Abstract
Vertebrate endoderm development has recently become the focus of intense investigation. In this report, we first show that the zebrafish bonnie and clyde (bon) gene plays a critical early role in endoderm formation. bon mutants exhibit a profound reduction in the number of sox17-expressing endodermal precursors formed during gastrulation, and, consequently, a profound reduction in gut tissue at later stages. The endodermal precursors that do form in bon mutants, however, appear to differentiate normally indicating that bon is not required at later steps of endoderm development. We further demonstrate that bon encodes a paired-class homeodomain protein of the Mix family that is expressed transiently before and during early gastrulation in both mesodermal and endodermal progenitors. Overexpression of bon can rescue endodermal gene expression and the formation of a gut tube in bon mutants. Analysis of a newly identified mutant allele reveals that a single amino acid substitution in the DNA recognition helix of the homeodomain creates a dominant interfering form of Bon when overexpressed. We also show through loss- and gain-of-function analyses that Bon functions exclusively downstream of cyclops and squint signaling. Together, our data demonstrate that Bon is a critical transcriptional regulator of early endoderm formation.
Keywords: Paired-class homeodomain, sox17, cyclops, squint, gut
One of the fundamental problems in developmental biology is to understand how the three germ layers—ectoderm, mesoderm, and endoderm—are established. The molecular mechanisms of ectoderm and mesoderm formation have been analyzed extensively and our understanding of their development is fairly advanced. In contrast, the molecular analysis of endoderm formation has only begun relatively recently, but already several Xenopus and zebrafish genes have been implicated in this process.
In Xenopus, several related paired-class homeodomain proteins, the Mix and Bix proteins, have been proposed to have important roles in endoderm formation (Rosa et al. 1989; Henry and Melton 1998; Mead et al. 1998; Tada et al. 1998). The genes encoding these proteins are all expressed in the prospective endoderm during gastrulation; most are also expressed in marginal cells that will form mesoderm, with the exception of Mixer, which is expressed exclusively in the endoderm (Henry and Melton 1998). When overexpressed in animal caps, these genes exhibit different abilities to activate endodermal gene expression: Mixer and Bix2/Milk appear to promote endodermal gene expression strongly, and at the expense of mesodermal gene expression (Ecochard et al. 1998; Henry and Melton 1998); Bix1 overexpression at low and high levels induces mesodermal and endodermal gene expression, respectively (Tada et al. 1998); and Mix.1 is able to activate endodermal gene expression only when co-expressed with the dorso-vegetal homeobox gene siamois (Lemaire et al. 1998). Additionally, Bix4 is able to restore endodermal gene expression to VegT depleted embryos, which lack all endoderm and most mesoderm (Zhang et al. 1998b), indicating that Bix4 acts downstream of VegT in endoderm formation (Casey et al. 1999). Therefore, whereas the various Xenopus Mix and Bix genes can regulate endoderm development, their individual roles and degree of importance in this process remain unresolved.
In contrast to the situation in Xenopus, only a single Mix-like gene has been identified so far in other vertebrates. The chick CMIX gene is expressed in the epiblast of the posterior marginal zone of early chick embryos, and starting at the onset of gastrulation, along the entire primitive streak except for its most posterior part (Peale et al. 1998; Stein et al. 1998). In mouse, a Mix-like gene, Mml, has recently been identified (Pearce and Evans 1999); it is expressed in the visceral endoderm at embryonic day 5.5 (E5.5), and starting at E6.5 throughout the entire primitive streak except for its most anterior part. In zebrafish, a Mix-like gene has also been described and provisionally named mixer, because of its homology with Xenopus Mixer and ability to induce endodermal gene expression in wild-type and mutant zebrafish embryos (Alexander et al. 1999; Alexander and Stainier 1999).
The Xenopus genes Xsox17α, and Xsox17β (here referred to collectively as Xsox17), which encode proteins containing a high mobility group DNA-binding domain, are also implicated in endoderm development (Hudson et al. 1997). Expression of Xsox17 becomes restricted to the endoderm at the onset of gastrulation, and when overexpressed in animal caps, Xsox17 activates endodermal gene expression. Mixer induces Xsox17 expression but Xsox17 does not induce Mixer expression in animal caps, suggesting that Mixer acts upstream of Xsox17; however, Xsox17 expression appears 3 hr before that of Mixer during normal development (Henry and Melton 1998). Therefore, it may be that these genes initiate expression independently of each other, with Mixer then regulating the maintenance of Xsox17 expression (Henry and Melton 1998; Yasuo and Lemaire 1999). Additionally, coexpression of Mixer and a dominant interfering form of Xsox17 (Xsox17–enR) in animal caps blocks the induction of endodermal gene expression, whereas coexpression of Xsox17 and Mixer–enR does not, suggesting that Mixer functions through Xsox17 in endoderm formation (Henry and Melton 1998). In the absence of genetic analyses of the Mix, Bix, and Xsox17 genes, however, it remains difficult to describe precisely the relationships between these various genes.
Several mutations have recently been reported to affect endoderm development in zebrafish (Schier et al. 1997; Feldman et al. 1998; Alexander et al. 1999; Reiter et al. 1999). The cyclops (cyc) and squint (sqt) genes encode Nodal-related proteins, and embryos mutant for both of these genes lack all endodermal and most mesodermal derivatives (Feldman et al. 1998). The same phenotype is seen in embryos that lack both maternal and zygotic one-eyed pinhead (oep), a gene encoding a member of the EGF–CFC protein family essential for Nodal signaling (Zhang et al. 1998a; Gritsman et al. 1999), whereas zygotic oep mutants lack endoderm as well as the prechordal plate and ventral neuroectoderm. Nodal-related genes have also been implicated in Xenopus endoderm development (Clements et al. 1999; Osada and Wright 1999; Yasuo and Lemaire 1999).
casanova (cas) and faust (fau), two zebrafish mutants that exhibit bilateral hearts (cardia bifida), have also been shown to be essential for endoderm development. In cas mutants, the early endodermal expression of genes such as axial, sox17, and fkd2 does not initiate, but expression of the Mix-like gene is normal, suggesting that cas acts downstream of, or in parallel to, this Mix-like gene to promote endoderm formation (Alexander et al. 1999). The cas gene has not yet been isolated. The fau locus is essential for multiple aspects of heart and endoderm development; fau mutants exhibit reduced amounts of cardiac and endodermal tissue and abnormal morphogenesis of the heart, pharyngeal endoderm, and gut (Reiter et al. 1999). fau encodes Gata5, a zinc finger-containing transcription factor, and is expressed in the endodermal and some mesodermal progenitors before to the onset of gastrulation (Reiter et al. 1999).
The bonnie and clyde (bon) mutation, like oep, cas, and fau, causes cardia bifida (Stainier et al. 1996). In this report, we show that in bon mutants the number of sox17-expressing endodermal precursors formed during gastrulation is significantly reduced, and that the gut tube is almost entirely absent at later stages. The endodermal precursors that do form in bon mutants, however, appear to differentiate normally—indicating that bon is not required at later steps of endoderm development. We also show that bon corresponds to the Mix-like gene that we isolated previously (Alexander et al. 1999). We further examined the relationship between bon and Nodal signaling and found that bon can induce a few cells to express sox17 expression in cyc;sqt double mutants, which normally completely lack sox17 expression. In contrast, bon cannot induce ectopic cyc and sqt expression in wild-type embryos, indicating that bon functions exclusively downstream of these nodal-related genes. Our results provide the first genetic evidence that Mix genes are critical for early endoderm development, show that bon is required only at a very early step of endoderm formation (i.e., the generation of sox17-expressing endodermal precursors), and suggest that additional factors act downstream of cyc and sqt to generate the full complement of endodermal precursors.
Results
bon mutants show severe defects in endoderm development
The zebrafish bon locus is defined by two mutant alleles: bonm425 was identified in a large-scale screen for embryonic lethal mutations (Stainier et al. 1996), whereas the second allele, bons9, was identified in a mosaic F1 screen for cardiac mutations (Alexander et al. 1998). Both alleles segregate as fully penetrant, completely recessive mutations, and cause essentially identical phenotypes at the morphological level. The transheterozygous phenotype (bonm425/bons9) is also fully penetrant and indistinguishable from that of either homozygous mutant at the morphological level.
bonm425 was originally classified as a cardia bifida mutation as it blocks cardiac fusion and leads to the presence of bilateral myocardial tissue (Stainier et al. 1996). bon mutants also exhibit pericardial edema, collapsed brain ventricles, as well as an enlarged yolk and a thickened yolk extension that may be secondary to the pronounced pericardial edema or defects in the yolk syncytial layer (Fig. 1A,B). The hearts, though bifid, contain distinct atrial and ventricular tissues (Fig. 1C,D).
Figure 1.

The bon mutations affect gut tube formation and heart morphogenesis. Light microscopic and immunofluorescent images of wild-type and bon mutant embryos at 30 hpf (A–D) and four days-postfertilization (E,F). (A,B) Compared with wild-type siblings (A), bon mutants (B) show pericardial edema (arrowhead), collapsed brain ventricles (asterisk), an enlarged yolk and a thickened yolk extension (arrow). (C,D) Ventral views of wild-type (C) and bon mutant (D) embryos stained with MF20 (TRITC) and S46 (FITC) antibodies. Red fluorescence indicates MF20 staining of ventricular tissue, whereas yellow fluorescence indicates the overlap of MF20 and S46 staining in atrial tissue. Wild-type embryos have a single heart tube with the ventricle (red) anterior to the atrium (yellow). bon mutants display cardia bifida, with the two separate hearts exhibiting distinct atrial and ventricular tissues. (E,F) bon mutants generally lack a gut tube, although sometimes a small amount of peristaltically contracting gut tissue is present (arrow in F). For comparison, we show a silent heart (sih) mutant where a similar extent of edema allows easy visualization of the gut tube (arrow in E).
oep, cas, and fau were also classified as cardia bifida mutations (Chen et al. 1996; Stainier et al. 1996), but recent experiments have shown that all three also cause endoderm defects that may in fact lead to the cardia bifida phenotype (Schier et al. 1997; Peyriéras et al. 1998; Alexander et al. 1999; Reiter et al. 1999). These data prompted us to analyze endoderm development in bon mutants. Light microscopic observation at four days post-fertilization (dpf) reveals that most bon mutants lack a gut tube, although some display a small amount of peristaltically contracting gut tissue (Fig. 1E,F). We next examined early endodermal gene expression in bon mutants and found profound defects. Zebrafish sox17, a homolog of Xenopus Xsox17, is expressed in endodermal precursors as well as in the forerunner cells (Alexander and Stainier 1999). In bon mutants, the number of sox17-expressing endodermal precursors is dramatically reduced compared with wild-type, whereas the number of forerunner cells appears to be unaffected (Fig. 2A,B; Table 1). The number of axial-expressing endodermal precursors is similarly reduced in bon mutants (data not included). Quantitation of sox17-expressing endodermal precursors in both bon alleles demonstrates that bon is required for the generation of normal numbers of sox17-expressing endodermal precursors and not for their subsequent proliferation, and that the bons9 endodermal defect is stronger than the bonm425 defect (Table 1).
Figure 2.
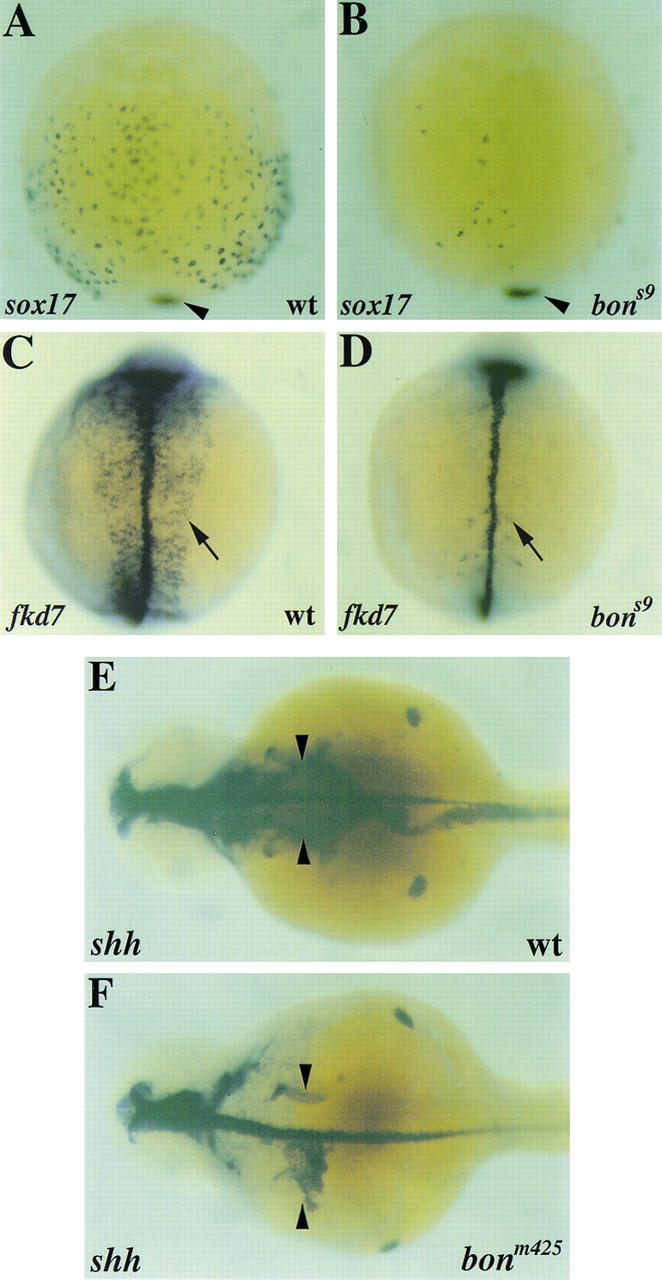
Endoderm development is defective in bon mutants. Wild-type (A,C,E) and bon mutant (B,D,F) embryos were examined for expression of sox17 at 90% epiboly (A,B), fkd7 at the 10-somite stage (C,D), and shh at 36 hpf (E,F). (A,B) During gastrulation sox17 is expressed in the endodermal precursors throughout the hypoblast as well as in the forerunner cells (arrowhead in A). Endodermal sox17 expression is greatly reduced in bon mutants (B), whereas sox17 expression in the forerunner cells is normal (arrowhead in B). (C,D) During mid-somitogenesis fkd7 is expressed in the endoderm (arrow in C) as well as in the floor plate and hypochord. In bon mutants (D), there are only a few fkd7-expressing endodermal cells (arrow in D), whereas expression in the floor plate and hypochord is normal. (E,F) Similarly, in 36 hpf wild-type embryos, shh is expressed throughout the endoderm (arrowheads in E); bon mutants exhibit small patches of shh-expressing endoderm (arrowheads in F). shh is also expressed in the floor plate, forebrain, and fin buds. All panels show dorsal views, with anterior to the top, except E and F, where anterior is to the left.
Table 1.
bons9 causes a stronger phenotype than bonm425
|
|
No. of sox17-expressing endodermal precursors
|
||
|---|---|---|---|
| shield (6 hpf)
|
75% epiboly (8 hpf)
|
90% epiboly (9 hpf)
|
|
| Wild type | 150 | 300 | 500 |
| bonm425 | 15.2 ± 1.1 | 39.0 ± 3.9 | 52.4 ± 3.0 |
| (n = 12) | (n = 9) | (n = 8) | |
| bons9 | 4.6 ± 0.9 | 18.5 ± 2.1 | 24.4 ± 4.7 |
| (n = 8) | (n = 11) | (n = 5) | |
The total number of sox17 expressing endodermal precursors was counted in bon mutants and estimated in wild-type embryos. P values (t-test) of the differences between bonm425 and bons9are 0.0007 (shield), 0.0035 (75% epiboly), and 0.0075 (90% epiboly).
At later stages, the forkhead-related gene fkd7 is expressed in the forming gut tube, as well as in the hypochord and floor plate (Odenthal and Nüsslein-Volhard 1998). In bon mutants, endodermal fkd7 expression is almost entirely absent, whereas hypochord and floor plate expression appears to be unaffected (Fig. 2C,D). We also examined the expression of sonic hedgehog (shh), which is expressed throughout the gut tube at larval stages (Krauss et al. 1993). Although shh expression in the floor plate, forebrain, and fin buds appears normal in bon mutants, endodermal shh expression is largely absent, although in some mutants, small clusters of shh-expressing endodermal cells can be seen (Fig. 2F). Together, these data indicate that bon has a critical role in the early events of endoderm development in zebrafish embryos.
The bon locus encodes a Mix-like paired-class homeodomain protein
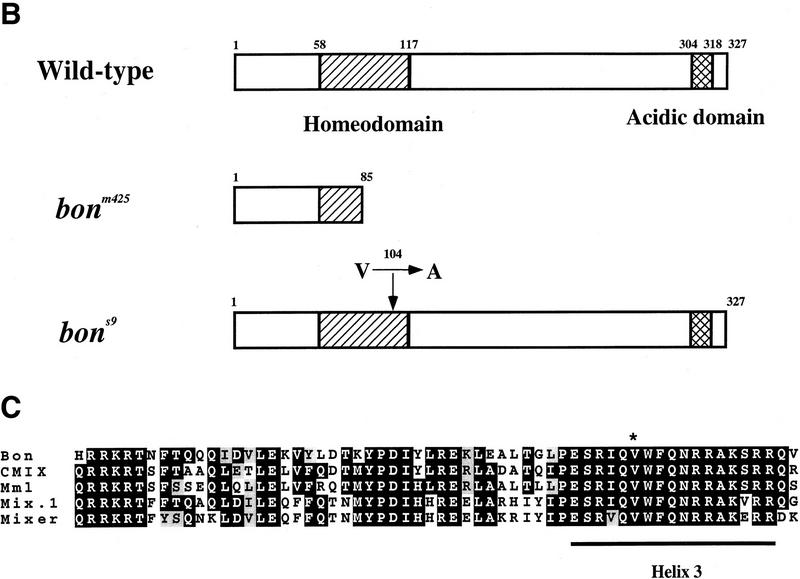
Using half-tetrad analysis (Johnson et al. 1995), we mapped bonm425 to LG20. Further studies revealed that bon is located ∼2.2 cM and 3.9 cM proximal to the simple sequence repeat marker Z13626 (Shimoda et al. 1999) and bmp2b, respectively, and ∼12 cM distal to Z20046 (Fig. 3A). Radiation hybrid mapping using the Goodfellow panel placed the zebrafish Mix-like gene (Alexander et al. 1999) 34.6 cR proximal to bmp2b, that is, in the same region as bon. Linkage analysis showed no recombination between bon and the Mix-like gene in 51 meiotic events (data not shown), and prompted us to sequence the Mix-like gene in both bonm425 and bons9 mutants. Sequencing the bonm425 allele revealed a T to A transversion (TAT → TAA) that introduces a premature translational stop at codon 86 and leads to a truncation in the homeodomain (Fig. 3B). In bons9, we found a T to C substitution (GTC → GCC) that introduces an alanine in place of a highly conserved valine at codon 104 (Fig. 3B) within the predicted third (DNA recognition) helix of the homeodomain (Kissinger et al. 1990; Wintjens and Rooman 1996).
Figure 3.
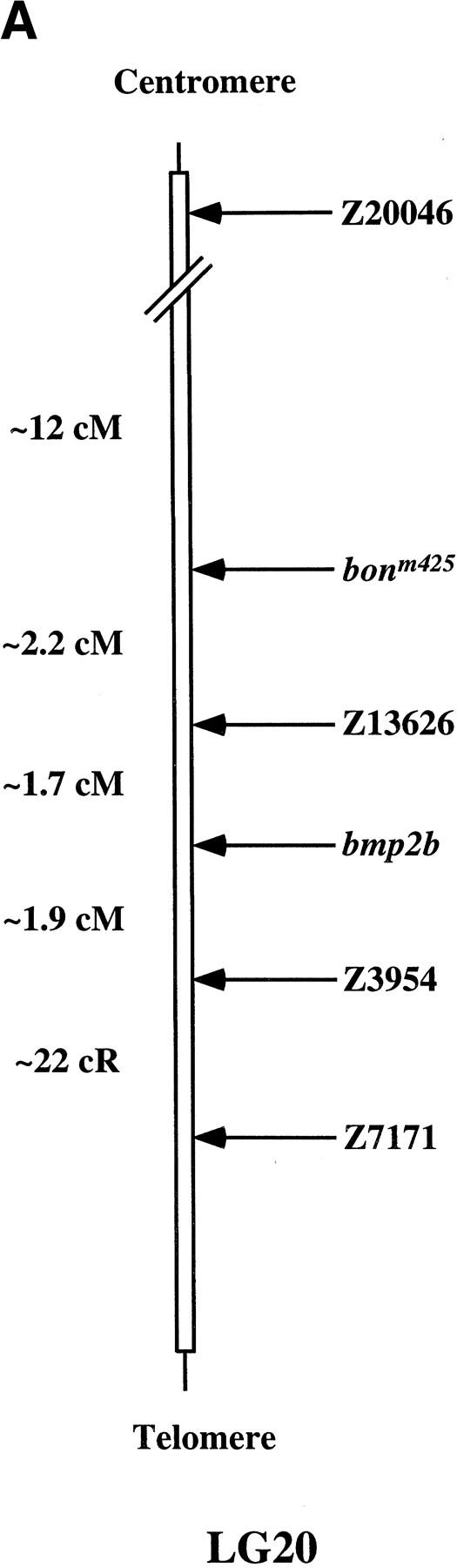
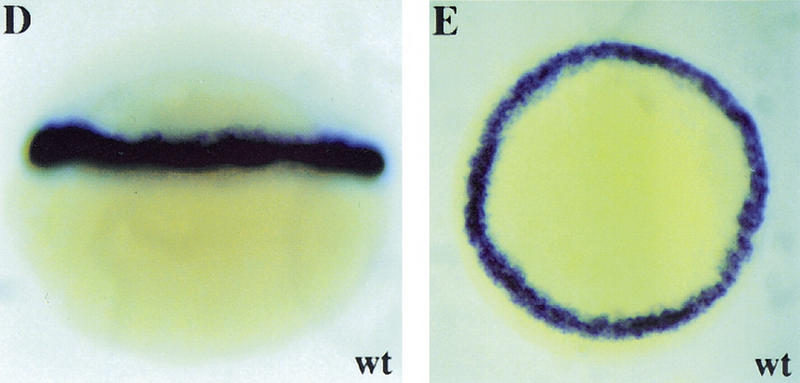
bon maps to LG20. (A) Schematic representation of part of LG20. bon maps ∼2.2 cM proximal to the SSR marker Z13626 (134 meioses), 3.9cM proximal to bmp2b (153 meioses), 5.8 cM proximal to Z3954 (134 meioses) and ∼12 cM distal to Z20046 (25 meioses). Radiation hybrid mapping using the Goodfellow panel placed Z7171 about 22 cR distal to Z3954. (B) Schematic representation of wild-type and mutant Bon proteins. The bonm425 mutation leads to a truncation in the homeodomain, whereas the bons9 mutation introduces a single amino acid substitution (valine-to-alanine) in the third (DNA recognition) helix of the homeodomain. (C) Comparison of the Bon, CMIX, Mml, Mix.1, and Mixer homeodomains. The Bon homeodomain shares the highest level of identity (70%) with the CMIX and Mml homeodomains. The asterisk indicates the highly conserved valine residue mutated in bons9. (D,E) Lateral view (D) and animal pole view (E) of wild-type embryos at 50% epiboly. bon is expressed in all cells of the blastoderm margin.
Using restriction fragment length polymorphisms to analyze linkage, we found that the mutant alleles bonm425 and bons9 segregated with the bon phenotype in 500 and 200 meiotic events, respectively. Together, the genetic data and identification of severe molecular lesions in this gene indicate that bon corresponds to the Mix-like gene we had provisionally named mixer (Alexander et al. 1999). According to the zebrafish nomenclature guidelines (Westerfield 1995), this gene will now be renamed bon.
As previously reported (Alexander et al. 1999), bon expression is first detected at sphere stage in a small group of dorsal cells, and by dome stage has spread throughout the marginal zone where it is expressed in both mesodermal and endodermal progenitors (Fig. 3D,E). By 60% epiboly bon expression is undetectable. bon expression is indistinguishable between wild-type, bonm425 and bons9 mutant embryos (data not shown), indicating that initiation and maintenance of bon expression does not require Bon function.
Overexpression of bon mRNA rescues sox17 expression and gut tube formation in bon mutants
To test further whether the bon phenotype is caused by the identified mutations, we injected synthetic wild-type bon mRNA (100 pg) into embryos derived from both bonm425/+ and bons9/+ heterozygote intercrosses. The injected embryos were processed for in situ hybridization at 75%–80% epiboly and the number of sox17-expressing endodermal precursors on the dorsal side was counted. As a control, we counted the number of sox17-expressing endodermal precursors in β-galactosidase (lacZ) mRNA-injected bon mutants. We found that the number of sox17-expressing endodermal precursors in bon mutants was significantly increased in both alleles following bon mRNA injection (Fig. 4A–D; Table 2). These data indicate that bon mRNA can rescue the early endodermal defect of bon mutants. To analyze whether this rescue was dose-dependent, we also injected 200 pg of bon mRNA and found that injection of a higher dose did not increase the average number of sox17-expressing endodermal precursors (data not shown).
Figure 4.
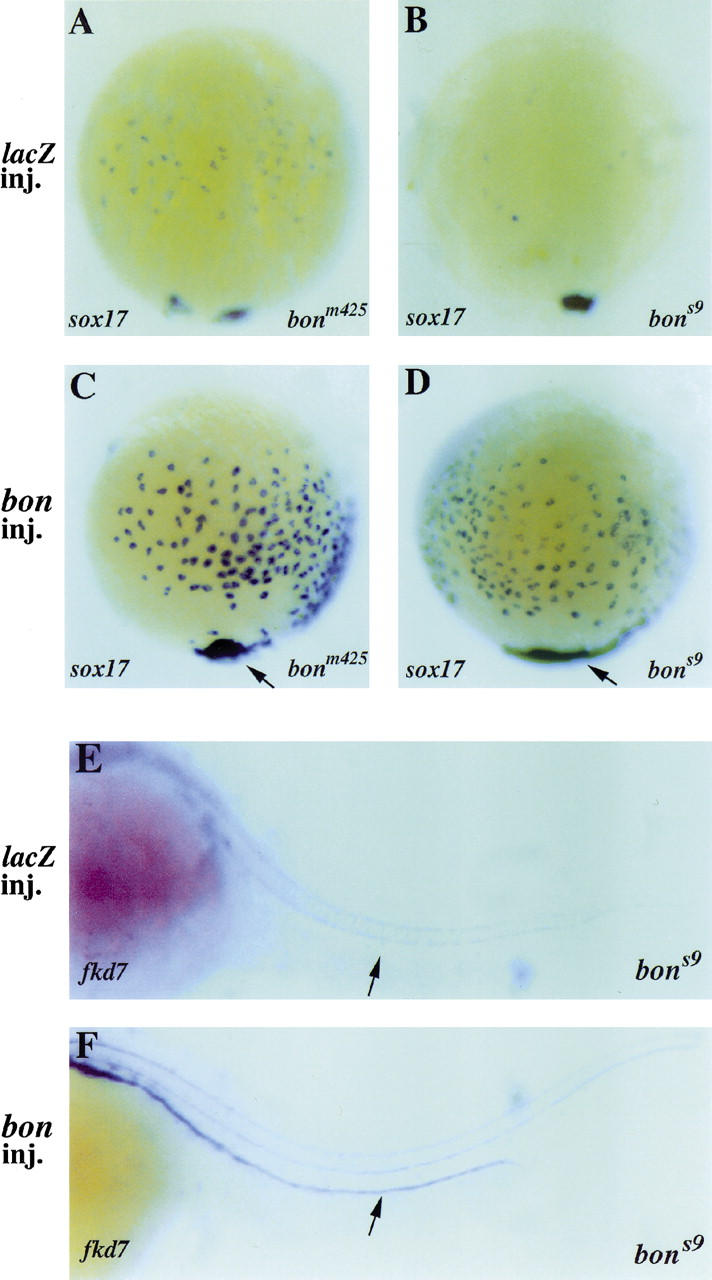
Overexpression of bon mRNA rescues endoderm formation in bon mutants. Embryos were injected with 100 pg of β-galactosidase (lacZ) or bon mRNA at the 1–4 cell stage (A–F). Gut tube formation was assessed by analyzing fkd7 expression in 24 hpf embryos (E,F). (A–D) Dorsal views. lacZ overexpression does not affect sox17 expression in bonm425 (A) or bons9 (B) mutants. The number of sox17-expressing endodermal precursors is significantly increased in bonm425 (C) and bons9 mutants (D) following bon overexpression. An enlargement of the most vegetal sox17-expressing population of cells is also seen in both alleles following bon overexpression (arrows in C and D). (E,F) Lateral views, anterior to the left and dorsal to the top. fkd7 expression in the gut tube, which is absent in lacZ-injected bons9 mutants (arrow in E), is restored following bon overexpression (arrow in F).
Table 2.
Overexpression of bon mRNA rescues sox17 expression in bon mutants
| Injected mRNA
|
Genotype
|
No. of sox17-expressing endodermal precursors on the dorsal side of the embryo
|
|---|---|---|
| lacZ | wild type | 164.1 ± 5.3 |
| (n = 10) | ||
| lacZ | bonm425 | 35.8 ± 3.0 |
| (n = 20) | ||
| bon | bonm425 | 103.7 ± 4.8 |
| (n = 14) | ||
| lacZ | bons9 | 7.5 ± 0.6 |
| (n = 15) | ||
| bon | bons9 | 80.1 ± 5.4 |
| (n = 16) |
The number of sox17-expressing endodermal precursors was counted at 75%–80% epiboly on the dorsal side of the embryo, which we determined to be representative of the whole embryos. P values (t-test) of the differences between bon and lacZ injections are 0.0001 for both sets of embryos (bonm425 and bons9).
To determine whether gut tube formation could also be rescued by bon mRNA injections, we injected 100 pg of bon or lacZ mRNA into bons9/+ heterozygote intercrosses, and examined fkd7 expression at 24 hpf. Out of a total of 105 injected embryos, we identified 23 bon mutants by genotyping, 18 of which showed a completely continuous gut tube as assessed by fkd7 expression (Fig. 4F). These successful rescue experiments provide further evidence that the bon phenotype is caused by the identified mutations.
The Bons9 protein acts in a dominant interfering fashion when overexpressed
The early endoderm defects in bons9 mutants are more severe than those in bonm425 mutants, yet the bons9 mutation causes a single amino acid substitution in the third helix of the homeodomain, whereas the bonm425 mutation causes a premature stop codon at the beginning of the homeodomain. As we would expect, the bonm425 mutation to be a null or near null mutation, we hypothesized that the Bons9 mutant protein functions as a dominant interfering form or antimorph. To test this hypothesis, we introduced an s9-type point mutation in wild-type bon by site-directed mutagenesis and analyzed the consequences of overexpressing bons9 mRNA in both wild-type and bonm425 mutant embryos. Overexpression of bons9 mRNA significantly reduced the average number of sox17-expressing endodermal precursors in both wild-type and bonm425 mutant embryos (Fig. 5A–D; Table 3). These data indicate that the Bons9 protein acts in a dominant interfering fashion when overexpressed.
Figure 5.

Overexpression of bons9 reduces the number of sox17-expressing endodermal precursors in wild-type and mutant embryos. Embryos were injected with 100 pg of lacZ or bons9 mRNA at the 1–4 cell stage. (A–D) Dorsal views. lacZ overexpression does not affect the number of sox17-expressing endodermal precursors in wild-type (A) or bonm425 mutant (B) embryos. The number of sox17-expressing endodermal precursors in wild-type (C) and bonm425 mutant (D) embryos is reduced following bons9 overexpression.
Table 3.
Bons9 acts in a dominant interfering fashion when overexpressed
| Injected mRNA
|
Genotype
|
No. of sox17-expressing endodermal precursors on the dorsal side of the embryo
|
|---|---|---|
| lacZ | wild type | 164.1 ± 5.3 |
| (n = 10) | ||
| bons9 | wild type | 119.4 ± 11.1 |
| (n = 10) | ||
| lacZ | bonm425 | 35.8 ± 3.0 |
| (n = 20) | ||
| bons9 | bonm425 | 8.5 ± 1.3 |
| (n = 24) |
The number of sox17-expressing endodermal precursors was counted at 75%–80% epiboly on the dorsal side of the embryo, which we determined to be representative of the whole embryo. P values (t-test) of the differences between bons9 and lacZ injection are 0.0009 (wild type) and 0.0001 (bonm425).
bon can induce sox17 expression in cyc;sqt double mutants
cyc;sqt double mutants lack all endodermal and most mesodermal derivatives and accordingly, lack endodermal sox17 and axial expression (Feldman et al. 1998; Alexander and Stainier 1999; Fig. 6B). In this report, we have shown through loss-of-function analyses that bon is necessary for the generation of sox17-expressing endodermal precursors. To analyze whether bon is sufficient to induce the formation of sox17-expressing endodermal precursors in the absence of Nodal signaling, we injected bon mRNA into cyc;sqt double mutants, in which bon expression is barely detectable (Alexander and Stainier 1999). We injected bon mRNA into 393 embryos from cyc/+, sqt/+ heterozygote intercrosses (the expected number of double mutants being 24 or 25) and genotyped 93 embryos that showed reduced or absent sox17 expression. We found that bon mRNA induced sox17 expression in 14 out of 24 cyc;sqt double mutants. The average number of sox17-expressing cells in these 14 embryos was very low (7.0 ± 1.6) (Fig. 6C,D), indicating that although bon can induce the formation of sox17-expressing endodermal precursors, its activity in the absence of Nodal signaling is quite weak.
Figure 6.
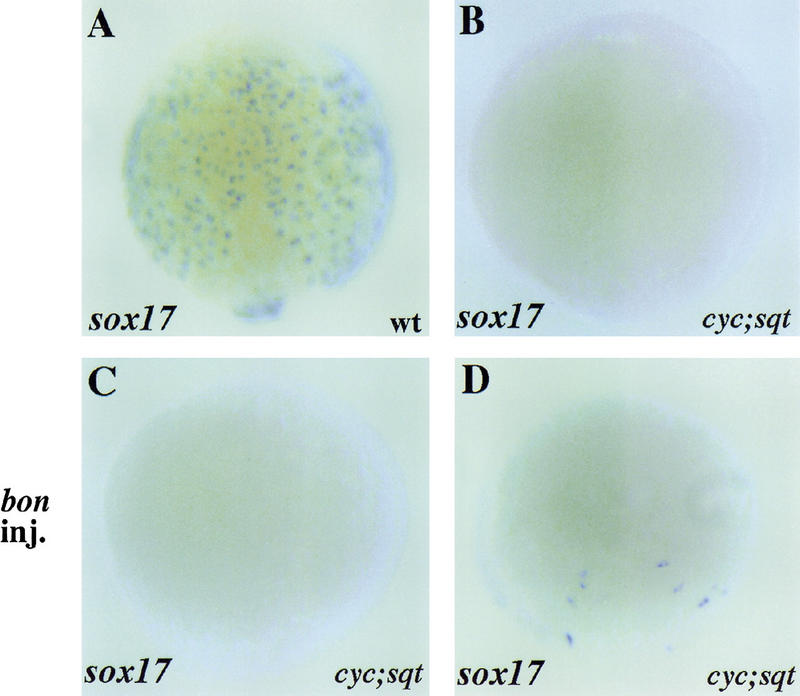
bon overexpression can induce sox17 expression in cyc;sqt double mutants. (A,B) Wild-type (A) and cyc;sqt double mutant (B) embryos were examined for sox17 expression at 90% epiboly. (A–D) Dorsal views. (A,B) sox17 expression is completely absent in cyc;sqt double mutants (B) as determined by genotyping (n = 10). (C,D) Embryos from cyc/+, sqt/+ heterozygote intercrosses were injected with 100 pg of bon mRNA at the 1–4 cell stage, examined for sox17 expression at 75%–80% epiboly and subsequently genotyped. Of 24 cyc;sqt double mutants, no sox17 expression was found in 10 (C), whereas some sox17 expression was observed in 14 (D).
Nodal-related signals have been shown to induce bon expression in zebrafish embryos (Alexander and Stainier 1999). To test whether bon may act reciprocally to promote or maintain cyc and sqt expression, we examined cyc and sqt expression in bon mRNA injected wild-type embryos. Overexpression of bon mRNA did not induce ectopic expression of cyc or sqt in wild-type embryos (Fig. 7C,D). Also, cyc and sqt expression is normal in bon mutants (data not included). These data, together with the fact that bon expression is barely detectable in cyc;sqt double mutants (Alexander and Stainier 1999), indicate that bon functions exclusively downstream of cyc and sqt and does not have a role in regulating their expression.
Figure 7.
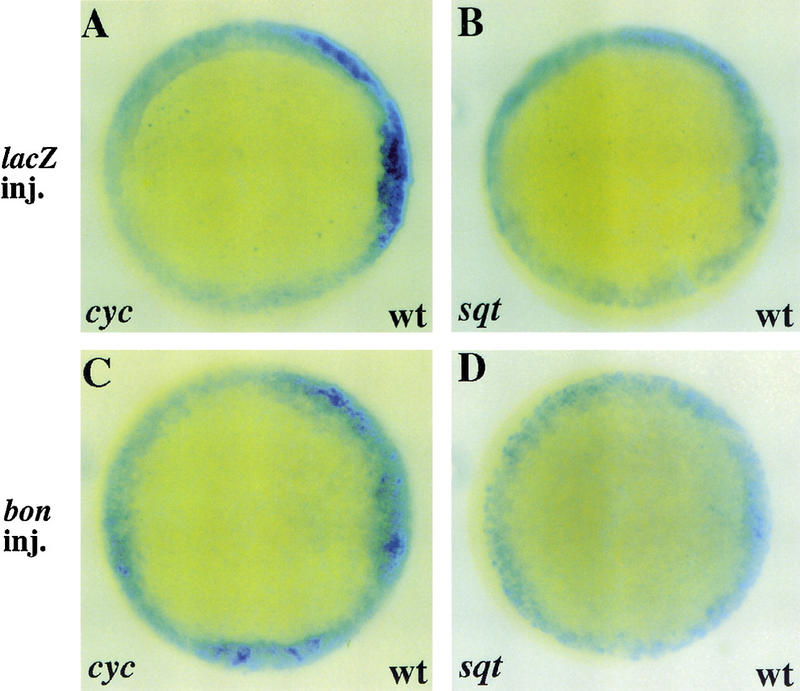
bon overexpression does not induce ectopic cyc or sqt expression in wild-type embryos. Wild-type embryos were injected with 100 pg of lacZ or bon mRNA and examined for cyc or sqt expression at 50% epiboly. (A–D) Animal pole views, dorsal to the right. lacZ overexpression does not affect cyc or sqt expression in wild-type embryos (A,B). Ectopic cyc or sqt expression was not observed in bon-injected wild-type embryos (C,D).
Discussion
bon is a critical regulator of early endoderm formation
In bon mutants, the number of sox17-expressing endodermal precursors is dramatically reduced during gastrulation, and later the gut tube is almost entirely missing. The number of sox17-expressing endodermal precursors does increase as development proceeds, however, suggesting that endodermal proliferation is not affected. Furthermore, the sox17-expressing endodermal precursors in bon mutants appear to differentiate normally, as they go on to express fkd7 and form peristaltically contracting gut tissue. Therefore, bon appears to regulate the initial formation of endodermal precursors but not their subsequent proliferation or differentiation. In Xenopus animal caps, overexpression of Mix and Bix family genes can induce the expression of endodermal genes and overexpression of Mixer–enR mRNA can block endodermal gene expression (Henry and Melton 1998; Mead et al. 1998; Tada et al. 1998; Casey et al. 1999), indicating that these genes are important determinants of endoderm development. The severe endoderm defects seen in bon mutants provide the first genetic evidence that a Mix-like gene, bon, is a critical determinant of early endoderm formation in vertebrates, and a detailed analysis of the bon phenotype brings us closer to understanding the role Mix genes have in endoderm development.
Whereas a small number of endodermal precursors develop in bon mutants, endodermal expression of sox17, axial, and fkd2 is absent completely in cas mutants (Alexander et al. 1999; Alexander and Stainier 1999). There are at least two possible explanations for the fact that some endodermal precursors remain in bon mutants. First, bonm425 and bons9 may not be null mutations. Alternatively, other factors may regulate the generation of sox17-expressing endodermal precursors. For example, the number of sox17- and axial-expressing endodermal precursors is also reduced in faust/gata5 mutants, and overexpression of gata5 mRNA can increase the number of endodermal precursors in wild-type and bon mutant embryos (Reiter et al., in prep.), indicating that at least one other transcription factor is important for early endoderm development in zebrafish embryos. gata5 expression is normal in bon mutants (Reiter et al., in prep.), and its activity may account for the residual endodermal development in these embryos. The cas gene product itself may also be expressed normally in bon mutants and account for the residual endodermal development.
bon encodes a zebrafish Mix-like protein
We present several lines of evidence that bon encodes a Mix-like protein. First, we found no recombination between the bon phenotype and the Mix-like gene in 751 meiotic events. Second, we found a premature stop codon at the beginning of the homeodomain in bonm425 mutants and a single amino acid substitution in a highly conserved residue in the DNA recognition helix of the homeodomain in bons9 mutants. Third, we were able to rescue sox17 expression and gut tube formation in both bonm425 and bons9 mutants by overexpressing wild-type bon mRNA. Together, these data convincingly demonstrate that bon encodes a Mix-like protein.
Bon appears to be a novel member of the Mix family
When comparing homeodomains, Bon is related more closely to chick CMIX (Peale et al. 1998; Stein et al. 1998) and mouse Mml (Pearce and Evans 1999) (70% identical) than to either Xenopus Mix.1 (Rosa 1989) (62% identical) or Mixer (Henry and Melton 1998) (55% identical) (Fig. 3C). The entire Bon protein, therefore, shows low identity with CMIX (24%), Mml (25%), Mix.1 (25%), and Mixer (24%). bon, CMIX, Mml, and Mix.1 appear to be expressed in progenitors of both the endoderm and mesoderm, in contrast to Mixer, which is expressed exclusively in the prospective endoderm. At the functional level, bon and Mixer, but not Mix.1, are able to induce sox17 expression, whereas the activities of the chick and mouse genes have not been reported. These sequence, expression pattern, and functional considerations suggest that bon is not the zebrafish homolog of a specific Xenopus Mix family member. As further information on human and mouse Mix genes is becoming available from genome sequencing data, the relationship between bon and mammalian Mix genes should become clearer.
A valine to alanine substitution in the DNA recognition helix of the homeodomain creates a dominant interfering form of Bon
The bons9 mutation leads to a valine-to-alanine substitution in the third (DNA recognition) helix of the homeodomain. The amino acid sequence of the third helix is highly conserved throughout the Mix and Bix family of proteins (Fig. 3C). Functional analyses have shown that paired-class homeodomains can bind cooperatively as homo- or heterodimers to the palindromic DNA sequence TAATYNRATTA (Y, C, or T; R, A, or G; N, any nucleotide) (Wilson et al. 1993). A high resolution crystal structure of a paired-class homeodomain bound to DNA demonstrates that the third helix is inserted into the major groove of the DNA, allowing two highly conserved amino acid residues (valine and asparagine) to make direct contacts with the DNA via hydrophobic/van der Waals interactions (Wilson et al. 1995). By changing this valine to an alanine, the bons9 mutation could affect the DNA binding ability or specificity of the third helix. Whereas Bons9 may have a reduced ability to bind DNA at its normal sites, it may still be able to interact with other factors of the transcriptional complex. Our results indicate that Bons9 acts in a dominant interfering fashion when overexpressed, consistent with a model whereby it sequesters other factors important for endoderm development. Such a model may also explain why the bons9 phenotype is stronger than the bonm425 one.
Mutations in the human paired-class homeodomain gene PAX3 can cause Waardenburg syndrome type I (DeStefano et al. 1998). In a specific family, a valine-to-phenylalanine substitution in the PAX3 homeodomain segregates with the syndrome (Lalwani et al. 1995). This mutation occurs in the same position of the homeodomain as that in Bons9, further supporting the hypothesis that the valine in the third helix is critical for the proper function of paired-class homeodomain proteins.
bon functions exclusively downstream of TGF-β signaling
bon expression is affected by mutations in nodal-related genes; it is reduced along the animal-vegetal axis and absent dorsally in sqt mutants, and barely detectable in cyc;sqt double mutants (Alexander and Stainier 1999). In addition, overexpression of a constitutively active form of the type I TGF-β receptor TARAM-A (Peyriéras et al. 1998) promotes widespread expression of bon (Alexander and Stainier 1999). Furthermore, overexpression of antivin/lefty1, a feedback inhibitor of Nodal signaling (Bisgrove et al. 1999; Meno et al. 1999; Thisse and Thisse 1999), eliminates bon expression (Alexander and Stainier 1999). These data indicate that Nodal signaling regulates bon expression. We have found that bon overexpression does not lead to ectopic cyc or sqt expression in wild-type embryos, and that cyc and sqt expression is normal in bon mutants. Together, these data demonstrate that bon functions exclusively downstream of Nodal signaling in the molecular pathway leading to endoderm formation in zebrafish.
Additional factors besides Bon regulate sox17 expression
Overexpression of bon increases the number of sox17- and axial-expressing endodermal precursors in zygotic oep mutants (Alexander and Stainier 1999), indicating that bon, by itself or in combination with putative cofactors, has the ability to activate endodermal gene expression in zygotic oep mutants. Here, we have shown that bon can induce the formation of some sox17-expressing endodermal precursors in cyc;sqt double mutants. Our results therefore suggest that Bon can induce endoderm formation in the absence of Nodal-related signaling, but that additional factors downstream of cyc and sqt are needed to generate the full complement of endodermal precursors. These factors may include other paired-class homeodomain proteins that heterodimerize with Bon.
Xenopus Mix.1 can bind to the palindromic DNA sequence (TAATTGAATTA) as a homodimer, and injection of Mix.1 mRNA transforms dorsal mesoderm to a ventral fate (Mead et al. 1996). Mix.1 can also form stable heterodimers with the paired-class homeodomain protein Siamois, and coinjection of Mix.1 and siamois mRNAs induces endodermal gene expression in animal caps (Mead et al. 1996; Tada et al. 1998). Therefore, cofactors of Mix and Bix proteins can have a critical role in specifying their function, raising the possibility that the identification of Bon cofactors will help to elucidate the molecular mechanisms that lead to vertebrate endoderm development.
Molecular events regulating endoderm formation in zebrafish
Previous work has demonstrated that Nodal-related signals encoded by the cyc and sqt genes, together with the putative extracellular Nodal cofactor Oep, are necessary for zebrafish endoderm formation (Feldman et al. 1998; Zhang et al. 1998a; Alexander and Stainier 1999; Gritsman et al. 1999). bon expression is dependent on Nodal signaling (Alexander and Stainier 1999), and in this report we have shown that bon works exclusively downstream of Nodal signaling, where it is necessary for the generation of the normal complement of endodermal precursors. bon expression closely parallels that of cyc and sqt temporally and spatially (Alexander and Stainier 1999), raising the possibility that bon expression is an immediate early response to Nodal signaling. sox17 expression in turn closely follows that of bon temporally. Only a subset of the bon-expressing cells go on to express sox17, however, and it remains an important challenge to understand how these cells are selected to become endoderm.
Materials and methods
Zebrafish strains
Adult zebrafish and embryos were maintained and staged as described in Westerfield (1995). Heterozygous carriers were intercrossed to obtain mutant embryos. Our mapping strain was constructed by crossing a bon heterozygote male to a WIK female. The following mutant alleles were used: cycb16 (Hatta et al. 1991), sqtcz35 (Feldman et al. 1998), sihtc300b (Chen et al. 1996).
Wholemount in situ hybridization and immunohistochemistry
Wholemount in situ hybridization and preparation of RNA probes were performed as described previously (Alexander et al. 1998).
Immunohistochemistry using the antibodies MF20 and S46 was performed as previously described (Alexander et al. 1998). MF20 was obtained from the Developmental Studies Hybridoma Bank, maintained by the Department of Biological Studies, University of Iowa under contract NO1-HD-2-3144 from the National Institute of Child Health and Development (NICHD).
Genetic mapping and single-strand conformational polymorphism (SSCP) analysis
Zebrafish bon was mapped on the Goodfellow zebrafish radiation hybrid panel (Research Genetics) by PCR with primers 5′-TACAAAACATGTCGGTGC-3′ and 5′-TCAAAGACTAAATGAAGC-3′ and the data were submitted for inclusion on the Tübingen map (see http://wwwmap.tuebingen.mpg.de/).
To extract genomic DNA from single embryos, 30 hpf bonm425/m425 and bons9/s9 embryos were digested overnight in 10 mm Tris-HCl at pH 7.5, 1 mm EDTA, 0.1% SDS, 100 mm NaCl, 20 μg/ml of proteinase K at 55°C. Twenty diploid mutant and 11 haploid mutant embryos were analyzed by SSCP (Orita et al. 1989) with primers from the 3′ untranslated region of bon: 5′-GAAAGAGACGCCATGTCTGG-3′ and 5′-TATTGAGATTTCGATGCTGG-3′.
Sequence analyses of bonm425 and bons9
Mutant genomic DNA was purified from 10 30 hpf embryos using the Nucleon ST Kits (Scotlab Bioscience). For sequencing the genomic locus, we performed three independent PCR reactions for each of six primer pairs designed to cover the bon locus, cloned the PCR fragments into pGEM-T (Promega), picked two independent clones from each PCR reaction, and sequenced the inserts using an ABI 377 DNA sequencer.
Linkage analysis
bon DNA fragments were amplified by PCR (for bonm425 5′-CTTCAGGACAGGATGAAGTTCC-3′ and 5′-GAATTCTCGATTCTGGCAAGCC-3′ and for bons9 5′-CAGCTCTGATCATTTCTGCAGC-3′ and 5′-CTTTGTAAGTTCTCCAGGCCGC-3′). The PCR conditions were 94°C for 5 min (1 cycle), then 94°C for 30 sec, 61°C for bonm425 and 64°C for bons9 for 30 sec and 72°C for 1 min (40 cycles), followed by 72°C for 5 min (1 cycle). Amplified products were digested by MseI for bonm425 and StuI for bons9 and resolved on 2% agarose gels. The bonm425 mutation creates a new MseI site and the bons9 mutation a new StuI site. Following in situ hybridization, embryos were genotyped as described above. Genotyping of cyc;sqt double mutants was done using published information (Feldman et al. 1998; Rebagliati et al. 1998).
mRNA injections
To introduce the s9 mutation in bon, we performed site-directed mutagenesis using QuickChange Site-Directed Mutagenesis Kit (Stratagene) according to the manufacturer's instructions and using the primer pair 5′-CCAGAATCGAGAATTCAGGC*CTGGTTCCAGAACCGAAGG-3′ and 5′-CCTTCGGTTCTGGAACCAGG*CCTGAATTCTCGATTCTGG-3′ (* indicates the site of the mutation). The coding region of bons9 was verified by sequencing both strands. Capped mRNA was transcribed in vitro from pCS2–bon or pCS2–bons9 using the mMESSAGE mMACHINE system (Ambion). For overexpression experiments, 100 pg or 200 pg of mRNA was microinjected at the 1–4 cell stage.
Acknowledgments
We thank Alex Navarro, Steve Waldron, and Hsiu-Fen Weeks for outstanding assistance; members of the Stainier lab for helpful discussions; Joseph De Young and Travis Taylor and the Program in Human Genetics Genomics Core Facility at UCSF for sequencing; Frank Stockdale for generously providing the S46 antibody; and Steve Waldron for help with the manuscript. Y.K. is a recipient of a long-term fellowship from the Human Frontier Science Program. J.F.R. and J.A. are members of the Medical Scientist Training Program of the National Institutes of Health. J.F.R. was also supported by an Achievement Rewards for College Scientists Fellowship and J.A. by an American Heart Association Predoctoral Fellowship. D.Y. was supported by an Amgen fellowship of the Life Sciences Research Foundation and is currently a recipient of a Career Award in the Biomedical Sciences from the Burroughs Wellcome Fund. This work was supported in part by grants from the American Heart Association, the Packard Foundation, the Sandler Foundation, and the Life and Health Insurance Medical Research Fund to D.Y.R.S.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL didier_stainier@biochem.ucsf.edu; FAX (415) 476-3892.
References
- Alexander J, Stainier DYR. A molecular pathway leading to endoderm formation in zebrafish. Curr Biol. 1999;9:1147–1157. doi: 10.1016/S0960-9822(00)80016-0. [DOI] [PubMed] [Google Scholar]
- Alexander J, Stainier DYR, Yelon D. Screening mosaic F1 females for mutations affecting zebrafish heart induction and patterning. Dev Genet. 1998;22:288–299. doi: 10.1002/(SICI)1520-6408(1998)22:3<288::AID-DVG10>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Alexander J, Rothenberg M, Henry GL, Stainier DYR. casanova plays an early and essential role in endoderm formation in zebrafish. Dev Biol. 1999;215:343–357. doi: 10.1006/dbio.1999.9441. [DOI] [PubMed] [Google Scholar]
- Bisgrove BW, Essner JJ, Yost HJ. Regulation of midline development by antagonism of lefty and nodal signaling. Development. 1999;126:3253–3262. doi: 10.1242/dev.126.14.3253. [DOI] [PubMed] [Google Scholar]
- Casey ES, Tada M, Fairclough L, Wylie CC, Heasman J, Smith JC. Bix4 is activated directly by VegT and mediates endoderm formation in Xenopus development. Development. 1999;126:4193–4200. doi: 10.1242/dev.126.19.4193. [DOI] [PubMed] [Google Scholar]
- Chen JN, Haffter P, Odenthal J, Vogelsang E, Brand M, van Eeden FJM, Furutani-Seiki M, Granato M, Hammerschmidt M, Heisenberg CP, et al. Mutations affecting the cardiovascular system and other internal organs in zebrafish. Development. 1996;123:293–302. doi: 10.1242/dev.123.1.293. [DOI] [PubMed] [Google Scholar]
- Clements D, Friday RV, Woodland HR. Mode of action of VegT in mesoderm and endoderm formation. Development. 1999;126:4903–4911. doi: 10.1242/dev.126.21.4903. [DOI] [PubMed] [Google Scholar]
- DeStefano AL, Cupples LA, Arnos KS, Asher JH, Jr, Baldwin CT, Blanton S, Carey ML, da Silva EO, Friedman TB, Greenberg J, et al. Correlation between Waardenburg syndrome phenotype and genotype in a population of individuals with identified PAX3 mutations. Hum Genet. 1998;102:499–506. doi: 10.1007/s004390050732. [DOI] [PubMed] [Google Scholar]
- Ecochard V, Cayrol C, Rey S, Foulquier F, Caillol D, Lemaire P, Duprat AM. A novel Xenopus Mix-like gene milk involved in the control of the endomesodermal fates. Development. 1998;125:2577–2585. doi: 10.1242/dev.125.14.2577. [DOI] [PubMed] [Google Scholar]
- Feldman B, Gates MA, Egan ES, Dougan ST, Rennebeck G, Sirotkin HI, Schier AF, Talbot WS. Zebrafish organizer development and germ-layer formation require nodal- related signals. Nature. 1998;395:181–185. doi: 10.1038/26013. [DOI] [PubMed] [Google Scholar]
- Gritsman K, Zhang J, Cheng S, Heckscher E, Talbot WS, Schier AF. The EGF-CFC protein one-eyed pinhead is essential for nodal signaling. Cell. 1999;97:121–132. doi: 10.1016/s0092-8674(00)80720-5. [DOI] [PubMed] [Google Scholar]
- Hatta K, Kimmel CB, Ho RK, Walker, C C. The cyclops mutation blocks specification of the floor plate of the zebrafish central nervous system. Nature. 1991;350:339–341. doi: 10.1038/350339a0. [DOI] [PubMed] [Google Scholar]
- Henry GL, Melton DA. Mixer, a homeobox gene required for endoderm development. Science. 1998;281:91–96. doi: 10.1126/science.281.5373.91. [DOI] [PubMed] [Google Scholar]
- Hudson C, Clements D, Friday RV, Stott D, Woodland HR. Xsox17α and β mediate endoderm formation in Xenopus. Cell. 1997;91:397–405. doi: 10.1016/s0092-8674(00)80423-7. [DOI] [PubMed] [Google Scholar]
- Johnson SL, Africa D, Horne S, Postlethwait JH. Half-tetrad analysis in zebrafish: Mapping the ros mutation and the centromere of linkage group I. Genetics. 1995;139:1727–1735. doi: 10.1093/genetics/139.4.1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissinger CR, Liu BS, Martin-Blanco E, Kornberg TB, Pabo CO. Crystal structure of an engrailed homeodomain-DNA complex at 2.8 Å resolution: A framework for understanding homeodomain-DNA interactions. Cell. 1990;63:579–590. doi: 10.1016/0092-8674(90)90453-l. [DOI] [PubMed] [Google Scholar]
- Krauss S, Concordet JP, Ingham PW. A functionally conserved homolog of the Drosophila segment polarity gene hh is expressed in tissues with polarizing activity in zebrafish embryos. Cell. 1993;75:1431–1444. doi: 10.1016/0092-8674(93)90628-4. [DOI] [PubMed] [Google Scholar]
- Lalwani AK, Brister JR, Fex J, Grundfast KM, Ploplis B, San Agustin TB, Wilcox ER. Further elucidation of the genomic structure of PAX3, and identification of two different point mutations within the PAX3 homeobox that cause Waardenburg syndrome type 1 in two families. Am J Hum Genet. 1995;56:75–83. [PMC free article] [PubMed] [Google Scholar]
- Lemaire P, Darras S, Caillol D, Kodjabachian L. A role for the vegetally expressed Xenopus gene Mix.1 in endoderm formation and in the restriction of mesoderm to the marginal zone. Development. 1998;125:2371–2380. doi: 10.1242/dev.125.13.2371. [DOI] [PubMed] [Google Scholar]
- Mead PE, Brivanlou IH, Kelley CM, Zon LI. BMP-4-responsive regulation of dorsal-ventral patterning by the homeobox protein Mix.1. Nature. 1996;382:357–360. doi: 10.1038/382357a0. [DOI] [PubMed] [Google Scholar]
- Mead PE, Zhou Y, Lustig KD, Huber TL, Kirschner MW, Zon LI. Cloning of Mix-related homeodomain proteins using fast retrieval of gel shift activities, (FROGS), a technique for the isolation of DNA-binding proteins. Proc Natl Acad Sci. 1998;95:11251–11256. doi: 10.1073/pnas.95.19.11251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meno C, Gritsman K, Ohishi S, Ohfuji Y, Heckscher E, Mochida K, Shimono A, Kondoh H, Talbot WS, Robertson EJ, et al. Mouse Lefty2 and zebrafish antivin are feedback inhibitors of nodal signaling during vertebrate gastrulation. Mol Cell. 1999;4:287–298. doi: 10.1016/s1097-2765(00)80331-7. [DOI] [PubMed] [Google Scholar]
- Odenthal J, Nüsslein-Volhard C. fork head domain genes in zebrafish. Dev Genes Evol. 1998;208:245–258. doi: 10.1007/s004270050179. [DOI] [PubMed] [Google Scholar]
- Orita M, Iwahana H, Kanazawa H, Hayashi K, Sekiya T. Detection of polymorphisms of human DNA by gel electrophoresis as single-strand conformation polymorphisms. Proc Natl Acad Sci. 1989;86:2766–2770. doi: 10.1073/pnas.86.8.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osada SI, Wright CV. Xenopus nodal-related signaling is essential for mesendodermal patterning during early embryogenesis. Development. 1999;126:3229–3240. doi: 10.1242/dev.126.14.3229. [DOI] [PubMed] [Google Scholar]
- Peale FV, Jr, Sugden L, Bothwell M. Characterization of CMIX, a chicken homeobox gene related to the Xenopus gene mix.1. Mech Dev. 1998;75:167–170. doi: 10.1016/s0925-4773(98)00089-6. [DOI] [PubMed] [Google Scholar]
- Pearce JJH, Evans MJ. Mml, a mouse mix-like gene expressed in the primitive streak. Mech Dev. 1999;87:189–192. doi: 10.1016/s0925-4773(99)00135-5. [DOI] [PubMed] [Google Scholar]
- Peyriéras N, Strähle, U. U, Rosa F. Conversion of zebrafish blastomeres to an endodermal fate by TGF-β related signaling. Curr Biol. 1998;8:783–786. doi: 10.1016/s0960-9822(98)70303-3. [DOI] [PubMed] [Google Scholar]
- Rebagliati MR, Toyama R, Haffter P, Dawid IB. cyclops encodes a nodal-related factor involved in midline signaling. Proc Natl Acad Sci. 1998;95:9932–9937. doi: 10.1073/pnas.95.17.9932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter JF, Alexander J, Rodaway A, Yelon D, Patient R, Holder N, Stainier DYR. Gata5 is required for the development of the heart and endoderm in zebrafish. Genes & Dev. 1999;13:2983–2995. doi: 10.1101/gad.13.22.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa FM. Mix.1, a homeobox mRNA inducible by mesoderm inducers, is expressed mostly in the presumptive endodermal cells of Xenopus embryos. Cell. 1989;57:965–974. doi: 10.1016/0092-8674(89)90335-8. [DOI] [PubMed] [Google Scholar]
- Schier AF, Neuhauss SC, Helde KA, Talbot WS, Driever W. The one-eyed pinhead gene functions in mesoderm and endoderm formation in zebrafish and interacts with no tail. Development. 1997;124:327–342. doi: 10.1242/dev.124.2.327. [DOI] [PubMed] [Google Scholar]
- Shimoda N, Knapik EW, Ziniti J, Sim C, Yamada E, Kaplan S, Jackson D, de Sauvage F, Jacob H, Fishman MC. Zebrafish genetic map with 2000 microsatellite markers. Genomics. 1999;58:219–232. doi: 10.1006/geno.1999.5824. [DOI] [PubMed] [Google Scholar]
- Stainier DYR, Fouquet B, Chen JN, Warren KS, Weinstein BM, Meiler SE, Mohideen MAPK, Neuhauss SCF, Solnica-Krezel L, Schier AF, et al. Mutations affecting the formation and function of the cardiovascular system in the zebrafish embryo. Development. 1996;123:285–292. doi: 10.1242/dev.123.1.285. [DOI] [PubMed] [Google Scholar]
- Stein S, Roeser T, Kessel M. CMIX, a paired-type homeobox gene expressed before and during formation of the avian primitive streak. Mech Dev. 1998;75:163–165. doi: 10.1016/s0925-4773(98)00088-4. [DOI] [PubMed] [Google Scholar]
- Tada M, Casey ES, Fairclough L, Smith JC. Bix1, a direct target of Xenopus T-box genes, causes formation of ventral mesoderm and endoderm. Development. 1998;125:3997–4006. doi: 10.1242/dev.125.20.3997. [DOI] [PubMed] [Google Scholar]
- Thisse C, Thisse B. Antivin, a novel and divergent member of the TGFβ superfamily, negatively regulates mesoderm induction. Development. 1999;126:229–240. doi: 10.1242/dev.126.2.229. [DOI] [PubMed] [Google Scholar]
- Westerfield M. The Zebrafish Book: A guide for the laboratory use of zebrafish. Eugene, OR: University of Oregon Press; 1995. [Google Scholar]
- Wilson D, Sheng G, Lecuit T, Dostatni N, Desplan C. Cooperative dimerization of paired class homeo domains on DNA. Genes & Dev. 1993;7:2120–2134. doi: 10.1101/gad.7.11.2120. [DOI] [PubMed] [Google Scholar]
- Wilson DS, Guenther B, Desplan C, Kuriyan J. High resolution crystal structure of a paired (Pax) class cooperative homeodomain dimer on DNA. Cell. 1995;82:709–719. doi: 10.1016/0092-8674(95)90468-9. [DOI] [PubMed] [Google Scholar]
- Wintjens R, Rooman M. Structural classification of HTH DNA-binding domains and protein-DNA interaction modes. J Mol Biol. 1996;262:294–313. doi: 10.1006/jmbi.1996.0514. [DOI] [PubMed] [Google Scholar]
- Yasuo H, Lemaire P. A two-step model for the fate determination of presumptive endodermal blastomeres in Xenopus embryos. Curr Biol. 1999;9:869–879. doi: 10.1016/s0960-9822(99)80391-1. [DOI] [PubMed] [Google Scholar]
- Zhang J, Talbot WS, Schier AF. Positional cloning identifies zebrafish one-eyed pinhead as a permissive EGF-related ligand required during gastrulation. Cell. 1998a;92:241–251. doi: 10.1016/s0092-8674(00)80918-6. [DOI] [PubMed] [Google Scholar]
- Zhang J, Houston DW, King ML, Payne C, Wylie C, Heasman J. The role of maternal VegT in establishing the primary germ layers in Xenopus embryos. Cell. 1998b;94:515–524. doi: 10.1016/s0092-8674(00)81592-5. [DOI] [PubMed] [Google Scholar]