Abstract
Background
Analysis of genomic sequence allows characterization of genome content and organization, and access beyond gene-coding regions for identification of functional elements. BAC libraries, where relatively large genomic regions are made readily available, are especially useful for species without a fully sequenced genome and can increase genomic coverage of phylogenetic and biological diversity. For example, no butterfly genome is yet available despite the unique genetic and biological properties of this group, such as diversified wing color patterns. The evolution and development of these patterns is being studied in a few target species, including Bicyclus anynana, where a whole-genome BAC library allows targeted access to large genomic regions.
Methodology/Principal Findings
We characterize ∼1.3 Mb of genomic sequence around 11 selected genes expressed in B. anynana developing wings. Extensive manual curation of in silico predictions, also making use of a large dataset of expressed genes for this species, identified repetitive elements and protein coding sequence, and highlighted an expansion of Alcohol dehydrogenase genes. Comparative analysis with orthologous regions of the lepidopteran reference genome allowed assessment of conservation of fine-scale synteny (with detection of new inversions and translocations) and of DNA sequence (with detection of high levels of conservation of non-coding regions around some, but not all, developmental genes).
Conclusions
The general properties and organization of the available B. anynana genomic sequence are similar to the lepidopteran reference, despite the more than 140 MY divergence. Our results lay the groundwork for further studies of new interesting findings in relation to both coding and non-coding sequence: 1) the Alcohol dehydrogenase expansion with higher similarity between the five tandemly-repeated B. anynana paralogs than with the corresponding B. mori orthologs, and 2) the high conservation of non-coding sequence around the genes wingless and Ecdysone receptor, both involved in multiple developmental processes including wing pattern formation.
Introduction
Accumulation of genomic sequence data for different species is allowing an in-depth understanding of genome properties and evolution. Analysis of whole genomes of target species enables a detailed characterization of genome content and structure, and comparative analysis of genomic sequence across species provides insights about different aspects of genome dynamics and evolution (e.g., [1]). Widening phylogenetic representation of genomic data has allowed in silico identification of new protein-coding and miRNA genes, and regulatory sequence (e.g., [2], [3]). However, and despite the increasing number of eukaryotic assembled genomes in the public depository [4], we are still far from representative coverage of biological diversity. This is especially so for groups with larger genomes whose full sequencing still requires a significant investment, and/or where repetitive or polymorphic sequence renders genome assembly a bioinformatic challenge.
Bacterial Artificial Chromosomes (BACs), where large (typically around 150 Kb) fragments of genomic DNA are cloned and can be accessed individually, are a valuable resource for species where a complete genome sequence is not (yet) available. They allow focus on particular genomic regions (often around genes of interest), and have been used successfully for different ends such as sequence annotation (including access to gene coding and regulatory regions, physical mapping, development of genetic markers, analysis of synteny, or to assist whole genome assembly (e.g., [5], [6]). BAC libraries are available for many species, including different lepidopterans (the insect order of butterflies and moths) which have relatively large and typically repetitive genomes [7]. This is one of the most diverse groups of animals and includes many agricultural pests and one of only two domesticated insects, the silkworm Bombyx mori.
The Lepidoptera have an unusual set of genetic properties, combining holocentric chromosomes, heterogametic females, and male-restricted meiotic recombination, whose consequences for genome evolution remain largely unexplored. The genome of B. mori is completed [8], [9] and provides an invaluable reference for comparative genomics in this group (e.g., [10], [11]). However, this is the only genome publicly available for this relatively vast and ancient group, with more than 150,000 described species [12], [13]. Butterflies have diverged from moths some 140 MYA [14] and, despite growth in genomic resources [15], no full genome sequence has yet been made available for any species in this group. Butterflies have interesting biological properties (such as color vision and novel wing color patterns) and include many textbook examples of studies in ecology and evolution – for example, long distance migrations of monarchs [16], mimicry in Papilio and Heliconius [17], [18], [19], mutualistic relationships between lycaenids and ants [20], and wing pattern plasticity and evo-devo in Bicyclus and Junonia [21]. Bicyclus anynana has been established as a butterfly model in the study of the evolution and development of wing color pattern elements called eyespots [21], [22], [23], [24]. This species has a large collection of expressed gene sequences and the densest gene-based linkage map available to date for any butterfly species [25], [26]. A BAC library available for B. anynana [27] allows access beyond the coding regions of genes of interest, including genes involved in wing pattern formation.
Here, we analyze large genomic regions in BAC clones selected for containing 11 genes expressed during B. anynana wing development, at stages relevant for color pattern formation [25]. The selected genes include those encoding signaling molecules proposed as candidate morphogens in the induction of eyespots (Decapentaplegic, Dpp, and Wingless, Wg; [23]) as well as some of their regulators possibly responsible for pattern variation (APC-like, APC, and Naked cuticle, Nkd; [28]), transcription factors implicated in eyespot ring patterning (Distal-less, Dll, and Engrailed, En; [29], [30], [31]) as well as other transcription regulators (Apterous, Ap, and DP transcription factor, Dp), enzyme Vermilion (V) presumably involved in pigment synthesis on developing wings [32], Ecdysone receptor (EcR) involved in wing pattern plasticity [33], [34], and the antioxidation gene Superoxide dismutase 2 (Sod2; [35]). Our annotation of the BAC sequences enabled the identification of repetitive DNA and transposable genetic elements, and prediction of putative protein-coding genes, including the 11 target genes as well as the genes around them. The comparative analysis to orthologous regions in other lepidopteran species allowed us to assess fine-scale conservation of gene order (synteny) and also of nucleotide sequence in predicted protein coding and non-coding DNA.
Results and Discussion
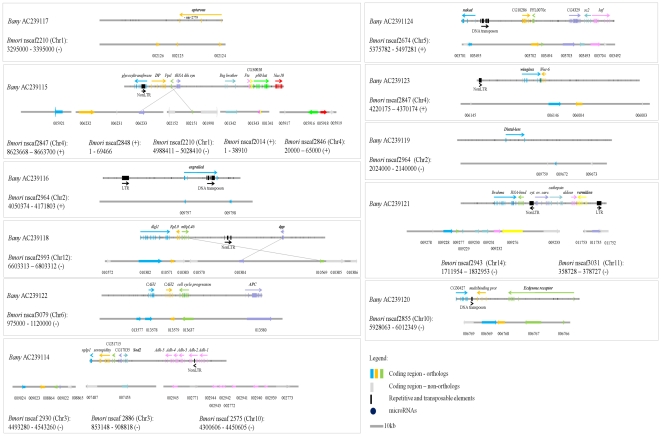
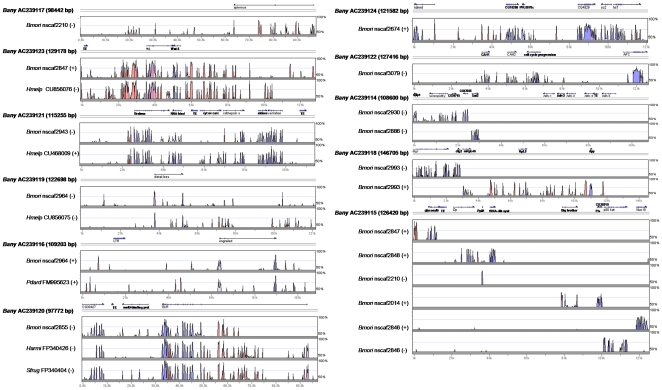
We analyzed ∼1.3 Mb of genomic sequence for the butterfly Bicyclus anynana, an emerging model in the study of wing pattern evolution and development [36]. This sequence was part of 11 BAC clones selected (from a library available for the species [27]) for containing 11 genes (Table 1) expressed in developing wings during the stages relevant for color pattern formation [25]. Assembled BAC sequences were characterized and annotated in relation to a number of criteria (see Methods), including detection of repetitive elements and prediction of protein-coding genes (Table 1, Figure 1). Comparison of gene content with the available gene-based linkage map of B. anynana [26], shows that the BACs analyzed correspond to regions on nine different chromosomes (Table 1). The annotated genomic regions were used for a comparative analysis of gene order in relation to the lepidopteran reference genome (Figure 1), and for a comparative analysis of nucleotide sequence in relation to this and other lepidopteran species with relevant sequence available (Figure 2).
Table 1. Main characteristics of the B. anynana BACs analyzed.
| BAC ID1 | Target Gene2 | Bany LG3 | Length | GC content | % repeats | TE type5 | KAIKO6 | Validated7 | # Unigenes8 | Sequence Annotation9 |
| (NCBI | CUGI | JGI) | (bp) | (%) | (S+LC)4 | BAC/cds | (%) | |||||
| AC239117 | 39A22 | 6132 | ap | Z | 98,442 | 36.1 | 0.3+1.9 | - | 35 (90) | 1(182) | 172/0 | 0.6/33.5/65.9 |
| AC239122 | 69H15 | 6133 | APC | 6 | 127,416 | 35.7 | 0.4+1.9 | - | 32 (177) | 4(678) | 163/3 | 6.5/16.9/76.6 |
| AC239115 | 19O01 | 6134 | DP | 4 | 126,420 | 34.7 | 1.2+2.6 | NonLTR/RTE-3 | 36 (183) | 10(449) | 141/23 | 10.6/33.7/55.7 |
| AC239118 | 39L19 | 6135 | dpp | 12 | 146,705 | 36.6 | 0.6+1.8 | NonLTR/DMRT1 | 52 (123) | 5(418) | 132/9 | 4.3/19.9/75.8 |
| AC239120 | 4N12 | 6136 | EcR | 10 | 97,772 | 36.3 | 0.4+2.0 | DNA/Mariner | 30 (122) | 4(401) | 180/10 | 4.9/65.9/29.2 |
| AC239116 | 23M04 | 6137 | en | 2 | 109,203 | 36.7 | 0.6+2.7 | LTR/BEL; Tc1-IS630-Pogo1 | 26 (166) | 2(1582) | 149/6 | 6.2/23.1/70.8 |
| AC239119 | 48N20 | 6138 | Dll | 2 | 122,698 | 35.4 | 0.6+2.8 | - | 45 (82) | 1(173) | 153/3 | 0.4/11.7/87.8 |
| AC239124 | 85J10 | 6139 | nkd | 5 | 121,582 | 35.3 | 0.3+2.2 | - | 28 (232) | 6(595) | 167/5 | 8.8/29.9/61.3 |
| AC239114 | 18H03 | 6140 | Sod2 | 3 | 108,600 | 36.1 | 0.6+3.0 | NonLTR/RTE-3 | 42 (147) | 11(275) | 161/16 | 7.7/32.1/60.3 |
| AC239121 | 68O14 | 6141 | v | 14 | 115,255 | 37.5 | 0.9+1.9 | NonLTR/CR1; LTR/Gypsy | 28 (225) | 8(601) | 120/22 | 12.5/42.3/45.2 |
| AC239123 | 84B11 | 6142 | Wg | 4 | 129,178 | 36.7 | 0.5+2.2 | NonLTR/RTE-1 | 44 (127) | 3(480) | 163/9 | 3.4/12.3/84.3 |
General characteristics observed for each BAC clone, including a summary of the annotation parameters discussed in the text.
BAC ID including NCBI accession number, BAC clone name from library at CUGI [27], and BAC sequence name from assembly by JGI. The CUGI and JGI names are used in the custom database [47];
Genes used to select the BACs for sequencing (see Introduction for abbreviations);
B. anynana linkage group based on mapping of the target genes (LG; [26]);
% repetitive sequence corresponding to single repeats (S) and low complexity (LC) regions as identified by RepeatMasker [41];
type of TEs, according to CENSOR [74] classification;
predicted peptides by Kaikogaas: number and (average aminoacid length);
predicted peptides after manual validation: number and (average aminoacid length);
Number of B. anynana UniGenes matching each BAC, and number corresponding to the validated predicted genes in Figure 1, cf. [47];
Percent sequence annotated as corresponding to protein-coding, intronic and intergenic DNA. Details in [47] and the supplementary files.
Figure 1. Annotation of B. anynana genomic regions and fine-scale synteny with B. mori.
Each B. anynana BAC sequence is represented, with the corresponding scaffold in B. mori (including information on chromosomal location). Each putative gene is represented by a different color: B. anynana gene names in bold correspond to those on which BAC selection was based (Table 1), and B. mori gene names reflect SilkDB annotation (e.g., 010572 is SilkDB gene BGIBMGA010572). Exons are explicitly annotated for B. anynana as stripes of the same color (darker shade for duplicated exons). Arrows indicate the direction of transcription of each gene, and fine lines are used for highlighting chromosomal rearrangements. The figure contains a legend for the representation of sequence length, and for the protein-coding genes, repetitive sequence, transposable elements, and microRNA identified in this study. Details on all B. anynana predicted peptides can be found in Table S3.
Figure 2. Conservation of DNA sequence in relation to other lepidopterans.
VISTA plots of all BAC sequences against B. mori and, when available, other lepidopterans (moths Bombyx mori, Helicoverpa armigera, Spodoptera frugipera, and butterflies Papilio dardanus, Heliconius melpomene). Regions more than 70% conserved in a 100 bp window (VISTA default settings) appear as peaks with blue corresponding to annotated protein-coding regions and red to conserved non-coding sequence. Figure S2 shows close-up and extended analysis of regions around genes wingless and Ecdysone receptor.
CG content, repetitive sequence, and mobile elements
We used a combination of web-available and custom-designed bioinformatic tools and extensive manual curation to characterize different aspects of the target genomic sequence (details in Methods). Similar to observations in other lepidopterans [10], [37], [38], the GC content was ∼36.1% for the total sequence analyzed (with some variation between regions; Table 1) and ∼45.4% for the 55 validated predicted protein-coding genes (see below). This is consistent with studies in Drosophila where functional (coding) regions exhibit higher GC content than presumably less constrained regions [39], possibly relating to the fact that preferred codons often end in C or G [40].
We used RepeatMasker [41] to identify and characterize repetitive regions, including the type and extent of different repetitive elements. We identified a total of 857 repeats larger than 20 bp, corresponding to ∼2.73% (35567 bp) of the sequence analyzed (Table 1 and S1). The majority (721) of those repetitive elements were characterized as low complexity (i.e. poly-purine/poly-pyrimidine stretches or regions of >87% AT or >89% GC), and ∼0.49% of the total genomic sequence corresponded to simple repeats (duplications of, typically, 1–5 bp). While the overall ∼2.73% estimated repetitiveness for B. anynana is lower than the >20% estimate for Heliconius butterflies [10], [37], the proportion of that corresponding to low complexity repeats is higher than estimates for other lepidopteran species and the % of simple repeats is comparable between all [10], [37], [38]. Because our estimates are based on BACs selected for carrying specific genes, rather than sequence from randomly-selected BACs, it avoids gene-less regions and might under-estimate the extent of repetitiveness in the whole genome. Aside the repetitive elements identified by RepeatMasker, we also specifically looked for the nine novel types of repeated elements identified in other model butterflies [10] (see Methods). We found sequence similar to two of these in the available B. anynana genomic sequence (Table 1), and also in nucleotide sequences available for other lepidopteran species in NCBI's sequence depository (Table S2).
Using a combination of tools (RepeatMasker, Kaikogaas, CENSOR and manual BLAST; see Methods), we identified sequences related to transposable elements (TEs; Table 1). These included DNA transposons (Tc1-IS630-Pogo and DNA/Mariner), retroelements encoding for a reverse transcriptase (three NonLTR/RTEs, one NonLTR/DMRT and one NonLTR/CR1), and two LTR-retrotransposons (one LTR/BEL and one LTR/Gypsy) (see Table 1 and Figure 1). Only one of these nine TEs (Tc1-IS630-Pogo in BAC AC239116) was identified by RepeatMasker, and was, thus, within the sequence that was masked before further annotation (see Methods). This element is, thus, not in Figure 1 or Table S3. Some of the TEs identified are located inside introns (e.g., in glycosyltransferase in AC239115, and Adh-4 in AC239114; Figure 1), and some are located near areas where synteny between B. anynana and B. mori is disrupted (e.g., inversion in AC239118, and transposition of cytosolic ovarian carcinoma antigen 1 in AC239121; Figure 1). This is especially interesting because TEs are thought to play an important role in genome evolution, including contributing to chromosomal rearrangements [42] and to the appearance of new exons and introns (albeit possibly to a lesser degree in invertebrates [43]).
In silico annotation and manual curation of protein-coding genes
For the in silico gene prediction we chose to use Kaikogaas [44], a web-available tool designed for annotation of genomic sequence of Bombyx mori, the lepidopteran reference [45]. This resulted in a total of 398 predicted peptides (Table 1 and S3). Of these, fewer than 10% (38) had any type of annotation (i.e. a putative gene name or function) beyond “hypothetical protein” (HP). To identify potential false positives and other issues with the in silico predictions, we manually curated the list of 398 Kaikogaas-derived genes extensively (details in Methods). In our conservative validation procedure we started by dismissing 111 predicted peptides shorter than 60 amino acids. We then used BLAST to check the remainder for sequence similarity in relation to relevant publicly-available gene collections. Only 55 predicted peptides (including the eight TEs not identified by RepeatMasker; see above) had significant similarity with proteins on NCBI and were kept for further analysis (Table 1, Figure 1 and Table S3). The curated 55 predicted genes in the 11 BAC sequences correspond to an average of 4.2 genes per 100 Kb which, despite our very conservative manual curation, falls well within published estimates for other insects (Figure S1), and is greater than that for other lepidopterans (∼2.5 genes/100 Kb in Heliconius butterflies [10] and ∼3.4 genes/100 Kb in the silkworm B. mori [46]). Note that gene density in the B. mori scaffolds orthologous to the available B. anynana sequence (the regions represented in Figure 1) is 4.5 genes/100 Kb, which is very close to that in B. anynana. Our manual curation strategy, designed to dismiss false positives at the expense of possibly generating false negatives, is expected to generate a conservative estimate of gene number. On the other hand, gene density in genomic regions selected for containing specific protein coding genes is probably higher than that in the whole genome as selection of gene-containing BACs avoids possible “gene deserts”.
The manual curation also allowed a more in-depth annotation of the predicted protein-coding genes. By annotating individual predicted exons we: 1) established that 41 of the 55 predicted genes had the complete putative coding sequences, 2) identified errors with the automated annotation process whereby exons of the same gene had been identified as different genes (e.g., Kaikogaas' HP12 gene in AC239120 corresponds to one of the exons of Ecdysone receptor; Table S3), and 3) highlighted instances of exon duplications, including cases of duplicated exons with (exons 11–15 of p80 katanin in AC239115, and Brahma in AC239121) and without (exons 2 and 3 of Sod2 in AC239114, and exons 5 and 6 of cell cycle progression in AC239122) non-sense mutations (Table S3).
Validation of in silico gene prediction using expressed sequence data
To match in silico gene predictions with expression data (see Methods), we used all B. anynana UniGenes (unique genes) from the available assembly of EST sequences [25], [26]. The results of this analysis are displayed graphically in a custom web-available database [47]. This approach allowed us not only to assess our annotation of protein-coding sequence, but also to assess EST assembly. We identified EST-derived UniGenes for most of our predicted peptides (Table S3), but also many UniGenes matching genomic regions with no predicted peptide (see [47]). We identified EST-derived UniGenes for 44 of the 55 predicted protein-coding genes; 17 with a single UniGene match and 27 with two to seven. In 23 of the 27 cases with more than one UniGene corresponding to the same predicted peptide, these UniGenes were at least partly overlapping. This reflects under-assembly of the ESTs, possibly due to polymorphisms in these sequences [25], [26], [48]. On the other hand, UniGenes in regions with no predicted peptides presumably correspond to false negatives of our conservative annotation. For example, Kaikogaas-predicted genes HP7 and HP35 in AC235115 and HP13 in AC239121 were discarded in the manual curation process (including one case under the 60-aminoacids long threshold). One UniGene corresponding to a putative transposable element not identified by Kaikogaas was detected in two BAC clones (AC239116 and AC239124). Still, the majority of UniGenes that did not match any predicted gene seemed to fall in repetitive regions identified and masked by RepeatMasker.
Alcohol dehydrogenase expansion and sequence similar to lepidopteran miRNAs
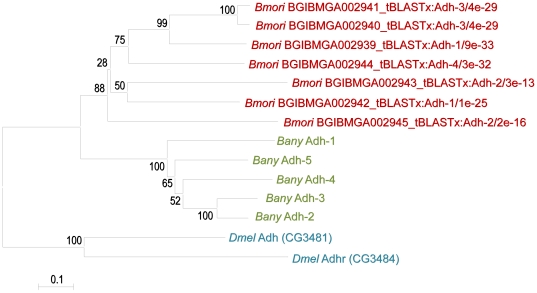
We identified an interesting case of gene duplication. Among the predicted genes, we identified five putative Alcohol dehydrogenase (Adh) genes in tandem in BAC AC239114 (Figure 1). BLAST analysis (see Methods) allowed us to identify seven orthologs in chromosome 10 (BGIBMGA002939-45; minimal e-value 1e-30) of the B. mori lepidopteran reference genome, also annotated as Adh genes [49]. Phylogenetic analysis of the 12 corresponding Adh proteins, and including the Adh and Adh-related proteins of D. melanogaster as outgroups (see Methods), showed that all B. anynana paralogs cluster together and separated from the cluster of B. mori paralogs (Figure 3). This pattern of higher similarities within than between species may result from either of two types of scenarios: 1) independent duplications of Adh having occurred after the separation of the lineages of B. mori and B. anynana, or 2) duplications having occurred prior to the split of the lineages with subsequent concerted evolution of paralogs (gene conversion; see [50]). Unlike what has been shown for converted duplicates in other species [51], CG content in the five B. anynana Adh genes (average ± standard for entire loci is 36.1%±2.3%) and in the remaining 50 predicted genes (37.6%±6.2%) is not significantly different (t-test t = 0.521, df = 53, p = 0.60). This, however, does not distinguish between the scenarios above. More and other types of data are necessary for such purpose, and also for unravelling the ecological value of this gene expansion [52].
Figure 3. Phylogenetic tree of Adh genes.
Neighbour-joining, unrooted tree reconstructed with MEGA 4 using the aminoacid sequence of the putative Adh genes in B. anynana (Bany, in green), together with the corresponding paralogs from chromosome 10 in B. mori (Bmori, in red, showing Silkdb gene accessions and BLAST results) and D. melanogaster (Dmel, in blue, showing FlyBase gene accessions). Numbers are bootstrap values for 1000 replicates.
We used BLAST to search for B. anynana genomic sequence similar to mature miRNA sequences (∼22 nt) from B. mori available in miRBase [53], [54] and from H. melpomene [55] (see Methods). We identified a putative B. anynana miRNA similar to one of the 487 B. mori miRNAs (in BAC AC239117, Figure 1). Its sequence was 95% and 100% identical to that of its B. mori and H. melpomene counterparts, respectively. Prediction of miRNAs based on sequence conservation remains of limited value and needs experimental validation. The extent to which miRNAs are conserved is only now starting to be characterized. For example, only 68 of 257 B. mori miRNAs were found to be conserved with other species (23 with other vertebrates and invertebrates, 13 limited to invertebrates and 32 limited to insects [54]), and 430 of 447 chicken miRNAs were considered to be exclusive to the avian lineage [56]. In Drosophila, where species from the Sophophora and Drosophila subgenus diverged ∼62 MYA [57], only 28 of 59 D. melanogaster miRNA were conserved throughout the phylogeny of those species with fully sequenced genomes [58].
Comparative analysis of gene order: fine-scale synteny with B. mori
The comparative analysis of gene order in the target B. anynana genomic regions (protein-coding gene annotation obtained as explained above) and the corresponding B. mori orthologous regions over 18 scaffolds (annotation available from SilkDB [49]) is represented in Figure 1. Note that both species have the same number of chromosomes and that B. anynana linkage groups were numbered following orthology with B. mori [26]. Some of the predicted 55 genes were excluded for a quantification of the synteny conservation: 1) three genes found isolated in three of the B. anynana BACs (AC239117, AC239116 and AC239119), 2) two genes whose B. mori orthologs were isolated in one scaffold (note that you need at least three gene pairs consecutive genes to assess conservation of order), and 3) the eight transposable elements in Figure 1. Out of 42 orthologous pairs analyzed, 36 genes (∼86%) are in the same order in both species. We also identified three B. mori genes (gray arrows in Figure 1) not represented in the corresponding B. anynana region (confirmed by running tBLASTn and tBLASTx of the B. mori sequence against the B. anynana BAC). One of these B. mori genes (BGIBMGA006231) presumably encodes a 57 amino acid protein with no SilkDB annotation [49] and might be a “false positive”. The other two (BGIBMGA010383 and BGIBMGA010570), however, are annotated and presumably encode longer peptides (776 and 304 amino acids, respectively). We used different BLAST algorithms to confirm that their absence in the corresponding B. anynana BAC was not the result of loss during our stringent manual curation. Interestingly, all three genes are associated with chromosomal rearrangements; one transposition in AC239115 and one inversion in AC239118 (Figure 1).
Previous results comparing genetic or physical maps had shown that chromosomal gene composition and gene order is highly conserved between butterflies and moths, but also revealed instances of chromosomal rearrangements [26], [59]. A fine-scale comparison of genomic sequence in ca. 420 Kb sequence between the butterfly Heliconius erato and the moth B. mori showed conserved gene order and distances for ca. 90% of the annotated protein-coding genes [10]. Here, in ca. 1303 Kb of sequence compared between B. anynana and B. mori, we detected smaller levels of conservation and identified 1) small scale inversions: inverted order of different genes (e.g., mRpL46 and dpp in AC239118) and inverted direction of single genes (e.g., p80 katanin in AC239115), and 2) transpositions: with a number of genes assigned to non-orthologous chromosomes (e.g., cytosolic ovarian carcinoma antigen 1 gene in AC239121 is in chromosome 11 in B. anynana and chromosome 14 in B. mori) and with the orthologs for the nine putative genes in AC239115 (B. anynana chromosome 1) distributed over five B. mori scaffolds, including regions in chromosome 4 and regions not yet assigned to a B. mori chromosome. Inversions and transpositions have been well documented between the sequenced genomes of Drosophila species [60], [61], [62] and are known to have played an important role in the evolution of their chromosomes.
Comparative sequence analysis highlights conserved non-coding regions
Of the ca. 1,303 Kb of B. anynana genomic sequence analyzed, 6% corresponded to estimated coding sequence (cds), while 28% and 66% corresponded to predicted intronic and intergenic regions, respectively (Table 1). Using VISTA [63], [64], we compared the genomic sequence available for B.anynana with that of orthologous regions (identified as described in the Methods section) of B. mori and other Lepidoptera. The results are displayed in Figure 2. Note that for the putative Adh genes, even though the B anynana and B. mori copies showed conservation at the protein level (Figure 3), the VISTA software was unable to identify nucleotide sequence conservation in the corresponding regions. For all B. anynana regions compared, VISTA estimated a total of 4.3% nucleotide identity with B. mori (and 1.3% with butterflies Papilio dardanus, and 6.4% with Heliconius melpomene, for available sequence), with 2.8% in predicted coding region, 0.4% in introns, and 1.1% in intergenic regions (Table 1). Conserved sequence regions correspond largely to the location of putative exons (blue areas in Figure 2), but, in some instances, also to putative non-protein coding regions (red areas). Non-protein-coding DNA forms the majority of the genomes of many multicellular eukaryotes (e.g. ∼80% of noncoding DNA in Drosophila [65]), and ∼99% in humans [66]), and is known to be functionally important in many respects (e.g. for the regulation of gene expression and chromosome packaging). Conservation of non-coding sequence is often taken as a sign of possible functional importance. Different studies found considerable levels of conservation of non-coding sequence (e.g. between pairs of Drosophila species [67], [68], diverged no more than 62 MYA [57]) and constraints on intergenic regions have been estimated to possibly be as high as 60% [69].
The regions of significant conservation of non-coding sequence are heterogeneously distributed in the neighborhood of different genes (Figure 2). In fact, we see high levels of conservation around some (notably, wingless and Ecdysone receptor) targeted “developmental genes”, but not all (e.g. around Distal-less and engrailed). Like has been suggested for Drosophila where levels of selective constrain (and putative functional role) appear to correlate with intron length [70], B. anynana wingless and Ecdysone receptor have relatively large introns (∼13 Kb and ∼50 Kb, respectively), compared to the average of ∼6.5 Kb for the other 46 non-intronless predicted genes (their intron length ranging from 50 bp in the putative DNA/Mariner element to ∼33 Kb for the gene encoding transcription factor apterous). The high degree of conservation of non-coding sequence around wingless and Ecdysone receptor between B. anynana and other lepidopterans (∼78 MYA for B. anynana and H. melpomene [71]), and more than 140 MYA for B. anynana and B. mori [14]) is lost when comparing B. anynana to species in other insect orders (Figure S2). It is noteworthy that wingless and Ecdysone receptor are highly pleiotropic genes involved in a multitude of developmental processes across in multiple species, and which, in B. anynana are thought to be associated to the formation of characteristic wing color pattern elements [28] and seasonal polyphenism [72]. It will be interesting to extend this analysis to more lepidopteran species, including closer relatives with comparable wing pattern properties, and to explore the functional role of the conserved non-coding sequence experimentally.
Overview and conclusions
We analyzed 11 BAC clones of the butterfly Bicyclus anynana, selected for the presence of key genes expressed during wing development at stages relevant for wing color pattern formation [25]. We have identified different genes in these regions, corresponding to gene densities similar to other lepidopterans. Among the genes identified, we discovered five tandemly arranged genes similar to Alcohol dehydrogenase (Adh), which potentially represent an expansion in the Lepidoptera. Comparative studies of sequence, expression and function of these genes are necessary to shed light onto their evolutionary history and ecological importance.
Our comparative analysis of the B. anynana genomic regions with the orthologous regions of the lepidopteran reference genome allowed assessment of conservation of fine-scale gene order and of DNA sequence. We detected strong synteny but 1) also multiple events where it was disrupted by different chromosomal rearrangements, including inversions and transpositions not detected with a previous comparative analysis of B. anynana versus B. mori linkage maps [26], and 2) lower proportion of genes in conserved order that a previous smaller-scale, fine-resolution analysis comparing Heliconius erato BAC-derived sequence with B. mori scaffolds [10]. We also detected instances of unusual high conservation of non-coding regions, in particular, around the genes encoding for Ecdysone Receptor and Wingless, between species diverged some 140 MYA. Understanding the functional significance of these regions will allow for a better understanding of the evolution and diversification of living organisms.
Materials and Methods
BAC sequences and target genes
We analyzed over 1,303,271 bp of nucleotide sequence from 11 B. anynana BAC clones deposited on GenBank (AC239114–AC239124). These BAC clones were obtained from the 9× coverage B. anynana BAC library available at Clemson University Genomics Institute (CUGI [27]) and were Sanger sequenced at 8–9× depth and their sequences assembled into 11 individual scaffolds by the Joint Genome Institute (JGI [73]), as part of the Community Sequencing Program FY2006. The sequenced BAC clones were originally selected by screening the BAC library filters with radioactively-labeled probes against 11 genes of interest (Table 1), expressed in developing wings [25].
Annotation of genomic sequence
For the annotation of the BAC sequences aimed at characterizing different aspects of their genomic composition, we used a combination of web-available and custom-designed bioinformatic tools, as well as manual curation. We also used sequence information from orthologous regions in other species, with emphasis on the silkworm B. mori, the lepidopteran genomic reference.
RepeatMasker [41] with CrossMatch search algorithm (default settings) was used to identify repetitive regions, including the type and extent of different repetitive elements. The repetitive regions identified in this way were masked before all further analysis. Also, using BLASTn (e-value cut-off of 1e-5) against the B. anynana BAC sequences, we searched for nine transposable element families recently identified in another butterfly [10]. CENSOR [74] was used for classifying the identified putative transposable elements in the different classes (nonLTRs, LTRs, and DNA transposons) and, when possible, families.
In silico gene prediction was done for each complete BAC sequence using Kaikogaas (a web-available tool customized for B. mori genomic data; [44]). Kaikogaas' output is a graphical display (Figure S3) and a list of all putative proteins identified (Table S3) in each target BAC. This list was extensively manually curated for validation of Kaikogaas' predictions and for further annotation. Manual curation involved a sequence of steps: 1) hypothetical proteins shorter than 60 amino acids were dismissed; 2) predicted peptides longer than 60 amino acid were used to search for similarity with the complete collection of non-redundant insect protein sequences (this collection consisted of 913470 entries when our analysis was run in 2010) using BLASTp (e-value cut-off of 1e-10) and best hits were used for gene identification; 3) the amino acid sequences corresponding to these proteins were then used for running BLASTp (default settings) against the Bombyx mori genome in SilkDB [49], from where the corresponding scaffolds were downloaded (assembly of August 2010) and used for later comparison of genomic sequence around predicted genes (see below); 4) to attempt to confirm the putative exons of each hypothetical gene, we used homologs from both B. mori (from SilkDB [49]) and D. melanogaster (from Flybase [75]) to run tBLASTx (default settings) against the B. anynana BAC sequences.
BAC annotation was also done using a custom-built platform [47] which, aside repetitive regions and orthologs to genes in relevant public databases, also uses the complete list of UniGenes identified from the assembly of a large collection of B. anynana ESTs [25], [26]. BLAST results comparing the sequences of each complete BAC to the UniGene collection were deposited in a database queried by the GBrowse interface [76]. This analysis allowed, on the one hand, validation of the in silico prediction of exons, and, on the other, an assessment of EST assembly.
For the identification of conserved putative microRNAs (miRNAs), we ran BLASTn analysis (e-value cut-off of 1E-3; word size of 15) using as query against the B. anynana BAC sequences, the mature sequence (∼22 nt) of the 487 miRNA sequences of B. mori (available at miRBase [53], [54]) and the recently identified miRNAs from Heliconius melpomene butterflies [55].
Comparative analysis of gene order and genomic sequence
To compare fine-scale physical linkage and gene order between B. anynana and B. mori, we used our annotation of the B. anynana BAC sequences (obtained as explained above) and the available annotation (SilkDB; [49]) for the corresponding B. mori scaffolds (identified as explained above, based on BLAST of predicted peptides during the manual curation).
To investigate nucleotide sequence conservation between B. anynana and other lepidopterans, we used VISTA (default settings: 70% identity, 100 bp window; [63], [64]) comparing whole B. anynana BACs with the corresponding genomic regions of other species. For B. mori these were obtained from the SilkDB scaffolds based on sequence similarity with the predicted B. anynana genes. For other species, they were obtained from BAC sequences available on NCBI, based on discontinuous megablast (default settings) of the complete B. anynana BAC sequence. Pairwise alignments between sequence for B. anynana sequence and each of the available corresponding sequences obtained were performed on the mVISTA program using the Avid alignment algorithm (default settings), which globally aligns DNA sequences of arbitrary length [77]. For the quantification of sequence conservation, we considered only the regions comprised between the first and last orthologous genes in each BAC that matched a single scaffold in the other species. The genomic regions around EcR, wg and Wnt-6 (plus 15 Kb upstream and downstream) from Drosophila melanogaster, Apis mellifera and Tribolium castaneum were downloaded from Flybase [75], Beebase [78] and BeetleBase [79], [80], respectively. They were used for comparison with the same regions in B. anynana (AC239120 and AC239123) using VISTA (Figure S2).
Phylogenetic analysis of Adh genes
Putative Adh genes from B. anynana (AC239114) were used for running tBLASTn and tBLASTx analysis against B. mori nucleotide and protein collection in SilkDB. The first hits corresponded to annotated Adh genes located at chromosome 10 of the silkworm. The corresponding proteins were downloaded, together with the ADH and ADHR (Adh-related) proteins of D. melanogaster from Flybase. These were used, together with the amino acid sequence of the putative B. anynana genes, for phylogenetic reconstruction using MEGA version 4 [81] (Neighbor-joining with pairwise deletion and bootstrap).
Supporting Information
Quantification of repetitive sequence in target B. anynana BACs. Identification and characterization of repeated regions was done with RepeatMasker (see Methods). Simple Repeats correspond to duplications of, typically 1–5bases. Low Complexity Sequence corresponds to poly-purine/poly-pyrimidine stretches or regions of >87% AT or >89% GC.
(PDF)
Heliconius repetitive elements identified in the target B. anynana BACs. Two of the novel repetitive elements identified in Heliconius [10] appear to be present in our target B. anynana genomic sequence, as well as in publicly available nucleotide sequence for other lepidopterans (cf. BLASTn analysis; see Methods).
(PDF)
List of Kaikogaas predicted genes and subsequent manual curation.
(TXT)
Gene density and genome size in insects. Gene density (number of genes per 100 Kb; cf. [10], [46], [82], [83], [84], [85], [86], [87], [88] in relation to genome size for different insect species (cf. [89]). Circles correspond to species where gene densities were estimated based on sequenced genomes – note that the size of assembled genome can differ from the estimates in this Figure. Other symbols correspond to species where gene density was estimated based on a few BAC clone sequences – including this paper for B. anynana.
(PDF)
Comparative analysis of the EcR and wg/Wnt-6 genomic regions. VISTA plots of the genomic regions comprising the genes EcR (BAC AC239120) and wg/Wnt-6 (BAC AC239123) with the orthologous regions in other insects with relevant sequence available: Heliconius melpomene, Bombyx mori, Helicoverpa armigera, Spodoptera frugiperda, Drosophila melanogaster, Tribolium castaneum, and Apis mellifera.
(PDF)
Kaikogaas graphical output of BAC annotation.
(PDF)
Acknowledgments
We thank Nicolien Pul, Surya Chhun, and Suzanne Saenko for technical help in the earlier stages of BAC selection, Antónia Monteiro for providing the probes for BAC selection of engrailed, Clemson University Genomics Institute for access to BAC library filters, and the Joint Genomes Institute for the sequencing and assembly of the BAC clones.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by research funds to PB from the Portuguese Foundation for Science and Technology FCT (http://alfa.fct.mctes.pt; reference PTDC/BIA-BDE/65295/2006) and the Dutch Scientific Organization NWO (http://www.nwo.nl/nwohome.nsf/pages/SPPD_5R2QE7_Eng; reference VIDI 864.08.010). The sequencing of the 11 BAC clones was performed under the auspices of the United States Department of Energy's Office of Science, Biological and Environmental Research Program, and by the University of California, Lawrence Berkeley National Laboratory under contract no. DE-AC02-05CH11231; Lawrence Livermore National Laboratory under contract no. DE-AC52-07NA27344; and Los Alamos National Laboratory under contract no. DE-AC02-06NA25396 (http://www.jgi.doe.gov/sequencing/why/3112.html). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Clark AG, Eisen MB, Smith DR, Bergman CM, Oliver B, et al. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- 2.Kellis M, Patterson N, Endrizzi M, Birren B, Lander ES. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423:241–254. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- 3.Stark A, Lin MF, Kheradpour P, Pedersen JS, Parts L, et al. Discovery of functional elements in 12 Drosophila genomes using evolutionary signatures. Nature. 2007;450:219–232. doi: 10.1038/nature06340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.NCBI National Center for Biotechnology Information. Genome sequencing projects statistics. Available: http://www.ncbi.nlm.nih.gov/genomes/static/gpstat.html. Accessed 2011 Aug 14.
- 5.Yamamoto K, Nohata J, Kadono-Okuda K, Narukawa J, Sasanuma M, et al. A BAC-based integrated linkage map of the silkworm Bombyx mori. Genome Biol. 2008;9:R21. doi: 10.1186/gb-2008-9-1-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Song X, Goicoechea JL, Ammiraju JS, Luo M, He R, et al. The 19 genomes of Drosophila: a BAC library resource for genus-wide and genome-scale comparative evolutionary research. Genetics. 2011;187:1023–1030. doi: 10.1534/genetics.111.126540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang DX. Lepidopteran microsatellite DNA: redundant but promising. Trends Ecol Evol. 2004;19:507–509. doi: 10.1016/j.tree.2004.07.020. [DOI] [PubMed] [Google Scholar]
- 8.Mita K, Kasahara M, Sasaki S, Nagayasu Y, Yamada T, et al. The genome sequence of silkworm, Bombyx mori. DNA Res. 2004;11:27–35. doi: 10.1093/dnares/11.1.27. [DOI] [PubMed] [Google Scholar]
- 9.Xia QY, Zhou ZY, Lu C, Cheng DJ, Dai FY, et al. A draft sequence for the genome of the domesticated silkworm (Bombyx mori). Science. 2004;306:1937–1940. doi: 10.1126/science.1102210. [DOI] [PubMed] [Google Scholar]
- 10.Papa R, Morrison CM, Walters JR, Counterman BA, Chen R, et al. Highly conserved gene order and numerous novel repetitive elements in genomic regions linked to wing pattern variation in Heliconius butterflies. BMC Genomics. 2008;9:345. doi: 10.1186/1471-2164-9-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wahlberg N, Wheat CW. Genomic outposts serve the phylogenomic pioneers: designing novel nuclear markers for genomic DNA extractions of Lepidoptera. Syst Biol. 2008;57:231–242. doi: 10.1080/10635150802033006. [DOI] [PubMed] [Google Scholar]
- 12.Roe AD, Weller SJ, Baixeras-Almela J, Brown J, Cummings MP, et al. Evolutionary framework for Lepidoptera model systems. In: Goldsmith M, Marec F, editors. Genetics and Molecular Biology of Lepidoptera. Boca Raton: Taylor and Francis; 2010. pp. 1–24. [Google Scholar]
- 13.Kristensen NP, Skalski AW. Paleontology and phylogeny. In: Kristensen NP, editor. Lepidoptera, Moths and Butterflies 1. Walter de Gruyter, Berlin & New York: Handbuch der Zoologie/Handbook of Zoology IV/35; 1998. pp. 7–25. [Google Scholar]
- 14.Timetree, The Timescale of Life. Available: http://www.timetree.org/time_query.php?taxon_a=7090Bombyx&taxon_b=110367. Accessed 2011 Aug 14.
- 15.Beldade P, McMillan WO, Papanicolaou A. Butterfly genomics eclosing. Heredity. 2008;100:150–157. doi: 10.1038/sj.hdy.6800934. [DOI] [PubMed] [Google Scholar]
- 16.Brower LP. Monarch butterfly orientation: Missing pieces of a magnificent puzzle. Journal of Experimental Biology. 1996;199:93–103. doi: 10.1242/jeb.199.1.93. [DOI] [PubMed] [Google Scholar]
- 17.Nijhout HF. Polymorphic mimicry in Papilio dardanus: mosaic dominance, big effects, and origins. Evolution & Development. 2003;5:579–592. doi: 10.1046/j.1525-142x.2003.03063.x. [DOI] [PubMed] [Google Scholar]
- 18.Counterman BA, Araujo-Perez F, Hines HM, Baxter SW, Morrison CM, et al. Genomic hotspots for adaptation: the population genetics of Mullerian mimicry in Heliconius erato. PLoS Genet. 2010;6:e1000796. doi: 10.1371/journal.pgen.1000796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baxter SW, Nadeau NJ, Maroja LS, Wilkinson P, Counterman BA, et al. Genomic hotspots for adaptation: the population genetics of Mullerian mimicry in the Heliconius melpomene clade. PLoS Genet. 2010;6:e1000794. doi: 10.1371/journal.pgen.1000794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Youngsteadt E, Devries PJ. The effects of ants on the entomophagous butterfly caterpillar Feniseca tarquinius, and the putative role of chemical camouflage in the Feniseca-ant interaction. J Chem Ecol. 2005;31:2091–2109. doi: 10.1007/s10886-005-6079-2. [DOI] [PubMed] [Google Scholar]
- 21.Beldade P, Brakefield PM. The genetics and evo-devo of butterfly wing patterns. Nat Rev Genet. 2002;3:442–452. doi: 10.1038/nrg818. [DOI] [PubMed] [Google Scholar]
- 22.Saenko SV, Marialva MS, Beldade P. Involvement of the conserved Hox gene Antennapedia in the development and evolution of a novel trait. Evodevo. 2011;2:9. doi: 10.1186/2041-9139-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Monteiro A, Glaser G, Stockslager S, Glansdorp N, Ramos D. Comparative insights into questions of lepidopteran wing pattern homology. BMC Dev Biol. 2006;6:52. doi: 10.1186/1471-213X-6-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beldade P, French V, Brakefield PM. Developmental and genetic mechanisms for evolutionary diversification of serial repeats: eyespot size in Bicyclus anynana butterflies. J Exp Zoolog B Mol Dev Evol. 2008;310:191–201. doi: 10.1002/jez.b.21173. [DOI] [PubMed] [Google Scholar]
- 25.Beldade P, Rudd S, Gruber JD, Long AD. A wing Expressed Sequence Tag resource for Bicyclus anynana butterflies, an evo-devo model. BMC Genomics. 2006;7:130. doi: 10.1186/1471-2164-7-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beldade P, Saenko SV, Pul N, Long AD. A gene-based linkage map for Bicyclus anynana butterflies allows for a comprehensive analysis of synteny with the lepidopteran reference genome. PLoS Genet. 2009;5:e1000366. doi: 10.1371/journal.pgen.1000366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.CUGI Ordering System. Available: https://www.genome.clemson.edu/cgi-bin/orders?&page=productGroup&service=bacrc&productGroup=421. Accessed 2011 Aug 14.
- 28.Saenko SV, Brakefield PM, Beldade P. Single locus affects embryonic segment polarity and multiple aspects of an adult evolutionary novelty. BMC Biol. 2010;8:111. doi: 10.1186/1741-7007-8-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brakefield PM, Gates J, Keys D, Kesbeke F, Wijngaarden PJ, et al. Development, plasticity and evolution of butterfly wing patterns. Nature. 1996;384:236–242. doi: 10.1038/384236a0. [DOI] [PubMed] [Google Scholar]
- 30.Keys DN, Lewis DL, Selegue JE, Pearson BJ, Goddrich LV, et al. Recruitment of a hedgehog regulatory circuit in butterfly eyespot evolution. Science. 1999;283:532–534. doi: 10.1126/science.283.5401.532. [DOI] [PubMed] [Google Scholar]
- 31.Brunetti CR, Selegue JE, Monteiro A, French V, Brakefield PM, et al. The generation and diversification of butterfly eyespot color patterns. Curr Biol. 2001;11:1578–1585. doi: 10.1016/s0960-9822(01)00502-4. [DOI] [PubMed] [Google Scholar]
- 32.Beldade P, Brakefield PM, Long AD. Generating phenotypic variation: prospects from “evo-devo” research on Bicyclus anynana wing patterns. Evolution & Development. 2005;7:101–107. doi: 10.1111/j.1525-142X.2005.05011.x. [DOI] [PubMed] [Google Scholar]
- 33.Koch PB, Brakefield PM, Kesbeke F. Ecdysteroids control eyespot size and wing color pattern in the polyphenic butterfly Bicyclus anynana (Lepidoptera: Satyridae). Journal of Insect Physiology. 1996;42:223–230. [Google Scholar]
- 34.Koch PB, Merk R, Reinhardt R, Weber P. Localization of ecdysone receptor protein during colour pattern formation in wings of the butterfly Precis coenia (Lepidoptera : Nymphalidae) and co-expression with Distal-less protein. Development Genes and Evolution. 2003;212:571–584. doi: 10.1007/s00427-002-0277-5. [DOI] [PubMed] [Google Scholar]
- 35.Pijpe J, Pul N, van Duijn S, Brakefield PM, Zwaan BJ. Changed gene expression for candidate ageing genes in long-lived Bicyclus anynana butterflies. Exp Gerontol. 2011;46:426–434. doi: 10.1016/j.exger.2010.11.033. [DOI] [PubMed] [Google Scholar]
- 36.Brakefield PM, Beldade P, Zwaan BJ. The African butterfly Bicyclus anynana: a model for evolutionary genetics and evolutionary developmental biology. In: Behringer RR, Johnson AD, Krumlauf RE, editors. Emerging Model Organisms: A Laboratory Manual. 2010/02/12 ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2009. 604. [DOI] [PubMed] [Google Scholar]
- 37.Wu C, Proestou D, Carter D, Nicholson E, Santos F, et al. Construction and sequence sampling of deep-coverage, large-insert BAC libraries for three model lepidopteran species. BMC Genomics. 2009;10:283. doi: 10.1186/1471-2164-10-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.d'Alencon E, Sezutsu H, Legeai F, Permal E, Bernard-Samain S, et al. Extensive synteny conservation of holocentric chromosomes in Lepidoptera despite high rates of local genome rearrangements. Proc Natl Acad Sci U S A. 2010;107:7680–7685. doi: 10.1073/pnas.0910413107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Singh ND, Arndt PF, Clark AG, Aquadro CF. Strong evidence for lineage and sequence specificity of substitution rates and patterns in Drosophila. Mol Biol Evol. 2009;26:1591–1605. doi: 10.1093/molbev/msp071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marais G, Mouchiroud D, Duret L. Does recombination improve selection on codon usage? Lessons from nematode and fly complete genomes. Proc Natl Acad Sci U S A. 2001;98:5688–5692. doi: 10.1073/pnas.091427698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smit AFA, Hubley R, Green P. 1996–2004. RepeatMasker Open-3.0 at http://repeatmasker.org.
- 42.González J, Petrov DA. The adaptive role of transposable elements in the Drosophila genome. Gene. 2009;448:124–133. doi: 10.1016/j.gene.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sela N, Kim E, Ast G. The role of transposable elements in the evolution of non-mammalian vertebrates and invertebrates. Genome Biol. 2010;11:R59. doi: 10.1186/gb-2010-11-6-r59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shimomura M, Minami H, Suetsugu Y, Ohyanagi H, Satoh C, et al. KAIKObase: an integrated silkworm genome database and data mining tool. BMC Genomics. 2009;10:486. doi: 10.1186/1471-2164-10-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goldsmith MR. Recent progress in silkworm genetics and genomics. In: Goldsmith MR, Marec F, editors. Molecular Biology and Genetics of the Lepidoptera. Boca Raton: CRC Press; 2010. pp. 25–47. [Google Scholar]
- 46.Consortium ISG. The genome of a lepidopteran model insect, the silkworm Bombyx mori. Insect Biochem Mol Biol. 2008;38:1036–1045. doi: 10.1016/j.ibmb.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 47.Bicyclus anynana BAC Database. Available: http://cstern.bio.uci.edu/cgi-bin/gbrowse/BanyBACsFinal/. Accessed 2011 Aug 14.
- 48.Long AD, Beldade P, Macdonald SJ. Estimation of population heterozygosity and library construction-induced mutation rate from expressed sequence tag collections. Genetics. 2007;176:711–714. doi: 10.1534/genetics.106.063610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang J, Xia Q, He X, Dai M, Ruan J, et al. SilkDB: a knowledgebase for silkworm biology and genomics. Nucleic Acids Res. 2005;33:D399–402. doi: 10.1093/nar/gki116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nei M, Rooney AP. Concerted and birth-and-death evolution of multigene families. Annu Rev Genet. 2005;39:121–152. doi: 10.1146/annurev.genet.39.073003.112240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Casola C, Ganote CL, Hahn MW. Nonallelic gene conversion in the genus Drosophila. Genetics. 2010;185:95–103. doi: 10.1534/genetics.110.115444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ashburner M. Speculations on the subject of alcohol dehydrogenase and its properties in Drosophila and other flies. BioEssays. 1998;20:949–954. doi: 10.1002/(SICI)1521-1878(199811)20:11<949::AID-BIES10>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 53.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu S, Li D, Li Q, Zhao P, Xiang Z, et al. MicroRNAs of Bombyx mori identified by Solexa sequencing. BMC Genomics. 2010;11:148. doi: 10.1186/1471-2164-11-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Surridge AK, Lopez-Gomollon S, Moxon S, Maroja LS, Rathjen T, et al. Characterisation and expression of microRNAs in developing wings of the neotropical butterfly Heliconius melpomene. BMC Genomics. 2011;12:62. doi: 10.1186/1471-2164-12-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Glazov EA, Cottee PA, Barris WC, Moore RJ, Dalrymple BP, et al. A microRNA catalog of the developing chicken embryo identified by a deep sequencing approach. Genome Res. 2008;18:957–964. doi: 10.1101/gr.074740.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tamura K, Subramanian S, Kumar S. Temporal patterns of fruit fly (Drosophila) evolution revealed by mutation clocks. Mol Biol Evol. 2004;21:36–44. doi: 10.1093/molbev/msg236. [DOI] [PubMed] [Google Scholar]
- 58.Ruby JG, Stark A, Johnston WK, Kellis M, Bartel DP, et al. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res. 2007;17:1850–1864. doi: 10.1101/gr.6597907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pringle EG, Baxter SW, Webster CL, Papanicolaou A, Lee SF, et al. Synteny and chromosome evolution in the lepidoptera: evidence from mapping in Heliconius melpomene. Genetics. 2007;177:417–426. doi: 10.1534/genetics.107.073122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ranz JM, Casals F, Ruiz A. How malleable is the eukaryotic genome? Extreme rate of chromosomal rearrangement in the genus Drosophila. Genome Res. 2001;11:230–239. doi: 10.1101/gr.162901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bhutkar A, Schaeffer SW, Russo SM, Xu M, Smith TF, et al. Chromosomal rearrangement inferred from comparisons of 12 Drosophila genomes. Genetics. 2008;179:1657–1680. doi: 10.1534/genetics.107.086108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Conceição IC, Aguadé M. High incidence of interchromosomal transpositions in the evolutionary history of a subset of or genes in Drosophila. J Mol Evol. 2008;66:325–332. doi: 10.1007/s00239-008-9071-y. [DOI] [PubMed] [Google Scholar]
- 63.Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 2004;32:W273–279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mayor C, Brudno M, Schwartz JR, Poliakov A, Rubin EM, et al. VISTA : visualizing global DNA sequence alignments of arbitrary length. Bioinformatics. 2000;16:1046–1047. doi: 10.1093/bioinformatics/16.11.1046. [DOI] [PubMed] [Google Scholar]
- 65.Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 66.Consortium IHGS. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 67.Bergman CM, Kreitman M. Analysis of conserved noncoding DNA in Drosophila reveals similar constraints in intergenic and intronic sequences. Genome Res. 2001;11:1335–1345. doi: 10.1101/gr.178701. [DOI] [PubMed] [Google Scholar]
- 68.Halligan DL, Eyre-Walker A, Andolfatto P, Keightley PD. Patterns of evolutionary constraints in intronic and intergenic DNA of Drosophila. Genome Research. 2004;14:273–279. doi: 10.1101/gr.1329204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Andolfatto P. Adaptive evolution of non-coding DNA in Drosophila. Nature. 2005;437:1149–1152. doi: 10.1038/nature04107. [DOI] [PubMed] [Google Scholar]
- 70.Haddrill PR, Charlesworth B, Halligan DL, Andolfatto P. Patterns of intron sequence evolution in Drosophila are dependent upon length and GC content. Genome Biol. 2005;6:R67. doi: 10.1186/gb-2005-6-8-r67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wahlberg N, Leneveu J, Kodandaramaiah U, Pena C, Nylin S, et al. Nymphalid butterflies diversify following near demise at the Cretaceous/Tertiary boundary. Proc Biol Sci. 2009;276:4295–4302. doi: 10.1098/rspb.2009.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Beldade P, Mateus AR, Keller RA. Evolution and molecular mechanisms of adaptive developmental plasticity. Mol Ecol. 2011;20:1347–1363. doi: 10.1111/j.1365-294X.2011.05016.x. [DOI] [PubMed] [Google Scholar]
- 73. http://www.jgi.doe.gov/sequencing/why/CSP2006/butterfly.html.
- 74.Jurka J, Klonowski P, Dagman V, Pelton P. CENSOR–a program for identification and elimination of repetitive elements from DNA sequences. Comput Chem. 1996;20:119–121. doi: 10.1016/s0097-8485(96)80013-1. [DOI] [PubMed] [Google Scholar]
- 75.Tweedie S, Ashburner M, Falls K, Leyland P, McQuilton P, et al. FlyBase: enhancing Drosophila Gene Ontology annotations. Nucleic Acids Res. 2009;37:D555–559. doi: 10.1093/nar/gkn788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stein LD, Mungall C, Shu S, Caudy M, Mangone M, et al. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bray N, Dubchak I, Pachter L. AVID: A global alignment program. Genome Res. 2003;13:97–102. doi: 10.1101/gr.789803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Munoz-Torres MC, Reese JT, Childers CP, Bennett AK, Sundaram JP, et al. Hymenoptera Genome Database: integrated community resources for insect species of the order Hymenoptera. Nucleic Acids Res. 2011;39:D658–662. doi: 10.1093/nar/gkq1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kim HS, Murphy T, Xia J, Caragea D, Park Y, et al. BeetleBase in 2010: revisions to provide comprehensive genomic information for Tribolium castaneum. Nucleic Acids Res. 2010;38:D437–442. doi: 10.1093/nar/gkp807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang L, Wang S, Li Y, Paradesi MS, Brown SJ. BeetleBase: the model organism database for Tribolium castaneum. Nucleic Acids Res. 2007;35:D476–479. doi: 10.1093/nar/gkl776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 82.Arensburger P, Megy K, Waterhouse RM, Abrudan J, Amedeo P, et al. Sequencing of Culex quinquefasciatus establishes a platform for mosquito comparative genomics. Science. 2010;330:86–88. doi: 10.1126/science.1191864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Consortium HGS. Insights into social insects from the genome of the honeybee Apis mellifera. Nature. 2006;443:931–949. doi: 10.1038/nature05260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Consortium IAG. Genome sequence of the pea aphid Acyrthosiphon pisum. PLoS Biol. 2010;8:e1000313. doi: 10.1371/journal.pbio.1000313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nene V, Wortman JR, Lawson D, Haas B, Kodira C, et al. Genome sequence of Aedes aegypti, a major arbovirus vector. Science. 2007;316:1718–1723. doi: 10.1126/science.1138878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Werren JH, Richards S, Desjardins CA, Niehuis O, Gadau J, et al. Functional and evolutionary insights from the genomes of three parasitoid Nasonia species. Science. 2010;327:343–348. doi: 10.1126/science.1178028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Holt RA, Subramanian GM, Halpern A, Sutton GG, Charlab R, et al. The genome sequence of the malaria mosquito Anopheles gambiae. Science. 2002;298:129–+. doi: 10.1126/science.1076181. [DOI] [PubMed] [Google Scholar]
- 88.Richards S, Gibbs RA, Weinstock GM, Brown SJ, Denell R, et al. The genome of the model beetle and pest Tribolium castaneum. Nature. 2008;452:949–955. doi: 10.1038/nature06784. [DOI] [PubMed] [Google Scholar]
- 89.Animal Genome Size Database. Available: http://www.genomesize.com. Accessed 2011 Aug 14.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Quantification of repetitive sequence in target B. anynana BACs. Identification and characterization of repeated regions was done with RepeatMasker (see Methods). Simple Repeats correspond to duplications of, typically 1–5bases. Low Complexity Sequence corresponds to poly-purine/poly-pyrimidine stretches or regions of >87% AT or >89% GC.
(PDF)
Heliconius repetitive elements identified in the target B. anynana BACs. Two of the novel repetitive elements identified in Heliconius [10] appear to be present in our target B. anynana genomic sequence, as well as in publicly available nucleotide sequence for other lepidopterans (cf. BLASTn analysis; see Methods).
(PDF)
List of Kaikogaas predicted genes and subsequent manual curation.
(TXT)
Gene density and genome size in insects. Gene density (number of genes per 100 Kb; cf. [10], [46], [82], [83], [84], [85], [86], [87], [88] in relation to genome size for different insect species (cf. [89]). Circles correspond to species where gene densities were estimated based on sequenced genomes – note that the size of assembled genome can differ from the estimates in this Figure. Other symbols correspond to species where gene density was estimated based on a few BAC clone sequences – including this paper for B. anynana.
(PDF)
Comparative analysis of the EcR and wg/Wnt-6 genomic regions. VISTA plots of the genomic regions comprising the genes EcR (BAC AC239120) and wg/Wnt-6 (BAC AC239123) with the orthologous regions in other insects with relevant sequence available: Heliconius melpomene, Bombyx mori, Helicoverpa armigera, Spodoptera frugiperda, Drosophila melanogaster, Tribolium castaneum, and Apis mellifera.
(PDF)
Kaikogaas graphical output of BAC annotation.
(PDF)