Abstract
Perfluoroalkyl acids (PFAAs) are ubiquitously present in human blood samples and the effects of these compounds on human health are not fully characterized. This study was conducted in order to investigate the applicability of peripheral blood gene expressions for exploring the impact of perfluorooctane sulfonate (PFOS), perfluorooctanoate (PFOA) and perfluorohexane sulfonate (PFHxS) exposure on the general population. PFOS, PFOA and PFHxS were analyzed in blood samples from a representative group of 270 healthy, postmenopausal Norwegian women (48-62 years). Gene expression was measured in the same samples using the Applied Biosystems microarray platform. Forty-eight different gene sets, all previously linked to PFAA exposure were explored in relation to the selected PFAAs. Two gene sets, both related to the citric acid cycle, were differentially expressed between the “PFOS high” (>30ng/ml, n=42) and the “PFOS low” (<30ng/ml, n=228) group. Based on the results of this study we believe that blood gene signatures have a large potential for elucidating which biological pathways are being affected by environmental pollutants. To the best of our knowledge, this study is the first assessment of the impact of PFAAs on blood gene expressions in humans from the general population.
Keywords: PFOS, gene expression, PFAA, peripheral blood, pollutants
Introduction
Over the last decade, perfluoroalkyl acids (PFAAs) have received increased attention due to their ubiquitous presence in the environment [1] and in human blood [2]. PFAAs possess unique properties of repelling both water and oil and have been widely used in industrial applications, e.g. as constituents of surface treatment products and as processing aids in the production of fluoropolymers [3]. Perfluorooctane sulfonate (PFOS) is usually the most abundant PFAA in human blood samples, but perfluorooc-tanoate (PFOA) and perfluorohexane sulfonate (PFHxS) are also frequently detected [2].
Potential health effects of PFAAs have been thoroughly evaluated by others using in vitro experiments or animal model studies. Among others, changes in blood lipid levels and gene expressions related to the fatty acid metabolism have been observed in rats and chickens exposed to PFOS and PFOA [4, 5]. Alterations in cell membrane fluidity, increased liver weight and increased mortality among newborn rats have also been associated with PFOS and PFOA exposure [6]. The toxicity of PFHxS has not been evaluated thoroughly, but reduced serum cholesterol levels and increased liver weight was, among others, recently observed in PFHxS exposed rodents [7].
Several epidemiological investigations of health effects among highly exposed workers in fluoropolymer industries have also been performed [8-11], but only a limited number of studies have looked into the potential health effects of PFAAs among the general population. Occupationally exposed workers in fluoropolymer industries have been studied in relation to morbidity, self-reported medical conditions and bladder cancer [8-10]. No association between place of work (as an exposure marker) and these endpoints was found. However, the results suggested a positive, although inconsistent, association between PFOA exposure, prostate cancer and diabetes mortality [11]. A positive relationship between serum PFOS concentrations, blood insulin levels, β-cell function and insulin resistance status was reported in a general population, suggesting that some PFAAs are associated with the metabolic syndrome [12]. Nelson et al. [13] found on the other hand, no association between insulin resistance, body size and PFOS, PFOA, PFHxS or PFNA among the general population from the United States (n=860). Recently, an association between increased PFOS and PFOA concentrations and increased total cholesterol and LDL-cholesterol was confirmed among children and adolescents previously exposed to PFOA contaminated drinking water [14]. Increased PFOA concentrations have also been linked to increased liver enzyme levels [15] and PFOS and PFOA have been associated with thyroid disease among 3974 adults from the general U.S population [16].
The effect of low-dose long term exposure to pollutants is often hard to investigate as the mechanism of action in humans is not characterized. The causal relationship may in addition be difficult to evaluate due to a long period between exposure and outcome, large normal variability in outcome measures or the outcome being a complex disease (e.g. cancer). Thus, there is a need for sensitive methods to investigate the effects of background pollutant concentrations on human health.
Gene expression signatures of human blood or tissues may have large implications in epidemiology as early biomarkers of disease or through investigations of the etiology of diseases. Expression profiles in peripheral human blood cells have been successfully used to assess the impact of environmental exposures, such as smoking [17], metal fumes [18], ionizing radiation [19], dioxin and benzene exposure [20, 21]. Despite that, using blood for gene expression analysis is complicated by inter-individual variations in blood cell distributions and the risk of gene expression changes due to technical variables such as batch number, amplification date, collection and storage time [22-24]. However, Dumeaux et al. [25] showed recently that body mass index (BMI), smoking, fasting status, hormone therapy (HT) and other medication use were mirrored in blood of the women included in the current analysis after adjustment for the significant effect of technical variables.
This study was undertaken to assess the applicability of blood gene signatures as a tool for exploring the effects of PFOS, PFOA and PFHxS exposure on the general population.
Materials and methods
Study participants and collection of blood samples
The women taking part in the current study are all participants in the Norwegian Women And Cancer Study (NOWAC) [26], which consists of more than 170 000 women who have answered one to three detailed questionnaires regarding their diet and lifestyle. From the original cohort, more than 50 000 women (born between 1943 and 1957) were randomly recruited in batches of 500 to the NOWAC postgenome study [27]. In addition to answer a two-page questionnaire regarding lifestyle and defined exposures, these women also donated a blood sample. Of a randomly selected batch of 500 women, 270 (blood drawn in 2005) fulfilled the inclusion criteria for the current analysis. Criteria for being included were defined as having postmenopausal status, successful blood delivery in one PAX gene Blood RNA tube (Preanalytix, Qiagen, Hilden, Germany) and in one blood collection tube containing citrate buffer. The blood sample had also to be frozen within three days after collection. In addition, sufficient RNA quantity, integrity or purity was demanded, at least 40% of the microarray probes had to have signal to noise ratio (S/N) ≥3, no use of diabetes medication were allowed and the recovery of the PFAA analysis had to be above 60%. Investigations on sex hormones and blood gene expression were performed on the same samples; therefore the study group was limited to postmenopausal women only.
Mean age among the 270 women included in the current study was 56 years, 26% were current smokers, 18% used HT and 57% used some other medication. Mean body mass index (BMI) was 25.5 kg/m2 and 9% of the women were fasting before blood delivery.
PFAA levels and the relationship to dietary intake in the larger study group have previously been assessed by Rylander et al [28]. HT use, sex hormone levels and detailed information about the blood collection procedures have been reported by Waaseth et al. [29].
Chemical analysis
The plasma analyses of PFAAs have been described in detail by Rylander et al. [30]. In brief, plasma concentrations of PFAAs were determined using sonication facilitated liquid-liquid extraction, activated charcoal clean-up and analysis on HPLC-QTOF-MS. The quality of the analysis was assured through repetitive analyses of blank samples and reference samples obtained from previous international comparison programs. For each batch of 30 samples, one reference material and two blank samples were prepared. Three times each year, our laboratory also participates in the AMAP Ringtest for Persistent Organic Pollutants in Human Serum, an international comparison program, organized by Institut National de Santé Publique du Québec, Canada. Results from interlaboratory comparisons indicate that the uncertainties of our analysis are well within +/- 30% of the assigned values. Due to the use of mass labeled internal standards, there was no need for recovery corrections in the samples. The recovery was, however, calculated for quality assurance purposes and varied between 60% and 150%. The values above 100% were a result of matrix induced ion suppression of the recovery standard signal. The method detection limit (LOD) was automatically calculated by the quantification software and accounted for individual matrix effects. PFOS, PFOA and PFHxS were detected in <10% of the blank samples. If the concentration of these compounds in the blank samples were larger than the software determined LOD for that batch of samples, LOD was determined from three times the concentration of analytes in the blanks.
Data defined as PFOS, PFOA and PFHxS are the sum of the linear and the dominating branched isomers.
The plasma concentrations of fatty acids were analyzed at the National Institute of Nutrition and Seafood Research in Bergen, Norway. The methods used have been described elsewhere [28].
RNA isolation, quality control, data capturing and preprocessing of data
All methods for RNA analysis, data capturing and preprocessing of data is described in detail by Dumeaux et al [25]. Microarray analysis was performed on the 270 samples using the Applied Biosystems expression array system (Foster City, Louisiana, USA). Briefly, 500 ng of total RNA was amplified and labeled using the NanoAmp RT-IVT Labeling Kit for one round of amplification. 10 μg of DIG-labeled cRNA was fragmented and hybridized to AB Human Genome Survey Microarray V2.0, in accordance with the Chemiluminescence Detection Kit Protocol. The AB Expression System software was used to export signal intensities, signal to noise ratios (S/N), and flagging values. Gene-wise intensities were adjusted for technical variability i.e. batch number, RNA extraction date and time between blood collection and storage [25].
Statistical analysis
The freely available software R version 2.8.1 (www.cran.r-project.org) with the Bioconductor packages was used for the statistical analysis. Study participants were divided into two groups (“high” and “low”) according to their concentrations of PFOS, PFOA and PFHxS. All contaminant data were right-skewed and samples following the normal quantile-quantile plot (normal qq-plot) were defined as the “low” group while samples with high, non-normal values were defined as the “high” group. The rationale for this grouping was the opportunity to compare the most extreme group to the rest. The cut-off values were determined to 30 ng/ml, 7.2 ng/ml and 1.7 ng/ml for PFOS, PFOA and PFHxS, respectively. Gene-wise linear models were used for evaluating differences in single gene expressions between groups. Enrichment of 48 gene sets were evaluated for PFOS, PFOA and PFHxS, respectively, using the global test [31]. All tested gene sets had previously been linked to PFAA exposure and were curated from the literature, the Kyoto Encyclopedia of Genes and Genomes (KEGG) [32] and Gene Ontology (GO) (Table S1 in the Supplemental material). The global test was adjusted for multiple testing using false discovery rates (FDR) [33]. Comparative p-values were calculated for each gene set and indicate the proportion of random gene sets of the same size as the tested gene lists being significant by chance.
Differences in age, BMI, fasting status, HT use, use of other medication, smoking and the ratio of n-6/n-3 fatty acids between the “high” and “low” groups were evaluated using linear models and chi square tests. Variables that were significantly different between the two groups were adjusted for in the gene set enrichment analysis. The gene plot from the global test was used to select core genes that were most important for explaining the differences between groups. Core genes were defined as genes with a standard deviation > 1.5 above the expected value under the null hypothesis of no association between gene set expression and exposure group. Genes that were strongly correlated to the core genes (r >0.75) were further evaluated using functional clustering in the Database for Annotation, Visualization and Integrated Discovery (DAVID) [34] to investigate groups of molecular pathways or processes related to PFAA exposure.
Results
In the current study group, the dominating PFAA was PFOS (median 19 ng/ml), followed by PFOA (4.4 ng/ml) and PFHxS (0.97 ng/ml) (Table 1). Two single genes- cytochrome C oxidase subunit VIb polypeptide 2 (COX6B2) (p=1.1e-5, FDR=0.14) and chromosome 17 open reading frame 74 (MGC17624) (p=1.7e-5, FDR=0.14) -were differentially expressed when comparing the “PFOS high” to the “PFOS low” group using gene-wise linear models. No significant single genes were differentially expressed according to PFOA and PFHxS concentrations.
Table 1.
Plasma concentrations of PFAAs in the study group
| Concentration (ng/ml) | Median | AM | Range | LOD | %>LOD |
|---|---|---|---|---|---|
| PFOS | |||||
| Total sample N=270 | 19 | 21 | 5.7-84 | 0.01-2.1 | 100 |
| High group N=42 | 37 | 40 | 31-84 | ||
| Low group N=228 | 18 | 18 | 5.4-30 | ||
| PFOA | |||||
| Total sample N=270 | 4.4 | 5.1 | 0.79-21 | 0.11-1.6 | 100 |
| High group N=32 | 9.2 | 11 | 7.3-21 | ||
| Low group N=238 | 2.9 | 2.8 | 0.79-7.2 | ||
| PFHxS | |||||
| Total sample N=270 | 0.97 | 1.3 | 0.15-13 | 0.01-1.2 | 94 |
| High group N=46 | 2.7 | 3.4 | 1.7-13 | ||
| Low group N=224 | 0.87 | 0.9 | 0.15-1.6 | ||
PFOS, Perfluorooctane sulfonate; PFOA, perfluorooctanoate; PFHxS, perfluorohexane sulfonate; AM, Arithmetic mean; LOD, Method limit of detection; %>LOD, Proportion of samples with concentrations > LOD
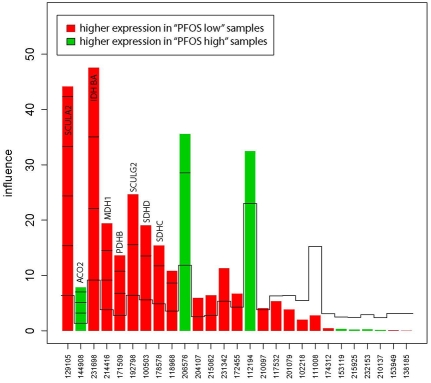
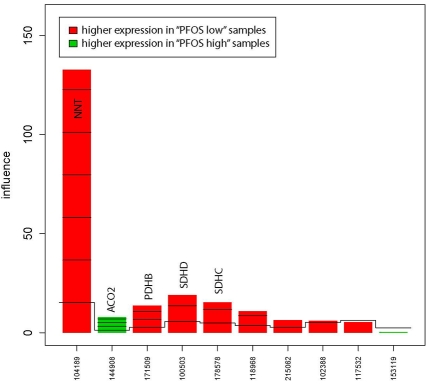
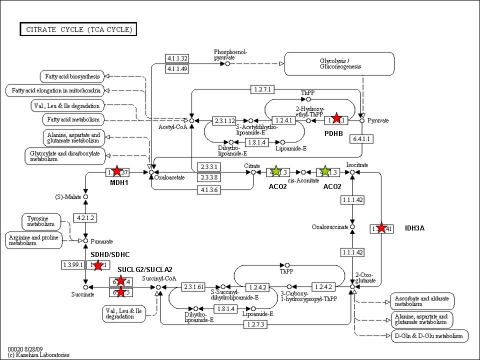
Two gene sets (gene set 39 & 46 in Table S1 in the Supporting informaion), both related to the citric acid cycle, were differentially expressed between the “PFOS high” and “PFOS low” group (Table 2). Figures 1 and 2 show the gene plot for each of the significant pathways. There were eight core genes (NNT, PDHB, SDHD, SDHC, SUCLA2, IDH3A, MDH1, SUCLG2) that were down-regulated in the “PFOS high” group and one gene (ACO2) that was up-regulated (Table 2, Figure 3). The core genes expressed in the citrate cycle pathway were strongly correlated to 58 identified single genes (Table S2 in the Supplemental material). Within these 58 genes, a functional cluster of three metabolic processes was identified (median FDR=2.3%, Table 3).
Table 2.
Significant gene sets associated with PFOS exposure
| No. probes tested | p-value | FDR adjusted | Comparative p-value | Core genes upregulated in the PFOS high group | Core genes down-regulated in the PFOS high group | |
|---|---|---|---|---|---|---|
| Citric acid cycle | 27 | 0.0393 | 0.37 | 0.18 | ACO2 | SUCLA2, IDH3A, MDH1, PDHB, SUCLG2, SDHD, SDHC |
| Citric acid cycle | 10 | 0.0504 | 0.3 | 0.30 | ACO2 | NNT, PDHB, SDND, SDHC |
PFOS, perfluorooctane sulfonate; FDR, false discovery rate
Figure 1.
Gene plot of the influence of “high” vs. “low” PFOS on the citric acid cycle (gene set 39). Core genes are identified by official gene symbols. The x-axis shows the probe IDs and the y-axis shows the influence of each gene on the test result.
Figure 2.
Gene plot of the influence of “high” vs. “low” PFOS on the citric acid cycle (gene set 46). Core genes are identified by official gene symbols. The x-axis shows the probe IDs and the y-axis shows the influence of each gene on the test result.
Figure 3.
The citric acid cycle pathway (reprinted from KEGG [33]). Up-regulated core genes in the “PFOS high” group are marked with a green star. Down-regulated core genes are marked with a red star.
Table 3.
Functional cluster of three metabolic processes, related to the 58 genes highly correlated to the core genes in the citrate cycle.
| Process (No. of genes in process) | p-value | Fold enrichment | FDR (%) |
|---|---|---|---|
| Macromolecule metabolic process (29) | 0.000064 | 1.7 | 0.1 |
| Cellular metabolic process (29) | 0.0012 | 1.5 | 2.3 |
| Primary metabolic process (29) | 0.0013 | 1.5 | 2.4 |
FDR, false discovery rate
The women in the “PFOS high” group were significantly older and had lower ratio of n-6/n-3 fatty acids than the women in the “PFOS low” group (Table 4). There were no differences in smoking status, BMI, fasting, medication or HT use between the two groups (Table 4). When adjusting for age and the ratio of n-6/n-3 fatty acids, the genes within the citrate cycle pathway remained differentially expressed between the “PFOS high” and “PFOS low” group. None of the tested gene sets were significantly enriched for PFOA or PFHxS.
Table 4.
Characteristics of the two PFOS groups
| PFOS high (N=42) | PFOS low (n=228) | p-value | |
|---|---|---|---|
| Age (years) | 57 | 55.5 | 0.01 |
| Body mass index (kg/m2) | 25.4 | 25.5 | 0.92 |
| Smoking (Y/N) | 7/35 | 63/164 | 0.19 |
| Hormone therapy use (Y/N) | 8/32 | 40/185 | 0.91 |
| Medication use (Y/N) | 21/20 | 130/94 | 0.52 |
| Fasting (Y/N) | 5/35 | 19/196 | 0.66 |
| n-6/n-3 fatty acid ratio (mg/ml)a | 5.2 | 6.2 | 0.006 |
PFOS, perfluorooctane sulfonate;
Include linolenic acid (LA)18:2n-6; eicosadienoic acid 20:2n-6; arachidonic acid (AA) 20:4n-6; dihomo-gamma-linolenic acid (DGLA) 20:3n-6; 16:3n-3; 16:4n-3; alpha-linolenic acid (ALA) 18:3n-3; stearidonic acid 18:4n-3; eicosatrienoic acid (ETE) 20:3n-3; eicosatetraenoic acid (ETA) 20:4n-3; eicosapentaenoic acid (EPA) C20:5n-3; docosapentaenoic acid (DPA) C22:5n-3 and docosahexaenoic acid (DHA) C22:6n-3 The p-value indicates if there was a significant difference between the two groups.
Discussion
To the best of our knowledge, this is the first population based study investigating the impact of organic pollutants on blood gene signatures in humans. We have successfully used gene expressions in peripheral blood cells for exploring the effects of PFAAs on the general population. Our results suggest gene profiles in human blood have large potential for elucidating which biological pathways that are being affected by long-term, low-dose exposure to contaminants and could therefore serve as a more public health relevant alternative to already established toxicological methods. In addition, blood gene signatures could be used for identifying a unique set of genes, differentially expressed by the specific pollutants or mixtures of contaminants, which could be further used as a diagnostic tool for detecting early effects of pollutants. However, before that is possible, large amounts of gene signature data from healthy individuals is needed in order to explore normal variability. The scope of this work adds new and important information to this newly developed research field.
It is important to emphasize that in the current work technical noise was present and largely corrected for as described in detail by Dumeaux et al. [25]. It is evident that the microarray platforms currently go through rapid development. Future platforms will be less sensitive to technical variables which in turn will provide improved gene expression results and wider applications for gene signatures. Hence, high through put assays open new research fields to investigate the effects of pollutants on human health.
Despite that a large proportion of the gene expression variability was explained by technical variables, the previous mentioned corrections made it possible to identify gene sets differentially expressed between the two PFOS groups. Genes coding for enzymes within the citric acid cycle were differentially expressed between women with “high” PFOS concentrations (>30 ng/ml) and women with “low” PFOS concentrations (<30 ng/ml). The result indicates that the glucose metabolism is affected by background concentrations of PFOS among the general population. The “high” PFOS group was significantly older and had lower ratio of n6/n3 fatty acids than the “low” group. The lower n6/n3 fatty acid ratio indicated dietary differences and a possible higher consumption of fatty fish in the “high” PFOS group. There was no difference in smoking or fasting status or BMI between the “high and “low” PFOS groups. When adjusting the analyses for age and fatty acid ratio, the citric acid cycle remained differentially expressed between the groups. Lin et al. [12] reported recently that PFOS (mean 24 ng/ml) and PFNA (0.8 ng/ml) were associated with indicators of metabolic syndrome in a group of adults from the general population (n=969), which supports our finding. Nelson et al. [13] found, on the other hand, little association between PFOS, PFOA, PFHxS, PFNA and body size and insulin resistance in a group of 860 people from the general population in the United States. They did, however, find a positive association between PFOS, PFOA and PFNA and cholesterol levels. In addition to epidemiological indications, there are also some toxicological findings suggesting that PFAAs may interfere with the carbohydrate metabolism [35] in rodents, which also converges in the citric acid cycle.
A set of 58 genes were identified as co-varying with the core genes in the citric acid cycle pathway. A cluster of three metabolic processes were identified within that gene list, indicating that genes involved in the metabolism were differentially expressed in the “PFOS high” group. Eight of the nine core genes in the citrate cycle gene sets encode central enzymes within that pathway. All, except for ACO2, were down-regulated in the “high” group. This could be a result of several different processes; PFOS may for example interfere with the citric acid cycle itself or it could be a result of a feedback mechanism induced by PFOS. The mitochondrion could also be affected and thereby affecting metabolic processes. The effect of the direction of the expressed genes has to be elucidated in future research.
Our results indicate an association between environmental PFOS exposure and chages in ezyme activities within the citric acid cycle. A number of diseases, e.g. type II diabetes and Alzheimer's disease [36] have been linked to citric acid cycle disorders, emphasizing the need for better understanding of the mechanism of action for PFOS. Additionally, increased mortality of diabetes was observed among workers “probably” exposed to APFO (precursor to PFOA) although that study was not consistent as there was no death from diabetes in the “definite” exposure group [11]. Four women were excluded from the analysis in the current study as they were using type II diabetes medicine. The median PFOS concentration among those women was 24 ng/ml (mean 20 ng/ml) whereas the corresponding median concentration was 19 ng/ml (mean 21 ng/ml) for the total study group (n=270). Although the diabetes group was too small for statistical purposes, the potential difference in PFOS concentration between diabetes patients and healthy individuals should be further assessed. Currently, it is too early to conclude if changes in enzyme activities within the citric acid cycle will have any consequences for public health. None of the epidemiological studies on occupationally exposed workers have revealed any strong associations between severe health effects and exposure to PFAAs. Nevertheless, there has been an enormous increase in type II diabetes incidence during the same periods as the PFOS exposure has increased considerable. It is important to emphasize that the results from the current study should be confirmed or refuted in an independent data set using improved microarray techniques. What is evident is that human blood gene signatures have an enormous and as yet unexplored potential in the interdisciplinary field of epidemiology and toxicology.
There are some limitations with the current study. As PFAAs were present in the blood of all participants in the current study group, it was not possible to divide them into sub-groups of “exposed” and “non-exposed” individuals. Differences between the “high” and “low” groups will therefore be small and there will be low chances of detecting differentially expressed single genes. Based on that, our analyses were focused on gene set enrichment instead of single genes analyses. Surprisingly, two single genes (COX6B2 and MGC17624) were differentially expressed between the “PFOS high” and the “PFOS low” group from the gene-wise linear analysis. This is an interesting finding, but could also be a result of chance.
The significantly affected pathways had false discovery rates of 37% and 25%. Breitling et al. recommended that gene sets with FDR ≤ 10% should be considered interesting [37]. In the current study, we accepted higher FDR values since our gene sets were curated from previous publications and thereby supported by toxicological/ epidemiological findings. In addition, none of the tested gene sets had comparative p-values high enough to raise concerns for false positive results.
We found no significantly enriched pathways for PFOA or PFHxS in this study group, which could be a result of low and uniform concentrations of the analytes within the population studied. Although PFOA and PFOS have been identified as potent peroxisome proliferators in rodents [6], the PPAR pathway was not enriched for any of the investigated PFAAs. A number of factors, including species-specific differences, could be the reason for the lack of differentially expressed genes within that biological pathway. Additionally, effects of PFOS and PFOA are mainly seen in the liver among test animals, and less pronounced effects in blood cells are therefore expected.
Acknowledgments
We gratefully acknowledge all participating women in NOWAC, the medical personnel helping out with data collection and Bente Augdal and Merete Albertsen for administrative work during the collection of data. Livar Frøyland has contributed with fatty acid analysis. This work was supported by grants from the Norwegian Research Council and the University of Tromsø and The Royal Ministry of Fisheries and Coastal Affairs.
Supplementary Data
Table S1.
Gene sets tested in gene set enrichment analysis
| Gene set ID number | Description | Number of genes | Reference |
|---|---|---|---|
| 1 | Beta oxidation of fatty acids | 46 | KEGG |
| 2 | Fatty acid beta oxidation | 24 | GO |
| 3 | Genes linked to PFOS and PFOA exposure in chickens | 36 | [1] |
| 4 | Gap junction intracellular communication | 96 | KEGG |
| 5 | Genes linked to oxidative stress in hepatocytes from freshwater tilapia after PFOS and PFOA exposure | 17 | [2] |
| 6 | Oxidative stress | 94 | GO |
| 7 | Regulation of fatty acid oxidation | 18 | GO |
| 8 | Steroid metabolism | 11 | KEGG |
| 9 | Cholesterol biosynthesis | 9 | KEGG |
| 10 | Cholesterol metabolism | 44 | GO |
| 11 | Steroid biosynthesis | 17 | KEGG |
| 12 | Cholesterol biosynthesis | 9 | GO |
| 13 | Genes linked to PFOS exposure in rat | 19 | [3] |
| 14 | Genes linked to PFOS and PFOA exposure in rat | 13 | [3] |
| 15 | Xenobiotic metabolic process | 13 | GO |
| 16 | Xenobiotic metabolism | 70 | KEGG |
| 17 | Synthesis and degradation of ketone bodies | 9 | KEGG |
| 18 | Fatty acid elongation | 10 | KEGG |
| 19 | Fatty acid metabolic process | 134 | GO |
| 20 | Unsaturated fatty acid biosynthesis | 22 | KEGG |
| 21 | Fatty acid biosynthesis | 7 | KEGG |
| 22 | Fatty acid biosynthesis and regulation | 19 | GO |
| 23 | Apoptosis | 88 | KEGG |
| 24 | Genes linked to PFOS exposure in zebrafish | 6 | [4] |
| 25 | Bile acid biosynthesis | 16 | KEGG |
| 26 | Bile acid metabolic process | 11 | GO |
| 27 | Genes linked to PFOS exposure in rat liver | 24 | [5] |
| 28 | Genes linked to PFOS exposure in hepatocytes from Atlantic salmon | 27 | [6] |
| 29 | Genes linked to PFOS exposure in carp | 20 | [7] |
| 30 | Genes linked to PFOS exposure in chicken embryo hepatocytes | 6 | [8] |
| 31 | Genes linked to PFOS exposure in bottlenose dolphin | 8 | [9] |
| 32 | Genes linked to PFOS exposure in rats | 4 | [10] |
| 33 | Genes linked to PFOS exposure in mouse | 8 | [11] |
| 34 | Glycolysis | 63 | KEGG |
| 35 | Glucose metabolic process | 57 | GO |
| 36 | Diabetes 2 | 44 | KEGG |
| 37 | Genes linked to PFOA exposure in rat | 76 | [3] |
| 38 | Leukocyte transendothelial migration | 116 | KEGG |
| 39 | Citric cycle | 32 | KEGG |
| 40 | Genes linked to PFOA exposure in mouse liver | 4 | [12] |
| 41 | Retinol metabolism | 64 | KEGG |
| 42 | PPAR | 69 | KEGG |
| 43 | Insulin signaling | 138 | KEGG |
| 44 | Glucose homeostasis | 22 | GO |
| 45 | Retinol metabolic process | 7 | GO |
| 46 | Citric acid cycle | 7 | GO |
| 47 | Insulin receptor signaling pathway | 33 | GO |
| 48 | Lipid transport | 95 | GO |
Table S2.
Genes highly correlated (r >0.75) to the core genes from gene set 39 and 46 (citric acid cycle).
| Official gene symbol |
|---|
| CYCS, PNRC2, HMGN1, ARPC5, GRPEL1, SLBP, SLTM, HNRPH3, SMT3, SPCS2, C9orf156, DDX1, CRLF3, CDKN1B, RAB1A, CALM2, HNRPA1, ACTR1B, CCAR1, FLJ20647, RBBP7, PPP1CC, PPCS, PSMC2, HMGN1, COX5A, GTF2A2, HMGN3, LMBRD1, MORF4L2, NDNL2, MGC12981, VDAC2, SELT, ARL2BP, MIS12, CEBPZ, XPA, GOLGA7, BXDC5, MGC4767, NIF3L1, SFRS10, CNOT8, C1orf108, OCIAD1, IGBP1, RPP38, PHGDHL1, SSB, PMPCB, PDHA1, TMED10, ZNF9, YWHAZ, C2orf25, MORF4L1, HNRPA1P4 |
References
- 1.Yeung LWY, Guruge KS, Yamanaka N, Miyazaki S, Lam PKS. Differential expression of chicken hepatic genes responsive to PFOA and PFOS. Toxicology. 2007;237:111–125. doi: 10.1016/j.tox.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 2.Liu C, Yu K, Shi X, Wang J, Lam PKS, Wu RSS, Zhou B. Induction of oxidative stress and apoptosis by PFOS and PFOA in primary cultured hepatocytes of freshwater tilapia (Oreochromis niloticus) Aquat Toxicol. 2007;82:135–143. doi: 10.1016/j.aquatox.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 3.Guruge KS, Yeung LWY, Yamanaka N, Miyazaki S, Lam PKS, Giesy JP, Jones PD, Yamashita N. Gene expression profiles in rat liver treated with perfluorooctanoic acid (PFOA) Toxicol Sci. 2006;89:93–107. doi: 10.1093/toxsci/kfj011. [DOI] [PubMed] [Google Scholar]
- 4.Shi X, Du Y, Lam PKS, Wu RSS, Zhou B. Developmental toxicity and alteration of gene expression in zebrafish embryos exposed to PFOS. Toxicol Appl Pharmacol. 2008;230:23–32. doi: 10.1016/j.taap.2008.01.043. [DOI] [PubMed] [Google Scholar]
- 5.Bjork JA, Lau C, Chang SC, Butenhoff JL, Wallace KB. Perfluorooctane sulfonate-induced changes in fetal rat liver gene expression. Toxicology. 2008;251:8–20. doi: 10.1016/j.tox.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 6.Krøvel AV, Søfteland L, Torstensen B, Olsvik PA. Transcriptional effects of PFOS in isolated hepatocytes from Atlantic salmon Salmo salar L. Comp Biochemi Physiol C. 2008;148:14–22. doi: 10.1016/j.cbpc.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 7.Hagenaars A, Knapen D, Meyer IJ, van der Ven K, Hoff P, De Coen W. Toxicity evaluation of perfluorooctane sulfonate (PFOS) in the liver of common carp (Cyprinus carpio) Aquat Toxicol. 2008;88:155–163. doi: 10.1016/j.aquatox.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 8.Cwinn MA, Jones SP, Kennedy SW. Exposure to perfluorooctane sulfonate or fenofibrate causes PPAR-[alpha] dependent transcriptional responses in chicken embryo hepatocytes. Comp Biochem Physiol C. 2008;148:165–171. doi: 10.1016/j.cbpc.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 9.Mollenhauer MAM, Carter BJ, Peden-Adams MM, Bossart GD, Fair PA. Gene expression changes in bottlenose dolphin, Tursiops truncatus, skin cells following exposure to methylmercury (MeHg) or perfluorooctane sulfonate (PFOS) Aquat Toxicol. 2009;91:10–18. doi: 10.1016/j.aquatox.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 10.Chang S-C, Ehresman DJ, Bjork JA, Wallace KB, Parker GA, Stump DG, Butenhoff JL. Gestational and lactational exposure to potassium perfluorooctanesulfonate (K+PFOS) in rats: Toxicokinetics, thyroid hormone status, and related gene expression. Reproduct Toxicol. 2009;27:387–399. doi: 10.1016/j.reprotox.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 11.Rosen MB, Schmid JE, Das KP, Wood CR, Zehr RD, Lau C. Gene expression profiling in the liver and lung of perfluorooctane sulfonate-exposed mouse fetuses: Comparison to changes induced by exposure to perfluorooctanoic acid. Reproduc Toxicol. 2007;27:278–288. doi: 10.1016/j.reprotox.2009.01.007. 2009. [DOI] [PubMed] [Google Scholar]
- 12.Ren HZ, Vallanat B, Nelson DM, Yeung LWY, Guruge KS, Lam PKS, Lehman-McKeeman LD, Corton JC. Evidence for the involvement of xenobiotic-responsive nuclear receptors in transcriptional effects upon perfluoroalkyl acid exposure in diverse species. Reproduc Toxicol. 2009;27:266–277. doi: 10.1016/j.reprotox.2008.12.011. [DOI] [PubMed] [Google Scholar]
References
- 1.Powley CR, George SW, Russell MH, Hoke RA, Buck RC. Polyfluorinated chemicals in a spatially and temporally integrated food web in the Western Arctic. Chemosphere. 2008;70:664–672. doi: 10.1016/j.chemosphere.2007.06.067. [DOI] [PubMed] [Google Scholar]
- 2.Calafat AM, Wong LY, Kuklenyik Z, Reidy JA, Needham LL. Polyfluoroalkyl chemicals in the US population: Data from the National Health and Nutrition Examination Survey (NHANES) 2003-2004 and comparisons with NHANES 1999-2000. Environ Health Perspect. 2007;115:1596–1602. doi: 10.1289/ehp.10598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fromme H, Tittlemier SA, Völkel W, Wilhelm M, Twardella D. Perfluorinated compounds -Exposure assessment for the general population in western countries. Int J Hyg Environ Health. 2009;212:239–270. doi: 10.1016/j.ijheh.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 4.Yeung LWY, Guruge KS, Yamanaka N, Miyazaki S, Lam PKS. Differential expression of chicken hepatic genes responsive to PFOA and PFOS. Toxicology. 2007;237:111–125. doi: 10.1016/j.tox.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 5.Curran I, Hierlihy SL, Liston V, Pantazopoulos P, Nunnikhoven Ae, Tittlemier S, Barker M, Trick K, Bondy G. Altered Fatty Acid Homeostasis and Related Toxicologic Sequelae in Rats Exposed to Dietary Potassium Perfluorooctanesul-fonate (PFOS) J Toxicol Environ Health A. 2008;71:1526–1541. doi: 10.1080/15287390802361763. [DOI] [PubMed] [Google Scholar]
- 6.Lau C, Anitole K, Hodes C, Lai D, Pfahles-Hutchens A, Seed J. Perfluoroalkyl acids: A review of monitoring and toxicological findings. Toxicol Sci. 2007;99:366–394. doi: 10.1093/toxsci/kfm128. [DOI] [PubMed] [Google Scholar]
- 7.Butenhoff JL, Chang S-C, Ehresman DJ, York RG. Evaluation of potential reproductive and developmental toxicity of potassium per-fluorohexanesulfonate in Sprague Dawley rats. Repro Toxicol. 2009;27:331–341. doi: 10.1016/j.reprotox.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 8.Alexander BH, Olsen GW. Bladder Cancer in Perfluorooctanesulfonyl Fluoride Manufacturing Workers. Ann Epidemiol. 2007;17:471–478. doi: 10.1016/j.annepidem.2007.01.036. [DOI] [PubMed] [Google Scholar]
- 9.Alexander BH, Olsen GW, Burris JM, Mandel JH, Mandel JS. Mortality of employees of a perfluorooctanesulphonyl fluoride manufacturing facility. Occup Environ Med. 2003;60:722–729. doi: 10.1136/oem.60.10.722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grice MM, Alexander BH, Hoffbeck R, Kampa DM. Self-reported medical conditions in perfluorooctanesulfonyl fluoride manufacturing workers. J Occup Environ Med. 2007;49:722–729. doi: 10.1097/JOM.0b013e3180582043. [DOI] [PubMed] [Google Scholar]
- 11.Lundin JI, Alexander BH, Olsen GW, Church TR. Ammonium Perfluorooctanoate Production and Occupational Mortality. Epidemiology. 2009;20:921–928. doi: 10.1097/EDE.0b013e3181b5f395. [DOI] [PubMed] [Google Scholar]
- 12.Lin CY, Chen PC, Lin YC, Lin LY. Association Among Serum Perfluoroalkyl Chemicals, Glucose Homeostasis, and Metabolic Syndrome in Adolescents and Adults. Diabetes Care. 2009;32:702–707. doi: 10.2337/dc08-1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nelson JW, Hatch EE, Webster TF. Exposure to Polyfluoroalkyl Chemicals and Cholesterol, Body Weight, and Insulin Resistance in the General US Population. Environ Health Perspect. 2010;118:197–202. doi: 10.1289/ehp.0901165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frisbee SJ, Shankar A, Knox SS, Steenland K, Savitz DA, Fletcher T, Ducatman AM. Per-fluorooctanoic Acid, Perfluorooctanesulfonate, and Serum Lipids in Children and Adolescents: Results From the C8 Health Project. Arch Pedi-atr Adolesc Med. 2010;164:860–869. doi: 10.1001/archpediatrics.2010.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin CY, Lin LY, Chiang CK, Wang WJ, Su YN, Hung KY, Chen PC. Investigation of the Associations Between Low-Dose Serum Perfluorinated Chemicals and Liver Enzymes in US Adults. Am J Gastroenterol. 2010;105:1354–1363. doi: 10.1038/ajg.2009.707. [DOI] [PubMed] [Google Scholar]
- 16.Melzer D, Rice N, Depledge MH, Henley WE, Galloway TS. Association between Serum Per-fluorooctanoic Acid (PFOA) and Thyroid Disease in the US National Health and Nutrition Examination Survey. Environ Health Perspect. 2010;118:686–692. doi: 10.1289/ehp.0901584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lampe JW, Stepaniants SB, Mao M, Radich JP, Dai HY, Linsley PS, Friend SH, Potter JD. Signatures of environmental exposures using peripheral leukocyte gene expression: Tobacco smoke. Cancer Epidemiol Biomark Prev. 2004;13:445–453. [PubMed] [Google Scholar]
- 18.Wang ZX, Neuburg D, Li C, Su L, Kim JY, Chen JC, Christiani DC. Global gene expression profiling in whole-blood samples from individuals exposed to metal fumes. Environ Health Perspect. 2005;113:233–241. doi: 10.1289/txg.7273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amundson SA, Do KT, Shahab S, Bittner M, Meltzer P, Trent J, Fornace AJ. Identification of potential mRNA biomarkers in peripheral blood lymphocytes for human exposure to ionizing radiation. Rad Res. 2000;154:342–346. doi: 10.1667/0033-7587(2000)154[0342:iopmbi]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 20.McHale CM, Zhang L, Hubbard AE, Zhao X, Baccarelli A, Pesatori AC, Smith MT, Landi MT. Microarray analysis of gene expression in peripheral blood mononuclear cells from dioxinexposed human subjects. Toxicology. 2007;229:101–113. doi: 10.1016/j.tox.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 21.McHale CM, Zhang LP, Lan Q, Li GL, Hubbard AE, Forrest MS, Vermeulen R, Chen J, Shen M, Rappaport SM, Yin SN, Smith MT, Rothman N. Changes in the peripheral blood transcriptome associated with occupational benzene exposure identified by cross-comparison on two microarray platforms. Genomics. 2009;93:343–349. doi: 10.1016/j.ygeno.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Radich JP, Mao M, Stepaniants B, Biery M, Castle J, Ward T, Schimmack G, Kobayashi S, Carleton M, Lampe J, Linsley PS. Individualspecific variation of gene expression in peripheral blood leukocytes. Genomics. 2004;83:980–988. doi: 10.1016/j.ygeno.2003.12.013. [DOI] [PubMed] [Google Scholar]
- 23.Tanner MA, Berk LS, Felten DL, Blidy AD, Bit SL, Ruff DW. Substantial changes in gene expression level due to the storage temperature and storage duration of human whole blood. Clin Lab Haematol. 2002;24:337–341. doi: 10.1046/j.1365-2257.2002.00474.x. [DOI] [PubMed] [Google Scholar]
- 24.Eady JJ, Wortley GM, Wormstone YM, Hughes JC, Astley SB, Foxall RJ, Doleman JF, Elliott RM. Variation in gene expression profiles of peripheral blood mononuclear cells from healthy volunteers. Physiol Genomics. 2005;22:402–411. doi: 10.1152/physiolgenomics.00080.2005. [DOI] [PubMed] [Google Scholar]
- 25.Dumeaux V, Olsen KS, Nuel G, Paulssen RH, Børresen-Dale A-L, Lund E. Deciphering normal blood gene expression - The NOWAC postgenome study. PLoS Genetics. 2010:e1000873. doi: 10.1371/journal.pgen.1000873. doi: 10.1371journal.pgen.1000873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lund E, Dumeaux V, Braaten T, Hjartaker A, Engeset D, Skeie G, Kumle M. Cohort profile: The Norwegian women and cancer study - NOWAC - Kvinner og kreft. Int J Epidemiol. 2008;37:36–41. doi: 10.1093/ije/dym137. [DOI] [PubMed] [Google Scholar]
- 27.Dumeaux V, Borresen-Dale AL, Frantzen JO, Kumle M, Kristensen VN, Lund E. Gene expression analyses in breast cancer epidemiology: the Norwegian Women and Cancer postgenome cohort study. Breast Cancer Res. 2008;10:9. doi: 10.1186/bcr1859. doi: 10.1186/bcr1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rylander C, Sandanger TM, Froyland L, Lund E. Dietary Patterns and Plasma Concentrations of Perfluorinated Compounds in 315 Norwegian Women: The NOWAC Postgenome Study. Environ Sci Technol. 2010;44:5225–5232. doi: 10.1021/es100224q. [DOI] [PubMed] [Google Scholar]
- 29.Waaseth M, Bakken K, Dumeaux V, Olsen KS, Rylander C, Figenschau Y, Lund E. Hormone replacement therapy use and plasma levels of sex hormones in the Norwegian Women and Cancer postgenome cohort - a cross-sectional analysis. BMC Women Health. 2008;8:1. doi: 10.1186/1472-6874-8-1. doi: 10.1186/1472-6874-8-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rylander C, Brustad M, Falk H, Sandanger TM. Dietary predictors and plasma concentrations of perfluorinated compounds in a coastal population from northern Norway. J Environ Public health. 2009 doi: 10.1155/2009/268219. 2009, Article ID 268219, doi: 10.1155/2009/268219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goeman JJ, van de Geer SA, de Kort F, van Houwelingen HC. A global test for groups of genes: testing association with a clinical outcome. Bioinformatics. 2004;20:93–99. doi: 10.1093/bioinformatics/btg382. [DOI] [PubMed] [Google Scholar]
- 32.Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Benjamini Y, Hochberg Y. Controlling the false discovery rate – a practical and powerful approach to multiple testing. J R Stat Soc Ser BMethodol. 1995;57:289–300. [Google Scholar]
- 34.Dennis G, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biology. 2003;4:11. [PubMed] [Google Scholar]
- 35.Guruge KS, Yeung LWY, Yamanaka N, Miyazaki S, Lam PKS, Giesy JP, Jones PD, Yamashita N. Gene expression profiles in rat liver treated with perfluorooctanoic acid (PFOA) Toxicol Sci. 2006;89:93–107. doi: 10.1093/toxsci/kfj011. [DOI] [PubMed] [Google Scholar]
- 36.Brooks WM, Lynch PJ, Ingle CC, Hatton A, Emson PC, Faull RLM, Starkey MP. Gene expression profiles of metabolic enzyme transcripts in Alzheimer's disease. Brain Res. 2007;1127:127–135. doi: 10.1016/j.brainres.2006.09.106. [DOI] [PubMed] [Google Scholar]
- 37.Breitling R. Biological microarray interpretation: The rules of engagement. Biochim Biophys Acta. 2006;1759:319–327. doi: 10.1016/j.bbaexp.2006.06.003. [DOI] [PubMed] [Google Scholar]