Abstract
MiRNAs are a widespread class of small non-coding RNAs that have the ability to silence gene expression through sequence complementarity to their targets. We describe their initial discovery in the nematode C.elegans and review what is currently known about their biogenesis. The regulation of expression and processing of miRNAs, and the mechanisms through which miRNAs locate their correct targets are not yet fully understood. MiRNAs are involved in a multitude of developmental and pathological processes leading to an explosion of research in disparate subject areas. In this review we draw attention to placentally expressed miRNAs that can be detected in the maternal plasma; and we discuss their potential use as biomarkers in non-invasive prenatal diagnostics.
Keywords: Small non-coding RNAs, miRNAs, circulating nucleic acids, maternal plasma biomarkers, prenatal diagnostics
Discovery and function of miRNAs
When RNA is extracted from either tissues or cells a substantial pool of small RNA molecules is observed. For many years it was assumed that much of the small RNA species were simply products of RNA degradation as a result of RNA extraction procedures. It is now clear that several small non-coding RNA molecules are an integral part of the cellular machinery of gene expression. Well-known examples include the snRNAs (small nuclear RNAs involved in pre-mRNA splicing) and the snoRNAs (small nucleolar RNAs involved in ribosomal RNA processing). However, these classes of RNAs do not account for all of the small non-coding RNAs. We now know that in fact there are additional classes of small non-coding RNAs that are involved in the silencing of gene expression. These are subdivided into three types: the siRNAs (short interfering RNAs) which target mRNAs for destruction; other siRNAs which target chromatin for epigenetic modification; and the miRNAs (or microRNAs). MiRNAs are generally thought to regulate mRNA translation; but it is important to note that they have also been involved in epigenetic regulation in plants, and perhaps elsewhere.
The first miRNA was described in the nematode C.elegans by the Ambros group at Harvard University in 1993 [1]. Ambros and colleagues were screening C.elegans for mutants that affect the timing of the switching of cell fate in development. Two genes were identified, lin-4 and let-7. Much to their surprise, these genes did not encode proteins, but instead small RNAs which were subsequently called miRNAs (or miRNAs).
The target of lin-4 miRNA is the 3'UTR (untranslated region) of lin-14 mRNA. They found that lin-4 has strong base sequence complementarity to lin-14. The lin-4 miRNA works during larval development to regulate the expression of lin-14; lin-14 encodes a nuclear protein involved in regulating the temporal switch between early and late cell fates in C.elegans development. Lin-4 regulates Lin-14 expression by repressing mRNA translation. In C.elegans, the let-7 miRNA is required for the timing of cell fate determination. Let-7 is now known to be part of a large, evolutionarily conserved family of miRNAs that have been identified in a wide range of species. Its best known targets are the oncogenes H-Ras, N-Ras and K-Ras [2].
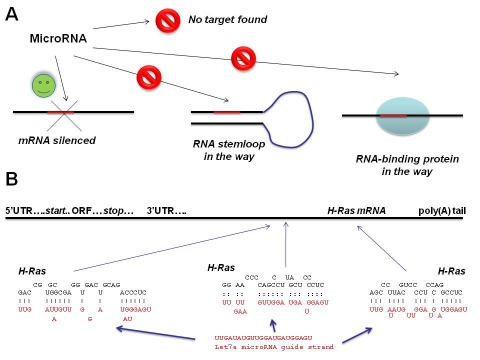
At the time of writing, over 15,000 miRNAs have been identified and catalogued in the public database miRBase (www.mirbase.org). MiRNAs have been identified in animals, plants and viruses. There are several ways to identify novel miRNAs. The first of these is to use traditional genetic screens, in which the gene responsible for a phenotype is identified and cloned - this is how Lin-4 and Let-7 were discovered. The second approach is direct cloning - in which small RNA molecules are physically isolated, for example out of an acrylamide gel, ligated to adaptors and cloned or amplified by PCR. This type of analysis is facilitated nowadays by the use of next generation sequencing technology, in which thousands of small RNAs can be sequenced in a short time frame - this is now the preferred approach in miRNA identification A third approach is to use bioinformatics: potential miRNAs can be inferred from genetic sequences. This is because miRNAs are transcribed as precursors with self-complementarity, so that they can be predicted to fold into a hairpin RNA. Figure 1 illustrates the structure of a typical miRNA: mammalian let -7a and its target sites in the 3' UTR of H-RAS mRNA.
Figure 1.
Target recognition by a typical miRNA. (A) A given miRNA may have several targets in an mRNA which could be masked by an RNA stemloop structure or by RNA-binding proteins. (B) Three target sequences of the human miRNA let-7a are shown in the 3'UTR of H-Ras mRNA [2]. Note the partial and differing level of sequence complementarity between the miRNA and its target sites. Note that in RNA the base T is represented by U (uracil) and that G can base pair with U.
The biological functions of miRNAs are increasingly wide-ranging [3]. It is now clear that they are involved in normal development and physiology in both animals and plants. In plants they control floral development and timing, leaf patterning and shape, vascular development and fertility, seed biology and the response to environmental stress [4-6]. In animals they are involved in several developmental processes including cardiovascular [7] and skeletal muscle development and disease [8]; in the response to stress [9] and in the control of cell proliferation and apoptosis [10]. MiRNAs have attracted much attention due to their involvement in cancer, where they can act as both oncogenes and tumour suppressors depending on their targets. MiRNAs are now biomarkers of specific cancers [11] and may point to novel therapeutic strategies [12]. Synthetic miRNAs can be used to mimic the effect of tumour suppressor miRNAs; conversely, synthetic “antagomirs” can also be designed to inactivate oncogenic miRNAs. The emerging roles of miRNAs in disease are not restricted to cancer, Some of the others diseases in which miRNAs have been implicated include type 2 diabetes [13]; and diseases of the central nervous system including Alzheimers, Parkinson's disease, Huntington's disease, schizophrenia and autism amongst several others [14]. Of particular relevance to the focus of this review, a recent study has shown that patterns of miRNA expression in human placentas appear to be altered in preeclampsia and preterm labour [15].
Biogenesis of miRNAs and target recognition
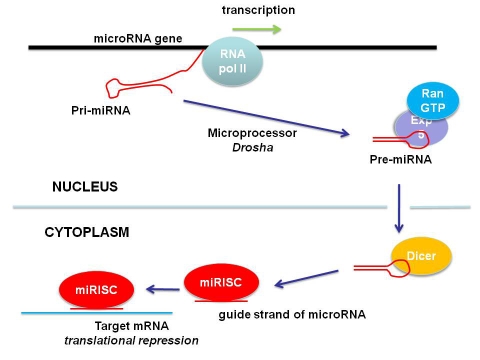
The biogenesis of miRNAs follows three sequential steps: (i) the transcription, generally by RNA polymerase II, of a primary transcript (pri-miRNA); followed by (ii) its partial processing into a precursor miRNA in the nucleus (pre-miRNA) and (iii) its final maturation into a functional miRNA in the cytoplasm [16]. The primary transcript (pri-miRNA) can be relatively long and is characterised by the formation of stem loop structures. In the nucleus, a macromolecular machinery known as the microprocessor then trims the pri-miRNA precursor through the activity of an enzyme called Drosha. This results in a pre-miRNA that is usually between 60-100 nucleotides long. The pre-miRNA is exported from the nucleus in a RanGTP-dependent process after binding with exportin-5. In the cytoplasm, pre-miRNA is further trimmed into a 22-nucleotide miRNA by the enzyme Dicer. The same happens in RNA interference, in which long dsRNA molecules are also trimmed by Dicer in the cytoplasm into 20-22 nucleotide siRNAs (short interfering RNAs). The processing of miRNAs in plants, however, is further complicated by the existence of several Dicer-like enzymes; thus Dicer-like 1 (DCL1) specifically processes microRNAs in the nucleus.
Thus a miRNA is effectively a genome-encoded siRNA. The distinction between siRNAs and miRNAs is to some extent blurred; however there are some notable differences. Small interfering RNAs are generally associated with the defence against viruses or transposons, whereas microRNAs target mRNAs. In plants, there is an additional class of siRNAs, the transacting siRNAs (or tasiRNAs). TasiRNAs are transcribed from the genome as polyadenylated, dsRNA precursors, producing a set of phased siRNAs that can effectively cooperate to silence the expression of target genes.
The two strands of a miRNA (or siRNA) are then separated; the ‘guide’ strand ends up in the RISC complex (RNA-induced silencing complex), leaving the ‘passenger’ strand behind. The guide strand is the strand which can then bind to its intended target through sequence complementarity. However the passenger strand is not always discarded, but can instead become part of a RISC complex itself; in this case it is indicated with an asterisk, the “star” miRNA. MiRNA*s may well have significant biological functions; for example in mammals miR155 and miR155* cooperatively regulate type I interferon production [17]. Furthermore, the paired use of miRNAs and miRNA*s can be tissue specific, adding another level of complexity to miRNA function [18]. A RISC complex with a miRNA guide strand is known as a miRISC complex. The miRISC complex, in the main, results in the translational repression of bound mRNAs by sterically hindering ribosome progress. However, as is the case with RNA interference, cleavage can occur through the “slicer” activity of Argonaute (Ago) proteins that are part of the RISC complex. The process of miRNA biogenesis is summarised in Figure 2.
Figure 2.
MiRNA biogenesis. A miRNA gene is transcribed by RNA polymerase II. The resultant primary transcript is termed the pri-miRNA. In the nucleus the pri-miRNAs is trimmed into a 60-100 nucleotide pre-miRNA by Drosha in the microprocessor complex. Bound by exportin 5 it is then exported via RanGTP into the cytoplasm. In the cytoplasm the nuclease Dicer further trims the pre-miRNA into a mature 22 nucleotide miRNA which becomes incorporated into a miRISC complex that retains the “guide strand”. The guide strand, perhaps through its “seed” sequence, then recognised the intended target sites frequently but not exclusively located in the 3' UTR of target mRNAs. Target mRNAs often have multiple target sites for any given miRNA; and a given miRNA may target several mRNAs.
The mechanisms through which miRNAs find their intended targets are not yet fully understood [19]. Whether or not a miRISC complex represses translation or induces active mRNA degradation can depend not only on the degree of sequence complementarity, but also on the nature of the specific Ago protein (which can be cleaving or non-cleaving). Whatever the mechanism involved, a miRNA will attenuate (i.e. silence) the expression of a target gene through the miRISC complex. Partial complementarity can in itself be thermodynamically stable; however the interaction between miRNA and target is likely to be modulated and faciltated by other factors, such as target-specific RNA binding proteins, or even secondary structures in the target RNA, or co-localisation of miRNAs and target mRNA in specific subcellular compartments. Whether or not a given miRNA binds to a putative target in vivo will depend on the accessibility of the target sequence. The target sequence can be masked by virtue of itself being part of a duplex; or because the target sequence is covered by an RNA-binding protein (Figure 1). Several lines of evidence point to the nucleotides at positions 2-8 at the 5' end of the miRNA as being particularly important for achieving target specificity (these are known as the seed sequence). However the role of the seed sequence in all target recognition events remains unclear as it is also possible to find a target without a strong seed sequence. In regards to the location of the miRNA targets in the mRNAs; their position does vary, although the vast majority have been reported in the 3'UTR. The 3'UTR is required, in many cases, for efficient translation; and there are also elements in the 3'UTR than can regulate mRNA stability and localisation - potentially, therefore, miRNAs can affect all of these processes. It is likely that 3'UTR sequences are favoured because they are not translated - therefore the translation machinery cannot displace associated miRNPs. It is also important to note that the translation of a single mRNA might be regulated by several miRNAs; and that a single miRNA might interact with and regulate the translation of several target mRNAs.
Regulation of miRNA expression
MiRNAs are encoded by genes; and as such, their expression is regulated. There are several ways in which their expression could be regulated: through either the regulation of transcription or processing [20]. Other layers of regulation are likely to emerge. For example the nuclear export of pre-miRNA precursors could be regulated; as could their localisation to specific compartments of the cells (mRNAs are localised to specific cytoplasmic compartments - so why not miRNAs?), their ability to find their target (variations in miRNP structure could affect this); their association with cytoplasmic compartments associated with the regulation of mRNA stability and RNA interference (P granules), and their stability and half-life could all be regulated. As miRNAs are now widely associated with developmental and disease processes, the question of what regulates them is topical.
An important issue to consider in terms of transcriptional regulation of miRNA expression is the position of the miRNA genes in the genome. Several miRNAs are found within the introns of other genes [21]. Therefore, they are co-transcribed with the gene that contains them; in other words, they are co-ordinately expressed. Thus the presence of miRNAs within the introns of specific sets of genes is because they are involved in the same biological pathways of the genes that contain them (their host genes). The processing of miRNA precursors out of introns, however, is not a trivial matter; and is in fact affected by the secondary structure of the intronic RNA and by RNA-binding proteins. On the other hand, several miRNAs are located in intergenic regions and are likely encoded by separate transcription units. In general, primary miRNA transcripts are transcribed by RNA poly-merase II. Like all genes, transcription is initiated via a promoter; and the promoters of miRNA genes are not yet well understood. However several transcription factors have been associated with the regulation of miRNA expression as illustrated by the following examples. The tumour suppressor p53 is a multifunctional transcription factor whose targets now include the miRNAs miR-17-92, miR-34, miR-145, miR-192 and miR-215. Many of these miRNAs are, like p53, also involved in the regulation of apoptosis and cell proliferation - thus these miRNAs can be considered to be effectors of p53 function [22]. MiR-21 is a widely expressed, reportedly oncogenic miRNA whose expression is regulated by several transcription factors including the androgen receptor (AR) [23]. AR binds to the miR-21 promoter directly and activates its transcription; in turn, miR-21 enhances prostate tumour growth in vivo [24].
Another important layer of miRNA regulation is posttranscriptional. That this should occur is illustrated by the fact that changes in the levels of miRNA precursors do not necessarily correspond to changes in levels of mature miRNAs [25]. HnRNP A1 is an abundant nuclear RNA-binding protein involved in several steps of pre-mRNA processing including packaging and alternative splicing. It promotes the production of miR-18a by inducing conformational changes in the pri-miRNA that enhance its processing by Drosha [26]. One of the first miRNAs ever identified, the let-7 family, is itself regulated posttran-scriptionally. Lin-28, also first discovered in C.elegans, encodes an RNA-binding protein that blocks the processing of the precursor of let-7 miRNA in embryonic cells [27]. In humans, lin-28 and let-7 are now implicated in cancer, stem cell ageing and the control of pluripotency. The manipulation of Lin-28 expression in transgenic mice results in significant changes in body size, crown-rump length and delayed onset of puberty [28]. ERα represses several miRNAs; it does so by hindering their biogenesis presumably by regulating the expression of genes involved in miRNA processing [29].
Use of miRNAs in non-invasive prenatal diagnostics
A novel application of miRNA science is its potential use in prenatal diagnostics. Conventional prenatal diagnostics methods, notably chorionic villus sampling and amniocentesis, are both invasive and risky. There is therefore a drive to develop non-invasive approaches. One approach is to isolate and analyse fetal cells that are present in the maternal circulation; but the problem is that circulating fetal cells are very rare [30]. The detection of cell-free fetal circulating nucleic acids (CNAs) is a potential alternative approach [31]. In the late 1990s, fetal DNA was detected in the cell-free fraction of maternal blood [32]. Fetal DNA presents the obvious advantage of allowing genotyping; and DNA is more stable than RNA. However, the advantage of RNA as a biomarker over DNA is the fact that DNA is of low copy number (generally two copies per cell). On the other hand there are several classes of RNAs that are very abundant (rRNA, tRNA, snRNAs, mRNAs corresponding to highly expressed genes; and potentially miRNAs). MiRNAs are in fact now considered to be among the available CNA biomarkers [33]. MiRNAs could potentially aid the early detection of clinical conditions associated with pregnancy or fetal development as follows.
The mammalian placenta aids fetal development by providing the interface between the mother and the fetus. The developing fetus obtains nutrients and gases through the placenta. Abnormalities in placentas can lead to medical problems including preeclampsia. Within the placenta the trophoblasts are cells that play a critical role in embryo implantation and in the ability of the placenta to interact with the maternal uterus. The placenta is covered by a layer of multinucleated syncytiotrophoblasts - which grow into the uterine endometrium. A recent study has shown that placental trophoblasts express miRNAs from a ∼100kb miRNA gene cluster on chromosome 19 in an area of the genome that is required for normal placental and embryonic development [34]. Placentally-expressed miRNAs have been detected in maternal plasma [35, 36]. The concentration of several miRNAs in the maternal plasma is increases during the various stages of pregnancy and then decreases postnatally. Some miRNAs, such as miR-141, are particularly stable when compared to CSH1, a typical placentally-expressed mRNA. This suggests that miRNAs could be used in the monitoring of physiological processes associated with pregnancy. How do placental miRNAs enter the maternal plasma? Perhaps they accumulate in the maternal plasma as a by-product of the remnants of dead cells; but there is an intriguing alternative. In situ hybridisation performed on a trophoblast cell line showed that the miRNA miR-517b is secreted via an exosome-mediated pathway [37]. This finding raises the intriguing possibility that miRNAs could be involved in extracellular signalling pathways.
Thus miRNAs could be playing a role in fetal-maternal communication [38] and genetic lesions that affect miRNA expression might be associated with pathologies of pregnancy. For example preeclampsia is associated with abnormally high expression of pro-angiogenic growth factors. The expression of pro-angiogenic growth factors is potentially regulated by miRNAs; and abnormal miRNA levels have recently been observed in placental tissue from patients with severe preeclampsia [39]. Alterations in miRNA expression are associated with several congenital defects including neural tube defects [40] or with teratogenic effects associated with ethanol exposure [41]. Clearly then the accurate measurement of miRNA levels could be used to detect several congenital defects as part of traditional invasive sampling methods. However there may also be contexts in which fetal miRNAs in the maternal plasma could be used to detect congenital defects [42].
There is a substantial incidence of congenital defects that lead to malformations of the heart and associated blood vessels (CHD) with an incidence of up to 8 per 1000 live births [43]. These defects are associated with up to 40% of perinatal and 20% of postnatal deaths [44]. If detected early it is possible in many cases to plan an early surgical intervention. Existing methods for the detection of CHDs include fetal echocardiography - but the required equipment is not easily available and the accuracy of the method is somewhat limited and influenced by the experience of the operating personnel [44]. As some miRNAs that are associated with CHD are also expressed in the placenta [45], it is conceivable that a complementary approach to the prenatal diagnosis of CHD could include the detection of relevant miRNA biomarkers.
Concluding remarks
When gene expression was first investigated by the pioneers of the field, it was presumed to occur mostly at a transcriptional level. Instead it is now apparent that epigenetic processes including chromatin modification and cotranscriptional and posttranscriptional processes contribute very significantly to the regulation of gene expression. The recent emergence of miRNAs as regulators of gene expression has added to the mix and opened up several new areas of research. MiRNAs have the clear potential to be used as biomarkers. The detection of placentally-expressed miRNAs in the maternal plasma suggests their potential use in non-invasive prenatal diagnostics.
References
- 1.Lee RC, Feinbaum RL, Ambros V. The C.elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 2.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 miRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 3.Huang Y, Shen ZJ, Zou Q, Wang SP, Tang SM, Zhang GZ. Biological functions of microR-NAs: a review. J Physiol Biochem. 2011;67:129–139. doi: 10.1007/s13105-010-0050-6. [DOI] [PubMed] [Google Scholar]
- 4.Chen X. Small RNAs and their roles in plant development. Annu Rev Cell Dev Biol. 2009;25:21–44. doi: 10.1146/annurev.cellbio.042308.113417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones-Rhoades MW, Bartel DP, Bartel B. MiRNAS and their regulatory roles in plants. Annu Rev Plant Biol. 2006;57:19–53. doi: 10.1146/annurev.arplant.57.032905.105218. [DOI] [PubMed] [Google Scholar]
- 6.Martin RC, Liu PP, Goloviznina NA, Nonogaki H. MiRNA, seeds, and Darwin?: diverse function of miRNA in seed biology and plant responses to stress. J Exp Bot. 2010;61:2229–2234. doi: 10.1093/jxb/erq063. [DOI] [PubMed] [Google Scholar]
- 7.da Costa Martins PA, Leptidis S, Salic K, De Windt LJ. MiRNA regulation in cardiovascular disease. Curr Drug Targets. 2010;11:900–906. doi: 10.2174/138945010791591322. [DOI] [PubMed] [Google Scholar]
- 8.Güller I, Russell AP. MiRNAs in skeletal muscle: their role and regulation in development, disease and function. J Physiol. 2010;588:4075–4087. doi: 10.1113/jphysiol.2010.194175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang C. Novel functions for small RNA molecules. Curr Opin Mol Ther. 2009;11:641–651. [PMC free article] [PubMed] [Google Scholar]
- 10.Leung AK, Sharp PA. MiRNA functions in stress responses. Mol Cell. 2010;40:205–215. doi: 10.1016/j.molcel.2010.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ruan K, Fang X, Ouyang G. MiRNAs: novel regulators in the hallmarks of human cancer. Cancer Lett. 2009;285:116–126. doi: 10.1016/j.canlet.2009.04.031. [DOI] [PubMed] [Google Scholar]
- 12.Li C, Feng Y, Coukos G, Zhang L. Therapeutic miRNA strategies in human cancer. AAPS J. 2009;11:747–757. doi: 10.1208/s12248-009-9145-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferland-McCollough D, Ozanne SE, Siddle K, Willis AE, Bushell M. The involvement of miRNAs in Type 2 diabetes. Biochem Soc Trans. 2010;38:1565–1570. doi: 10.1042/BST0381565. [DOI] [PubMed] [Google Scholar]
- 14.De Smaele E, Ferretti E, Gulino A. MiRNAs as biomarkers for CNS cancer and other disorders. Brain Res. 2010;1338:100–111. doi: 10.1016/j.brainres.2010.03.103. [DOI] [PubMed] [Google Scholar]
- 15.Mayor-Lynn K, Toloubeydokhti T, Cruz AC, Chegini N. Expression profile of microRNAs and mRNAs in human placentas from pregnancies complicated by preeclampsia and preterm labor. Repr Sci. 2011;18:46–56. doi: 10.1177/1933719110374115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beezhold KJ, Castranova V, Chen F. Microprocessor of miRNAs: regulation and potential for therapeutic intervention. Mol Cancer. 2010;9:134–143. doi: 10.1186/1476-4598-9-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou H, Huang X, Cui H, Luo X, Tang Y, Chen S, Wu L, Shen N. miR155 and its star-form partner miR-155* cooperatively regulate type I interferon production by human plasmacytoid dendritic cells. Blood. 2010;116:5885–5994. doi: 10.1182/blood-2010-04-280156. [DOI] [PubMed] [Google Scholar]
- 18.Ro S, Oark C, Young D, Saunders KM, Yan W. Tissue-dependent paired expression of miRNAs. Nuc Acids Res. 2007;35:5944–5953. doi: 10.1093/nar/gkm641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bartel DP. MiRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang X, Zeng Y. Regulation of mammalian miRNA expression. J Cardiovasc Trans Res. 2010;3:197–203. doi: 10.1007/s12265-010-9166-x. [DOI] [PubMed] [Google Scholar]
- 21.Golan D, Levy C, Friedman B, Shomron N. Biased hosting of intronic micro RNA genes. Bioinformatics. 2010;26:992–995. doi: 10.1093/bioinformatics/btq077. [DOI] [PubMed] [Google Scholar]
- 22.Feng Z, Zhang C, Wu R, Hu W. Tumour suppressor p53 meets microRNAs. J Mol Cell Biol. 2011;3:44–50. doi: 10.1093/jmcb/mjq040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ribas J, Ni X, Haffner M, Wentzel EA, Salmasi AH, Chowdhury WH, Kudrolli TA, Yegnasubramanian S, Luo J, Rodriguez R, Mendell JT, Lu-pold SE. miR-21: an androgen receptor-regulated miRNA that promotes hormone-dependent and hormone-independent prostate cancer growth. Cancer Res. 2009;69:7165–7169. doi: 10.1158/0008-5472.CAN-09-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ribas J, Lupold SE. The transcriptional regulation of miR-21, its multiple transcripts, and their implication in prostate cancer. Cell Cycle. 2010;9:923–929. doi: 10.4161/cc.9.5.10930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Obernosterer G, Leuschner PJ, Alenius M, Martinez J. Post-transcriptional regulation of miRNA expression. RNA. 2006;12:1161–1167. doi: 10.1261/rna.2322506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guil S, Cáceres JF. The multifunctional RNA-binding protein hnRNP A1 is required for processing of miR-18a. Nat Struct Mol Biol. 2007;14:591–596. doi: 10.1038/nsmb1250. [DOI] [PubMed] [Google Scholar]
- 27.Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of miRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu H, Shah S, Shyh-Chang N, Shinoda G, Einhorn WS, Viswanathan SR, Takeuchi A, Grasemann C, Rinn JL, Lopez MF, Hirschhorn JN, Palmert MR, Daley GQ. Lin28a transgenic mice manifest size and puberty phenotypes identified in human genetic association studies. Nat Genet. 2010;42:626–630. doi: 10.1038/ng.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamagata K, Fujiyama S, Ito S, Ueda T, Murata T, Naitou M, Takeyama K, Minami Y, O'Malley BW, Kato S. Maturation of miRNA is hormonally regulated by a nuclear receptor. Mol Cell. 2009;36:340–347. doi: 10.1016/j.molcel.2009.08.017. [DOI] [PubMed] [Google Scholar]
- 30.Hamada H, Arinami T, Kubo T. Hamaguchi H, Iwasaki H. Fetal nucleated cells in maternal peripheral blood: frequency and relationship to gestational age. Hum Genet. 1993;91:427–432. doi: 10.1007/BF00217766. [DOI] [PubMed] [Google Scholar]
- 31.Hung EC, Chiu RW, Lo YM. Detection of circulating fetal nucleic acids: a review of methods and applications. J Clin Pathol. 2009;62:308–313. doi: 10.1136/jcp.2007.048470. [DOI] [PubMed] [Google Scholar]
- 32.Lo YM, Corbetta N, Chamberlain PF, Rai V, Sargent IL, Redman CW, Wainscoat JS. Presence of fetal DNA in maternal plasma and serum. Lancet. 1997;350:485–487. doi: 10.1016/S0140-6736(97)02174-0. [DOI] [PubMed] [Google Scholar]
- 33.Gilad S, Meiri E, Yogev Y, Benjamin S, Lebanony D, Yerushalmi N, Benjamin H, Kushnir M, Cholakh H, Melamed N, Bentwich Z, Hod M, Goren Y, Chajut A. Serum miRNAs are promising novel biomarkers. PLoS One. 2008;3:e3148. doi: 10.1371/journal.pone.0003148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miura K, Miura S, Yamasaki K, Higashijima A, Kinoshita A, Yoshiura K, Masuzaki H. Identification of pregnancy-associated miRNAs in maternal plasma. Clin Chem. 2010;56:1767–1771. doi: 10.1373/clinchem.2010.147660. [DOI] [PubMed] [Google Scholar]
- 35.Chim SS, Shing TK, Hung EC, Leung TY, Lau TK, Chiu RW, Lo YM. Detection and characterization of placental miRNAs in maternal plasma. Clin Chem. 2008;54:482–490. doi: 10.1373/clinchem.2007.097972. [DOI] [PubMed] [Google Scholar]
- 36.Kotlabova K, Doucha J, Hromadnikova I. Placental-specific microRNA in maternal circulation - identification of appropriate pregnancyassociated microRNAs with diagnostic potential. J Rep Immunol. 2011;89:185–191. doi: 10.1016/j.jri.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 37.Luo SS, Ishibashi O, Ishikawa G, Ishikawa T, Katayama A, Mishima T, Takizawa T, Shigihara T, Goto T, Izumi A, Ohkuchi A, Matsubara S, Takeshita T, Takizawa T. Human villous trophoblasts express and secrete placentaspecific miRNAs into maternal circulation via exosomes. Biol Reprod. 2009;81:717–729. doi: 10.1095/biolreprod.108.075481. [DOI] [PubMed] [Google Scholar]
- 38.Chiu RWK, Lo YMD. Pregnancy-associated miRNAs in maternal plasma: a channel for fetal-maternal communication? Clin Chem. 2010;65:1656–1657. doi: 10.1373/clinchem.2010.153684. [DOI] [PubMed] [Google Scholar]
- 39.Noack F, Ribbat-Idel J, Thorns C, Chiriac A, Axt-Fliedner R, Diedrich K, Feller AC. miRNA expression profiling in formalin-fixed and paraffinembedded placental tissue samples from pregnancies with severe preeclampsia. J Perinat Med. 2011 doi: 10.1515/jpm.2011.012. (in press) [DOI] [PubMed] [Google Scholar]
- 40.Shookhoff JM, Gallicano GI. A new perspective on neural tube defects: folic acid and miRNA misexpression. Genesis. 2010;48:282–294. doi: 10.1002/dvg.20623. [DOI] [PubMed] [Google Scholar]
- 41.Wang LL, Zhang Z, Li Q, Yang R, Pei X, Xu Y, Wang J, Zhou SF, Li Y. Ethanol exposure induces differential miRNA and target gene expression and teratogenic effects which can be suppressed by folic acid supplementation. Human Rep. 2009;24:562–579. doi: 10.1093/humrep/den439. [DOI] [PubMed] [Google Scholar]
- 42.Yu Z, Han S, Hu P, Zhu C, Wang X, Qian L, Guo X. Potential role of maternal serum miRNAs as a biomarker for fetal congenital heart defects. Med Hypotheses. 2011;76:424–426. doi: 10.1016/j.mehy.2010.11.010. [DOI] [PubMed] [Google Scholar]
- 43.Jone PN, Schowengerdt KO. Prenatal diagnosis of congenital heart disease. Pediatr Clin North Am. 2009;56:709–715. doi: 10.1016/j.pcl.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 44.Trojnarska O, Grajek S, Katarzynski S, Kramer I. Predictors of mortality in adult patients with congenital heart disease. Cardiol J. 2009;16:341–347. [PubMed] [Google Scholar]
- 45.Liang Y, Ridzon D, Wong I, Chen C. Characterization of miRNA expression profiles in normal human tissues. BMC Genomics. 2007;8:166. doi: 10.1186/1471-2164-8-166. [DOI] [PMC free article] [PubMed] [Google Scholar]