Abstract
Helicobacter pylori BabA adhesin metastability could yield variants with potential for periodic activation and deactivation of their mediated adherence. babA/B or babB/A chimeras could play an important role in translational regulation. We investigated the frequency of different bab gene profiles in paired isolates from antrum and corpus recovered from patients with chronic gastritis. Isolates from 174 biopsies from 34 patients were included, and bab genes at the three common chromosomal loci were investigated. Inter-micro-niche variation was found in 1/4 patients, counting duplicate copies of babA or babB, babB/A or babA/B chimeras, opposite location of babA and babB or babC and babB, and absence of babB ATG translational codon. Truncated BabA was identified in 2/34 patients without inter-micro-niche variation. Isolates from 12/34 patients harbored babA/B or babB/A chimeras -either in one, several or all micro-niches indicating that chimera formation is a common mechanism to control BabA expression. To note, babA gene was absent in 11/34 patients, and in this population, babA/B chimeras which lack expression predominated over babB/A, able to exhibit Leb binding phenotype.
Keywords: H. pylori, babA/B chimera, babB/A chimera
Introduction
Genetic diversification can aid in the persistence of microorganisms that continue to replicate during chronic infection. In addition, H. pylori genetic variation may help to adapt to different micro-niches within a single host and to the changing conditions in the host over time [1]. One example of how H. pylori's genetic variability helps it adapt to new environments involves its adhesin genes [1]. BabA adhesin recognizes both H-type-1 and Lewis b (Leb) blood-group antigens expressed on normal gastric mucosa of secretor individuals [2]. It was suggested that BabA metastability results in clones having high potential for periodic activation and deactivation of mucosal binding, appropriate for the intensity of the host response to infection [3, 4]. babA, babB, and babC genes are members of the paralogous hop family of outer membrane proteins. Considering the flanking regions, bab genes can be located in at least three different H. pylori chromosomal loci referred to as locus A, B and C (downstream of the hypD gene, the s18 gene, and the conserved hypothetical protein gene, hp0318 in H. pylori 26695, respectively). In addition to an extensive genotypic diversity in babA, babB and babC within the mid-region across inter-patients clinical isolates, individual strains with duplicate copies of babA, frameshift mutations, chimeric babA/B (resulting in loss of BabA expression), and chimeric babB/A (that subject protein expression to phase variation), have been described [5, 6, 7, 8]. Homologous recombination of bab genes has been observed in clinical isolates from different patients [6, 7, 8]. Besides, in experimental infections with babA functional strains, recombination has also been reported as a likely mechanism for generating Leb non-binding babA output isolates [9, 10]. However, Ohno et al., [11] demonstrated that the lack of BabA expression after 6 months of infection of mongolian gerbils with a functional babA was mainly attributable to nucleotide changes within the babA gene that resulted in a truncated protein. Nevertheless, these authors did not exclude the possible presence of recombinant allelic variants in the output population assuming that recombination occurs at low frequencies [11].
This study was aimed to investigate the frequency of different bab gene profiles in paired isolates from antrum and corpus from patients with chronic gastritis, to evaluate the results of microevolution during persistent colonization.
Materials and methods
Patients and samples
A total of 174 isolates from 34 patients (20 from a previous study [12] and 14 referred to the Gastroenterology Service of an Ambulatory Care Centre, Clínica Bazterrica, Buenos Aires, Argentina for upper gastric endoscopies) were included. All patients agreed to participate in the study by signing an informed consent. Six biopsy specimens were obtained from each patient (three from antrum: a1, a2, a3, and three from corpus: c1, c2, c3 [12]) in the same endoscopic session.
Bacterial isolation, DNA extraction and strain delineation
Biopsy specimens were cultured separately as previously described [12]. DNA was extracted from confluent cultures with fewer than three “in vitro” passages by standard protocols using a pool of colonies from the isolation plate of each biopsy (microniche) and from the subculture of single-colony of this plate. When results suggested intra-microniche diversity, expansion of extra single-colonies (n: 10) was done by sub-culturing sweep of bacteria from the isolation plate conserved at -80°C. Strain delineation was achieved by lspA-glmM RFLP and RAPD fingerprints as previously described [12].
bab genes analysis
bab genes location was investigated by PCR using consensus primers with hypD (locus A) and s18 (locus B) combined with primers for babA and babB [7]; LC-F1 and LC-R1 primers were used to analyze locus C [6]. babA, babB or babC at each locus, and locus C empty-site (absence of bab genes) [6] were confirmed by sequence analysis. bab genes promoter regions for sequencing were amplified with one of the following primers: LocA-1 (5’GGCTCATAACCCAA AGGTC-3’), LocA-2 (5’-GTTTTGGTCCTGGCATTC-3’), LocB-1 (5’-GGATAGCCCTTTAAAACAGC-3’), or LocB-2 (5’-GAAGTAGCGATCAAAAGAG-3’); and the babA and babB specific primers used by Colbeck et al. [7]. PCR amplicons were purified using Wizard-PCR Preps (Promega, Madison, WI, USA) according to the manufacturer's instructions. Sequencing was performed on both strands using an ABI 373 DNA sequencer (Applied Biosystems). Nucleotide sequence data were then compared with nucleotide sequences deposited in the GenBank (Blast software at the National Center for Biotechnology Information) in order to identify the most closely related sequences.
Nucleotide sequence accession numbers
GenBank accession numbers JF922315-JF922350 were assigned to bab genes sequences newly determined in this study.
Results
According to lspA-glmM RFLP and RAPD fingerprints, 33/34 (97%) of the patients included in the study were colonized with a single strain [12]. bab genes inter-micro-niche variation was observed in 9/34 (26.47%) patients. This variation included the co-existence of isolates with: a) single or double copy of babA, b) inverse location of babA and babB, d) single and double copy of babB, e) presence and absence of babC at locus A, f) presence and absence of the chimeric babB/A gene; and g) babB gene with and without ATG translational start codon (Figure 1). Inter-micro-niche variation was not commonly associated with isolates from antrum or corpus. Intra-micro-niche variation could be demonstrated in 2/34 patients, but in only one of four or six H. pylori-positive biopsies from each of them; including co-existence of isolates with babA and isolates with babB at locus B and babA at locus A, or isolates babA and isolates with chimeric babA/B at locus A and babB at locus B, respectively (Figure 1). In both patients, PCR with pooled colonies DNA exhibited amplification with babA and babB primers at loci A or B respectively, then expansion of single-colony revealed the co-existence of isolates with different bab genes at each locus, confirmed by sequence analysis. Figure 2 shows bab gene profiles of the other 23/34 patients. In the patient colonized with two different H. pylori strains (one recovered from antrum and the other from corpus biopsies), isolates from both strains harbored a double copy of babB gene with 100% identity among loci and strains.
Figure 1.
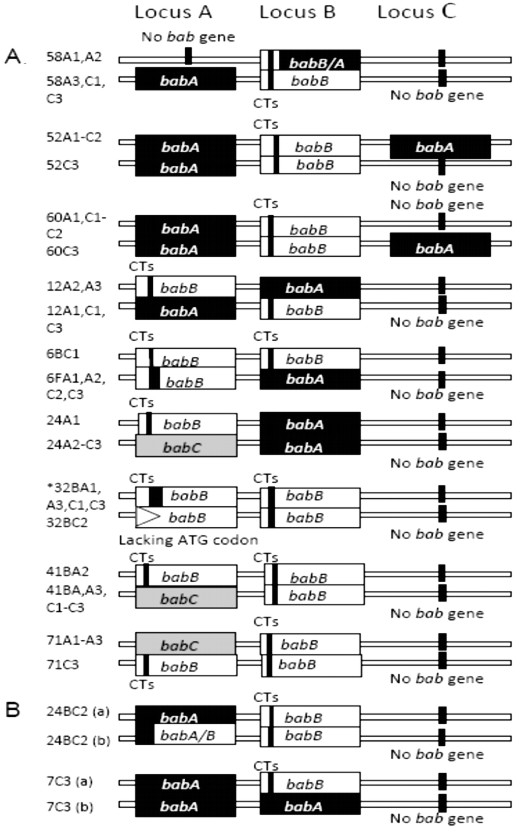
Patients with co-existence of isolates with different bab genes profiles: A. Inter-micro-niche variation. In patients numbered 32B, isolates from a1, a3, c1 y c3 showed babB gene recombined with babA promoter region at locus A and with babC promoter region at locus B. After initial translational ATG codon babB sequences at loci A and B are identical except for the CT repeats number. In addition, in isolates from c2, babB at locus A lacked the ATG start codon. B. Intra-micro-niche variation. a1, a2 and a3: antrum biopsies, c1, c2 and c3: corpus.
Figure 2.
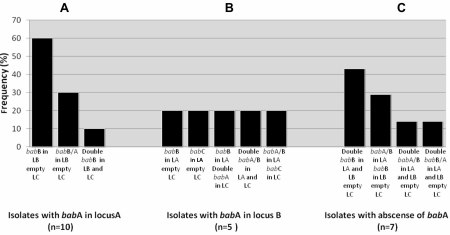
Frequency of bab genes profiles at the three common chromosomal sites in patients without inter- or intra-micro-niche variations. A. Isolates with babA in locus A. B. Isolates with babA in locus B. C. Isolates with absence of babA. Locus A: downstream of hypD gene, Locus B downstream s18 gene, Locus C: downstream the conserved hypothetical protein gene, hp0318 in H. pylori 26695. LA: Locus A, LB: Locus B, LC: Locus C.
To note, isolates from 11/34 (33%) patients lacked babA gene at the three common chromosomal sites. In these individuals, the chimeric babA/B predominated over babB/A. Considering both chimeric recombinant variants: babA/B or babB/A, these were observed in 12/34 (35%) individuals either at loci A, B or C, even in the double copy (babB/A at loci A and B; babA/B at loci A and C and at loci A and B) (Figure 1 and Figure 2). babA/B was identified in 7/34 (20%) patients, more frequently in locus A and mainly in patients without inter-micro-niche variation (Figure 2). babB/A was identified in 5/34 (15%) patients and conversely to babA/B, more frequently in locus B (Figure 2). babB/A carried a variable number of CT repeats in isolates from different patients and also at different loci of the same isolate harboring a double copy of this recombinant variant. Recombination upstream of the ATG translational start codon of bab genes could be identified in isolates from 5/34 (14%) patients. babA gene was preceded by the babB promoter region at locus B in isolates from one patient; babB gene was preceded by the babA promoter region at locus A in isolates from 3 patients, and at locus B in isolates from one patient. Among these five patients, from the numbered 32B with inter-micro-niche variation (Figure 1), H. pylori was recovered from 5 micro-niches: two from antrum and three from corpus (a1, a3, c1, c2 and c3), and all isolates harbored a double copy of babB. Isolates from a1, a3, c1, and c3 showed babA sequences upstream of the babB ATG start codon at locus A, while at locus B recombination seemed to occur with the babC promoter region. After the ATG, babB sequences at loci A and B were identical except for the CT repeat number. In addition, from c2, isolates with the absence of babB ATG start codon were recovered.
The babA gene promoter region showed high inter-patient diversity. Poly-adeninetract within the -10 to -35 varied from 5 to 16 adenine number, and between Shine-Dalgarno sequences and ATG start codon, it varied from 3 to 7 repeats. Only isolates recovered from one patient showed inter-microniche variation in the number of adenine repeats between the -10 and the -35.
Isolates with truncated BabA were recovered from two patients only. babA gene with nonsense mutation (substitution A→ C at position 656) leading to a stop codon at BabA amino acid 219 was observed in one of them, and frameshift mutation (insertion of G at position 229, and deletion of GG at position 247-248 towards a stop codon at BabA amino acid 92) was present in the other patient.
Discussion
Micro- and macro-diversity among H. pylori isolates act as an important driving force for adaptation to the hostile gastric environment and the variable living conditions during the inflammation process [13]. The initial observation of loss of BabA expression during experimental infection with H. pylori strains expressing adhesine due to the emergence and co-existence of isolates with a second copy of babB and isolates with babA harboring CT repeats preceding the 5’ signal peptide sequence, led to the hypothesis that recombination events might reflect a response to selective pressure that may also be apparent in human clinical isolates [7, 9, 10]. Ohno et al. [11] also found the loss of BabA expression post experimental infection, although this loss seemed to be attributable to nucleotide changes within the babA. Concerning clinical isolates, variation of bab genes location at the three common chromosomal loci as well as single or double copy of one of them, or the presence of chimeric variants have been reported among those from different patients [4, 5, 6, 7]. All together, these results suggested that the variation of bab genes during chronic colonization is highly dynamic. Nevertheless, babA gene status or BabA level of expression has been related with gastric disease outcome [14, 15]. In this study, microevolution of bab genes defined as inter- micro-niche variation in paired isolates of the same host could be clearly demonstrated with a frequency of 1/4 patients colonized with a single strain. However, the only patient from whom two different strains were recovered, all isolates harbored identical babB at loci A and B. Considering the great diversity of bab gene profiles among clinical strains, the last result suggested that the presence of one bab gene profile or one bab gene variant could be related to variable conditions during the colonization process rather than the characteristic of a specific strain. In the two patients showing mixed bab genotypes at locus A or B by PCR with pooled colonies DNA (babA and babB at locus B, or babA and babB at locus A, respectively) the expansion of single colonies demonstrated co-existence of isolates with different bab genes at each locus.
To note, babA gene was absent in 1/3 patients. Moreover, in two patients with inter-niche variation, isolates lacking babA were confined to specific micro-niches. Concerning chimeric variants, the frequency of patients with isolates carrying babB/A was close to that previously reported [6, 7], whereas in patients with isolates harboring babA/B, it was higher [5, 15]. In none of our isolates babC showed CT repeats at the 5’ end like in H. pylori strain HPAG-1. However, in one patient, sequences upstream of the babB ATG start codon showed the highest similarity with the babC promoter region, supporting that identity in babB and babC can also promote recombination [2, 15].
The absence of a babA translational initiation codon was not observed in any of our studied isolates, and the sequence analysis of the promoter region of this gene did not clearly support the suggested babA1 and babA2 allele differentiation [4, 15]. Conversely, isolates from one patient with a double copy of babB, lacked the ATG start codon at the copy in locus A. Therefore, although rare, the absence of the translational initiation codon may be a common mechanism of lack of expression of babA and babB genes.
In this study population, nucleotide changes within the babA gene generating a truncated protein were not a common event, supporting the finding of Henning et al. [6].
In the present study, bab gene variation across a single host gastric mucosa at a specific point in time was demonstrated in about 1/4 patients, including the duplicate copy of babA or babB, presence of babB/A or babA/B chimeras, opposite location of babA and babB or babC and babB. Therefore, the intra-patient diversity could be equal and even higher than that found inter-patient. These variations may represent the H. pylori adaptation during chronic colonization. Therefore, inter-micro-niche variation should be considered when genes or the protein status is used to correlate with disease progression.
Acknowledgments
This study was supported by grants BID 1728 OC-AR PICT 2006-0458 provided by Agencia Nacional de Promoción Científica y Tecnológica and by M064 from Universidad de Buenos Aires (UBACyT 2010-2012).
Declaration of conflict of interests
There is no conflict of interests among authors.
References
- 1.Dorer MS, Talarico S, Salama NR. Helicobacter pylori's unconventional role in health and disease. PLoS Pathog. 2009:e1000544. doi: 10.1371/journal.ppat.1000544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boren T, Falk P, Roth KA, Larson G, Normark S. Attachment of Helicobacter pylori to human gastric epithelium mediated by blood group antigens. Science. 1993;262:1892–1895. doi: 10.1126/science.8018146. [DOI] [PubMed] [Google Scholar]
- 3.Marcos NT, Magalhães A, Ferreira B, Oliveira MJ, Carvalho AS, Mendes N, Gilmartin T, Head SR, Figueiredo C, David L, Santos-Silva F, Reis CA. Helicobacter pylori induces beta3GnT5 in human gastric cell lines, modulating expression of the SabA ligand sialyl-lewis x. J Clin Invest. 2008;118:2325–2336. doi: 10.1172/JCI34324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bäckström A, Lundberg C, Kersulyte D, Berg DE, Boren T, Arnqvist A. Metastability of Helicobacter pylori bab adhesion genes and dynamics in Lewis b antigen binding. Proc Natl Acad Sci USA. 2004;101:16923–16928. doi: 10.1073/pnas.0404817101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pride DT, Meinersmann RJ, Blaser MJ. Allelic variation within Helicobacter pylori babA and babB. Infect Immun. 2001;69:1160–1171. doi: 10.1128/IAI.69.2.1160-1171.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hennig EE, Allen JM, Cover TL. Multiple chromosomal loci for the babA gene in Helicobacter pylori. Infect Immun. 2006;74:3046–3051. doi: 10.1128/IAI.74.5.3046-3051.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colbeck JC, Hansen LM, Fong JM, Solnick JV. Genotypic profile of the outer membrane proteins BabA and BabB in clinical isolates of Helicobacter pylori. Infect Immun. 2006;74:4375–4378. doi: 10.1128/IAI.00485-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pride DT, Blaser MJ. Concerted evolution between duplicated genetic elements in Helicobacter pylori. J Mol Biol. 2002;316:629–642. doi: 10.1006/jmbi.2001.5311. [DOI] [PubMed] [Google Scholar]
- 9.Solnick JV, Hansen LM, Salama NR, Boonjakuakul JK, Syvanen M. Modification of Helicobacter pylori outer membrane protein expression during experimental infection of rhesus macaques. Proc Natl Acad Sci U S A. 2004;101:2106–2111. doi: 10.1073/pnas.0308573100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Styer CM, Hansen LM, Cooke CL, et al. Expression of the BabA adhesin during experimental infection with Helicobacter pylori. Infect Immun. 2010;78:1593–1600. doi: 10.1128/IAI.01297-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ohno T, Vallstrom A, Rugge M, et al. Effects of blood group antigen-binding adhesin expression during Helicobacter pylori infection of Mongolian gerbils. J Infect Dis. 2011;203:726–735. doi: 10.1093/infdis/jiq090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matteo MJ, Granados G, Perez CV, Olmos M, Sanchez C, Catalano M. Helicobacter pylori cag pathogenicity island genotype diversity within the gastric micro-niche of a single host. J Med Microbio. 2007;56:664–669. doi: 10.1099/jmm.0.46885-0. [DOI] [PubMed] [Google Scholar]
- 13.Odenbreit S, Swoboda K, Barwig I, Ruhl S, Borén T, Koletzko S, Haas R. Outer membrane protein expression profile in Helicobacter pylori clinical isolates. Infect Immun. 2009;77:3782–3790. doi: 10.1128/IAI.00364-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fujimoto S, Olaniyi Ojo O, Arnqvist A, et al. Helicobacter pylori BabA expression, gastric mucosal injury, and clinical outcome. Clin Gastroenterol Hepatol. 2007;5:49–58. doi: 10.1016/j.cgh.2006.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamaoka Y. Roles of Helicobacter pylori BabA in gastroduodenal pathogenesis. World J Gastroenterol. 2008;14:4265–4272. doi: 10.3748/wjg.14.4265. [DOI] [PMC free article] [PubMed] [Google Scholar]