Abstract
P-TEFb is a key regulator of the process controlling the processivity of RNA polymerase II and possesses a kinase activity that can phosphorylate the carboxy-terminal domain of the largest subunit of RNA polymerase II. Here we report the cloning of the small subunit of Drosophila P-TEFb and the finding that it encodes a Cdc2-related protein kinase. Sequence comparison suggests that a protein with 72% identity, PITALRE, could be the human homolog of the Drosophila protein. Functional homology was suggested by transcriptional analysis of an RNA polymerase II promoter with HeLa nuclear extract depleted of PITALRE. Because the depleted extract lost the ability to produce long DRB-sensitive transcripts and this loss was reversed by the addition of purified Drosophila P-TEFb, we propose that PITALRE is a component of human P-TEFb. In addition, we found that PITALRE associated with the activation domain of HIV-1 Tat, indicating that P-TEFb is a Tat-associated kinase (TAK). An in vitro transcription assay demonstrates that the effect of Tat on transcription elongation requires P-TEFb and suggests that the enhancement of transcriptional processivity by Tat is attributable to enhanced function of P-TEFb on the HIV-1 LTR.
Keywords: P-TEFb, transcription elongation factor, RNA polymerase II, HIV-1 Tat, transactivation
The intricate pattern of gene expression exhibited by all eukaryotic organisms is accomplished, in part, through control of the steps required to generate mature mRNAs. It is increasingly clear that one of the major regulatory processes affects the elongation potential of RNA polymerase II after initiation (Spencer and Groudine 1990; Kerppola and Kane 1991; Wright 1993; Bentley 1995). Negative factors instigate this elongation control process by limiting the processivity of RNA polymerase II, leading to abortive elongation, a process characterized by premature termination (Marshall and Price 1992). One such factor, Drosophila factor 2, causes transcript release (Xie and Price 1996). Positive factors, exemplified by Drosophila positive transcriptional elongation factor b (P-TEFb; Marshall and Price 1995), trigger the escape into productive elongation and allow the generation of transcripts that extend beyond the 3′ ends of mature mRNAs. Polymerases that have made the transition into productive elongation are further affected by elongation factors, such as S-II (Guo and Price 1993), TFIIF (Kephart et al. 1994), ELL (Shilatifard et al. 1996), and elongin (Aso et al. 1995); these factors increase the efficiency of transcription elongation (Reines et al. 1996). Whereas the effects of the latter factors are important, the transition from abortive to productive elongation is the major regulated step (Wright 1993).
The carboxy-terminal domain (CTD) of the largest subunit of RNA polymerase II plays an important role in the elongation control process. It was first noticed that phosphorylation of the CTD occurred at the same time as the transition into productive elongation (O’Brien et al. 1994; Dahmus 1996). It was then found that an intact CTD was required for the elongation control process (Chun and Jeang 1996; Marshall et al. 1996). A connection between elongation control and CTD phosphorylation came with the discovery that Drosophila P-TEFb had CTD kinase activity (Marshall and Price 1995; Marshall et al. 1996). The CTD kinase activity and transcriptional function of P-TEFb are blocked by low levels of 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole (DRB), a nucleoside analog that inhibits CTD phosphorylation (Egyhazi et al. 1996) and the production of mRNAs (Tamm and Kikuchi 1979) in vivo.
HIV-1 Tat greatly stimulates gene expression from the viral promoter located in the long terminal repeat (LTR) of the human immunodeficiency virus (HIV) genome (Jones and Peterlin 1994). The primary effect of Tat is to increase the processivity of RNA polymerases that otherwise would prematurely terminate after the synthesis of short nascent transcripts (Kao et al. 1987; Laspia et al. 1989; Feinberg et al. 1991; Marciniak and Sharp 1991; Laspia et al. 1993). The elongation effect requires an RNA element, transactivation response (TAR), that forms a stem and loop structure with which Tat can associate (Jones and Peterlin 1994). Consistent with its effect on elongation Tat has been found to be an integral component of activated elongation complexes (Keen et al. 1996). The function of Tat in elongation is similar to the function of P-TEFb in that the transcriptional stimulation by Tat is sensitive to DRB (Braddock et al. 1991; Marciniak and Sharp 1991) and requires the CTD of the largest subunit of RNA polymerase II (Chun and Jeang 1996; Parada and Roeder 1996; Yang et al. 1996). In vitro (Herrmann and Rice 1993, 1995; Chun and Jeang 1996) and in vivo (Yang et al. 1996) Tat specifically associates with a serine/threonine kinase through its activation domain. Like P-TEFb, this Tat-associated kinase (TAK) is sensitive to DRB (Yang et al. 1996). It has been suggested that Tat might function by recruiting TAK to phosphorylate the CTD (Yang et al. 1996). Tat has also been shown to affect initiation (Laspia et al. 1989; Veschambre et al. 1995). Tat can associate with transcription preinitiation complexes and this association does not require the TAR element (Garcia-Martinez et al. 1997a). Consistent with a function in initiation, recent results have shown that Tat can interact with the RNA polymerase II holoenzyme in the absence of TAR (Cujec et al. 1997).
Besides the involvement of P-TEFb in elongation control (Marshall and Price 1995; Marshall et al. 1996), TFIIH has also been implicated in the process. There is a correlation between activators that stimulate elongation and their ability to associate with TFIIH (Blau et al. 1996; Tsuchiya et al. 1996). In addition, antibodies against subunits of TFIIH inhibited elongation of transcription of the c-myc gene when both were injected into Xenopus oocytes (Yankulov et al. 1996). Parada and Roeder (1996) and Garcia-Martinez et al. (1997b), but not Yang et al. (1996), found that TFIIH could associate with Tat and was required for Tat-stimulated transcription. Strong evidence links TFIIH with a late event in initiation, promoter escape (Goodrich and Tjian 1994), whereas P-TEFb has been shown to function during elongation (Marshall and Price 1995). It is possible that both factors are involved, and they act independently or one may regulate the activity of the other.
In this paper we report the cloning of the kinase subunit of Drosophila P-TEFb and show that a previously known protein, PITALRE, is the homologous human P-TEFb catalytic subunit. We provide several lines of evidence indicating that P-TEFb associates with the HIV-1 Tat. Furthermore, we show that human P-TEFb is required for Tat-mediated stimulation of transcription elongation. Our results suggest that the ability of Tat to increase the processivity of RNA polymerase II is mediated by its ability to enhance the function of the elongation control factor, P-TEFb.
Results
Cloning of the kinase subunit of Drosophila P-TEFb
To further our understanding of the function of P-TEFb we cloned a cDNA encoding the small subunit of the Drosophila factor. The two subunits of purified P-TEFb were separated by gel electrophoresis, and the small subunit was excised and subjected to protein sequencing. The peptide sequence information was used in the cloning of full-length cDNA, as described in Materials and Methods. The deduced amino acid sequence (Fig. 1) identified the small subunit of Drosophila P-TEFb as a member of the Cdc2-like cyclin dependent kinase family with >40% identity to Schizosaccharomyces pombe Cdc2. A search of the protein database revealed a human protein, PITALRE (Grana et al. 1994), that exhibits 72% identity and 83% similarity to the Drosophila protein (Fig. 1). The high level of sequence similarity indicated that PITALRE is a potential homolog of the small subunit of Drosophila P-TEFb and therefore may be a component of human P-TEFb. Two kinases from Saccharomyces cerevisiae, SGV1 (Irie et al. 1991) and CTK1 (Sterner et al. 1995), each share 43% identity with PITALRE and the small subunit of Drosophila P-TEFb. Although sequence similarity does not allow the prediction of a potential yeast homolog, CTK1 has recently been demonstrated to increase the elongation efficiency of RNA polymerase II (Lee and Greenleaf 1997).
Figure 1.

The small subunit of Drosophila P-TEFb is similar to PITALRE. (Dm) Small subunit of Drosophila P-TEFb; (Hu) human PITALRE. Black reverse shading indicates identity.
PITALRE is a component of human P-TEFb
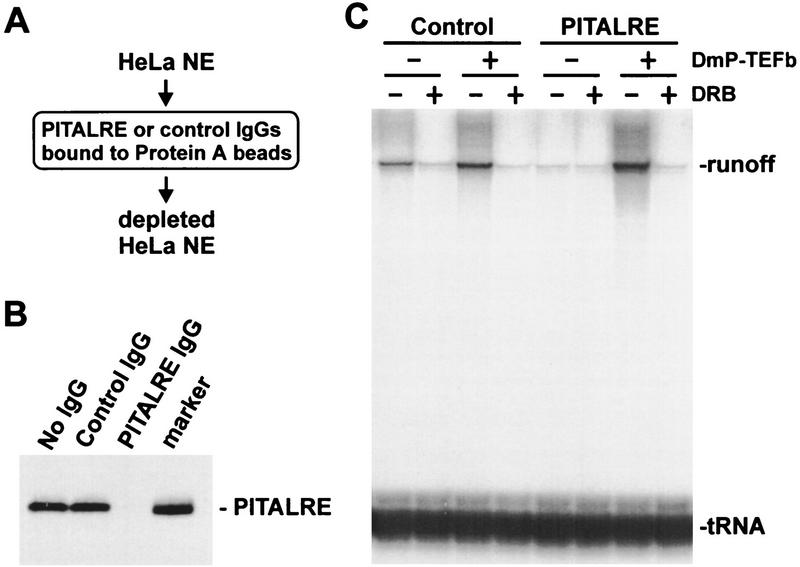
Drosophila P-TEFb functions during elongation by increasing the processivity of RNA polymerase II, allowing the synthesis of long transcripts (Marshall and Price 1995; Marshall et al. 1996). P-TEFb function is indicated by the generation of DRB-sensitive runoff transcripts during transcription in vitro. If PITALRE is the functional homolog of the small subunit of Drosophila P-TEFb, removal of PITALRE from HeLa nuclear extract (HNE) should eliminate DRB-sensitive runoff transcripts. To test this we immunodepleted PITALRE from HNE with antibodies directed against the last 20 amino acids of PITALRE that are not shared with other known kinases (Fig. 2A). Western blot analysis indicated that PITALRE was removed by anti-PITALRE antibodies to levels below detection but not by control antibodies (Fig. 2B). The depleted HNE was unable to generate DRB-sensitive 633-nucleotide runoff transcripts from an HIV-1 LTR template in a pulse/chase transcription reaction (Fig. 2C). Addition of pure Drosophila P-TEFb to the depleted extract restored DRB-sensitive transcription (Fig. 2C). These results indicate that depletion of PITALRE abolished human P-TEFb activity, supporting the hypothesis that PITALRE is a component of human P-TEFb.
Figure 2.
Human P-TEFb is required for the generation of DRB-sensitive runoff transcripts. (A) Immunodepletion. HNE was passed over two successive protein A columns containing either affinity-purified PITALRE–CT IgG or affinity-purified rabbit anti-goat IgG (control) as diagramed. (B) Western blot of HNE depleted with indicated antibodies probed with PITALRE–CT antibodies. (Marker) Bacterially expressed PITALRE. (C) Transcriptional activity of human P-TEFb. The pulse–chase protocol is described in Materials and Methods. (DmP-TEFb) Drosophila P-TEFb; (tRNA) tRNA labeled with [32P]CTP during transcription reaction (recovery control).
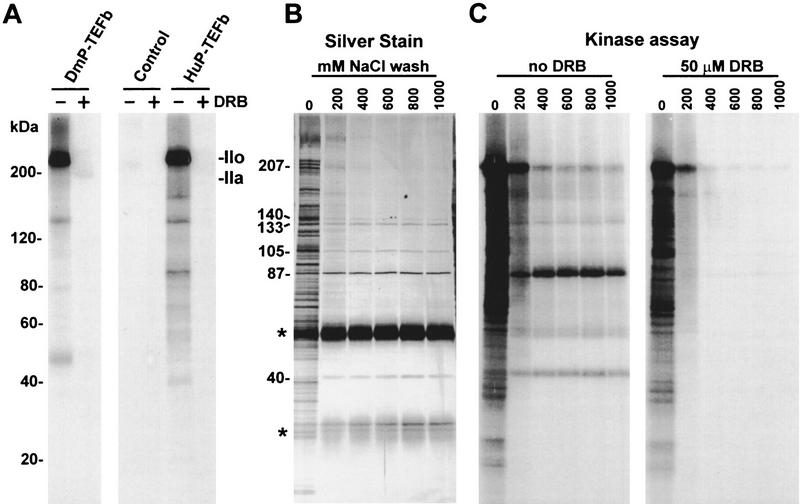
PITALRE was not known previously to be a CTD kinase. Therefore, we examined the material immunoprecipitated during the depletion of HNE for CTD kinase activity. The antibody-loaded beads containing PITALRE were washed extensively with high salt and subjected to a CTD kinase assay. Similar to Drosophila P-TEFb, beads containing PITALRE (together with any other strongly associated proteins) were able to convert the largest subunit of Drosophila RNA polymerase II to the hyperphosphorylated IIo form (Fig. 3A). Control beads were inactive. As expected, all phosphorylation was sensitive to 50 μm DRB. In the control reaction with Drosophila P-TEFb autophosphorylation of both subunits (43 and 124 kD) was seen. In the reaction with beads containing PITALRE antibodies, the 40-kD PITALRE, a band of similar size to the large subunit of Drosophila P-TEFb, and several other bands were phosphorylated.
Figure 3.
Immunoprecipitation of human P-TEFb. (A) CTD kinase assay. Purified Drosophila RNA polymerase II (Price et al. 1987; Marshall and Price 1995) was used as substrate. (DmP-TEFb) Drosophila P-TEFb; (Control) beads after depletion of HNE by affinity-purified rabbit anti-goat IgG; (HuP-TEFb) beads after depletion of HNE by PITALRE–CT IgG. Products were analyzed by 6%–15% gradient SDS-PAGE. Selected size standards from the 10-kD ladder are indicated and apply to all gels. (B) Silver-stained SDS-PAGE of proteins bound to PITALRE–CT IgG beads after washing with a buffer containing 20 mm HEPES (pH 7.6), 0.5% NP-40, 1% Triton X-100, and 5 mm DTT and the indicated amount of NaCl. Sizes of proteins remaining after high salt wash are indicated. Bands marked with an asterisk (*) are from IgG. (C) Kinase assay with only endogenous substrates of the fractions analyzed in B either without or with 50 μm DRB, as indicated.
To examine the association of other proteins with PITALRE, immunoprecipitates were washed with buffer containing nonionic detergents and increasing amounts of salt. The proteins associated with the beads were analyzed by SDS–PAGE, followed by silver staining (Fig. 3B). When no salt was present many proteins were retained on the beads. Most of these proteins were removed by washing with 200 mm NaCl. No changes occurred in the proteins visible after a wash with buffer containing NaCl higher than 400 mm. Besides immunoglobulin heavy and light chain and PITALRE (40 kD), proteins with sizes 87, 105, 133, and 140 kD were found. When rabbit anti-goat IgG control beads were used no other proteins except for the immunoglobulins were seen after high salt washes (data not shown). The immunoprecipitates were incubated with [γ-32P]ATP to determine which proteins became phosphorylated (Fig. 3C). The beads washed without salt carried out extensive phosphorylation of many protein substrates with little DRB sensitivity. After washing with 400 mm or higher concentration of NaCl, only a few proteins were labeled and all phosphorylation was DRB sensitive. Of the major proteins associated with PITALRE, only the 105-kD protein was not phosphorylated. At all salt concentrations the partially DRB-sensitive phosphorylation of a 207-kD protein was observed. Except for PITALRE the identity of the other proteins is unknown. The sizes of these proteins do not correlate with subunits of other known basal transcription factors. The other proteins could be constituents of a larger complex containing P-TEFb or different complexes containing PITALRE.
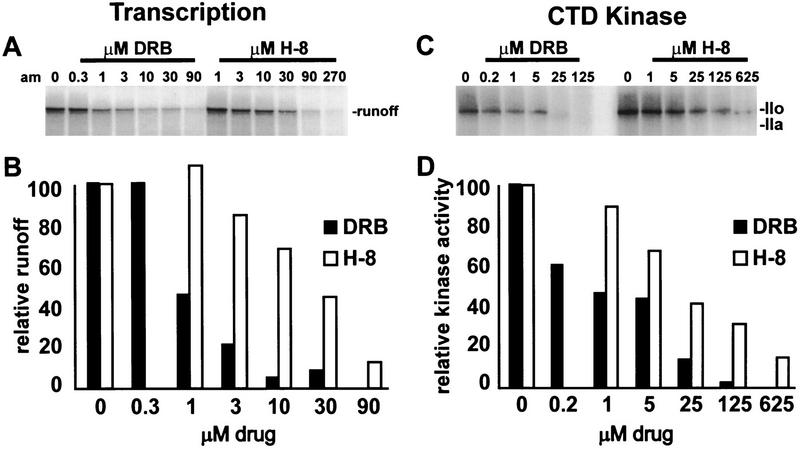
A distinguishing characteristic of P-TEFb is its sensitivity to the kinase inhibitors DRB and H-8. In vitro transcription and the CTD kinase activity of Drosophila P-TEFb are both inhibited by these two compounds, and in both assays DRB is 10-fold more potent than H-8 (Marshall and Price 1995; Marshall et al. 1996). The effect of DRB and H-8 were determined on transcription in the HNE (Fig. 4A,B) and on kinase assays using the immunoprecipitated human P-TEFb (Figure 4C,D). In both assays DRB was the more effective inhibitor. DRB and H8 compete with ATP for binding to the kinase active site; therefore, it is inappropriate to compare directly the 50% inhibition points under different conditions, especially if different concentrations of ATP are used (Marshall et al. 1996). However, the ratio of 50% points for different compounds under identical conditions can be compared. This ratio is more likely to be independent of assay condition. The ratio of 50% inhibition points (H-8/DRB) was 23 μm/1 μm = 23 for the transcription assay and 16 μm/0.65 μm = 25 for the CTD kinase assay. These ratios are similar to each other, suggesting that the same kinase, namely human P-TEFb, is being inhibited in both assays. Considering the sequence similarity between the small subunit of Drosophila P-TEFb and PITALRE, the functional similarity between Drosophila P-TEFb and the activity removed from HNE by PITALRE antibodies, and the presence of other potential subunits in the immunoprecipitates, we conclude that PITALRE is a component of human P-TEFb.
Figure 4.
DRB and H-8 inhibition of transcription and human P-TEFb activity. (A,B) Transcription of HIV LTR template (633-nucleotide runoff); (C,D) CTD kinase assay using immunoprecipitated human P-TEFb and Drosophila RNA polymerase II as substrate, as in Fig. 3A. (B,D) Plot of radioactivity in runoff or polymerase IIo after quantitation using a Packard InstantImager and normalization to the starting amount (100).
P-TEFb specifically associates with the activation domain of HIV Tat
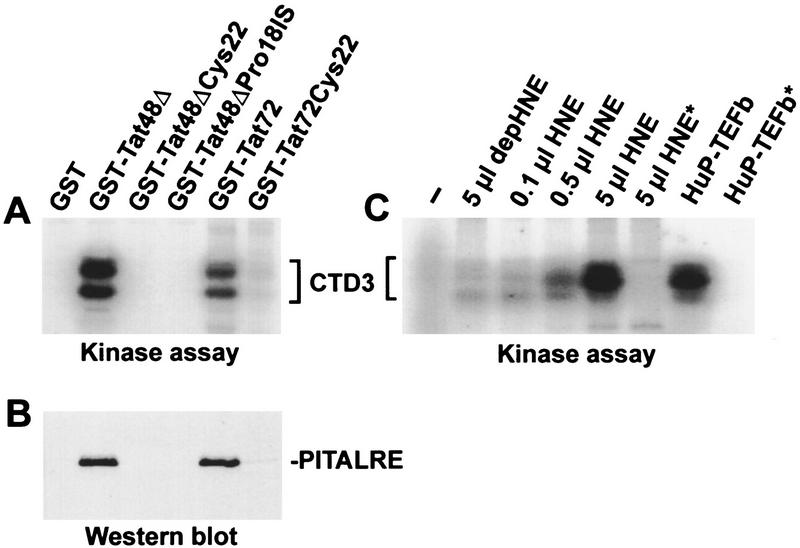
Several lines of evidence led us to hypothesize that P-TEFb might associate with the viral transactivator Tat. Tat transactivation is sensitive to DRB (Braddock et al. 1991; Marciniak and Sharp 1991) and requires the CTD (Chun and Jeang 1996; Parada and Roeder 1996; Yang et al. 1996), and Tat associates with a DRB-sensitive CTD kinase (TAK) (Herrmann and Rice 1993, 1995; Chun and Jeang 1996; Yang et al. 1996). The hypothesis was tested by ascertaining whether the human P-TEFb kinase associates with Tat during incubation with HeLa extracts. Glutathione beads containing various glutathione S-transferase (GST)–Tat fusion proteins were incubated with HeLa extract and washed extensively. Proteins associated with Tat constructs containing an intact activation domain (Tat72 and Tat48Δ) were able to phosphorylate the synthetic peptide CTD3 (Fig. 5A), as well as RNA polymerase II (data not shown). GST–Tat fusions containing mutations in the activation domain that abolish Tat transactivation (Rice and Carlotti 1990; Herrmann and Rice 1995) were not able to pull down TAK (Fig. 5A). The proteins associated with the Tat constructs were probed with anti-PITALRE antibody by Western blot analysis (Fig. 5B). PITALRE was detected only when the constructs contained an intact Tat transactivation domain. This suggests that P-TEFb is a TAK.
Figure 5.
PITALRE associates with the activation domain of HIV-1 Tat. (A) TAK activity assay using CTD3 peptide as substrate. The indicated GST Tat or Tat mutant proteins were used. Both Tat72 and Tat48Δ have intact transactivation domains, whereas other constructs have mutations that abolish transactivation. (Tat72) HIV-1 Tat containing residues 1–72; (Tat48Δ) the activation domain of HIV-1 Tat containing residues 1–48; (Cys22) mutant Tat containing Gly instead of Cys at position 22; (Pro18IS) Tat mutant containing Glu and Phe insertion after Pro-18. (B) Western blot of the same samples using antibodies against PITALRE–CT. (C) TAK activity assays using GST–Tat48Δ were performed with whole or depleted (depHNE) HNE. In the kinase assay CTD3 peptide was used as substrate. (−) No kinase added; (*) no CTD3 peptide added. Human P-TEFb (HuP-TEFb) was immunoprecipitated from the equivalent of 1 μl of HNE.
When human P-TEFb was depleted from HNE by antibodies to PITALRE, TAK activity (assayed by GST–Tat48Δ pull-down) was reduced to <2% of that found in the intact extract (Fig. 5C, cf. the lane using 5 μl of depleted extract and that using 0.1 μl of whole extract). This strongly suggests that under the conditions used human P-TEFb is the predominant CTD kinase that associates with Tat. Others have shown that TFIIH can associate with Tat (Parada and Roeder 1996; Garcia-Martinez et al. 1997b), but under the conditions used, we did not detect any p62 subunit of TFIIH bound to Tat nor did we detect a reduction in the amount of the subunit in the PITALRE-depleted extract. In addition, when glutathione beads containing TAK were probed with antibodies to all three subunits of CAK (CDK7, cyclin H, Mat1; supplied by D. Morgan, University of California, San Diego) we found no evidence of the TFIIH-associated kinase (data not shown).
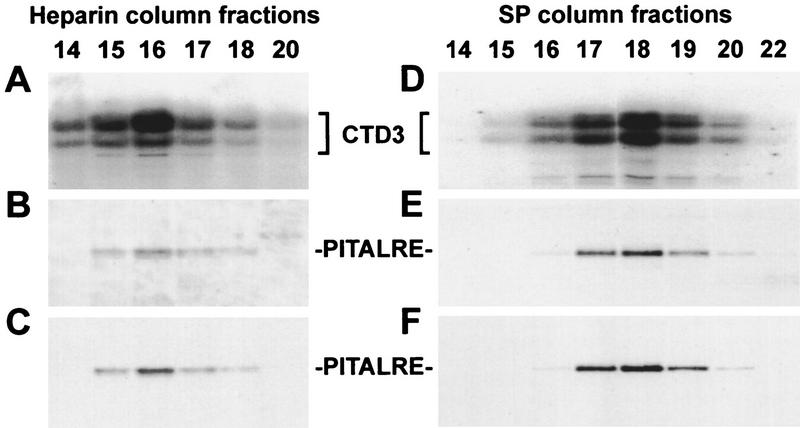
To further confirm that human P-TEFb can associate with Tat, TAK was partially purified from HeLa cells by sequential chromatography on DEAE, heparin, and SP resins. Fractions eluted from the second and third columns, Heparin and SP were assayed for TAK activity and probed for human P-TEFb (Fig. 6). In the eluate of both columns, TAK activity assayed by GST–Tat48Δ pull-down (Fig. 6A,D) correlated with P-TEFb determined by Western analysis using two different preparations of PITALRE antibodies (Fig. 6B,C,E,F). A GST construct with a mutation in the Tat activation domain did not become associated with CTD kinase activity when incubated with the same column fractions (data not shown). In addition, we compared the drug sensitivity of TAK and human P-TEFb. As expected, TAK activity was inhibited by DRB and H-8 in a manner similar to P-TEFb (data not shown).
Figure 6.
PITALRE coelutes with TAK activity. (A–C) Heparin column fractions; (D–F) SP column fractions. TAK activity was assayed with GST–Tat48Δ across both columns using CTD3 as the kinase substrate (A,D); PITALRE was detected in Western blots using antibodies against whole PITALRE (B,E) or PITALRE–CT, (C,F).
Human P-TEFb is required for Tat-stimulated elongation
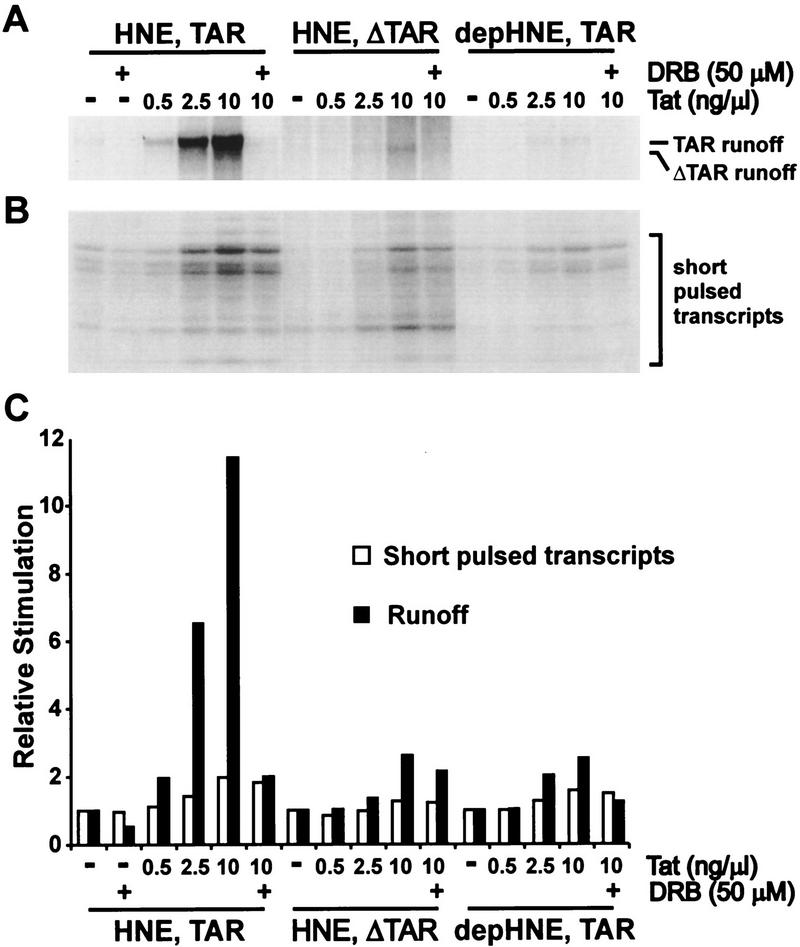
To investigate the function of P-TEFb in Tat transactivation directly, we compared the effect of Tat on initiation and elongation in whole or PITALRE-depleted extracts (Fig. 7). Two different DNA templates were used. The TAR template contained HIV-1 LTR sequences from −475 to +76, whereas the ΔTAR template lacked the sequence encoding TAR and contained HIV-1 LTR sequences from −475 to +19. With HNE (not depleted) and a continuous labeling protocol (Fig. 7A), 10 ng/μl of Tat stimulated the generation of runoff ∼12-fold (Fig. 7C, solid bars). This effect of Tat was mostly inhibited by 50 μm DRB and required the TAR sequence. The DRB sensitivity of Tat-stimulated runoff was similar to that determined for the low level of runoff from the HIV LTR in the absence of Tat shown in Figure 4 (data not shown). When PITALRE-depleted extract was used, the majority of the stimulatory effect of Tat was abolished. This result strongly suggests that human P-TEFb is required for efficient transactivation by Tat. Add-back of Drosophila P-TEFb to the depleted extract stimulated runoff in the presence or absence of Tat, as seen in Figure 2, but did not restore the ability of Tat to specifically enhance elongation (data not shown). This suggests that other required factors were removed by the depletion of PITALRE or that Drosophila P-TEFb lacks appropriate domains required for interaction with Tat or other cofactors. Addition of the high salt-washed PITALRE immunoprecipitate had no effect on runoff in the presence or absence of Tat (data not shown), indicating that immobilization had a negative impact on the function of P-TEFb. Although consistent with an effect on elongation, the experiment shown in Figure 7A did not rule out the possibility that the major effect of Tat was on initiation.
Figure 7.
P-TEFb is required for Tat transactivation in vitro. (A) In vitro Tat transactivation using a continuous labeling protocol with the indicated templates and extracts. Runoff transcripts were analyzed in a 6% TBE/urea gel. (B) Similar reaction mixtures as in A were subjected to a 2-min pulse, and the short transcripts generated (<30 nucleotides) were analyzed in an 18% TBE/urea gel. (C) Quantitation data for A and B. The amount of runoff or short transcripts for each template/extract combination was normalized to the corresponding lane with no DRB or Tat added.
From the autoradiograph (Figure 7A) and the normalized quantitation data graphed in Figure 7C (solid bar) it was obvious that Tat had a modest effect on transcription (about twofold increase of runoff) in the presence of 50 μm DRB, in the presence of ΔTAR template, or even in the absence of human P-TEFb. To further analyze the effect of Tat on transcription, we performed a similar set of experiments, as shown in Figure 7A, except that a pulse-only transcription protocol was used instead of a continuous labeling protocol (Fig. 7B). Quantitation of the short pulsed transcripts (<20 nucleotides in length) gives the efficiency of initiation. Under these conditions it was clear that increasing Tat had the effect of increasing transcription initiation (Fig. 7B and open bars in C). At the highest level of Tat used (10 ng/μl) initiation increased about twofold compared to initiation when no Tat was added. This effect on initiation did not require TAR (Fig. 7B,C). Moreover, the effect of Tat on initiation was not sensitive to DRB and did not require P-TEFb. The effect of Tat on initiation can be used to explain the slight effect of Tat on the generation of runoff in the presence of DRB, ΔTAR template, or depleted HNE (Fig. 7A). The two assays taken together indicate that the major effect of Tat is on elongation, although initiation is slightly affected. Most importantly, P-TEFb is required for the effect of Tat on elongation but not for its effect on initiation.
Discussion
We have cloned the small subunit of Drosophila P-TEFb and identified it as the counterpart of two human protein kinases whose mutual identity was not recognized previously. Sequence similarity suggested that the small subunit of Drosophila P-TEFb was the homolog of PITALRE, a Cdc2-related kinase of hitherto unknown function. This was confirmed when Drosophila P-TEFb was able to functionally replace the transcriptional activity removed from HeLa nuclear extract with PITALRE antibodies. Using immunological and biochemical tests we found that human P-TEFb was a TAK. Furthermore, we showed that the effect of Tat on transcription elongation was dependent on P-TEFb. These results provide a foundation for understanding the mechanism of action of Tat in HIV transcriptional transactivation and should facilitate the development of defined in vitro Tat transactivation systems and of drugs aimed at blocking Tat transactivation.
All three of these activities, P-TEFb, PITALRE, and TAK, previously were known to be associated with a 42- to 43-kD phosphoprotein that is now characterized as the kinase subunit of the human P-TEFb complex. In Drosophila this complex contains two subunits of 43 and 124 kD (Marshall and Price 1995), whereas the human P-TEFb complex is less well defined. The small subunit is found in association with several high molecular weight polypeptides (Fig. 3; Grana et al. 1994; Garriga et al. 1996a). The roles of the additional polypeptides are unknown, but it is possible that they interact with other components of the transcriptional machinery and/or regulate kinase activity. Because PITALRE has similarity to other cyclin-dependent kinases it is likely that one of these associated proteins is a cyclin subunit. PITALRE with its associated proteins is a proline-directed serine/threonine kinase (Garriga et al. 1996b) that, like its Drosophila counterpart, is able to hyperphosphorylate the CTD of RNA polymerase II (Figs. 3 and 5; Herrmann and Rice 1995; Marshall et al. 1996). Although proteins such as the retinoblastoma protein pRB (but not histone H1) can serve as substrates (Grana et al. 1994), at least in vitro, the link between transcription elongation and RNA polymerase II phosphorylation makes it likely that the CTD is a major substrate for the P-TEFb kinase in vivo.
CTD hyperphosphorylation is associated with processive elongation (Dahmus 1996). Correspondingly, the CTD is essential for P-TEFb-mediated transition to productive elongation in vitro (Marshall et al. 1996) and for Tat transactivation in vivo and in vitro (Parada and Roeder 1996; Yang et al. 1996). Drosophila P-TEFb acts at an early stage of elongation to allow transcription complexes to make the transition to productive elongation (Marshall et al. 1996), and results presented here extend this finding to the human transcription system (Fig. 2). Similarly, Tat binds to short nascent transcripts (Jones and Peterlin 1994) and overcomes promoter–proximal termination (Kao et al. 1987; Laspia et al. 1989, 1990). A priori, the properties of human P-TEFb make it an attractive candidate for the critical agent recruited to the elongating complex by Tat. However, Tat has been reported to associate with numerous components of the transcription apparatus, as well as other proteins (Jones and Peterlin 1994).
Several lines of evidence indicate that P-TEFb is involved in the stimulation of processivity brought about by Tat. P-TEFb binds specifically to the activation domain of Tat, and the interaction is eliminated by mutations that abrogate transactivation (Fig. 5; Herrmann and Rice 1993). The interaction of TAK with Tat survives exhaustive washing under stringent conditions and has been observed in vitro (Herrmann and Rice 1993, 1995; Chun and Jeang 1996) and in vivo (Yang et al. 1996). Furthermore, TAK (P-TEFb) binds to the Tat proteins of HIV-2 and equine infectious anemia virus (Herrmann and Rice 1993, 1995), which both appear to function through the same cofactor as HIV-1 Tat (Carroll et al. 1992; Madore and Cullen 1993). A second line of support derives from experiments with the nucleoside analog DRB. The enhanced processivity brought about by P-TEFb and by Tat are both selectively sensitive to low levels of DRB (Figs. 4 and 7; Braddock et al. 1991; Marciniak and Sharp 1991; Marshall and Price 1992). Correspondingly, the CTD kinase activities of Drosophila P-TEFb and TAK are sensitive to low levels of DRB and related inhibitors (Fig. 4; Herrmann and Rice 1995; Egyhazi et al. 1996; Marshall et al. 1996). Perhaps the most compelling argument for the role of P-TEFb in Tat transactivation comes from depletion experiments (Figs. 2, 3, 5, and 7). Antibodies directed against a 20-residue peptide from the carboxyl terminus of PITALRE simultaneously removed TAK activity and rendered the HeLa nuclear extract dependent on P-TEFb for efficient elongation. P-TEFb could be detected in the immunoprecipitates by silver staining and CTD kinase assay. Most importantly, HNE depleted of P-TEFb was unable to support Tat transactivation (Fig. 7A).
We observed that Tat stimulated transcriptional initiation as well as elongation in our in vitro transcription assay. This has been observed before (Laspia et al. 1989; Veschambre et al. 1995) and is consistent with the fact that Tat interacts with the RNA polymerase II holoenzyme (Cujec et al. 1997) and associates with transcription preinitiation complexes (Garcia-Martinez et al. 1997a). However, Tat stimulated initiation only ∼2-fold, whereas its runoff transcripts increased >10-fold (Fig. 7), consistent with previous results (Kao et al. 1987; Laspia et al. 1989; Feinberg et al. 1991) that the major effect of Tat is on elongation. The fact that TAR and P-TEFb are only required for the effect on elongation further differentiate the effect of Tat on initiation and elongation. The stimulation of initiation by Tat may be mediated through its interaction with SP1 (Jeang et al. 1993), TATA-binding protein (TBP) (Kashanchi et al. 1994; Veschambre et al. 1995), the p62 subunit of TFIIH (Parada and Roeder 1996), or RNA polymerase II (Mavankal et al. 1996). The effect of Tat on elongation is likely mediated by the interaction of TAR RNA with a complex containing Tat and P-TEFb. The complex may contain other cofactors such as Tat–SF1 (Zhou and Sharp 1996), which is required for Tat transactivation. Supporting this idea, Tat–SF1 has been shown to associate with a kinase that is also required for Tat transactivation (Zhou and Sharp 1996). A model integrating the effects on initiation and elongation is difficult to formulate. However, work of Hernandez and coworkers has indicated that Tat might affect the way RNA polymerase II initiates at the HIV LTR under different conditions because of the presence of a DNA element around the start point of transcription that induces short transcripts (IST) (Cullen 1993; Sheldon et al. 1993; Pendergrast and Hernandez 1997). The different types of early elongation complexes could have different receptivity to P-TEFb action.
Although P-TEFb is required for normal elongation control and Tat transactivation, an important question is whether Tat enhances the function of P-TEFb specifically during transcription of the HIV LTR. The association of P-TEFb with the activation domain of Tat suggests that an enhancement of activity is possible. The results in Figure 7 also strongly suggest that Tat allows P-TEFb to function more effectively. If P-TEFb merely performed its normal function the amount of DRB-sensitive runoff generated would be related to the number of initiation events. Because Tat stimulated initiation about twofold from the LTR either with or without TAR, the effect on DRB-sensitive runoff should have been twofold. However, when the TAR sequence was present Tat increased DRB-sensitive runoff transcripts ∼12-fold. The most likely explanation is that Tat enhanced the function of P-TEFb. Because P-TEFb is limiting in nuclear extracts (Marshall and Price 1992, 1995) Tat may allow the available P-TEFb to be recruited to early elongation complexes containing TAR RNA. Consistent with the idea that the enhancement of P-TEFb function by Tat is attributable to increased local concentration of P-TEFb, we have observed a stimulation of DRB-sensitive runoff by increasing the concentration of P-TEFb in the extract in the absence of Tat (see Fig. 2). One caveat to the interpretation of our results is that the method used to measure initiation only looked in a 2-min window. It is possible that the effect of Tat on initiation during the 20-min continuous labeling period used to examine Tat transactivation might have been different. Further experiments will be needed to resolve this and other uncertainties about the mechanism of the enhancement of P-TEFb function.
Although we have shown that P-TEFb plays a role in elongation control, others have implicated TFIIH in the process. In our work with P-TEFb we have not uncovered a specific function for TFIIH in elongation control, although it is present in all reactions because of its initiation requirement. Drosophila P-TEFb has been shown to increase the processivity of RNA polymerase II when added to in vitro reactions after the initiation phase was complete (Marshall et al. 1996). In the same study purified Drosophila TFIIH was unable to functionally substitute for P-TEFb even though it possessed a similar level of CTD kinase activity (Marshall et al. 1996). In comparing work from a number of laboratories, a point of confusion is the sensitivity of the two kinases to the competitive inhibitors DRB and H8. Although it has been suggested that TFIIH is the DRB-sensitive kinase (Yankulov et al. 1995), we showed in the Drosophila system that P-TEFb was significantly more sensitive to DRB than was TFIIH (Marshall et al. 1996). A comparison of the DRB sensitivity of human TFIIH and P-TEFb kinase activity has not been made directly using identical conditions; however, using a standard CTD kinase assay (10 μm ATP) DRB had a 50% inhibition point of <1 μm against P-TEFb, which is significantly lower than the 10–50 μm reported for TFIIH under similar (7.5 μm ATP) conditions (Yankulov et al. 1995). Therefore, we suggest that caution should be exercised when interpreting experiments utilizing kinase inhibitors such as DRB and H8. A second point of confusion is the association of the CTD kinases with Tat. Neither we (T. Pe’ery, unpubl.) nor Yang et al. (1996) found TFIIH in association with Tat. However, Parada and Roeder (1996) as well as Garcia-Martinez et al. (1997b) reported that TFIIH associated with Tat and was required for Tat-stimulated transcription. Unlike our results in which P-TEFb associated with Tat in the presence of a crude nuclear extract, the latter group found that TFIIH only associated with Tat after partial purification (Garcia-Martinez et al. 1997b). Parada and Roeder (1996) only found hyperphosphorylation of the CTD when a crude fraction of TFIIH containing other factors was used. Despite the confusion, strong evidence for the involvement of TFIIH in elongation control comes from experiments that do not involve kinase inhibitors but, rather, antibodies against subunits of TFIIH (Yankulov et al. 1996). Yankulov et al. (1996) showed that injection of antibodies against TFIIH had an effect on transcription similar to DRB. Our previous results and those presented here do not rule out a significant role for the kinase activity of TFIIH in elongation control or Tat transactivation, but rather, indicate only that another kinase, P-TEFb, is required. Now that the kinase subunit of P-TEFb has been identified, examining the potential interplay between TFIIH and P-TEFb is an exciting possibility.
Materials and methods
Cloning of the small subunit of Drosophila P-TEFb
Drosophila P-TEFb was purified as described (Marshall and Price 1995). The small subunit was excised after SDS-PAGE and subjected to peptide sequencing (Keck Facility, Yale University, New Haven, CT). Based on peptide sequences, two degenerate PCR primers (GGAATTCNATGYTNCARCARCC and AACTGCAGTCCARAARAARTCRTGRTT) were synthesized and used to amplify cDNA from Drosophila Kc cell total cDNA made from poly(A)+ RNA. A 1.1-kb cDNA fragment was amplified by PCR using Taq DNA polymerase. After digestion with PstI (an internal site in the PCR product and a site designed in the 3′ end), the 0.7-kb fragment from the 3′ portion of the PCR product was cloned into the Bluescript II SK+ vector (Stratagene). The 3′ region of the full-length cDNA sequence was obtained using a GIBCO 3′ RACE kit with two specific primers from the 0.7-kb fragment (5′-TGTCAAGGATCAAACCGGCTGTGAT and 5′-CGAATTCCAAGAAACGCATCGATGC). The 5′ region of the full-length cDNA was obtained using a GIBCO 5′ RACE kit with three gene-specific primers from the 0.7-kb fragment (5′-AGACCTGCCAAATCGTGT, 5′-AGAAGGTGGATCTGTAACCATTCGT, and 5′-GGAATTCAGATCTCGATCAGATTCA). The entire 1.2-kb coding region of the small subunit was cloned using a GIBCO reverse transcription PCR kit. First, the cDNA encoding the small subunit was generated in a reverse transcription with a designed primer (5′-TTACTACTCGAGCTACCAAACCCGGTC) and purified template (Drosophila embryonic mRNA). Second, the coding sequence was produced in a PCR reaction using Vent DNA polymerase, two primers (5′-TAAGCAAGCTTCTATGGCGCACATGTCC and 5′-TTACTACTCGAGCTACCAAACCCGGTC), and the cDNA as the template. Third, the coding sequence was digested with HindIII and XhoI and cloned into pET-21a vector.
Generation of antibodies to PITALRE
PITALRE–CT antibodies were affinity-purified rabbit IgG directed to the carboxy-terminal 20 amino acids of PITALRE (Santa Cruz Biotechnology). Antibodies against the whole PITALRE were generated using purified recombinant protein as antigen. First, two primers (5′-GCAGGATCCAGAATTCCATATGGCAAAGCAGTACGACTCGG and 5′- pGATCTCGAGCCCTCAGAAGACGCGCTCAAA) were used in a PCR reaction to amplify the coding region of the cDNA of PITALRE. The human brain cDNA mix (Clontech Co.) was used as template. The PCR product was cloned into a pET21a vector (Novagen Co.). The final vector was amplified and transformed into DE3 (BL21)-competent cells for expression of PITALRE. The transformed DE3 cells were grown to an OD600 of 0.6 and induced with 1 mm IPTG. After 3 hr induction, the cells were collected and lysed by passing through a French press three times. The lysate was subjected to centrifugation at 15,000g for 30 min. The pellet was solubilized in 20 mm Tris (pH 7.5), 0.1 m NaCl, and 7 m urea and loaded onto a Mono Q column (Pharmacia Co.). The flowthrough fraction of the Mono Q column was loaded onto a Mono S column (Pharmacia Co.); the flowthrough fraction of the Mono S column, in turn, was subjected to dialysis against PBS. The dialyzed solution was centrifuged at 15,000g for 30 min. The pellet was suspended in the same buffer and sent for generation of antibodies in rabbit (Pocono Rabbit Farm & Laboratory, Inc.).
Immunodepletion of human P-TEFb
Immunodepletion was performed by passing protein A–Sepharose-precleared HNE (in 20 mm HEPES at pH 7.6, 15% glycerol, 165 mm KCl, 0.1 mm EDTA, 1 mm DTT, 0.1 mm PMSF) through two affinity columns made with protein A–Sepharose beads prebound with anti-PITALRE–CT antibodies or control IgGs (affinity-purified rabbit anti-goat IgG; Sigma). For every 50 μl of HNE, 10 μl of protein A beads containing 1 μg of bound IgG was used. After depletion of HNE, the antibody-containing beads were washed extensively with 100 times the bead volume of 20 mm HEPES (pH 7.6), 0.5% NP-40, 1% Triton X-100, 5 mm DTT, and 600 mm or indicated concentration of NaCl and then washed with 25 times the bead volume of 20 mm HEPES (pH 7.6) and 1 mm DTT. The amounts of the washed beads used for the kinase assays and silver staining were the equivalent of 1 and 10 μl of HNE, respectively.
CTD kinase assay
CTD kinase assays were performed in a 20-μl reaction containing 20 mm HEPES (pH 7.6), 10 μm ATP, 5 μCi [γ-32P]ATP, ∼10 ng of Drosophila RNA polymerase II, 5 mm MgCl2, and purified Drosophila P-TEFb or immunoprecipitated human P-TEFb. In the reactions containing DRB it was added to 50 μm unless other indicated amounts were used. The reactions were incubated at 30°C for 1 hr.
TAK activity assay
Preparation of Tat fusion proteins and the TAK pull-down assay were conducted as described by Herrmann and Rice (1993, 1995) with modifications. Nuclear extract, cytoplasmic extract, or fractions containing partially purified TAK were incubated with glutathione–Sepharose beads containing GST–Tat fusion proteins for 1 hr at 4°C with gentle rocking. DE3 (BL21) bacteria containing the GST–Tat expression vectors were obtained from NIH AIDS Research and Reference Reagent Program. For maximum sensitivity, GST–Tat48Δ was used unless specified otherwise. The beads (30 μl, 50% slurry) were washed six to eight times with 1 ml of EBCD buffer (50 mm Tris at pH 8.0, 120 mm NaCl, 0.5% NP-40, 5 mm DTT) containing 0.03% SDS, then two to four times with Tat kinase buffer (TKB/Mg: 50 mm Tris-HCl at pH 7.6, 5 mm DTT, 5 mm MnCl2, and 4 mm MgCl2) and brought to 50 μl with TKB/Mg buffer and kinase assay mix. The final reaction contained 2 μm ATP, 10 μCi [γ-32P]ATP (ICN, 3000 Ci/mmole), and 50–100 μm CTD trimer peptide CTD3 (ACSYSPTSPSYSPTSPSYSPTSPSKK). Reactions were incubated at 25°C for 40 min, stopped by boiling in Laemmli sample buffer, and resolved by electrophoresis in 15% polyacrylamide/bis-acrylamide (30%:0.15%) gels.
Partial purification of TAK
TAK was partially purified from a HeLa cell cytoplasmic fraction (2.6 grams of protein) prepared according to Ausubel et al. (1989), except that dialysis was omitted. Proteins precipitating between 10% and 40% saturation of ammonium sulfate (816 mg) were resuspended in DEAE buffer (25 mm HEPES at pH 7.6, 150 mm KCl, 0.1 mm EDTA, 1 mm DTT, 0.1 mm PMSF, 4 mm MgCl2, 1 μg/ml each of aprotinin, leupeptin, pepstatin A, and 10% glycerol), dialyzed against the same buffer, and applied to a 230-ml DEAE–Sepharose column equilibrated in DEAE buffer. The flowthrough fraction (350 mg of protein) was concentrated by 50% ammonium sulfate precipitation. The proteins were resuspended in HE buffer (same as DEAE buffer, except 25 mm HEPES at pH 6.9, and 100 mm KCl were used) and loaded onto a perfusion chromatography heparin affinity column (POROS 20 HE; PerSeptive Biosystems). The column was washed with the same buffer, and proteins were eluted with a linear gradient of 100–500 mm KCl in 15 column volumes of HE buffer. TAK activity eluted from 200 to 250 mm KCl. Active fractions were diluted with an equal volume of the same buffer, except that the pH was 8.0 and KCl was omitted, and loaded onto a perfusion chromatography cation exchange column (POROS 20 SP; PerSeptive Biosystems). The column was washed with SP buffer (the same as HE buffer, except that the buffer was 25 mm HEPES at pH 7.5) and eluted with a linear gradient of 100–500 mm KCl in 15 column volumes of SP buffer. The active fractions (1 mg of protein) eluted at ∼300 mm KCl.
Transcription assay
The transcription template (NcoI-digested pLTR-4/CAT vector containing the HIV-1 LTR from −153 to +80, provided by P. Luciw, University of California, Davis) was used in a pulse-chase transcription experiment (Fig. 2). Reactions [12 μl total; 20 mm HEPES at pH 7.6, 7 mm MgCl2, 20 μg/ml of DNA template, 3 μl of HNE, 64 mm KCl, with 50 μm DRB or 0.3 μl of Drosophila P-TEFb (Marshall et al. 1996), as indicated] were preincubated for 20 min at 30°C, pulsed for 2 min by adding ATP, GTP, and UTP to 600 μm each and 5 μCi [α-32P]CTP (∼0.1 μm), and chased for 5 min by adding CTP to 1.2 mm. Reactions were stopped, phenol extracted, and analyzed on a 6% polyacrylamide gel as described (Marshall and Price 1995). pLTR–TAR–Luc and pLTR–ΔTAR–Luc plasmids were digested with EcoRI to generate TAR and ΔTAR template that were used in the experiments shown in Figure 7. The TAR template contains HIV-1 LTR from −475 to +76, whereas the ΔTAR template contains HIV-1 LTR from −475 to +19. Transcription with TAR and ΔTAR templates will generate runoffs of 694 and 646 nucleotides, respectively. In Figure 7A, the transcription reaction mixture contained 20 mm HEPES (pH 7.6), 7 mm MgCl2, 60 mm KCl, 0.5 μl of HNE (or depHNE as indicated), 20 μg/ml of TAR template (or ΔTAR template as indicated), an indicated amount of HIV-1 Tat (86-residue HIV-1 Tat followed by a streptavidin binding tag at its carboxyl terminus, kindly provided by O. Flores, Tularik, Inc., South San Francisco, CA), and DRB. The reaction mix was preincubated for 15 min at 30°C, and the transcription was started by adding nucleotides to final concentrations of 50 μm ATP, 50 μm GTP, 50 μm UTP, 10 μmCTP, and 5 μCi [α-32P]CTP (∼0.1 μm) and continuously labeling for 20 min (Fig. 7A), or by adding nucleotides to final concentrations of 50 μmM ATP, 50 μm GTP, 50 μm UTP, and 5 μCi [α-32P]CTP (∼0.1 μm), and pulsed for 2 min (Fig. 7B). The reactions were stopped, and the reaction mixtures were phenol-extracted. The transcripts were then analyzed on a 6% polyacrylamide gel as described (Marshall and Price 1995).
Acknowledgments
We thank Ken Williams at the Keck Foundation Biotechnology Resource Laboratory at Yale University for providing an excellent protein sequencing service, Osvaldo Flores for providing Tat protein, and Ofer Gileadi for providing the CTD3 peptide. We also thank Izhak Haviv and Yosef Shaul for fruitful discussions and help with purification, and Pam Geyer for critical reading of the manuscript. This work was supported by National Institutes of Health grants GM35500, A131802, and TW00506.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL david-price@uiowa.edu; FAX (319) 335-9570.
References
- Aso T, Lane WS, Conaway JW, Conaway RC. Elongin (SIII): A multisubunit regulator of elongation by RNA polymerase II. Science. 1995;269:1439–1443. doi: 10.1126/science.7660129. [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current protocols in molecular biology. New York, NY: Greene Publishing Associates/Wiley-Intersience; 1989. [Google Scholar]
- Bentley DL. Regulation of transcriptional elongation by RNA polymerase II. Curr Opin Genet Dev. 1995;5:210–216. doi: 10.1016/0959-437x(95)80010-7. [DOI] [PubMed] [Google Scholar]
- Blau J, Xiao H, McCracken S, O’Hare P, Greenblatt J, Bentley D. Three functional classes of transcriptional activation domains. Mol Cell Biol. 1996;16:2044–2055. doi: 10.1128/mcb.16.5.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braddock M, Thorburn AM, Kingsman AJ, Kingsman SM. Blocking of Tat-dependent HIV-1 RNA modification by an inhibitor of RNA polymerase II processivity. Nature. 1991;350:439–441. doi: 10.1038/350439a0. [DOI] [PubMed] [Google Scholar]
- Carroll R, Peterlin BM, Derse D. Inhibition of human immunodeficiency virus type 1 Tat activity by coexpression of heterologous trans activators. J Virol. 1992;66:2000–2007. doi: 10.1128/jvi.66.4.2000-2007.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun RF, Jeang KT. Requirements for RNA polymerase II carboxyl-terminal domain for transcription of human retroviruses, human T-cell lymphotrophic virus, and HIV-1. J Biol Chem. 1996;271:27888–27894. doi: 10.1074/jbc.271.44.27888. [DOI] [PubMed] [Google Scholar]
- Cujec TP, Cho H, Maldonado E, Meyer J, Reinberg D, Peterlin BM. The human immunodeficiency virus transactivator Tat interacts with the RNA polymerase II holoenzyme. Mol Cell Biol. 1997;17:1817–1823. doi: 10.1128/mcb.17.4.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen BR. Does HIV-1 Tat induce a change in viral initiation rights? Cell. 1993;73:417–420. doi: 10.1016/0092-8674(93)90126-b. [DOI] [PubMed] [Google Scholar]
- Dahmus ME. Reversible phosphorylation of the c-terminal domain of RNA polymerase II. J Biol Chem. 1996;271:19009–19012. doi: 10.1074/jbc.271.32.19009. [DOI] [PubMed] [Google Scholar]
- Egyhazi E, Ossoinak A, Pigon A, Holmgren C, Lee JM, Greenleaf AL. Phosphorylation dependence of the initiation of productive transcription of Balbiani ring 2 genes in living cells. Chromosoma. 1996;104:422–433. doi: 10.1007/BF00352266. [DOI] [PubMed] [Google Scholar]
- Feinberg MB, Baltimore D, Frankel AD. The role of Tat in the human immunodeficiency virus life cycle indicates a primary effect on transcriptional elongation. Proc Natl Acad Sci. 1991;88:4045–4049. doi: 10.1073/pnas.88.9.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Martinez LF, Ivanov D, Gaynor RB. Association of Tat with purified HIV-1 and HIV-2 transcription preinitiation complexes. J Biol Chem. 1997a;272:6951–6958. doi: 10.1074/jbc.272.11.6951. [DOI] [PubMed] [Google Scholar]
- Garcia-Martinez LF, Mavankal G, Neveu JM, Lane WS, Ivanov D, Gaynor RB. Purification of a Tat-associated kinase reveals a TFIIH complex that modulates HIV-1 transcription. EMBO J. 1997b;16:2836–2850. doi: 10.1093/emboj/16.10.2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garriga J, Mayol X, Grana X. The CDC2-related kinase PITALRE is the catalytic subunit of active multimeric protein complexes. Biochem J. 1996a;319:293–298. doi: 10.1042/bj3190293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garriga J, Segura E, Mayol X, Grubmeyer C, Grana X. Phosphorylation site specificity of the CDC2-related kinase PITALRE. Biochem J. 1996b;320:983–989. doi: 10.1042/bj3200983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich JA, Tjian R. Transcription factors IIE and IIH and ATP hydrolysis direct promoter clearance by RNA polymerase II. Cell. 1994;77:145–156. doi: 10.1016/0092-8674(94)90242-9. [DOI] [PubMed] [Google Scholar]
- Grana X, De Luca A, Sang N, Fu Y, Claudio PP, Rosenblatt J, Morgan DO, Giordano A. PITALRE, a nuclear CDC2-related protein kinase that phosphorylates the retinoblastoma protein in vitro. Proc Natl Acad Sci. 1994;91:3834–3838. doi: 10.1073/pnas.91.9.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Price DH. Mechanism of DmS-II-mediated pause suppression by Drosophila RNA polymerase II. J Biol Chem. 1993;268:18762–18770. [PubMed] [Google Scholar]
- Herrmann CH, Rice AP. Specific interaction of the human immunodeficiency virus Tat proteins with a cellular protein kinase. Virology. 1993;197:601–608. doi: 10.1006/viro.1993.1634. [DOI] [PubMed] [Google Scholar]
- ————— Lentivirus Tat proteins specifically associate with a cellular protein kinase, TAK, that hyperphosphorylates the carboxyl-terminal domain of the large subunit of RNA polymerase II: Candidate for a Tat cofactor. J Virol. 1995;69:1612–1620. doi: 10.1128/jvi.69.3.1612-1620.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irie K, Nomoto S, Miyajima I, Matsumoto K. SGV1 encodes a CDC28/cdc2-related kinase required for a G alpha subunit-mediated adaptive response to pheromone in S. cerevisiae. Cell. 1991;65:785–795. doi: 10.1016/0092-8674(91)90386-d. [DOI] [PubMed] [Google Scholar]
- Jeang K-T, Chun R, Lin NH, Gatignol A, Glabe CG, Fan H. In vitro and in vivo binding of human immunodeficiency virus type 1 Tat protein and Sp1 transcription factor. J Virol. 1993;67:6224–6233. doi: 10.1128/jvi.67.10.6224-6233.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KA, Peterlin BM. Control of RNA initiation and elongation at the HIV-1 promoter. Annu Rev Biochem. 1994;63:717–743. doi: 10.1146/annurev.bi.63.070194.003441. [DOI] [PubMed] [Google Scholar]
- Kao S-Y, Calman AF, Luciw PA, Peterlin EM. Antitermination of transcription within the long terminal repeat of HIV by the tat gene product. Nature. 1987;330:489–493. doi: 10.1038/330489a0. [DOI] [PubMed] [Google Scholar]
- Kashanchi F, Piras G, Radonovich MF, Duvall JF, Fattaey A, Chiang CM, Roeder RG, Brady JN. Direct interaction of human TFIID with the HIV-1 transactivator tat. Nature. 1994;367:295–299. doi: 10.1038/367295a0. [DOI] [PubMed] [Google Scholar]
- Keen NJ, Gait MJ, Karn J. Human immunodeficiency virus type-1 Tat is an integral component of the activated transcription-elongation complex. Proc Natl Acad Sci. 1996;93:2505–2510. doi: 10.1073/pnas.93.6.2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kephart DD, Wang BQ, Burton ZF, Price DH. Functional analysis of Drosophila factor 5 (TFIIF), a general transcription factor. J Biol Chem. 1994;269:13536–13543. [PubMed] [Google Scholar]
- Kerppola TK, Kane CM. RNA polymerase: Regulation of transcript elongation and termination. FASEB J. 1991;5:2833–2842. doi: 10.1096/fasebj.5.13.1916107. [DOI] [PubMed] [Google Scholar]
- Laspia MF, Rice AP, Mathews MB. HIV-1 Tat protein increases transcriptional initiation and stabilizes elongation. Cell. 1989;59:283–292. doi: 10.1016/0092-8674(89)90290-0. [DOI] [PubMed] [Google Scholar]
- Laspia MF, Rice AP, Mathews MB. Synergy between HIV-1 Tat and adenovirus E1A is principally due to stabilization of transcriptional elongation. Genes & Dev. 1990;4:2397–2408. doi: 10.1101/gad.4.12b.2397. [DOI] [PubMed] [Google Scholar]
- Laspia MF, Wendel P, Mathews MB. HIV-1 Tat overcomes inefficient transcriptional elongation in vitro. J Mol Biol. 1993;232:732–746. doi: 10.1006/jmbi.1993.1427. [DOI] [PubMed] [Google Scholar]
- Lee JM, Greenleaf AL. Modulation of RNA polymerase II elongation efficiency by c-terminal heptapeptide repeat domain kinase I. J Biol Chem. 1997;272:10990–10993. doi: 10.1074/jbc.272.17.10990. [DOI] [PubMed] [Google Scholar]
- Madore SJ, Cullen BR. Genetic analysis of the cofactor requirement for human immunodeficiency virus type 1 Tat function. J Virol. 1993;67:3703–3711. doi: 10.1128/jvi.67.7.3703-3711.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marciniak RA, Sharp PA. HIV-1 Tat protein promotes formation of more-processive elongation complexes. EMBO J. 1991;10:4189–4196. doi: 10.1002/j.1460-2075.1991.tb04997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall NF, Price DH. Control of formation of two distinct classes of RNA polymerase II elongation complexes. Mol Cell Biol. 1992;12:2078–2090. doi: 10.1128/mcb.12.5.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J Biol Chem. 1995;270:12335–12338. doi: 10.1074/jbc.270.21.12335. [DOI] [PubMed] [Google Scholar]
- Marshall NF, Peng JM, Xie Z, Price DH. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J Biol Chem. 1996;271:27176–27183. doi: 10.1074/jbc.271.43.27176. [DOI] [PubMed] [Google Scholar]
- Mavankal G, Ou SHI, Oliver H, Sigman D, Gaynor RB. Transcription factors, and gene expression. Human immunodeficiency virus type 1 and 2 Tat proteins specifically interact with RNA polymerase II. Proc Natl Acad Sci. 1996;93:2089–2094. doi: 10.1073/pnas.93.5.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien T, Hardin S, Greenleaf A, Lis JT. Phosphorylation of RNA polymerase II C-terminal domain and transcriptional elongation. Nature. 1994;370:75–77. doi: 10.1038/370075a0. [DOI] [PubMed] [Google Scholar]
- Parada CA, Roeder RG. Enhanced processivity of RNA polymerase II triggered by Tat-induced phosphorylation of its carboxy-terminal domain. Nature. 1996;384:375–378. doi: 10.1038/384375a0. [DOI] [PubMed] [Google Scholar]
- Pendergrast PS, Hernandez N. RNA-targeted activators, but not DNA-targeted activators, repress the synthesis of short transcripts at the human immunodeficiency virus type 1 long terminal repeat. J Virol. 1997;71:910–917. doi: 10.1128/jvi.71.2.910-917.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price DH, Sluder AE, Greenleaf AL. Fractionation of transcription factors for RNA polymerase II from Drosophila Kc cell nuclear extracts. J Biol Chem. 1987;262:3244–3255. [PubMed] [Google Scholar]
- Reines D, Conaway JW, Conaway RC. The RNA polymerase II general elongation factors. Trends Biochem Sci. 1996;21:351–355. [PMC free article] [PubMed] [Google Scholar]
- Rice AP, Carlotti F. Structural analysis of wild-type and mutant human immunodeficiency virus type 1 Tat proteins. J Virol. 1990;64:6018–6026. doi: 10.1128/jvi.64.12.6018-6026.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon M, Ratnasabapathy R, Hernandez N. Characterization of the inducer of short transcripts, a human immunodeficiency virus type 1 transcriptional element that activates the synthesis of short RNAs. Mol Cell Biol. 1993;13:1251–1263. doi: 10.1128/mcb.13.2.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A, Lane WS, Jackson KW, Conaway RC, Conaway JW. An RNA polymerase II elongation factor encoded by the human ELL gene. Science. 1996;271:1873–1876. doi: 10.1126/science.271.5257.1873. [DOI] [PubMed] [Google Scholar]
- Spencer CA, Groudine M. Transcription elongation and eukaryotic gene regulation. Oncogene. 1990;5:777–786. [PubMed] [Google Scholar]
- Sterner DE, Lee JM, Hardin SE, Greenleaf AL. The yeast carboxyl-terminal repeat domain kinase CTDK-I is a divergent cyclin-cyclin-dependent kinase complex. Mol Cell Biol. 1995;15:5716–5724. doi: 10.1128/mcb.15.10.5716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamm I, Kikuchi T. Early termination of heterogeneous nuclear RNA transcripts in mammalian cells: accentuation by 5,6-dichloro 1-beta-D-ribofuranosylbenzimidazole. Proc Natl Acad Sci. 1979;76:5750–5754. doi: 10.1073/pnas.76.11.5750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya H, Iseda T, Hino O. Identification of a novel protein (VBP-1) binding to the vonHippel-Lindau (VHL) tumor suppressor gene product. Cancer Res. 1996;56:2881–2885. [PubMed] [Google Scholar]
- Veschambre P, Simard P, Jalinot P. Evidence for functional interaction between the HIV-1 Tat transactivator and the TATA box binding protein in vivo. J Mol Biol. 1995;250:169–180. doi: 10.1006/jmbi.1995.0368. [DOI] [PubMed] [Google Scholar]
- Wright S. Regulation of eukaryotic gene expression by transcriptional attenuation. Mol Biol Cell. 1993;4:661–668. doi: 10.1091/mbc.4.7.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Price DH. Purification of an RNA polymerase II transcript release factor from Drosophila. J Biol Chem. 1996;271:11043–11046. doi: 10.1074/jbc.271.19.11043. [DOI] [PubMed] [Google Scholar]
- Yang XZ, Herrmann CH, Rice AP. The human immunodeficiency virus tat proteins specifically associate with TAK in vivo and require the carboxy-terminal domain of RNA polymerase II for function. J Virol. 1996;70:4576–4584. doi: 10.1128/jvi.70.7.4576-4584.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yankulov K, Yamashita K, Roy R, Egly JM, Bentley DL. The transcriptional elongation inhibitor 5,6-dichloro-1-β-D-ribofuranosylbenzimidazole inhibits transcription factor IIH-associated protein kinase. J Biol Chem. 1995;270:23922–23925. doi: 10.1074/jbc.270.41.23922. [DOI] [PubMed] [Google Scholar]
- Yankulov KY, Pandes M, McCracken S, Bouchard D, Bentley DL. TFIIH functions in regulating transcriptional elongation by RNA polymerase II in Xenopus oocytes. Mol Cell Biol. 1996;16:3291–3299. doi: 10.1128/mcb.16.7.3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou QA, Sharp PA. TAT-SF1 - cofactor for stimulation of transcriptional elongation by HIV-1 Tat. Science. 1996;274:605–610. doi: 10.1126/science.274.5287.605. [DOI] [PubMed] [Google Scholar]