Abstract
Background
Genome-wide association studies are currently identifying new loci with potential roles in thrombosis and hemostasis: these loci include novel polymorphisms associated with platelet function traits and count. However, no genome-wide study performed on children has been reported to date, in spite of the potential that these subjects have in genetic studies, when compared to adults, given the minimal degree of confounders, i.e., acquired and environmental factors, such as smoking, physical activity, diet, and drug or hormone intake, which are particularly important in platelet function.
Design and Methods
To identify new genetic variants involved in platelet reactivity and count, we performed a genome-wide association study on 75 children (8.5±1.8 years) using the Illumina Sentrix Human CNV370-Quad BeadChip containing 320,610 single nucleotide polymorphisms. Functional analyses included assessment of platelet aggregation and granule secretion triggered by different agonists (arachidonic acid, collagen, epinephrine, ADP), as well as platelet count. Associations were selected based on statistical significance and physiological relevance for a subsequent replication study in a similar sample of 286 children.
Results
We confirmed previously established associations with plasma levels of factors XII, VII and VIII as well as associations with platelet responses to ADP. Additionally, we identified 82 associations with platelet reactivity and count with a P value less than 10−5. From the associations selected for further replication, we validated two single nucleotide polymorphisms with mildly increased platelet reactivity (rs4366150 and rs1787566) on the LPAR1 and MYO5B genes, encoding lisophosphatidic acid receptor-1 and myosin VB, respectively; and rs1937970, located on the NRG3 gene coding neuroregulin-3, associated with platelet count.
Conclusions
Our genome-wide association study performed in children, followed by a validation analysis, led us to the identification of new genes potentially relevant in platelet function and biogenesis.
Keywords: genome-wide associations, platelet function, platelet count, SNP, children
Introduction
Identification of genetic factors associated with thrombosis and hemostasis has been a common quest since the discovery of classical factors such as protein C, protein S and antithrombin deficiencies, ABO blood group, factor V Leiden and prothrombin G20210A.1 In recent years this search has increased drastically mainly because of: (i) the recognized multifactorial character of thrombotic diseases;2 (ii) the high level of heritability of these diseases2,3 and (iii) the availability of genome-wide association studies (GWAS) as a useful experimental instrument.4 GWAS allow the analysis of hundreds of thousands of genomic variations or single nucleotide polymorphisms (SNP) across the whole human genome, in a non-selected, rapid, and accurate manner although the technique is expensive. In the context of thrombosis and hemostasis, a few GWAS studies have provided promising results,5–10 although the genetic variants were associated with relatively small increments in risk.
Regarding platelet physiology, newly identified SNP have been associated with platelet responses.11 Associations between newly identified SNP/genes and P-selectin exposure and fibrinogen binding induced by ADP and collagen were found in a study based on candidate genes.12 A recent GWAS in a large number of subjects also showed associations between novel SNP and platelet count and volume.13 Moreover, the first whole-genome analysis involving direct measurement of platelet function, the Framingham Heart Study, recently identified seven loci associated with platelet aggregation induced by ADP, epinephrine and collagen.14 Certainly, these studies exemplified the potential of genomic instruments to discover new loci with a role in platelet biology.
To date, all GWAS have been conducted in populations formed by adult subjects, some of whom exceeded 70 years old. Adults are affected by both acquired factors such as atherosclerosis, hypertension, endothelial dysfunction, excessive weight (body mass index >25 Kg/m2) and environmental factors such as smoking, physical activity, diet, and drug or hormone intake. All these factors are known to affect platelet function, as previously described.15–17 We, therefore, postulated that studying a population less influenced by acquired and environmental factors would be a novel and promising approach for a whole-genome analysis of platelet function traits. We reasoned that children would be an ideal population with a minimal amount of confounders, if any, and hence, we performed the INGENIAHS study (Identification of New GENes Indirectly Affecting the Hemostatic System). We measured 75 hematologic variables, of which 21 were platelet-related, in a population of 75 healthy children aged 5 to 11 years old. In a subsequent step, we conducted a validation study of the selected, most relevant SNP in a larger number of subjects.
Design and Methods
Subjects
The INGENIAHS study included 75 non-related healthy children (39 males, 36 females; 8.5±1.8 years old), with no history of bleeding as assessed by a bleeding score. Subjects did not have known diseases and were not taking any medication at the time of the laboratory testing. The details of this population have been published previously.18 The subjects’ parents gave informed consent to the tests carried out on their children. All procedures were performed in accordance with the Ethics Committee of the Pontificia Universidad Católica de Chile and the Declaration of Helsinki, as amended in Edinburgh in 2000. Blood samples were collected for both isolation of DNA and functional assays.
For the replication analysis of selected SNP, a larger sample of children from the same origin was studied. Any potential control subject who was concurrently taking drugs, was pregnant, or had infections, other diseases, a platelet count less than 100×109/L, a hemoglobin concentration less than 9 g/dL, elevated concentrations of serum creatinine or transaminases (alanine transminase/aspartate transminase), or a C-reactive protein greater than 1 mg/dL were excluded. This sample consisted of 286 healthy children (129 males, 157 females) aged 12.3±6.6 years old. Exceptionally, for the validation study involving rs1937970, 433 subjects were included since their platelet count was already available by the time of the analysis.
Sample collection, hematologic tests and platelet assays
Blood was collected, platelets prepared and plasma stored as previously reported.18 We analyzed 75 laboratory parameters related to hemostasis. Coagulation-related traits included measurements of different clotting times, clot lysis assays, and plasma concentrations of coagulation, fibrinolytic and thrombophilic factors. Platelet parameters included platelet count, aggregation (determined by light aggregation) and secretion triggered by type III collagen (1 and 2 μg/mL), ADP (4 and 8 μM), arachidonic acid (1 mM), epinephrine (10 μM) and ristocetin (1.2 mg/mL), closure times determined using the platelet function analyzer (PFA-100™) with collagen-epinephrine and collagen-ADP cartridges, and ADP, ATP and serotonin platelet contents. These assays were performed as described in detail elsewhere.18,19
Genotyping
Samples were genotyped using the Illumina Sentrix Human CNV370-Quad BeadChip (Illumina, Essex, UK) containing 338,897 SNP and covering the whole human genome including high density SNP regions within 10 Kb of a referenced sequenced gene, highly conserved regions, regions of copy number variation, major histocompatibility complex regions, mega-satellite and SNP deserts. Genotyping was performed by the commercial service of the Centro Nacional de Genotipado (Madrid, Spain).
For the replication studies, selected SNP were genotyped by means of TaqMan® SNP Genotyping Assays from Applied Biosystems: C___2676047_10 for rs4366150 (LPAR1), C___8803065_10 for rs933880 (BTBD11), C__11686881_10 for rs1787566 (MYO5B) and C__12117859_10 for rs1937970 (NRG3).
Statistical analysis
Different quality-control filters were applied to DNA samples, SNP and laboratory tests included in the INGENIAHS. Samples with a call rate greater than 90% and a factor less than 95% in the identical-by-state test were selected. SNP with a call rate greater than 90%, a minor allele frequency greater than 0.04, a false discovery rate of less than 0.2 for detection of unacceptably high individual heterozygosity, and a P value greater than 0.05 for the Hardy-Weinberg equilibrium were included. Laboratory tests with a call rate greater than 0.4 were also selected. Consequently, the final statistical analysis was based on data from 70 non-related subjects, 320,610 SNP and 62 laboratory assays. The statistical analysis was performed using the generalized linear models from the GenABEL package of R statistical software. Parameters indicative of the strength of association between laboratory parameters and SNP were obtained: (i) the proportion, an empirical genome-wide significance P value between 0 and 1 which indicates the strength of association between a SNP and a functional trait, was calculated using 1,000 replicas and incorporating a multiple testing correction; (ii) minor allele frequency, which is inversely related to the risk of non-relevant or false association; and (iii) the P value, which indicates the statistical relevance of the association.
A t-test or a Mann Whitney U-test was performed, as appropriate, for the validation studies.
Results
Whole-genome analysis: the INGENIAHS study
This report is focused on associations with platelet phenotypes. As summarized in Table 1, the INGENIAHS study revealed 82 associations with quantitative parameters of platelet reactivity and platelet count, with P values ranging from 10−8 to 10−5. From among these, we selected four for further analysis based on the following criteria: (i) P value; (ii) potential of the SNP to influence platelet reactivity in more than one functional test and/or towards various agonists; and (iii) biological relevance of the identified locus in the processes of platelet activation and/or platelet biogenesis. Thus, four SNP were selected for the replication studies: rs4366150, rs933880, rs1787566 and rs1937970.
Table 1.
SNP identified in the INGENIAHS with lowest P values related to quantitative platelet parameters.
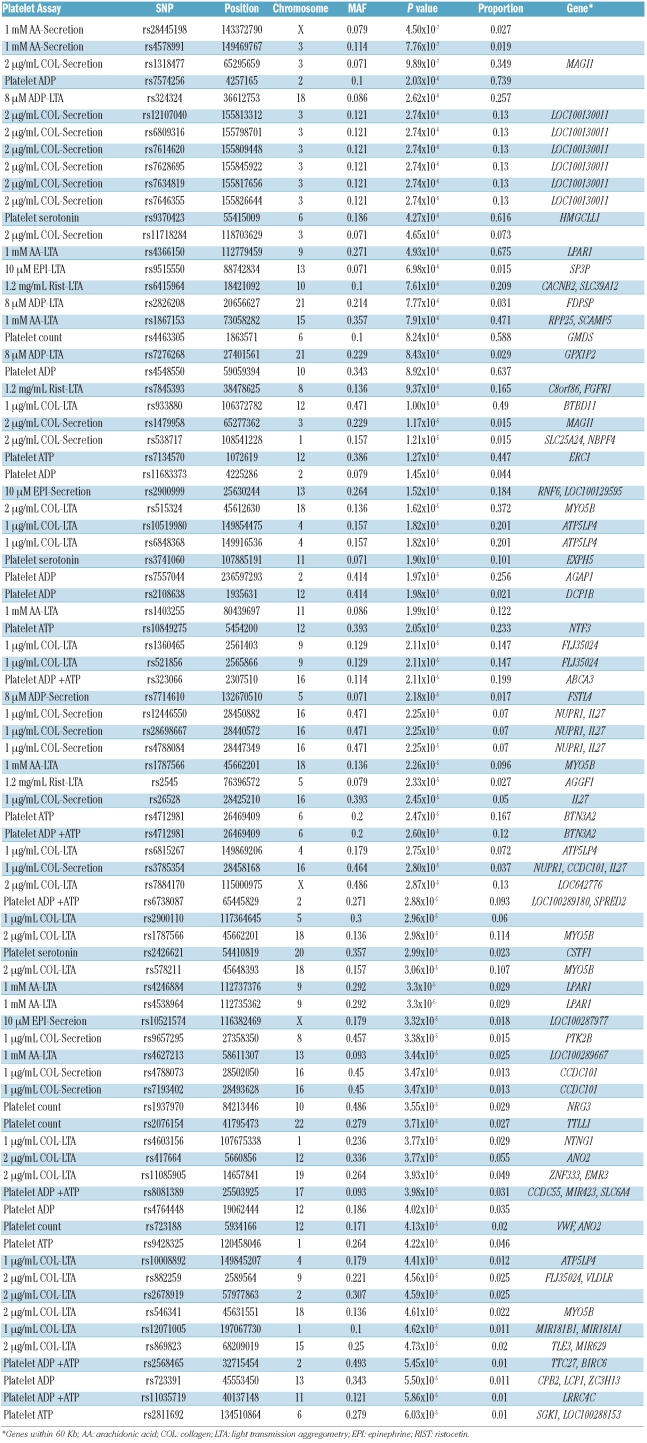
Firstly, rs4366150 with a minor allele frequency of 0.271, a P value of 4.93×10−6 and a proportion of 0.675, was associated with platelet aggregation induced by arachidonic acid. This SNP is located in intron 1 of the LPAR1 gene on chromosome 9. LPAR1 encodes lisophosphatidic acid receptor-1 (LPAR-1) involved in the platelet activation mediated by this phospholipid derivative.20 rs4366150 is in linkage disequilibrium with 19 other SNP with unknown roles (Online Supplementary Table S1) including rs4538964 and rs4246884, also present in our array (r2 >0.8) and showing similar statistical significance. In our population the allelic distribution of rs4366150 was 0.729 and 0.271 for alleles A and G, respectively. The presence of the polymorphic allele G, instead of the ancestral A, predisposed subjects to a mildly but significantly increased platelet aggregation response to arachidonic acid (77.0±4.6, 81.2±4.3 and 89.5±5.1 % for AA, AG and GG subjects, respectively, Figure 1A). Interestingly, this SNP also showed significant associations with platelet aggregation responses to other agonists used, with a common pattern of higher reactivity for carriers of the G allele (Figure 1A). Results of the platelet function tests performed are detailed in Online Supplementary Table S2.
Figure 1.
Effect of rs4366150 (LPAR1) on platelet aggregation and secretion assays. (A) Data from the INGENIAHS. In all cases P value <0.01. (B) Data from the replication population. *indicates a P value <0.05 when AA controls were compared to AG+GG individuals in the replication study. Results represent mean ± SD.
Secondly, rs933880 was associated with platelet aggregation induced by collagen. This SNP is located in intron 1 of the BTBD11 gene which encodes a BTB domain-containing protein. BTB domains are found in both actin-binding and DNA-binding proteins such as zinc-finger transcription factors,21 and are involved in the determination of cell fate and function in the hematopoietic system.22 Its relevance in platelet function is still unknown. This SNP was not validated in the further replication study performed. Nevertheless, results regarding this SNP can be seen in Online Supplementary Table S3.
Thirdly, rs1787566 was statistically associated with platelet aggregation triggered by two different platelet agonists, arachidonic acid and collagen, with a minor allele frequency of 0.136. A proportion of 0.096 and a P value of 2.26×10−5 were obtained with arachidonic acid-induced platelet aggregation, while a proportion of 0.114, and a P value of 2.98×10−5 were found with collagen-triggered aggregation. rs1787566 localizes in intron 22 of the MYO5B gene on chromosome 18 and encodes myosin VB, a member of the myosin superfamily. This family of proteins plays a relevant role in the dynamics of platelet cytoskeleton.23 Defects on myosin VA have been associated with Griscelli syndrome,24 characterized by a platelet dysfunction, among other abnormalities. However, as far as we know, the presence of myosin VB has not been evaluated in platelets. The allelic distribution of rs1787566 in our population was 0.864 and 0.136 for alleles A and G, respectively. The polymorphic allele G of rs1787566 was associated with a mildly increased platelet aggregation response (Figure 2A). Furthermore, the G allele was associated with higher platelet secretion induced by 1 mM arachidonic acid, 1 μg/mL collagen and 2 μg/mL collagen. rs1787566 is in linkage disequilibrium with fourteen SNP with unknown roles within the same gene (r2 >0.8) (Online Supplementary Table S4), including rs546341, which is also present in our array, showing similar results. Two other SNP within this gene, rs515324 and rs578211, partially linked with rs1787566 (r2 =0.618) also showed a statistical association with collagen (2 μg/mL)-induced platelet aggregation (Table 1). Results from all platelet functional tests are shown in Online Supplementary Table S5.
Figure 2.
Effect of rs1787566 (MYO5B) on platelet aggregation and secretion. (A) Data from the INGENIAHS. In all cases P value <0.01. (B) Results obtained in the validation population. *indicates a P value <0.05 when AA controls were compared to AG+GG individuals in the replication study. Results represent mean ± SD.
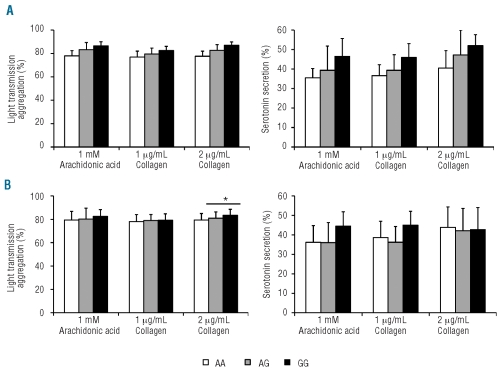
Finally, the INGENIAHS study also identified a strong association between rs1937970 and platelet count, showing a minor allele frequency of 0.485, a proportion of 0.029, and a P value of 3.55×10−5. rs1937970 is located in intron 2 of the NRG3 gene on chromosome 10, and is in linkage disequilibrium with fourteen intronic SNP on the same gene (r2 > 0.8) (Online Supplementary Table S6). The NRG3 gene encodes neuregulin-3, a ligand for the trans-membrane tyrosine kinase receptor ERBB4 that, upon activation, induces several cellular responses including proliferation, migration, differentiation, and survival or apoptosis.25 Neuregulin-3 is involved in neuron and mammary gland biology and also in diseases such as schizophrenia and schizoaffective disorders.26 To our knowledge, the potential role of neuregulin-3 in megakaryopoiesis and platelet formation is still unknown. In our population, the allelic frequency of rs1937970 was 0.515 and 0.485 for alleles A and G, respectively. Interestingly, the A allele was associated with higher platelet count (260.9±46.1×109/L in GG subjects, 289.3±56.6×109/L in AG subjects, and 347.3± 45.2×109/L in AA individuals, Figure 3A). No associations with other platelet parameters were detected in the INGE-NIAHS study (Online Supplementary Table S7).
Figure 3.
Effect of rs1937970 (NRG3) on platelet count. (A) Data from the INGENIAHS. P value = 3.55 x 10−5. (B) Results obtained in the validation population. *indicates a P value <0.05 when AA controls were compared to AG+GG individuals in the replication study. Results represent mean ± SD.
Validation analysis
In order to confirm the significant associations described above, we performed a validation study in 286 children from the same population. The findings for each SNP are described below.
rs4366150 (LPAR1)
In agreement with the INGENIAHS results, carriers of the G allele showed a mildly increased aggregation response to arachidonic acid (79.3±10.0, 79.7±5.4, and 82.4±5.9 % for AA, AG and GG individuals, respectively; Figure 1B), although differences did not reach statistical significance. Platelet aggregation induced by epinephrine (10 μM), and platelet secretion by both epinephrine and collagen (1 μg/mL) were significantly greater in carriers of the G allele (P<0.05) (Figure 1B). Results of all platelet functional assays performed in the replication population are detailed in Online Supplementary Table S2.
rs933880 (BTBD11)
As mentioned earlier, this SNP was not confirmed in the replication sample. Results can be seen in Online Supplementary Table S3.
rs1787566 (MYO5B)
Validation studies confirmed the association of this SNP with greater platelet reactivity (Figure 2B). As shown, carriers of the G allele had a greater platelet aggregation response to arachidonic acid (79.5±7.4, 80.2±9.3 and 82.5±5.8 % in AA, AG and GG subjects, respectively), and to 2 μg/mL collagen (79.4±5.6, 81.0±5.2, and 83.5±5.0 % for AA, AG and GG subjects, respectively, P<0.05) (Figure 2B). Values for all platelet parameters evaluated in this replication study are shown in Online Supplementary Table S5.
rs1937970 (NRG3)
The association of this SNP with platelet count was further validated in 433 subjects. In a similar fashion, we found higher platelet counts in carriers of the A allele (286.6±65.98×109, 272.35±66.12, and 267.6±54.4 platelets/L in AA, AG and GG subjects, respectively; P<0.05 when AA subjects were compared with AG subjects) (Figure 3B). As in the INGENIAHS study, rs1937970 was not associated with any platelet function variable in the additional subjects evaluated (Online Supplementary Table S7).
Discussion
In recent years, GWAS have become a powerful instrument for identifying new genetic factors involved in human diseases, including thrombosis. Different genes and their variants have been identified using a broad variety of GWAS designs. Studies aimed at elucidating the genetic predisposition to thrombosis are based on large case-control populations of affected and healthy subjects.5 Genetics of platelet function have also been explored using a single control population.13,14 As far as concerns the sample sizes of GWAS, there are robust studies containing thousands of subjects14 and others based on only several hundreds.12 Recently, a whole-genome expression assay from 37 subjects identified new loci regulating platelet function which were validated in subsequent steps.27 To date, all GWAS have been performed on adult subjects who usually have confounding factors such as being overweight, smoking, and taking drugs or hormones that could mask the identification of genetic loci involved in platelet biology. In order to diminish the influence of confounders, we performed a genetic study on a sample of healthy children (n=75).
In contrast to previous approaches assessing only a few platelet traits, we evaluated a wide range of parameters, including platelet aggregation and secretion elicited by several agonists at different concentrations. Considering that the performance and analysis of a whole set of platelet assays is a complex and laborious process requiring a high degree of standardization,28 we believe our study provides a unique source of genetic associations that can contribute to identify new loci relevant in platelet physiology. Statistically, the INGENIAHS revealed associations with P values in the 10−8–0−5 range similar to those previously reported for GWAS.13,27 Hence, taking into account our study design, we considered that selection of associations based only on the statistically stringent threshold could mask potentially relevant SNP with low minor allele frequency. For this reason, we based our selection not only on the P value but also on the biological relevance of the SNP identified and its correlation with other tests and agonists analyzed.
Of note, our study confirmed associations between previously reported SNP and hemostatic and platelet traits, indicating that our experimental design has the ability to identify true associations. First, rs2731672, in linkage disequilibrium (r2= 0.936) with the Kozak −4C>T polymorphism located in the F12 gene, was associated with plasma levels of factor XII;29 rs8176746 (ABO gene) was associated with factor VIII levels;30 and rs488703 (F7 gene) with factor VII levels31 (Online Supplementary Table S8). Second, although some SNP associated with platelet function were not validated in our study (Online Supplementary Table S9), our data seem to support the associations of variants in JAK2, LRRFIP1, COMMD7 and JMJD1C with platelet reactivity12,27 in the same direction (Online Supplementary Table S9). These results corroborate that these proteins might play a significant role in platelet reactivity. Further studies are required to characterize the pathways involved, as well as the mechanisms implicated in the functional effect and the potential relevance in diseases.
The INGENIAHS revealed 82 associations with platelet biology, with the lowest P values. Four of these were selected for further validation in 286 controls. rs933880 (BTBD11) was not further validated; however, associations between rs4366150 (LPAR1) and rs1787566 (MYO5B) with platelet hyper-reactivity and rs1937970 (NRG3) with platelet count were confirmed in the replication sample.
rs4366150 (LPAR1) was consistently associated with increased platelet reactivity to several agonists. As mentioned, LPAR1 codes for LPAR-1, a member of the G-protein coupled receptor family, which mediates platelet activation by lisophosphatidic acid. This phospholipid derivative is a component of the mildly-oxidized low-density lipoproteins and the lipid rich core of atherosclerotic plaques.32 To date, information on the role of lisophosphatidic acid in platelet function is limited and controversial. Platelets express mRNA from the seven members of the lisophosphatidic acid receptor family, LPAR-1 being the second lowest expressed.20 Nevertheless, LPAR-1 antagonists inhibit lisophosphatidic acid-induced platelet aggregation.33 Moreover, it was recently reported that 20% of healthy controls failed to respond to lisophosphatidic acid.34 Genetic variants of the genes encoding lisophosphatidic acid receptors, such as rs4366150 for LPAR-1, could be involved in this inter-individual heterogeneity, although this needs further studies.
rs1787566 (MYO5B) was associated with increased platelet response in our studies. The role of myosin super family proteins is widely known in cell biology, as they are motor proteins that move along actin filaments in an ATP-dependent manner.35 Megakaryocytes and platelets express myosin IIA, IIB, VA and very likely other types of non-filamentous myosin (from the book “Platelets” by A. Michaelson ISBN 13: 978-0-12-369367-9). Also, myosin deficiency is associated with rare bleeding disorders.24 In addition, with regards to the role of myosin VB in other cell types, it is known that MYO5B mutations cause microvillus inclusion disease and disrupt epithelial cell polarity in the bowel.36 Thus, although the expression of myosin VB in platelets is still unknown, the association identified in our assay deserves additional attention. Studies on the role of myosin VB in platelets will elucidate the relevance of this isoform in platelet biology as well as the importance of SNP on this gene as modulators of platelet function.
While an appropriate number of circulating platelets is crucial to maintain vascular integrity and to prevent bleeding, an excessive number of platelets predisposes to thrombosis.37–39 To date, the basis of the variability found in platelet count among normal individuals has remained elusive. Rare inherited syndromes characterized by abnormal megakaryopoiesis and platelet formation, added to studies using knockout mice and recent GWAS related to platelet count have shed some light on to this issue.40,41 In our study, previously reported associations between SNP and platelet number11 were not confirmed (Online Supplementary Table S10). However, a new identified polymorphism, rs1937970 (NRG3), was consistently associated with higher platelet count. NRG3 encodes a ligand for the transmembrane tyrosine kinase receptor ERBB4, relevant in cell differentiation and survival.25 This ligand was initially described in neurons, whereas information on its presence in platelets is lacking. Interestingly, neurons and platelets share similarities regarding secretory pathways and secretion products.42 Platelets and neurons store and release neuro-transmitters, e.g, 5-hydroxytryptamine, and drugs blocking the serotonin re-uptake affect both neuron and platelet replenishment with the amine.43 Moreover, mental disorders, e.g., schizophrenia, have also been associated with platelet dysfunction.26 All these data suggest that neuregulin-3 may play an important role in platelet biology which deserves further study.
As mentioned earlier, rs4366150, rs1787566 and rs1937970 localize within intronic regions of their respective genes and are in linkage disequilibrium with many other intronic SNP on the same gene (Online Supplementary Tables S1, S4 and S6). Further studies involving fine mapping and/or additional functional analyses are needed to find the causative SNP for the described test. At present, the fact that only 12% of SNP associated with traits are located in protein-coding regions of genes has drawn attention to the potential roles of intronic and intergenic regions in regulating gene expression.4
In conclusion, our study identified two novel loci potentially associated with a mild increase in platelet reactivity (rs4366150 and rs1787566) as well as a novel locus (rs1937970) potentially influencing platelet count in a population of children. The results obtained in this GWAS must be interpreted with caution because of the small number of subject analyzed (n=75), although we also performed a replication analysis in 286 subjects for platelet reactivity and 433 controls for platelet count. The finding that the SNP identified in our study exert only a mild effect on platelet function is, in some way, predictable since similar modest effects have been found in other genome-wide studies.4,13,14 However, the identification of the SNP may highlight the biological relevance of these novel genes in platelet physiology. It is also plausible that other less frequent polymorphisms not included in our array, or even rare mutations affecting these genes may have stronger functional effect or, may even play a relevant pathological role in bleeding or thrombotic disorders. Additionally, these genes could constitute targets for novel therapies. Further studies in other populations may be helpful to elucidate the physiological role of each of these genes in platelet physiology.
Footnotes
The online version of this article has a Supplementary Appendix.
Authorship and Disclosures
The information provided by the authors about contributions from persons listed as authors and in acknowledgments is available with the full text of this paper at www.haematologica.org.
Financial and other disclosures provided by the authors using the ICMJE (www.icmje.org) Uniform Format for Disclosure of Competing Interests are also available at www.haematologica.org.
Funding: this study was supported partially by PI10/02594 (Fondo de Investigaciones Sanitarias), FEHH and Novo Nordisk Pharma, S.A., and grants 04515/GERM/06 (Fundación Seneca), SAF2009-08993 (MCYT & FEDER), RD06/0014/0039 (ISCIII & FEDER), PI-08/0420 and PI-08/0756. This study was also supported by FOONDECYT, Chile 1090170 to DM. JAG is supported by a Sara Borrell postdoctoral fellowship. CM is an investigator from “Fundación para la Formación e Investigación Sanitarias de la Región de Murcia”. JMS was supported by “Programa d’Estabilització d’Investigadors de la Direcció d’Estrategia i Coordinació del Departament de Salut” (Generalitat de Catalunya).
References
- 1.Reitsma PH. How to identify new genetic risk factors for VTE? Thromb Res. 2009;123(Suppl 4):S22–S24. doi: 10.1016/S0049-3848(09)70138-0. [DOI] [PubMed] [Google Scholar]
- 2.Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461(7265):747–53. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Lange M, Snieder H, Ariens RA, Spector TD, Grant PJ. The genetics of haemostasis: a twin study. Lancet. 2001;357(9250):101–5. doi: 10.1016/S0140-6736(00)03541-8. [DOI] [PubMed] [Google Scholar]
- 4.Manolio TA. Genomewide association studies and assessment of the risk of disease. N Engl J Med. 2010;363(2):166–76. doi: 10.1056/NEJMra0905980. [DOI] [PubMed] [Google Scholar]
- 5.Bezemer ID, Bare LA, Doggen CJ, Arellano AR, Tong C, Rowland CM, et al. Gene variants associated with deep vein thrombosis. JAMA. 2008;299(11):1306–14. doi: 10.1001/jama.299.11.1306. [DOI] [PubMed] [Google Scholar]
- 6.Tregouet DA, Heath S, Saut N, Biron-Andreani C, Schved JF, Pernod G, et al. Common susceptibility alleles are unlikely to contribute as strongly as the FV and ABO loci to VTE risk: results from a GWAS approach. Blood. 2009;113(21):5298–303. doi: 10.1182/blood-2008-11-190389. [DOI] [PubMed] [Google Scholar]
- 7.Yang Q, Kathiresan S, Lin JP, Tofler GH, O’Donnell CJ. Genome-wide association and linkage analyses of hemostatic factors and hematological phenotypes in the Framingham Heart Study. BMC Med Genet. 2007;8(Suppl 1):S12. doi: 10.1186/1471-2350-8-S1-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soranzo N, Spector TD, Mangino M, Kuhnel B, Rendon A, Teumer A, et al. A genome-wide meta-analysis identifies 22 loci associated with eight hematological parameters in the HaemGen consortium. Nat Genet. 2009;41(11):1182–90. doi: 10.1038/ng.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Malarstig A, Buil A, Souto JC, Clarke R, Blanco-Vaca F, Fontcuberta J, et al. Identification of ZNF366 and PTPRD as novel determinants of plasma homocysteine in a family-based genome-wide association study. Blood. 2009;114(7):1417–22. doi: 10.1182/blood-2009-04-215269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buil A, Tregouet DA, Souto JC, Saut N, Germain M, Rotival M, et al. C4BPB/C4BPA is a new susceptibility locus for venous thrombosis with unknown protein S-independent mechanism: results from genome-wide association and gene expression analyses followed by case-control studies. Blood. 2010;115(23):4644–50. doi: 10.1182/blood-2010-01-263038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kunicki TJ, Nugent DJ. The genetics of normal platelet reactivity. Blood. 2010;116(15):2627–34. doi: 10.1182/blood-2010-04-262048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones CI, Bray S, Garner SF, Stephens J, de Bono B, Angenent WG, et al. A functional genomics approach reveals novel quantitative trait loci associated with platelet signaling pathways. Blood. 2009;114(7):1405–16. doi: 10.1182/blood-2009-02-202614. [DOI] [PubMed] [Google Scholar]
- 13.Soranzo N, Rendon A, Gieger C, Jones CI, Watkins NA, Menzel S, et al. A novel variant on chromosome 7q22.3 associated with mean platelet volume, counts, and function. Blood. 2009;113(16):3831–7. doi: 10.1182/blood-2008-10-184234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson AD, Yanek LR, Chen MH, Faraday N, Larson MG, Tofler G, et al. Genome-wide meta-analyses identifies seven loci associated with platelet aggregation in response to agonists. Nat Genet. 2010;42(7):608–13. doi: 10.1038/ng.604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schafer AI. Genetic and acquired determinants of individual variability of response to antiplatelet drugs. Circulation. 2003;108(8):910–1. doi: 10.1161/01.CIR.0000088843.52678.8A. [DOI] [PubMed] [Google Scholar]
- 16.Fusegawa Y, Hashizume H, Okumura T, Deguchi Y, Shina Y, Ikari Y, et al. Hypertensive patients with carotid artery plaque exhibit increased platelet aggregability. Thromb Res. 2006;117(6):615–22. doi: 10.1016/j.thromres.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 17.Zangari M, Guerrero J, Cavallo F, Prasad HK, Esseltine D, Fink L. Hemostatic effects of bortezomib treatment in patients with relapsed or refractory multiple myeloma. Haematologica. 2008;93(6):953–4. doi: 10.3324/haematol.12522. [DOI] [PubMed] [Google Scholar]
- 18.Quiroga T, Goycoolea M, Panes O, Aranda E, Martinez C, Belmont S, et al. High prevalence of bleeders of unknown cause among patients with inherited mucocutaneous bleeding. A prospective study of 280 patients and 299 controls. Haematologica. 2007;92(3):357–65. doi: 10.3324/haematol.10816. [DOI] [PubMed] [Google Scholar]
- 19.Martinez C, Anton AI, Corral J, Quiroga T, Panes O, Lozano ML, et al. Genotype-phenotype relationship for six common polymorphisms in genes affecting platelet function from 286 healthy subjects and 160 patients with mucocutaneous bleeding of unknown cause. Br J Haematol. 2009;146(1):95–103. doi: 10.1111/j.1365-2141.2009.07713.x. [DOI] [PubMed] [Google Scholar]
- 20.Khandoga AL, Fujiwara Y, Goyal P, Pandey D, Tsukahara R, Bolen A, et al. Lysophosphatidic acid-induced platelet shape change revealed through LPA(1–5) receptor-selective probes and albumin. Platelets. 2008;19(6):415–27. doi: 10.1080/09537100802220468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perez-Torrado R, Yamada D, Defossez PA. Born to bind: the BTB protein-protein interaction domain. Bioessays. 2006;28(12):1194–202. doi: 10.1002/bies.20500. [DOI] [PubMed] [Google Scholar]
- 22.Bilic I, Ellmeier W. The role of BTB domain-containing zinc finger proteins in T cell development and function. Immunol Lett. 2007;108(1):1–9. doi: 10.1016/j.imlet.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 23.Nachmias VT, Kavaler J, Jacubowitz S. Reversible association of myosin with the platelet cytoskeleton. Nature. 1985;313(5997):70–2. doi: 10.1038/313070a0. [DOI] [PubMed] [Google Scholar]
- 24.Gunay-Aygun M, Huizing M, Gahl WA. Molecular defects that affect platelet dense granules. Semin Thromb Hemost. 2004;30(5):537–47. doi: 10.1055/s-2004-835674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang D, Sliwkowski MX, Mark M, Frantz G, Akita R, Sun Y, et al. Neuregulin-3 (NRG3): a novel neural tissue-enriched protein that binds and activates ErbB4. Proc Natl Acad Sci USA. 1997;94(18):9562–7. doi: 10.1073/pnas.94.18.9562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang YC, Chen JY, Chen ML, Chen CH, Lai IC, Chen TT, et al. Neuregulin 3 genetic variations and susceptibility to schizophrenia in a Chinese population. Biol Psychiatry. 2008;64(12):1093–6. doi: 10.1016/j.biopsych.2008.07.012. [DOI] [PubMed] [Google Scholar]
- 27.Goodall AH, Burns P, Salles I, Macaulay IC, Jones CI, Ardissino D, et al. Transcription profiling in human platelets reveals LRRFIP1 as a novel protein regulating platelet function. Blood. 2010;116(22):4646–56. doi: 10.1182/blood-2010-04-280925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quiroga T, Goycoolea M, Matus V, Zuniga P, Martinez C, Garrido M, et al. Diagnosis of mild platelet function disorders. Reliability and usefulness of light transmission platelet aggregation and serotonin secretion assays. Br J Haematol. 2009;147(5):729–36. doi: 10.1111/j.1365-2141.2009.07890.x. [DOI] [PubMed] [Google Scholar]
- 29.Endler G, Exner M, Mannhalter C, Meier S, Ruzicka K, Handler S, et al. A common C-- >T polymorphism at nt 46 in the promoter region of coagulation factor XII is associated with decreased factor XII activity. Thromb.Res. 2001;101(4):255–60. doi: 10.1016/s0049-3848(00)00404-7. [DOI] [PubMed] [Google Scholar]
- 30.Smith NL, Chen MH, Dehghan A, Strachan DP, Basu S, Soranzo N, et al. Novel associations of multiple genetic loci with plasma levels of factor VII, factor VIII, and von Willebrand factor: The CHARGE (Cohorts for Heart and Aging Research in Genome Epidemiology) Consortium. Circulation. 2010;121(12):1382–92. doi: 10.1161/CIRCULATIONAHA.109.869156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ken-Dror G, Drenos F, Humphries SE, Talmud PJ, Hingorani AD, Kivimaki M, et al. Haplotype and genotype effects of the F7 gene on circulating factor VII, coagulation activation markers and incident coronary heart disease in UK men. J Thromb Haemost. 2010;8(11):2394–403. doi: 10.1111/j.1538-7836.2010.04035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu YJ, Aziz OA, Bhugra P, Arneja AS, Mendis MR, Dhalla NS. Potential role of lysophosphatidic acid in hypertension and atherosclerosis. Can J Cardiol. 2003;19(13):1525–36. [PubMed] [Google Scholar]
- 33.Rother E, Brandl R, Baker DL, Goyal P, Gebhard H, Tigyi G, et al. Subtype-selective antagonists of lysophosphatidic acid receptors inhibit platelet activation triggered by the lipid core of atherosclerotic plaques. Circulation. 2003;108(6):741–7. doi: 10.1161/01.CIR.0000083715.37658.C4. [DOI] [PubMed] [Google Scholar]
- 34.Pamuklar Z, Lee JS, Cheng HY, Panchatcharam M, Steinhubl S, Morris AJ, et al. Individual heterogeneity in platelet response to lysophosphatidic acid: evidence for a novel inhibitory pathway. Arterioscler Thromb Vasc Biol. 2008;28(3):555–61. doi: 10.1161/ATVBAHA.107.151837. [DOI] [PubMed] [Google Scholar]
- 35.Nambiar R, McConnell RE, Tyska MJ. Myosin motor function: the ins and outs of actin-based membrane protrusions. Cell Mol Life Sci. 2010;67(8):1239–54. doi: 10.1007/s00018-009-0254-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muller T, Hess MW, Schiefermeier N, Pfaller K, Ebner HL, Heinz-Erian P, et al. MYO5B mutations cause microvillus inclusion disease and disrupt epithelial cell polarity. Nat Genet. 2008;40(10):1163–5. doi: 10.1038/ng.225. [DOI] [PubMed] [Google Scholar]
- 37.Geddis AE. Megakaryopoiesis. Semin Hematol. 2010;47(3):212–9. doi: 10.1053/j.seminhematol.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thon JN, Italiano JE. Platelet formation. Semin Hematol. 2010;47(3):220–6. doi: 10.1053/j.seminhematol.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Skoda RC. Thrombocytosis. Hematology Am Soc Hematol Educ Program. 2009:159–67. doi: 10.1182/asheducation-2009.1.159. [DOI] [PubMed] [Google Scholar]
- 40.Nurden P, Nurden AT. Congenital disorders associated with platelet dysfunctions. Thromb Haemost. 2008;99(2):253–63. doi: 10.1160/TH07-09-0568. [DOI] [PubMed] [Google Scholar]
- 41.Biino G, Balduini CL, Casula L, Cavallo P, Vaccargiu S, Parracciani D, et al. Analysis of 12,517 inhabitants of a Sardinian geographic isolate reveals that predispositions to thrombocytopenia and thrombocytosis are inherited traits. Haematologica. 2011;96(1):96–101. doi: 10.3324/haematol.2010.029934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Da Prada M, Cesura AM, Launay JM, Richards JG. Platelets as a model for neurones? Experientia. 1988;44(2):115–26. doi: 10.1007/BF01952193. [DOI] [PubMed] [Google Scholar]
- 43.Bismuth-Evenzal Y, Roz N, Gurwitz D, Rehavi M. N-methyl-citalopram: a quaternary selective serotonin reuptake inhibitor. Biochem Pharmacol. 2010;80(10):1546–52. doi: 10.1016/j.bcp.2010.07.035. [DOI] [PubMed] [Google Scholar]