Abstract
Two conserved genomic fragments viz. 289bp of ORF1a and 449bp of ORF2 amplified by RT-PCR showed emergence of interesting recombinant strains representing new and novel genetic variants (n=5) within eight different genotypes of astroviruses known to date. HAstV-positive cases with ORF1a [HAstV genotype G2 or G8] and ORF2 [HAstV genotype G1, G2, or G3] were detected as sole or mixed infection among infants, children and adults in Kolkata with severe illness owing to acute gastroenteritis that required hospitalization for treatment between 2007 and 2009. The twelve interesting recombinants were of type HAstV _ ORF1a _ ORF2 as HAstV _ G8_ G2 (n=1), HAstV _ G8_ G1 (n=10) and HAstV _ G2_ G3 (n=1).
Keywords: Human astrovirus inter-genotype recombinants, unique emerging variants for ORF1a and ORF2, acute gastroenteritis, genotyping
Introduction
Human astroviruses (HAstVs) are gaining importance as etiological agents of acute gastroenteritis (AGE) among infants, children and adults [1-3]. Astrovirus infection has also been detected in other hosts, viz: turkeys, chicken, cattle, sheep, dogs, cats, cheetahs, ducks and bats and immuno-compromised patients [4-6]. Incidence of HAstV infection ranges from approx. 0.3 to 10% in children worldwide [7-8]. HAstV infection is normally transmitted through the fecal-oral route. Clinical symptoms observed include mild and self-limiting diarrhea, vomiting, fever, anorexia or abdominal pain that typically lasts for 2-3days [2].
Till date, eight genotypes of HAstVs have been detected that are genetically distinct from one another. Recently, the highly divergent astrovirus strain (Ast-MLB1) was reported from USA and India [9-10]. In the course of this study, twelve recombinant-like astroviruses with three different inter-genotype combinations were found to be associated with acute viral gastroenteritis among hospitalized infants, children and adults in Kolkata, India. Five cases showed unique, hitherto unreported amino acid changes within the conserved stretch of ORF1a and ORF2 fragments, indicating the emergence of HAstV variants in our setting that were different from all the eight different genotypes of astroviruses known to date.
Materials and methods
Molecular detection of astrovirus and other co-infecting enteropathogens
The etiological role of human astroviruses was evaluated during a surveillance study from 2007 to 2009 comprising infants, children and adults hospitalized with symptoms of acute watery diarrhea and vomiting or fever at the Infectious Disease and Beliaghata General Hospital (ID&BGH), Kolkata. Fecal samples were simultaneously screened for viral, bacterial and parasitic pathogens. Viral pathogens screened by Reverse Transcriptase Polymerase Chain Reaction (RT-PCR) were Astrovirus (HAstV), Sapovirus, Norovirus (NoVGI and NoVGII). Rotavirus and enteric Adenovirus (type 40 and 41) were detected with a lateral immunochromatography-based dual-detection kit. Bacterial enteropathogens such as Vibrio cholerae, Vibrio parahaemolyticus, Campylobacter jejuni, Aeromonas spp, ETEC group, EAEC and Shigella were detected by a combination of bacterial culture and PCR-techniques. Parasitic pathogens Cryptosporidium spp, Giardia lamblia and Entamoeba histolytica were detected using conventional microbiological and PCR techniques, respectively [11].
Molecular characterization of human astroviruses
Viral RNA was extracted from fecal samples using QIAamp® Viral RNA Mini Kit (QIAGEN, GmbH, Hilden, Germany). cDNA was synthesized by reverse transcription using random primers. Next, PCR with published primers Mon (+) 340: (5'-C GTCATTATTTGTTGTCATAC T-3') and Mon 348 (-): (ACATGTGCTGCTGTTA C T A T G-3') generated 289bp amplicons within highly conserved ORF1a (encoding serine protease). A second primer pair - Mon 269 (+): (5'-CAACTCAGGAAACAGGGTGT-3') and Mon 270 (-): (5'-TCAGATGCATTGTCATTGGT-3') - was used to amplify the 449bp fragment of the ORF2 (encoding capsid gene) of HAstVs, strongly conserved within specific genotypes [12-13].
All the appropriate-sized PCR products were purified with QIAquick PCR Purification Kit (QIAGEN, GmbH, Hilden, Germany) and sequenced using the ABI PRISM Big-Dye Terminator Cycle Sequencing Ready Reaction Kit version 3.1 in an automated DNA sequencer Model 3730 (Applied Biosystems, Foster City, CA). All sequences were read using FinchTV (v.1.4.0) and sequence data obtained was compared with other reference sequences in the DNA databases, using BLAST [14].
The deduced amino acid sequences were obtained using DNAsis software and aligned with the hitherto reported amino acid sequences from reference strains of astroviruses, available in GenBank, with ClustalW [15]. MEGA (Version 4.0) [16] was used for constructing phylogenetic tree. The bootstrapped phylogenetic tree (bootstrap of 1000 replicates) was constructed using Neighbor-Joining method [17] following Juke-Cantor's parameter. Percentage bootstrap support is indicated by the values at each node.
Results
Status of illness and microbial infection in the astrovirus positive cases
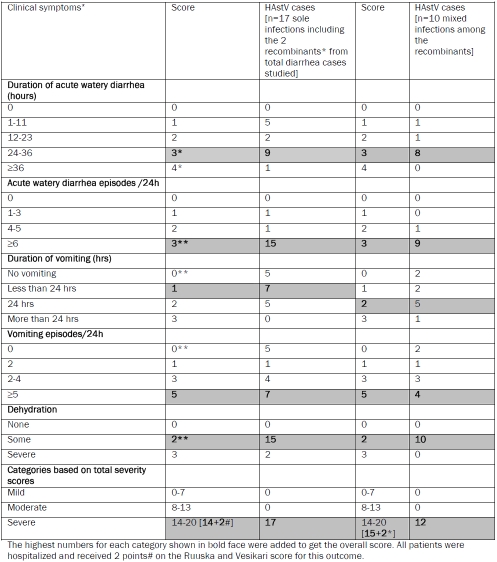
The in-depth analysis of clinical details, nature of infection by different diarrheagenic pathogens and molecular characterization to understand the genotype nature of partial ORF1a (289bp) and ORF2 (449bp) indicated that 12 HAstV positives comprising infants, children and adults were potential inter-genotype recombinants. HAstV sole infection was detected in 2 adult females (45 years and 65 years respectively). Among five HAstV positive cases, co-infection with Rotavirus was detected as the most common co-pathogen in infants and children. HAstV infection was also observed in children below two years with co-infection of Adenovirus (n=1), Sapovirus (n=1) or combination of Rotavirus, Adenovirus and Norovirus (n=1). In one child, HAstV infection with Rotavirus and three bacteria viz. V.choleraeO1, C.jejuni and Entero Adherent Escherichia coli [EAEC] was observed. In another child, parasitic co-infection with Cryptosporidium spp was observed. Abdominal pain and vomiting were associated clinical symptoms recorded besides loose stool or acute watery diarrhea, during severe illness with dehydration, requiring hospitalization for treatment (Table 1). The severity of gastroenteritis was estimated with the numerical scoring system by the Ruuska and Vesikari model [18] as shown in Table 2. The duration of diarrhea was 1-2 days in most cases with ≥6 episodes/day. There were no vomiting episodes in some cases. Some dehydration was detected in most cases. In a single case of adult diarrhea, the patient had diarrhea and abdominal pain. Of the 12 cases, 6 were male infants or children, 4 were female infants or children along with 2 adult females. The overall condition was severe illness (14-20 score according to different clinical symptoms observed during hospitalization for acute gastroenteritis).
Table 1.
Clinical details of cases infected by astrovirus recombinants and nature of the conserved 289bp fragment of ORF1a and 449bp fragment of ORF2 detected by RT-PCR in Kolkata, India. The five unique HAstV variants of ORF1a and three of ORF2 are shown in boldface.
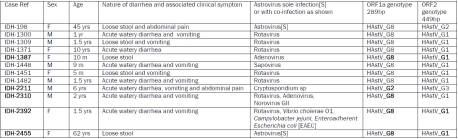 |
Table 2.
Comparison of clinical symptoms associated with sole or mixed infection of astrovirus according to their severity scores.
 |
Comparison of ORF1a sequence with other HAstVs
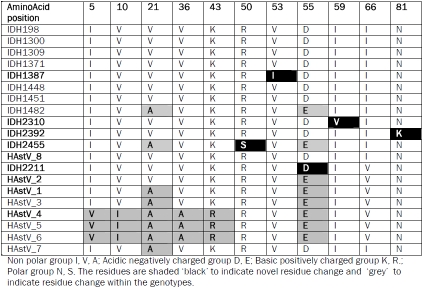
The comparison of deduced amino acid sequences of conserved partial ORF1a fragments indicated close homology for one strain IDH2211 to the HAstV2 Oxford strain L13745 reported from USA with one change in aa55 (E to D*). The remaining eleven HAstVs showed close homology to HAstV8 strains reported from Mexico and India with interesting amino acid changes described below. The strain IDH1482* showed two amino acid changes from the reference strain of HAstV8 (Yuc8, AF260508; from Mexico) at aa21 (V to A*) and aa55 (D to E*). The strain IDH2455* showed three amino acid differences when compared to the reference strain HAstV8 (Yuc8, AF260508) at aa21 (V to A*), aa50 (R basic, non-polar to S* polar) and aa55 (D to E*). Next, the following observations indicate that novel amino acid changes occurring within the ORF1a fragment in HAstVG8 strains (n=4) detected during this study has set them apart from all hitherto reported reference strains of astroviruses for 289bp conserved sequence of ORF1a (Table 3). The strain IDH1387* showed an interesting amino acid change at aa53 position (V to I*). The strain IDH2310* showed another interesting amino acid variation at aa59 (I to V*). The strain IDH2392* showed an interesting amino acid variation at aa81 (N polar to K* basic, non-polar). The strain IDH2455* showed an interesting amino acid variation at aa50 (R basic, non-polar to S* polar) Sequence alignment and comparison of partial ORF1a region of twelve HAstVs detected during the study with the eight known genotypes of HAstVs indicated that four HAstVs were novel variants of genotype HAstV8 at ORF1a while one was a novel variant of genotype HAstV2 at ORF1a.
Table 3.
Comparison of conserved amino acids and amino acid changes in positions indicated within the 289bp ORF1a fragment of astrovirus positives detected during the study and representative strains of eight different genotypes of astroviruses.
 |
Phylogenetic analysis of the ORF1a (289bp) region of the 12 recombinant HAstV strains showed that in eleven instances (2 adults, 9 infants or children) they closely clustered with reference strain of HAstV8 (Yuc 8, AF260508) reported from Mexico and in one instance (child) with HAstV2 Oxford strain L13745 from USA (Figure 1).
Figure 1.
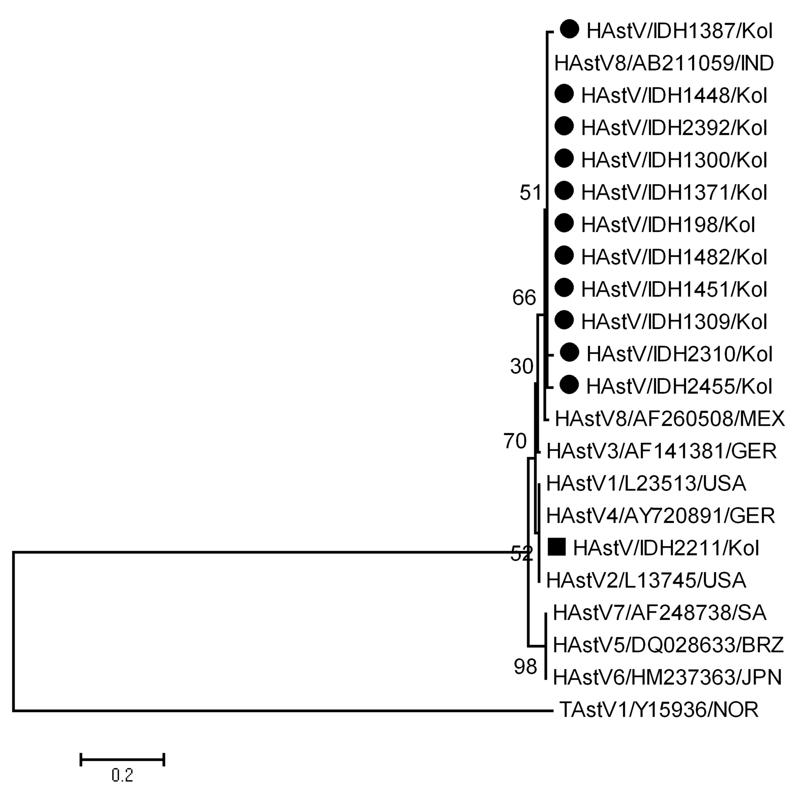
Phylogenetic analysis of deduced amino acid sequences (from 289bp fragment of partial ORF1a encoding serine protease) of human astrovirus strains detected in Kolkata, India. The Kolkata strains of HAstVs are indicated by black symbols. Scale bar indicates amino acid substitution per site. Reference sequences were obtained from GenBank under accession nos.HAstV1 (L23513/USA), HAstV2 (L13745/USA), HAstV3 (AF141381/GER), HAstV4 (AY720891/GER), HAstV5 (DQ028633/BRZ), HAstV6 (HM237363/JPN), HAstV7 (AF248738/SA) and HAstV8 (AF260508/MEX). The nucleotide sequences of 289bp ORF1a fragments of the HAstVs from Kolkata (variants marked with bold face) were deposited in DDBJ under accession nos. IDH198/AB607960, IDH1300/AB551381; IDH1309/AB551382; IDH1371/AB551383; IDH1387/AB551384; IDH1448/AB551385; IDH1451/AB607961; IDH1482/AB551386; IDH2211/ AB551387; IDH2310/ AB551388; IDH2392/AB607962;IDH2455/AB607963.
Comparison of ORF2 sequence with other HAstVs
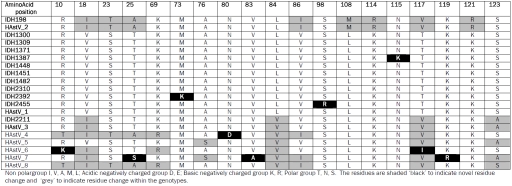
The analysis of deduced amino acid sequence of ORF2 fragments from the HAstV positives showed that ten closely matched genotype 1 Oxford strain, HAstV1 L23513 reported from USA; moreover, three strains showed interesting amino acid changes viz. at aa73 (M, non-polar, acidic to K* basic) in IDH2392, aa98 (S, polar, acidic to R* non-polar, basic) in IDH2455 and aa115 (N polar, acidic to K*non-polar, basic) in IDH1387. These interesting amino acid changes within the ORF2 fragment in these HAstVG1 strains (n=3) detected during this study, indicates the emergence of new variants that are different from all hitherto reported astroviruses for 449bp conserved sequence of ORF2. The deduced amino acid sequences of two strains IDH198 and IDH2211, however, showed 100% similarity with the reference strains HAstV2 (Oxford strain, L13745 reported from USA) and HAstV3 (Berlin strain, AF141381 reported from Germany) respectively (Table 4). Sequence alignment and comparison of partial ORF2 region with the eight known genotypes of HAstVs indicated that partial ORF2 fragments showed close identity to HAstV1 (n=10), HAstV2 (n=1) and HAstV3 (n=1) genotypes of astroviruses.
Table 4.
Comparison of conserved amino acids and amino acid changes in positions indicated within the 449bp fragment of astrovirus positives detected during the study and representative strains of eight different genotypes of astroviruses.
 |
Thus, of the twelve HAstV positives, eleven were potential inter-genotype recombinants showing HAstV genotype 8 for ORF1a in combination with either HAstV genotype 1 (n=10) or HAstV genotype 2 (n=1) for ORF2. One HAstV positive was another inter-genotype recombinant with HAstV genotype 2 for ORF1a and HAstV genotype 3 for ORF2. Phylogenetic analysis of the ORF2 fragment (deduced amino acids corresponding to the 449bp stretch conserved within specific genotypes) of the 12 recombinant HAstV strains showed that they clustered closely with HAstV1 Oxford strain L23513 reported from USA for ten cases (1 adult, nine infants or children); with HAstV2 Oxford strain L13745 reported from USA (for an adult) and with HAstV3 strain (Berlin strain, AF141381) reported from Germany for a child (Figure 2).
Figure 2.

Phylogenetic analysis of deduced amino acid sequences (from 449bp fragment of the partial ORF2 encoding capsid gene) of human astrovirus strains detected in Kolkata, India. The Kolkata strains are indicated by black symbols. Scale bar indicates amino acid substitution per site. Reference sequences were obtained from GenBank under accession nos. HAstV1 (L23513/ USA), HAstV2 (L13745/USA), HAstV3 (AF141381/GER), HAstV4 (AY720891/GER), HAstV5 (DQ028633/BRZ), HAstV6 (HM237363/ JPN), HAstV7 (AF248738/SA), HAstV8 (AF260508/MEX), HAstVMLB1 (FJ402983/USA), HAstV1 (AB000301/PAK), HAstVVA1 (FJ973602/ USA) and TAstV1 (Y15936/NOR). The nucleotide sequences of 449bp ORF2 fragments of the HAstVs from Kolkata (variants marked with bold face) were deposited in DDBJ under accession nos. IDH198/AB551371, IDH1300/ AB540662; IDH1309/ AB548400; IDH1371/ AB548401; IDH1387/AB551372; IDH1448/AB548402; IDH1451/AB548403; IDH1482/AB551373; IDH2211/ AB548404; IDH2310/AB548405; IDH2392/AB551374; IDH2455/AB551375.
Discussion
Twelve interesting recombinants of human astroviruses (HAstV _ ORF1a _ ORF2) are reported herewith as HAstV _ G8_ G2 (n=1), HAstV _ G8_ G1 (n=10) and HAstV _ G2_ G3 (n=1) that were detected in course of the surveillance study in Kolkata, India. A novel recombinant strain of HAstV was reported earlier in children [19]. This study brings new evidence of inter-genotype recombinant-like astrovirus strains associated with acute gastroenteritis in adults involving different genotypes (ORF1a_G8 with ORF2_G2, ORF1a_G8 with ORF2 _G1). The recombinants among infants or children from Kolkata were ORF1a_G8 with ORF2 _G1 and ORF1a_G2 with ORF2_G3. For the first time to the best of our knowledge from Kolkata, India, the conserved stretch of amino acids spanning the 289bp fragment of ORF1a (in 4 strains of genotype G8 and one strain of genotype G2) and 449bp fragment of ORF2 (in 3 strains) showed unique amino acid changes indicating emergence of unique variants of HAstVs, hitherto unreported among the eight recognized genotypes of astroviruses. The ongoing surveillance to track emergence of interesting variants of astroviruses and study their association with acute viral gastroenteritis among adults, infants or children, as sole or mixed infections has yielded interesting data and indicates that continuous monitoring is essential for better understanding of the severity of astrovirus infections. Further, molecular characterization of astroviruses is of utmost importance in the context of increasing our awareness and understanding their genetic diversity.
Acknowledgments
We sincerely thank all surveillance staff of NICED Unit at Infectious Diseases and Beliaghata General Hospital (ID&BGH) for sample collection and clinical information from all the enrolled cases. The study was funded by Okayama University Program of Founding Research Center for Emerging and Reemerging Infectious Diseases, Ministry of Education, Culture, Sports, Science and Technology of Japan. We would like to thank Prof. Yoshifumi Takeda and Dr G. Balakrish Nair for constant encouragement. We are thankful to our research trainee Parna Banerjee and staff members Md. Mussaraf Hossain, Bimal Bera and Khokon Sen for invaluable technical support. We are thankful to Ainul Haque for data analysis. M. Pativada and Nataraju S.M are supported by Senior Research Fellowships from Indian Council of Medical Research (ICMR).
Conflict of interest
None of the authors have a commercial or other association that might pose a conflict of interest.
Ethical clearance
This study was reviewed and approved by the institutional ethics committee of National Institute of Cholera & Enteric Diseases
References
- 1.Chen SY, Chang YC, Lee YS, Chao HC, Tsao KC, Lin TY, Ko TY, Tsai CN, Chiu CH. Molecular epidemiology and clinical manifestation of viral gastroenteritis in hospitalized pediatric patients in Northern Taiwan. J Clin Microbiol. 2007;45:2054–2057. doi: 10.1128/JCM.01519-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhattacharya R, Sahoo GC, Nayak MK, Ghosh S, Dutta P, Bhattacharya MK, Mitra U, Gangopadhyay D, Dutta S, Niyogi SK, Saha DR, Naik TN, Bhattacharya SK, Krishnan T. Molecular epidemiology of human astrovirus infection in Kolkata, India. Infect Genet Evol. 2006;6:425–435. doi: 10.1016/j.meegid.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 3.Belliot G, Lavern H, Monroe SS. Outbreak of gastroenteritis in military recruits associated with serotype 3 astrovirus infection. J Med Virol. 1997;51:101–106. doi: 10.1002/(sici)1096-9071(199702)51:2<101::aid-jmv3>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 4.Koci MD, Schultz-Cherry S. Avian astroviruses. Avian Pathol. 2002;31:213–227. doi: 10.1080/03079450220136521. [DOI] [PubMed] [Google Scholar]
- 5.Chu DK, Poon LL, Guan Y, Peiris JS. Novel astroviruses in insectivorous bats. J Virol. 2008;82:9107–9114. doi: 10.1128/JVI.00857-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Quan PL, Wagner TA, Briese T, Torgerson TR, Hornig M, Tashmukhamedova A, Firth C, Palacios G, Baisre-De-Leon A, Paddock CD, Hutchison SK, Egholm M, Zaki SR, Goldman JE, Ochs HD, Lipkin WI. Astrovirus encephalitis in boy with X-linked Agammaglobulinemia. Emerg Infect Dis. 2010;16:918–925. doi: 10.3201/eid1606.091536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Grazia S, Glammanco GM, Colomba C, Cascio A, Arista S. Molecular epidemiology of astrovirus infection in Italian children with gastroenteritis. Clin Microbiol Infect. 2004;10:1025–1029. doi: 10.1111/j.1469-0691.2004.00995.x. [DOI] [PubMed] [Google Scholar]
- 8.Liu MQ, Yang BF, Peng JS, Zhou DJ, Tang L, Wang B, Liu Y, Sun SH, Ho WZ. Molecular epidemiology of astrovirus infection in infants in Wuhan, China. J Clin Microbiol. 2007;45:1308–1309. doi: 10.1128/JCM.00010-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finkbeiner SR, Le BM, Holtz LR, Storch GA, Wang D. Detection of newly described astrovirus MLB1 in stool samples from children. Emerg Infect Dis. 2009;15:441–444. doi: 10.3201/1503.081213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Finkbeiner SR, Holtz LR, Jiang Y, Rajendran P, Franz CJ, Zhao G, Kang G, Wang D. Human stool contains a previously unrecognized diversity of novel astroviruses. Virology Journal. 2009;8:161–165. doi: 10.1186/1743-422X-6-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nair GB, Ramamurthy T, Bhattacharya MK, Krishnan T, Ganguly S, Saha DR, Rajendran K, Manna B, Ghosh M, Okamoto K, Takeda Y. Emerging trends in the etiology of enteric pathogens as evidenced from an active surveillance of hospitalized diarrheal patients in Kolkata, India. Gut Pathogens. 2010;2:4. doi: 10.1186/1757-4749-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Noel JS, Lee TW, Kurtz JB, Glass RI, Monroe SS. Typing of human astroviruses from clinical isolates by enzyme immunoassay and nucleotide sequencing. J Clin Microbiol. 1995;33:797–801. doi: 10.1128/jcm.33.4.797-801.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gabby YB, Chamone CB, Nakamura LS, Oliveira DS, Abreu SF, Cavalcante-Pepino EL, Mascarenhas JD, Leite JP, Linhares AC. Characterization of an astrovirus genotype 2 strain causing an extensive outbreak of gastroenteritis among Maxakali Indians, South Brazil. J Clin Virol. 2006;37:287–292. doi: 10.1016/j.jcv.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 14.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis(MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 17.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 18.Ruuska T, Vesikari T. Rotavirus disease in Finnish children: use of numerical scores for clinical severity of diarrhoel episodes. Scand J Infect Dis. 1990;22:259–267. doi: 10.3109/00365549009027046. [DOI] [PubMed] [Google Scholar]
- 19.Walter JE, Brings J, Guerrero ML, Matson DO, Pickering LK, Ruiz-Palacios G, Berke T, Mitchell DK. Molecular characterization of a novel recombinant strain of human astrovirus associated with gastroenteritis in children. Arch Virol. 2001;146:2357–2367. doi: 10.1007/s007050170008. [DOI] [PMC free article] [PubMed] [Google Scholar]


