Abstract
The cellular components required to form the 3′ ends of small nuclear RNAs are unknown. U5 snRNA from Saccharomyces cerevisiae is found in two forms that differ in length at their 3′ ends (U5L and U5S). When added to a yeast cell free extract, synthetic pre-U5 RNA bearing downstream genomic sequences is processed efficiently and accurately to generate both mature forms of U5. The two forms of U5 are produced in vitro by alternative 3′-end processing. A temperature-sensitive mutation in the RNT1 gene encoding RNase III blocks accumulation of U5L in vivo. In vitro, alternative cleavage of the U5 precursor by RNase III determines the choice between the two multistep pathways that lead to U5L and U5S, one of which (U5L) is strictly dependent on RNase III. These results identify RNase III as a trans-acting factor involved in 3′-end formation of snRNA and show how RNase III might regulate alternative RNA processing pathways.
Keywords: snRNP, Saccharomyces cerevisiae, endonuclease, spliceosomal snRNA, RNT1
In addition to its role in mRNA synthesis, RNA polymerase II transcribes many of the genes encoding small nuclear RNAs (snRNAs). Whereas the 3′ ends of most mRNAs are generated by endonucleolytic cleavage and polyadenylation, the mechanism by which snRNAs acquire their 3′ ends is much less well understood. In vertebrates, proper snRNA 3′-end formation requires expression from snRNA promoters, because transcription of snRNAs using mRNA promoters results in aberrant processing of the transcript by the mRNA cleavage and polyadenylation machinery (Ciliberto et al. 1986; Hernandez and Weiner 1986; Neuman de Vegvar et al. 1986). Thus, the primary events that lead to 3′-end formation of snRNA in vertebrates are linked to transcription initiation. Whether snRNA 3′-end formation is a result of termination or processing, and how the promoter identity influences the primary events on the nascent snRNA transcript, is not known.
Secondary processing events that mature the ends of snRNA appear to be linked to snRNP biogenesis. Precursors of vertebrate snRNAs that contain up to 16 extra nucleotides at their 3′ ends can be detected (Madore et al. 1984a,b; Yuo et al. 1985; Neuman de Vegvar and Dahlberg 1990). Most of this extension is removed in the cytoplasm (Madore et al. 1984a; Kleinschmidt and Pederson 1987; Neuman de Vegvar and Dahlberg 1990), where Sm protein binding to the snRNA and cap hypermethylation also occurs (for review, see Mattaj 1988; Nagai and Mattaj 1994). In Saccharomyces cerevisiae and Schizosaccharomyces pombe, mutations leading to defects in snRNP biogenesis are correlated with the accumulation of 3′ extended snRNAs (Potashkin and Frendewey 1990; Noble and Guthrie 1996b). Thus, proper maturation of the snRNA 3′ end may be an important step in snRNP biogenesis.
In vitro systems have been developed to study cytoplasmic 3′ trimming of exogenous precursors of vertebrate snRNAs containing an extension of a few nucleotides (Yuo et al. 1985; Kleinschmidt and Pederson 1987), but these systems are unable to process species longer than 10 nucleotides. Furthermore, no trans-acting factors involved in snRNA 3′-end formation have been identified yet. We have addressed this question by studying 3′-end formation of yeast U5. We show that a precursor containing a long downstream genomic sequence can be processed efficiently and accurately in vitro to produce the two forms of U5 that are found in vivo (Patterson and Guthrie 1987; Frank et al. 1994). This processing occurs in at least two steps, the first of which involves endonucleolytic cleavage. Based on genetic results in S. pombe (Potashkin and Frendewey 1990; Rotondo et al. 1995), and the observation of a defect in U5 synthesis in vivo in an RNase III mutant strain, we tested the involvement of yeast RNase III in this process in vitro. We show that RNase III cleaves pre-U5 RNA at two different sites. The choice of the site of cleavage by RNase III determines the form into which U5 will be finally processed. These results identify a trans-acting factor involved in snRNA 3′-end formation and reveal how RNase III cleavage site selection can influence alternative RNA processing.
Results
In vitro processing of yeast U5 snRNA precursor
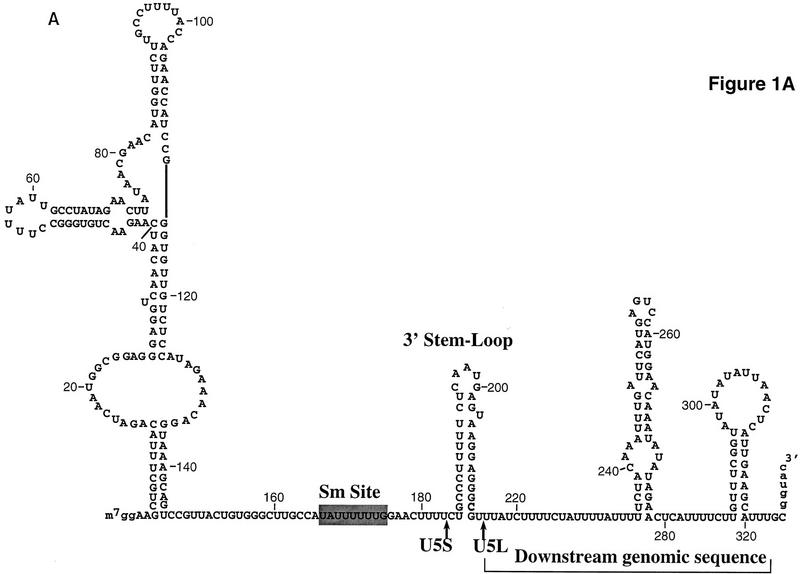
To develop an in vitro system to study snRNA 3′-end formation in yeast, a U5 precursor bearing 116 nucleotides of downstream genomic sequences (Fig. 1A) was transcribed in vitro and incubated in a yeast whole-cell extract (Umen and Guthrie 1995; Fig. 1B; see also Materials and Methods). This precursor was processed efficiently to give rise to four shorter RNAs. Two species, ∼270 and 240 nucleotides long (σ and λ; Fig. 1B), are produced very rapidly, after <1 min of incubation in the extract. The two shorter species, whose sizes are consistent with those of the two forms of mature U5 found in vivo (U5S and U5L; Fig. 1B) appear later, after the formation of σ and λ. This processing is strictly dependent on the addition of extract and magnesium (data not shown).
Figure 1.
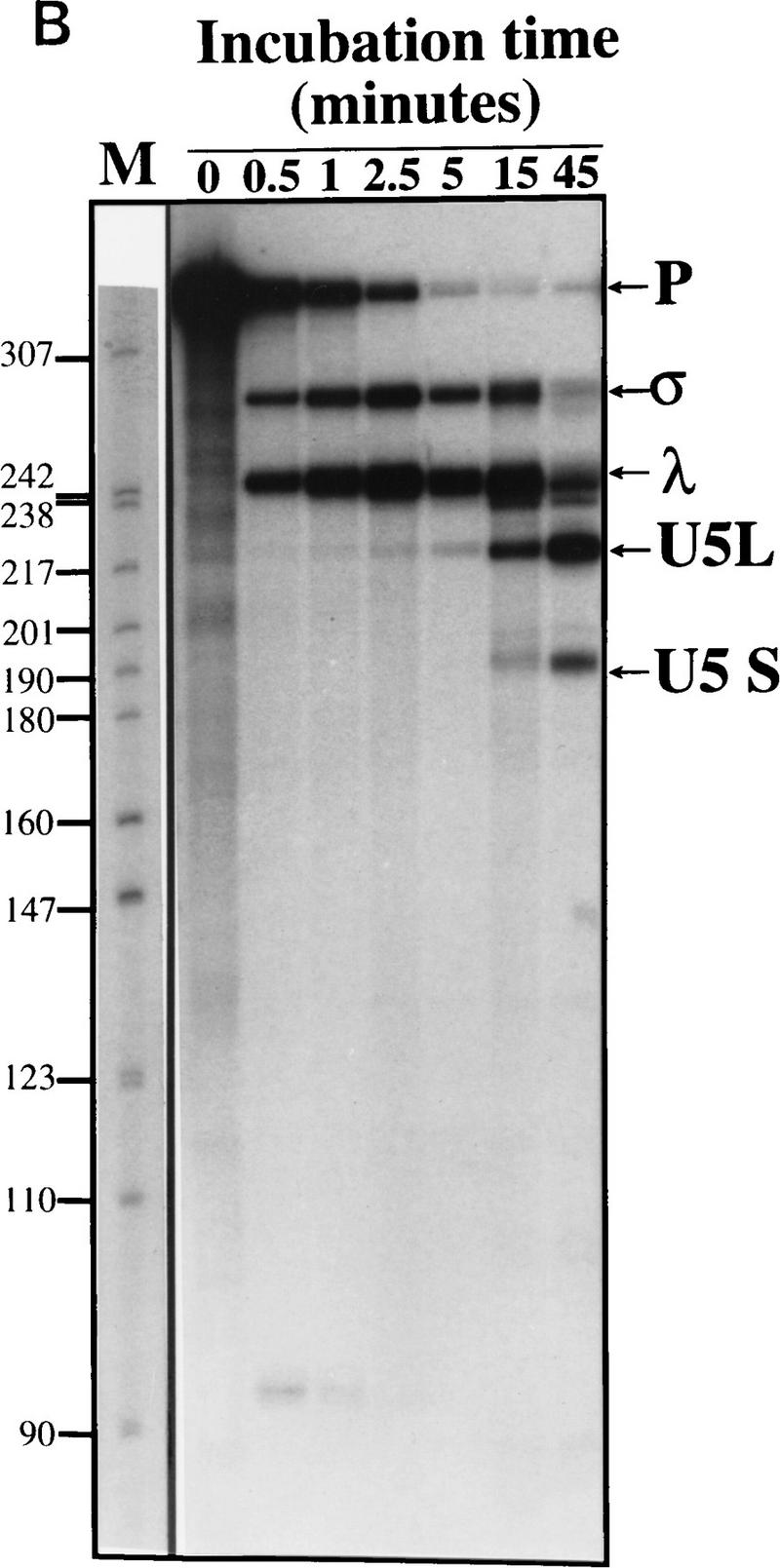
In vitro processing of a model pre-U5 transcript. (A) Sequence and a secondary structure model of the precursor used in this study. The secondary structure of mature U5 is drawn from Frank et al. (1994). The most stable potential secondary structure of the downstream genomic sequence calculated using Mfold (Zuker 1994) is shown here but has not been proved experimentally nor phylogenetically. Sequences not derived from the U5 gene are indicated in lowercase. They include two additional guanosines at the 5′ end of the molecule to facilitate transcription by T7 RNA polymerase and part of a BamHI restriction site at the 3′ end. (B) Time course of processing. Precursor-U5 (P) was incubated in a whole-cell extract for the times indicated. Shown is an autoradiograph of a 6% gel. σ and λ are generated rapidly and have the characteristics of intermediates, whereas U5L and U5S have characteristics of products (see text). The molecular weight marker (M) is a pBR322 plasmid digested with MspI and labeled with [γ-32P]ATP.
The processing reaction produces new 3′ ends
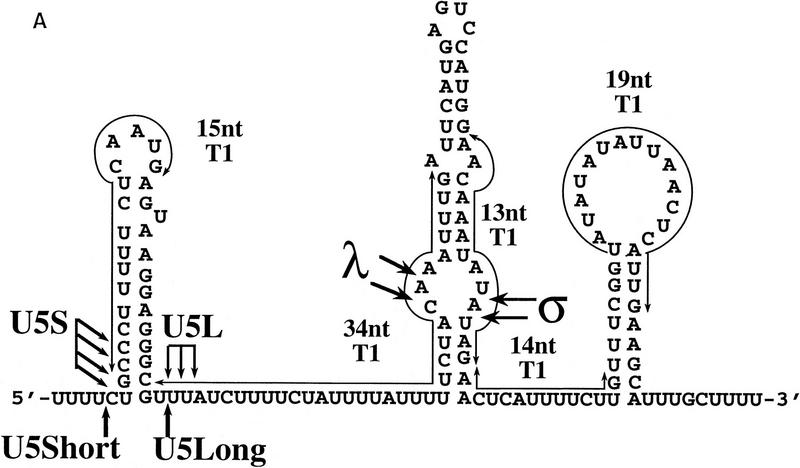
To characterize the products of the processing reaction, we first mapped them by digestion with RNase T1. We took advantage of several large, unique T1 fragments derived from the 3′ part of the precursor (Fig. 2A). Digestion of the input precursor by RNase T1 yielded the expected fragments of 34, 19, 15, 14, and 13 nucleotides long (Fig. 2B). RNA species were produced in the in vitro reaction, gel-purified, and subjected to digestion by RNase T1. Absence of the 19-, 14-, and 13-nucleotide fragments diagnostic of the 3′ end indicated that this part of all the species tested had been removed during processing. Digestion of the σ product still yielded the 34-nucleotide fragment, indicating that the 3′ end of this species is located between the 34- and 13-nucleotide fragments, or within the latter. Digestion of the λ species produces a 27- to 28-nucleotide-long fragment that must be derived from the 34-nucleotide fragment. The U5L digestion yields the 15-nucleotide fragment, indicating that the 3′ stem–loop is intact in U5L, which is absent in the U5S product (Fig. 2B). These results show that the processing reaction removes sequences from the 3′ end of pre-U5, generating a set of U5 RNA species with different 3′ ends.
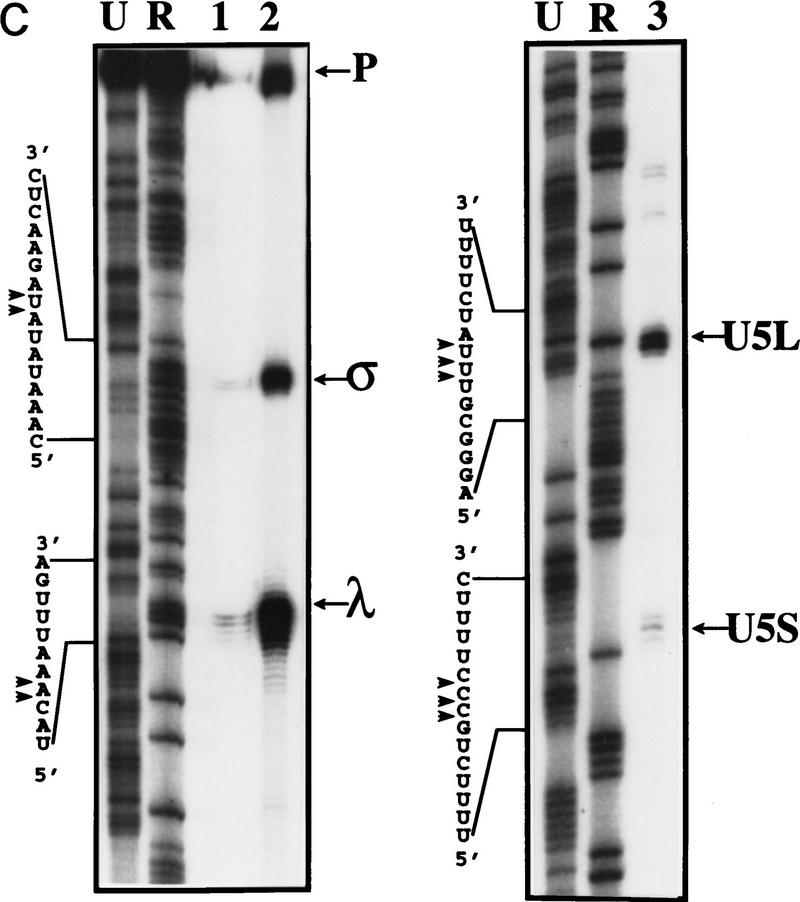
Figure 2.
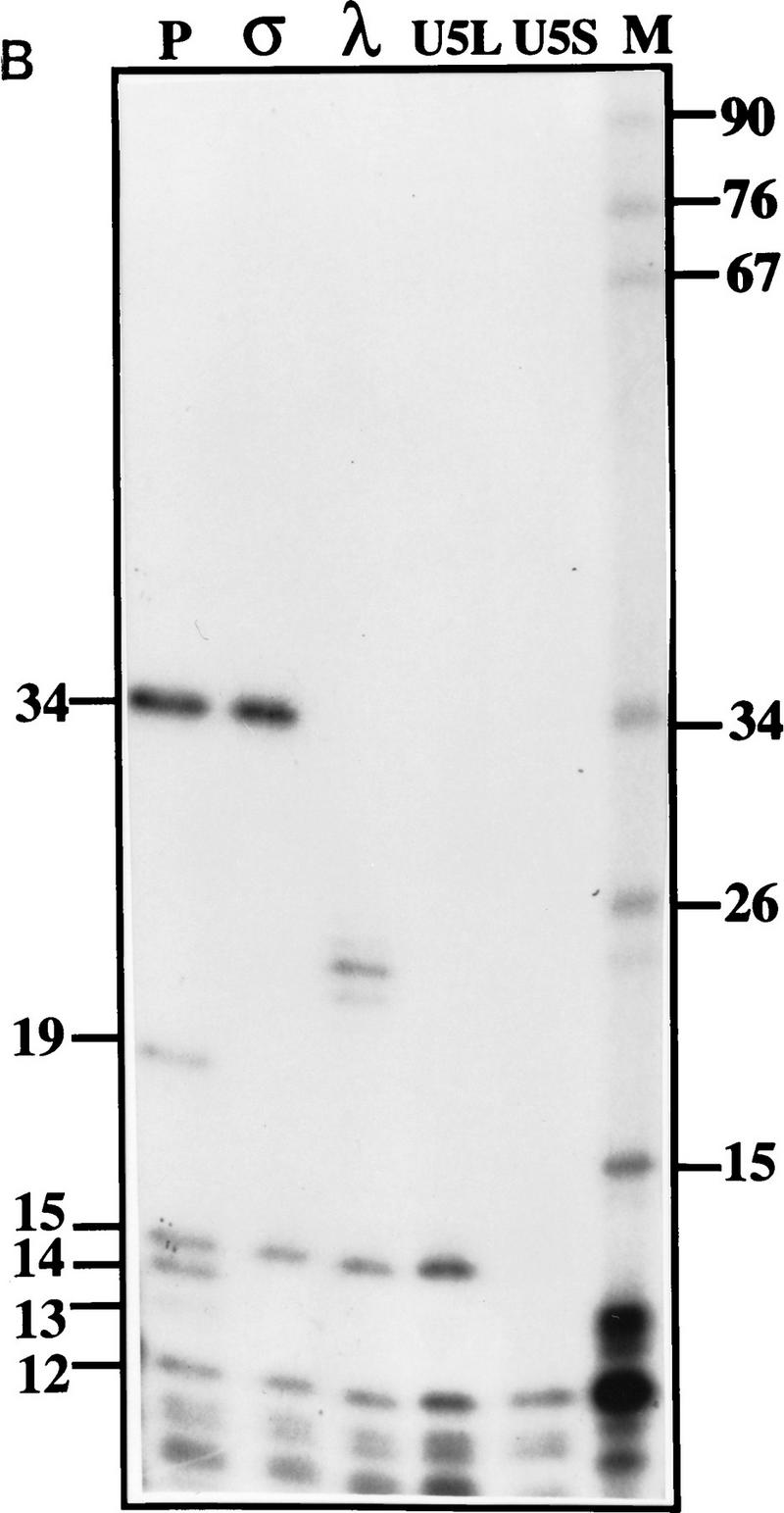
Mapping of 3′ ends of transcripts produced by in vitro processing. (A) Structure of the 3′ downstream genomic sequence. Shown are the diagnostic RNase T1 fragments, as well as the 3′ ends of putative intermediates, and products, deduced from the experiments shown in B and C and in Figs. 3B and 5C. Arrows from U5L and U5S indicate 3′ ends of the products of the in vitro reaction, whereas U5Long and U5Short indicate the 3′ ends of the long and short forms of U5 mapped in vivo. No diagnostic T1 fragment is found in the remaining part of the precursor; therefore, this part of the molecule is not shown. (B) RNase T1 mapping. Shown is an autoradiograph of a 10% acrylamide gel. Each of the species (precursor, intermediates and products) was subjected to total digestion by RNase T1 (see Materials and Methods). Legends and marker as in Fig. 1B. Marker is shown as an indication of molecular weight but is composed of double-stranded DNA, whereas the mapped species are single-stranded RNA. Therefore, direct comparison of the sizes of low molecular weight fragments is not possible. (C) Mapping cleavage sites to the U5 flanking sequence. A 5′-labeled precursor was incubated for 2 min (lanes 1,2) or 45 min (lane 3) to give rise to intermediates and products and was loaded on a 5% acrylamide sequencing gel, in parallel with a sequence generated from the same precursor (see Materials and Methods), substituted with purine phosphorothioates (R) or uridine (U), and cleaved with iodine. Lanes 1 and 2 are loadings of different amounts of the same reaction.
To map precisely the 3′ end of each RNA, a 5′-end-labeled precursor was incubated 2 min to yield the σ and λ products, or 45 min to yield the U5L and U5S forms. The lengths of these species were compared with a sequence ladder obtained in parallel from the same 5′-labeled precursor substituted with phosphorothioate nucleotides and cleaved with iodine (Fig. 2C). The 3′ ends of σ and λ (Fig. 2A,C) are located on either side of an internal loop of a putative stem–loop structure within the precursor, in good agreement with the RNase T1 mapping experiment. The 3′ end of the U5L and U5S products are indicated in Figure 2A. The 3′ ends of U5L in vitro is consistent with the S1 protection experiment used to map the long form of U5 in vivo (Patterson and Guthrie 1987; Frank et al. 1994). The 3′ end of U5S generated in the extract is 3–4 nucleotides longer than reported for the in vivo short form (Patterson and Guthrie 1987; Frank et al. 1994), a discrepancy that may be attributable to competition with processing to U5L (see below). Thus, incubation of synthetic pre-U5 in the extract leads to formation of two different 3′-truncated transcripts with the kinetic properties of intermediates (σ and λ; Fig. 1B), and two transcripts with authentic U5 3′ ends.
U5L and U5S are generated by alternative 3′-processing reactions through distinct intermediates
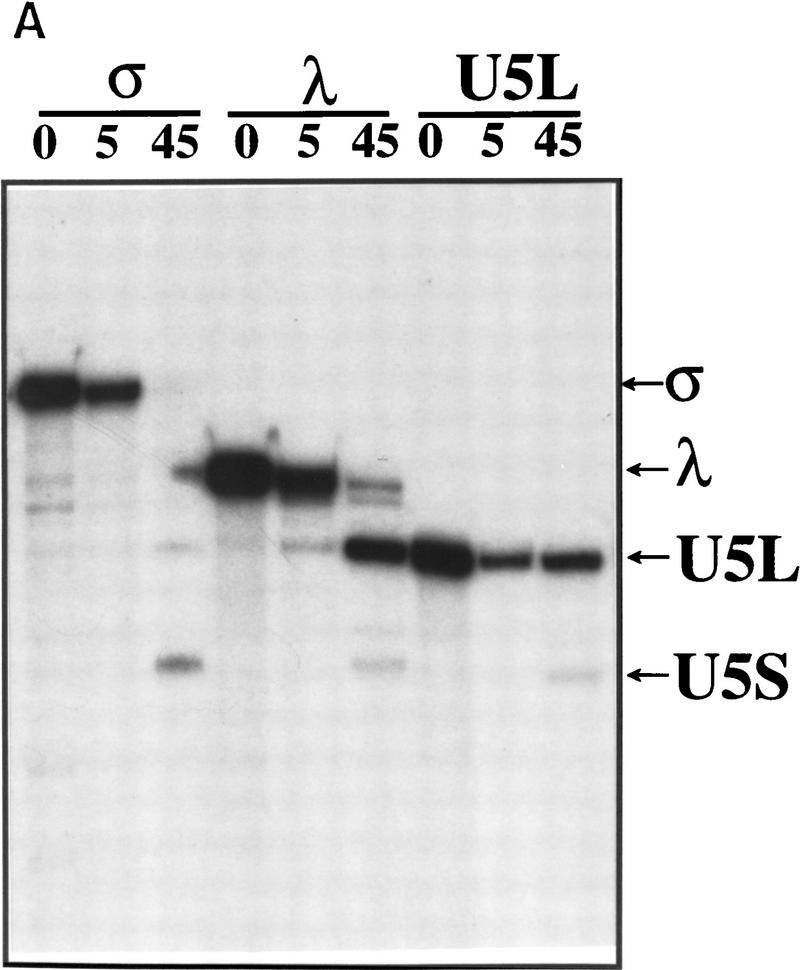
To determine directly whether a precursor-to-product relationship exists between the rapidly generated transcripts σ and λ, and mature U5S and U5L, the σ and λ transcripts generated in the extract were gel-purified on a denaturing acrylamide gel and reincubated in extract (Fig. 3A). Incubation of σ gave rise in majority to U5S, whereas λ produced mostly U5L. Incubation of gel-purified U5L showed that this product did not generate a significant amount of U5S, indicating that U5L is not a precursor of U5S. These observations show that the in vitro processing reaction occurs via two pathways, each of which has two steps. In one pathway, the pre-U5 substrate gives rise to λ, which is further processed to U5L. In the other pathway, pre-U5 gives rise to σ, which is further processed to U5S. Thus, U5S and U5L are the products of alternative 3′-end processing reactions that generate distinct intermediates with different fates.
Figure 3.
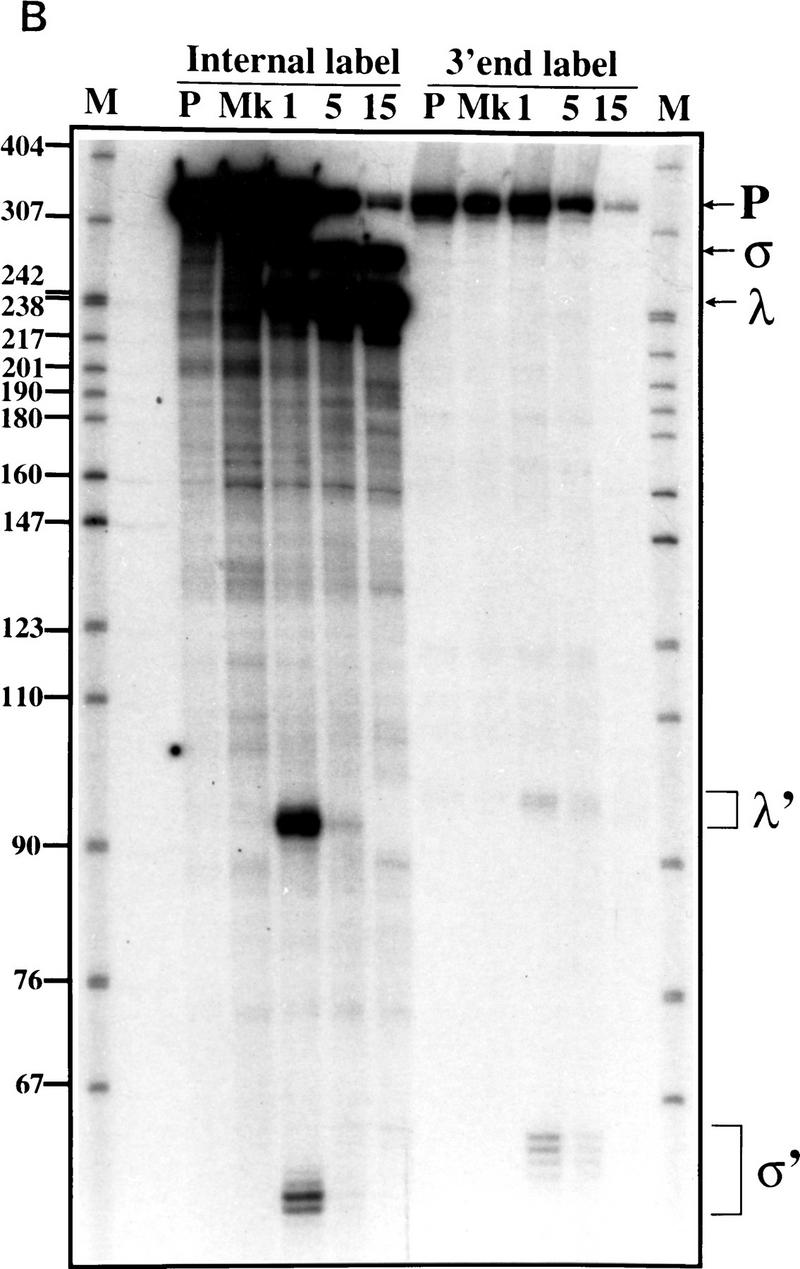
U5L and U5S arise from distinct intermediates generated by endonucleolytic cleavage. (A) The long and the short forms of U5 arise from different intermediates. Gel-purified σ, λ, and U5L species were incubated in extracts for the amount of time indicated and fractionated on a 6% acrylamide gel. Legends as in Figure 1B. (B) Time course of 3′ processing of internally labeled or 3′-labeled precursors. Legends as in Fig. 1B. In the mock incubation points (Mk), samples were incubated for 15 min at 30°C with extract, buffer, and 25 mm EDTA. The sums of σ′ (60 nucleotides) plus σ (270 nucleotides), or λ′ (90 nucleotides) plus λ (240 nucleotides) are about the same as the total length of pre-U5 (332 nucleotides), supporting the interpretation that the short unstable RNAs are the downstream products of endonucleolytic cleavages that generate the intermediates.
Intermediates in U5 3′-end formation are generated by endonucleolytic cleavage
To determine whether the intermediates are generated by endo- or exonucleolytic cleavage, we attempted to identify possible downstream cleavage products. We incubated 3′-end-labeled pre-U5 RNA in extracts and separated the processed RNA on a denaturing gel (Fig. 3B). At 1 min of incubation, fragments of low molecular weight (labeled σ′ and λ′; Fig. 3B) are detected using 3′ end-labeled (or internally labeled) pre-U5 substrate. These disappear on longer incubations presumably because of their instability in extracts. The lengths of these two fragments are consistent with those expected for the downstream products of an endonucleolytic cleavage that would generate σ and λ (see Fig. 2). We conclude that the maturation of U5 3′ ends begins with cleavage of a long pre-U5 RNA by an endonuclease.
Sm proteins associate with U5 intermediates and products during 3′-end formation
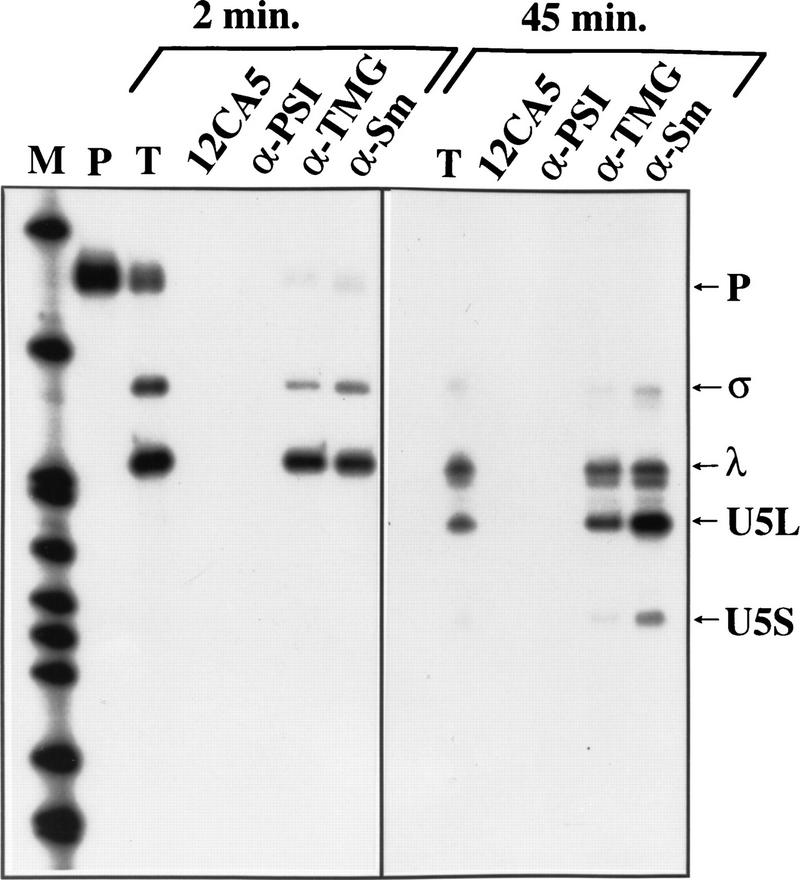
To determine which processed species might be incorporated into snRNPs, we performed immunoprecipitation on in vitro processing reactions using anti-Sm antibodies (Fig. 4; Siliciano et al. 1987). Incubation of pre-U5 synthesized with a monomethyl guanosine cap followed by immunoprecipitation showed that the intermediates and products of the in vitro reaction, but not the pre-U5, are efficiently immunoprecipitated by anti-Sm human sera (Fig. 4), as well as by antitrimethylguanosine monoclonal antibody (Krainer 1988). In contrast, they were not immunoprecipitated by the 12CA5 monoclonal antibody, nor by anti-PSI (P element somatic inhibitor) crude ascites from mouse (Siebel et al. 1994). This result suggests that the intermediates and products of the processing reaction, but not the precursor, associate with Sm proteins and that the monomethylguanosine cap becomes hypermethylated. Association of Sm proteins and cap hypermethylation are hallmark events in snRNP biogenesis (Mattaj 1988), suggesting that snRNA 3′-end formation occurs concomitantly with snRNP assembly in vitro.
Figure 4.
Intermediates and products of the processing reaction associate with Sm proteins. After 2 or 45 min of incubation, RNAs were immunoprecipitated with various antibodies (see text and Materials and Methods). T (total) represents  of the amount of RNAs used in each reaction. The other lanes contain RNAs isolated from immunoprecipitates. Shown is an autoradiograph of a 6% acrylamide gel. Legends as in Fig. 1B.
of the amount of RNAs used in each reaction. The other lanes contain RNAs isolated from immunoprecipitates. Shown is an autoradiograph of a 6% acrylamide gel. Legends as in Fig. 1B.
Altered levels of U5L and U2 snRNA in RNase III mutant cells
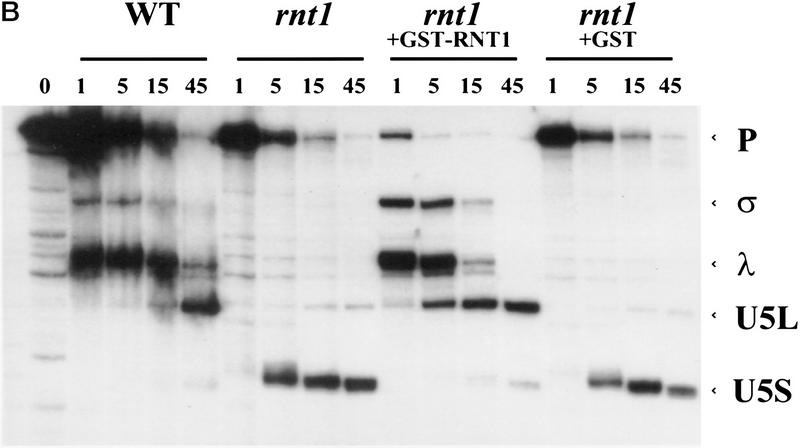
In S. pombe, snm1-1, a mutant defective in snRNA biosynthesis and 3′-end formation, can be suppressed by overexpression of pac1, the S. pombe homolog of RNase III (Potashkin and Frendewey 1990; Rotondo et al. 1995), suggesting involvement of RNase III in the snRNA biosynthesis pathway. We observed a defect in U5 accumulation in vivo in an RNase III-defective mutant strain of S. cerevisiae (rnt1; Abou Elela et al. 1996) at the restrictive temperature, in a general screen for RNA processing defects in this mutant. Total RNA was extracted from wild-type and rnt1 yeast strains grown at permissive temperature or shifted to restrictive temperature, fractionated on a denaturing polyacrylamide gel and probed for snRNAs. Consistent with our initial observation, the long form of U5 was found to be absent in rnt1 strain, even at the permissive temperature (Fig. 5A). A shift to nonpermissive temperature did not cause a significant decrease in the level of U5S (Fig. 5A), nor of U1, U4, or U6 snRNAs. In contrast, U2 snRNA levels are reduced in the mutant at permissive temperature, and decrease slightly on shift to nonpermissive temperature (Fig. 5A). We conclude that RNase III influences U5 and U2 snRNA metabolism in S. cerevisiae, and seems essential for the synthesis of U5L, but not U5S, in vivo. Because RNase III activity in preribosomal RNA processing is greatly reduced by a temperature shift (Abou Elela et al. 1996), we suspected that U5S production could be supported by an RNase III independent pathway in vivo.
Figure 5.
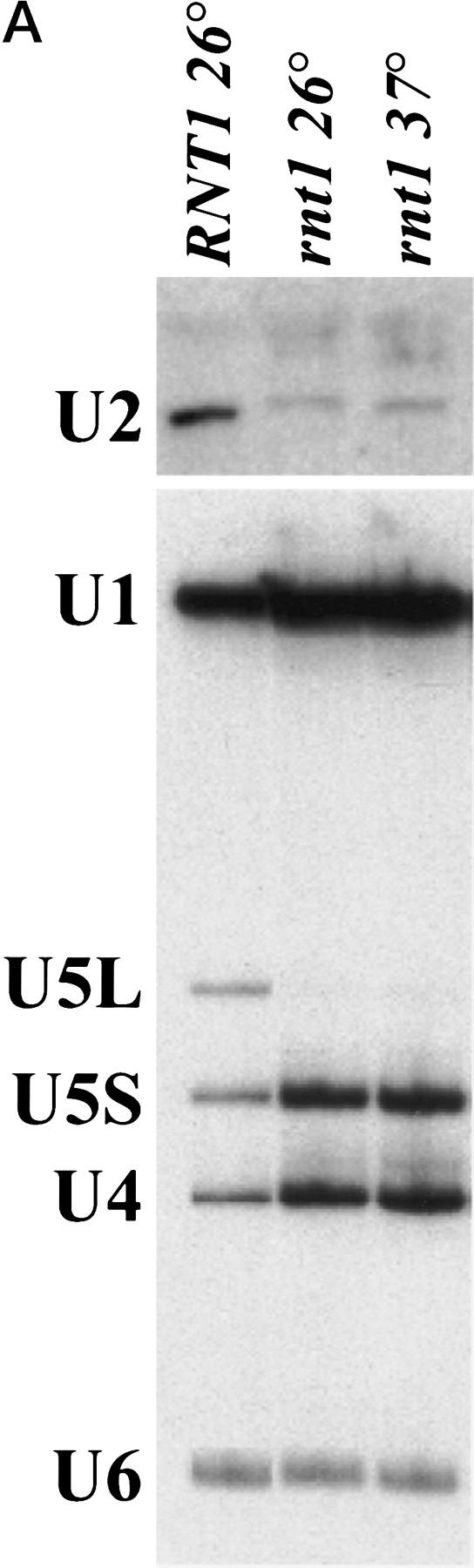
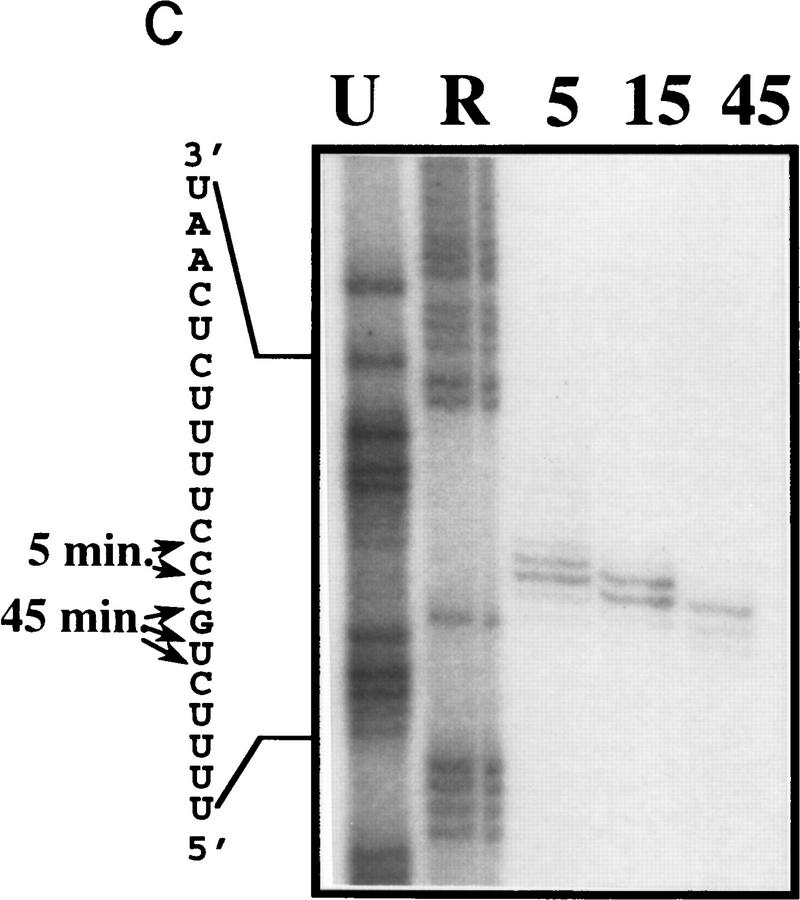
RNase III is required for correct U5 3′-end processing. (A) The rnt1 mutant strain does not produce the long form of U5 in vivo. Total RNAs were extracted from wild-type or rnt1 strains, grown at 26°C or shifted to 37°C for 4 hr. The RNAs were loaded onto a denaturing 6% acrylamide gel, transferred to a nylon membrane, and hybridized to DNA fragments complementary to snRNAs. (B) Extracts from rnt1 cells are deficient in the production of intermediates and U5L. Internally-labeled transcript was incubated in extracts made from wild-type (WT) or mutant rnt1 strains for the times indicated, and products were loaded in a denaturing 6% acrylamide gel. Legends as in Fig. 1B. (C) 3′-end trimming of the short form of U5 in rnt1 extracts. A 5′-labeled transcript was incubated for 5, 15, or 45 min in rnt1 extract and loaded on a 5% sequencing gel, in parallel with a phosphorothioate generated sequence with purines (R) and uridine (U) as in Fig. 3C. Arrowheads on the sequence indicate the 3′ ends after 5 and 45 min of incubation. Note that the trimming must be at the 3′ end, because the 5′ end of the molecule is labeled.
Extracts from RNase III-deficient cells fail to generate U5 intermediates or the U5L product in vitro
To assess the possibility of a direct role of RNase III in U5 3′-end processing, we incubated pre-U5 RNA in extracts prepared from wild-type and RNase III defective rnt1 strains grown at permissive temperature. The kinetics of U5 3′-end processing in vitro are strikingly different in the rnt1 mutant extract as compared with the wild-type control (Fig. 5B). Neither the σ nor λ intermediate is observed, and only U5S was produced in rnt1 extracts. Early in the reaction this form comigrates with U5S generated in the wild-type extract (data not shown). As the reaction proceeds in the mutant extract, U5S is further trimmed at its 3′ end (Fig. 5C). The mutant phenotype in vitro is strikingly similar to the mutant phenotype in vivo, where only the short form of U5 is produced (Fig. 5A). Taken together, these observations suggest that in addition to the two major pathways that give rise to U5L and U5S via the λ and σ intermediates, a bypass pathway ensures production of mature U5S in the absence of RNase III. Components of the bypass pathway may partly or completely overlap with those that generate U5S from σ in wild-type cells (Fig. 3A).
To show that reduced processing in rnt1 extracts is attributable to reduced RNT1 protein activity, we tried to complement the mutant extract by adding back a glutathione S-transferase (GST)–RNT1 fusion protein produced in Escherichia coli (Abou Elela et al. 1996). Addition of this protein to the extract restores the production of the intermediates and of the long form of U5, whereas GST alone has no effect (Fig. 5B). These results indicate a direct involvement of RNT1 in U5 3′-end processing in vitro, and suggest a catalytic role for this protein in the first endonucleolytic cleavages of both major processing pathways.
Purified RNase III cleaves pre-U5 to generate both intermediates in the absence of extract
To determine whether RNase III catalyzes cleavage at the σ and λ sites, we incubated U5 precursor with GST–RNT1 or with GST, in the absence of extract (Fig. 6). After 5 min of incubation internally labeled or 5′-end-labeled precursors are efficiently cleaved, whereas incubation with GST has no effect. The 5′-end-labeled substrate is cleaved to produce two species the sizes of the σ and λ intermediates, whereas the internally labeled substrate produces four species, σ and λ as well as the σ′ and λ′ downstream fragments (Fig. 6). This indicates that GST–RNT1 alone cleaves pre-U5 at positions identical to that observed in complete wild-type extracts. Together these results argue that RNT1 is responsible for the first catalytic step of the 3′ processing of U5, but is not sufficient for the second step of the reaction.
Figure 6.

RNase III cleaves pre-U5 to produce intermediates in the absence of other factors. 5′-Labeled or internally-labeled pre-U5 transcripts were incubated for 5 and 45 min with GST–RNT1 or for 45 min with GST alone (Mk). Markers are the phosphorothioate-generated sequence with purines (R) and pyrimidines (Y) as in Fig. 3C.
Discussion
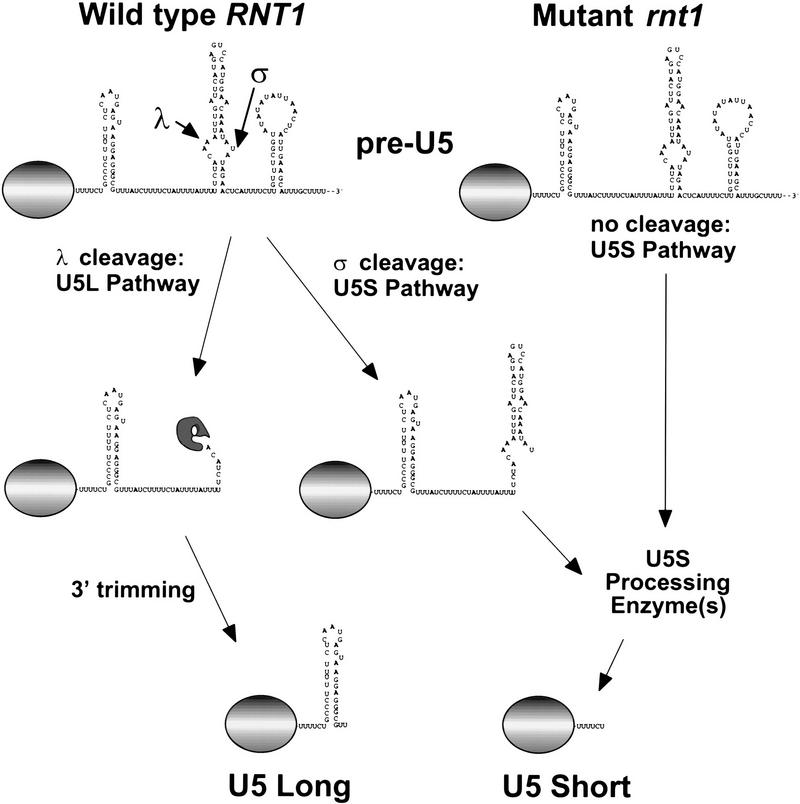
From its single U5 gene, yeast produces two forms of U5 RNA that differ at their 3′ ends (Patterson and Guthrie 1987; Frank et al. 1994). We have found that the two different 3′ ends of U5 are formed by RNA processing (Figs. 1 and 2) through two distinct intermediates along different pathways (Figs. 2 and 3, summarized in Fig. 7). Sm protein binding and cap hypermethylation, two events associated with snRNP biogenesis (Mattaj 1988), occur during 3′-end formation (Fig. 4), suggesting that snRNA processing may be coordinated with snRNP assembly. The requirement for RNase III in U5L and U2 accumulation in vivo (Fig. 5A) and for U5L processing in vitro (Fig. 5B) identifies this ribonuclease as an important trans-acting factor in snRNA metabolism. Production of U5S when RNase III activity is reduced may include factors that act on the products of RNase III cleavage during normal processing (Fig. 7). These results extend the known substrates of the ubiquitous RNase III-like processing enzymes to include snRNAs.
Figure 7.
RNA processing pathways for generating the 3′ ends of yeast U5 snRNA. The 5′ part of U5 snRNA is represented by the shaded ovals.
Trans-acting factors in snRNA 3′ end formation
The role of RNase III as a trans-acting factor in U5 snRNA 3′ end formation is limited to the generation of intermediates (Fig. 6), indicating that other factors are required for 3′ end formation in vivo. These additional steps can be uncoupled from RNase III activity as demonstrated by correct processing of gel-purified λ and σ intermediates added to a rnt1 extract (data not shown). The factors required for the second step of each processing reaction are still unknown, but in the case of the λ to U5L pathway, an exonuclease is probably responsible, because a ladder of bands can sometimes be observed between the λ intermediate and the final product U5L (data not shown). The reaction stops at the bottom of the 3′ stem–loop of U5L, which is GC-rich and may constitute the block to further processing (Figs. 1A and 7).
The factors required for production of the short form are more mysterious. By incubating a 3′-labeled transcript in rnt1 extracts, we tried to obtain evidence for a second endonuclease that helps process U5S in the RNase III bypass pathway (Fig. 5C). Unfortunately, this activity (if it exists) is weaker than that which degrades downstream cleavage products (see Fig. 3B). Thus, we are unable to conclude whether an endonuclease or a highly processive exonuclease is responsible for production of U5S in the RNase III-independent pathway. The 3′ trimming that completes processing of U5S (Fig. 5C) suggests that an exonuclease is involved in the final stages of U5S processing. The 3′ end of U5S is very close to the Sm-binding site (boxed in Fig. 1A; Frank et al. 1994), suggesting that Sm proteins may block this exonuclease. This hypothesis is supported by in vitro data showing that Sm proteins bind rapidly to the U5 intermediates (Fig. 1C), and by in vivo data showing that some mutations in the U5 Sm-binding site result in transcripts shorter than U5S (Haltiner-Jones and Guthrie 1990). These observations argue that in addition to RNT1, at least one exonuclease (for the U5L pathway), at least one other nuclease (for the U5S pathways), Sm proteins, and possibly snRNP assembly events are also important for correct U5 3′-end formation in yeast (Fig. 7).
A temperature-sensitive mutation in the S. pombe snm1 gene shows defects in snRNA metabolism similar to those we observe in the rnt1 strain (Potashkin and Frendewey 1990; Rotondo et al. 1995). The snm1 mutation can be suppressed by extra copies of pac1 (which encodes S. pombe RNase III), suggesting that snm1 is a cofactor with RNase III in 3′-end formation or is itself an RNase III-like enzyme. If other proteins are required to help guide RNase III to the nascent transcript in vivo, to generate the correctly folded substrate, or to coordinate these activities, then the overexpression of RNase III may rescue conditional mutations in such components. Alternatively, a second RNase III homolog exists in S. pombe (Rotondo and Frendewey 1996). If this protein is primarily responsible for snRNA 3′-end formation and the snm1 mutation reduces its activity, increased pac1 expression could suppress the defect. We have not found a potential open reading frame encoding a second RNase III homolog in the S. cerevisiae genome. Perhaps genetic screens with rnt1 will reveal cofactors in snRNA 3′-end formation.
We also investigated whether the yeast pre-mRNA cleavage and polyadenylation machinery contributes to the U5 3′-end processing reaction by preparing extracts inactivated (by heat treatment or immunodepletion) for Rna14, Rna15, or Brr5/Ysh1, essential protein components of the yeast pre-mRNA cleavage and polyadenylation machinery (Minvielle-Sebastia et al. 1994; Chanfreau et al. 1996; Jenny et al. 1996). No obvious defect in U5 processing could be observed (data not shown), indicating that the machinery involved in U5 3′-end formation is not likely to include these cleavage and polyadenylation factors.
Uncoupling snRNA 3′-end formation from transcription in yeast
Yeast U5 3′-end formation can be completely uncoupled from transcription (Fig. 1). Our model pre-U5 transcript does not correspond to any known natural precursor resulting from transcription termination. We have tried without success to detect natural U5 precursors and intermediates in vivo. The steady-state levels of such precursors and intermediates may be too low to detect by our RNase protection experiments (not shown), possibly because they are processed to the mature forms at a high rate in vivo. In the absence of data concerning the U5 transcription unit in yeast, the efficient processing of the precursor used in our study shows that this transcript contains sequences necessary for correct 3′-end formation in vitro.
In contrast, 3′-end formation of vertebrate snRNAs is dependent not only on transcription, but also on snRNA promoter-specific elements (Ciliberto et al. 1986; Hernandez and Weiner 1986; Neuman de Vegvar et al. 1986). In yeast, however, substitution of snRNA promoters with elements of mRNA promoters seems not to perturb 3′-end formation of snRNAs (Patterson and Guthrie 1987; Seraphin and Rosbash 1989; Hughes and Ares 1991; Miraglia et al. 1991; Seraphin et al. 1991; Noble and Guthrie 1996b). These mechanistic differences may reflect the different genomic architecture and demands for snRNA in these organisms. Yeast snRNAs are generally encoded by single-copy genes (Wise et al. 1983; Guthrie and Patterson 1988), whereas vertebrate snRNAs are encoded by repeated gene families (for review, see Dahlberg and Lund 1988). Special measures may be necessary to ensure the high transcriptional output of 2 × 106 to 3 × 106 snRNA transcripts per vertebrate cell. Such measures may not be needed in yeast where snRNA levels are three to four orders of magnitude lower (Wise et al. 1983).
The specialization of snRNA promoters as a unique class of RNA polymerase II promoters (for review, see Dahlberg and Lund 1988) may have carried with it specialization in 3′-end formation. In the case of mRNA 3′-end processing, factors associate with RNA polymerase II (McCracken et al. 1997). If vertebrate snRNA promoters direct 3′-end formation by loading factors on polymerase at the promoter, the factors must dissociate rapidly, as extending the distance between the promoter and 3′-end formation signals greatly reduces snRNA 3′-end formation and increases cleavage and polyadenylation (Ramamurthy et al. 1996). Our findings help explain why yeast snRNAs can be large and vertebrate snRNAs are small: Vertebrate snRNA 3′-end formation is constrained to occur near the promoter (Ramamurthy et al. 1996), whereas yeast snRNA 3′ end formation is an RNA processing event that can be uncoupled from transcription (Fig. 1B), allowing yeast snRNAs to tolerate insertions and expand. Whether vertebrate snRNA 3′-end formation occurs by termination or processing remains to be determined.
Involvement of RNase III in alternative and redundant RNA processing pathways
How does RNase III cleavage site choice result in alternative 3′-end formation? Bacterial RNase III has been shown to cut at a single site (see Court 1993), or in a concerted fashion at two sites on the opposite strands of an RNA duplex-containing structure (see Bram et al. 1980). In the case of pre-U5 processing, RNase III cleaves at two sites opposite each other on the same potential stem–loop to generate the λ and σ intermediates (Fig. 7). The enzyme most likely generates the two intermediates by alternative cleavage of a single folded form, rather than by efficient concerted cleavage at both sites. The λ intermediate can be generated by a single cleavage event, as indicated by the appearance of λ′ (Figs. 3 and 6) or by cleavage at both sites, as indicated by the appearance of a fragment of the size expected for a fragment comprised between the λ and σ cleavage sites (data not shown; see Fig. 7). In the case of σ, single cleavage at this site is an obligatory event. Although we cannot strictly rule out that concerted cleavage is the preferred pathway in vivo, we observe a significant rate of production of both intermediates and of their corresponding downstream cleavage fragments (λ′ and σ′) in vitro, suggesting that single cleavage must also occur. Finally, we cannot rule out the possibility that cleavage events at λ and σ occur on transcripts that are alternatively folded.
Because most substrate molecules are cleaved only once at one or the other site, and because each of the distinct intermediates is efficiently converted into a different final product (Fig. 3A), the choice between cleavage at λ or σ commits the transcript to the pathway leading to U5L or U5S (see Fig. 7). Commitment to the U5L pathway is probably a result of the fact that the λ intermediate lacks most of the 3′ downstream sequence and is therefore rapidly trimmed down to the U5L end. In the case of U5S, the σ intermediate may be in a stable configuration that favors the action of the enzymes responsible for production of U5S. In particular, cleavage at σ inhibits subsequent cleavage by RNase III at the λ site (Fig. 3A) probably because the transcript lacks the bottom of the putative recognition stem. Therefore, in a wild-type context, cleavage at σ sequesters the transcript out of the U5L pathway. However, this cleavage is not strictly required for production of U5S, because U5S can be efficiently produced when RNase III function is debilitated both in vivo and in vitro (the RNase III bypass pathway; Fig. 7). Finally, alternative RNA folding of the pre-U5 transcript in wild-type cells could prevent formation of the RNase III cleavage site, promoting the bypass pathway leading to U5S in wild-type cells. These multiple pathways afford numerous opportunities for regulation by modulating substrate folding, cleavage site choice, and RNase III activity.
The functional significance of these redundant pathways, as well as of the two forms of U5, remains enigmatic. Both U5L and U5S are incorporated into snRNPs (Madhani et al. 1990) and U5L is dispensable in vivo (Haltiner-Jones and Guthrie 1990; Frank et al. 1994). In addition, some yeast species very close to S. cerevisiae have lost the long form of U5, whereas in vertebrates, only the long form exists (Frank et al. 1994). The two forms of U5 in S. cerevisiae may simply result from the presence of redundant processing machineries that would protect the cell from loss of one of the processing pathways. Such redundancy is reminiscent of yeast tRNA 3′-end formation, which relies on two independent pathways: an endonucleolytic pathway requiring the La protein, and an exonucleolytic processing pathway (Yoo and Wolin 1997). The species differences in U5 3′ ends can be explained by loss of one or another pathway, or by loss of cis-acting U5 substrate features, in different species.
Finally, it seems likely that the targets of RNase III in eukaryotic cells are not restricted to rRNA and snRNAs. The presence of regulatory sequences in the 5′ or 3′ untranslated region is a common feature of mRNAs, and it is conceivable that structures recognized by RNT1 are present in some mRNAs. For example, overexpression of RNase III in S. pombe inhibits mating and sporulation, probably by degrading specific mRNA(s) required for sexual development (Xu et al. 1990; Iino et al. 1991). Given the variety of targets of RNase III in prokaryotic cells, and the effect of RNase III cleavage site choice on alternative U5 3′-end formation, it seems possible that eukaryotic cells have developed additional uses for this enzyme.
Materials and methods
Plasmids and strains
Yeast manipulation was done as described (Guthrie and Fink 1991). The wild-type strain used for extract preparation is YGS2, a derivative of S288C (Noble and Guthrie 1996a). Wild-type and rnt1 mutant strains used for the experiments in Figure 5 are derived from a strain carrying a chromosomal disruption of RNT1 by insertion of a HIS3 fragment (Abou Elela et al. 1996) and the wild-type RNT1 gene on a LEU2 plasmid (wild type), or no plasmid (rnt1). Although RNT1 scores as an essential gene by tetrad dissection (Abou Elela et al. 1996), long-term incubation of the disrupted strain complemented by a URA3–RNT1 plasmid on 5-fluoro-orotic acid medium (which selects for cells that lack the URA3 gene; Guthrie and Fink 1991) reproducibly gives rise to very slow growing (generation time ∼6–7 hr at 25°C) and temperature-sensitive cells carrying only the disrupted allele. This phenotype is rescued by a plasmid-borne RNT1 gene (data not shown). Because the disruption retains the potential to express a pet56–rnt1 fusion protein, the residual growth at 25°C may be attributable to a low level of activity of this fusion. U5 sequences were amplified from genomic DNA using Pfu Polymerase (Stratagene) and primers T7U5 (5′-GCGAATTCTAATACGACTCACTATAGGGAAGCAGCTTTACAG) and U5DS (5′-CGGCATCCGCAAATGCTTCAATGAG). The amplified fragment was cut with BamHI and EcoRI and ligated into PUC19 to create pT7U5P.
In vitro 3′-end processing reaction
Yeast whole-cell extracts were prepared as described (Umen and Guthrie 1995). RNA synthesis and labeling was done as described (Chanfreau and Jacquier 1996), using pT7U5p digested with BamHI as a template, except that cap nucleotide (1 mm) was included in the transcriptions (except for RNAs destined to be 5′ end-labeled). Internal labeling was done by the addition of [α-32P]ATP, GTP, or UTP in the in vitro transcription reaction. Phosphorothioate incorporation and iodine cleavage were done as described (Christian and Yarus 1992). In vitro reactions were usually done in 10 μl containing 30%–60% extract (vol/vol), 100 mm K-acetate (pH 7.2), 2.5 mm Mg-acetate, 3% (vol/vol) polyethylene glycol (average molecular weight 8000), 2 mm ATP, 0.5 mm CaCl2, 1.5 mm dithiothreitol. The role of ATP in the processing reaction is unclear. Although it is not required for the first step of the processing, replacement of ATP by GTP moderately inhibits the conversion from the λ intermediate to the U5 long form, and production of the short form. ATP depletion from the extract has not been assayed. Presence of dithiothreitol and CaCl2 increased the production of U5S without changing the rate of formation of other species. No difference in processing efficiency was noticed between capped and uncapped transcripts (data not shown). After incubation, reactions were quenched on ice and by addition of 400 μl of STOP buffer (0.2 m NaCl, 25 mm EDTA at pH 8.0, 1% Na-dodecylsulfate). RNAs were extracted with phenol–chloroform (1:1) and precipitated with 1 ml of ethanol, and pellets were resuspended in formamide loading buffer (95% deionized formamide, 10 mm Tris at pH 7.5, 5 mm EDTA at pH 8.0) and loaded on acrylamide gels. Gels were fixed 20 min in 20% ethanol and 10% acetic acid (vol/vol), dried and exposed for autoradiography or PhosphorImager scanning.
RNase T1 digestion
Gel-purified RNAs were digested 1 hr at 37°C, in 6-μl reactions with 1000 units of RNase T1 (Boehringer Mannheim), in 10 mm Tris-HCl at pH 7.5, 10 mm EDTA at pH 8.0, 1 mg/ml of E. coli tRNA. Digestion reactions were loaded directly on 10% acrylamide gels after addition of 2 volumes of formamide loading buffer.
Northern blot
Northern blots were done as described (Good et al. 1994). Briefly, 4 μg of total RNA extracted from cells carrying RNT1 or rnt1-1 allele were loaded on 6% acrylamide denaturing gels, and transferred to a nylon membrane (Hybond-N, Amersham). The membrane was hybridized to probes prepared by random priming (Megaprime Kit, Amersham) in 5× SSPE, 0.1% SDS, 2× Denhardt’s solution, and 50% formamide at 50°C. Filters were washed in a solution containing 2× SSPE and 0.1% SDS at room temperature and exposed for autoradiography.
Immunoprecipitations
Protein A–Sepharose beads (Pharmacia) were washed extensively in IP buffer (50 mm Tris-HCl at pH 7.5, 300 mm NaCl, 0.05% NP40, 5 mm EDTA, 0.1 mg/ml of PMSF, 1 μg/ml of leupeptin, 1 mm benzamidine). Eighty microliters of this slurry were incubated with 50 μl of crude serum or ascites in 600 μl of IP buffer with 0.1 mg/ml of bovine serum albumin (BSA), and nutated for 1 hr at 4°C. Antibodies bound to protein A beads were washed seven times with IP buffer. Scaled-up 3′-end processing reactions (10×) were added to antibodies bound to protein A beads, in a total volume of 800 μl of IP buffer, with 0.3 mg/ml of E. coli tRNA and 0.1 mg/ml of BSA to prevent nonspecific RNA and protein binding, and nutated for 30 min at 4°C. Beads were washed five times with IP buffer, and bound nucleic acids were phenol-extracted, ethanol-precipitated, and loaded on a denaturing 6% polyacrylamide gel.
In vitro cleavage by RNase III
Fifty femtomoles of U5 precursor (5′-labeled or internally labeled) were incubated at 30°C in the presence of GST or GST–RNT1, prepared as described (Abou Elela et al. 1996) from a transformed E. coli strain mutant for bacterial RNase III (generously provided by A. Nicholson, Wayne State University, Detroit, MI), in 30 mm Tris-HCl at pH 7.5, 10 mm MgCl2, 5 mm spermidine, 30 mm KCl, E. coli tRNA (0.1 mg/ml) in a total volume of 10 μl. Reactions were stopped by adding 400 μl of STOP buffer, the RNAs extracted with phenol–chloroform and ethanol-precipitated, and loaded on a denaturing 6% acrylamide gel.
Acknowledgments
We are greatly indebted to G. Rotondo for suggesting the involvement of RNase III in U5 3′-end processing. We thank C. Siebel for anti PSI ascites; A. Krainer for anti-TMG monoclonal antibody; A. Nicholson for RNase III− E. coli strain; A. Perrin and A. Jacquier for help with MFold; J. Abelson, A. Jacquier, G. Rotondo, and J. Staley for stimulating discussions; T. Esperas, C. Pudlow, and H. Roiha for wonderful technical assistance; and D. Frendewey, A. Jacquier, P. Legrain, G. Rotondo, and J. Staley for critical reading of the manuscript. G.C. was supported by a long-term fellowship from Human Frontier Science Program Organization. G.C. is Chargé de Recherches at the CNRS (URA 1300) and thanks A. Jacquier and P. Legrain for generous support during the late steps of this work. This work was supported by National Institutes of Health grants GM21119 (to C.G.) and GM55557 (to M.A.). C.G. is an American Cancer Society Research Professor of Molecular Genetics.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL guthrie@cgl.ucsf.edu; FAX (415) 502-5306.
References
- Abou Elela SA, Igel H, Ares M., Jr RNase III cleaves eukaryotic preribosomal RNA at a U3 snoRNP-dependent site. Cell. 1996;85:115–124. doi: 10.1016/s0092-8674(00)81087-9. [DOI] [PubMed] [Google Scholar]
- Bram RJ, Young RA, Steitz JA. The ribonuclease III site flanking 23S sequences in the 30S ribosomal precursor RNA of E. coli. Cell. 1980;19:393–401. doi: 10.1016/0092-8674(80)90513-9. [DOI] [PubMed] [Google Scholar]
- Chanfreau G, Jacquier A. An RNA conformational change between the two chemical steps of group II self-splicing. EMBO J. 1996;15:3466–3476. [PMC free article] [PubMed] [Google Scholar]
- Chanfreau G, Noble SM, Guthrie C. Essential yeast protein with unexpected similarity to subunits of mammalian cleavage and polyadenylation specificity factor (CPSF) Science. 1996;274:1511–1514. doi: 10.1126/science.274.5292.1511. [DOI] [PubMed] [Google Scholar]
- Christian EL, Yarus M. Analysis of the role of phosphate oxygens in the group I intron from Tetrahymena. J Mol Biol. 1992;228:743–758. doi: 10.1016/0022-2836(92)90861-d. [DOI] [PubMed] [Google Scholar]
- Ciliberto G, Dathan N, Frank R, Philipson L, Mattaj IW. Formation of the 3′ end on U snRNAs requires at least three sequence elements. EMBO J. 1986;5:2931–2937. doi: 10.1002/j.1460-2075.1986.tb04589.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Court D. RNase III: A double-strand processing enzyme. In: Brawerman G, Belasco J, editors. Control of mRNA stability. New York, NY: Academic Press; 1993. pp. 70–116. [Google Scholar]
- Dahlberg JE, Lund E. The genes and transcription of the major small nuclear RNAs. In: Birnstiel M-L, editor. Structure and function of major and minor small nuclear ribonucleoprotein particles. New York, NY: Springer-Verlag; 1988. pp. 38–70. [Google Scholar]
- Frank DN, Roiha H, Guthrie C. Architecture of the U5 small nuclear RNA. Mol Cell Biol. 1994;14:2180–2190. doi: 10.1128/mcb.14.3.2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good L, Abou Elela S, Nazar RN. Tetrahymena ribozyme disrupts rRNA processing in yeast. J Biol Chem. 1994;269:22169–22172. [PubMed] [Google Scholar]
- Guthrie C, Fink GR. Guide to yeast genetics and molecular biology. San Diego, CA: Academic Press; 1991. [Google Scholar]
- Guthrie C, Patterson B. Spliceosomal snRNAs. Annu Rev Genet. 1988;22:387–419. doi: 10.1146/annurev.ge.22.120188.002131. [DOI] [PubMed] [Google Scholar]
- Haltiner-Jones M, Guthrie C. Unexpected flexibility in an evolutionarily conserved protein-RNA interaction: Genetic analysis of the Sm binding site. EMBO J. 1990;9:2555–2561. doi: 10.1002/j.1460-2075.1990.tb07436.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez N, Weiner AM. Formation of the 3′ end of U1 snRNA requires compatible snRNA promoter elements. Cell. 1986;47:249–258. doi: 10.1016/0092-8674(86)90447-2. [DOI] [PubMed] [Google Scholar]
- Hughes JM, Ares M., Jr Depletion of U3 small nucleolar RNA inhibits cleavage in the 5′ external transcribed spacer of yeast pre-ribosomal RNA and impairs formation of 18S ribosomal RNA. EMBO J. 1991;10:4231–4239. doi: 10.1002/j.1460-2075.1991.tb05001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iino Y, Sugimoto A, Yamamoto M. S. pombe pac1+, whose overexpression inhibits sexual development, encodes a ribonuclease III-like RNase. EMBO J. 1991;10:221–226. doi: 10.1002/j.1460-2075.1991.tb07939.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenny A, Minvielle-Sebastia L, Preker PJ, Keller W. Sequence similarity between the 73-kilodalton protein of mammalian CPSF and a subunit of yeast polyadenylation factor I. Science. 1996;274:1514–1517. doi: 10.1126/science.274.5292.1514. [DOI] [PubMed] [Google Scholar]
- Kleinschmidt AM, Pederson T. Accurate and efficient 3′ processing of U2 small nuclear RNA precursor in a fractionated cytoplasmic extract. Mol Cell Biol. 1987;7:3131–3137. doi: 10.1128/mcb.7.9.3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krainer AR. Pre-mRNA splicing by complementation with purified human U1, U2, U4/U6 and U5 snRNPs. Nucleic Acids Res. 1988;16:9415–9429. doi: 10.1093/nar/16.20.9415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhani HD, Bordonne R, Guthrie C. Multiple roles for U6 snRNA in the splicing pathway. Genes & Dev. 1990;4:2264–2277. doi: 10.1101/gad.4.12b.2264. [DOI] [PubMed] [Google Scholar]
- Madore SJ, Wieben ED, Pederson T. Intracellular site of U1 small nuclear RNA processing and ribonucleoprotein assembly. J Cell Biol. 1984a;98:188–192. doi: 10.1083/jcb.98.1.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madore SJ, Wieben ED, Kunkel GR, Pederson T. Precursors of U4 small nuclear RNA. J Cell Biol. 1984b;99:1140–1144. doi: 10.1083/jcb.99.3.1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattaj IW. U snRNP assembly and transport. In: Birnstiel M-L, editor. Structure and function of major and minor small nuclear ribonucleoprotein particles. New York, NY: Springer-Verlag; 1988. pp. 100–114. [Google Scholar]
- McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson SD, Wickens M, Bentley DL. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature. 1997;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- Minvielle-Sebastia L, Preker PJ, Keller W. RNA14 and RNA15 proteins as components of a yeast pre-mRNA 3′-end processing factor. Science. 1994;266:1702–1705. doi: 10.1126/science.7992054. [DOI] [PubMed] [Google Scholar]
- Miraglia L, Seiwert S, Igel AH, Ares M., Jr Limited functional equivalence of phylogenetic variation in small nuclear RNA: Yeast U2 RNA with altered branchpoint complementarity inhibits splicing and produces a dominant lethal phenotype. Proc Natl Acad Sci. 1991;88:7061–7065. doi: 10.1073/pnas.88.16.7061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai K, Mattaj IW. RNA–protein interactions in the splicing snRNPs. In: Nagai K, Mattaj IW, editors. RNA–protein interactions. Oxford, UK: IRL Press; 1994. pp. 150–177. [Google Scholar]
- Neuman de Vegvar HE, Dahlberg JE. Nucleocytoplasmic transport and processing of small nuclear RNA precursors. Mol Cell Biol. 1990;10:3365–3375. doi: 10.1128/mcb.10.7.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman de Vegvar HE, Lund E, Dahlberg JE. 3′ end formation of U1 snRNA precursors is coupled to transcription from snRNA promoters. Cell. 1986;47:259–266. doi: 10.1016/0092-8674(86)90448-4. [DOI] [PubMed] [Google Scholar]
- Noble SM, Guthrie C. Identification of novel genes required for yeast pre-mRNA splicing by means of cold-sensitive mutations. Genetics. 1996a;143:67–80. doi: 10.1093/genetics/143.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Transcriptional pulse-chase analysis reveals a role for a novel snRNP-associated protein in the manufacture of spliceosomal snRNPs. EMBO J. 1996b;15:4368–4379. [PMC free article] [PubMed] [Google Scholar]
- Patterson B, Guthrie C. An essential yeast snRNA with a U5-like domain is required for splicing in vivo. Cell. 1987;49:613–624. doi: 10.1016/0092-8674(87)90537-x. [DOI] [PubMed] [Google Scholar]
- Potashkin J, Frendewey D. A mutation in a single gene of Schizosaccharomyces pombe affects the expression of several snRNAs and causes defects in RNA processing. EMBO J. 1990;9:525–534. doi: 10.1002/j.1460-2075.1990.tb08139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramamurthy L, Ingledue TC, Pilch DR, Kay BK, Marzluff WF. Increasing the distance between the snRNA promoter and the 3′ box decreases the efficiency of snRNA 3′-end formation. Nucleic Acids Res. 1996;24:4525–4534. doi: 10.1093/nar/24.22.4525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotondo G, Frendewey D. Purification and characterization of the Pac1 ribonuclease of Schizosaccharomyces pombe. Nucleic Acids Res. 1996;24:2377–2386. doi: 10.1093/nar/24.12.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotondo G, Gillespie M, Frendewey D. Rescue of the fission yeast snRNA synthesis mutant snm1 by overexpression of the double-strand-specific Pac1 ribonuclease. Mol Gen Genet. 1995;247:698–708. doi: 10.1007/BF00290401. [DOI] [PubMed] [Google Scholar]
- Seraphin B, Rosbash M. Identification of functional U1 snRNA–pre-mRNA complexes committed to spliceosome assembly and splicing. Cell. 1989;59:349–358. doi: 10.1016/0092-8674(89)90296-1. [DOI] [PubMed] [Google Scholar]
- Seraphin B, Abovich N, Rosbash M. Genetic depletion indicates a late role for U5 snRNP during in vitro spliceosome assembly. Nucleic Acids Res. 1991;19:3857–3860. doi: 10.1093/nar/19.14.3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebel CW, Kanaar R, Rio DC. Regulation of tissue-specific P-element pre-mRNA splicing requires the RNA-binding protein PSI. Genes & Dev. 1994;8:1713–1725. doi: 10.1101/gad.8.14.1713. [DOI] [PubMed] [Google Scholar]
- Siliciano PG, Jones MH, Guthrie C. Saccharomyces cerevisiae has a U1-like small nuclear RNA with unexpected properties. Science. 1987;237:1484–1487. doi: 10.1126/science.3306922. [DOI] [PubMed] [Google Scholar]
- Umen JG, Guthrie C. A novel role for a U5 snRNP protein in 3′ splice site selection. Genes & Dev. 1995;9:855–868. doi: 10.1101/gad.9.7.855. [DOI] [PubMed] [Google Scholar]
- Wise JA, Tollervey D, Maloney D, Swerdlow H, Dunn EJ, Guthrie C. Yeast contains small nuclear RNAs encoded by single copy genes. Cell. 1983;35:743–751. doi: 10.1016/0092-8674(83)90107-1. [DOI] [PubMed] [Google Scholar]
- Xu HP, Riggs M, Rodgers L, Wigler M. A gene from S. pombe with homology to E. coli RNase III blocks conjugation and sporulation when overexpressed in wild-type cells. Nucleic Acids Res. 1990;18:5304. doi: 10.1093/nar/18.17.5304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo CJ, Wolin SL. The yeast La protein is required for the 3′ endonucleolytic cleavage that matures tRNA precursors. Cell. 1997;89:393–402. doi: 10.1016/s0092-8674(00)80220-2. [DOI] [PubMed] [Google Scholar]
- Yuo CY, Ares M, Jr, Weiner AM. Sequences required for 3′ end formation of human U2 small nuclear RNA. Cell. 1985;42:193–202. doi: 10.1016/s0092-8674(85)80115-x. [DOI] [PubMed] [Google Scholar]
- Zuker M. Prediction of RNA secondary structure by energy minimization. Methods Mol Biol. 1994;25:267–294. doi: 10.1385/0-89603-276-0:267. [DOI] [PubMed] [Google Scholar]