Abstract
In this article we define vaccinomics as the integration of immunogenetics and immunogenomics with systems biology and immune profiling. Vaccinomics is based on the use of cutting edge, high-dimensional (so called “omics”) assays and novel bioinformatics approaches to the development of next-generation vaccines and the expansion of our capabilities in individualized medicine. Vaccinomics will allow us to move beyond the empiric “isolate, inactivate, and inject” approach characterizing past vaccine development efforts, and toward a more detailed molecular and systemic understanding of the carefully choreographed series of biological processes involved in developing viral vaccine-induced “immunity.” This enhanced understanding will then be applied to overcome the obstacles to the creation of effective vaccines to protect against pathogens, particularly hypervariable viruses, with the greatest current impact on public health. Here we provide an overview of how vaccinomics will inform vaccine science, the development of new vaccines and/or clinically relevant biomarkers or surrogates of protection, vaccine response heterogeneity, and our understanding of immunosenescence.
The Developing Science of Vaccinomics
Undoubtedly among the hallmarks of 21st century medicine will be the application of increasingly high dimensional genetic, proteomic, and other assays, combined with novel bioinformatics approaches in the development and clinical utility of individualized or “precision” medicine. Individualized drug therapies are now beginning to be routinely informed by the application of knowledge gleaned from the science base of pharmacogenomics and pharmacogenetics. Similar to this work with drugs, we have published work defining the term “vaccinomics” by which we mean the application of these same scientific fields of study when applied to understanding immunologic mechanisms behind vaccine response heterogeneity (Poland, 2007; Poland et al. 2007). In this article we expand upon this definition by including the integration of immunogenetics and immunogenomics with systems biology and immune profiling approaches to understanding immune responses to vaccines and provide examples of such work in regards to vaccines against viral diseases. Vaccinomics seeks to understand variations in individual's immune responses to vaccines by combining the strengths of immunogenetics and immunogenomics including both single nucleotide polymorphism (SNP) discovery and functional validation with immune profiling and systems biology approaches. Such information is useful in the design of new vaccine candidates, in advancing our understanding of how immune responses and adverse reactions develop, and perhaps as biomarkers in vaccine clinical trials, among others. Although this article focuses on our work with vaccines against viral diseases, the concepts involved are more broadly applicable to prophylactic and therapeutic vaccines against other infectious and noninfectious diseases. We have, in part, previously discussed some of these examples in other publications and reviews of the field of vaccinomics (Haralambieva and Poland, 2010; Ovsyannikova and Poland, 2011; Poland, 2007; Poland et al. 2007, 2009a).
Large studies of naïve individuals receiving live viral vaccines lead to “archetype” phenotypes of immune response such as: “nonresponders,” “normal responders,” and “hyperresponders” as used by current serologic methods of defining humoral antibody response to vaccines. We also recognize that such archetypes can be expanded (consistent with the science) into archetypes based on measures of cell-mediated immunity, and combinations of humoral and cell-mediated immunity such as: nonresponders [low antibody (Ab) and low cell-mediated immunity (CMI)], hyperresponder (high Ab and high CMI), individuals with skewed responses (high Ab but low CMI, high CMI but low Ab), or even “early” or “late” responders with differences in immune pathway activation kinetics. But for the purposes of illustration we will illustrate in this article the concepts we wish to discuss using the three simple archetypes of non-, normal-, and hyperresponders.
Although clinicians recognize that antidepressant, anticancer (azithioprine, for example), antihypertensive, and antimicrobial (isoniazid, for example) drugs result in significant variations in therapeutic effects, and therefore require different dosing regimens and result in different side-effect profiles for each individual, it is only recently that science has recognized that the mechanisms for these variations are genetically based. We propose that similar mechanisms explain heterogeneity of biologic immune responses to, and adverse effects from, vaccines (Poland et al., 2009b).
As we will discuss, we have defined associations between genetic polymorphisms and significant heterogeneity in immune responses to several live, attenuated viral vaccines (Poland, 2007; Poland et al., 2007, 2009a). We believe such phenotype–genotype data, when combined with systems biology approaches and high throughput next-generation sequencing and bioinformatics, will substantially influence and drive both vaccinomics and personalized vaccinology. In turn, these areas will assist in the understanding and immune responses and the prediction of adverse events to vaccines, allowing for informed development of next-generation vaccines. This approach is a logical extension of current medical practice to diagnose an individual patient's situation and tailor a personalized medical intervention suited to their needs. Examples of such approaches already known to clinicians include increased hepatitis B antigen doses to patients with either renal failure or who are immunocompromised resulting in improved seroconversion and protection. Infants receive a different dose and numbers of doses of influenza vaccine compared to older children and adults. From an immunogenetics point of view, an extended HLA haplotype associated with nonresponse to hepatitis B vaccine has been defined (Kruskall et al., 1992). These individuals may benefit from multiple doses or vaccines containing increased amounts of antigen.
Problems with Empiric Vaccine Development
We are calling for a new directed approach that would lead to the informed development of next-generation vaccines. This approach contrasts with an empiric approach that has served us well since the late 18th century with Jenner's work with cowpox in developing a vaccine against smallpox. In the empirical approach, the investigators first isolate the organism that causes the infectious disease or alternatively results in immunity to the disease. The investigators then work to modify candidate agents to both improve their success in generating immunity as well as improve their safety and tolerability as an immunizing agent; the immunity generated prevents either infection from the agent and thus the pathology that would develop (Johnston and Fauci, 2008). Of note, the empirical approach does not require that the investigators understand the details of the immunologic response to the immunizing agent (Reed et al., 2006). Nonetheless, the empirical approach has resulted in the array of vaccines that make up our routine childhood and adult vaccine schedule. Together they have been hailed as one of the 20th Century's most important public health achievements (Centers for Disease Control and Prevention, 1999a, 1999b). Nonetheless, barriers to the empirical approach have left many important infectious diseases (DeMarco and Verjovski-Almeida, 2009; Reed et al., 2006)—including a number of viral infections, such as respiratory syncytial virus (Murata, 2009; Varga, 2009), human immunodeficiency virus (Johnston and Fauci, 2008), and herpes simplex (Mascola, 1999)—without safe and effective vaccines. These barriers include the following:
• Individuals do not appear to acquire long-lasting, effective immunity from an infection (Johnston and Fauci, 2008; Mascola, 1999)
• Individuals require repeated infections to generate immunity (Reed et al. 2006)
• The infectious agent rapidly mutates resulting in either numerous or sequentially different antigenic exposures (Murata, 2009; Rappuoli, 2007)
• Individuals can develop natural immunity from infection, but remain contagious (Johnston and Fauci, 2008)
• Neutralizing antibodies alone do not effectively prevent infection (Rappuoli, 2007)
• Worse, antibodies formed build immune complexes causing harm (Varga, 2009)
• Individuals fail to respond immunologically to the infection, perhaps because of developmental immaturity of the immune system or because of interference from passively acquired maternal immunity (Murata, 2009)
• Nature lacks benign agents that result in mild infection but produce sufficient immunity
• The infectious agent cannot be attenuated without impairing the immunogenic qualities
• The attenuated form, although immunogenic, suffers from cold-chain issues preventing its distribution in parts of the world that cannot maintain a cold chain
• The attenuated form reverts to its more dangerous wild type
• The attenuated form still presents a danger despite attenuation to the immune compromised
Where these barriers exist, we would propose a directed approach, moving away from the empirical approach that begins with the isolation of the immunizing agent followed by its modification. Instead, we propose moving toward the study of the immunogenetic processes and deploying the new tools of genomics and proteomics, and we call that directed approach vaccinomics.
How Vaccinomics Informs New Vaccine Development and Vaccine Science
For an individual to develop a protective immune response to an antigen, a complex series of biologic, molecular, genetic, physiologic, and other processes must be activated (and perhaps in some cases suppressed). Antigen recognition, processing, presentation, and activation of innate, adaptive, and cell-mediated immune responses must occur. Protective immunity requires the activation/suppression of specific genes as well as protein transcription, expression, secretion, and function. The “immune response gene network” theory seeks to provide a framework for the above described interactive and carefully regulated processes that result in protective responses against pathogen threats (Poland et al., 2007). Genes involved in pathogen recognition, binding and cell entry, the processing and/or presentation of antigen to T or B cells, innate and adaptive effector function, immunoregulation, and many others are critical and necessary components to host survival. This theory also recognizes that epigenetic modifications, complementation, and other important events also impact the process of developing immune responses.
Vaccinomics offers advances in vaccine science in at least two primary dimensions: that of scientific deliverables, and of clinical deliverables. Proceeding from this perspective, vaccinomics has much to offer vaccine development. Below we elaborate and give specific examples relating to these concepts.
Vaccinomics and New Scientific Understandings
From the scientific deliverable aspect, the increasing availability of high-dimensional sequencing and arrays such as transcriptomics, in combination with increasingly sophisticated bioinformatic approaches, will allow ever more detailed genotype:phenotype information and informed understanding of the roles of gene pathways, epigenetics, and complementation. Understanding how, for example, genetic heterogeneity (such as SNPs/haplotypes) in a specific dominant gene involved in producing the receptor for a virus is effected by different polymorphic forms of this gene, and in turn affects the receptor's ability to bind and recognize the viral antigen, could profoundly inform and allow directed development of a vaccine antigen/antigens that could be universally bound by the known spectrum of polymorphic receptors, or include directed and targeted adjuvants that can overcome genetic restrictions.
Vaccinomics serves to identify these mechanisms and pathways by which innate and adaptive immune responses occur. At first, attention will be, and has been, given to discovering dominant polymorphic effects whereby single gene polymorphisms demonstrate significant effects on immune response. Although these types of analyses are useful, it is more likely, as illustrated by our work over the last 20 years, that the immune response is the summative totality of a series (or pathways) of polymorphisms and gene effects that individually contribute small effects, but that in toto might explain and predict a much larger proportion of the overall immune response(s) (Pankratz et al., 2010; Poland et al., 2007). We are currently undertaking the important tasks of replicating our immunogenetic findings in independent cohorts and pursuing mechanistic studies aimed at elucidating the downstream consequences of these genetic associations. These current and future studies are a necessary stepping stone in the development of a solid scientific foundation upon which to develop new vaccines and medical interventions.
The concept of vaccinomics is constantly evolving in advancing our understanding of how the immune system works and will likely soon encompass new challenging areas in vaccine research (not addressed currently) including pathogen evolution and immune escape, coinfection, prior exposure to infection, and therapeutic vaccination. Similarly, pathogen (not just host) variability can be used in developing new vaccines. For example, genomic analysis of pandemic influenza A/H1N1 isolates reveals associations between specific hemagglutinin mutations and subsequent human disease severity (Glinsky, 2010). This information, in combination with information regarding the genetic basis for variations in immune response to these identified mutations, could inform new candidate vaccine design.
The Case of Immune Response Heterogeneity to Vaccines
Studies demonstrate that vaccination induces heterogeneous innate and adaptive (humoral and cellular) immune responses, including vaccine failure, in individuals treated identically. Some vaccines, such as measles, mumps, rubella, hepatitis B, and others, fail to induce life-long protective levels of antibody in approximately 5 to 10% of healthy recipients, resulting in the inability to protect against infectious diseases and the accumulation of vaccine failures resulting in eventual outbreaks (Anders et al. 1996; Centers for Disease Control and Prvention, 2010; Poland and Jacobson, 1994). For this reason, among others, outbreaks continue to take place worldwide, as well as in highly vaccinated populations (Chen et al., 1989; Poland and Jacobson, 1994). For example, in the mumps outbreak that began in the summer of 2009 in an upstate New York summer camp and spread to Brooklyn and four counties in New York and New Jersey, the majority of cases were indeed vaccinated; in fact, among 1,521 cases, only 9.1% lacked a previous mumps vaccination (Centers for Disease Control and Prevention, 2010). The factors associated with vaccine failure and extensive heterogeneity of immune responses to vaccines in these specific outbreaks are unclear; however, we have conducted a twins study to demonstrate, when controlling for environmental factors, that almost 89, 39, and 46% of the variation in measles, mumps, and rubella humoral immunity (as measured by IgG antibody levels), respectively, following vaccination can be attributed to genetic factors rather than chance (Tan et al., 2001). This study, along with a number of other twins and siblings studies that examine the heritability of vaccine response and reactivity profiles, strongly supports the rationale for a genomics approach to interindividual variations in vaccine immune response (Jacobson et al., 2007; Klein et al., 2007; Newport et al., 2004; Tan et al., 2001).
The concept of immune response “archetypes,” defined earlier as nonresponder, hyperresponder, and responder, assist in studies of the mechanisms for such archetypes. Multiple host genetic factors contribute to infectious disease susceptibility and variations in adaptive immune responses to vaccination. Among immune response genes, human leukocyte antigen (HLA) genes are critical genes in host immunity and vaccine immune responses. Several findings in regard to measles, mumps, rubella, influenza, and vaccinia vaccine antigens and HLA genetics were previously reported, demonstrating that HLA genes are important determinants of variability in vaccine response (Ovsyannikova et al., 2004a, 2004b, 2005a, 2006a, 2007a; Poland et al., 2001, 2008a). Our research indicates that specific HLA class I and class II gene polymorphisms, including haplotypes and supertypes, are associated with variations in MMR vaccine-induced antibody levels, demonstrating that individual genetic variation or immunogenetic profiling is important in explaining variations in protective antibody levels following vaccination (Ovsyannikova et al., 2004a, 2004b, 2005a, 2006b, 2007a, 2009). These immunogenetic studies demonstrate that humoral responses to vaccines are influenced by polymorphisms of the HLA genes. By identifying naturally processed epitopes, combined with knowledge of HLA supertypes, one could create adjuvanted vaccines with the combinations of those peptides most likely to be optimally immunogenic (Ovsyannikova et al., 2007b). As an example, using a mass spectrometry approach, 13 naturally processed and presented measles virus peptides were identified from the class II HLA-DRB1 peptide binding groove of human cells (Johnson et al., 2005; Ovsyannikova et al., 2003). From these peptides, promiscuous peptides capable of binding across common population HLA supertypes could be utilized in the construct of new candidate vaccines.
HLA heterozygosity has been associated with variations in vaccine-generated immunity to a variety of pathogens; however, at both individual and population levels, HLA homozygosity may adversely affect immune responses to vaccines. For example, HLA homozygosity is associated with lower measles virus-specific IgG levels after a single dose of MMR vaccine (Sauver et al., 2002); however, this HLA homozygosity disadvantage can, in some cases, be overcome by additional doses of measles vaccine in many, but not all, individuals (Ovsyannikova et al., 2007c; St. Sauver et al., 2005). In the case of rubella vaccine, however, following two doses of rubella-containing vaccine, homozygosity within the HLA-DPB1 locus was associated with increased levels of rubella virus-specific IgG, an effect driven by a common DPB1*0401 allele (Kennedy et al., 2010).
We have also demonstrated that HLA genes significantly influence CMI responses following MMR vaccination, but do not explain all of the variance in immune responses seen within the population (Ovsyannikova et al., 2005b, 2005c, 2006a, 2007d). For example, we have estimated that the HLA genes explain approximately 9% of the variance observed in rubella antibody levels. Because the heritability for rubella was 46%, this indicates that variation in HLA loci accounted for approximately 20% of the overall genetic variation in humoral immunity to rubella (Ovsyannikova et al., 2009). Thus, the immune response to vaccines is controlled by a multiple genetic loci, which is consistent with our immune response network theory.
For example, recent studies demonstrate that other candidate immune response gene polymorphisms may play a role in vaccine-induced immunity (Jin and Wang, 2003). Our studies have found significant associations between polymorphisms in cytokine, cytokine receptors, and Toll-like receptor (TLRs) genes and measures of humoral and CMI responses to MMR vaccine (Dhiman et al., 2008, 2010). These data further confirmed the influence of immunogenetics on vaccine response variability. Specifically, cytokine and cytokine receptor polymorphisms may be important contributing factors to vaccine responsiveness, and data suggest a possible involvement of genetic variation in the interleukin (IL)2, IL10, and IL12B genes in measles-specific immunity (Dhiman et al., 2007). Significant associations were also demonstrated between SNPs in the IL10RA, IL12RB1, and IL12RB2 cytokine receptor genes and variations in antibody and mumps-specific lymphoproliferative responses to mumps vaccine (Ovsyannikova et al., 2008). In particular, allele DQB1*0303 was associated with lower mumps-specific antibody titers, and the occurrence of a minor allele T for an intronic SNP (rs2201584) in the IL12RB2 gene was associated with an allele dose-related decrease in mumps antibodies (Ovsyannikova et al., 2008). Such data could lead to the development of a novel mumps vaccine consisting of a “peptide cocktail” with cytokine adjuvants that could overcome these immunogenetic limitations (Poland, 2007; Poland et al., 2007).
Our data also provide support for associations of polymorphisms in the TLR3 gene with both measles and rubella vaccine immune responses. Specifically, the heterozygous genotype for SNP rs5743305 in the TLR3 gene was significantly associated with: rubella-induced granulocyte macrophage-colony stimulating factor (GM-CSF) secretion, lower measles-specific antibody titer, and lower lymphoproliferation to measles vaccine. Further, the major alleles of coding SNPs in the TLR2 (rs3804100) and TLR4 (rs5030710) genes were both associated with variations in measles-vaccine induced humoral immunity in allele dose-dependent manner (Ovsyannikova et al., 2011). As an example of this application, specific adjuvants could be used to differentially activate TLRs (including TLR4/MyD88) to boost B cell and T cell immunity) (Mata-Haro et al., 2007). Similarly, increased occurrence of a minor allele T for nonsynonymous SNP rs3796504 in the SLAM gene resulted in a substantial decrease in measles-specific antibody levels. In addition, several specific polymorphisms in CD46 (rs11118580 and rs2724384) gene demonstrated allele dose-dependent reductions in measles antibody levels (Dhiman et al., 2007). Importantly, an association of the CD46 SNP rs2724384 with measles vaccine-specific antibodies was validated in a separate replication study (unpublished data). Understanding how genetic variation affects viral antigen recognition by the SLAM (and CD46) receptor could inform and allow directed development of a novel vaccine antigen universally bound by SLAM regardless of whether specific polymorphisms are present in the exons of gene(s) that encode for this receptor.
We also identified novel genetic associations between variability in host genes and variations in immune outcomes following rubella vaccination. Innate immunity also takes part in the response to viral pathogens, playing an important role in the initiation and modulation of adaptive immunity. For this reason, SNP associations were identified between the vitamin A (RARA, RARB, and RARG), RIG-I/DDX58, and TRIM genes and rubella vaccine-specific immunity (Ovsyannikova et al., 2010a, 2010b). Notably, the coding TRIM5 SNPs rs3740996 (previously found to affect TRIM5 antiviral activity) (Goldschmidt et al., 2006; Javanbakht et al., 2006; Sawyer et al., 2006; van Manen et al., 2008), rs3740996, and rs10838525 were associated with variations in rubella-specific humoral immunity, tumor necrosis factor (TNF)-α secretion, and IL-2/GM-CSF production, respectively. Further, we studied polymorphisms in inteferon (IFN) response genes and identified a number of associations between SNPs in the 2′-5′-oligoadenylate synthetase (OAS) gene and rubella virus-specific IL-2, IL-10, IL-6 production, and antibody levels (Haralambieva et al., 2010). Three OAS1 SNPs (rs3741981/Ser162Gly, rs1051042/Thr361Arg, rs2660) were associated with higher IL-2 secretion.
Finally, we recently demonstrated the value of examining both HLA genes and other genes in the class III region, including lymphotoxin alpha (LTA), tumor necrosis factor (TNF) and leukocyte-specific transcript-1 (LST1) genes, as part of the extended haplotypes useful in better understanding genomic drivers regulating vaccine immune response outcomes. In this regard, we examined the association between rubella vaccine-induced immune responses and extended class I-class II-class-III haplotypes (62). A number of associations were discovered between haplotypes extending across the HLA class I region, SNP haplotypes, and the HLA class II region (i.e., A-C-B-LTA-TNF-LST1-DRB1-DQA1-DQB1-DPA1-DPB1) and rubella vaccine-induced antibodies. Associations were also found between both extended A*02-C*03-B*15-AAAACGGGGC-DRB1*04-DQA1*03-DQB1*03-DPA1*01-DPB1*04 and HLA-only A*02-C*03-B*15-DRB1*04-DQA1*03-DQB1*03-DPA1*01-DPB1*04 haplotypes and higher rubella antibody levels. The class II HLA-only haplotype DRB1*13-DQA1*01-DQB1*06-DPA1*01-DPB1*04 lacking LTA-TNF-LST1 SNPs was associated with lower rubella antibody responses (Ovsyannikova et al., 2010c). Such data illustrate the need for more population-based genetic vaccine studies to elucidate the influence of population genetics on response to vaccines.
As discussed, numerous investigations have now established that genetic factors are involved in vaccine-induced immunity, including measles, mumps, and rubella. We have demonstrated that adaptive immune responses to viral vaccines are significantly associated with HLA alleles and SNPs in multiple classes of immune response genes, but these associations do not account for all immune responses variation at the population level. Whether or not and how these immune responses may be influenced by other human genes are important questions and areas for further investigation.
The Case of Immunosenescence
Both infants and the elderly are more susceptible to infections and have ineffective immune responses, and therefore have specific immunization requirements. As discussed earlier in the article, the proposed concept of immune response “archetypes” includes the low responder “archetype,” which in turn may integrate common genetic and physiological conditions associated with immune dysfunction such as immune immaturity and immunosenescence. With the perspective of rapidly growing elderly populations throughout the developed world, there is a particular public and scientific interest in preventive strategies that improve protection and vaccination against infectious diseases in older subjects.
Immunosenescence is the decline in the body's ability to mount an adequate immune response to either infection or vaccination (Doria and Frasca, 2000; Smith et al., 2006; Targonski et al., 2007b). Critical for vaccinology is the comprehensive understanding of the functional capacity of the aged immune system, which affects both innate and adaptive immunity (Allman and Miller, 2005a, 2005b; Gomez et al., 2005, 2008; Weng, 2006). The elderly exhibit abnormal cytokine secretion and a chronic inflammatory state that has been called “inflamm-aging,” as well as diminished T cell, B cell, and antigen presenting cell function, leading to dysfunctional immunity and impaired memory formation (Allman and Miller, 2005a, 2005b; Aw et al., 2007; Cancro et al., 2009; Chen et al., 2009; Franceschi et al., 2000; Katz et al., 2004; Riley et al., 2005).
In an attempt to define better correlates of protection in the elderly and potentially new predictive biomarkers for vaccine efficacy, we and others have demonstrated that CD28 expression on cytotoxic T cells, telomerase activity, and T cell receptor rearrangement excision circles (TREC) may be reasonable phenotypic markers of immunosenescence and the first two markers have been associated with the capacity of the elderly to develop vaccine-induced protection against influenza (Effros et al., 2005; Geenen et al., 2003; Goronzy et al., 2001; Targonski et al., 2007a, 2007b; Weng et al., 1996; Xie and McElhaney, 2007). Some of these markers, such as the TREC levels, were found to be modulated using IL-7 and thus enhance response to influenza vaccination (Aspinall et al., 2007). The importance of cytotoxic T lymphocytes (CTL) as a key defense mechanism against influenza and other viral infections has long been recognized (Bangham, 2009; Brien et al., 2009; Brown and Kelso, 2009; Ishii and Koziel, 2008; O'Connell et al., 2009), mediated mainly through Granzyme B (Johnson et al., 2003; Lawrence et al., 2005). Previous vaccine studies provide evidence that correlates of protection may differ between age groups. For example, T cell responses have been proposed by some investigators as better markers of influenza vaccine-induced protection than hemagglutination inhibition assay (HAI) antibodies in the elderly, and granzyme B was demonstrated to correlate with protection and increased CTL response in this age group (McElhaney et al., 1988, 1996, 1998, 2001, 2006, 2009).
As mentioned, it is well established that immunosenescence substantially contributes to the decreased efficacy of vaccines in elderly persons, and that vaccine efficacy tends to be age-dependent as well as infectious agent/vaccine-dependent, although the underlying mechanisms are not completely understood (Weinberger et al., 2008). For example, vaccine efficacy estimates against influenza is only 29–46% in persons ≥75 years, compared with 41–58% in persons 60–74 years of age (Weinberger et al., 2008). Vaccine efficacy against shingles is 64% in 60–70 year olds, but only 18% in persons ≥80 years old, whereas vaccine efficacy of live attenuated yellow fever vaccine in elderly persons is believed to be 100% (Weinberger et al., 2008). Building on our immunogenetics/immunogenomics expertise, our new vaccinomics concept combines a systems biology approach, immune profiling, and functional studies with immunogenetics/immunogenomics to better comprehensively elucidate the causes/metrics of immune function, immune dysfunction, or alteration leading to decreased immune responses to preventive vaccination. This could potentially provide new insights into age-related vaccine failures and impact the development of new strategies that confer improved prevention and infection control in the elderly.
It is well documented that aging exerts significant effects on key components of the innate immune system expression of key pattern recognition receptors and other important host defense molecules (Gomez et al., 2008; Katz et al., 2004; van Duin and ShAw, 2007). Our immunogenetic studies provide compelling evidence for the contribution of a variety of innate immunity genes (including TLRs and related signaling molecules MyD88 and MD2, RIG-I/DDX58, TRIM5, VISA, IRF9, OAS, MX1, etc.) to immune response variations after measles and rubella vaccines (Dhiman et al., 2010; Haralambieva et al., 2010; Ovsyannikova et al., 2010a, 2010b). Knowledge of such polymorphisms and/or other causative polymorphisms in the innate signaling pathways, in combination with multiple, layered, interactive systems studies—functional studies, high dimensional transcriptomics studies, immune profiling studies, and novel bioinformatic approaches—might allow us to better understand the mechanisms by which such genetic variants influence innate and adaptive immune responses. In turn, such knowledge could revolutionize our understanding of immune activation and provide the scientific framework for the directed and rational design of new generation vaccines and vaccination strategies targeting the general population or specific populations with known innate immunity defects (the elderly). Examples include the implementation of new molecular adjuvants that utilize TLRs or cytoplasmic pattern recognition receptors (NOD proteins, RIG-I, MDA-5) and/or modulate critical innate pathways to increase the ability of the immune system to circumvent age-related immune restrictions (such as TLR agonists and ligands: CpG-containing oligodeoxynucleotides, poly(I:C), imidazoquinoline compounds: R-848, monophosphoryl lipid A, bacterial flagellins, etc.) (Kornbluth and Stone, 2006; Pichichero, 2008; Rosenthal, 2006). Other strategies that may enhance vaccine immunogenicity/efficacy in the elderly include: increased doses of antigen, new adjuvants and delivery systems, novel mucosal vaccines, “early prime-later boost” immunizations, T cell rejuvenation therapies such as stem cell interventions, androgen blockade, IL-7 treatment, or manipulation of other key cytokines and/or costimulatory molecules (Brien et al., 2009; Rosenthal, 2006; Siegrist and Aspinall, 2009).
Regardless, it is clear that a more substantial and focused research approach, such as our vaccinomics approach, is required to provide deeper insights into the underlying mechanisms of the diminished and inadequate vaccine-related protection against illness and address the elevated risk for adverse infectious outcomes that are frequently observed in the elderly, and/or to point out effective ways for immune function restoration.
Vaccinomics and New Vaccine Products
From the clinical deliverable (product) point of view, our work, as briefly reviewed above, has illustrated significant associations between genetic polymorphisms and immune response heterogeneity to live viral vaccines (Dhiman et al., 2007, 2008, 2010; Poland et all, 2001; Ovsyannikova et al., 2004a, 2005a, 2008; Sauver et al., 2002). Population-based immunogenetic studies inform and elucidate the mechanisms of protective immunity and provide a foundation for future vaccine development.
Our laboratory has identified naturally processed and presented viral epitopes from a number of HLA molecules (Johnson et al., 2009; Ovsyannikova et al., 2007b). The current smallpox vaccine containing live vaccinia virus causes life-threatening adverse events and has significant contraindications in up to 30% of the population and safe, effective smallpox vaccines are urgently needed. Several national governments, including the United States, the United Kingdom, and Canada are actively procuring smallpox vaccine stockpiles as biodefense measures, and have expressed the desire for nonreplicating and/or subunit vaccines. We have identified, using mass spectrometry methods, 116 novel vaccinia-derived peptides eluted from the HLA peptide binding groove of some common HLA class I molecules (HLA-A*0201, B*1501, and C*03) (Johnson et al., 2009). Preliminary testing has shown that vaccine recipients have immune responses specifically directed against many of these peptides, and we are currently evaluating the immunogenicity and protective efficacy of these peptides in a mouse model. If the results are promising, advanced animal studies might then be undertaken to satisfy the FDA's animal rule as a prerequisite for eventual GMP production and clinical trials.
Thus, our ability to identify critical poxvirus immune epitopes and demonstrate efficacy in appropriate animal models are significant first steps in developing a peptide-based smallpox vaccine—identified through our vaccinomics approach. A cocktail of identified and tested peptides could then be constructed around known HLA allele prevalence in the general population, or even within specific subpopulations such as those with specifically defined genetic restrictions preventing expression of certain antigenic epitopes.
The Case of Vaccine Clinical Testing Biomarkers
The goal of vaccine development is to produce products that prevent illness and disease in the vaccine recipients. The earliest measure of vaccine efficacy was disease prevention as illustrated by Jenner's vaccination of James Phipps and subsequent exposure to smallpox (Henderson et al., 2008) Pasteur's use of weakened anthrax cultures in chickens and sheep (Geison, 1995; Pasteur et al., 2002; Scorpio et al., 2006) or the first use of the rabies vaccine on Joseph Meister (Plotkin et al., 2008). Early immunologists identified antibodies as the protective component of humoral immunity and measurements of serum antibody titers were quickly adopted as correlates of protection. Further investigation led to a greater understanding of the role of antibodies in neutralization, opsonization, hemagglutination, and complement fixation. For many pathogens these functional activities are now thought to better correlate with disease protection (Amanna et al., 2008; Feng et al., 2009; Mascola and Montefiori, 2010; McElhaney, 2008).
Similarly, the prominent role of cellular immunity in resolving infections is now being examined. The control of latent viral infections [including cytomegalovirus (CMV), varicella zoster virus (VZV), Epstein-Barr virus (EBV), and Herpes Simplex virus (HSV)] is associated with cell-mediated immune responses, indicating that measures of cellular immunity should make ideal correlates of protection for these diseases. Advances in critical elements of vaccinomics (pathogen biology, epitope identification, and host response) have furthered our understanding of key targets of protective immunity, critical gene and pathways activated during immune responses, and appropriate endpoints and measures of clinical efficacy, thereby opening the door to improving current vaccines and creating next-generation vaccines against difficult pathogens such as tuberculosis, francisella, plague, malaria, subunit-based vaccines and, just as importantly, improved measures of protection and vaccine efficacy (Amanna and Slifka, 2009; de Boer et al., 2010; Vaine et al., 2010).
As these examples demonstrate, as our understanding of host–pathogen interactions and immune response pathways grow, so too does our ability to harness this knowledge and develop better correlates of protection, surrogate endpoints reached soon after treatment, and enhanced biomarkers for vaccine development. These might include: Elispot or flow cytometric analysis of specific T or B cell subsets, quantification of T or B cell reactivity to key epitopes, or expression analysis of critical innate pathways (TLR engagement, IFN response, and others). Vaccinomics provides this enhanced understanding and can also be used in developing “go, no go” decisions in early stages of vaccine development. Through the use of deep immune profiling we will soon understand what genes or gene products must be activated/suppressed/transcribed in order for effective immune responses to occur. Patterns of gene activation (transcriptomics), T cell receptor immunophenotypes, epigenetic modifications, and protein expression profiles can be used to understand the molecular and genetic basis for different vaccine response archetypes. Critical pathways can then be defined which, if present (“go”), lead to protective immune responses and, if absent (“no-go”), lead to nonprotective immune responses. Thus, a vaccine candidate could be inexpensively and quickly studied to determine whether predetermined genetic/molecular archetype markers are elicited and used to determine the potential for efficacy of the vaccine candidate, and hence whether further investment in development is warranted.
With a population, there is always a wide range of response to disease. It may be possible to identify additional genetic markers of disease clearance or susceptibility. An excellent example is the CCR5 Δ32 deletion that results in almost complete protection in HIV+ patients (Liu et al., 1996). This knowledge may, in turn, lead to the development of next-generation therapeutics (Telenti, 2009). Another important point to realize is that we cannot fully capture the benefits and risks of a given drug or vaccine with just a single endpoint. For example, levels of serum cytokines (IL-10, CSF-3) after receipt of the smallpox vaccine may serve as biomarkers of excess inflammation, a side effect that measuring only neutralizing antibody titers or T cell IFN-γ ELISPOT responses would be unable to identify (Reif et al., 2009). The simultaneous use of multiple biomarkers therefore allows a more complete and accurate assessment of various facets of vaccine intervention.
We have previously articulated what we see as the “next steps” in personalized vaccinology (Poland et al., 2008b) These include:
• Properly powered gene association studies to detect clinically meaningful associations (Burgner et al., 2006; Cordell and Clayton, 2005; Gordon and Finch, 2005; Hattersley and McCarthy, 2005; Hirschhorn and Daly, 2005; Majunder and Ghosh, 2005; Motsinger et al., 2007)
• Studies providing maximal information content (i.e., pilot studies of candidate genes are less promising than genome wide association studies)
• Follow-up validation studies to confirm true associations (Chanock et al., 2007), an increased emphasis and funding of such studies
• As well as databases and biobanks allowing open access to study results, biological specimens, and protocols, thereby facilitating additional genotype:phenotype association studies expeditiously and inexpensively
• Reliable, reproducible, rapid, and inexpensive genetic tests
• Innovative bioinformatics analysis tools that rely on a sophisticated understanding of genetics, immunology, and immunogenetics (Poland, 2007; Poland et al., 2007, 2009a)
Vaccinomics and How It Can Overcome the Barriers to Empiric Vaccine Development
Given the barriers to empiric vaccine development listed earlier, we point out three of these as further examples of how vaccinomics might approach their solutions. The first occurs when natural disease does not provide durable and effective/protective immunity. Here investigators might identify individuals who appear to have a natural resistance to the disease during outbreaks or when exposed. Immunogenetic investigations and pathway/gene set analyses might uncover genetic differences in immune response leading to potential strategies for novel vaccine candidates.
A second barrier to empiric vaccine development occurs when humoral immunity alone cannot control the infection (e.g., requires T cell immunity). Here immunogenetic studies of response to infection, including immunogenomic, transcriptomic, and proteomic studies, might identify pathways toward cellular immunity. Epitope identification algorithms may allow us to target immune responses to critical proteins, while understanding the type of cellular response, the interplay of cytokines and the role of genetic restriction may uncover the optimal type of cellular immune response necessary for protection. Knowing the critical pathways to cellular immunity can help us overcome our lack of a useful antibody-biomarker for vaccine development. Another barrier occurs when passively transmitted maternal immunity interferes with vaccine response. Actually, this is the basis for our own investigations with the measles vaccine. Here we start with identifying the genetic and genomic associations with immune response to the current vaccine given after the age when maternal immunity no longer interferes. We identify which measles virus peptides appear to bind with which key HLA genes or HLA supertypes while pursuing which cytokines and their receptors are up- and downregulated. We envision from this would come a cocktail of peptides and cytokines that would form a vaccine that could be administered at birth to override the maternal antibody that neutralizes the live viral vaccine when administered before 12 months of life, long after many infants are exposed to measles.
Conclusion
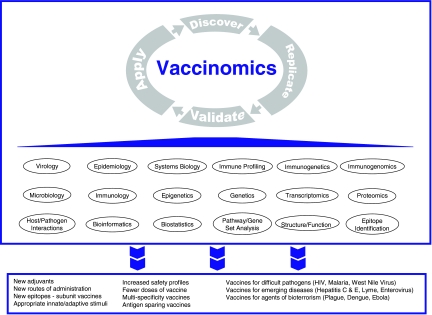
Vaccinomics represents a new paradigm in vaccine development. This paradigm moves the field away from the predominant “isolate, inactivate/attenuate, inject” empiric pathway of the past and toward one of directed vaccine development that we have characterized as “Discover–Replicate–Validate/Characterize–Apply.” In turn, this new paradigm is informed by a deep systems biology approach toward the genetic and immunologic mechanisms and drivers of antigen-activated innate and adaptive immune responses (Fig. 1). In this article we have illustrated the concept of vaccinomics as the integration of immunogenetics/immunogenomics with a system biology and immune profiling approach, and provided examples of how discovering new scientific insights can lead to novel vaccine candidates.
FIG. 1.
Vaccinomics approach to new vaccine development. Vaccinomics is the application of multidimensional science to the understanding of biologic and immunologic vaccine response and variability in immune responses. Detailed studies of the mechanisms underlying host responses to pathogens coupled with high dimensional analysis of the epigenetic, genetic, transcriptomic, and proteomic, events that culminate in protective immunity, is used to design rational vaccine approaches. These insights lead to the discovery of new knowledge, which, after replication and validation, is then applied to the development of novel vaccine candidates, and in turn, provides new insights into biological processes that further extend scientific knowledge and discovery.
It has been stated, “Just as pharmacogenetics has suggested ways of designing drugs to minimize population variability, understanding mechanisms of immunogenetic variation may lead to new vaccines designed specifically to minimize immunogenetically based vaccine failure”(Spielberg, 1998). We believe the 21st century will ultimately prove vaccinomics to be an innovative and transformative driver of new vaccine design and utilization. Such work will allow for the development of additional vaccine candidates capable of eliminating/minimizing viral vaccine nonresponse, overcoming immunosenescence, and overcoming genetic restriction.
Acknowledgments
This work was supported by NIH Grants AI 48793, AI 33144, AI 89859, HHSN272201000025C, and 5UL1RR024150-03 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health, and the NIH Roadmap for Medical Research. Its contents are solely the responsibility of the authors and do not necessarily represent the official view of NCRR or NIH.
Author Disclosure Statement
Dr. Poland is the chair of a safety evaluation committee for novel vaccines undergoing clinical studies by Merck Research Laboratories. Dr. Poland offers consultative advice on new vaccine development to Merck & Co., Inc., Avianax, Theraclone Sciences (formally Spaltudaq Corporation), MedImmune LLC, Liquidia Technologies, Inc., Emergent BioSolutions, Novavax, Dynavax, EMD Serono, Inc., Novartis Vaccines, and Therapeutics and PAXVAX, Inc. Dr. Jacobson serves on a Safety Review Committee for two postlicensure studies of Gardasil [Human Papillomavirus Quadrivalent (Types 6, 11, 16, 18) Vaccine, Recombinant] for Kaiser-Permanente.
References
- Allman D. Miller J.P. B cell development and receptor diversity during aging. Curr Opin Immunol. 2005a;17:463–467. doi: 10.1016/j.coi.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Allman D. Miller J.P. The aging of early B-cell precursors. Immunol Rev. 2005b;205:18–29. doi: 10.1111/j.0105-2896.2005.00269.x. [DOI] [PubMed] [Google Scholar]
- Amanna I.J. Slifka M.K. Wanted, dead or alive: new viral vaccines. Antiviral Res. 2009;84:119–130. doi: 10.1016/j.antiviral.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amanna I.J. Messaoudi I. Slifka M.K. Protective immunity following vaccination: how is it defined? Hum Vaccin. 2008;4:316–319. doi: 10.4161/hv.4.4.5751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders J.F. Jacobson R.M. Poland G.A. Jacobsen S.J. Wollan P.C. Secondary failure rates of measles vaccines: a metaanalysis of published studies. Pediatr Infect Dis J. 1996;15:62–66. doi: 10.1097/00006454-199601000-00014. [DOI] [PubMed] [Google Scholar]
- Aspinall R. Pido-Lopez J. Imami N. Henson S.M. Ngom P.T. Morre M., et al. Old rhesus macaques treated with interleukin-7 show increased TREC levels and respond well to influenza vaccination. Rejuvenat Res. 2007;10:5–17. doi: 10.1089/rej.2006.9098. [DOI] [PubMed] [Google Scholar]
- Aw D. Silva A.B. Palmer D.B. Immunosenescence: emerging challenges for an ageing population. Immunology. 2007;120:435–446. doi: 10.1111/j.1365-2567.2007.02555.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bangham C.R. CTL quality and the control of human retroviral infections. Eur J Immunol. 2009;39:1700–1712. doi: 10.1002/eji.200939451. [DOI] [PubMed] [Google Scholar]
- Brien J.D. Uhrlaub J.L. Hirsch A. Wiley C.A. Nikolich-Zugich J. Key role of T cell defects in age-related vulnerability to West Nile virus. J Exp Med. 2009;206:2735–2745. doi: 10.1084/jem.20090222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown L.E. Kelso A. Prospects for an influenza vaccine that induces cross-protective cytotoxic T lymphocytes. Immunol Cell Biol. 2009;87:300–308. doi: 10.1038/icb.2009.16. [DOI] [PubMed] [Google Scholar]
- Burgner D. Jamieson S.E. Blackwell J.M. Genetic susceptibility to infectious diseases: big is beautiful, but will bigger be even better? Lancet Infect Dis. 2006;6:653–663. doi: 10.1016/S1473-3099(06)70601-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancro M.P. Hao Y. Scholz J.L. Riley R.L. Frasca D. Dunn-Walters D.K., et al. B cells and aging: molecules and mechanisms. Trends Immunol. 2009;30:313–318. doi: 10.1016/j.it.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention. Achievements in public health, 1900–1999 impact of vaccines universally recommended for children—United States, 1990–1999. MMWR. 1999a;48:243–248. [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention. Ten great public health achievments—United States, 1900–1999. MMWR. 1999b;48:241–264. [Google Scholar]
- Centers for Disease Control and Prevention. Update: mumps outbreak—New York and New Jersey, June 2009–January 2010. MMWR. 2010;59:125–129. [PubMed] [Google Scholar]
- Chanock S.J. Manolio T. Boehnke M. Boerwinkle E. Hunter D.J. Thomas G., et al. Replicating genotype–phenotype associations. Nature. 2007;447:655–660. doi: 10.1038/447655a. [DOI] [PubMed] [Google Scholar]
- Chen R.T. Goldbaum G.M. Wassilak S.G.F. Markowitz L.E. Orenstein W.A. An explosive point-source measles outbreak in a highly vaccinated population. Modes of transmission and risk factors for disease. Am J Epidemiol. 1989;129:173–182. doi: 10.1093/oxfordjournals.aje.a115106. [DOI] [PubMed] [Google Scholar]
- Chen W.H. Kozlovsky B.F. Effros R.B. Grubeck-Loebenstein B. Edelman R. Sztein M.B. Vaccination in the elderly: an immunological perspective. Trends Immunol. 2009;30:351–359. doi: 10.1016/j.it.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordell H.J. Clayton D.G. Genetic association studies. Lancet. 2005;366:1121–1131. doi: 10.1016/S0140-6736(05)67424-7. [DOI] [PubMed] [Google Scholar]
- de Boer S.M. Kortekaas J. Antonis A.F. Kant J. van Oploo J.L. Rottier P.J., et al. Rift Valley fever virus subunit vaccines confer complete protection against a lethal virus challenge. Vaccine. 2010;28:2330–2339. doi: 10.1016/j.vaccine.2009.12.062. [DOI] [PubMed] [Google Scholar]
- DeMarco R. Verjovski-Almeida S. Schistosomes—proteomics studies for potential novel vaccines and drug targets. Drug Discov Today. 2009;14:472–478. doi: 10.1016/j.drudis.2009.01.011. [DOI] [PubMed] [Google Scholar]
- Dhiman N. Cunningham J.M. Jacobson R.M. Vierkant R.A. Wu Y. Ovsyannikova I.G., et al. Variations in measles vaccine-specific humoral immunity by polymorphisms in SLAM and CD46 measles virus receptors. J Allergy Clin Immunol. 2007a;120:666–672. doi: 10.1016/j.jaci.2007.04.036. [DOI] [PubMed] [Google Scholar]
- Dhiman N. Ovsyannikova I.G. Cunningham J.M. Vierkant R.A. Kennedy R.B. Pankratz V.S., et al. Associations between measles vaccine immunity and single nucleotide polymorphisms in cytokine and cytokine receptor genes. J Infect Dis. 2007b;195:21–29. doi: 10.1086/510596. [DOI] [PubMed] [Google Scholar]
- Dhiman N. Ovsyannikova I.G. Vierkant R.A. Ryan J.E. Pankratz V.S. Jacobson R.M., et al. Associations between SNPs in toll-like receptors and related intracellular signaling molecules and immune responses to measles vaccine: preliminary results. Vaccine. 2008;26:1731–1736. doi: 10.1016/j.vaccine.2008.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhiman N. Haralambieva I.H. Kennedy R.B. Vierkant R.A. O'Byrne M.M. Ovsyannikova I.G., et al. SNP/haplotype associations in cytokine and cytokine receptor genes and immunity to rubella vaccine. Immunogenetics. 2010;62:197–210. doi: 10.1007/s00251-010-0423-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doria G. Frasca D. Genetic factors in immunity and aging. Vaccine 2000. 2000;18:1591–1595. doi: 10.1016/s0264-410x(99)00491-0. [DOI] [PubMed] [Google Scholar]
- Effros R.B. Dagarag M. Spaulding C. Man J. The role of CD8+ T-cell replicative senescence in human aging. Immunol Rev. 2005;205:147–157. doi: 10.1111/j.0105-2896.2005.00259.x. [DOI] [PubMed] [Google Scholar]
- Feng J. Gulati U. Zhang X. Keitel W.A. Thompson D.M. James J.A., et al. Antibody quantity versus quality after influenza vaccination. Vaccine. 2009;27:6358–6362. doi: 10.1016/j.vaccine.2009.06.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschi C. Bonafe M. Valensin S. Olivieri F. De Luca M. Ottaviani E., et al. Inflamm-aging. An evolutionary perspective on immunosenescence. Ann N Y Acad Sci. 2000;908:244–254. doi: 10.1111/j.1749-6632.2000.tb06651.x. [DOI] [PubMed] [Google Scholar]
- Geenen V. Poulin J.F. Dion M.L. Martens H. Castermans E. Hansenne I., et al. Quantification of T cell receptor rearrangement excision circles to estimate thymic function: an important new tool for endocrine-immune physiology. J Endocrinol. 2003;176:305–311. doi: 10.1677/joe.0.1760305. [DOI] [PubMed] [Google Scholar]
- Geison G.L. The Private Science of Louis Pasteur. Princeton University Press; Princeton, NJ: 1995. [DOI] [PubMed] [Google Scholar]
- Glinsky G.V. Genomic analysis of pandemic (H1N1) 2009 reveals association of increasing disease severity with emergence of novel hemagglutinin mutations. Cell Cycle. 2010;9:958–970. doi: 10.4161/cc.9.5.10913. [DOI] [PubMed] [Google Scholar]
- Goldschmidt V. Bleiber G. May M. Martinez R. Ortiz M. Telenti A. Role of common human TRIM5alpha variants in HIV-1 disease progression. Retrovirology. 2006;3:54. doi: 10.1186/1742-4690-3-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez C.R. Boehmer E.D. Kovacs E.J. The aging innate immune system. Curr Opin Immunol. 2005;17:457–462. doi: 10.1016/j.coi.2005.07.013. [DOI] [PubMed] [Google Scholar]
- Gomez C.R. Nomellini V. Faunce D.E. Kovacs E.J. Innate immunity and aging. Exp Gerontol. 2008;43:718–728. doi: 10.1016/j.exger.2008.05.0168.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D. Finch S.J. Factors affecting statistical power in the detection of genetic association. J Clin Invest. 2005;115:1408–1418. doi: 10.1172/JCI24756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goronzy J.J. Fulbright J.W. Crowson C.S. Poland G.A. O'Fallon W.M. Weyand C.M. Value of immunological markers in predicting responsiveness to influenza vaccination in elderly individuals. J Virol. 2001;75:12182–12187. doi: 10.1128/JVI.75.24.12182-12187.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haralambieva I.H. Poland G.A. Vaccinomics, predictive vaccinology and the future of vaccine development. Future Microbiol. 2010;5:1757–1760. doi: 10.2217/fmb.10.146. [DOI] [PubMed] [Google Scholar]
- Haralambieva I.H. Dhiman N. Ovsyannikova I.G. Vierkant R.A. Pankratz V.S. Jacobson R.M., et al. Oligoadenylate synthetase single-nucleotide polymorphisms and haplotypes are associated with variations in immune responses to rubella vaccine. Hum Immunol. 2010;71:383–391. doi: 10.1016/j.humimm.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattersley A.T. McCarthy M.I. What makes a good genetic association study? Lancet. 2005;366:1315–1323. doi: 10.1016/S0140-6736(05)67531-9. [DOI] [PubMed] [Google Scholar]
- Henderson D.A. Borio L.L. Grabenstein J.D. Smallpox and vaccinia. In: Plotkin S.A., editor; Orenstein W.A., editor; Offit P.A., editor. Vaccines. 5th. Elsevier; China: 2008. pp. 773–804. [Google Scholar]
- Hirschhorn J.N. Daly M.J. Genome-wide association studies for common diseases and complex traits. Genetics. 2005;6:95–108. doi: 10.1038/nrg1521. [DOI] [PubMed] [Google Scholar]
- Ishii S. Koziel M.J. Immune responses during acute and chronic infection with hepatitis C virus. Clin Immunol. 2008;128:133–147. doi: 10.1016/j.clim.2008.03.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson R.M. Ovsyannikova I.G. Targonski P.V. Poland G.A. Studies of twins in vaccinology. Vaccine. 2007;25:3160–3164. doi: 10.1016/j.vaccine.2007.01.048. [DOI] [PubMed] [Google Scholar]
- Javanbakht H. An P. Gold B. Petersen D.C. O'Huigin C. Nelson G.W., et al. Effects of human TRIM5alpha polymorphisms on antiretroviral function and susceptibility to human immunodeficiency virus infection. Virology. 2006;354:15–27. doi: 10.1016/j.virol.2006.06.031. [DOI] [PubMed] [Google Scholar]
- Jin P. Wang E. Polymorphism in clinical immunology—from HLA typing to immunogenetic profiling. J Transl Med. 2003;1:8. doi: 10.1186/1479-5876-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson B.J. Costelloe E.O. Fitzpatrick D.R. Haanen J.B. Schumacher T.N. Brown L.E., et al. Single-cell perforin and granzyme expression reveals the anatomical localization of effector CD8+ T cells in influenza virus-infected mice. Proc Natl Acad Sci USA. 2003;100:2657–2662. doi: 10.1073/pnas.0538056100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K.L. Ovsyannikova I.G. Poland G. Muddiman D.C. Identification of class II HLA-DRB1*03-bound measles virus peptides by 2D-liquid chromatography tandem mass spectrometry. J Proteome Res. 2005;4:2243–2249. doi: 10.1021/pr0501416. [DOI] [PubMed] [Google Scholar]
- Johnson K.L. Ovsyannikova I.G. Mason C.J. Bergen H.R., III Poland G.A. Discovery of naturally processed and HLA-presented class I peptides from vaccinia virus infection using mass spectrometry for vaccine development. Vaccine. 2009;28:38–47. doi: 10.1016/j.vaccine.2009.09.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M.I. Fauci A.S. An HIV vaccine—challenges and prospects. N Engl J Med. 2008;359:888–890. doi: 10.1056/NEJMp0806162. [DOI] [PubMed] [Google Scholar]
- Katz J.M. Plowden J. Renshaw-Hoelscher M. Lu X. Tumpey T.M. Sambhara S. Immunity to influenza: the challenges of protecting an aging population. Immunol Res. 2004;29:113–124. doi: 10.1385/IR:29:1-3:113. [DOI] [PubMed] [Google Scholar]
- Kennedy R.B. Ovsyannikova I.G. Vierkant R.A. Jacobson R.M. Poland G.A. Effect of human leukocyte antigen homozygosity on rubella vaccine-induced humoral and cell-mediated immune responses. Hum Immunol. 2010;71:128–135. doi: 10.1016/j.humimm.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein N.P. Fireman B. Enright A. Ray P. Black S. Dekker C.L. A role for genetics in the immune response to the varicella vaccine. Pediatr Infect Dis J. 2007;26:300–305. doi: 10.1097/01.inf.0000257454.74513.07. [DOI] [PubMed] [Google Scholar]
- Kornbluth R.S. Stone G.W. Immunostimulatory combinations: designing the next generation of vaccine adjuvants. J Leukoc Biol. 2006;80:1084–1102. doi: 10.1189/jlb.0306147. [DOI] [PubMed] [Google Scholar]
- Kruskall M.S. Alper C.A. Awdeh Z. Yunis E.J. Marcus-Bagley D. The immune response to hepatitis B vaccine in humans: inheritance patterns in families. J Exp Med. 1992;175:495–502. doi: 10.1084/jem.175.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence C.W. Ream R.M. Braciale T.J. Frequency, specificity, and sites of expansion of CD8+ T cells during primary pulmonary influenza virus infection. J Immunol. 2005;174:5332–5340. doi: 10.4049/jimmunol.174.9.5332. [DOI] [PubMed] [Google Scholar]
- Liu R. Paxton W.A. Choe S. Ceradini D. Martin S.R. Horuk R., et al. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell. 1996;86:367–377. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- Majumder P.P. Ghosh S. Mapping quantitative trait loci in humans: achievements and limitations. J Clin Invest. 2005;115:1419–1424. doi: 10.1172/JCI24757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascola J.R. Herpes simplex virus vaccines—Why don't antibodies protect. JAMA. 1999;282:379–380. doi: 10.1001/jama.282.4.379. [DOI] [PubMed] [Google Scholar]
- Mascola J.R. Montefiori D.C. The role of antibodies in HIV vaccines. Annu Rev Immunol. 2010;28:413–444. doi: 10.1146/annurev-immunol-030409-101256. [DOI] [PubMed] [Google Scholar]
- Mata-Haro V. Cekic C. Martin M. Chilton P.M. Casella C.R. Mitchell T.C. The vaccine adjuvant monophosphoryl lipid A as a TRIF-biased agonist of TLR4. Science. 2007;316:1628–1632. doi: 10.1126/science.1138963. [DOI] [PubMed] [Google Scholar]
- McElhaney J.E. Influenza vaccination in the elderly: seeking new correlates of protection and improved vaccines. Aging Health. 2008;4:603–613. doi: 10.2217/1745509X.4.6.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElhaney J.E. Gravenstein S. Upshaw C.M. Hooton J.W. Krause P. Drinka P. Immune response to influenza vaccination in institutionalized elderly: effect on different T-cell subsets. Vaccine. 1988;16:403–409. doi: 10.1016/s0264-410x(97)80918-8. [DOI] [PubMed] [Google Scholar]
- McElhaney J.E. Pinkoski M.J. Upshaw C.M. Bleackley R.C. The cell-mediated cytotoxic response to influenza vaccination using an assay for granzyme B activity. J Immunol Methods. 1996;190:11–20. doi: 10.1016/0022-1759(95)00235-9. [DOI] [PubMed] [Google Scholar]
- McElhaney J.E. Gravenstein S. Krause P. Hooton J.W. Upshaw C.M. Drinka P. Assessment of markers of the cell-mediated immune response after influenza virus infection in frail older adults. Clin Diagn Lab Immunol. 1998;5:840–844. doi: 10.1128/cdli.5.6.840-844.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElhaney J.E. Gravenstein S. Upshaw C.M. Hooton J.W. Krause P. Drinka P., et al. Granzyme B: a marker of risk for influenza in institutionalized older adults. Vaccine. 2001;19:3744–3751. doi: 10.1016/s0264-410x(01)00087-1. [DOI] [PubMed] [Google Scholar]
- McElhaney J.E. Xie D. Hager W.D. Barry M.B. Wang Y. Kleppinger A., et al. T cell responses are better correlates of vaccine protection in the elderly. J Immunol. 2006;176:6333–6339. doi: 10.4049/jimmunol.176.10.6333. [DOI] [PubMed] [Google Scholar]
- McElhaney J.E. Ewen C. Zhou X. Kane K.P. Xie D. Hager W.D., et al. Granzyme B: correlates with protection and enhanced CTL response to influenza vaccination in older adults. Vaccine. 2009;27:2418–2425. doi: 10.1016/j.vaccine.2009.01.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motsinger A.A. Haas D.W. Hulgan T. Ritchie M.D. Human genomic association studies: a primer for the infectious diseases specialist. J Infect Dis. 2007;195:1737–1744. doi: 10.1086/518247. [DOI] [PubMed] [Google Scholar]
- Murata Y. Respiratory syncytial virus vaccine development. Clin Lab Med. 2009;29:725–739. doi: 10.1016/j.cll.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newport M.J. Goetghebuer T. Weiss H.A. The MRC Gambia Twin Study Group. Whittle H. Siegrist C.A., et al. Genetic regulation of immune responses to vaccines in early life. Genes Immun. 2004;5:122–129. doi: 10.1038/sj.gene.6364051. [DOI] [PubMed] [Google Scholar]
- O'Connell K.A. Bailey J.R. Blankson J.N. Elucidating the elite: mechanisms of control in HIV-1 infection. Trends Pharmacol Sci. 2009;30:631–637. doi: 10.1016/j.tips.2009.09.005. [DOI] [PubMed] [Google Scholar]
- Ovsyannikova I.G. Poland G.A. Vaccinomics: Current Findings, Challenges and Novel Approaches for Vaccine Development. American Association of Pharmaceutical Scientists; Arlington, VA: 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovsyannikova I.G. Johnson K.L. Naylor S. Muddiman D.C. Poland G.A. Naturally processed measles virus peptide eluted from class II HLA-DRB1*03 recognized by T lymphocytes from human blood. Virology. 2003;312:495–506. doi: 10.1016/s0042-6822(03)00281-2. [DOI] [PubMed] [Google Scholar]
- Ovsyannikova I.G. Jacobson R.M. Vierkant R.A. Pankratz S.V. Jacobsen S.J. Poland G.A. Associations between human leukocyte antigen (HLA) alleles and very high levels of measles antibody following vaccination. Vaccine. 2004a;22:1914–1920. doi: 10.1016/j.vaccine.2003.11.016. [DOI] [PubMed] [Google Scholar]
- Ovsyannikova I.G. Jacobson R.M. Vierkant R.A. Jacobsen S.J. Pankratz V.S. Poland G.A. The contribution of HLA class I antigens in immune status following two doses of rubella vaccination. Hum Immunol. 2004b;65:1506–1515. doi: 10.1016/j.humimm.2004.07.001. [DOI] [PubMed] [Google Scholar]
- Ovsyannikova I.G. Jacobson R.M. Vierkant R.A. Jacobsen S.J. Pankratz V.S. Poland G.A. Human leukocyte antigen class II alleles and rubella-specific humoral and cell-mediated immunity following measles-mumps-rubella-II vaccination. J Infect Dis. 2005a;191:515–519. doi: 10.1086/427558. [DOI] [PubMed] [Google Scholar]
- Ovsyannikova I.G. Jacobson R.M. Ryan J.E. Vierkant R.A. Pankratz V.S. Jacobsen S.J., et al. HLA class II alleles and measles virus-specific cytokine immune response following two doses of measles vaccine. Immunogenetics. 2005b;56:798–807. doi: 10.1007/s00251-004-0756-0. [DOI] [PubMed] [Google Scholar]
- Ovsyannikova I.G. Ryan J.E. Vierkant R.A. Pankratz V.S. Jacobson R.M. Poland G.A. Immunologic significance of HLA class I genes in measles virus-specific IFN-gamma and IL-4 cytokine immune responses. Immunogenetics. 2005c;57:828–836. doi: 10.1007/s00251-005-0061-6. [DOI] [PubMed] [Google Scholar]
- Ovsyannikova I.G. Jacobson R.M. Ryan J.E. Vierkant R.A. Pankratz V.S. Poland G.A. Human leukocyte antigen and interleukin 2, 10 and 12p40 cytokine responses to measles: is there evidence of the HLA effect? Cytokine. 2006a;36:173–179. doi: 10.1016/j.cyto.2006.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovsyannikova I.G. Pankratz S.V. Vierkant R. Jacobson R.M. Poland G.A. Human leukocyte antigen haplotypes in the genetic control of immune response to measles-mumps-rubella vaccine. J Infect Dis. 2006b;193:655–663. doi: 10.1086/500144. [DOI] [PubMed] [Google Scholar]
- Ovsyannikova I.G. Jacobson R.M. Vierkant R.A. Pankratz V.S. Poland G.A. HLA supertypes and immune responses to measles-mumps-rubella viral vaccine: Findings and implications for vaccine design. Vaccine. 2007a;25:3090–3100. doi: 10.1016/j.vaccine.2007.01.020. [DOI] [PubMed] [Google Scholar]
- Ovsyannikova I.G. Johnson K.L. Bergen H.R., III Poland G.A. Mass spectrometry and peptide-based vaccine development. Clin Pharmacol Ther. 2007b;82:644–652. doi: 10.1038/sj.clpt.6100389. [DOI] [PubMed] [Google Scholar]
- Ovsyannikova I.G. Dhiman N. Jacobson R.M. Vierkant R.A. Pankratz V.S. Poland G.A. HLA homozygosity does not adversely effect measles vaccine-induced cytokine responses. Virology. 2007c;364:87–94. doi: 10.1016/j.virol.2007.02.028. [DOI] [PubMed] [Google Scholar]
- Ovsyannikova I.G. Jacobson R.M. Ryan J.E. Dhiman N. Vierkant R.A. Poland G.A. Relationship between HLA polymorphisms and gamma interferon and interleukin-10 cytokine production in healthy individuals after rubella vaccination. Clin Vaccine Immunol. 2007d;14:115–122. doi: 10.1128/CVI.00247-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovsyannikova I.G. Jacobson R.M. Dhiman N. Vierkant R.A. Pankratz V.S. Poland G.A. Human leukocyte antigen and cytokine receptor gene polymorphisms associated with heterogeneous immune responses to mumps viral vaccine. Pediatrics. 2008;121:e1091–e1099. doi: 10.1542/peds.2007-1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovsyannikova I.G. Jacobson R.M. Vierkant R.A. O'Byrne M.M. Poland G.A. Replication of rubella vaccine population genetic studies: validation of HLA genotype and humoral response associations. Vaccine. 2009;27:6926–6931. doi: 10.1016/j.vaccine.2009.08.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovsyannikova I.G. Dhiman N. Haralambieva I.H. Vierkant R.A. O'Byrne M.M. Jacobson R.M., et al. Rubella vaccine-induced cellular immunity: evidence of associations with polymorphisms in the Toll-like, vitamin A and D receptors, and innate immune response genes. Hum Genet. 2010s;127:207–221. doi: 10.1007/s00439-009-0763-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovsyannikova I.G. Haralambieva I.H. Dhiman N. O'Byrne M.M. Pankratz V.S. Jacobson R.M., et al. Polymorphisms in the vitamin A receptor and innate immunity genes influence the antibody response to rubella vaccination. J Infect Dis. 2010b;201:207–213. doi: 10.1086/649588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovsyannikova I.G. Vierkant R.A. Pankratz V.S. Jacobson R.M. Poland G.A. Extended LTA, TNF, LST1 and HLA gene haplotypes and their association with rubella vaccine-induced immunity. PLoS ONE. 2010c;5:e11806. doi: 10.1371/journal.pone.0011806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovsyannikova I.G. Haralambieva I.H. Vierkant R.A. Pankratz V.S. Poland G.A. The role of polymorphisms in Toll-like receptors and their associated intracellular signaling genes in measles vaccine immunity. Hum Genet. 2011 doi: 10.1007/s00439-011-0977-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankratz V.S. Vierkant R.A. O'Byrne M.M. Ovsyannikova I.G. Poland G.A. Associations between SNPs in candidate immune-relevant genes and rubella antibody levels: a multigenic assessment. BMC Immunol. 2010;11:48. doi: 10.1186/1471-2172-11-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasteur L. Chamberland Roux. Summary report of the experiments conducted at Pouilly-le-Fort, near Melun, on the anthrax vaccination, 1881. Yale J Biol Med. 2002;75:59–62. [PMC free article] [PubMed] [Google Scholar]
- Pichichero M.E. Improving vaccine delivery using novel adjuvant systems. Hum Vaccin. 2008;4:262–270. doi: 10.4161/hv.4.4.5742. [DOI] [PubMed] [Google Scholar]
- Plotkin S.A. Koprowski H. Rupprecht C.E. Rabies vaccine. In: Plotkin S.A., editor; Orenstein W.A., editor; Offit P.A., editor. Vaccines. 5th. Elsevier; China: 2008. pp. 687–714. [Google Scholar]
- Poland G.A. Pharmacology, vaccinomics, and the 2nd golden age of vaccinology. Clin Pharmacol Ther. 2007;82:623–626. doi: 10.1038/sj.clpt.6100379. [DOI] [PubMed] [Google Scholar]
- Poland G.A. Jacobson R.M. Failure to reach the goal of measles elimination. Apparent paradox of measles infections in immunized persons. Arch Intern Med. 1994;154:1815–1820. [PubMed] [Google Scholar]
- Poland G.A. Ovsyannikova I.G. Jacobson R.M. Vierkant R.A. Jacobsen S.J. Pankratz V.S., et al. Identification of an association between HLA class II alleles and low antibody leve.ls after measles immunization. Vaccine. 2001;20:430–438. doi: 10.1016/s0264-410x(01)00346-2. [DOI] [PubMed] [Google Scholar]
- Poland G.A. Ovsyannikova I.G. Jacobson R.M. Smith D.I. Heterogeneity in vaccine immune response: the role of immunogenetics and the emerging field of vaccinomics. Clin Pharmacol Ther. 2007;82:653–664. doi: 10.1038/sj.clpt.6100415. [DOI] [PubMed] [Google Scholar]
- Poland G.A. Ovsyannikova I.G. Jacobson R.M. Immunogenetics of seasonal influenza vaccine response. Vaccine. 2008a;26S:D35–D40. doi: 10.1016/j.vaccine.2008.07.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poland G.A. Ovsyannikova I.G. Jacobson R.M. Personalized vaccines: the emerging field of vaccinomics. Expert Opin Biol Ther. 2008b;8:1659–1667. doi: 10.1517/14712598.8.11.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poland G.A. Ovsyannikova I.G. Jacobson R.M. Application of pharmacogenomics to vaccines. Pharmacogenomics. 2009a;10:837–852. doi: 10.2217/PGS.09.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poland G.A. Ovsyannikova I.G. Jacobson R.M. Adversomics: the emerging field of vaccine adverse event immunogenetics. Pediatr Infect Dis J. 2009b;28:431–432. doi: 10.1097/INF.0b013e3181a6a511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poland G.A. Ovsyannikova I.G. Jacobson R.M. Vaccinomics and Personalized Vaccinology (Jordan Report) 2011.
- Rappuoli R. Bridging the knowledge gaps in vaccine design. Nat Biotechnol. 2007;25:1361–1366. doi: 10.1038/nbt1207-1361. [DOI] [PubMed] [Google Scholar]
- Reed Z.H. Friede M. Kieny M.P. Malaria vaccine development: progress and challenges. Curr Mol Med. 2006;6:231–245. doi: 10.2174/156652406776055195. [DOI] [PubMed] [Google Scholar]
- Reif D.M. Motsinger-Reif A.A. McKinney B.A. Rock M.T. Crowe J.E., Jr. Moore J.H. Integrated analysis of genetic and proteomic data identifies biomarkers associated with adverse events following smallpox vaccination. Genes Immun. 2009;10:112–119. doi: 10.1038/gene.2008.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley R.L. Blomberg B.B. Frasca D. B cells, E2A, and aging. Immunol Rev. 2005;205:30–47. doi: 10.1111/j.0105-2896.2005.00268.x. [DOI] [PubMed] [Google Scholar]
- Rosenthal K.L. Tweaking innate immunity: the promise of innate immunologicals as anti-infectives. Can J Infect Dis Med Microbiol. 2006;17:307–314. doi: 10.1155/2006/195957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauver J.L. Ovsyannikova I.G. Jacobson R.M. Jacobsen S.J. Vierkant R.A. Schaid D.J., et al. Associations between human leukocyte antigen homozygosity and antibody levels to measles vaccine. J Infect Dis. 2002;185:1545–1549. doi: 10.1086/340573. [DOI] [PubMed] [Google Scholar]
- Sawyer S.L. Wu L.I. Akey J.M. Emerman M. Malik H.S. High-frequency persistence of an impaired allele of the retroviral defense gene TRIM5alpha in humans. Curr Biol. 2006;16:95–100. doi: 10.1016/j.cub.2005.11.045. [DOI] [PubMed] [Google Scholar]
- Scorpio A. Blank T.E. Day W.A. Chabot D.J. Anthrax vaccines: Pasteur to the present. Cell Mol Life Sci. 2006;63:2237–2248. doi: 10.1007/s00018-006-6312-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegrist C.A. Aspinall R. B-cell responses to vaccination at the extremes of age. Nat Rev Immunol. 2009;9:185–194. doi: 10.1038/nri2508. [DOI] [PubMed] [Google Scholar]
- Smith N.M. Bresee J.S. Shay D.K. Uyeki T.M. Cox N.J. Strikas R.A. Prevention and control of influenza: recommendations of the Advisory Committee on Immunization Practices (ACIP) MMWR Recomm Rep. 2006;55:1–42. [PubMed] [Google Scholar]
- Spielberg S.P. Therapeutics and toxicology. Curr Opin Pediatr. 1998;10:201–202. doi: 10.1097/00008480-199804000-00015. [DOI] [PubMed] [Google Scholar]
- St. Sauver J.L. Dhiman N. Ovsyannikova I.G. Jacobson R.M. Vierkant R.A. Pankratz S.V., et al. Extinction of the human leukocyte antigen homozygosity effect after two doses of the measles–mumps–rubella vaccine. Hum Immunol. 2005;66:788–798. doi: 10.1016/j.humimm.2005.03.008. [DOI] [PubMed] [Google Scholar]
- Tan P.L. Jacobson R.M. Poland G.A. Jacobsen S.J. Pankratz S.V. Twin studies of immunogenicity—determining the genetic contribution to vaccine failure. Vaccine. 2001;19:2434–2439. doi: 10.1016/s0264-410x(00)00468-0. [DOI] [PubMed] [Google Scholar]
- Targonski P.V. Caldwell C.R. Strausbauch M. Wettstein P. Poland G.A. Tangalos E.G. White blood cell telomerase activity and incident respiratory illness among community-dwelling elderly vaccinated against seasonal influenza. Clin Pharmacol Ther. 2007a;82:694–699. doi: 10.1038/sj.clpt.6100410. [DOI] [PubMed] [Google Scholar]
- Targonski P.V. Jacobson R.M. Poland G.A. Immunosenescence: role and measurement in influenza vaccine response among the elderly. Vaccine. 2007b;25:3066–3069. doi: 10.1016/j.vaccine.2007.01.025. [DOI] [PubMed] [Google Scholar]
- Telenti A. Safety concerns about CCR5 as an antiviral target. Curr Opin HIV AIDS. 2009;4:131–135. doi: 10.1097/COH.0b013e3283223d76. [DOI] [PubMed] [Google Scholar]
- Vaine M. Wang S. Hackett A. Arthos J. Lu S. Antibody responses elicited through homologous or heterologous prime-boost DNA and protein vaccinations differ in functional activity and avidity. Vaccine. 2010;28:2999–3007. doi: 10.1016/j.vaccine.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Duin D. Shaw A.C. Toll-like receptors in older adults. J Am Geriatr Soc. 2007;55:1438–1444. doi: 10.1111/j.1532-5415.2007.01300.x. [DOI] [PubMed] [Google Scholar]
- van Manen D. Rits M.A. Beugeling C. van Dort K. Schuitemaker H. Kootstra N.A. The effect of Trim5 polymorphisms on the clinical course of HIV-1 infection. PLoS Pathog. 2008;4:e18. doi: 10.1371/journal.ppat.0040018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga S.M. Fixing a failed vaccine. Nat Med. 2009;15:21–22. doi: 10.1038/nm0109-21. [DOI] [PubMed] [Google Scholar]
- Weinberger B. Herndler-Brandstetter D. Schwanninger A. Weiskopf D. Grubeck-Loebenstein B. Biology of immune responses to vaccines in elderly persons. Clin Infect Dis. 2008;46:1078–1084. doi: 10.1086/529197. [DOI] [PubMed] [Google Scholar]
- Weng N.P. Aging of the immune system: how much can the adaptive immune system adapt? Immunity. 2006;24:495–499. doi: 10.1016/j.immuni.2006.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng N.P. Levine B.L. June C.H. Hodes R.J. Regulated expression of telomerase activity in human T lymphocyte development and activation. J Exp Med. 1996;183:2471–2479. doi: 10.1084/jem.183.6.2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie D. McElhaney J.E. Lower GrB + CD62Lhigh CD8 TCM effector lymphocyte response to influenza virus in older adults is associated with increased CD28null CD8 T lymphocytes. Mech Ageing Dev. 2007;128:392–400. doi: 10.1016/j.mad.2007.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]