Abstract
Gene expression profiling studies have distinguished diffuse large B-cell lymphomas (DLBCLs) by cell of origin, with distinct pathogenetic mechanisms and prognosis. We attempted to identify DLBCL molecular subtypes in an epidemiologic study of 214 DLBCL patients diagnosed during 1998-2000 with archival tissues to investigate etiology. Immunohistochemical staining for CD10, BCL6, LMO2, MUM1/IRF4, and BCL2 and fluorescence in situ hybridization for t(14;18) were conducted, with ≥93% blinded duplicate agreement. CD10, LMO2, and BCL2 expression was similar to previous reports (32%, 44%, and 44% of DLBCLs, respectively), but BCL6 and MUM1/IRF4 expression was lower than expected (29% and 5%, respectively). We classified 112/214 (52%) cases as germinal center B-cell-like DLBCL (GCB-DLBCL; Hans et al., Blood 2004; CD10+ or CD10-/BCL6+/MUM1-), with no difference in prognosis compared with non-GCB-DLBCL (Cox regression, P=0.48). Comparing other GCB correlates, LMO2 expression and t(14;18) were more common but not exclusive to GCB-DLBCL as defined in our study, whereas BCL2 expression did not differ between DLBCL molecular subtypes. We could not confidently identify patients with GCB-DLBCL using these immunohistochemistry-based markers on archival tissues.
Keywords: diffuse large B-cell lymphoma, germinal center, molecular epidemiology, immunohistochemistry
Introduction
Diffuse large B-cell lymphoma (DLBCL), the most common non-Hodgkin lymphoma (NHL) subtype, represents a heterogeneous group of malignancies [1]. Although DLBCL is potentially curable, with approximately 50% average five-year overall survival, the clinical course varies widely, and the International Prognostic Index (IPI) remains the best clinical prognostic predictor available [2-3].
Studies of gene expression profiles have distinguished DLBCL subtypes by cell of origin, demonstrating that patients with germinal center B-cell-like DLBCL (GCB-DLBCL) have a substantially better prognosis than patients with non-GCB-DLBCL [4]. Additional research has shown that the disease subtypes are characterized by distinct oncogenic events [e.g., t(14;18) in GCB-DLBCL and constitutive NFkB activation in non-GCB-DLBCL] [5-6], leading us to hypothesize that the disease subtypes may also have distinct etiologies.
Identification of GCB- versus non-GCB DLBCL in epidemiologic studies to investigate etiology requires a biomarker that can be applied to fixed archival tissue specimens, since few epidemiologic studies collect fresh tissues. Using immunohistochemical (IHC) staining, Hans et al. distinguished DLBCL subtypes based on expression of three key markers of B-cell differentiation (CD10, BCL6, and MUM1/IRF4), by directly comparing results from IHC and gene expression for the same cases [7]. However, other IHC-based studies of protein expression and DLBCL prognosis using these or other combinations of markers have yielded inconsistent results [8-19], possibly due to small sample sizes, use of convenience clinical samples with variability in patient populations, methodological differences in IHC staining and interpretation, and varying ability of IHC to recapitulate the GEP signature [20-21].
In order to identify molecular subtypes of DLBCL in a population-based epidemiologic study with archival tissue, we systematically evaluated expression of CD10, BCL6, and MUM1/IRF4 as well as other GCB correlates [LMO2 and BCL2 expression, t(14;18)].
Materials and methods
Study population
The study population derived from the National Cancer Institute-Surveillance Epidemiology and End Results (NCI-SEER) NHL population-based case-control and prognosis studies, described in detail previously [22-23]. Briefly, the population-based case-control study included 1321 NHL cases (participation rate 76%) diagnosed during 1998-2000, aged 20-74 years, without known HIV infection, identified using rapid case ascertainment among residents of four SEER registries (Iowa, Detroit, Los Angeles, Seattle). Demographic data were obtained from patient interviews, and clinical data (date of diagnosis, stage, presence of B-symptoms, first course of therapy, date of last follow-up, and vital status) were obtained for 99% of cases from linkage to the SEER registries most recently in early 2008. Institutional Review Boards at the National Cancer Institute and each SEER center approved the study protocol. Participants provided written, informed consent prior to completing the interview.
Pathology
All 1321 NHL cases were initially histologically confirmed as NHL and coded according to the International Classification of Diseases for Oncology, 2nd Edition (ICD-O-2) by the local diagnosing pathologist. Updated ICD-O-2/ICD-O-3 codes based on local pathology review were received during the 2008 SEER record linkage. Pathology reports were obtained for 1215/1321 (92%) patients for review by an expert hematopathologist (Mohammad A. Vasef, University of New Mexico), who classified cases according to the World Health Organization classification for lymphoid neoplasms and assigned a confidence score to the subtype diagnosis (≥90% versus <90%). For all cases with low confidence in the NHL subtype classification as well as a 10% random sample of cases with high confidence in the NHL subtype classification, additional immunostaining was conducted to establish the NHL subtype for those patients with available pathology material (N=472). All cases were then assigned a final diagnosis based on the review of tumor specimens (N=472) or pathology reports (N=743), updated ICD-O-2 code from the SEER record linkage if pathology review data were not available (N=76), or original ICD-O-2 code from SEER at the time of case identification if updated data were not available (N=30).
Cases assigned a final diagnosis of DLBCL (ICD-O-2: 9680-84, 9688, 9712; ICD-O-3: 9678-80, 9684; N=417, of which 381 were confirmed by pathology report and/or specimen review) were eligible for this analysis. Sufficient archived, unstained slides (5-micron sections) from formalin-fixed, paraffin-embedded tumors were available for 240/417 (58%) DLBCL patients for laboratory analysis; tumor specimens were not available for the remaining patients (N=177, 42%).
Laboratory methods
Immunohistochemistry (IHC) staining was conducted for five markers (CD10, BCL6, MUM1/ IRF4, LMO2, and BCL2) according to previously published methods [7, 16]. Samples were evaluated by expert hematopathologists, and those with ≥30% tumor cells stained were considered positive. Successful staining was achieved for ≥94% of slides for all markers. The final analytic study population included 214 DLBCL patients with IHC data for all five markers and available follow-up and clinical data. Duplicate slides from 28 randomly selected individuals were interspersed and blinded from the laboratory and hematopathologists, with ≥93% agreement for all markers. Additional information on the antibodies and approach to interpretation is available upon request.
We also identified tumors with the t(14;18) chromosomal translocation, the most common cytogenetic abnormality in NHL, using fluorescence in situ hybridization according to previously published methods (Bhavana J. Dave, Smrati Jain, University of Nebraska) [24].
Statistical Analysis
The prognostic significance of marker expression was evaluated using hazard ratios (HRs) and 95% confidence intervals (CIs) derived from multivariate Cox proportional hazards regression, adjusting for age, demographics (sex, race, study center, years of education), and clinical factors (stage, presence of B-symptoms, type of initial therapy) using a continuous risk score (SAS v9.1, SAS Institute, Inc., Cary, NC) [23]. Characteristics of the DLBCL molecular subtypes were compared using the Pearson chi-square statistic.
Results
The study population was predominantly non-Hispanic white (90%) and male (59%) with a median age at DLBCL diagnosis of 59 years (Table 1). Characteristics of the present DLBCL case subset were comparable to all DLBCL cases from the parent case-control study. Clinically, 28% of patients had B-symptoms, and 86% received initial chemotherapy (pre-rituximab era). During follow-up, 69 (32%) patients died. The median follow-up of living patients was 7.7 years (range, 2.3-9.0).
Table 1.
Clinical and demographic characteristics among all DLBCL patients and by DLBCL molecular subtype*
| DLBCL patients in the present analysis | ||||
|---|---|---|---|---|
| All DLBCL patients | Total | GCB-DLBCL | Non-GCB-DLBCL | |
| Characteristic | N (%) | N (%) | N (%) | N (%) |
| Total | 417 (100.0) | 214 (100.0) | 112 (100.0) | 102 (100.0) |
| Age at DLBCL diagnosis | ||||
| <45 years | 96 (23.0) | 45 (21.0) | 22 (19.6) | 23 (22.5) |
| 45-64 | 196(47.0) | 96 (44.9) | 52 (46.4) | 44 (43.1) |
| 65+ | 125 (30.0) | 73(34.1) | 38 (33.9) | 35 (34.3) |
| Sex | ||||
| Male | 236 (56.6) | 125 (58.4) | 68 (60.7) | 57 (55.6) |
| Female | 181 (43.4) | 89 (41.6) | 44 (39.3) | 45 (44.1) |
| Race | ||||
| White | 360 (86.3) | 186 (86.9) | 97 (86.6) | 89 (87.3) |
| Non-white | 57 (13.7) | 28(13.1) | 15 (13.4) | 13 (12.7) |
| Education | ||||
| <12 years | 42(10.1) | 24 (11.2) | 11 (9.8) | 13 (12.7) |
| 12-15 | 250(60.1) | 129 (60.3) | 67 (59.8) | 62 (60.8) |
| 16+ | 124 (29.8) | 61(28.5) | 34 (30.4) | 27 (26.5) |
| Study center | ||||
| Detroit | 100 (24.0) | 37 (17.3) | 23 (20.5) | 14 (13.7) |
| Iowa | 119 (28.5) | 81(37.9) | 39 (34.8) | 42 (41.2) |
| Los Angeles | 99(23.7) | 43(20.1) | 24 (21.4) | 19 (18.6) |
| Seattle | 99(23.7) | 53 (24.8) | 26 (23.2) | 27 (26.5) |
| B-symptoms | ||||
| No | 148 (35.5) | 90(42.1) | 47 (42.0) | 43 (42.2) |
| Yes | 112 (26.9) | 60 (28.0) | 29 (25.9) | 31 (30.4) |
| Unknown | 157 (37.6) | 64 (29.9) | 36 (32.1) | 28 (27.5) |
| Stage | ||||
| Local | 137 (32.9) | 70(32.7) | 39 (34.8) | 31 (30.4) |
| Regional | 96 (23.0) | 56 (26.2) | 27 (24.1) | 29 (28.4) |
| Distant | 164 (39.3) | 83 (38.8) | 44 (39.3) | 39 (38.2) |
| Unknown | 20 (4.8) | 5 (2.3) | 2 (1.8) | 3 (2.9) |
| Initial chemotherapy | ||||
| No | 55 (13.2) | 29 (13.6) | 18 (16.1) | 11 (10.8) |
| Yes | 355(85.1) | 185 (86.4) | 94 (83.9) | 91 (89.2) |
| Unknown | 7(1.7) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| t(14;18) translocation | ||||
| Negative | 120 (28.8) | 120(56.1) | 56 (50.0) | 64 (62.7) |
| Positive | 92(22.1) | 92 (43.0) | 55 (49.1) | 28 (36.3) |
| Unknown | 205 (49.2) | 2 (0.9) | 1 (0.9) | 1 (1.0) |
Abbreviations: diffuse large B-cell lymphoma (DLBCL), germinal center B-cell (GCB).
DLBCL molecular subtype assigned using Hans et al.[7] (GCB-DLBCL: CD10+ or CD10-/BCL6+/MUM1-).
CD10, LMO2, and BCL2 were expressed in 32%, 44%, and 44% of DLBCLs, respectively, similar to previous reports, but expression of BCL6 and MUM1/IRF4 was lower than expected (29% and 5%, respectively). Of the five markers evaluated, the strongest individual predictors of overall survival were BCL2 (44% positive, HR=2.15, 95%CI 1.33-3.50, P=0.0019) and LMO2 (44% positive, HR=0.62, 95%CI 0.38-1.03, P=0.062; Table 2).
Table 2.
Prognostic significance of immunohistochemical markers, individually and in combination, in 214 patients with histologically confirmed DLBCL from a population-based study
| HR (95% CI) |
||||||
|---|---|---|---|---|---|---|
| N (%) | % Patients Deceased | Unadjusted | P | Adjusted* | P | |
| Individual markers | ||||||
| BCL6 | ||||||
| Negative | 151(70.6) | 35.1 | 1.00 (referent) | 1.00 (referent) | ||
| Positive | 63 (29.4) | 25.4 | 0.70 (0.40-1.22) | 0.20 | 0.77 (0.44-1.35) | 0.36 |
| CD10 | ||||||
| Negative | 146 (68.2) | 35.1 | 1.00 (referent) | 1.00 (referent) | ||
| Positive | 68 (31.8) | 33.8 | 1.15(0.70-1.90) | 0.58 | 1.12 (0.68-1.85) | 0.65 |
| LMO2 | ||||||
| Negative | 119 (55.6) | 37.8 | 1.00 (referent) | 1.00 (referent) | ||
| Positive | 95 (44.4) | 25.3 | 0.59 (0.36-0.97) | 0.038 | 0.62 (0.38-1.03) | 0.062 |
| MUM1/IRF4 | ||||||
| Negative | 204 (95.3) | 31.9 | 1.00 (referent) | 1.00 (referent) | ||
| Positive | 10(4.7) | 40 | 1.33 (0.48-3.65) | 0.58 | 1.17 (0.42-3.23) | 0.77 |
| BCL2 | ||||||
| Negative | 120(56.1) | 22.5 | 1.00 (referent) | 1.00 (referent) | ||
| Positive | 94 (43.9) | 44.7 | 2.26 (1.39-3.67) | 0.0010 | 2.15 (1.33-3.50) | 0.0019 |
| Combinations of markers | ||||||
| Hans et al.[7]† | ||||||
| Non-GCB-DLBCL | 102(47.7) | 36.3 | 1.00 (referent) | 1.00 (referent) | ||
| GCB-DLBCL | 112 (52.3) | 28.6 | 0.78 (0.49-1.26) | 0.31 | 0.84 (0.52-1.35) | 0.48 |
Abbreviations: confidence interval (CI), diffuse large B-cell lymphoma (DLBCL), germinal center B-cell (GCB), hazard ratio (HR)
Adjusted for age, demographic, and clinical factors.
GCB-DLBCL: CD10+ or CD10-/BCL6+/MUM1-, non-GCB-DLBCL: CD10-/BCL6- or CD10-/BCL6-/MUM1+
Based on the combination of markers recommended by Hans et al. [7], we classified 112 patients with GCB-DLBCL (CD10+ or CD10-/ BCL6+/MUM1-), and while these patients had somewhat better overall survival than patients with non-GCB-DLBCL (HR=0.84, 95%CI 0.52-1.35; Table 2), this was not statistically significant (P=0.48). To further evaluate the molecular classification of the DLBCLs, we compared expression of other GCB correlates. LMO2 expression and the t(14;18) were more common in GCB- than non-GCB-DLBCL [LMO2: 61% versus 26%, P<0.0001; t(14;18): 49% versus 36%, P=0.06), whereas BCL2 expression did not differ between the DLBCL molecular subtypes (51% versus 44%, P=0.32). Other clinical and demographic characteristics also were similar among patients with GCB- and non-GCB-DLBCL (Table 1).
Discussion
We used IHC staining and fluorescence in situ hybridization for six key markers of B-cell differentiation to attempt to discriminate DLBCL molecular subtypes in a population-based epidemiologic study with archival tissues. Using Hans et al. [7], we classified 112 (52%) cases as GCB-DLBCL (CD10+ or CD10-/BCL6+/MUM1-), but patterns of additional molecular characteristics in our data suggested that we could not be confident in our classification. We observed the t(14;18) in 37% of non-GCB-DLBCL, whereas other series have reported a prevalence of <5% [6]. This observation suggests misclassification of the GCB-DLBCL subtype in our cases because of the high reliability of using fluorescence in situ hybridization to identify the t(14;18). Further supporting the likelihood of misclassification, expression of the GC marker LMO2 was absent in 39% of GCB-DLBCL and present in 26% of non-GCB-DLBCL, whereas expression of the activated B-cell marker BCL2 was present in 51% of GCB-DLBCL and absent in 56% of non-GCB-DLBCL. Although both LMO2 and BCL2 are expressed in a subset of lymphocytes at other stages of differentiation [16, 25], their lack of clear clustering with the DLBCL molecular subtypes in our data suggests that we cannot rule out the possibility of misclassification of the GCB-DLBCL subtype in our cases.
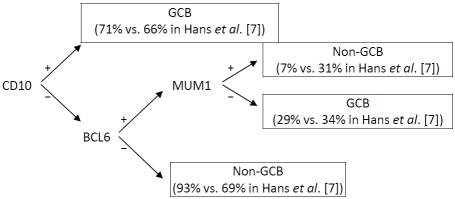
Our experience is illustrative of the promise and the challenges of molecular subtyping in epidemiologic studies. Our duplicate quality control samples indicated ≥93% agreement for all markers, supporting the internal consistency of our results. However, previous research has demonstrated that variability in IHC staining and interpretation can markedly affect results, particularly for BCL6 and MUM1/IRF4 [26]. This variability and degradation of the antigens on our stored slides may account for the lower than expected percentage of cases staining positive for each of these markers in our study and would contribute to misclassification of the DLBCL molecular subtypes. Specifically, a substantially larger proportion of our cases compared with Hans et al. [7] were classified as non -GCB-DLBCL (93% vs. 69%) due to a lack of BCL6 expression (Figure 1).
Figure 1.
Comparison of the assignment of GCB- versus non-GCB DLBCL in our cases versus Hans et al.[7]
Although our study was population-based and used rapid reporting, we were unable to enroll cases with rapidly fatal disease. Observed survival in our study was therefore better than expected in the general population, but consistent with survival estimates for DLBCL patients surviving one year following diagnosis [23]. Exclusion of cases with rapidly fatal disease may have limited our ability to assess the prognostic significance of certain markers and decreased the proportion of non-GCB-DLBCLs, which have worse prognosis, in our population. Additionally, we were unable to obtain tumor tissue for all cases, though patient characteristics were similar for those with and without tissue. Detailed treatment and clinical data were not available, but our demographic and clinical risk score has a level of predictability similar to the IPI [5, 23].
Finally, we did not evaluate GCET1 and FOXP1, which were recently shown to modestly improve identification of GCB-DLBCL compared with Hans et al.[27].
Discrimination of DLBCL molecular subtypes in epidemiologic studies requires identification of valid and reliable markers that can be used on archival tissues, are robust to variability in patient populations (e.g., by treatment, race, or comorbid conditions such as AIDS),[12, 14-15, 17-19] and have high inter-laboratory agreement [26]. Establishment of such markers will also facilitate widespread adoption of DLBCL molecular subtyping in routine clinical care. Although IHC staining is a standard approach, methodological differences in IHC staining and interpretation for some of the current key B-cell markers suggest the need for new markers, further optimization of current markers for archival tissues, or the pursuit of alternative approaches [28].
Acknowledgments
We gratefully acknowledge the contributions of the staff and scientists at the SEER centers of Iowa, Los Angeles, Detroit, and Seattle for the conduct of the study's field effort; Freda Selk (University of Iowa) for coordination of tumor specimens; and Matthew Butcher, Mary McAdams, Pete Hui, Lonn Irish, and Michael Stagner (Information Management Services, Inc.) for programming support.
Disclosures
The authors have no conflicts of interest to disclose.
References
- 1.Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J, Vardiman JW. Lyon: IARC Press; 2008. World Health Organization Classification of Tumours of Haematopoietic and Lymphoid Tissues. [Google Scholar]
- 2.The International Non-Hodgkin's Lymphoma Prognostic Factors Project. A predictive model for aggressive non-Hodgkin's lymphoma. New Engl J Med. 1993;329:987–994. doi: 10.1056/NEJM199309303291402. [DOI] [PubMed] [Google Scholar]
- 3. Surveillance, Epidemiology, and End Results (SEER) Program ( www.seer.cancer.gov) SEER*Stat Database: Incidence - SEER 13 Regs Limited-Use, Nov 2008 Sub (1992-2006). National Cancer Institute, DCCPS, Surveillance Research Program, Cancer Statistics Branch, released April 2009, based on the November 2008 submission, 2009.
- 4.Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, Powell JI, Yang L, Marti GE, Moore T, Hudson J, Jr, Lu L, Lewis DB, Tibshirani R, Sherlock G, Chan WC, Greiner TC, Weisenburger DD, Armitage JO, Warnke R, Levy R, Wilson W, Grever MR, Byrd JC, Botstein D, Brown PO, Staudt LM. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 5.Rosenwald A, Wright G, Chan WC, Connors JM, Campo E, Fisher RI, Gascoyne RD, Muller-Hermelink HK, Smeland EB, Giltnane JM, Hurt EM, Zhao H, Averett L, Yang L, Wilson WH, Jaffe ES, Simon R, Klausner RD, Powell J, Duffey PL, Longo DL, Greiner TC, Weisenburger DD, Sanger WG, Dave BJ, Lynch JC, Vose J, Armitage JO, Montserrat E, Lopez-Guillermo A, Grogan TM, Miller TP, LeBlanc M, Ott G, Kvaloy S, Delabie J, Holte H, Krajci P, Stokke T, Staudt LM, the Lymphoma/Leukemia Molecular Profiling P The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. New Engl J Med. 2002;346:1937–1947. doi: 10.1056/NEJMoa012914. [DOI] [PubMed] [Google Scholar]
- 6.Leich E, Hartmann E, Burek C, Ott G, Rosenwald A. Diagnostic and prognostic significance of gene expression profiling in lymphomas. APMIS. 2007;115:1135. doi: 10.1111/j.1600-0463.2007.apm_867.xml.x. [DOI] [PubMed] [Google Scholar]
- 7.Hans CP, Weisenburger DD, Greiner TC, Gascoyne RD, Delabie J, Ott G, Muller-Hermelink HK, Campo E, Braziel RM, Jaffe ES, Pan Z, Farinha P, Smith LM, Falini B, Banham AH, Rosenwald A, Staudt LM, Connors JM, Armitage JO, Chan WC. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103:275–282. doi: 10.1182/blood-2003-05-1545. [DOI] [PubMed] [Google Scholar]
- 8.Colomo L, Lopez-Guillermo A, Perales M, Rives S, Martinez A, Bosch F, Colomer D, Falini B, Montserrat E, Campo E. Clinical impact of the differentiation profile assessed by immunophenotyping in patients with diffuse large B-cell lymphoma. Blood. 2003;101:78–84. doi: 10.1182/blood-2002-04-1286. [DOI] [PubMed] [Google Scholar]
- 9.Berglund M, Thunberg U, Amini RM, Book M, Roos G, Erlanson M, Linderoth J, Dictor M, Jerkeman M, Cavallin-Stahl E, Sundstrom C, Rehn-Eriksson S, Backlin C, Hagberg H, Rosenquist R, Enblad G. Evaluation of immunophenotype in diffuse large B-cell lymphoma and its impact on prognosis. Mod Pathol. 2005;18:1113–1120. doi: 10.1038/modpathol.3800396. [DOI] [PubMed] [Google Scholar]
- 10.Zinzani PL, Dirnhofer S, Sabattini E, Alinari L, Piccaluga PP, Stefoni V, Tani M, Musuraca G, Marchi E, Falini B, Baccarani M, Pileri SA. Identification of outcome predictors in diffuse large B-cell lymphoma. Immunohistochemical profiling of homogenously treated de novo tumors with nodal presentation on tissue micro-arrays. Hematol J. 2005;90:341–347. [PubMed] [Google Scholar]
- 11.De Paepe P, De Wolf-Peeters C. Diffuse large B-cell lymphoma: a heterogeneous group of non-Hodgkin lymphomas comprising several distinct clinicopathological entities. Leukemia. 2006;21:37–43. doi: 10.1038/sj.leu.2404449. [DOI] [PubMed] [Google Scholar]
- 12.Haarer CF, Roberts RA, Frutiger YM, Grogan TM, Rimsza LM. Immunohistochemical classification of de novo, transformed, and relapsed diffuse large B-cell lymphoma into germinal center B-cell and nongerminal center B-cell subtypes correlates with gene expression profile and patient survival. Arch Pathol Lab Med. 2006;2006:1819. doi: 10.5858/2006-130-1819-ICODNT. [DOI] [PubMed] [Google Scholar]
- 13.Muris JJF, Meijer C, Vos W, van Krieken JHJM, Jiwa NM, Ossenkoppele GJ, Oudejans JJ. Immunohistochemical profiling based on Bcl-2, CD10 and MUM1 expression improves risk stratification in patients with primary nodal diffuse large B cell lymphoma. J Pathol. 2006;208:714–723. doi: 10.1002/path.1924. [DOI] [PubMed] [Google Scholar]
- 14.Nyman H, Adde M, Karjalainen-Lindsberg M-L, Taskinen M, Berglund M, Amini R-M, Blomqvist C, Enblad G, Leppa S. Prognostic impact of immunohistochemically defined germinal center phenotype in diffuse large Bcell lymphoma patients treated with immunochemotherapy. Blood. 2007;109:4930–4935. doi: 10.1182/blood-2006-09-047068. [DOI] [PubMed] [Google Scholar]
- 15.Fu K, Weisenburger DD, Choi WWL, Perry KD, Smith LM, Shi X, Hans CP, Greiner TC, Bierman PJ, Bociek RG, Armitage JO, Chan WC, Vose JM. Addition of rituximab to standard chemotherapy improves the survival of both the germinal center B-cell-like and nongerminal center B-cell-like subtypes of diffuse large B-cell lymphoma. J Clin Oncol. 2008;26:4587–4594. doi: 10.1200/JCO.2007.15.9277. [DOI] [PubMed] [Google Scholar]
- 16.Natkunam Y, Farinha P, Hsi ED, Hans CP, Tibshirani R, Sehn LH, Connors JM, Gratzinger D, Rosado M, Zhao S, Pohlman B, Wongchaowart N, Bast M, Avigdor A, Schiby G, Nagler A, Byrne GE, Levy R, Gascoyne RD, Lossos IS. LMO2 protein expression predicts survival in patients with diffuse large B-cell lymphoma treated with anthracycline-based chemotherapy with and without rituximab. J Clin Oncol. 2008;26:447–454. doi: 10.1200/JCO.2007.13.0690. [DOI] [PubMed] [Google Scholar]
- 17.Anderson JJ, Fordham S, Overman L, Dignum H, Wood K, Proctor SJ, Crosier S, Angus B, Culpin RE, Mainou-Fowler T. Immunophenotyping of diffuse large B-cell lymphoma (DLBCL) defines multiple sub-groups of germinal centre-like tumours displaying different survival characteristics. Int J Oncol. 2009;35:961–971. doi: 10.3892/ijo_00000409. [DOI] [PubMed] [Google Scholar]
- 18.Chadburn A, Chiu A, Lee JY, Chen X, Hyjek E, Banham AH, Noy A, Kaplan LD, Sparano JA, Bhatia K, Cesarman E. Immunophenotypic analysis of AIDS-related diffuse large B-cell lymphoma and clinical implications in patients from AIDS Malignancies Consortium Clinical Trials 010 and 034. J Clin Oncol. 2009;27:5039–5048. doi: 10.1200/JCO.2008.20.5450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Y, Han T, Iqbal J, Irons R, Chan WC, Zhu X, Fu K. Diffuse large B-cell lymphoma in Chinese patients. Am J Clin Pathol. 2010;133:305–313. doi: 10.1309/AJCP4H6ADGYDZMOA. [DOI] [PubMed] [Google Scholar]
- 20.Gutierrez-Garcia G, Cardesa-Salzmann T, Climent F, Gonzalez-Barca E, Mercadal S, Mate JL, Sancho JM, Arenillas L, Serrano S, Escoda L, Martinez S, Valera A, Martinez A, Jares P, Pinyol M, Garcia-Herrera A, Martinez-Trillos A, Gine E, Villamor N, Campo E, Colomo L, Lopez-Guillermo A. Gene-expression profiling and not immunophenotypic algorithms predicts prognosis in patients with diffuse large B-cell lymphoma treated with immunochemotherapy. Blood. 2011;117:4836–4843. doi: 10.1182/blood-2010-12-322362. [DOI] [PubMed] [Google Scholar]
- 21.Meyer PN, Fu K, Greiner TC, Smith LM, Delabie J, Gascoyne RD, Ott G, Rosenwald A, Braziel RM, Campo E, Vose JM, Lenz G, Staudt LM, Chan WC, Weisenburger DD. Immunohistochemical methods for predicting cell of origin and survival in patients with diffuse large B-cell lymphoma treated with rituximab. J Clin Oncol. 2011;29:200–207. doi: 10.1200/JCO.2010.30.0368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chatterjee N, Hartge P, Cerhan JR, Cozen W, Davis S, Ishibe N, Colt J, Goldin L, Severson RK. Risk of non-Hodgkin's lymphoma and family history of lymphatic, hematologic, and other cancers. Cancer Epidemiol Biomarkers Prev. 2004;13:1415–1421. [PubMed] [Google Scholar]
- 23.Habermann TM, Wang SS, Maurer MJ, Morton LM, Lynch CF, Ansell SM, Hartge P, Severson RK, Rothman N, Davis S, Geyer SM, Cozen W, Chanock SJ, Cerhan JR. Host immune gene polymorphisms in combination with clinical and demographic factors predict late survival in diffuse large B-cell lymphoma patients in the pre-rituximab era. Blood. 2008;112:2694–2702. doi: 10.1182/blood-2007-09-111658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chiu BCH, Dave BJ, Blair A, Gapstur SM, Zahm SH, Weisenburger DD. Agricultural pesticide use and risk of t(14;18)-defined subtypes of non-Hodgkin lymphoma. Blood. 2006;108:1363–1369. doi: 10.1182/blood-2005-12-008755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iqbal J, Neppalli VT, Wright G, Dave BJ, Horsman DE, Rosenwald A, Lynch J, Hans CP, Weisenburger DD, Greiner TC, Gascoyne RD, Campo E, Ott G, Muller-Hermelink HK, Delabie J, Jaffe ES, Grogan TM, Connors JM, Vose JM, Armitage JO, Staudt LM, Chan WC. BCL2 expression is a prognostic marker for the activated B-cell-like type of diffuse large B-cell lymphoma. J Clin Oncol. 2006;24:961–968. doi: 10.1200/JCO.2005.03.4264. [DOI] [PubMed] [Google Scholar]
- 26.de Jong D, Xie W, Rosenwald A, Chhanabhai M, Gaulard P, Klapper W, Lee A, Sander B, Thorns C, Campo E, Molina T, Hagenbeek A, Horning S, Lister A, Raemaekers J, Salles G, Gascoyne RD, Weller E. Immunohistochemical prognostic markers in diffuse large B-cell lymphoma: validation of tissue microarray as a prerequisite for broad clinical applications (a study from the Lunenburg Lymphoma Biomarker Consortium) J Clin Pathol. 2009;62:128–138. doi: 10.1136/jcp.2008.057257. [DOI] [PubMed] [Google Scholar]
- 27.Choi WW, Weisenburger DD, Greiner TC, Piris MA, Banham AH, Delabie J, Braziel RM, Geng H, Iqbal J, Lenz G, Vose JM, Hans CP, Fu K, Smith LM, Li M, Liu Z, Gascoyne RD, Rosenwald A, Ott G, Rimsza LM, Campo E, Jaffe ES, Jaye DL, Staudt LM, Chan WC. A new immunostain algorithm classifies diffuse large Bcell lymphoma into molecular subtypes with high accuracy. Clin Cancer Res. 2009;15:5494–5502. doi: 10.1158/1078-0432.CCR-09-0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rimsza LM, LeBlanc ML, Unger JM, Miller TP, Grogan TM, Persky DO, Martel RR, Sabalos CM, Seligmann B, Braziel RM, Campo E, Rosenwald A, Connors JM, Sehn LH, Johnson N, Gascoyne RD. Gene expression predicts overall survival in paraffin-embedded tissues of diffuse large B-cell lymphoma treated with R-CHOP. Blood. 2008;112:3425–3433. doi: 10.1182/blood-2008-02-137372. [DOI] [PMC free article] [PubMed] [Google Scholar]