not dissociate from the template in our cell-free system.
In this paper we describe how Nun induces transcription arrest at defined template sites in vitro. We demonstrate that paused or stalled RNA polymerase (RNAP) complexes respond to Nun in distinct ways. These data suggest that Nun blocks the translocation of RNAP at a defined point in the elongation process.
Results
Effects of Nun on RNA polymerization
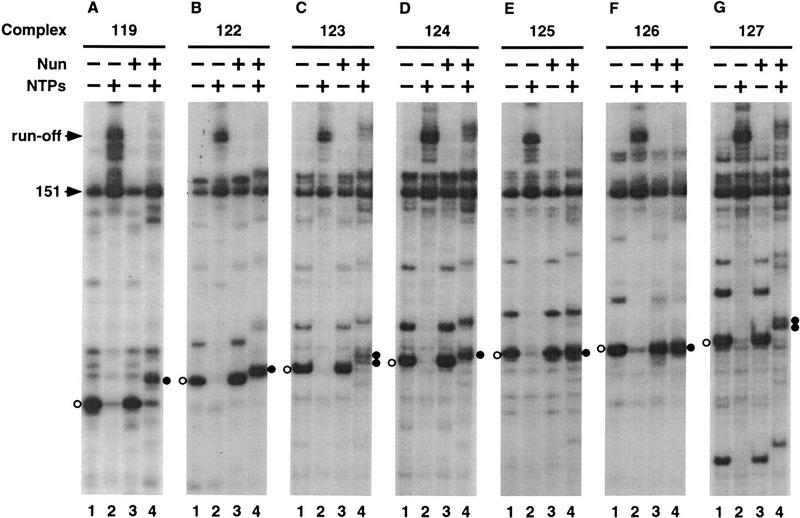
Nun arrests elongating RNAP at or near intrinsic pause sites (Hung and Gottesman 1995). This may indicate that Nun recognizes specific structural features of RNAP paused at such sites. Alternatively, Nun modification of elongating RNAP may be a slow process, so that Nun only arrests transcription complexes that dwell for an extended period at any template site. We distinguished between these possibilities using an interrupted elongation strategy. Transcription elongation on a λ pL operon template was permitted for a brief period and then interrupted by the removal of NTPs (see Materials and Methods for details). Under these conditions, many of the complexes paused at the 119-nucleotide site, that is, contained a 119-nucleotide nascent transcript (Fig. 1A, lane 1). The paused RNAP was then “walked” by repeated addition and removal of the appropriate NTP subsets to generate complexes stalled at selected downstream sites (Fig. 1B–G, lanes 1; see Materials and Methods for details). The stalled complexes were elongation proficient, as addition of the complete set of NTPs generated principally the 161-nucleotide and 162-nucleotide runoff transcripts (Fig. 1A–G, lanes 2). In addition, a 151-nucleotide transcript, present in a spontaneously arrested complex, formed in all transcription assays (Fig. 1A–G; see below).
Figure 1.
Nun-induced arrest of transcription elongation complexes. Transcription elongation complexes located at various positions on a λ pL operon template were prepared and designated as described in Materials and Methods. Each panel shows the transcript of a complex located at a particular position (lanes 1) and the RNA product(s) after the complex was chased with NTPs (lanes 2), incubated with Nun (lanes 3), and incubated with Nun and then chased with NTPs (lanes 4). In this and subsequent figures, the + or − sign in the lane description denotes the addition or omission, respectively, of the reagent shown to the far left of the sign. The descending order of lane descriptions corresponds to the order of treatments of the transcription complex. The pointers with run-off and 151 labels locate the runoff transcript and the 151-nucleotide transcript of the spontaneously arrested complex, respectively. In each panel, the transcript of the starting complex is located by ○ on the left, and the transcript(s) of the Nun-arrested complex(es) thus derived is located • on the right.
To determine whether these stalled complexes were susceptible to arrest, they were first preincubated with Nun for 5 min and then chased with four NTPs. Note that preincubation with Nun did not alter the size of the transcript over this time period (Fig. 1A–G, lanes 1,3). With the addition of the four NTPs, all of the Nun-pretreated complexes arrested (Fig. 1A–G, lanes 2,4). With the exception of the 122-nucleotide arrested complex, which derives from the paused complex 119, none of the arrested complexes had been observed in an “uninterrupted” elongation assay (data not shown). This result indicates that Nun can act on stalled or paused complexes and rules out the notion that the latter represent a particular Nun-sensitive class of RNAPs. Most complexes arrested after the incorporation of one or a few nucleotides. Complexes 125 and 126, however, arrested without further transcript elongation. The concentrations of chase NTPs affected neither the efficiency of arrest nor the number of nucleotides incorporated by the Nun-treated complex prior to arrest (data not shown).
We next determined the kinetics of Nun interaction with RNAP. Nun was preincubated with complexes 122 and 126 for different periods of time prior to the addition of NTPs (Fig. 2A,B). Little or no arrest could be detected when Nun and NTPs were added simultaneously (lanes 3). The efficiency of arrest increased with the time of preincubation with Nun and reached a plateau after >1 min of preincubation (lanes 4–7). These data confirm that Nun can arrest elongating RNAP at any site, but the dwell time of the transcription complex at that site must be sufficient to allow interaction with Nun.
Figure 2.
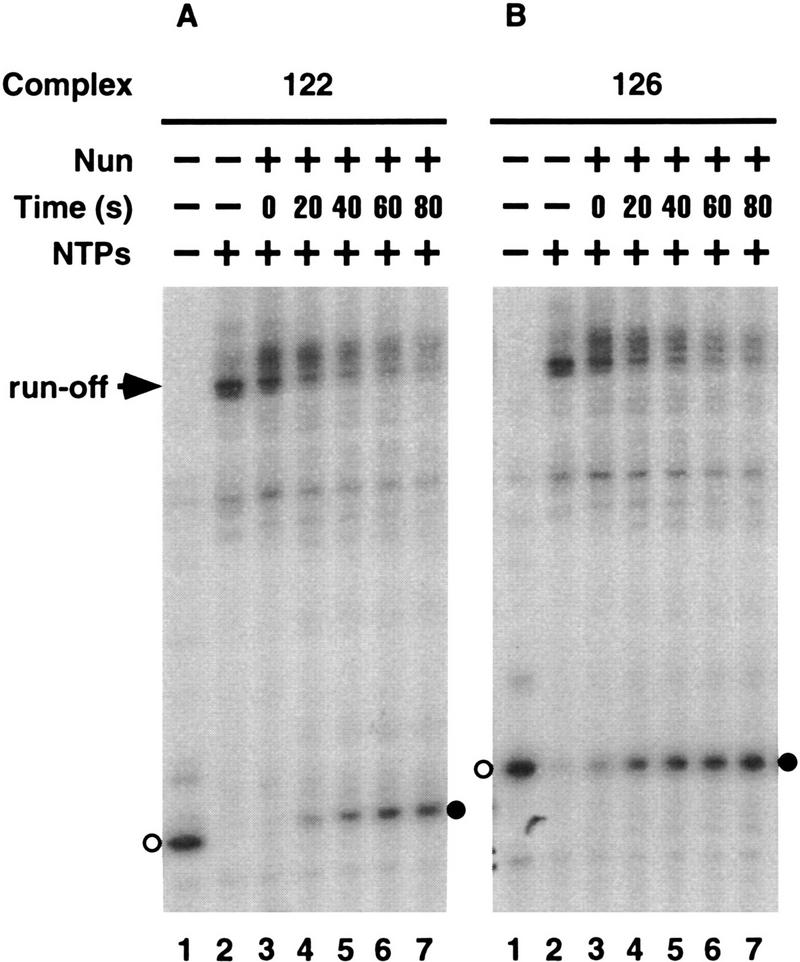
Time requirement of complex modification by Nun transcription complexes 122 and 126 (lanes 1) were incubated with Nun (lanes 3–7) for the indicated periods of time (Time) and were then chased with NTPs (lanes 2–7).
Nun-induced transcription arrest and GreB-stimulated transcript cleavage
Transcription complexes located at certain sites tend to arrest spontaneously (Arndt and Chamberlin 1990). Such complexes are particularly sensitive to endonucleolytic transcript cleavage stimulated by the E. coli GreB protein (Borukhov et al. 1993). Transcript cleavage reactivates spontaneously arrested complexes, allowing elongation to continue from the new 3′-OH terminal nucleotide (Borukhov et al. 1993; Nudler et al. 1994). We asked if complexes arrested by Nun could also be reactivated by GreB.
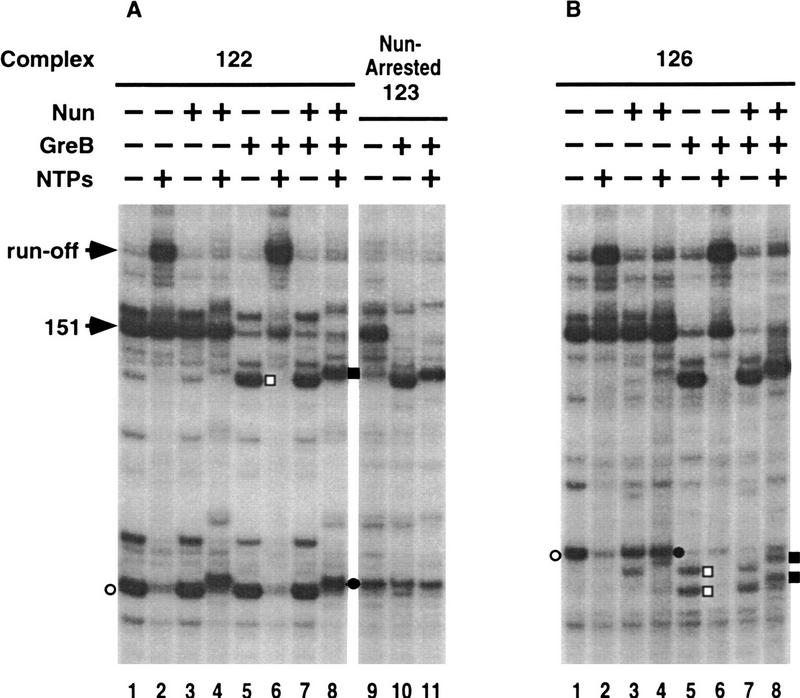
Elongation complexes were incubated with approximately equimolar concentrations of GreB for 5 min (Fig. 3). Under these conditions, only the 126 and 151 complexes were GreB-sensitive. The 126-nucleotide transcript was cleaved to 122 and 124 nucleotides (lane 12). Recall that complex 126 was arrested by Nun without transcript elongation (Fig. 1F). All other GreB-sensitive complexes likewise arrested without nucleotide addition (data not shown).
Figure 3.
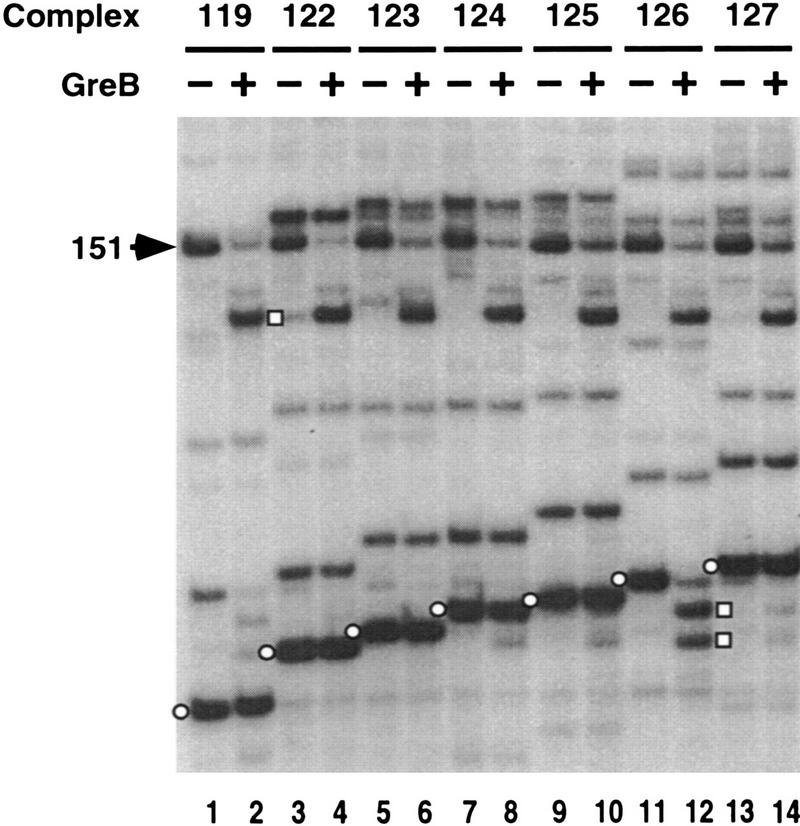
GreB-stimulated transcript cleavage of transcription complexes. Transcription complexes (odd-numbered lanes) were incubated with approximately equimolar (50 nm) of GreB (even-numbered lanes). The cleaved transcript is indicated □ on the right.
Transcription reactions performed with Nun and GreB demonstrate that GreB does not block Nun-mediated arrest and that the initial sensitivity or resistance of a transcript to GreB-induced cleavage is retained in the Nun-arrested complex. For example, the GreB-resistant complex 122 arrested after the incorporation of a single nucleotide in the presence or absence of GreB (Fig. 4A, lanes 1,4,8). The 123-nucleotide transcript was stable when the Nun-arrested complex was incubated with GreB (lane 10) and did not elongate when the complex was subsequently incubated with NTPs (lane 11). The GreB-sensitive 126 complex remained sensitive after exposure to Nun (Fig. 4B, lanes 1,5,7). However, the cleaved 122- and 124-nucleotide transcripts, unlike the original 126-nucleotide transcript, added a single nucleotide prior to Nun arrest (Fig. 4B, lane 8). The same pattern of transcription elongation and arrest was seen when the order of addition of Nun and GreB was reversed or when both proteins were added simultaneously (data not shown). Recall that the stalled complexes 122 and 124 also arrested after the incorporation of a single nucleotide (Fig. 1B,D). Figure 4B also shows that Nun has weak GreB-like activity. A portion of the transcript in the 126 complex was cleaved to 124 nucleotide nt after 15 min incubation with Nun (cf. lanes 1 and 3).
Figure 4.
Effects of GreB on Nun-induced transcription arrest. Transcription complexes 122 and 126 (lanes 1) were incubated with different combinations of Nun, GreB, and NTPs, as detailed in the lane descriptions (lanes 2–8). The Nun-arrested complex with a 123-nucleotide transcript (Nun-Arrested 123) used in the reactions shown in A (lanes 9–11), was prepared from the reaction shown in A (lane 4) after removal of unincorporated NTPs. The complex was incubated with GreB (lanes 10–11) and then chased with NTPs (lane 11). (▪) The transcript of an arrested complex after treatment with Nun and GreB.
The effect of Nun on the spontaneously arrested 151 complex is also shown in Figure 4. The 151-nucleotide transcript fails to elongate either in the presence or absence of Nun (lanes 2,4). That the transcript is still engaged in the complex is shown by its efficient cleavage when exposed to GreB (lanes 5). Transcript cleavage reactivated the complex, and the shortened transcripts elongated upon incubation with NTPs (lanes 6). The 151 complex was sensitive to GreB-stimulated transcript cleavage in the presence of Nun (lanes 7). As shown above for the 126 complex, the cleaved transcripts incorporated a single nucleotide prior to Nun-mediated arrest (lanes 8). We conclude that Nun modification persists after transcript cleavage stimulated by GreB and limits subsequent elongation to a single nucleotide.
Effects of Nun modification on RNA pyrophosphorolysis
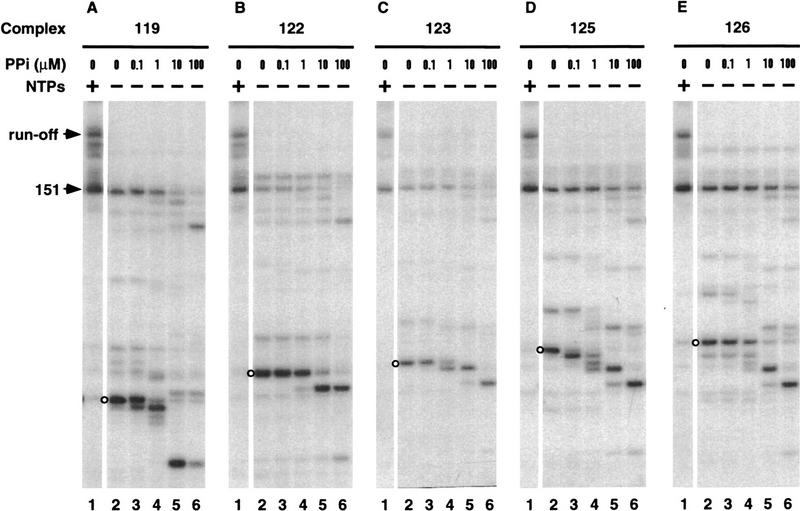
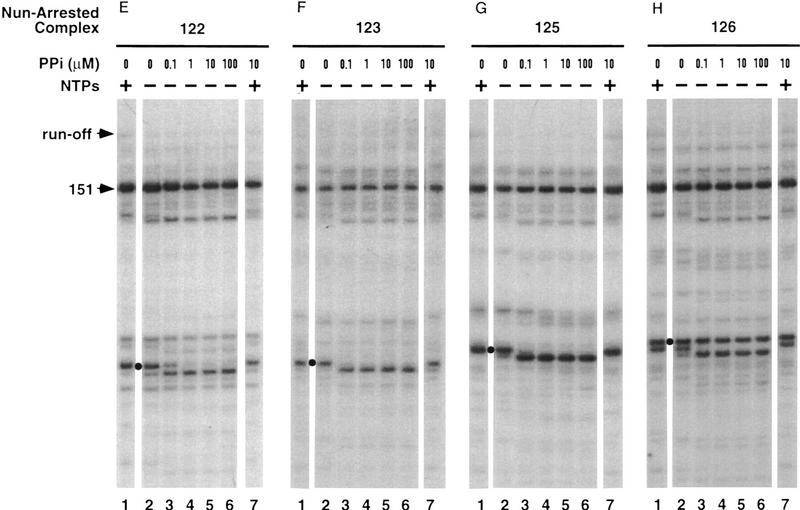
We wondered whether, in addition to blocking RNA polymerization, Nun also inhibited the reverse reaction, RNA pyrophosphorolysis. Elongation complexes were incubated with pyrophosphate over a broad range of concentrations (0.1–100 μm) to determine the sensitivity of their nascent transcripts to pyrophosphorolysis (Fig. 5). Complex 125 was the most sensitive; the terminal nucleotide of the 125-nucleotide transcript was cleaved at 0.1 μm pyrophosphate (Fig. 5D, lane 3). Other complexes were cleaved only at ⩾1 μm pyrophosphate. The sensitivity of a complex to GreB was unrelated to its sensitivity to pyrophosphate. At >10 μm pyrophosphate, all nascent transcripts underwent multiple rounds of pyrophosphorolysis (Fig. 5A–E, lanes 5,6).
Figure 5.
RNA pyrophosphorolysis of transcription complexes. Transcription complexes (lanes 2) were chased with NTPs (lanes 1) or incubated with sodium pyrophosphate (PPi) at the indicated concentrations for 15 min (lanes 3–6).
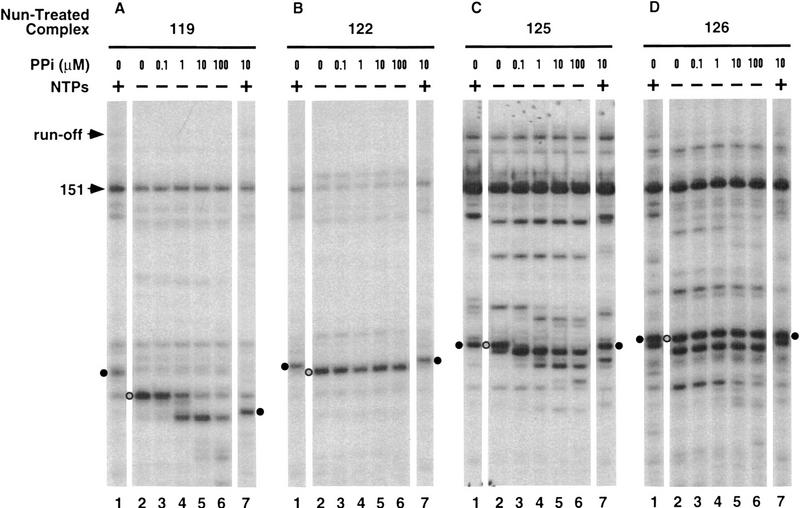
We next determined how exposure to Nun, in the absence of NTPs, affected the pyrophosphate sensitivity of several stalled or paused transcription complexes (Fig. 6A–D). The response of each of these Nun-modified complexes to pyrophosphate was unique.
Figure 6.
Effects of Nun on RNA pyrophosphorolysis. (A–D) Nun-treated complexes 119, 122, 125, and 126 (lanes 2) were prepared by preincubating the corresponding elongation complexes with Nun for 5 min. The transcript of such a complex is indicated by a shaded circle on the left. (E–H) Nun-arrested complexes 122, 123, 125, and 126 (lanes 2) were prepared from Nun-treated complexes 119, 122, 125 and 126, respectively, by chasing with NTPs and then removing unincorporated NTPs. The complexes were then incubated with sodium pyrophosphate (PPi) at the indicated concentrations for 15 min (lanes 3–7) and/or chased with NTPs (lanes 1,7).
Incubation of complex 119 with Nun did not protect it from repeated transcript cleavage (Fig. 6A). At 10 μm pyrophosphate or below, the treated complex was limited to three rounds of pyrophosphorolysis (lane 5). That the complex was arrested was indicated by the incorporation of only one nucleotide upon subsequent addition of NTPs (lane 7). The efficiency of arrest at this site was reduced at higher pyrophosphate concentrations (lane 6).
Nun modification protected complex 122 against pyrophosphorolysis (Fig. 6B, lanes 2–6). The modified complex added a single nucleotide prior to arrest, either in the presence or absence of pyrophosphate (Fig. 6B, lanes 1,7).
Complex 125 retained its exquisite sensitivity to pyrophosphate after exposure to Nun. Although the terminal nucleotide was still removed at 0.1 μm pyrophosphate, there was no further transcript cleavage, even at 100 μm pyrophosphate (Fig. 5D and 6C, cf. lanes 3–6). The single nucleotide removed by pyrophosphorolysis was reincorporated when NTPs were subsequently added (Fig. 6C, lane 7). Recall that complex 125 arrested without nucleotide addition (Fig. 6C, lane 1).
The GreB-sensitive complexes 126 and 151 were completely refractory to pyrophosphorolysis after Nun modification (Fig. 6D, lanes 3–6). Incubation of complex 126 with Nun resulted in partial cleavage of the transcript, principally to a 124-nucleotide transcript (Fig. 6D, lane 2) as described above (Fig. 4, lane 3). The 124-nucleotide transcript was likewise resistant to pyrophosphate cleavage. When incubated with NTPs, the cleaved complex arrested after incorporating one nucleotide (lane 1), resembling the Nun-treated elongation complex 124 (Fig. 1D).
The properties of the different complexes before and after Nun treatment are summarized in Table 1.
Table 1.
A summary of the properties of λpL transcription complexes observed in this study
| Complex
|
Nun
|
Nucleotide incorporationa
|
Pyrophosphorolysisa
|
GreB sensitivityb
|
Preferred configurationc
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| 0.1 μm PPi
|
10 μm PPi
|
|||||||||
| −
|
+
|
−
|
+
|
−
|
+
|
−
|
+
|
|||
| 119 | U | 3 | 0 | 0 | U | 3 | R | R | paused | |
| 122 | U | 1 | 0 | 0 | U | 0 | R | R | poly. (I) | |
| 125 | U | 0 | 1 | 1 | U | 1 | R | R | pyro. (II) | |
| 126 | U | 0 | 0 | 0 | U | 0 | S | S | endo. (III) | |
| 151 | 0 | 0 | 0 | 0 | U | 0 | S | S | endo. (III) | |
Each complex is identified by the number of nucleotides in its transcript. The properties of each complex with (+) or without (−) preincubation with Nun are shown.
(U) An unlimited reaction. When a limited reaction occurs, the number of nucleotides incorporated or removed is indicated.
(R or S) GreB resistant or GreB sensitive, respectively.
Poly., pyro., and endo. are abbreviations for polymerization-, pyrophosphorolysis-, or endonucleolysis-ready configuration, respectively; the bracketed Roman numeral refers to the structure in Figure 7 that depicts the corresponding configuration. The preferred configuration of the paused complex 119 cannot be determined by the observed properties and is thus labeled paused. (See text for a detailed discussion.)
Pyrophosphate sensitivity of Nun-arrested complexes
We then examined the pyrophosphate sensitivity of complexes arrested in the presence of Nun and NTPs. Arrested complexes 122, 123, 125, and 126 were obtained by incubating elongation complexes 119, 122, 125, and 126, respectively, with Nun and NTPs and then removing unreacted NTPs by gel filtration. Arrest was confirmed by demonstrating that the complexes could not elongate upon the addition of NTPs (Fig. 6E–H, lanes 1). The transcript of arrested complex 126 was totally resistant to pyrophosphate (Fig. 6H, lanes 3–6). In contrast, the terminal nucleotides in the transcripts of arrested complexes 122, 123, or 125 were efficiently cleaved by pyrophosphate at 0.1 μm (Fig. 6E–G, lanes 3). The shortened transcript was then completely refractory to pyrophosphorolysis (Fig. 6E–G, lanes 3–6). After pyrophosphorolysis, the arrested complexes reincorporated only the cleaved nucleotide (Fig. 6E–G, lanes 7). These responses to pyrophosphate and NTPs were identical in all arrested GreB-resistant complexes (data not shown). Thus, the ultimate nucleotide incorporated in the presence of Nun is sensitive to pyrophosphorolysis, but all other nucleotides are resistant. How this unique pattern of pyrophosphorolysis may reflect the special conformation of the arrested complex is discussed below.
Discussion
Elongation complexes vary in their response to Nun
Using an interrupted elongation strategy, we have shown that the HK022 Nun protein inhibits RNA polymerization by RNAP elongation complexes paused at or walked to and stalled at various positions on a λ DNA template. We have also demonstrated that there is a slow step in the Nun-induced reaction. This explains why Nun arrest sites are associated with intrinsic pause sites in a continuous elongation assay (Hung and Gottesman 1995). Nun-induced transcription arrest is not specific to paused complexes. In fact, RNAP located at a pause site may be resistant to full modification by Nun (see below).
Nun also inhibits the reverse reaction of RNA polymerization; transcripts in arrested complexes are resistant to extensive pyrophosphorolysis. We suggest that these properties reflect a common feature of the Nun reaction, namely that Nun blocks both forward and backward translocation of the nascent transcript in the elongation complex. The complexes that we have analyzed differ with respect to the sensitivity of their transcripts to pyrophosphorolysis and to cleavage induced by the E. coli GreB protein. They also vary in how they respond to Nun. Based on these characteristics, we suggest that the various paused and stalled complexes represent different and structurally distinct intermediates in the elongation cycle.
Structure and properties of a transcription elongation complex
Although direct visualization of RNAP is only now becoming possible (Polyakov et al. 1995), the analysis of its enzymatic properties has yielded considerable structural information. Transcribing RNAP interacts with the 3′-proximal nucleotides of the nascent transcript through the product-binding (P) site of the protein (Krakow and Fronk 1969; see Chan and Landick 1994). The RNAP active center includes a nucleotide-binding (N) site that can accommodate either a nucleotide in the nascent transcript or the nucleoside monophosphate moiety of the substrate NTP. Phosphodiester bonds are formed or disrupted 5′ to the N site. These RNAP sites and the nascent transcript in an elongating complex are depicted in Figure 7.
Figure 7.
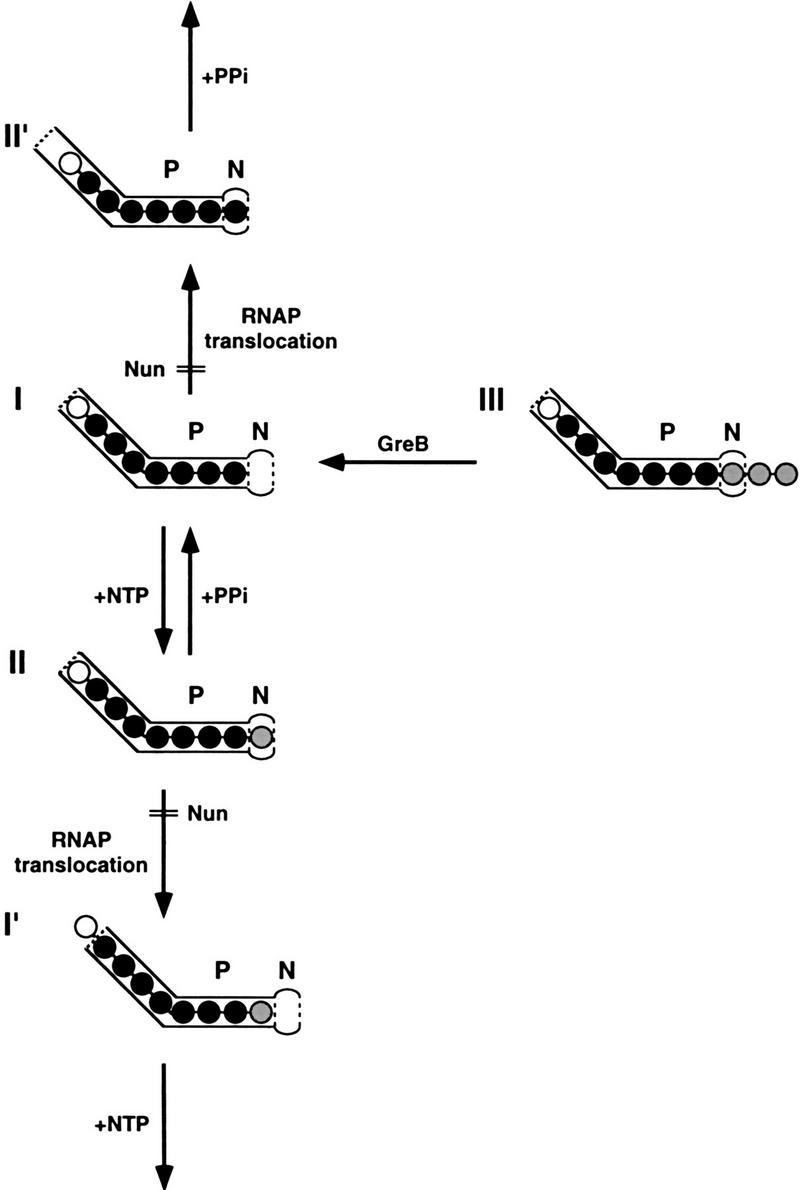
A model for the action of Nun on transcription complexes. Each numbered structure represents a transcription complex at a particular transitional stage during elongation. An arrow between two structures indicates a transition. Transitions that involve RNAP translocation are indicated, as is the substrate/factor required for a transition. A transition inhibited by Nun is shown by double lines labeled Nun. In each structure, the bent rectangle represents the product-binding (P) site of RNAP. The oval represents the nucleotide-binding (N) site of the active center of RNAP. The circles represent nucleotides in the nascent transcript. To help visualize the translocation of RNAP, some circles are shaded differently to identify particular nucleotides in the transcript. The number of nucleotides accommodated by the product-binding site is arbitrarily depicted. (See text for detailed description.)
To support RNA polymerization, the N site must be located immediately downstream to the 3′-hydroxyl of the nascent transcript. Structure I depicts such a polymerization-ready RNAP, in which the N site is vacant. The appropriate NTP then enters the N site and forms a phosphodiester bond with the 3′-hydroxyl of the nascent transcript, yielding structure II. Further nucleotide incorporation requires downstream translocation of the RNAP, which places the new 3′-hydroxyl nucleotide upstream of the N site (structure I′; which is configurationally equivalent to structure I). Processive RNA polymerization occurs by repeated nucleotide incorporation and translocation (Fig. 7, downward-pointing arrows).
Pyrophosphorolysis is the reverse reaction of RNA polymerization. Both reactions are thought to be carried out by the same active center (Krakow and Fronk 1969). In contrast to elongation, pyrophosphorolysis requires that the 3′-terminal nucleotide of the nascent transcript be located in the N site, as shown in structure II (the pyrophosphorolysis-ready form). Pyrophosphate cleaves the 3′-most phosphodiester bond by a nucleophilic attack on the phosphorus atom (Rozovskaya et al. 1984). Removal of the NTP by-product yields structure I. A second round of pyrophosphorolysis occurs when upstream translocation of RNAP feeds the newly created 3′-terminal nucleotide into the N site, producing structure II′, which is configurationally equivalent to structure II. Processive pyrophosphorolysis entails repeated bond breakage and translocation (Fig. 7, upward-pointing arrows) and may require higher pyrophosphate concentrations than the removal of the initial 3′-terminal nucleotide (Kassavetis et al. 1986).
Endonucleolytic hydrolysis of nascent transcripts has been demonstrated in both prokaryotic and eukaryotic systems (Surratt et al. 1991; see Reines 1994). An RNAP-associated factor, such as GreA or GreB for E. coli RNAP (Borukhov et al. 1992, 1993), or TFIIS for RNAPII (Izban and Luse 1992; Reines 1992; Guo and Price 1993) stimulates hydrolytic cleavage at a phosphodiester bond two or more nucleotides from the transcript 3′ terminus. The small oligonucleotide product is released, whereas the upstream portion of the transcript remains engaged and can be elongated. Although efficient cleavage occurs in vitro only in the presence of a stimulatory factor, hydrolytic transcript cleavage is an intrinsic activity of the RNAP (Orlova et al. 1994; Rudd et al. 1994) and is believed to be promoted by the RNAP active center (Rudd et al. 1994). The active center can approach the scissile phosphodiester bond by upstream sliding of RNAP along the transcript (Komissarova and Kashlev 1997; Nudler et al. 1997), as shown in Figure 7, structure III (the endonucleolysis-ready form). Structure III is incapable of transcript elongation. Cleavage and the release of the downstream oligonucleotide product regenerates structure I and permits resumption of RNA polymerization.
With the exception of complex 151, all of the complexes in this study were elongation proficient. All of the nascent transcripts were cleaved at high concentrations of pyrophosphate or GreB. Our data are consistent with the idea that a particular transcription complex is in equilibrium among various structures, although one structure may predominate. Presumably, sensitivity to pyrophosphate, GreB, or Nun reflects the population distribution of these various structures, although other possibilities have not been excluded. Thus, complex 125, which is particularly sensitive to pyrophosphate, exists largely in the pyrophosphorolysis-ready form (structure II). Complex 126 is in equilibrium between an elongation proficient form and a GreB-sensitive, presumably inactive, form (structure III). Complex 119 or 122, which is relatively resistant to both transcript cleavage reactions, may be in equilibrium between the polymerization-ready form (structure I) and another configuration.
How Nun induces transcription arrest
Our data show that Nun acts by inhibiting the translocation of RNAP. The 3′-terminal nucleotide of a GreB-resistant complex arrested by Nun is highly susceptible to pyrophosphorolysis, arguing that the arrested complex is in the pyrophosphorolysis-ready form (Fig. 7, structure II). However, the complex is totally resistant to a second round of pyrophosphorolysis. Moreover, the arrested complex can reincorporate only the single cleaved nucleotide. The arrested complex is therefore limited to making and breaking the same phosphodiester bond. This indicates that the active center of the Nun-arrested complex retains its catalytic activities but cannot translocate relative to the nascent transcript.
Translocation of RNAP entails the shifting of the active center and the nucleic acid-binding channels relative to RNA and DNA, the unwinding and rewinding of DNA at the promoter–distal (downstream) and promoter–proximal (upstream) edges of the transcription bubbles, respectively, and the unwinding of the RNA–DNA heteroduplex near the upstream edge of the transcription bubble. Inhibition of any one of these activities would prevent translocation of RNAP. It is not yet known which of these activities is/are blocked in a Nun-modified RNAP.
As predicted from this model of Nun action, complex 125, which is in the pyrophosphorolysis-ready form, with the N site occupied, is arrested without nucleotide incorporation. Complex 122 is in the polymerization-ready form (structure I). By preventing translocation of RNAP upstream, Nun blocks the filling of the N site with the transcript 3′-OH terminus, and pyrophosphorolysis cannot take place. Because complex 122 has a vacant N site, it can incorporate one, but only one, nucleotide after Nun modification.
This mechanism also explains the response of a GreB-sensitive complex to Nun. We assume that Nun modifies a GreB-sensitive complex after upstream translocation of the RNAP. This configuration (structure III) is maintained in the modified complex, preventing both elongation and pyrophosphorolysis. GreB-stimulated transcript cleavage converts the modified complex to structure I, and it can now incorporate one nucleotide prior to arrest.
The spontaneously paused complex 119 differs from the stalled complexes described above. After exposure to Nun, this complex can undergo multiple rounds of nucleotide incorporation or pyrophosphorolysis prior to arrest. It appears that Nun is unable to modify fully RNAP paused at position 119 and that translocation arrest takes effect only after the RNAP has moved to an upstream or downstream position. Interestingly, most of the complexes isolated directly from kinetically interrupted elongation reactions undergo more than one round of nucleotide incorporation or pyrophosphorolysis after incubation with Nun. The special configuration(s) of these paused complexes (Chan et al. 1997) may render them resistant to full modification by Nun.
Spontaneous and Nun-mediated elongation arrest
Borukhov et al. (1993) suggest that spontaneously arrested complexes arise when the RNAP active center and 3′ terminus of the nascent transcript misalign. GreB-stimulated cleavage removes the protruding 3′ nucleotides and permits further elongation. The response of the spontaneously arrested 151 complex to Nun is consistent with this model. Complex 151 is unable to elongate prior to treatment with GreB, although it undergoes transcript cleavage at high pyrophosphate concentrations. The 151 RNAP is therefore capable of some upstream movement. By blocking the translocation of RNAP, Nun renders complex 151 entirely resistant to pyrophosphate. As expected, the Nun-modified complex remains sensitive to GreB and is reactivated by GreB-induced cleavage. The reactivated complex, however, adds only a single nucleotide, implying that GreB-stimulated cleavage generates a complex of the type I structure that remains modified by Nun.
Materials and methods
Nun protein was purified as described (Chattopadhyay et al. 1995). RNAP was purified according to Burgess and Jendrisak (1975) from E. coli strain AD8571, which carries disrupted greA and greB genes (Orlova et al. 1995). GreB protein was a generous gift from Dr. S. Borukhov (State University of New York Health Science Center, Brooklyn). [α-32P]NTPs and unlabeled ultrapure NTPs were purchased from Amersham and Pharmacia, respectively.
Transcription complexes are identified by the lengths of their nascent transcripts, for example, a complex containing a nascent transcript of 100 nucleotides is denoted as complex 100. The DNA template used for all experiments presented in this paper contained the phage λ sequence from the coordinates 35422–35660, including the pL promoter and the nutL sequence; the runoff transcript is 161 nucleotides. The template was prepared as described (Hung and Gottesman 1995), except the following oligonucleotides were used as primers for PCR: 5′-CATACAGATAACCATCTGCGGTG-3′ and 5′-CCCCGCGATTGGCACATTCGGAGC-3′.
The transcription buffer and the synthesis of the minus UTP starter 15 complex were essentially as described (Hung and Gottesman 1995), except that the DNA template, RNAP, and GTP were used at 100 nm, 300 nm, and 150 μm, respectively. GTP concentration was increased to suppress transcript slippage at the λ pL promoter (Severinov and Goldfarb 1994). To obtain paused complex 119, a mixture of rifampicin, ATP, and other NTPs was added to complex 15 so that the final concentrations of the respective components were 10 μg/ml, 3 μm, and 150 μm, respectively. Elongation was permitted for 1 min and halted by adding EDTA to 50 mm. To remove NTPs and EDTA from the transcription complex, the bovine serum albumin content of the reaction was first adjusted to 1.25 mg/ml, and a portion (<30μl) of the mixture was loaded onto a BioSpin 30 gel filtration spin column (Bio-Rad) pre-equilibrated with transcription buffer. The loaded column was spun at 1000g for 15 min at 4°C, and the filtrate, which contained the transcription complex, was saved. To walk to various downstream template sites, the complex was incubated with appropriate combinations of NTPs for 10 min at 12°C. The final concentration of NTP used to walk a transcription complex from an intrinsic pause site was 5 μm; to walk from other sites, the NTP concentration was 0.5 μm. After each walk, NTPs were removed by gel filtration chromatography as described above. Counting from the transcription start site of the λ pL operon, the nontemplate sequence from 120 to 128 nucleotides is AAGTGCGAT. Thus, complexes 122 and 124 were obtained by walking complex 119 with ATP + GTP and ATP + GTP + UTP, respectively; complex 123 by walking complex 122 with UTP; complexes 125, 126, and 127 by walking complex 124 with CTP, CTP + GTP, and ATP + CTP + GTP, respectively. To complete transcription elongation, that is, to chase a particular complex, all four NTPs were added to 50 μm each. Where indicated, Nun was added at 500 nm. After each addition of protein factor or reagent, samples were incubated at 32°C for 5 min prior to the next step, unless otherwise indicated. The transcription products were treated as described (Hung and Gottesman 1995) and electrophoresed on a 40 × 35 × 0.4-cm 6% polyacrylamide/8.3 m urea gel at a constant power of 100 W until the xylene cyanol FF dye had migrated for 35 cm. Radiolabeled transcripts were detected by autoradiography.
In most assays a spontaneously arrested complex at 151 nucleotides was detected. Arrest at 151 nucleotides is template, but not sequence, specific. The 151 complex was not observed in transcription assays using the template described above but carrying additional downstream DNA (data not shown). We suggest that spontaneous arrest at 151 nucleotides was most likely caused by the loss of RNAP–DNA contact because of the proximity of the elongating RNAP to the end of the template (Izban et al. 1995).
To improve the efficiency of the Nun reaction to ∼100%, we used high Nun concentrations. At these levels, Nun activity is independent of the λ nut site (data not shown). We believe, however, that our results do reveal the mechanism of Nun action, as high Nun concentrations stimulate but do not affect the essential characteristics of the Nun reaction. Thus, the sites of arrest and the susceptibility of the arrested complexes to transcript cleavage shown here are also seen at low Nun levels, when λ nut is required (data not shown). As discussed previously (Chattopadhyay et al. 1995), it is likely that the λ nut sequence in the nascent transcript serves only to increase the local Nun concentration in the vicinity of RNAP and can be compensated for by high Nun concentrations. Similarly, overexpression of Nun is known to enhance termination on mutant λ nut sequences in vivo (Baron and Weisberg 1992).
Acknowledgments
We thank Dr. S. Borukhov for providing GreB protein and Dr. A. Das for providing E. coli strain AD8571. We are grateful to Drs. M. Kashlev, E. Nudler, and K. Severinov for technical advice and R.S. Watnick for reading the manuscript. This work was supported by National Institutes of Health grant GM37219.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL gottesma@cuccfa.ccc.columbia.edu; FAX (212) 305-1741.
References
- Arndt KM, Chamberlin MJ. RNA chain elongation by Escherichia coli RNA polymerase. Factors affecting the stability of elongating ternary complexes. J Mol Biol. 1990;213:79–108. doi: 10.1016/S0022-2836(05)80123-8. [DOI] [PubMed] [Google Scholar]
- Baron J, Weisberg RA. Mutations of the phage λ nutL region that prevent the action of Nun, a site-specific transcription termination factor. J Bacteriol. 1992;174:1983–1989. doi: 10.1128/jb.174.6.1983-1989.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borukhov S, Polyakov A, Nikiforov V, Goldfarb A. GreA protein: A transcription elongation factor from Escherichia coli. Proc Natl Acad Sci. 1992;89:8899–8902. doi: 10.1073/pnas.89.19.8899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borukhov S, Sagitov V, Goldfarb A. Transcript cleavage factors from E. coli. Cell. 1993;72:459–466. doi: 10.1016/0092-8674(93)90121-6. [DOI] [PubMed] [Google Scholar]
- Burgess RR, Jendrisak JJ. A procedure for the rapid, large-scale purification of Escherichia coli DNA-dependent RNA polymerase involving Polymin P precipitation and DNA-cellulose chromatography. Biochemistry. 1975;14:4634–4638. doi: 10.1021/bi00692a011. [DOI] [PubMed] [Google Scholar]
- Chan CL, Landick R. New perspectives on RNA chain elongation and termination by E. coli RNA polymerase. In: Conaway RC, Conaway JW, editors. Transcription: Mechanisms and regulation. New York, NY: Raven Press; 1994. pp. 297–321. [Google Scholar]
- Chan CL, Wang D, Landick R. Multiple interactions stabilize a single paused transcription intermediated in which hairpin to 3′ end spacing distinguishes pause and termination pathway. J Mol Biol. 1997;268:54–68. doi: 10.1006/jmbi.1997.0935. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay S, Hung SC, Stuart AC, Palmer III AG, Garcia-Mena J, Das A, Gottesman ME. Interaction between the phage HK022 Nun protein and the nut RNA of phage λ. Proc Natl Acad Sci. 1995;92:12131–12135. doi: 10.1073/pnas.92.26.12131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Price DH. Mechanism of DmS-II-mediated pause suppression by Drosophila RNA polymerase II. J Biol Chem. 1993;268:18762–18770. [PubMed] [Google Scholar]
- Hung SC, Gottesman ME. Phage HK022 Nun protein arrests transcription on phage λ DNA in vitro and competes with the phage λ N antitermination protein. J Mol Biol. 1995;247:428–442. doi: 10.1006/jmbi.1994.0151. [DOI] [PubMed] [Google Scholar]
- Izban MG, Luse DS. Factor-stimulated RNA polymerase II transcribes at physiological elongation rates on naked DNA but very poorly on chromatin templates. J Biol Chem. 1992;267:13647–13655. [PubMed] [Google Scholar]
- Izban MG, Samkurashvili I, Luse DS. RNA polymerase II ternary complexes may become arrested after transcribing to within 10 bases of the end of linear templates. J Biol Chem. 1995;270:2290–2297. doi: 10.1074/jbc.270.5.2290. [DOI] [PubMed] [Google Scholar]
- Kassavetis GA, Zentner PG, Geiduschek EP. Transcription at bacteriophage T4 variant late promoters. An application of a newly devised promoter-mapping method involving RNA chain retraction. J Biol Chem. 1986;261:14256–14265. [PubMed] [Google Scholar]
- Komissarova N, Kashlev M. RNA polymerase switches between inactivated and activated states translocating back and forth along the DNA and the RNA. J Biol Chem. 1997;272:15329–15338. doi: 10.1074/jbc.272.24.15329. [DOI] [PubMed] [Google Scholar]
- Krakow JS, Fronk E. Azotobacter vinelandii ribonucleic acid polymerase. 8. Pyrophosphate exchange. J Biol Chem. 1969;244:5988–5993. [PubMed] [Google Scholar]
- Nudler E, Goldfarb A, Kashlev M. Discontinuous mechanism of transcription elongation. Science. 1994;265:793–796. doi: 10.1126/science.8047884. [DOI] [PubMed] [Google Scholar]
- Nudler E, Mustaev M, Lukhtanov E, Goldfarb A. The RNA-DNA hybrid maintains the register of transcription by preventing backtracking of RNA polymerase. Cell. 1997;89:33–41. doi: 10.1016/s0092-8674(00)80180-4. [DOI] [PubMed] [Google Scholar]
- Orlova M, Newlands J, Das A, Goldfarb A, Borukhov S. Intrinsic transcript cleavage activity of RNA polymerase. Proc Natl Acad Sci. 1995;92:4596–4600. doi: 10.1073/pnas.92.10.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polyakov A, Severinova E, Darst SA. Three-dimensional structure of E. coli core RNA polymerase: Promoter binding and elongation conformations of the enzyme. Cell. 1995;83:365–373. doi: 10.1016/0092-8674(95)90114-0. [DOI] [PubMed] [Google Scholar]
- Reines D. Elongation factor-dependent transcript shortening by template-engaged RNA polymerase II. J Biol Chem. 1992;267:3795–3800. [PMC free article] [PubMed] [Google Scholar]
- ————— . Nascent RNA cleavage by transcription elongation complexes. In: Conaway RC, Conaway JW, editors. Transcription: Mechanisms and regulation. New York, NY: Raven Press; 1994. pp. 263–278. [Google Scholar]
- Robert J, Sloan SB, Weisberg RA, Gottesman ME, Robledo R, Harbrecht D. The remarkable specificity of a new transcription termination factor suggests that the mechanisms of termination and antitermination are similar. Cell. 1987;51:483–492. doi: 10.1016/0092-8674(87)90644-1. [DOI] [PubMed] [Google Scholar]
- Robledo R, Gottesman ME, Weisberg RA. λ nutR mutations convert HK022 Nun protein from a transcription termination factor to a suppressor of termination. J Mol Biol. 1990;212:635–643. doi: 10.1016/0022-2836(90)90226-c. [DOI] [PubMed] [Google Scholar]
- Robledo R, Atkinson BL, Gottesman ME. Escherichia coli mutations that block transcription termination by phage HK022 Nun protein. J Mol Biol. 1991;220:613–619. doi: 10.1016/0022-2836(91)90104-e. [DOI] [PubMed] [Google Scholar]
- Rozovskaya TA, Rechinsky VO, Bibilashvili RS, Karpeisky M, Tarusova NB, Khomutov RM, Dixon HB. The mechanism of pyrophosphorolysis of RNA by RNA polymerase. Endowment of RNA polymerase with artificial exonuclease activity. Biochem J. 1984;224:645–650. doi: 10.1042/bj2240645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd MD, Izban MG, Luse LS. The active site of RNA polymerase II participates in transcript cleavage within arrested ternary complexes. Proc Natl Acad Sci. 1994;91:8057–8061. doi: 10.1073/pnas.91.17.8057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severinov K, Goldfarb A. Topology of the product binding site in RNA polymerase revealed by transcript slippage at the phage λ PL promoter. J Biol Chem. 1994;269:31701–31705. [PubMed] [Google Scholar]
- Sloan SB, Weisberg RA. Use of a gene encoding a suppressor tRNA as a reporter of transcription: Analyzing the action of the Nun protein of bacteriophage HK022. Proc Natl Acad Sci. 1993;90:9842–9846. doi: 10.1073/pnas.90.21.9842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surratt CK, Milan SC, Chamberlin MJ. Spontaneous cleavage of RNA in ternary complexes of Escherichia coli RNA polymerase and its significance for the mechanism of transcription. Proc Natl Acad Sci. 1991;88:7983–7987. doi: 10.1073/pnas.88.18.7983. [DOI] [PMC free article] [PubMed] [Google Scholar]