Abstract
Regulated mRNA turnover is a highly important process, but its mechanism is poorly understood. Using interleukin-2 (IL-2) mRNA as a model, we described a role for the JNK-signaling pathway in stabilization of IL-2 mRNA during T-cell activation, acting via a JNK response element (JRE) in the 5′ untranslated region (UTR). We have now identified two major RNA-binding proteins, nucleolin and YB-1, that specifically bind to the JRE. Binding of both proteins is required for IL-2 mRNA stabilization induced by T-cell activation signals and for JNK-induced stabilization in a cell-free system that duplicates essential features of regulated mRNA decay. Nucleolin and YB-1 are required for formation of an IL-2 mRNP complex that responds to specific mRNA stabilizing signals.
Keywords: Nucleolin, JNK, T-cell activation, stabilization, trans-acting factors
Much knowledge has accumulated regarding the mechanisms by which signal transduction pathways regulate gene expression at the transcriptional level (Hunter and Karin 1992; Hill and Treisman 1995; Karin and Hunter 1995). In comparison, the mechanistic basis for regulation of mRNA turnover, which also plays a very important role in control of gene expression, remains nebulous. The stability of different mRNAs varies considerably and in many cases is regulated in response to extracellular stimuli. Expression of immediate early genes and cytokines is usually transient because their mRNAs are inherently unstable. Rapid mRNA turnover is mediated by cis-acting elements that are distributed throughout the mRNA molecule (Sachs 1993; Ross 1995). A common destabilizing element found in the 3′ untranslated region (UTR) of short-lived mRNAs is the AU-rich element (ARE), consisting of multiple copies of the pentanucleotide, AUUUA, and a high content of U residues (Caput et al. 1986; Chen and Shyu 1995). Although an ARE or an UUAUUUAUU nonamer can direct rapid mRNA turnover by promoting deadenylation followed by rapid decay of the mRNA body (Shaw and Kamen 1986; Shyu et al. 1989; Lagnado et al. 1994; Zubiaga et al. 1995; Xu et al. 1997), the mechanism underlying this activity is largely unknown. In addition to ARE-mediated mRNA decay, unstable mRNAs can be targeted for rapid decay by other pathways (Shyu et al. 1991; Wisdom and Lee 1991; Brown et al. 1996; Peng et al. 1996).
ARE-directed mRNA decay may be positively or negatively regulated by extracellular stimuli (Iwai et al. 1993; Stoecklin et al. 1994; Ross 1995; Kishore et al. 1999). Moreover, distinct ARE-containing mRNAs can be differentially regulated within the same cell, underlining the complexity of mRNA degradation. For example, in a monocytic cell line, the 3′ UTRs of c-fos or c-myc destabilize a reporter mRNA, whereas the 3′ UTR of granulocyte-macrophage colony-stimulating factor (GM-CSF) does not (Schuler and Cole 1988). Activation of quiescent T-cells with antibodies to the T-cell receptor (TCR)–CD3 complex and the CD28 auxiliary receptor stabilizes several cytokine mRNAs, whereas c-fos and c-myc mRNAs remain labile (Lindsten et al. 1989). Such differential regulation may be due to different mRNA decay pathways. For instance, c-fos and c-myc mRNAs may be targeted to rapid decay by ARE-independent pathways, whereas the ARE-directed pathway is inhibited in response to cell stimulation (Shyu et al. 1991; Wisdom and Lee 1991). Alternatively, specific cis elements present in some cytokine mRNAs, but not others, are recognized by signal-responsive trans-acting factors that modulate or determine ARE-directed decay. The signaling pathways that regulate ARE-directed mRNA decay have not been widely explored, and the relevant cis elements that mediate signal-induced mRNA stabilization are largely unknown. The AREs of GM-CSF or IL-3 are sufficient for regulation of mRNA turnover in response to calcium signals (Iwai et al. 1993; Stoecklin et al. 1994). However, phorbol ester (TPA)-induced stabilization of GM-CSF mRNA requires an additional region upstream to its ARE (Iwai et al. 1991). Ca2+ signals do not stabilize a reporter mRNA containing the 3′ UTRs of granulocyte colony-stimulating factor (G-CSF), IL-2, or IL-6, because an mRNA decay pathway specified by a novel instability element, in addition to AREs, is insensitive to Ca2+-induced mRNA stabilization (Brown et al. 1996). mRNA stabilization may require cooperation between multiple RNA elements (Yang et al. 1996; Dibbens et al. 1999), suggesting that either proper mRNA folding is necessary for signal-induced stabilization or that interactions between different trans-acting factors can modulate ARE-directed mRNA decay.
Although many ARE-binding proteins were identified (Hamilton et al. 1993; Zhang et al. 1993; Nagy and Rigby 1995; Nakagawa et al. 1995; Myer et al. 1997; Lai et al. 1999), with a few exceptions, their functional significance remains enigmatic. Overexpression of a human homolog of Drosophila ELAV (embryonic lethal, abnormal vision), HuR, stabilizes ARE-containing mRNAs (Fan and Steitz 1998; Peng et al. 1998). ELAV proteins also stabilize mRNA in vitro (Ford et al. 1999). Mice lacking the tristetraprolin protein (TTP) exhibit decreased TNFα mRNA turnover (Carballo et al. 1998). TTP binds the AREs of TNFα and other cytokine mRNAs and promotes deadenylation and decay (Lai et al. 1999). AUF1, another ARE-binding protein, promotes mRNA decay in vitro (Brewer 1991). AUF1 is also involved in heat shock-induced stabilization of ARE-containing mRNA (Laroia et al. 1999) and in ARE-mediated mRNA destabilization in vivo (Loflin et al. 1999). Whether any of these ARE-binding proteins are involved in signal-regulated mRNA stabilization remains unknown. In addition, it is unlikely that general ARE-binding proteins account for differential regulation of mRNA stability. Most likely, other trans-acting factors, recognizing cis elements throughout the mRNA molecule, can modulate the activity of ARE-binding proteins and thereby confer differential mRNA stabilization.
To understand how extracellular stimuli regulate mRNA stability, we chose IL-2 mRNA as a model. Like other short-lived cytokine mRNAs, IL-2 mRNA contains several AREs in the 3′ UTR, which may be responsible for its short half-life (t1/2 = 30–60 min) in resting T cells (Chen et al. 1998). Upon T-cell activation, the t1/2 of IL-2 mRNA is prolonged (Lindsten et al. 1989). The signal transduction pathways and the cis elements responsible for IL-2 mRNA stabilization in activated T cells remained elusive for many years. Recently, IL-2 mRNA stabilization in response to CD28 signaling was shown to require multiple cis elements (Ragheb et al. 1999). We found that stabilization of IL-2 mRNA in response to T-cell activation signals is mediated via at least two cis elements (Chen et al. 1998). One of the pathways that leads to IL-2 mRNA stabilization is the JNK mitogen-activated protein kinase (MAPK) cascade, which targets a 5′ JNK response element (JRE), that needs to interact with undefined element within the 3′ UTR. Whereas the IL-2 3′ UTR contains both stabilizing and destabilizing elements, some of which may overlap, the 5′ JRE only mediates mRNA stabilization in response to JNK activation (Chen et al. 1998). Deletion or inactivation of the 5′ JRE has no effect either on IL-2 mRNA stability in nonstimulated T cells or its response to other stabilizing signals. Due to its well defined and specific function, we focused our effort on understanding how the 5′ JRE functions. We identified two proteins that specifically recognize the 5′ JRE and are required for JNK- and mitogen-induced IL-2 mRNA stabilization both in vivo and in a cell-free system, in which mRNA decay can be regulated by physiological signals, including activated JNK.
Results
Identification of cytoplasmic 5′ JRE-binding proteins
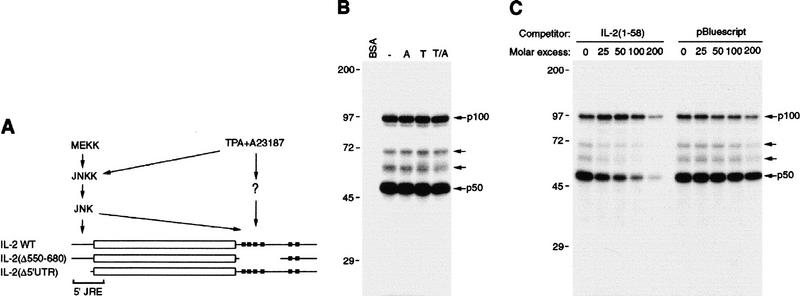
IL-2 mRNA contains multiple cis elements required for stabilization in activated T cells (Fig. 1A). JNK-induced stabilization requires at least two cis elements; one within the 5′ UTR and the other within the 3′ UTR (Chen et al. 1998). The 5′ JRE was defined previously by two deletion mutations Δ(1–40) and Δ(58–105), both of which abolished JNK-mediated stabilization. Nucleotides 1–120 are sufficient for conferring JNK-dependent stabilization on a chimeric transcript containing chloramphenicol acetyltransferase (CAT)-coding region and IL-2 3′ UTR (Chen et al. 1998). However, the 5′ sequences required for JNK-mediated stabilization were not further defined.
Figure 1.
Identification of JRE-binding proteins and critical sequences. (A) Schematic representation of wild-type and mutant IL-2 transcripts. (Open box) Coding region; (thin line) 5′ and 3′ UTRs; (solid boxes) AUUUA motifs. The signaling pathways involved in regulation of IL-2 mRNA stability are shown with their cis element targets. (B) UV cross-linking assay. Cytoplasmic extracts of Jurkat cells either unstimulated or stimulated with TPA (T), A23187 (A), or both (T/A) were incubated with 32P-labeled IL-2(1–58) RNA. Following UV cross-linking, RNA–protein complexes were analyzed by denaturing SDS-PAGE and are indicated by arrows. (C) Specificity of RNA–protein complex formation. IL-2(1–58) RNA probe was coincubated with extracts of unstimulated Jurkat cells and the indicated molar excess of either homologous or heterologous RNA competitors (nonspecific 90-nucleotide transcript produced from the polylinker of pBluescript). (D) Sequences of the 5′ UTRs of wild-type and mutant IL-2 RNAs and their half-lives. CAT reporter genes flanked by IL-2 5′ and 3′ UTRs were cotransfected with MEKK1 or an empty expression vector into Jurkat cells and their t1/2s were determined. The average t1/2 (on the basis of two separate transfection experiments done in duplicate) of each RNA in the absence or presence of MEKK1 is indicated at right.
To detect JRE-binding proteins, uniformly labeled RNAs containing nucleotides 1–130 or 1–58 were incubated with cytoplasmic extracts from unstimulated or mitogen-stimulated Jurkat cells, a human T-cell leukemia line. After initial incubation, binding mixtures were treated with RNase T1 to remove unprotected RNA. RNase-resistant RNA–protein complexes were UV cross-linked, treated with RNase A, and separated by SDS-PAGE under reducing conditions. Two major, 50-kD (p50) and 100-kD (p100), and two minor RNA–protein complexes were detected (Fig. 1B). The RNA–protein complexes detected with nucleotides 1–58 were identical to those detected with nucleotides 1–130 (data not shown). The abundance or the mobility of any of the complexes was not affected by cell stimulation (Fig. 1B). Formation of the p50–RNA complex was competed by excess homologous, but not heterologous, unlabeled RNA (Fig. 1C). Neither homologous nor heterologous RNAs competed for formation of the p100–RNA complex. These results suggest that p50 binds specifically to IL-2(1–58) RNA and the binding of p100 may either be nonspecific or not efficiently competed in the presence of p50 or other cytosolic proteins (see Fig. 2). The two less-abundant RNA–protein complexes appeared to be specific (Fig. 1C). These results also suggest that most of the protein binding occurred between nucleotides 1 and 58, whereas little if any binding of p50 and p100 occurred between nucleotides 59 and 130. To further define the 5′ JRE, we constructed four linker-scanning mutants, M1–M4, spanning the 5′ UTR. These, as well as the wild-type sequence were placed in front of a CAT-IL-2 3′ UTR chimera and examined for JNK responsiveness by cotransfection with an expression vector for MEKK1, a JNK kinase kinase. Whereas M1 and M2 lost the ability to respond to the JNK-activating signal, mutants M3 and M4 were as responsive as the wild-type reporter (Fig. 1D). Therefore, the 5′ JRE requires sequences between the cap site and nucleotide 22.
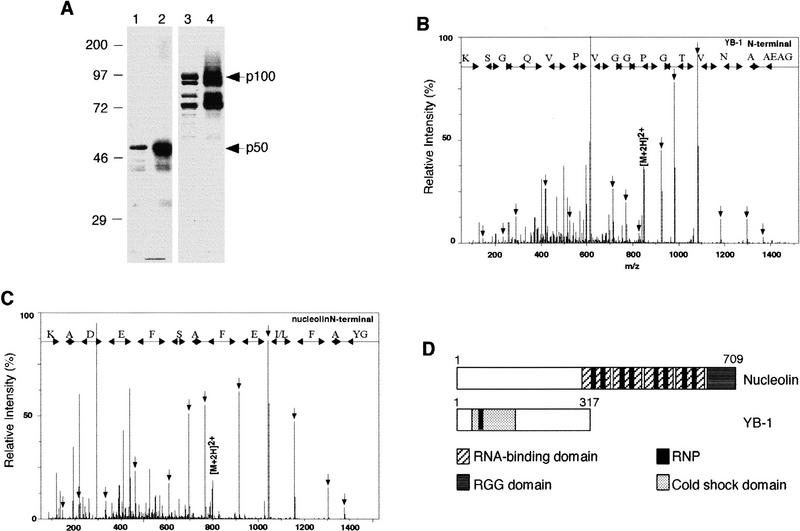
Figure 2.
Molecular identification of p50 and p100. (A) Coomassie blue staining of purified p50 (lane 1) or p100 (lane 3) and their respective RNA–polypeptide complexes (lanes 2,4) detected by UV cross-linking and autoradiography. (B,C) Identification of YB-1 and nucleolin by the nanoelectrospray tandem mass spectra of doubly charged peptides extracted from the 50- and 100-kD bands, respectively. Sequence tags were assembled from a series of fragment ions and used to search for a matching pattern. Fragment masses calculated for the retrieved YB-1 GAEAANVTGPGGVPVQGSK or the retrieved nucleolin GYAFI/LEFASFEDAK sequences were compared with the complete fragmentation spectrum to confirm the match. (D) Schematic representation of nucleolin and YB-1. (Hatched boxes) RNA-binding domain; (dark grey box) RGG domain; (black boxes) RNP; (stippled boxes) cold shock domain.
To purify p50 and p100, Jurkat cytoplasmic extracts were fractionated by ion-exchange chromatography. After washing, column-bound proteins were eluted with a linear salt gradient and their binding to IL-2(1–58) RNA measured by UV cross-linking. This procedure separated p50 from p100 (data not shown). Fractions containing p50 and p100 were pooled separately and fractionated by gel filtration, followed by affinity chromatography on a column containing immobilized IL-2(1–58) RNA. Bound proteins were step eluted and fractions containing p50 or p100 were collected, concentrated, and analyzed by RNA cross-linking and SDS-PAGE (Fig. 2A). This revealed that p50 was purified to near homogeneity, whereas p100 copurified with three other polypeptides. All polypeptides had very similar electrophoretic mobilities to their RNA cross-linked complexes after RNase treatment (Fig. 2A).
Polypeptide bands were excised from the gel and analyzed by nanoelectrospray mass spectrometry to derive peptide sequence tags (Wilm et al. 1996). The 50-kD band (Fig. 2B) contained a unique 19 amino acid peptide from human Y box-binding protein (YB-1; Didier et al. 1988). Tandem mass spectra of >15 peptides derived from the 100-kD band matched peptide sequences from human nucleolin (Srivastava et al. 1989). One of the peptides and its mass spectrum are shown in Figure 2C. The fast-migrating bands coeluting with p100 exhibited very similar spectra to p100 itself. Therefore, these bands contain proteolyzed nucleolin. Nucleolin and YB-1 are schematically described in Figure 2D. The amino-terminal portion of nucleolin consists of acidic stretches and is highly phosphorylated (Ginisty et al. 1999). Its carboxy-terminal portion has four RNA-binding domains (RBDs), each containing two highly conserved RNP-1 and RNP-2 motifs (Burd and Dreyfuss 1994). YB-1 can be divided into three putative domains, an amino-terminal region, the cold shock domain (CSD), which is responsible for nucleic acid binding and contains RNP-1 motif, and a carboxy-terminal tail, which may be responsible for protein–protein interactions (Wolffe et al. 1992).
Nucleolin and YB-1 bind IL-2 mRNA in vitro and in vivo
To confirm that the two JRE-binding proteins are YB-1 and nucleolin, UV cross-linking reaction mixtures were immunoprecipitated with anti-YB-1 or anti-nucleolin. The p50-JRE and p100-JRE complexes were specifically precipitated by anti-YB-1 and anti-nucleolin antibodies, respectively (Fig. 3A). Immunoblot analysis confirmed that the p50–JRE and p100–JRE complexes contain YB-1 and nucleolin, respectively. We examined binding of nucleolin and YB-1 to nucleotides 1–58 of wild-type IL-2 mRNA or mutants M1–M4. In agreement with the JNK responsiveness of these sequences (Fig. 1D), nucleolin and YB-1 in Jurkat cytosolic extracts bound the wild-type, M3, and M4 probes, but not the M1 and M2 probes (Fig. 3B). Similar specificity was displayed by nucleolin and YB-1 GST fusion proteins (Fig. 3C,D). Collectively, these results demonstrate that p50 and p100 are YB-1 and nucleolin, respectively, and show that they bind specifically to the 5′ JRE of IL-2 mRNA. We also found that nucleolin and YB-1 can bind simultaneously to the 5′ JRE to form a ternary complex (data not shown). Mutations that disrupt binding of nucleolin and YB-1 to the 5′ JRE abolish JNK-mediated IL-2 mRNA stabilization (Fig. 1D).
Figure 3.
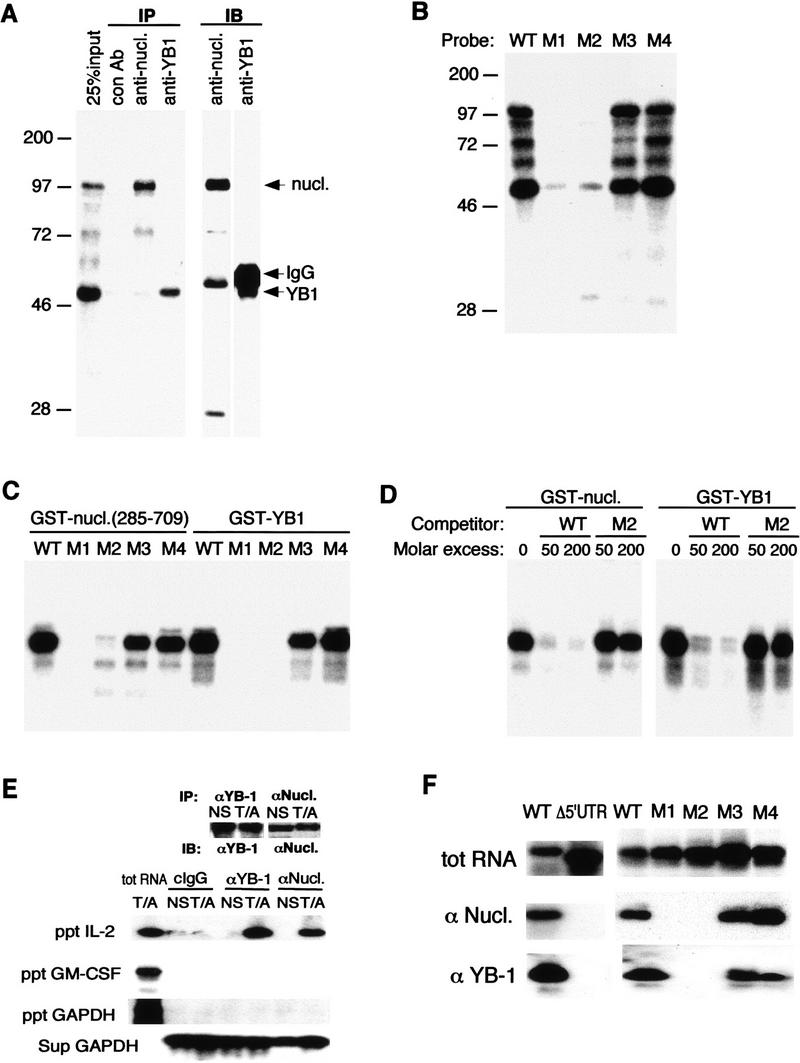
Binding of YB-1 and nucleolin to IL-2 RNA. (A) Jurkat cytoplasmic extracts were incubated with 32P-labeled IL-2(1–58) RNA, UV cross-linked, and immunoprecipitated with control antibodies [mouse IgG (lane 2) or rabbit IgG (not shown)], a monoclonal antibody against nucleolin (lane 3), or polyclonal antibodies against YB-1 (lane 4). The input (25%, lane 1) and the precipitates were fractionated by SDS-PAGE, transferred to a membrane, and autoradiographed. These membranes were then probed with anti-nucleolin (lane 5) or anti-YB-1 (lane 6) antibodies and visualized by ECL. (B) UV cross-linking of wild-type and mutant IL-2(1–58) RNAs incubated with Jurkat extracts. (C) Binding of recombinant nucleolin or YB-1 to wild-type and mutant IL-2(1–58) RNAs. Each probe was incubated with GST–nucleolin(285–709) (100 ng) or GST–YB-1 (25 ng) and analyzed by UV cross-linking. (D) Specific binding of GST–nucleolin(285–709) or GST–YB-1 to IL-2(1–58) RNA. Wild-type 32P-IL-2(1–58) RNA was incubated with nucleolin or YB-1 in the presence of molar excess of unlabeled wild-type or M2 IL-2(1–58) RNAs and analyzed by UV cross-linking. (E) In vivo association of IL-2 mRNA with nucleolin and YB-1. The proteins were immunoprecipitated from either unstimulated (NS) or stimulated (T/A) Jurkat cells. Portions of the precipitates were analyzed by immunoblotting (IB) with either anti-YB-1 or anti-nucleolin (top). RNA was extracted from the immune complexes (ppt) and analyzed by RT–PCR and Southern blotting. RNA was also extracted from supernatants (sup) and similarly analyzed. (F) In vivo association of nucleolin and YB-1 with wild-type or IL-2 mRNA mutants. Jurkat cells were transfected with vectors expressing either wild-type IL-2 mRNA or a mutant in which the IL-2 5′ UTR was removed (Δ5′-UTR) or CAT reporters, containing wild-type or mutant 5′ UTRs, described in Fig. 1D. After 48 hr, cells were stimulated, lysed, and immunoprecipitated with anti-nucleolin or anti-YB-1. The presence of reporter-derived transcripts in the immune complexes was analyzed by RT–PCR. (Top) Expression of each transcript in transfected cells; (middle, bottom) precipitated RNAs.
To determine whether nucleolin and YB-1 interact with IL-2 mRNA in vivo, we immunoprecipitated either protein from lysates of either unstimulated or stimulated Jurkat cells and examined the immune compex for presence of IL-2 mRNA by RT–PCR. Although the amounts of either nucleolin or YB-1 were unchanged upon T-cell stimulation, both proteins coprecipitated IL-2 mRNA only from stimulated cell extracts (Fig. 3E). Coprecipitation of IL-2 mRNA was specific, as no GAPDH or GM–CSF mRNAs could be detected in the immune complexes. We next investigated whether the 5′ JRE was required for in vivo interaction of nucleolin and YB-1 with IL-2 mRNA. Jurkat cells were transiently transfected with either a wild-type IL-2 reporter gene, a mutant lacking the 5′ JRE (Δ5′ UTR), or CAT reporters containing the M1–M4 versions of the 5′ UTR. The coprecipitation of exogenous transcripts with nucleolin or YB-1 was analyzed by RT–PCR. Both anti-nucleolin and anti-YB-1-precipitated wild-type IL-2, as well as CAT-IL-2 3′ UTR transcripts generated by the wild-type, M3, and M4 reporters, but no precipitation of transcripts derived from the JRE-deleted reporter or the M1 and M2 CAT-IL-2 3′ UTR reporters could be detected (Fig. 3F). These data strongly suggest that nucleolin and YB-1 specifically associate with IL-2 mRNA in vivo and this interaction requires the same 5′ UTR sequences needed for JNK-mediated IL-2 mRNA stabilization. On the basis of a RNase protection experiment, we estimate that >50% of IL-2 mRNA is associated with nucleolin and YB-1 (data not shown).
Establishment of a regulated in vitro mRNA decay system
The mechanisms underlying regulated mRNA turnover in mammalian cells are largely unknown. By-and-large this has been due to the lack of a reliable in vitro assay for studying regulated mRNA stability. An optimal in vitro system for studying regulation of mRNA stability should reproduce general aspects of mRNA turnover observed in vivo. For instance, labile mRNAs should be relatively unstable but stabilizable in response to the appropriate signals or factors. In addition, rapid decay should be ARE dependent. To establish such a system for studying the control of IL-2 mRNA stability, we adopted a recently described assay system in which rapid mRNA degradation was shown to be ARE dependent (Ford et al. 1999). Soluble cytoplasmic extracts (S100) were prepared from unstimulated or stimulated Jurkat cells. Capped and polyadenylated 32P-labeled full-length IL-2 RNA was synthesized by in vitro transcription and incubated with S100 extracts in the presence of an ATP-generating system, as rapid RNA degradation is ATP dependent (Ford et al. 1999). At various time points, IL-2 RNA was isolated and its abundance determined by gel electrophoresis and autoradiography. In unstimulated extracts, IL-2 RNA was degraded with t1/2 = 12 min, but the same RNA was considerably more stable (t1/2 = 45 min) in stimulated extracts (Fig. 4A). The fold increase in IL-2 mRNA t1/2 seen in vitro is similar to the extent of IL-2 mRNA stabilization detected in vivo (Chen et al. 1998). In contrast, polyadenylated and similarly produced GAPDH RNA was equally stable in both extracts (Fig. 4A,B). A deleted IL-2 mRNA lacking AREs from its 3′ UTR was quite stable in either extract with t1/2 similar to that of wild-type IL-2 mRNA in stimulated cell extracts (Fig. 4B).
Figure 4.
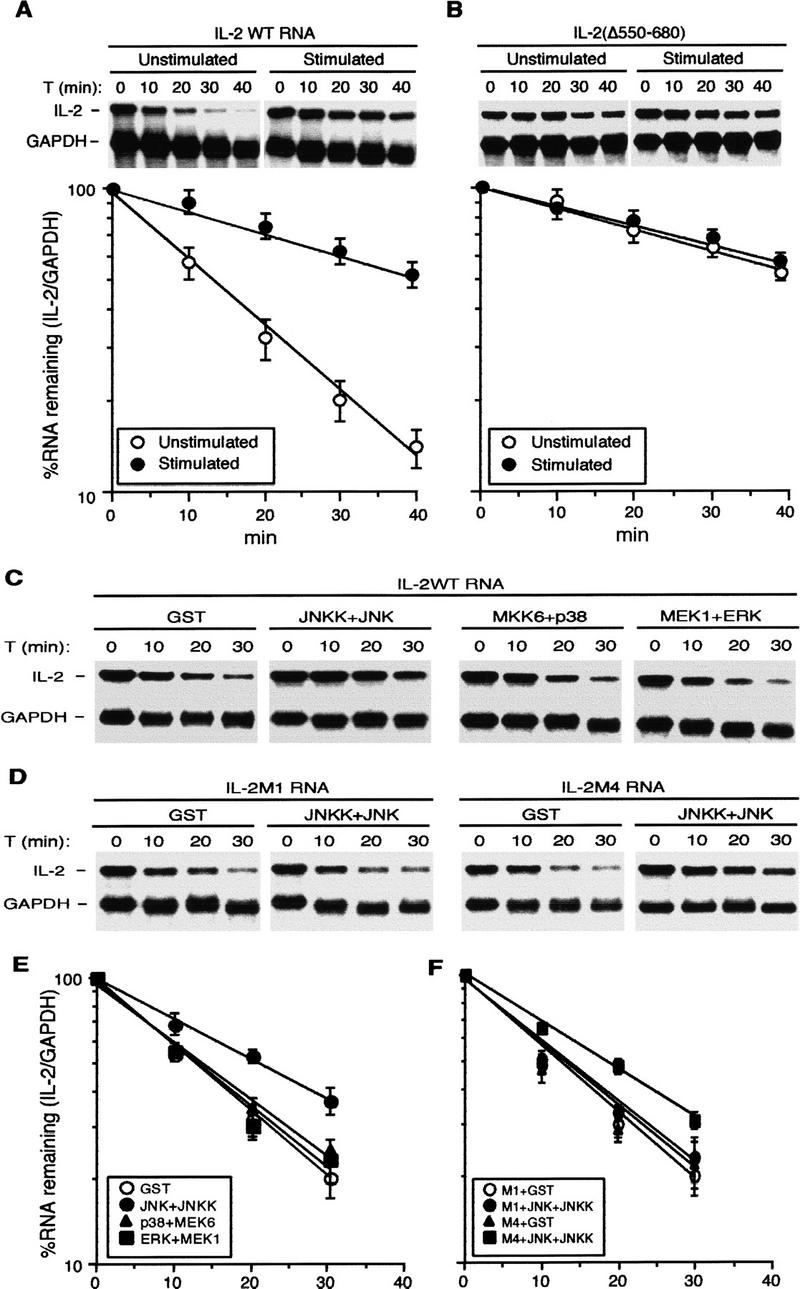
In vitro regulation of IL-2 mRNA stability. (A) Stability of IL-2 mRNA in nonstimulated and stimulated extracts. 32P-labeled, capped, and polyadenylated IL-2 and GAPDH RNAs were incubated with S100s from unstimulated or stimulated Jurkat cells at 37°C for various times. RNAs were isolated, resolved on a denaturing gel, and visualized by autoradiography. The amount of IL-2 RNAs was quantitated by phosphorimaging, normalized to the amount of GAPDH RNA, and plotted semilogarithmicly using a linear regression program. The values shown are averages of three independent experiments. (B) Stability of IL-2 RNA lacking nucleotides 550–680, including several AREs, in unstimulated or stimulated S100s was determined and quantitated as described as above. (○) Unstimulated; (●) stimulated. (C,D) In vitro IL-2 mRNA stabilization by JNK. S100 of unstimulated Jurkat cells was preincubated with recombinant activated JNK2, p38α, or ERK2, at 30°C for 20 min in the presence of ATP. RNA decay assays were performed as described above using wild-type IL-2 RNA (C) or IL-2 RNA containing linker scanning mutations (M1 or M4) within the 5′ UTR (D). (E,F) Quantitation of IL-2 RNA decay from three independent experiments identical to those shown in C and D, respectively. (E) (○) GST; (●) JNK + JNKK; (▴) p38 + MEK6; (█) ERK + MEK1. (F) (○) M1 + GST; (●) M1 + JNK + JNKK; (▴) M4 + GST; (█) M4 + JNK + JNKK.
We used this in vitro system to investigate the role of the JNK pathway in stabilization of IL-2 mRNA. We prepared recombinant MAPKs, including JNK2, p38α, and ERK2, and constitutively active MAPK kinases (MAPKKs), including JNKK2(DD), MKK6(EE), and MEK1(EE) (Mansour et al. 1994; Jiang et al. 1996; Wu et al. 1997). None of the MAPKs was active unless incubated with its respective MAPKK (data not shown). Unstimulated S100 was preincubated with each of the activated MAPKs, respectively, in the presence of the ATP and RNA decay was examined. IL-2 mRNA was considerably stabilized in the JNK2-pretreated S100, but not in the p38α or ERK2-pretreated S100s (Fig. 4C,E). Mutations that prevented binding of YB-1 and nucleolin to the 5′ JRE (Fig. 3), abolished JNK-induced in vitro mRNA stabilization (Fig. 4D,F). Whereas IL-2M1 RNA was no longer stabilized by preincubating the S100 with activated JNK2, IL-2M4 RNA, which still binds nucleolin and YB-1, was stabilized as efficiently as wild-type IL-2 RNA. Thus JNK-mediated IL-2 mRNA stabilization can be reproduced in vitro. As in vivo (Chen et al. 1998), the in vitro response to JNK depends on the 5′ JRE and accounts for ∼50% of the total increase in IL-2 mRNA stability (twofold vs. fourfold).
Nucleolin and YB-1 are required for JNK-induced IL-2 mRNA stabilization
The in vitro RNA decay system was used to determine whether YB-1 or nucleolin are required for JNK-induced IL-2 mRNA stabilization. YB-1 or nucleolin were immunodepleted from unstimulated S100 with antibodies to either protein immobilized on protein A-agarose beads (Fig. 5A). Although in crude cytoplasmic extracts, nucleolin and YB-1 are tightly coassociated with messenger ribonucleoprotein (mRNP) particles (data not shown), in the S100 fraction, the two proteins can be independently immunoprecipitated. The immunodepleted supernatants were preincubated with activated JNK2 and used to perform RNA decay assays. Whereas immunodepletion with an antibody against La protein, an RNA-binding protein associated with RNA polymerase III transcripts (Pruijn et al. 1995), did not impair JNK-induced IL-2 mRNA stabilization, removal of either YB-1 or nucleolin prevented JNK-induced stabilization (Fig. 5B,C), suggesting that they are necessary for JNK-mediated IL-2 mRNA stabilization. Addition of purified nucleolin or YB-1 to the depleted S100s restored JNK-induced IL-2 mRNA stabilization, but no significant effect was observed upon addition of nucleolin or YB-1 to the depeleted S100s in the absence of JNK (Fig. 5B,D).
Figure 5.
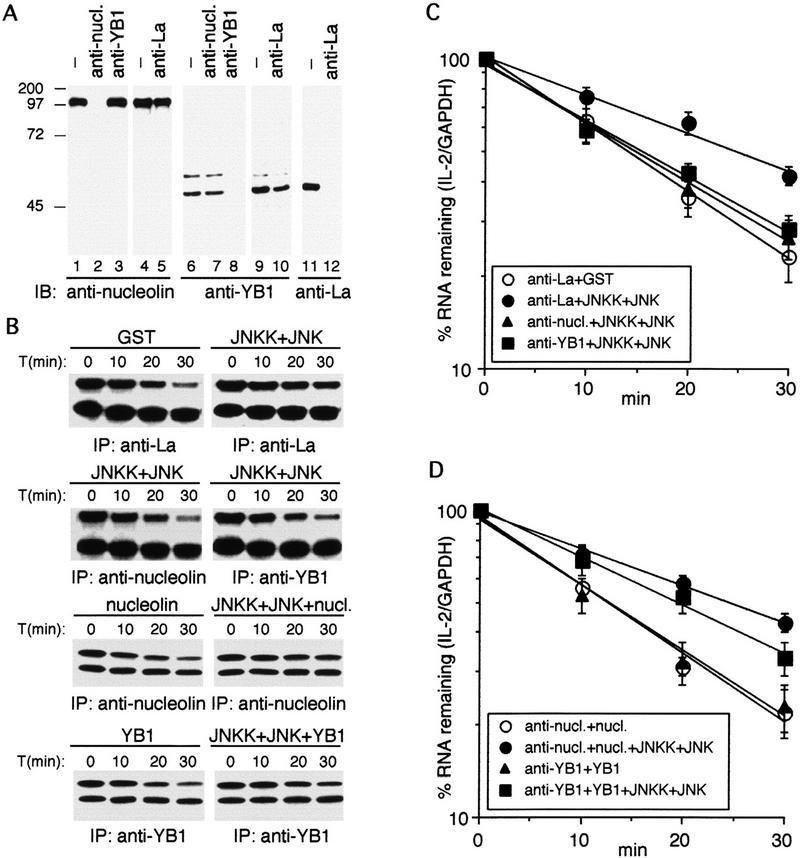
Nucleolin and YB-1 are required for JNK-induced IL-2 RNA stabilization in vitro. (A) Immunoblot analysis with anti-nucleolin (lanes 1–5), anti-YB-1 (lanes 6–10), or anti-La (lanes 11,12) of Jurkat S100 fraction that was untreated (lanes 1,4,6,9,11) or passed (three times) through protein A-agarose beads containing anti-nucleolin (lanes 2,7), anti-YB-1 (lanes 3,8), or anti-La (lanes 5,10,12). (B) The immunodepleted supernatants were preincubated with activated JNK2 (JNKK+JNK), purified nucleolin or YB-1, or activated JNK2 plus nucleolin or YB-1 (as indicated) and used to examine the decay of wild-type IL-2 RNA. (C,D) Quantitation of IL-2 RNA decay in three independent experiments identical to those shown in B. (C) (○) anti-La + GST; (●) anti-La + JNKK + JNK; (▴) anti-nucl. + JNKK + JNK; (▪) anti-YB1 + JNKK + JNK. (D) (○) anti-nucl. + nucl.; (●) anti-nucl. + nucl. + JNKK + JNK; (▴) anti-YB1 + YB1; (█) anti-YB1 + YB1 + JNKK + JNK.
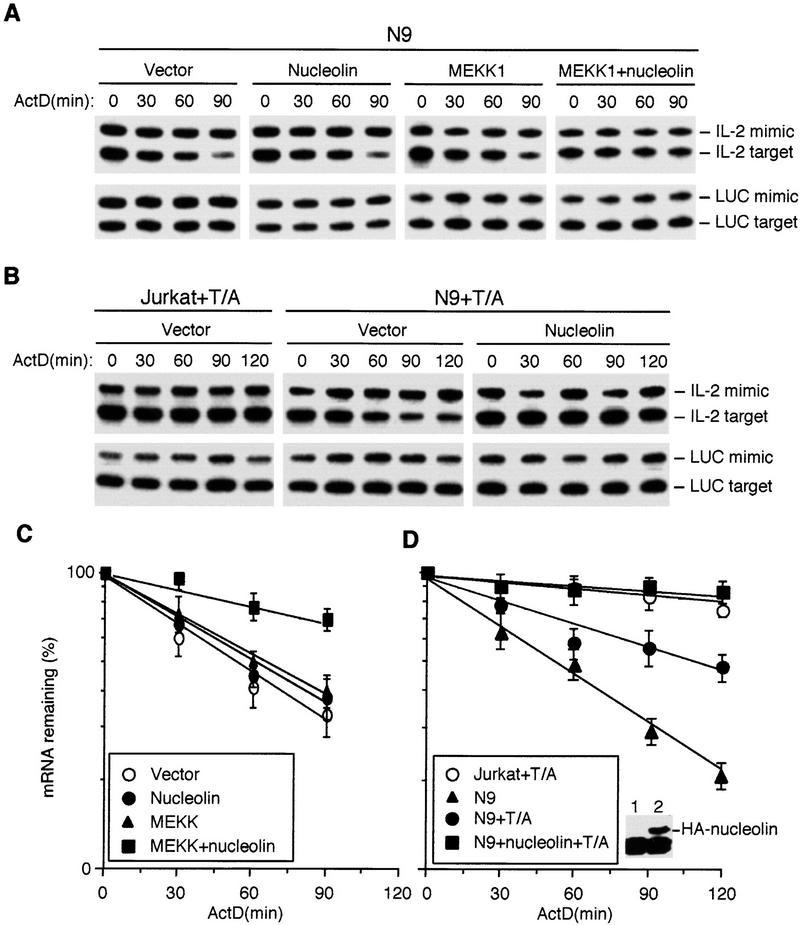
We next examined whether nucleolin or YB-1 are involved in stabilization of IL-2 mRNA in vivo. As YB-1 and nucleolin are relatively abundant and ubiquitous proteins, we chose to examine the effect of reduced expression rather than overexpression. Coexpression of antisense RNAs to either YB-1 or nucleolin partially inhibited MEKK1-induced IL-2 mRNA stabilization (Fig. 6A,D). Neither antisense RNA affected MEKK1 expression but did cause partial reduction in expression of nucleolin or YB-1 (data not shown). We also attempted to establish Jurkat cell clones stably expressing antisense RNAs for YB-1 or nucleolin. After selecting and screening many clones, we obtained several transfectants that expressed considerably lower levels of nucleolin, but could not identify clones expressing <50% of the parental levels of YB-1. One of the stable cell lines, N9, expressed ∼10% of the parental amount of nucleolin (Fig. 6B). Whereas in control Jurkat cells (stably transfected with an empty vector), stimulation with TPA + A23187 resulted in IL-2 mRNA stabilization, the stability of endogenous IL-2 mRNA was dramatically reduced in N9 cells (from t1/2 = 360 min to t1/2 = 45 min). Decreased nucleolin expression had no effect on the stability of actin mRNA (Fig. 6C).
Figure 6.
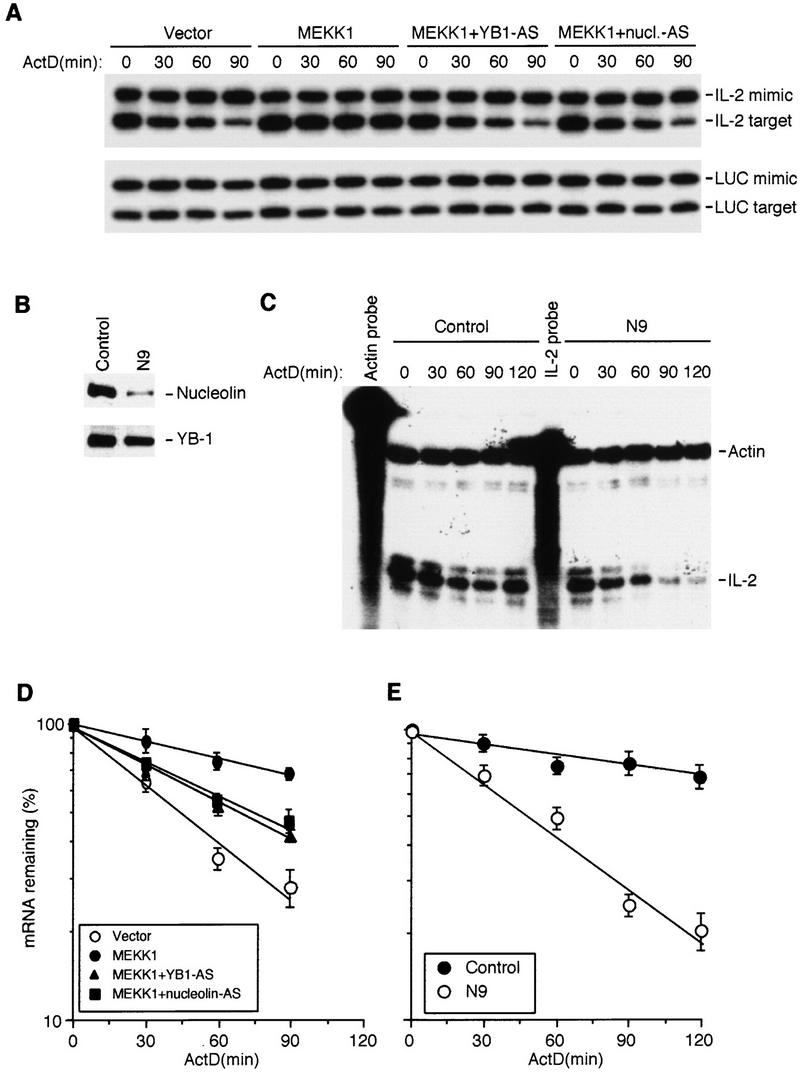
Nucleolin and YB-1 are involved in IL-2 mRNA stabilization in vivo. (A) Jurkat cells were transfected with an IL-2 reporter, a control plasmid expressing luciferase (LUC) mRNA, and either MEKK1 or an empty expression vector along with constructs expressing YB-1 or nucleolin anti-sense RNAs. After 36 hr, actinomycin D (ActD) was added, total RNA was isolated at the indicated times, and the amount of IL-2 mRNA was analyzed by competitive RT–PCR. IL-2 mimic is a PCR product derived from a DNA template added to each reaction to control for amplification efficiency. IL-2 target is derived from transfected IL-2 mRNA. LUC mimic and target were also amplified. Each mimic and target were amplified with the same primer sets. (B) The amounts of nucleolin and YB-1 in Jurkat cells stably transfected with an empty vector (control) or antisense nucleolin RNA (N9). (C) Stability of endogenous IL-2 mRNA in control or N9 cells after stimulation with TPA + A23187. Total RNA was isolated at the indicated times after addition of ActD and the levels of IL-2 and β-actin mRNAs was determined by RNase protection. (D,E) Quantitation of exogenous and endogenous IL-2 mRNA decay in experiments as those shown in A and C, respectively. The quantitated IL-2 signals were normalized to the LUC and β-actin signals and plotted as described above. Each point is the average of three independent experiments. (D) (○) Vector; (●) MEKK1; (▴) MEKK1 + YB1 − AS; (█) MEKK1 + nucleolin-AS. (E) (●) Control; (○) N9.
To demonstrate that destabilization of IL-2 mRNA in N9 cells was due to reduced nucleolin expression, we transiently transfected the IL-2 reporter and either an empty vector or a nucleolin expression vector into N9 cells and determined the stability of the transfected IL-2 mRNA in the absence or presence of MEKK1. Unlike parental Jurkat cells, no stabilization of transfected IL-2 mRNA by coexpressed MEKK1 was detected in N9 cells, but coexpression of nucleolin restored the response to MEKK1 (Fig. 7A,C). On its own, the nucleolin expression vector had no considerable effect on IL-2 mRNA stability. As shown for endogenous IL-2 mRNA, transfected IL-2 transcripts were also less stable in N9 cells stimulated with TPA + A23187 relative to the parental cells (Fig. 7B). Expression of exogenous nucleolin in N9 cells completely restored the response to TPA + A23187 (Fig. 7B,D). These results indicate that nucleolin is required for stabilization of IL-2 mRNA by either deliberate JNK activation or T-cell mitogens.
Figure 7.
Restoration of IL-2 mRNA stabilization in N9 cells by exogenous nucleolin. (A) N9 cells were cotransfected with the IL-2 reporter and expression vectors as indicated. The stability of IL-2 mRNA in the transfected cells was determined as described in Fig. 6A. (B) IL-2 mRNA stabilization in response to stimulation (T/A) of parental or N9 Jurkat cells transfected with either a nucleolin or an empty expression vector. (C,D) Quantitation of IL-2 mRNA decay in A and B. Each point is the average of two transfection experiments. Inset in D shows expression of HA-tagged nucleolin in transfected N9 cells. (C) (○) Vector; (●) nucleolin; (▴) MEKK; (█) MEKK + nucleolin. (D) (○) Jurkat + T/A; (▴) N9; (●) N9 + T/A; (█) N9 + nucleolin + T/A.
Discussion
Activation of T cells results in IL-2 mRNA stabilization. The JNK MAPK cascade plays an important role in this response (accounting for ∼50% of the total increase in t1/2) and its effects are transmitted via a specific cis element, the JRE, in the 5′ UTR of IL-2 mRNA (Chen et al. 1998). We have delineated the JRE to nucleotides 1–22 of IL-2 mRNA and purified two RNA-binding proteins that recognize this element. On the basis of peptide sequences and immunoreactivity, these proteins were identified as nucleolin and YB-1. The results described above indicate that both proteins are bound to endogenous IL-2 mRNA in intact Jurkat cells and this association, although not affected by T-cell mitogens or JNK activation, depends on integrity of the 5′ JRE. Through the use of an in vitro system for regulated RNA decay and expression of antisense RNA constructs in intact cells, we found that both nucleolin and YB-1 are required for JNK-mediated stabilization of IL-2 mRNA during T-cell activation. Thus, nucleolin and YB-1 are essential protein factors in a specific pathway of stimulus-dependent mRNA stabilization.
Nucleolin and YB-1 are multifunctional proteins thought to be involved in many cellular processes. Nucleolin was suggested to be involved in ribosome biogenesis and processing of ribosomal RNA (rRNA) by binding to a specific site within the rRNA precursor (Ginisty et al. 1998). Nucleolin was also implicated in transcriptional repression (Yang et al. 1994) and as a component of the B-cell-specific transcription factor and switch region binding protein, LR1 (Hanakahi et al. 1997). Nucleolin was also shown to specifically interact with the 3′ UTR of amyloid protein precursor (APP) mRNA, the region that regulates the stability of that mRNA (Zaidi and Malter 1995). Although there is no obvious sequence homology between the nucleolin-binding site within the rRNA precursor (Ghisolfi-Nieto et al. 1996) and the 5′ JRE of IL-2 mRNA, the 5′ half of the nucleolin-binding site in the 3′ UTR of APP mRNA, 5′-CUCUCUUUA-3′ (Zaidi et al. 1994), is identical to nucleotides 5–13 of IL-2 mRNA, which are required for nucleolin binding and JRE function. However, whether nucleolin modulates the stability of APP mRNA or other mRNAs was not examined previously.
YB-1 was first identified as a DNA-binding protein that interacts with the Y-box sequence in MHC class II promoters (Didier et al. 1988). However, YB-1, which belongs to a family of evolutionarily conserved proteins (Wolffe et al. 1992), is most likely an RNA-binding protein and not a double-stranded DNA-binding protein. A Xenopus Y-box protein, FRGY2, is a major mRNA storage protein in oocytes, sequestering mRNA from ribosomes (Ranjan et al. 1993). p50, a rabbit YB-1, is a major core protein of mRNP particles and the most abundant protein of inactive and active globin mRNPs (Evdokimova et al. 1995). Depending on the p50 to mRNA ratio, p50 may act either as a translational repressor or an activator (Evdokimova and Ovchinnikov 1999). An involvement of Y-box proteins in control of mRNA stability was heretofore not demonstrated.
Whereas some previous work suggested that nucleolin and YB-1 tend to interact nonspecifically with RNA, such conclusions were based on filter-binding assays, which reduce the specificity of most RNA-binding proteins (X.D. Fu, pers. comm.). The amino- and carboxy-terminal regions of the first two RBDs of nucleolin were shown to be required for specific RNA binding and specific nucleolin-binding sites were identified (Serin et al. 1997). Similarly, the conserved CSD of FRGY2 is required for sequence-specific interaction with RNA, which is facilitated by amino- and carboxy-terminal regions flanking the CSD (Bouvet et al. 1995). In all cases, where sequence-specific binding of nucleolin and YB-1 were demonstrated, either gel-shift or UV cross-linking assays, which include incubation with heparin sulfate and nonspecific RNA, and/or RNase treatment, were used. Such conditions reduce nonspecific binding and facilitate detection of sequence-specific RNA–protein interactions. Our results demonstrate that both proteins interact with the JRE in the 5′ UTR of IL-2 mRNA with considerable specificity in vitro and in vivo. Most importantly, using a series of linker-scanning mutants through the 5′ JRE, we found that binding of nucleolin and YB-1 to the 5′ UTR of IL-2 mRNA correlates perfectly with JNK responsiveness. Two nonresponsive mutants (M1 and M2) no longer bind nucleolin and YB-1 in vitro or in vivo, whereas two other mutants that retain JNK responsiveness (M3 and M4) bind both proteins in vitro and in vivo. The latter association was demonstrated via an RNA immunoprecipitation procedure that may detect only those mRNA species that bind with rather high affinity to the precipitated protein. Several other mRNA species, including both the stable GAPDH and the ARE-containing GM–CSF mRNAs did not coprecipitate with either nucleolin or YB-1. We also found that nucleolin and YB-1 could be coprecipitated with each other from crude cytoplasmic extracts of stimulated Jurkat cells, but this interaction was abolished by RNase treatment (data not shown). As nucleolin and YB-1 can bind simultaneously to the IL-2 5′ UTR (data not shown), such interaction may generate additional specificity and higher affinity than that provided by the individual proteins.
Using a cell-free system for regulated RNA decay, we reproduced several important aspects of regulated mRNA turnover in mammalian cells. As observed in a similar in vitro system derived from HeLa cells (Ford et al. 1999), exogenous IL-2 and GAPDH RNA substrates were deadenylated prior to degradation of the RNA body (data not shown) and rapid IL-2 RNA degradation was dependent on AREs in its 3′ UTR. Most importantly, IL-2 RNA was more stable (fourfold stabilization) in extracts of mitogen-stimulated Jurkat cells. This increase in stability is similar to what has been observed in vivo after Jurkat cell activation (Chen et al. 1998). Furthermore, as found previously in vivo, stabilization of IL-2 RNA was induced by incubating unstimulated S100 extracts with activated recombinant JNK, but not upon incubation with activated p38 or ERK. Both in vivo and in vitro, JNK activation increased mRNA stability by a similar factor (two- to threefold) and both responses required a functional JRE. Although much remains to be learned about the mechanism of regulated mRNA turnover in mammalian cells, the present results reveal that JNK-mediated phosphorylation is unlikely to directly affect the machinery responsible for ARE-directed mRNA decay. Most likely, through the 5′ JRE, JNK modulates the activity of factors that affect the recruitment of this ribonuclease to JRE- and ARE-containing IL-2 mRNA. Immunodepletion experiments confirm that nucleolin and YB-1 not only bind to the 5′ JRE, but are specifically required for JNK-dependent stabilization of JRE-containing IL-2 RNA in vitro. Dependence of JNK-induced IL-2 mRNA stabilization on nucleolin and YB-1 was also detected in intact cells. Transient expression of antisense YB-1 or nucleolin RNAs partially inhibited IL-2 mRNA stabilization and a more extensive inhibition was observed in a cell line that stably expresses nucleolin antisense RNA and as a result contains 10% of the normal amount of nucleolin. Expression of exogenous nucleolin restored IL-2 mRNA stabilization in these cells. However, as no complete elimination of YB-1 and nucleolin expression was achieved in these experiments, the observed inhibition was not as extensive as the one caused by the immunodepletion experiments.
Although essential components, several results suggest that neither nucleolin nor YB-1 is a direct target for JNK in the IL-2 mRNA stabilization pathway. First, we did not detect JNK-induced IL-2 mRNA stabilization in cell lines that are not of T-cell origin using either in vivo or in vitro assays (C.-Y. Chen, unpubl.). Therefore, this response probably requires a T-cell-specific cofactor, whereas nucleolin and YB-1 are present in all cell types. Furthermore, extracts from cell lines in which IL-2 mRNA is not stabilized by JNK produce the same pattern of JRE-binding activities detected in Jurkat extracts (C.-Y. Chen, unpubl.). Second, neither purified YB-1 nor nucleolin are efficiently phosphorylated by JNK in vitro in comparison with physiological JNK substrates, such as c-Jun (C.-Y. Chen, unpubl.). We therefore suggest that nucleolin and YB-1 serve as docking proteins that recruit a cell-type-specific cofactor whose ability to inhibit ARE-directed IL-2 mRNA decay depends on JNK-mediated phosphorylation. Alternatively, nucleolin and YB-1 act as RNA chaperons or unfolding proteins that allow binding of the JNK-responsive factor to another site, possibly within the 3′ UTR. In support of this model, both nucleolin and YB-1 were reported to exhibit RNA unfolding activity (for review, see Evdokimova and Ovchinnikov 1999; Ginisty et al. 1999). Although necessary, the 5′ JRE is not sufficient for conferring JNK-mediated stabilization upon heterologous mRNAs (Chen et al. 1998). This response also requires an element(s) within the IL-2 3′ UTR. Thus, binding of both proteins to the 5′ JRE of IL-2 mRNA may ensure formation of a proper mRNP particle that responds to JNK-generated signals.
In summary, we identified two RNA-binding proteins that are necessary for JNK-induced IL-2 mRNA stabilization. Although these proteins, nucleolin and YB-1, are likely to be multifunctional RNA-binding proteins, this work provides the first demonstration of their involvement in stimulus-regulated mRNA stability. The development of a stimulus-responsive in vitro system for studying regulated mRNA decay should allow the biochemical identification of additional factors that are involved in the nucleolin- and YB-1-dependent JNK-responsive mRNA stabilization pathway, as well as more general factors involved in ARE-directed mRNA decay.
Materials and methods
Plasmids
cDNAs used to produce RNA probes were subcloned into pBluescript II-KS (Stratagene). The 58-bp double-stranded linkers encompassing nucleotides 1–58 of IL-2 cDNA and corresponding to wild-type, M1, M2, M3, and M4 (Fig. 1D), were subcloned into the vector. cDNA fragments containing nucleotides 1–58, full-length IL-2, or nucleotides 590–1070 of human GAPDH, generated by PCR, were subcloned into different sites within the polylinker of pSP64poly(A) (Promega). In vitro transcription of EcoRI-linearized plasmids with SP6 RNA polymerase produces RNA copies of the inserted sequences that contain a 3′ poly(A) tail of 30 residues. Plasmids expressing full-length IL-2 mRNA with mutations in the 5′ UTR were constructed by standard procedures. Details of all cloning procedures will be provided upon request.
IL-2, luciferase, and CAT reporter genes were described previously (Chen et al. 1998) as was a nucleolin expression vector, pNtag4 (Hanakahi et al. 1997). To express antisense RNAs, pcDNA3-nucleolin-AS and pcDNA3-YB1-AS were constructed by subcloning an EcoRI fragment containing nucleotides 114–2239 of human nucleolin (Srivastava et al. 1989) and a NotI fragment containing nucleotides 21–1059 of human YB-1 (Didier et al. 1988) into pcDNA3 (Invitrogen) in the antisense orientation. GST–YB-1 expression vector was described (Li et al. 1997). GST–nucleolin (285–709) was constructed by subcloning a MscI–EcoRI fragment encoding the carboxy-terminal half of human nucleolin (amino acids 285–709) into pGEX-2T (Pharmacia).
Preparation of RNA substrates
Linearized plasmids were used as templates to produce 32P-labeled RNAs using a MAXIscript in vitro transcription kit (Ambion). Capped and polyadenylated 32P-labeled IL-2 and GAPDH RNAs were synthesized in a reaction containing 0.5 mm ATP and CTP, 0.2 mm GTP and UTP, 1 mm m7G5′ppp5′G (cap analog), and 50 μCi of [α-32P]UTP. Labeled RNAs were purified on CHROMA spin columns (Clontech). Large quantities of RNA were produced using the RiboMAX large-scale RNA production system (Promega).
Preparation of cytoplasmic extracts and RNA-binding assays
Jurkat cells were either unstimulated or stimulated with TPA (15 ng/ml) and A23187 (1 μg/ml) for 1 hr. Cytoplasmic extracts and RNA-binding reactions were conducted as described (Bohjanen et al. 1991) with minor modifications. Cytoplasmic extracts (20 μg protein) and 32P-labeled RNA (0.5 ng = 2 × 105 cpm) were incubated at room temperature for 20 min in a RNA-binding buffer (20 μl) containing 10 mm HEPES (pH 7.6), 3 mm MgCl2, 100 mm KCl, 2 mm DTT, 5% glycerol, 0.5% NP-40, yeast RNA (1 μg), and heparin (1 μg). For competition experiments, unlabeled RNAs were preincubated with lysates for 10 min before addition of RNA probe. Unbound RNA was digested with RNase T1 (200 units per reaction) for 15 min at 37°C. Reaction mixtures were transferred to a 96-well plate and irradiated at 4°C for 10 min with a UV Stratalinker (Stratagene) at a distance of 5 cm. After subsequent digestion with RNase A (200 ng per reaction) for 10 min at 37°C, samples were separated by electrophoresis on reducing 10% SDS–polyacrylamide gels. Gels were dried, and 32P-labeled proteins were visualized by autoradiography.
Protein purification and sequence determination
Cytoplasmic extracts were prepared from 5 × 109 Jurkat cells. Protein samples (250 mg) were loaded on a 5-ml HiTrap Q column (Pharmacia Biotech). After washing with 10-column volumes (CVs) of RNA-binding buffer, a 24-CV linear gradient to 1 m KCl was applied and 3-ml fractions collected. Samples (5 μl) from every other fraction were examined by UV cross-linking. Fractions containing peak p50 or p100 binding activities were separately pooled, diluted to 100 mm KCl, reloaded onto a 1-ml HiTrap Q column, washed as above, and eluted with 3 ml of RNA-binding buffer + 1 m KCl. Eluates containing p50 or p100 were dialyzed against 1 liter of RNA binding buffer + 100 mm KCl. Samples (0.5 ml) were chromatographed on a Superose 6B column (Pharmacia Biotech) and fractions containing peak RNA-binding activity were pooled. Pooled p50 fractions were further purified by RNA affinity chromatography. Pooled p100 fractions were loaded on a 1-ml HiTrap heparin-Sepharose column (Pharmacia Biotech). After washing, a 15-CV linear gradient to 500 mm KCl was applied and 1-ml fractions collected. Fractions containing p100 were identified, pooled, dialyzed, and further purified by RNA affinity chromatography. In vitro synthesized and polyadenylated IL-2(1–58) RNA (500 μg) was immobilized onto 0.5-ml poly(U)–Sepharose beads (Sigma) by incubating at 4°C for 1 hr. After washing, the RNA beads were incubated with samples containing p50 or p100 in RNA-binding buffer containing RNasin, heparin, and yeast RNA at 4°C for 1 hr. The mixtures were packed into a 10-ml column and washed with 20 CVs of RNA-binding buffer containing 5 mg/ml heparin and 20 CVs of RNA-binding buffer. Bound proteins were step eluted (0.5 ml of RNA-binding buffer + 0.5, 1.0, or 2.0 m KCl) and aliquots were tested for RNA binding after 10-fold dilution without KCl. Purified p50 or p100 were dialyzed and concentrated on a Centricon-30 device and analyzed by SDS-PAGE. Coomassie brilliant blue-stained protein bands were excised and subjected to mass spectroscopy.
Protein bands were reduced, S-alkylated, and digested in gel with an excess of trypsin. Peptides were eluted using a microcolumn with 1 μl of 60% methanol, 5% formic acid directly into a nanoelectrospray needle. Spectra were recorded on a quadrupole time-of-flight mass spectrometer (QSTAR, Perkin-Elmer Sciex, Toronto, Canada). Peptide-sequence tags were assembled from fragmentation spectra and searched against a nonredundant protein-sequence database (NRDB) maintained at the European BioInformatics Institute (EBI, Hinxton Park, England) using the PeptideSearch program (Wilm et al. 1996).
Recombinant proteins and antibodies
Purified recombinant p38α and active MKK6(EE) were kindly provided by Dr. L. Tong (Columbia University). GST–JNKK2(DD), GST–JNK2, GST–ERK2, GST–nucleolin(285–709), GST–YB-1, and histidine-tagged MEK1(EE) were produced in Escherichia coli and purified using GSH–Sepharose 4B (Pharmacia Biotech) or Ni-NTA (Qiagen) resin. Purity and concentration of the purified proteins were determined by a Bradford assay and Coomassie blue staining after gel electrophoresis. Monoclonal antibody against nucleolin, MS3 (Valdez et al. 1995) was described previously. Polyclonal antibodies against YB-1 (Jürchott et al., in prep.) will be described.
RNA immunoprecipitation assay
Jurkat cells were stimulated with TPA + A23187 for 4 hr and lysed in a buffer containing 50 mm Tris-HCl (pH 8.0), 150 mm NaCl, 100 mm NaF, 1 m Na3VO4, 1 mm EGTA, 1 mm EDTA, 1% Triton-X 100, 2 mm PMSF, 10 mm vanadyl ribonucleoside complex, and 1 mg/ml aprotinin for 15 min at 4°C. Lysates were spun at 14,000g for 15 min at 4°C and supernatants were incubated overnight with antibodies at 4°C under rotation. Protein A–Sepharose was added for 4 hr at 4°C, the precipitates were washed three times in lysis buffer, and collected. RNA in the precipitates or supernatants was extracted with RNAzol and ethanol precipitated. Equal amounts of RNA were reverse transcribed and subjected to PCR as described (Chen et al. 1998). Sequences of primers used for PCR will be provided upon request.
In vivo and in vitro RNA degradation assays
Analysis of in vivo RNA turnover was described previously (Chen et al. 1998). For in vitro analysis of RNA decay, S100 fractions were prepared from either unstimulated or stimulated Jurkat cells as described by Ford et al. (1999). RNA decay reactions were performed according to Brewer and Ross (1990) and Ford et al. (1999) with several modifications. The 5′-capped and 3′-polyadenylated 32P-labeled IL-2 (10 ng) and GAPDH (5 ng) RNAs were incubated with 20 μg of S100 protein in 25 μl of RNA decay buffer containing 100 mm KCH3COOH, 2 mm Mg(CH3COOH)2, 10 mm Tris-Cl (pH 7.6), 2 mm DTT, 10 mm creatine phosphate, 1 μg of creatine phosphokinase, 1 mm ATP, 0.4 mm GTP, 0.1 mm spermine, 4 units of RNasin, and 500 ng of poly(A) oligonucleotide (Pharmacia Biotech). Reactions were incubated at 37°C for the indicated times and stopped with 100 μl of stop buffer [400 mm NaCl, 25 mm Tris-Cl (pH 7.6), 0.1% SDS]. RNAs were phenol-chloroform extracted, ethanol precipitated, and analyzed on a 5% polyacrylamide gel containing 7 m urea. To examine IL-2 RNA stabilization by recombinant MAPKs, MAPKs (200 ng) were pre-incubated with active MAPKKs (100 ng) in kinase buffer (Wu et al. 1997) for 20 min at 30°C in the presence of 20 μm ATP, after which, S100 (20 μg) was added to the activated MAPKs in RNA decay buffer and further incubated for 20 min. Kinase-activated S100 was used for RNA degradation assays as described above.
Acknowledgments
We thank Drs. X.D. Fu and N. Sonenberg for critical comments and discussion, Dr. H. Busch for anti-nucleolin antibody, Dr. Pruijn for anti-La antibody, Dr. N. Maizels for nucleolin cDNA and HA-nucleolin plasmid, Dr. L. Tong for recombinant p38α and MKK6(EE), Dr. A. Lee for GST–YB-1 plasmid, and Dr. M. Ellisman for access to NIH-supported microscopy facility. C.-Y.C. was supported by fellowships from the California Tobacco-Related Disease Research Program and is a Special Fellow of The Leukemia & Lymphoma Society. R.G. is on leave from Istituto Nazionale per la Ricerca sul Cancro (Genova, Italy) and was partially supported by grants from AIRC and American-Italian Cancer Foundation. This work was supported by NIH grant nos. AI43447 and ES04151 and a grant from Boehringer Ingelheim.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL karinoffice@ucsd.edu; FAX (858) 534-8158.
References
- Bohjanen PR, Petryniak B, June CH, Thompson CB, Lindsten T. An inducible cytoplasmic factor (AU-B) binds selectively to AUUUA multimers in the 3′ untranslated region of lymphokine mRNA. Mol Cell Biol. 1991;11:3288–3295. doi: 10.1128/mcb.11.6.3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvet P, Matsumoto K, Wolffe AP. Sequence-specific RNA recognition by the Xenopus Y-box proteins. An essential role for the cold shock domain. J Biol Chem. 1995;270:28297–28303. doi: 10.1074/jbc.270.47.28297. [DOI] [PubMed] [Google Scholar]
- Brewer G. An A + U-rich element RNA-binding factor regulates c-myc mRNA stability in vitro. Mol Cell Biol. 1991;11:2460–2466. doi: 10.1128/mcb.11.5.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer G, Ross J. Messenger RNA turnover in cell-free extracts. Methods Enzymol. 1990;181:202–209. doi: 10.1016/0076-6879(90)81122-b. [DOI] [PubMed] [Google Scholar]
- Brown CY, Lagnado CA, Goodall GJ. A cytokine mRNA-destabilizing element that is structurally and functionally distinct from A+U-rich elements. Proc Natl Acad Sci. 1996;93:13721–13725. doi: 10.1073/pnas.93.24.13721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd CG, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- Caput D, Beutler B, Hartog K, Thayer R, Brown-Shimer S, Cerami A. Identification of a common nucleotide sequence in the 3′-untranslated region of mRNA molecules specifying inflammatory mediators. Proc Natl Acad Sci. 1986;83:1670–1674. doi: 10.1073/pnas.83.6.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carballo E, Lai WS, Blackshear PJ. Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science. 1998;281:1001–1005. doi: 10.1126/science.281.5379.1001. [DOI] [PubMed] [Google Scholar]
- Chen CY, Shyu AB. AU-rich elements: Characterization and importance in mRNA degradation. Trends Biochem Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- Chen CY, Del Gatto-Konczak F, Wu Z, Karin M. Stabilization of interleukin-2 mRNA by the c-Jun NH2-terminal kinase pathway. Science. 1998;280:1945–1949. doi: 10.1126/science.280.5371.1945. [DOI] [PubMed] [Google Scholar]
- Dibbens JA, Miller DL, Damert A, Risau W, Vadas MA, Goodall GJ. Hypoxic regulation of vascular endothelial growth factor mRNA stability requires the cooperation of multiple RNA elements. Mol Biol Cell. 1999;10:907–919. doi: 10.1091/mbc.10.4.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didier DK, Schiffenbauer J, Woulfe SL, Zacheis M, Schwartz BD. Characterization of the cDNA encoding a protein binding to the major histocompatibility complex class II Y box. Proc Natl Acad Sci. 1988;85:7322–7326. doi: 10.1073/pnas.85.19.7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evdokimova VM, Ovchinnikov LP. Translational regulation by Y-box transcription factor: Involvement of the major mRNA-associated protein, p50. Int J Biochem Cell Biol. 1999;31:139–149. doi: 10.1016/s1357-2725(98)00137-x. [DOI] [PubMed] [Google Scholar]
- Evdokimova VM, Wei CL, Sitikov AS, Simonenko PN, Lazarev OA, Vasilenko KS, Ustinov VA, Hershey JW, Ovchinnikov LP. The major protein of messenger ribonucleoprotein particles in somatic cells is a member of the Y-box binding transcription factor family. J Biol Chem. 1995;270:3186–3192. doi: 10.1074/jbc.270.7.3186. [DOI] [PubMed] [Google Scholar]
- Fan XC, Steitz JA. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J. 1998;17:3448–3460. doi: 10.1093/emboj/17.12.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford LP, Watson J, Keene JD, Wilusz J. ELAV proteins stabilize deadenylated intermediates in a novel in vitro mRNA deadenylation/degradation system. Genes & Dev. 1999;13:188–201. doi: 10.1101/gad.13.2.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghisolfi-Nieto L, Joseph G, Puvion-Dutilleul F, Amalric F, Bouvet P. Nucleolin is a sequence-specific RNA-binding protein: Characterization of targets on pre-ribosomal RNA. J Mol Biol. 1996;260:34–53. doi: 10.1006/jmbi.1996.0380. [DOI] [PubMed] [Google Scholar]
- Ginisty H, Amalric F, Bouvet P. Nucleolin functions in the first step of ribosomal RNA processing. EMBO J. 1998;17:1476–1486. doi: 10.1093/emboj/17.5.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginisty H, Sicard H, Roger B, Bouvet P. Structure and functions of nucleolin. J Cell Sci. 1999;112:761–772. doi: 10.1242/jcs.112.6.761. [DOI] [PubMed] [Google Scholar]
- Hamilton BJ, Nagy E, Malter JS, Arrick BA, Rigby WF. Association of heterogeneous nuclear ribonucleoprotein A1 and C proteins with reiterated AUUUA sequences. J Biol Chem. 1993;268:8881–8887. [PubMed] [Google Scholar]
- Hanakahi LA, Dempsey LA, Li MJ, Maizels N. Nucleolin is one component of the B cell-specific transcription factor and switch region binding protein, LR1. Proc Natl Acad Sci. 1997;94:3605–3610. doi: 10.1073/pnas.94.8.3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill CS, Treisman R. Transcriptional regulation by extracellular signals: Mechanisms and specificity. Cell. 1995;80:199–211. doi: 10.1016/0092-8674(95)90403-4. [DOI] [PubMed] [Google Scholar]
- Hunter T, Karin M. The regulation of transcription by phosphorylation. Cell. 1992;70:375–387. doi: 10.1016/0092-8674(92)90162-6. [DOI] [PubMed] [Google Scholar]
- Iwai Y, Bickel M, Pluznik DH, Cohen RB. Identification of sequences within the murine granulocyte-macrophage colony-stimulating factor mRNA 3′-untranslated region that mediate mRNA stabilization induced by mitogen treatment of EL-4 thymoma cells. J Biol Chem. 1991;266:17959–17965. [PubMed] [Google Scholar]
- Iwai Y, Akahane K, Pluznik DH, Cohen RB. Ca2+ ionophore A23187-dependent stabilization of granulocyte-macrophage colony-stimulating factor messenger RNA in murine thymoma EL-4 cells is mediated through two distinct regions in the 3′-untranslated region. J Immunol. 1993;150:4386–4394. [PubMed] [Google Scholar]
- Jiang Y, Chen C, Li Z, Guo W, Gegner JA, Lin S, Han J. Characterization of the structure and function of a new mitogen-activated protein kinase (p38beta) J Biol Chem. 1996;271:17920–17926. doi: 10.1074/jbc.271.30.17920. [DOI] [PubMed] [Google Scholar]
- Karin M, Hunter T. Transcriptional control by protein phosphorylation: signal transmission from the cell surface to the nucleus. Curr Biol. 1995;5:747–757. doi: 10.1016/s0960-9822(95)00151-5. [DOI] [PubMed] [Google Scholar]
- Kishore R, Tebo JM, Kolosov M, Hamilton TA. Cutting edge: Clustered AU-rich elements are the target of IL-10-mediated mRNA destabilization in mouse macrophages. J Immunol. 1999;162:2457–2461. [PubMed] [Google Scholar]
- Lagnado CA, Brown CY, Goodall GJ. AUUUA is not sufficient to promote poly(A) shortening and degradation of an mRNA: The functional sequence within AU-rich elements may be UUAUUUA(U/A)(U/A) Mol Cell Biol. 1994;14:7984–7995. doi: 10.1128/mcb.14.12.7984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai WS, Carballo E, Strum JR, Kennington EA, Phillips RS, Blackshear PJ. Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor alpha mRNA. Mol Cell Biol. 1999;19:4311–4323. doi: 10.1128/mcb.19.6.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laroia G, Cuesta R, Brewer G, Schneider RJ. Control of mRNA decay by heat shock-ubiquitin-proteasome pathway. Science. 1999;284:499–502. doi: 10.1126/science.284.5413.499. [DOI] [PubMed] [Google Scholar]
- Li WW, Hsiung Y, Wong V, Galvin K, Zhou Y, Shi Y, Lee AS. Suppression of grp78 core promoter element-mediated stress induction by the dbpA and dbpB (YB-1) cold shock domain proteins. Mol Cell Biol. 1997;17:61–68. doi: 10.1128/mcb.17.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsten T, June CH, Ledbetter JA, Stella G, Thompson CB. Regulation of lymphokine messenger RNA stability by a surface-mediated T cell activation pathway. Science. 1989;244:339–343. doi: 10.1126/science.2540528. [DOI] [PubMed] [Google Scholar]
- Loflin P, Chen CY, Shyu AB. Unraveling a cytoplasmic role for hnRNP D in the in vivo mRNA destabilization directed by the AU-rich element. Genes & Dev. 1999;13:1884–1897. doi: 10.1101/gad.13.14.1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour SJ, Matten WT, Hermann AS, Candia JM, Rong S, Fukasawa K, Vande Woude GF, Ahn NG. Transformation of mammalian cells by constitutively active MAP kinase kinase. Science. 1994;265:966–970. doi: 10.1126/science.8052857. [DOI] [PubMed] [Google Scholar]
- Myer VE, Fan XC, Steitz JA. Identification of HuR as a protein implicated in AUUUA-mediated mRNA decay. EMBO J. 1997;16:2130–2139. doi: 10.1093/emboj/16.8.2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy E, Rigby WF. Glyceraldehyde-3-phosphate dehydrogenase selectively binds AU-rich RNA in the NAD(+)-binding region (Rossmann fold) J Biol Chem. 1995;270:2755–2763. doi: 10.1074/jbc.270.6.2755. [DOI] [PubMed] [Google Scholar]
- Nakagawa J, Waldner H, Meyer-Monard S, Hofsteenge J, Jenö P, Moroni C. AUH, a gene encoding an AU-specific RNA binding protein with intrinsic enoyl-CoA hydratase activity. Proc Natl Acad Sci. 1995;92:2051–2055. doi: 10.1073/pnas.92.6.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng SS, Chen CY, Shyu AB. Functional characterization of a non-AUUUA AU-rich element from the c-jun proto-oncogene mRNA: Evidence for a novel class of AU-rich elements. Mol Cell Biol. 1996;16:1490–1499. doi: 10.1128/mcb.16.4.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng SS, Chen CY, Xu N, Shyu AB. RNA stabilization by the AU-rich element binding protein, HuR, an ELAV protein. EMBO J. 1998;17:3461–3470. doi: 10.1093/emboj/17.12.3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruijn GJM, Thijssen JPH, Smith PR, Williams DG, Van Venrooij WJ. Anti-La monoclonal antibodies recognizing epitopes within the RNA-binding domain of the La protein show differential capacities to immunoprecipitate RNA-associated La protein. Eur J Biochem. 1995;232:611–619. doi: 10.1111/j.1432-1033.1995.611zz.x. [DOI] [PubMed] [Google Scholar]
- Ragheb JA, Deen M, Schwartz RH. CD28-Mediated regulation of mRNA stability requires sequences within the coding region of the IL-2 mRNA. J Immunol. 1999;163:120–129. [PubMed] [Google Scholar]
- Ranjan M, Tafuri SR, Wolffe AP. Masking mRNA from translation in somatic cells. Genes & Dev. 1993;7:1725–1736. doi: 10.1101/gad.7.9.1725. [DOI] [PubMed] [Google Scholar]
- Ross J. mRNA stability in mammalian cells. Microbiol Rev. 1995;59:423–450. doi: 10.1128/mr.59.3.423-450.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs AB. Messenger RNA degradation in eukaryotes. Cell. 1993;74:413–421. doi: 10.1016/0092-8674(93)80043-e. [DOI] [PubMed] [Google Scholar]
- Schuler GD, Cole MD. GM-CSF and oncogene mRNA stabilities are independently regulated in trans in a mouse monocytic tumor. Cell. 1988;55:1115–1122. doi: 10.1016/0092-8674(88)90256-5. [DOI] [PubMed] [Google Scholar]
- Serin G, Joseph G, Ghisolfi L, Bauzan M, Erard M, Amalric F, Bouvet P. Two RNA-binding domains determine the RNA-binding specificity of nucleolin. J Biol Chem. 1997;272:13109–13116. doi: 10.1074/jbc.272.20.13109. [DOI] [PubMed] [Google Scholar]
- Shaw G, Kamen R. A conserved AU sequence from the 3′ untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell. 1986;46:659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- Shyu AB, Greenberg ME, Belasco JG. The c-fos transcript is targeted for rapid decay by two distinct mRNA degradation pathways. Genes & Dev. 1989;3:60–72. doi: 10.1101/gad.3.1.60. [DOI] [PubMed] [Google Scholar]
- Shyu AB, Belasco JG, Greenberg ME. Two distinct destabilizing elements in the c-fos message trigger deadenylation as a first step in rapid mRNA decay. Genes & Dev. 1991;5:221–231. doi: 10.1101/gad.5.2.221. [DOI] [PubMed] [Google Scholar]
- Srivastava M, Fleming PJ, Pollard HB, Burns AL. Cloning and sequencing of the human nucleolin cDNA. FEBS Lett. 1989;250:99–105. doi: 10.1016/0014-5793(89)80692-1. [DOI] [PubMed] [Google Scholar]
- Stoecklin G, Hahn S, Moroni C. Functional hierarchy of AUUUA motifs in mediating rapid interleukin-3 mRNA decay. J Biol Chem. 1994;269:28591–28597. [PubMed] [Google Scholar]
- Valdez BC, Henning D, Busch RK, Srivastava M, Busch H. Immunodominant RNA recognition motifs of human nucleolin/C23. Mol Immunol. 1995;32:1207–1213. doi: 10.1016/0161-5890(95)00093-3. [DOI] [PubMed] [Google Scholar]
- Wilm M, Shevchenko A, Houthaeve T, Breit S, Schweigerer L, Fotsis T, Mann M. Femtomole sequencing of proteins from polyacrylamide gels by nano-electrospray mass spectrometry. Nature. 1996;379:466–469. doi: 10.1038/379466a0. [DOI] [PubMed] [Google Scholar]
- Wisdom R, Lee W. The protein-coding region of c-myc mRNA contains a sequence that specifies rapid mRNA turnover and induction by protein synthesis inhibitors. Genes & Dev. 1991;5:232–243. doi: 10.1101/gad.5.2.232. [DOI] [PubMed] [Google Scholar]
- Wolffe AP, Tafuri S, Ranjan M, Familari M. The Y-box factors: A family of nucleic acid binding proteins conserved from Escherichia coli to man. New Biologist. 1992;4:290–298. [PubMed] [Google Scholar]
- Wu Z, Wu J, Jacinto E, Karin M. Molecular cloning and characterization of human JNKK2, a novel Jun NH2-terminal kinase-specific kinase. Mol Cell Biol. 1997;17:7407–7416. doi: 10.1128/mcb.17.12.7407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N, Chen CY, Shyu AB. Modulation of the fate of cytoplasmic mRNA by AU-rich elements: Key sequence features controlling mRNA deadenylation and decay. Mol Cell Biol. 1997;17:4611–4621. doi: 10.1128/mcb.17.8.4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Steussy CN, Fuhrer DK, Hamilton J, Yang YC. Interleukin-11 mRNA stabilization in phorbol ester-stimulated primate bone marrow stromal cells. Mol Cell Biol. 1996;16:3300–3307. doi: 10.1128/mcb.16.7.3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang TH, Tsai WH, Lee YM, Lei HY, Lai MY, Chen DS, Yeh NH, Lee SC. Purification and characterization of nucleolin and its identification as a transcription repressor. Mol Cell Biol. 1994;14:6068–6074. doi: 10.1128/mcb.14.9.6068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi SH, Malter JS. Nucleolin and heterogeneous nuclear ribonucleoprotein C proteins specifically interact with the 3′-untranslated region of amyloid protein precursor mRNA. J Biol Chem. 1995;270:17292–17298. doi: 10.1074/jbc.270.29.17292. [DOI] [PubMed] [Google Scholar]
- Zaidi SH, Denman R, Malter JS. Multiple proteins interact at a unique cis-element in the 3′-untranslated region of amyloid precursor protein mRNA. J Biol Chem. 1994;269:24000–24006. [PubMed] [Google Scholar]
- Zhang W, Wagner BJ, Ehrenman K, Schaefer AW, DeMaria CT, Crater D, DeHaven K, Long L, Brewer G. Purification, characterization, and cDNA cloning of an AU-rich element RNA-binding protein, AUF1. Mol Cell Biol. 1993;13:7652–7665. doi: 10.1128/mcb.13.12.7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubiaga AM, Belasco JG, Greenberg ME. The nonamer UUAUUUAUU is the key AU-rich sequence motif that mediates mRNA degradation. Mol Cell Biol. 1995;15:2219–2230. doi: 10.1128/mcb.15.4.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]