Abstract
Background
A synergistic effect resulting from a combination of BCL2 and MYC or MYC and CCND1 has been implicated in human B-cell lymphomas. Although the identification of other cooperative genes involved is important, our present understanding of such genes remains scant. The objective of this study was to identify the additional cooperative gene(s) associated with BCL2 and MYC or MYC and CCND1. First, we assessed whether Bcl2, Myc and Ccnd1 could cooperate. Next, we developed a synergism-based functional screening method for the identification of other oncogene(s) that act with Bcl2 and Myc.
Design and Methods
Growth in culture, colony formation and oncogenicity in vivo were assessed in mouse primary B cells exogenously expressing various combinations of Bcl2, Myc and Ccnd1. For the functional screening, Bcl2- and Myc-expressing primary B cells were infected with a retroviral cDNA library. Inserted cDNA of transformed cells in culture were then identified.
Results
Primary B cells exogenously expressing Bcl2, Myc and Ccnd1 showed factor-independent growth ability, enhanced colony-forming capability and aggressive oncogenicity, unlike the cases observed with the expression of any combination of only two of the genes. We identified CCND3 and NRAS as cooperative genes with Bcl2 and Myc through the functional screening.
Conclusions
Bcl2, Myc and Ccnd1 or Bcl2, Myc and CCND3 synergistically transformed mouse primary B cells into aggressive malignant cells. Our new synergism-based method is useful for the identification of synergistic gene combinations in tumor development, and may expand our systemic understanding of a wide range of cancer-causing elements.
Keywords: cooperative genes, malignant transformation, oncogenes, Bcl2, Myc, Ccnd1
Introduction
Intense efforts to examine genetic alterations in human cancer have provided a catalogue of cancer-causing genes. It is speculated that most cancers arise as a result of accumulation of altered cancer-causing genes. The study of functional synergism among cancer-causing genes is thus becoming one of the central issues in cancer research.1 However, it is difficult to determine which combinations of genes are involved in oncogenesis because of the large number of possible candidates. This accounts for the fact that our knowledge about synergistic combinations of cancer-causing genes has remained limited.
Genetic alterations of BCL2 (B-cell CLL/lymphoma 2), MYC [v-myc myelocytomatosis viral oncogene homolog (avian)] and CCND1 (cyclin D1) are the most frequently found alterations in human B-cell lymphomas. These genes are transcriptionally deregulated as the partner genes of IgH translocation, and are thus thought to perform crucial roles in human B-cell lymphomagenesis.2 A synergistic effect resulting from a combination of two genes from BCL2, MYC and CCND1 has been implicated in human B-cell lymphomagenesis. About 4% of cases of diffuse large B-cell cell lymphoma possess BCL2/MYC double translocations, suggesting a synergistic effect of BCL2 and MYC in lymphoma development.3–7 The synergistic effect of BCL2 and MYC has also been implicated in the histological and clinical transformation of indolent follicular lymphoma into a more aggressive lymphoma.8–12 Cases of CCND1/MYC double translocation are relatively frequent in mantle cell lymphoma.7 Importantly, it is believed that other hitherto unknown genes also play important roles in lymphomagenesis in addition to the synergistic effects of the aforementioned two oncogenes since human B-cell lymphomas often show a variety of genes subject to alterations and/or deregulated expression.7 That multiple genes are involved in human lymphoma formation has also been suggested by experimental mouse models. Cory et al. pointed out that additional genetic alterations were involved in lymphoma development in an Emu mouse model ectopically expressing Myc and Bcl2.13 The identification and clarification of the multiple cooperative genes implicated in human lymphoma formation is important, although our present understanding of such genes remains scant.
The objective of this study was to identify additional cooperative gene(s) associated with BCL2 and MYC or MYC and CCND1 in human B-cell lymphomagenesis. Importantly, human B-cell lymphoma cases with concurrent multiple translocations including BCL2, MYC and CCND1 have been reported.14,15 Given the different biological functions of BCL2, MYC and CCND1,16–20 we investigated the possibility that these three genes might act synergistically. In vitro assays demonstrated that ectopic expression of all three genes could transform mouse primary B-cells, unlike the cases observed following the expression of any combination of only two of the three genes. We also determined that this synergistic effect contributed to lethal tumor development in mice. Furthermore, we used these findings to develop a new functional screening method, with which we were able to identify other transforming gene combinations.
Design and Methods
Generation of retrovirus
Retroviral vector plasmids were transiently co-transfected with MCV-Ecopac vector21 (kindly provided by Dr. Richard Van Etten, Tufts-NEMC Cancer Center, Boston, MA, USA) into 293 T cells using the calcium phosphate precipitation method (Profection mammalian transfection system; Promega) or the FuGENE6 transfection reagent (Roche) according to the manufacturers’ instructions. Twenty-four hours following transfection, the culture medium was replaced with Feeder medium [Iscove’s modified Dulbecco’s medium (IMDM) supplemented with 2% fetal calf serum (FCS) containing 2- mercaptoethanol (5×10−5 M; Sigma, St. Louis, MO, USA) and Primatone RL (0.03% wt/vol; Cellgro) with interleukin-7 [IL-7; 5% of mouse IL-7-producing cell line J558 supernatant (kindly provided by Dr. Tariq Enver, University of Oxford, Oxford, UK)]. Cells were incubated at 32°C for 24 h before harvesting of the virus supernatants. The virus supernatants were filtered (0.45 μM) and then frozen at −80°C.
Preparation and retroviral infection of cells
On day 15 or 16 of gestation, fetal liver cells were enriched to produce pro B-cells by cell sorting for B220 and c-Kit expression, and cultured at 5×103 /mL in Feeder medium containing IL-7 (5%) on 15 Gy-irradiated ST-2 stromal cells. Following pro B-cell growth for 3–4 days, cells were used for retroviral infection. Cells (1.25 ×105) were plated onto ST-2 cells in a 6-well culture plate (Costar). After 24 h, cells were suspended in virus supernatant and spin infected at 2000 rpm (840g, 1.5 h) at room temperature. For drug selection, cells were serially infected with various retroviral vectors and then purified by serial selection using neomycin (1.2–2 mg/mL), hygromycin (0.3–1.5 mg/mL) and blasticidin (15–20 μg/mL). For cell sorting, cells were simultaneously infected with various retroviral vectors and sorted for green fluorescent protein (GFP) and huKO expression after 2 or 3 days. In Screening 2, cells were infected with MSCV-Bcl2-pgk-Myc-ires-GFP and sorted for GFP expression. The combination of retroviral vectors used for establishing the various stable integrants are detailed in Online Supplementary Figure S1.
In vitro liquid assays
Prior to commencement of the in vitro liquid assay without IL-7 and ST-2 cells, cell samples were washed twice in Feeder medium. Cells (1×105 or 5×105) were seeded into wells of a 24-well plate on day 0.
In vitro colony-forming assays
Cells were plated in triplicate onto 35-mm dishes in 1 mL of semisolid, 1% methylcellulose-based medium supplemented with FCS (30%), mouse stem cell factor (SCF, 5% of mouse SCF producing a cell line supernatant), mouse IL-7 (5%), and mouse FLT3 (FMS-like tyrosine kinase 3) ligand (10 ng/mL; 250–31L, Peprotech)
In vivo transplantation assays
Cells (1×107) were transplanted intravenously into SCID mice. Cell suspensions of spleen or lymph nodes were obtained by pressing the tissue gently between two glass microscope slides and then treating the sample with ACK (0.15 M NH4Cl, 1.0 mM KHCO3 and 0.1 mM EDTA) to lyse the red blood cells. Tissues were fixed in phosphate-buffered saline containing 10% formaldehyde, embedded in paraffin, and then stained with hematoxylin and eosin. Experiments were performed with the approval of the Institutional Ethical Committee for Animal Experiments of Aichi Cancer Center Research Institute.
Construction of the retroviral cDNA expression library and screening
Construction of the retroviral cDNA expression library was performed as previously described.22 In brief, a double-stranded cDNA longer than 1.0 kb obtained from the SU-DHL-6 cell line was ligated into a BstXI digested pMXs vector. These procedures allowed the use of an endogenous stop codon of each gene in this expression library system. Escherichia coli DH10B was transformed by electroporation, resulting in the production of about 1.0×106 clones. To obtain the results from functional screenings, the inserted cDNA were recovered by PCR-amplification using the retroviral-specific primers (S: 5′-GGTGGACCATCCTCTAGACT-3′, AS: 5′-CCCTTTTTCTGGAGACTAAAT-3′) from the retrovirus-integrated genomic DNA of proliferating cells. Anti-sense oriented cDNA clones were excluded from analysis in this study.
Statistical analysis
Mean values ± s.e.m. were used throughout. Data were analyzed using the Student’s t-test and Kaplan–Meier survival analysis was conducted using Statview software (SAS Institute).
Further details on the construction of vectors, flow cytometry, Southern blot analysis and analysis of expression microarray data are provided in the Online Supplementary Design and Methods.
Results
In vitro growth of primary B cells with deregulated expression of IgH translocation-associated oncogenes
A defining characteristic of the transformed cell in vitro is the lack of dependency on exogenous proliferating and/or survival signals. As BALB/c fetal liver-derived pro B-cells could be cultured for over 6 months in the presence of IL-7 on ST-2 stromal cells,23,24 our initial efforts focused on identifying oncogenes that impart B cells independence from IL-7 and ST-2 cells. We infected primary B cells with retrovirus containing one oncogene (Bcl2, Myc or Ccnd1) and a drug selection marker. Drug-selected cells with a single gene could not proliferate in the absence of IL-7 and ST-2 cells (data not shown). All combinations comprising two genes from Bcl2, Myc and Ccnd1 were also unable to transform primary B cells (Figure 1A, Experiment (EXP) 1, see also Online Supplementary Figure S1 for vector constructions and stable integrants). In marked contrast, primary B cells stably expressing Bcl2, Myc and Ccnd1 (Online Supplementary Figure S2A) showed stable growth in the absence of IL-7 and ST-2 cells (Figure 1A, EXP 1). These findings were reproducible (Figure 1A, EXP 2).
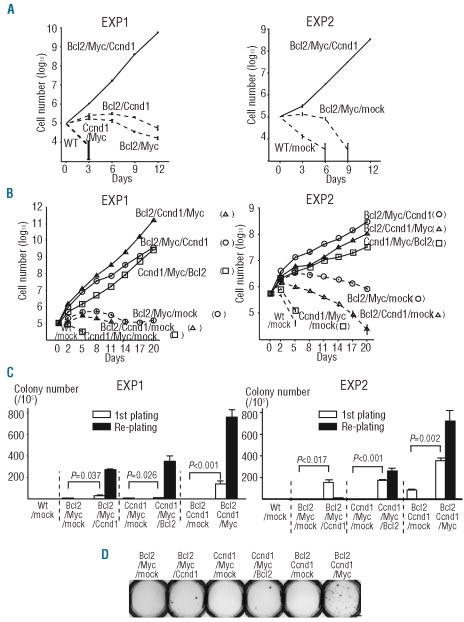
Figure 1.
Transformation of primary mouse B cells stably expressing exogenous Bcl2, Myc and Ccnd1 in vitro. (A and B) In vitro growth curves of various stable integrants established by the drug selection (A) or cell sorting (B) methods in the absence of IL-7 and ST-2 cells. Right and left panels show the results of Experiment 1 (EXP 1) and Experiment 2 (EXP 2), respectively. Data are the mean ± s.e.m. (triplicate). (C and D) Methylcellulose colony-forming abilities of various stable integrants established by the cell sorting method (C) and photograph of representative colonies in the first plating (D). Right and left panels in (C) show the results of EXP 1 and EXP 2, respectively. Data are the mean±s.e.m. (triplicate). P values are two-sided (Student’s test). Scale bar represent 10 mm.
To confirm these initial results, primary B cells stably expressing exogenous Bcl2, Myc and Ccnd1 were established with a non-drug selection method. We simultaneously infected primary B cells with two vectors. One vector co-expressed Bcl2, Myc and GFP (MSCV-Bcl2-pgk-Myc-ires-GFP)25 and the other vector co-expressed Ccnd1 and humanized Kusabira-Orange (huKO)26 (pGCDNsam-Ccnd1-ires-huKO) (Online Supplementary Figure S1). GFP and huKO double-positive cells were purified by cell sorting and designated as “Bcl2/Myc/Ccnd1”. Ectopic expression of BCL2, MYC and CCND1 proteins in Bcl2/Myc/Ccnd1 cells was confirmed by western blot analysis (Online Supplementary Figure S3). This procedure allows for faster purification of the stable integrant compared to the drug selection method, and hence minimizes uncontrollable factors that may have additive effects on cellular transformation. Bcl2/Myc/Ccnd1 cells could grow in the absence of IL-7 and ST-2 cells, unlike Bcl2/Myc/mock cells (Figure 1B, EXP 1, see also Online Supplementary Figure S1 for vector constructions and stable integrants). Other stable integrants stably expressing all three genes (“Ccnd1/Myc/Bcl2” and “Bcl2/Ccnd1/Myc”) were also established (see Online Supplementary Figure S1 for vector constructions and stable integrants and Online Supplementary Figure S3 for protein expression). As expected, both Ccnd1/Myc/Bcl2 and Bcl2/Ccnd1/Myc cells could grow in the absence of IL-7 and ST-2 cells, unlike Ccnd1/Myc/mock and Bcl2/Ccnd1/mock cells (Figure 1B, EXP 1). Reproducibility was demonstrated by an independent experiment (Figure 1B, EXP 2). These results confirmed that ectopic expression of Bcl2, Myc and Ccnd1 could synergistically transform primary B cells to gain independence from IL-7 and ST-2 cells.
Enhanced colony-forming efficiency of primary B cells with deregulated expression of Bcl2, Myc and Ccnd1 in a methylcellulose matrix
The ability to form colonies in semisolid media is another characteristic feature of transformed cells. As drug-selected stable integrants expressing the three genes formed more colonies than those expressing any combination comprising only two of the genes (Online Supplementary Figures S2B,C and S4), we confirmed these results using sorted stable integrants. In the first plating, Bcl2/Myc/Ccnd1, Ccnd1/Myc/Bcl2 and Bcl2/Ccnd1/Myc cells showed statistically significant more efficient colony formation than Bcl2/Myc/mock, Ccnd1/Myc/mock and Bcl2/Ccnd1/mock, respectively (Figure 1C, EXP 1, and D). In the re-plating, cells expressing any combination of the two genes did not show any colonies, while Bcl2/Myc/Ccnd1, Ccnd1/Myc/Bcl2 and Bcl2/Ccnd1/Myc cells formed a substantial number of colonies (Figure 1C, EXP 1). Similar results were obtained using an independent experiment (Figure 1C, EXP 2). These results indicate that the ectopic expression of Bcl2, Myc and Ccnd1 synergistically transforms primary B cells so that they acquire efficient colony-forming capability in a methylcellulose matrix. However, the colony-forming efficiency of the stable integrants with Bcl2, Myc and Ccnd1 was much lower than that of the human B-cell lymphoma cell lines SU-DHL-6 and Raji (Bcl2/Myc/Ccnd1 25/105 (0.025%); Ccnd1/Myc/Bcl2 5/105 (0.005%); Bcl2/Ccnd1/Myc 141/105 (0.141%); SU-DHL-6 21/103 (2.1%); Raji 864/103 (86.4%) in the first plating).
The contribution of deregulated expression of Bcl2, Myc and Ccnd1 in converting primary B cells into highly aggressive malignant cells
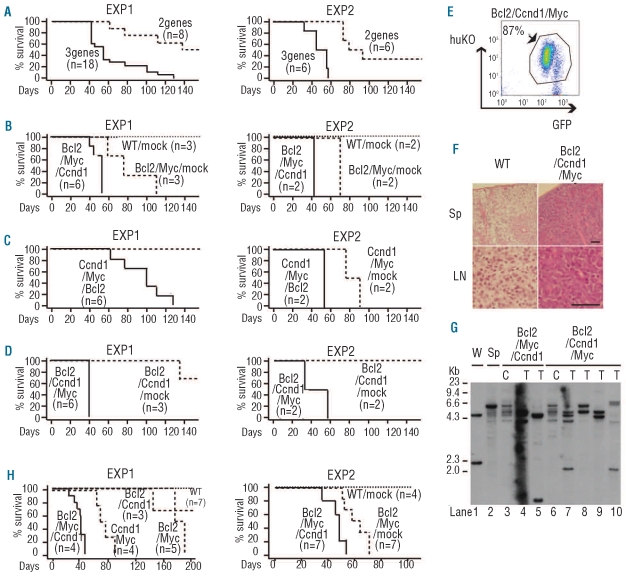
In an effort to examine whether the synergistic effect of Bcl2, Myc and Ccnd1 could contribute to tumor development, we transplanted 107 cells of the sorted stable integrants into non-irradiated SCID mice. SCID mice transplanted with Bcl2/Myc/Ccnd1, Ccnd1/Myc/Bcl2 or Bcl2/Ccnd1/Myc cells developed fatal leukemia/lymphoma more rapidly than mice transplanted with cells containing only two of the oncogenes [Figure 2A, EXP 1, average survival time: 63.5 days for the cells expressing three genes (Bcl2/Myc/Ccnd1, n=6; Ccnd1/Myc/Bcl2, n=6; and Bcl2/Ccnd1/Myc, n=6) versus 163.4 days for cells expressing two genes (Bcl2/Myc/mock, n=3; Ccnd1/Myc/mock, n=2 and Bcl2/Ccnd1/mock, n=3), P=0.0003; Figure 2B, EXP 1, average survival time: 51.3 days for Bcl2/Myc/Ccnd1 versus 83.7 days for Bcl2/Myc/mock, P=0.0128; Figure 2C, EXP 1, 97.0 days for Ccnd1/Myc/Bcl2 versus 243.5 days for Ccnd1/Myc/mock, P=0.0319; Figure 2D, EXP 1, 42.2 days for Bcl2/Ccnd1/Myc versus 189.7 days for Bcl2/Ccnd1/mock, P=0.0051]. Dissections of two mice with Bcl2/Myc/Ccnd1 and four mice with Bcl2/Ccnd1/Myc showed that GFP+huKO+ tumor cells had mainly infiltrated the enlarged lymph nodes, spleen and thymus (Figure 2E,F), and bone marrow was also saturated with tumor cells. Fluorescence-activated cell sorter (FACS) analysis of the surface markers of the malignant cells infiltrating lymph nodes indicated that they had an immature B-cell phenotype (pre B; c-Kit−, B220+, CD19+, IgM−). Southern blot analysis of genomic DNA extracted from enlarged lymph nodes showed that all cases comprised one to several clonal IgH rearrangement(s) and that the patterns of IgH rearrangements among these cases differed (Figure 2G, lanes 4,5,7–10). Reproducibility was determined using an independent experiment (Figure 2A-D, EXP 2). Consistent results were also obtained when drug-selected stable integrants were transplanted into SCID mice (Figure 2H). All of these findings obtained with our in vivo mouse model clearly indicate that ectopic expression of Bcl2, Myc and Ccnd1 can transform primary B cells so that they develop a highly aggressive malignant potential.
Figure 2.
Enhanced tumorigenic properties of primary mouse B-cells stably expressing exogenous Bcl2, Myc and Ccnd1. Stable integrants established by the cell sorting method were used. (A-D) Kaplan-Meier survival curves of SCID mice transplanted with various stable integrants established by the cell sorting method. Right and left panels show the results of EXP 1 and EXP 2, respectively. “3 genes” (n=18) in EXP 1 of (A) indicates Bcl2/Myc/Ccnd1 (n=6), Ccnd1/Myc/Bcl2 (n=6) and Bcl2/Ccnd1/Myc (n=6). “2 genes” (n=8) in EXP 1 of (A) indicates Bcl2/Myc/mock (n=3), Ccnd1/Myc/mock (n=2) and Bcl2/Ccnd1/mock (n=3). “3 genes” (n=6) in EXP 2 of (A) indicates Bcl2/Myc/Ccnd1 (n=2), Ccnd1/Myc/Bcl2 (n=2) and Bcl2/Ccnd1/Myc (n=2). “2 genes” (n=6) in EXP 2 of (A) indicates Bcl2/Myc/mock (n=2), Ccnd1/Myc/mock (n=2) and Bcl2/Ccnd1/mock (n=2). In EXP 2 of (A), three-gene-expressing cells versus two-gene-expressing cells, P=0.0006. In EXP 2 of (B), Bcl2/Myc/Ccnd1 versus Bcl2/Myc/mock, P=0.0833. In EXP 2 of (C), Ccnd1/Myc/Bcl2 versus Ccnd1/Myc/mock, P=0.0833. In EXP 2 of (D), Bcl2/Ccnd1/Myc versus Bcl2/Ccnd1/mock, P=0.0896. (E) The GFP and huKO expression pattern of cells infiltrating a lymph node of a representative Bcl2/Ccnd1/Myc mouse. (F) Hematoxylin/eosin staining of spleen (Sp) and lymph node (LN) in wild-type (WT) and representative Bcl2/Ccnd1/Myc mice. Scale bar represents 50 μm. (G) Southern blot analysis of IgH gene rearrangements in the stable integrants established by the cell sorting method. Genomic DNA from WEHI231 (murine B-cell lymphoma cell line) (W) and spleen cells (Sp) of wild-type BALB/c mouse were used as positive controls. ”C” in lanes 3 and 6 or ”T” in lanes 4,5,7–10 denotes genomic DNA extracted from in vitro cultured cells or tumor cells of neoplastic lymph nodes of SCID mice transplanted with stable integrants. (H) Kaplan-Meier survival curves of SCID mice transplanted with various stable integrants established by the drug selection method. Right and left panels show the results of EXP 1 and EXP 2, respectively. In EXP 1, Bcl2/Myc/Ccnd1 cells killed SCID mice more rapidly compared to Bcl2/Myc or Ccnd1/Myc cells (P=0.0013, log-rank test), or Bcl2/Ccnd1 cells. In EXP 2, Bcl2/Myc/Ccnd1 cells killed SCID mice more rapidly compared to Bcl2/Myc/mock cells (P=0.0012, log-rank test).
Screening a retroviral cDNA expression library to identify functional genes cooperating with Bcl2 and Myc in human B-cell lymphoma
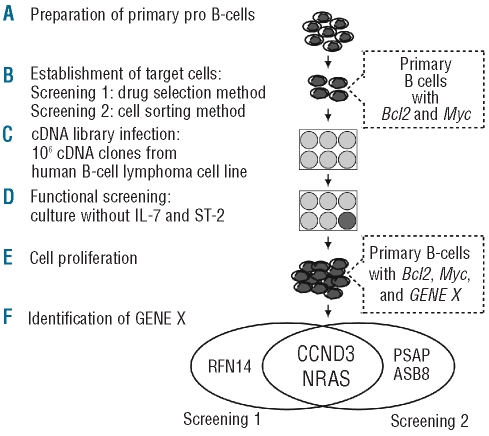
Having established that the synergistic effect of the IgH translocation-associated oncogenes Bcl2, Myc and Ccnd1 could transform mouse primary B cells both in vitro and in vivo, we then utilized this system in an effort to identify other oncogenes which might cooperate with Bcl2 and Myc in transforming mouse primary B cells in vitro. Bcl2-and Myc-expressing primary B cells newly established using the drug selection or cell sorting methods were used for Screening 1 or Screening 2, respectively (Figure 3A,B). Cells expressing Bcl2 and Myc were infected with a retro-viral library expressing cDNA from SU-DHL-6, a human B-cell lymphoma cell line with the BCL2/IgH translocation (Figure 3C), followed by initiation of functional screening, in which the infected cells were cultured in the absence of IL-7 and ST-2 cells (Figure 3D). Retroviral library-infected cells showed robust proliferation for about 10 days (Screening 1) and for 2 to 5 weeks (Screening 2) following library infection, unlike the mock-infected cells (Figure 3E). We recovered the inserted cDNA from retrovirus-integrated genomic DNA of the proliferating cells. This process resulted in the identification of CCND3, NRAS and RNF14 in Screening 1, and CCND3, NRAS, PSAP and ASB8 in Screening 2 (Figure 3F). Of special note is that CCND3 and NRAS were identified in both screenings. The recovered sequence of NRAS contained a missense mutation in codon 61 (Q61K), and we confirmed that SU-DHL-6 possessed the mutated NRAS allele.
Figure 3.
Diagram of the retroviral cDNA expression library screening protocol. (A) Mouse primary pro B-cells were purified from fetal liver of BALB/c mice. (B) Bcl2 and Myc-expressing target cells were established by the drug selection method for Screening 1 and the cell sorting method for Screening 2. (C) Target cells were infected with a retroviral expression library containing 106 cDNA from SU-DHL-6. (D) Functional screenings were performed under culture condition in the absence of IL-7 and ST-2 cells. (E) Proliferation of cells transduced with GENE X, which cooperates with Bcl2 and Myc. (F) Inserted cDNA were recovered by PCR-amplification using the retro-viral-specific primers from retrovirus-integrated genomic DNA of proliferating cells. CCND3 and NRAS were identified in both screenings.
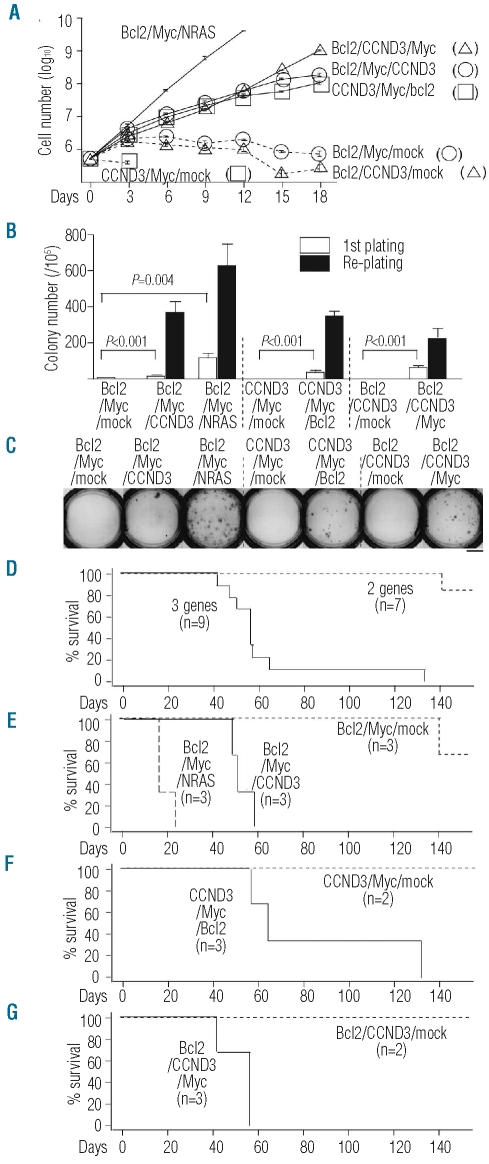
We also conducted another experiment to confirm these results. To this purpose, we established various stable integrants using the sorting method (see Online Supplementary Figure S1 for vector constructions and stable integrants). In the liquid culture assay without IL-7 and ST-2 cells, expression of CCND3 or NRAS (Q61K) led to the stable growth of primary B cells expressing combinations of Bcl2 and Myc (Figure 4A, Bcl2/Myc/CCND3 or Bcl2/Myc/NRAS), unlike the case with RNF14, PSAP or ASB8 (data not shown). Furthermore, cells expressing CCND3 and Myc (CCND3/Myc/mock) or Bcl2 and CCND3 (Bcl2/CCND3/mock) could not proliferate under the same culture conditions (Figure 4A). This led us to conclude that one of the genes (Bcl2, Myc or CCND3) is indispensable for the transformation of primary mouse B cells.
Figure 4.
Experiments confirming the synergistic effect of CCND3 or NRAS with Bcl2 and Myc. (A) In vitro growth curves of various stable integrants in the absence of IL-7 and ST-2 cells. Data are the mean ± s.e.m. (triplicate). (B) Methylcellulose colony-forming abilities of various stable integrants. Data are the mean ± s.e.m. (triplicate). P values are two-sided (Student’s t test). (C) Photograph of representative colonies of various integrants in the first plating. Scale bar represents 10 mm. (D-G) Kaplan-Meier survival curves of SCID mice transplanted with various stable integrants. “3 genes” (n=9) in (D) indicates Bcl2/Myc/CCND3 (n=3), CCND3/Myc/Bcl2 (n=3) and Bcl2/CCND3/Myc (n=3). “2 genes” (n=7) in (D) indicates Bcl2/Myc/mock (n=3), CCND3/Myc/mock (n=2) and Bcl2/CCND3/mock (n=2).
We then determined whether ectopic expression of Bcl2, Myc and CCND3 or Bcl2, Myc and NRAS (Q61K) could transform primary B cells in a colony-forming assay. In the first plating of the methylcellulose colony assay, Bcl2/Myc/CCND3, CCND3/Myc/Bcl2 and Bcl2/CCND3/Myc cells showed more efficient colony formation than Bcl2/Myc/mock, CCND3/Myc/mock and Bcl2/CCND3/mock, respectively (Figure 4B,C). On re-plating, Bcl2/Myc/mock, CCND3/Myc/mock and Bcl2/CCND3/mock cells did not show any colony formation, while cells with the three genes expressing stable integrants (Bcl2/Myc/CCND3, CCND3/Myc/Bcl2 and Bcl2/CCND3/Myc) showed a higher number of colonies (Figure 4B). Moreover, Bcl2/Myc/NRAS cells formed colonies at a higher frequency than Bcl2/Myc/mock (Figure 4B). We also examined the synergistic effects of these gene combinations in the SCID transplantation model and found that Bcl2/Myc/CCND3, CCND3/Myc/Bcl2 and Bcl2/CCND3/Myc cells caused B-cell leukemia/lymphoma with shorter survival times than cells containing only two of the oncogenes [Figure 4D, cells expressing three genes (Bcl2/Myc/CCND3, n=3; CCND3/Myc/Bcl2 n=3; and Bcl2/CCND3/Myc, n=3) versus cells expressing two genes (Bcl2/Myc/mock, n=3; CCND3/Myc/mock, n=2; and Bcl2/CCND3/mock, n=2), P≤0.0001; Figure 4E, Bcl2/Myc/CCND3 versus Bcl2/Myc/mock, P=0.0246; Figure 4F, CCND3/Myc/Bcl2 versus CCND3/Myc/mock, P=0.0643; Figure 4G, Bcl2/CCND3/Myc versus Bcl2/CCND3/mock, P=0.0645]. Bcl2/Myc/NRAS cells also killed SCID mice with an associated shorter survival time than Bcl2/Myc/mock cells (P=0.0224, Figure 4E). These findings confirmed that ectopic expression of Bcl2, Myc and CCND3 or Bcl2, Myc and NRAS (Q61K) can function synergistically to transform primary B cells into highly aggressive malignant cells. The available expression microarray data obtained from a public database also suggested that BCL2, MYC and CCND3 function synergistically and contribute to the histological transformation of some cases of human follicular lymphoma (Online Supplementary Figure S5).
Expression microarray analysis to identify significant up- or down-regulated signaling pathways in cells expressing three genes compared to those expressing two genes
In order to investigate the deregulated pathways in the cells expressing three genes, expression microarray was employed using the stable integrants established by the cell sorting method in EXP 1 and the GSEA program with gene sets of canonical pathways. Twenty-six and 125 gene sets were significantly up- or down-regulated, respectively, in Bcl2/Myc/Ccnd1 cells (nominal P value <0.05 and false discovery rate q-value <0.25) relative to Bcl2/Myc/mock cells (Online Supplementary Table 2A,B). The five gene sets with the most up-regulated normalized enrichment score (NES) included three muscle contraction-associated gene sets (Online Supplementary Table S2A, REACTOME_SMOOTH_MUSCLE_CONTRACTION, KEGG_VASCULAR_SMOOTH_MUSCLE_CONTRACTION and REACTOME_MUSCLE_CONTRACTION). The five gene sets with the most down-regulated NES included four translation-associated gene sets (Online Supplementary Table S2B; KEGG_RIBOSOME, REACTOME_PEPTIDE_CHAIN_ELONGATION, REACTOME_FORMATION_OF_A_POOL_OF_FREE_40S_SUB UNITS and REACTOME_VIRAL_MRNA_TRANSLATION). Fifty-seven and 78 gene sets were significantly up-or down-regulated, respectively, in Ccnd1/Myc/Bcl2 cells relative to Ccnd1/Myc/mock cells (Online Supplementary Table S2C, D). All of the five gene sets with the most up-regulated NES comprised translation-associated gene sets (Online Supplementary Table S2C). The five gene sets with the most down-regulated NES included three cell cycle-associated gene sets (Online Supplementary Table S2D; REACTOME_MITOTIC_M_M_G1_PHASES, REACTOME_CELL_CYCLE_MITOTIC and REACTOME_SCF_BETA_TRCP_MEDIATED_DEGRADATION_OF_EMI1). One hundred and fifty-two and 83 gene sets were significantly up- or down-regulated, respectively, in Bcl2/Ccnd1/Myc cells relative to Bcl2/Ccnd1/mock cells (Online Supplementary Table S2E,F). The five gene sets with the most up-regulated NES comprised translation-associated gene sets (Online Supplementary Table S2E). The five gene sets with the most down-regulated NES comprised cell surface receptor-associated gene sets (Online Supplementary Table S2F). This analysis did not show any gene sets which were commonly up- or down-regulated in all variants of the cells expressing three genes (Online Supplementary Table S2).
In addition to the mRNA analysis, the protein expression of some selected genes was examined. It is known that a failure to induce senescence underlies the transformation process of some mouse primary cells. We performed western blot analysis to assess expression of senescence-associated proteins, including p53, p21, p16 and p27 and cytochemical staining of senescence-associated β galactosidase. We were not, however, able to show any differences between cells expressing three or two genes (data not shown).
Discussion
In this study, we demonstrated for the first time that the IgH translocation junction B-cell lymphoma oncogenes Bcl2, Myc and Ccnd1 function synergistically to transform mouse primary B cells into highly aggressive malignant cells. The transformation of primary mouse B cells could be induced in vitro by the exogenous introduction of all three genes. Transplanted cells ectopically expressing all three genes killed recipient mice more rapidly than those expressing any combination of two of the three genes. These findings provided direct evidence that the synergic effect contributed to lethal tumor development in mice, thus increasing our understanding of human B-cell lymphoma development. Importantly, there are reports of some cases of human B-cell lymphoma possessing concurrent BCL2, MYC and CCND1 translocations.14,15 Our results strongly support the notion that BCL2, MYC and CCND1 can operate synergistically in the malignant processes related to these cases.
Although cases involving a triple translocation of BCL2, MYC and CCND1 are rare, concurrent double translocations, especially of BCL2 and MYC, have been occasionally identified in cases of human B-cell lymphoma.3–7 It is possible that lymphoma cases with deregulated expression of BCL2 and MYC might have a cooperative genetic alteration which can substitute for CCND1. Based on such a hypothesis, we performed functional screening for genes cooperating with Bcl2 and Myc using 106 candidate cDNA clones of the retroviral SU-DHL-6 cDNA expression library. Through the screening, we not only identified mutated NRAS, one of the best-known oncogenes, but also CCND3.
It was previously reported that CCND3 is recurrently, but infrequently, targeted by IgH translocations.27,28 Moreover, it should be emphasized that we previously reported that CCND3 was the putative oncogene at the 6p21 gain/amplification region frequently found in human B-cell lymphomas.29 Although many other reports have proposed that CCND3 is involved in human B-cell lymphomagenesis,30–34 no direct evidence for its involvement has been provided. To the best of our knowledge, our study has provided the first direct evidence that CCND3 can participate in the process of malignant transformation of primary B cells. Since it is well known that CCND1 and CCND3 share a redundant role,35,36 our findings may imply that any of several genes involved in cell cycle regulation could has a synergistic effect in combination with Bcl2 and Myc on human B-cell lymphomagenesis.
Our screenings picked up the NRAS cDNA sequence containing the missense mutation in codon 61 (Q61K). We could confirm the presence of the mutated NRAS allele in SU-DHL-6. Mutated Nras has been implicated in a minority of E-mu-myc mouse lymphomas.37 However, mutated NRAS is unlikely to be the third cooperative gene acting with BCL2 and MYC in human B-cell lymphoma since it is well known that human B-cell lymphomas rarely have such mutations.38
As shown in Figure 2G, the transplantation of polyclonal integrants expressing Bcl2, Myc and Ccnd1 into mice resulted in tumor development with one to several clonal IgH rearrangements. Figure 1C shows that only a limited cell population of integrants expressing Bcl2, Myc and Ccnd1 could form colonies, suggesting that an additional unknown gene or genes cooperating with Bcl2, Myc and Ccnd1 may further contribute to B-cell transformation. This notion has some credence given that human cases reported to possess BCL2, MYC and CCND1 translocations also harbored additional BCL6 translocations.14,15 It is important to determine whether BCL6 and/or the other cooperative genes are necessary for human lymphoma formation.
The stromal microenvironment plays an important role in lymphoma formation in vivo.39–41 Lymphoma cells escaping the influence of the microenvironment in vivo could mimic the synergism which we observed under the culture condition without IL-7 and ST-2 stromal cells in vitro, while it remains to be determined whether lymphoma cells under the influence of the microenvironment in vivo could mimic the synergism. In this context, it is important to note that we were able to identify some cases of histologically transformed follicular lymphoma which showed increased expression of MYC and CCND3 (Online Supplementary Figure S5). Further studies are needed to determine whether co-activation of BCL2, MYC and CCND3 affect the relationship between follicular lym-phoma cells and their microenvironment.
GSEA was utilized in an effort to identify deregulated pathways which might be implicated in the synergistic effect of Bcl2, Myc and Ccnd1. Many pathways were deregulated in Bcl2/Myc/Ccnd1, Ccnd1/Myc/Bcl2 and Bcl2/Ccnd1/Myc cells relative to Bcl2/Myc/mock, Ccnd1/Myc/mock and Bcl2/Ccnd1/mock cells, respectively. Translation-associated pathways were significantly down-regulated in Bcl2/Myc/Ccnd1 cells and up-regulated in Ccnd1/Myc/Bcl2 and Bcl2/Ccnd1/Myc cells. We were unable to identify gene sets which were commonly deregulated in all variants of the cells expressing three genes. This may suggest that the synergistic effect of Bcl2, Myc and Ccnd1 is not regulated by changes in mRNA expression but by changes in protein stability and/or modification. Further studies should provide important insights into the downstream processes associated with the synergistic effect of Bcl2, Myc and Ccnd1.
The studied cells expressing three genes showed some minor variations in their response to the cell growth assays without IL-7 and ST-2, and considerable differences in their response to the colony-forming and in vivo assays. These variant cell types were established using different retroviral vectors, and thus the cells ectopically expressed different amounts of BCL2, MYC and CCND1 proteins (Online Supplementary Figure S3). These differences in protein expression may have affected the results of the in vitro and in vivo assays. However, it is important to note that all cell variants expressing three genes showed a malignant phenotype in every assay, relative to the respective control cells expressing two genes.
In this study, we were able to identify cooperative oncogenes that act with Bcl2 and Myc on the basis of SU-DHL-6 retroviral cDNA library screening. The use of other cDNA libraries may facilitate the identification of other cooperative oncogenes and/or microRNA. The use of siRNA or shRNA libraries could enable the identification of cooperative suppressor oncogenes. Our synergism-based screening strategy could be useful in both in vitro and in vivo studies. In our preliminary study, we recurrently identified the TCL1A gene as a gene that cooperates with Bcl2 and Myc (data not shown). Our synergism-based functional analysis method is useful not only for the identification of hitherto unknown genes, but also for the identification of oncogenic synergisms with previously identified cancer-causing genes and microRNA, which may expand our systemic understanding of a wide range of cancer-causing elements. However, the method used in our study also selected pseudo-positive genes, suggesting that the method requires further improvement.
Acknowledgments
The authors would like to thank Dr. Toshio Kitamura for providing pMXs and pMXs-neo, Dr. Masafumi Onodera for providing pGCDNsam-ires-huKO, Dr. Mel Greaves for providing the mouse JH subcloned vector, Dr. Shigetaka Kitajima for providing the pGEM-T-mouse-Myc vector, Dr. Richard Van Etten for providing the MCV-ecopac vector, Dr. Minetaro Ogawa for providing ST-2 stromal cells, and Dr. Tariq Enver for providing mouse IL-7-and SCF-producing cell lines. The outstanding technical assistance of Mss. Yumiko Kasugai, Kyoko Hirano and Seiko Sato is very much appreciated, and we are grateful to the other members of the laboratory staff for their encouragement throughout this study.
Footnotes
The online version of this article has a Supplementary Appendix.
Authorship and Disclosures
The information provided by the authors about contributions from persons listed as authors and in acknowledgments is available with the full text of this paper at www.haematologica.org.
Financial and other disclosures provided by the authors using the ICMJE (www.icmje.org) Uniform Format for Disclosure of Competing Interests are also available at www.haematologica.org.
Funding: this work was supported in part by Grants-in-Aid from the Ministry of Health, Labor and Welfare of Japan; from the Ministry of Education, Culture, Sports, Science and Technology of Japan; and from the Japanese Society for the Promotion of Science.
References
- 1.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10(8):789–99. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 2.Kuppers R, Dalla-Favera R. Mechanisms of chromosomal translocations in B cell lymphomas. Oncogene. 2001;20(40):5580–94. doi: 10.1038/sj.onc.1204640. [DOI] [PubMed] [Google Scholar]
- 3.Johansson B, Mertens F, Mitelman F. Cytogenetic evolution patterns in non-Hodgkin’s lymphoma. Blood. 1995;86(10):3905–14. [PubMed] [Google Scholar]
- 4.Niitsu N, Okamoto M, Miura I, Hirano M. Clinical significance of 8q24/c-MYC translocation in diffuse large B-cell lymphoma. Cancer Sci. 2009;100(2):233–7. doi: 10.1111/j.1349-7006.2008.01035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson NA, Savage KJ, Ludkovski O, Ben-Neriah S, Woods R, Steidl C, et al. Lymphomas with concurrent BCL2 and MYC translocations: the critical factors associated with survival. Blood. 2009;114(11):2273–9. doi: 10.1182/blood-2009-03-212191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kluin PM, Harris NL, Stein H, Leoncini L. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. ed 4. Lyon, France: IARC; 2008. B-cell lymphoma, unclassifiable, with features intermediate between diffuse large B-cell lymphoma and Burkitt lymphoma; pp. 265–266. Chapter 10. [Google Scholar]
- 7.Aukema SM, Siebert R, Schuuring E, van Imhoff GW, Kluin-Nelemans H, Boerma EJ, et al. Double hit B-cell lymphomas. Blood. 2011;117(8):2319–31. doi: 10.1182/blood-2010-09-297879. [DOI] [PubMed] [Google Scholar]
- 8.De Jong D, Voetdijk BM, Beverstock GC, van Ommen GJ, Willemze R, Kluin PM. Activation of the c-myc oncogene in a pre-cursor-B-cell blast crisis of follicular lymphoma, presenting as composite lymphoma. N Engl J Med. 1988;318(21):1373–8. doi: 10.1056/NEJM198805263182106. [DOI] [PubMed] [Google Scholar]
- 9.Lee JT, Innes DJ, Jr, Williams ME. Sequential bcl-2 and c-myc oncogene rearrangements associated with the clinical transformation of non-Hodgkin’s lymphoma. J Clin Invest. 1989;84(5):1454–9. doi: 10.1172/JCI114320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thangavelu M, Olopade O, Beckman E, Vardiman JW, Larson RA, McKeithan TW, et al. Clinical, morphologic, and cytogenetic characteristics of patients with lymphoid malignancies characterized by both t(14;18)(q32;q21) and t(8;14)(q24;q32) or t(8;22)(q24;q11) Genes Chromosomes Cancer. 1990;2(2):147–58. doi: 10.1002/gcc.2870020211. [DOI] [PubMed] [Google Scholar]
- 11.Yano T, Jaffe ES, Longo DL, Raffeld M. MYC rearrangements in histologically progressed follicular lymphomas. Blood. 1992;80(3):758–67. [PubMed] [Google Scholar]
- 12.Lossos IS, Alizadeh AA, Diehn M, Warnke R, Thorstenson Y, Oefner PJ, et al. Transformation of follicular lymphoma to diffuse large-cell lymphoma: alternative patterns with increased or decreased expression of c-myc and its regulated genes. Proc Natl Acad Sci USA. 2002;99(13):8886–91. doi: 10.1073/pnas.132253599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cory S, Vaux DL, Strasser A, Harris AW, Adams JM. Insights from Bcl-2 and Myc: malignancy involves abrogation of apoptosis as well as sustained proliferation. Cancer Res. 1999;59(7 Suppl):1685s–1692s. [PubMed] [Google Scholar]
- 14.Kawakami K, Miyanishi S, Sonoki T, Nakamura S, Nomura K, Taniwaki M, et al. Case of B-cell lymphoma with rearrangement of the BCL1, BCL2, BCL6, and c-MYC genes. Int J Hematol. 2004;79(5):474–9. doi: 10.1532/ijh97.03105. [DOI] [PubMed] [Google Scholar]
- 15.Bacher U, Haferlach T, Alpermann T, Kern W, Schnittger S, Haferlach C. Several lymphoma-specific genetic events in parallel can be found in mature B-cell neoplasms. Genes Chromosomes Cancer. 2011;50(1):43–50. doi: 10.1002/gcc.20831. [DOI] [PubMed] [Google Scholar]
- 16.Danial NN, Korsmeyer SJ. Cell death: critical control points. Cell. 2004;116(2):205–19. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- 17.Cory S, Adams JM. The Bcl2 family: regulators of the cellular life-or-death switch. Nat Rev Cancer. 2002;2(9):647–56. doi: 10.1038/nrc883. [DOI] [PubMed] [Google Scholar]
- 18.Pelengaris S, Khan M, Evan G. c-MYC: more than just a matter of life and death. Nat Rev Cancer. 2002;2(10):764–76. doi: 10.1038/nrc904. [DOI] [PubMed] [Google Scholar]
- 19.Dominguez-Sola D, Ying CY, Grandori C, Ruggiero L, Chen B, Li M, et al. Non-transcriptional control of DNA replication by c-Myc. Nature. 2007;448(7152):445–51. doi: 10.1038/nature05953. [DOI] [PubMed] [Google Scholar]
- 20.Massague J. G1 cell-cycle control and cancer. Nature. 2004;432(7015):298–306. doi: 10.1038/nature03094. [DOI] [PubMed] [Google Scholar]
- 21.Li S, Ilaria RL, Jr, Million RP, Daley GQ, Van Etten RA. The P190, P210, and P230 forms of the BCR/ABL oncogene induce a similar chronic myeloid leukemia-like syndrome in mice but have different lymphoid leukemogenic activity. J Exp Med. 1999;189(9):1399–412. doi: 10.1084/jem.189.9.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kitamura T, Koshino Y, Shibata F, Oki T, Nakajima H, Nosaka T, et al. Retrovirus-mediated gene transfer and expression cloning: powerful tools in functional genomics. Exp Hematol. 2003;31(11):1007–14. [PubMed] [Google Scholar]
- 23.Rolink A, Kudo A, Karasuyama H, Kikuchi Y, Melchers F. Long-term proliferating early pre B cell lines and clones with the potential to develop to surface Ig-positive, mitogen reactive B cells in vitro and in vivo. Embo J. 1991;10(2):327–36. doi: 10.1002/j.1460-2075.1991.tb07953.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ogawa M, Nishikawa S, Ikuta K, Yamamura F, Naito M, Takahashi K, et al. B cell ontogeny in murine embryo studied by a culture system with the monolayer of a stromal cell clone, ST2: B cell progenitor develops first in the embryonal body rather than in the yolk sac. Embo J. 1988;7(5):1337–43. doi: 10.1002/j.1460-2075.1988.tb02949.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsuzuki S, Seto M, Greaves M, Enver T. Modeling first-hit functions of the t(12;21) TEL-AML1 translocation in mice. Proc Natl Acad Sci USA. 2004;101(22):8443–8. doi: 10.1073/pnas.0402063101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kamiya A, Kakinuma S, Onodera M, Miyajima A, Nakauchi H. Prospero-related homeobox 1 and liver receptor homolog 1 coordinately regulate long-term proliferation of murine fetal hepatoblasts. Hepatology. 2008;48(1):252–64. doi: 10.1002/hep.22303. [DOI] [PubMed] [Google Scholar]
- 27.Sonoki T, Harder L, Horsman DE, Karran L, Taniguchi I, Willis TG, et al. Cyclin D3 is a target gene of t(6;14)(p21.1;q32.3) of mature B-cell malignancies. Blood. 2001;98(9):2837–44. doi: 10.1182/blood.v98.9.2837. [DOI] [PubMed] [Google Scholar]
- 28.Wlodarska I, Dierickx D, Vanhentenrijk V, an Roosbroeck K, Pospisilova H, Minnei F, et al. Translocations targeting CCND2, CCND3, and MYCN do occur in t(11;14)-negative mantle cell lymphomas. Blood. 2008;111(12):5683–90. doi: 10.1182/blood-2007-10-118794. [DOI] [PubMed] [Google Scholar]
- 29.Kasugai Y, Tagawa H, Kameoka Y, Morishima Y, Nakamura S, Seto M. Identification of CCND3 and BYSL as candidate targets for the 6p21 amplification in diffuse large B-cell lymphoma. Clin Cancer Res. 2005;11(23):8265–72. doi: 10.1158/1078-0432.CCR-05-1028. [DOI] [PubMed] [Google Scholar]
- 30.Doglioni C, Chiarelli C, Macri E, Dei Tos AP, Meggiolaro E, Dalla Palma P, et al. Cyclin D3 expression in normal, reactive and neoplastic tissues. J Pathol. 1998;185(2):159–66. doi: 10.1002/(SICI)1096-9896(199806)185:2<159::AID-PATH73>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 31.Suzuki R, Kuroda H, Komatsu H, Hosokawa Y, Kagami Y, Ogura M, et al. Selective usage of D-type cyclins in lymphoid malignancies. Leukemia. 1999;13(9):1335–42. doi: 10.1038/sj.leu.2401485. [DOI] [PubMed] [Google Scholar]
- 32.Filipits M, Jaeger U, Pohl G, Stranzl T, Simonitsch I, Kaider A, et al. Cyclin D3 is a predictive and prognostic factor in diffuse large B-cell lymphoma. Clin Cancer Res. 2002;8(3):729–33. [PubMed] [Google Scholar]
- 33.Rosenwald A, Wright G, Wiestner A, Chan WC, Connors JM, Campo E, et al. The proliferation gene expression signature is a quantitative integrator of oncogenic events that predicts survival in mantle cell lymphoma. Cancer Cell. 2003;3(2):185–97. doi: 10.1016/s1535-6108(03)00028-x. [DOI] [PubMed] [Google Scholar]
- 34.Fu K, Weisenburger DD, Greiner TC, Dave S, Wright G, Rosenwald A, et al. Cyclin D1-negative mantle cell lymphoma: a clinico-pathologic study based on gene expression profiling. Blood. 2005;106(13):4315–21. doi: 10.1182/blood-2005-04-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ciemerych MA, Kenney AM, Sicinska E, Kalaszczynska I, Bronson RT, Rowitch DH, et al. Development of mice expressing a single D-type cyclin. Genes Dev. 2002;16(24):3277–89. doi: 10.1101/gad.1023602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kozar K, Ciemerych MA, Rebel VI, Shigematsu H, Zagozdzon A, Sicinska E, et al. Mouse development and cell proliferation in the absence of D-cyclins. Cell. 2004;118(4):477–91. doi: 10.1016/j.cell.2004.07.025. [DOI] [PubMed] [Google Scholar]
- 37.Alexander WS, Bernard O, Cory S, Adams JM. Lymphomagenesis in E mu-myc transgenic mice can involve ras mutations. Oncogene. 1989;4(5):575–81. [PubMed] [Google Scholar]
- 38.Neri A, Knowles DM, Greco A, McCormick F, Dalla-Favera R. Analysis of RAS oncogene mutations in human lymphoid malignancies. Proc Natl Acad Sci USA. 1988;85(23):9268–72. doi: 10.1073/pnas.85.23.9268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dave SS, Wright G, Tan B, Rosenwald A, Gascoyne RD, Chan WC, et al. Prediction of survival in follicular lymphoma based on molecular features of tumor-infiltrating immune cells. N Engl J Med. 2004;351(21):2159–69. doi: 10.1056/NEJMoa041869. [DOI] [PubMed] [Google Scholar]
- 40.Farinha P, Al-Tourah A, Gill K, Klasa R, Connors JM, Gascoyne RD. The architectural pattern of FOXP3-positive T cells in follicular lymphoma is an independent predictor of survival and histologic transformation. Blood. 2010;115(2):289–95. doi: 10.1182/blood-2009-07-235598. [DOI] [PubMed] [Google Scholar]
- 41.Lenz G, Wright G, Dave SS, Xiao W, Powell J, Zhao H, et al. Stromal gene signatures in large-B-cell lymphomas. N Engl J Med. 2008;359(22):2313–23. doi: 10.1056/NEJMoa0802885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakagawa M, Nakagawa-Oshiro A, Karnan S, Tagawa H, Utsunomiya A, Nakamura S, et al. Array comparative genomic hybridization analysis of PTCL-U reveals a distinct subgroup with genetic alterations similar to lymphoma-type adult T-cell leukemia/lymphoma. Clin Cancer Res. 2009;15(1):30–8. doi: 10.1158/1078-0432.CCR-08-1808. [DOI] [PubMed] [Google Scholar]