Abstract
We have purified and characterized a Gcn5-independent nucleosomal histone H3 HAT complex, NuA3 (Nucleosomal Acetyltransferase of histone H3). Peptide sequencing of proteins from the purified NuA3 complex identified Sas3 as the catalytic HAT subunit of the complex. Sas3 is the yeast homolog of the human MOZ oncogene. Sas3 is required for both the HAT activity and the integrity of the NuA3 complex. In addition, NuA3 contains the TBP- associated factor, yTAFII30, which is also a component of the TFIID, TFIIF, and SWI/SNF complexes. Sas3 mediates interaction of the NuA3 complex with Spt16 both in vivo and in vitro. Spt16 functions as a component of the yeast CP (Cdc68/Pob3) and mammalian FACT (facilitates chromatin transcription) complexes, which are involved in transcription elongation and DNA replication. This interaction suggests that the NuA3 complex might function in concert with FACT–CP to stimulate transcription or replication elongation through nucleosomes by providing a coupled acetyltransferase activity.
Keywords: HAT, NuA3, Sas3, TAFII30, Spt16, FACT
The association of DNA with histones results in the formation of repressive structures that are largely incompatible with basic nuclear processes such as replication and transcription. Numerous cellular activities have been identified that can overcome the repressive effects of chromatin (Workman and Kingston 1998). For example, there is a class of chromatin remodeling activities including SWI/SNF (Kwon et al. 1994; Wang et al. 1996), NURF (Tsukiyama and Wu 1995), CHRAC (Varga-Weisz et al. 1997), ACF (Ito et al. 1997), RSC (Cairns et al. 1996b), and RSF (LeRoy et al. 1998), which use the energy of ATP hydrolysis to alter nucleosomal structure so that the underlying DNA becomes more accessible to DNA-binding proteins. In addition, nucleosomal histones can be modified by histone acetyltransferases (HATs). The conserved amino-terminal tails of histones are the primary sites of acetylation and serve as targets for HATs. A rich history of studies have linked acetylation of core histones and transcriptional regulation (Allfrey et al. 1964; Loidl 1994).
HATs can be broadly categorized into two groups: the Type A HATs that can acetylate nucleosomal histones and the Type B HATs that can only acetylate free histones (not DNA associated). Type A HATs are typically associated with transcriptional processes (Brownell et al. 1996; Grant et al. 1997) whereas Type B HATs are involved in the modification of histones targeted for deposition during replication (Kleff et al. 1995; Parthun et al. 1996).
A consequence of histone acetylation is the neutralization of the positively charged lysines thereby changing the overall charge distribution of the histone tails (Kleff et al. 1995; Parthun et al. 1996; Workman and Kingston 1998). While acetylation of histone tails does not appear to result in gross changes in nucleosome structure, observed consequences include: enhanced DNase I cleavage (Simpson et al. 1985), increased transcription factor accessibility (Vettese-Dadey et al. 1996), and enhanced transcription (Nightingale et al. 1998; Steger et al. 1998). Indeed, chromosomal domains that are actively transcribed tend to be enriched in hyperacetylated histones while transcriptionally repressed domains are enriched in hypoacetylated histones (Turner and O'Neill 1995). The level of histone acetylation is counterbalanced by the action of histone deacetylases (HDACs), which are typically associated with co-repressor complexes (Struhl 1998).
The key observation that co-activator proteins like Gcn5 possess intrinsic HAT activity provided a molecular link between histone acetylation and transcriptional activation (Brownell and Allis 1995; Brownell et al. 1996). Gcn5 belongs to a diverse superfamily of genes (GNATs) that share multiple domains of homology including a region involved in acetyl-CoA binding (HAT domain) (Neuwald and Landsman 1997; Wang et al. 1998; Lin et al. 1999). Recently, other co-activator proteins have been shown to possess histone acetyltransferase activity. A partial list includes p300/CBP (Bannister and Kouzarides 1996), p/CAF (Yang et al. 1996), ACTR (Chen et al. 1997), Src-1 (Spencer et al. 1997), TAFII250 (Mizzen et al. 1996), and TFIIIC (Kundu et al. 1999).
Members of the MYST family of proteins share homology with Gcn5 and other HATs. This group includes Sas2, Sas3, Esa1, MOZ, and Tip60 (Kamine et al. 1996; Kimura and Horikoshi 1998; Reifsnyder et al. 1996; Smith et al. 1998), all of which share remarkable homology with each other, especially in the conserved HAT domain. Sas3, Esa1, and Tip60 have been shown to possess histone acetylase activity, but are unable to acetylate nucleosomal substrates (Kimura and Horikoshi 1998; Smith et al. 1998; Takechi and Nakayama 1999). Mutations in either SAS2 or SAS3 restore silencing to derepressed HMR loci, but also enhance silencing defects at the HML silent mating loci. Therefore, it appears that Sas2 and Sas3 can act as either positive or negative regulators of transcription depending on the specific gene context (Reifsnyder et al. 1996). Similarly, gcn5 mutants have been shown to both activate and repress gene expression (Holstege et al. 1998; Spellman et al. 1998).
Co-activator/HAT proteins have been found to function as part of multiprotein complexes that represent a novel class of transcription regulators (Grant and Berger 1999). Analysis of these complexes has revealed important insights into the biochemical functions of several gene products implicated in transcriptional regulation (Winston and Sudarsanam 1998). Four distinct high-molecular-weight HAT complexes (ADA, NuA4, NuA3, and SAGA) have been identified in yeast (Grant et al. 1997). NuA4 (1.2 MD) acetylates histone H4 on nucleosomal substrates and contains the essential yeast protein Esa1 (Essential SAS2-related acetyltransferase; Clarke et al. 1999; Smith et al. 1998) as its catalytic subunit and Tra1, a protein that may interact with transcriptional activators (J. Cote, pers. comm.). ADA (0.8 MD), SAGA (1.8 MD) and NuA3 all acetylate primarily histone H3 on nucleosomal substrates (Grant et al. 1997). The ADA and SAGA complexes both contain the catalytic subunit, Gcn5, and the adaptor proteins Ada2 and Ada3 (Grant et al. 1997). However, the SAGA and ADA complexes also contain unique subunits. The AHC1 gene product is an essential component of the ADA complex (Eberharter et al. 1999). SAGA has been found to contain the TBP group of Spt proteins, a subset of TAFs (yTAFII90, yTAFII68, yTAFII60, and yTAFII17) ,and Tra1 (Grant et al. 1998).
Here, we describe the purification and characterization of the first Gcn5-independent histone H3 HAT complex from yeast. Using a combination of biochemical and genetic approaches, we show that the Something About Silencing protein, Sas3, is the sole catalytic subunit of the NuA3 complex. In addition, the complex is shown to contain the TBP associated factor, yTAFII30 (hereafter referred to as TAF30). Although NuA3 has been shown to stimulate transcription from chromatin templates in an acetyl-CoA-dependent fashion, it has not been found to interact directly with acidic activation domains as does SAGA and NuA4 (Ikeda et al. 1999; Steger et al. 1998). In this report, we show that, instead, the NuA3 complex interacts with the amino terminus of Spt16 both in vitro and in vivo. Spt16, an abundant and essential yeast protein, is a component of CP (Cdc68/Pob3) and FACT (facilitates chromatin transcriptions), regulatory complexes implicated in transcriptional and replication processes (Brewster et al. 1998; Orphanides et al. 1998; Wittmeyer and Formosa 1997).
Results
Identification of Sas3 and TAF30 as components of the NuA3 complex
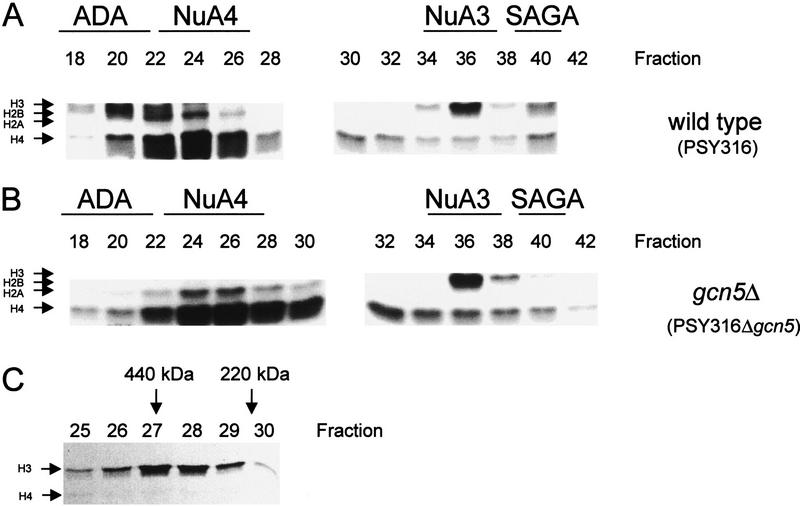
Previously, we have identified four native histone acetyltransferase complexes from yeast that efficiently acetylate nucleosomal histones (Grant et al. 1997). These complexes are separated on a MonoQ column, which is the second column in their purification (Fig. 1). The ADA, NuA3, and SAGA complexes acetylate primarily H3 on nucleosomal templates (Fig. 1A, fractions 20, 36, and 40, respectively). The Esa1-dependent H4 HAT (NuA4) targets primarily H4 and H2A (Fig. 1A, fractions 22–26) on nucleosomal templates. In a gcn5 deletion strain (PSY316Δgcn5) the ADA and SAGA complexes are inactive (Fig. 1, cf. A and B, fractions 20 and 40) whilst the NuA4 and NuA3 activities (Fig. 1, cf. A and B, fractions 22–26 and fractions 34–36) remain unaffected. This result suggested that NuA3, while modifying H3, just like the ADA and SAGA HATs, contains a novel Gcn5-independent catalytic subunit. Furthermore, fractionation of NuA3 over a sizing column suggested that this activity, like the other HATs, was present in cells as a high-molecular-weight complex (0.4–0.5 MD, Fig. 1C).
Figure 1.
NuA3 is a high-molecular-weight Gcn5-independent complex. Separation of distinct HAT complexes with differing acetylation specificities on nucleosomal templates. Whole-cell extracts were prepared and bound to nickel–agarose resin. The nickel eluate was loaded onto a MonoQ column. Four HATs are identified in this fractionation scheme. The names of these complexes are indicated above the fractions where they elute. The ADA complex typically elutes in fractions 18–20, NuA4 in 22–28, NuA3 in 34–36 and SAGA at 40. (Arrows) Migration position of the four core histones. (A) Fluorogram using 1 μl of MonoQ fractions from a wild-type strain (PSY316). (B) Fluorogram using 1 μl of MonoQ fractions from a gcn5 deletion strain (PSY316Δgcn5). Note the absence of ADA and SAGA complexes in this deletion. NuA4 and NuA3 complexes are unaffected.(C) Pooled NuA3 fractions (fractions 35–37) from A were concentrated and put over a Superose 6 sizing column. NuA3 elutes in fractions 27 and 28 with a predicted molecular weight of 400–500 kD. The molecular weights of protein standards are indicated by arrows above the appropriate fraction.
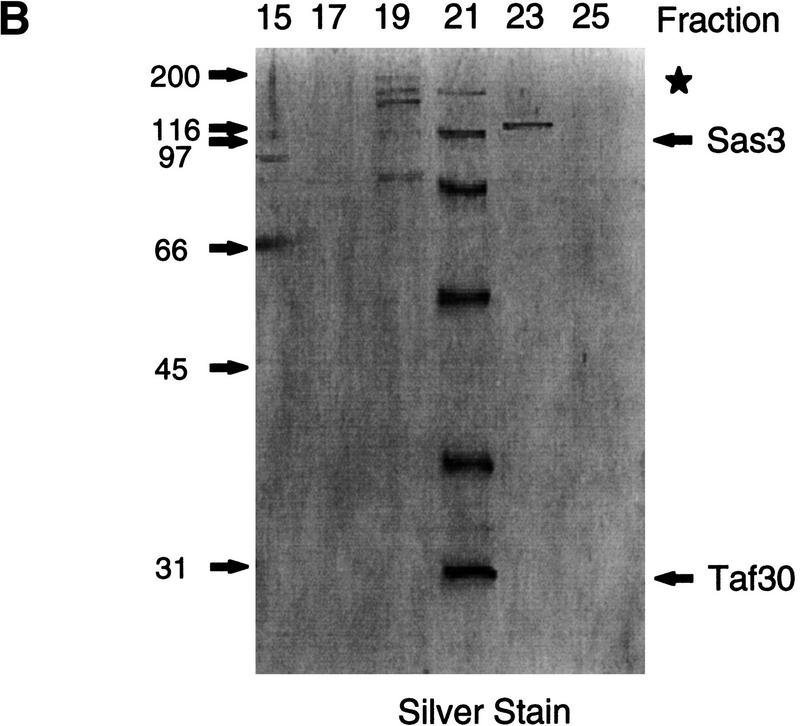
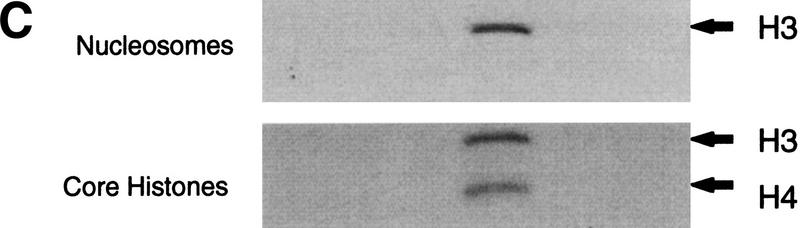
To investigate the composition and functions of NuA3 we undertook purification of the NuA3 activity as outlined in Figure 2A. As no subunits of this complex were known, NuA3 was purified on the basis of its activity in acetylating histone H3 in nucleosomes (see Materials and Methods). Figure 2B illustrates the protein composition after the eighth and final column. Five clearly visible proteins coelute with HAT activity and range in molecular weight from >100-kD to ∼30-kD (Fig. 2B, fraction 21). The identity of two of these bands was determined by peptide sequencing. The band migrating at >100 kD yielded peptide sequences to Sas3 (predicted molecular weight, 97 kD), and the 30 kD protein was identified as yTAFII30 (for peptide sequence, see Materials and Methods). Peptide sequencing was also used to tentatively identify the remaining proteins within the complex. Peptides were identified from the following ORFs; YHR197w, YNL061w, YJR002w, YER049w, YGR145w, YPL101w, and YHR187W. Experiments are in progress to confirm whether these proteins are bona fide subunits of the NuA3 complex. The acetylation specificity of the peak Mini Q fraction is illustrated in Figure 2C. NuA3 acetylates H3 on nucleosomal substrates but, in addition, can also acetylate H4 on free histones.
Figure 2.
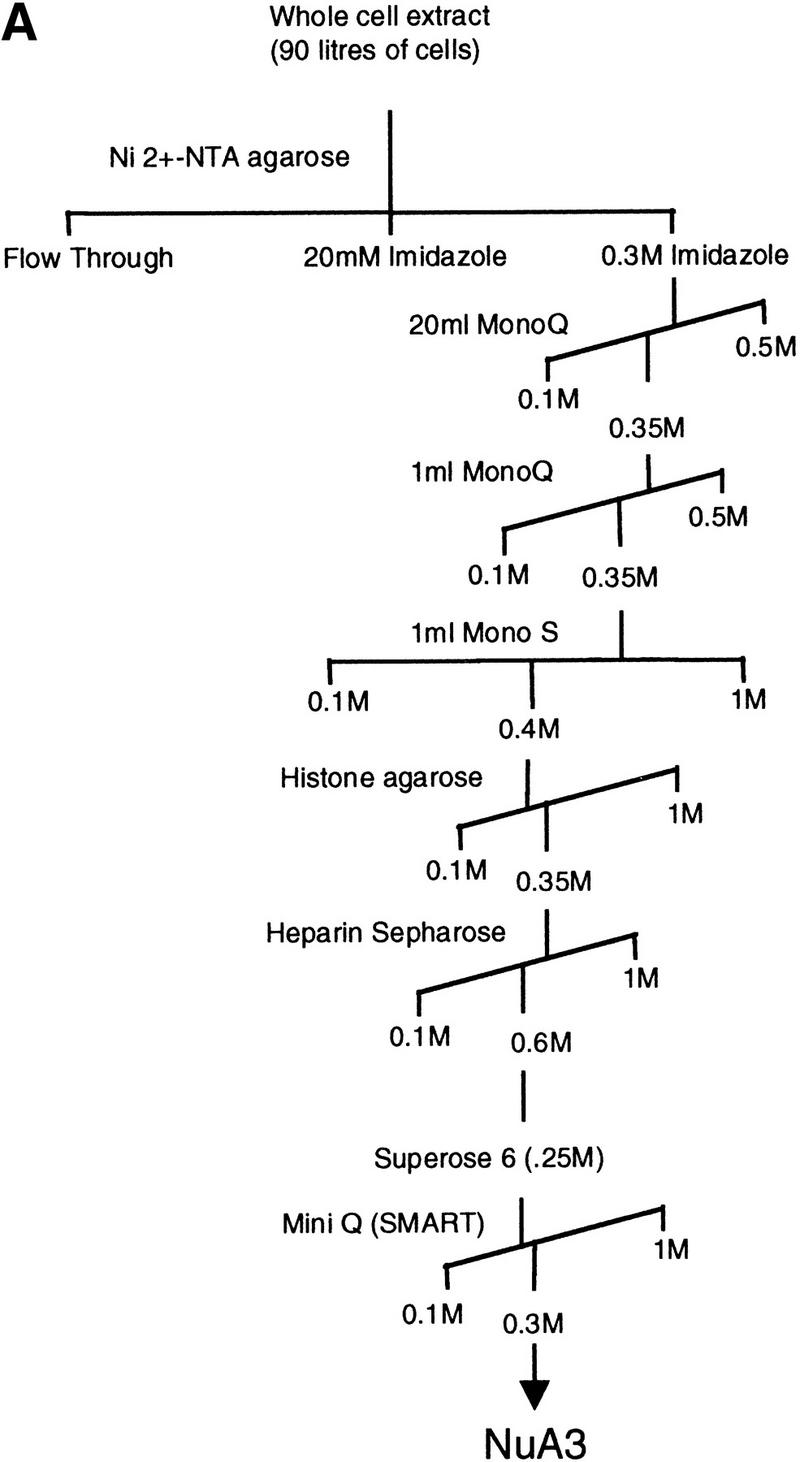
Purification scheme of the yeast NuA3 complex. Whole-cell extract from 90 liters of yeast strain CY396 were prepared for the biochemical purification of the NuA3 complex. (A) Purification scheme for the 0.4–0.5 MD nucleosomal HAT complex, NuA3 (see Materials and Methods for details). (B) Silver-stained gel from the final MiniQ column. (Arrows) Bands in the peak fraction (fraction 21) that yielded peptide sequences to Sas3 and TAF30. The identity of the other bands in fraction 21 is unconfirmed. (Star) Contaminating band that is also present in fraction 19. (C) Fluorogram of HAT assays on nucleosomes and core histones using 0.25 μl of MiniQ fractions show that the specificity of NuA3 is primarily histone H3 on nucleosomes and H3 and weakly H4 on core histones. The peak HAT activity is coincident with the protein peak. (Arrows) Positions of histones H3 and H4.
Sas3 is a component of the NuA3 complex
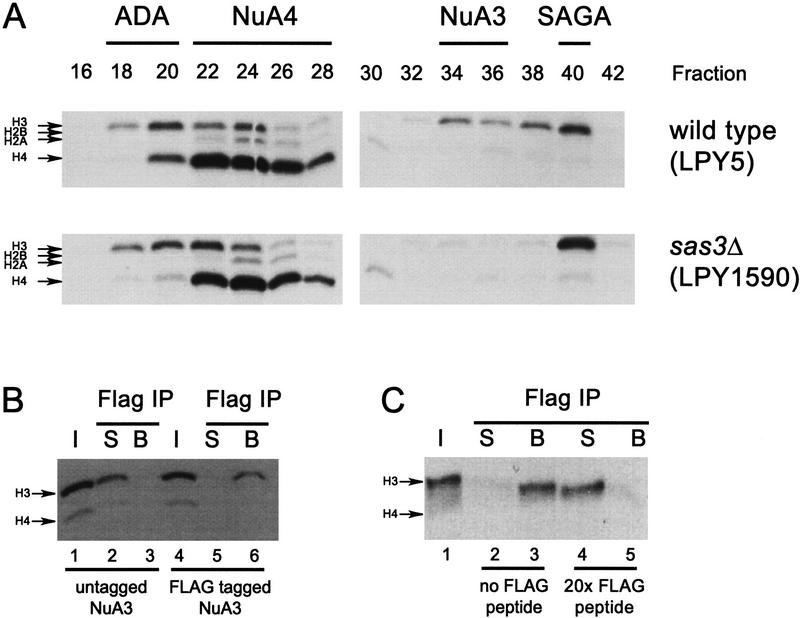
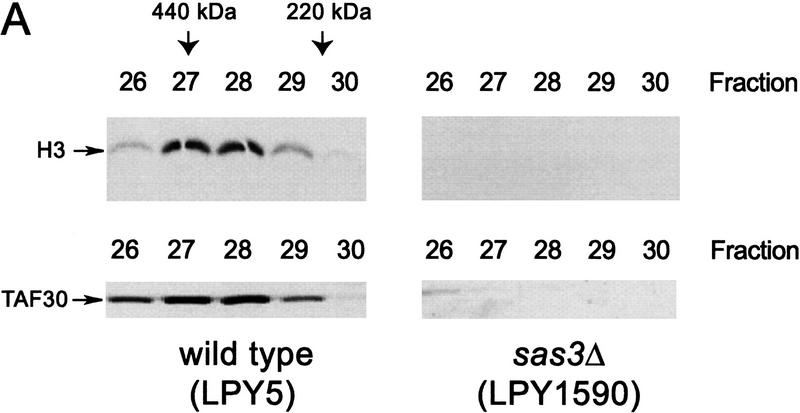
To provide support for the protein sequencing data suggesting that Sas3 is a component of the NuA3 complex, a sas3 deletion strain (LPY1590) and the corresponding isogenic wild-type strain (LPY5) were used to test for any dependence of NuA3 on the SAS3 gene product (see Materials and Methods). Extracts were made from each strain and then subjected to the initial chromatographic steps illustrated in Figure 2A. MonoQ fractions from both strains were assayed for HAT activity and are shown in Figure 3A. A comparison of the four HAT complexes indicates that while ADA, NuA4, and SAGA activities are unchanged in extracts prepared from a sas3-deleted yeast strain, no HAT activity on either nucleosomes (cf. fractions 34–36) or core histones (data not shown) were observed for NuA3. To implicate further Sas3 as a component of the NuA3 complex, we transformed the sas3 deletion strain with a CEN/ARS plasmid bearing the wild-type SAS3 gene (yeast strain YJW115) under the control of its own promoter (1 kb of upstream sequence) and with a C terminal Flag epitope tag. Partially purified NuA3 fractions from this strain were then immunoprecipitated with the Flag M2 antibody (see Materials and Methods), and the supernatant and beads from the immunoprecipitation were assayed for HAT activity. As shown in Figure 3B (lanes 4–6), the tagged version of NuA3 is quantitatively depleted onto Flag M2 beads. The specificity of the immunoprecipitation is demonstrated by either using the endogenous (untagged) NuA3 complex in a control immunoprecipitation or by incubating the tagged HAT complex with Flag antibody in the presence of Flag peptide. In both cases (Fig. 3B, lanes 1–3 and 3C, lanes 4–5), the NuA3 activity resides in the supernatant.
Figure 3.
Sas3 is a component of the NuA3 complex. Whole-cell extracts were prepared from wild type and sas3 deletion strains and fractionated as described (see Materials and Methods). (A) Fluorogram using 1 μl of MonoQ fractions from a wild-type strain (LPY5) and from an sas3 deletion strain (LPY1590) on oligonucleosomes. (B) Fluorogram of supernatant and beads of Flag immunoprecipitates (IP) using Superose 6 fractions of untagged NuA3 (lanes 1–3) and Flag-tagged NuA3 (lanes 4–6). Only Flag-tagged NuA3 fractions can be depleted by Flag antibody. I, Input; S, supernatant; B, beads. (C) Fluorogram of supernatant and beads of Flag immunoprecipitates using Superose 6 Flag-tagged NuA3 in the absence (lanes 2,3) or presence (lanes 4,5) of Flag peptide. The presence of excess Flag peptide prevents Flag-tagged NuA3 from interacting with Flag antibody. Abbreviations as in B.
TAF30 is a component of the NuA3 complex
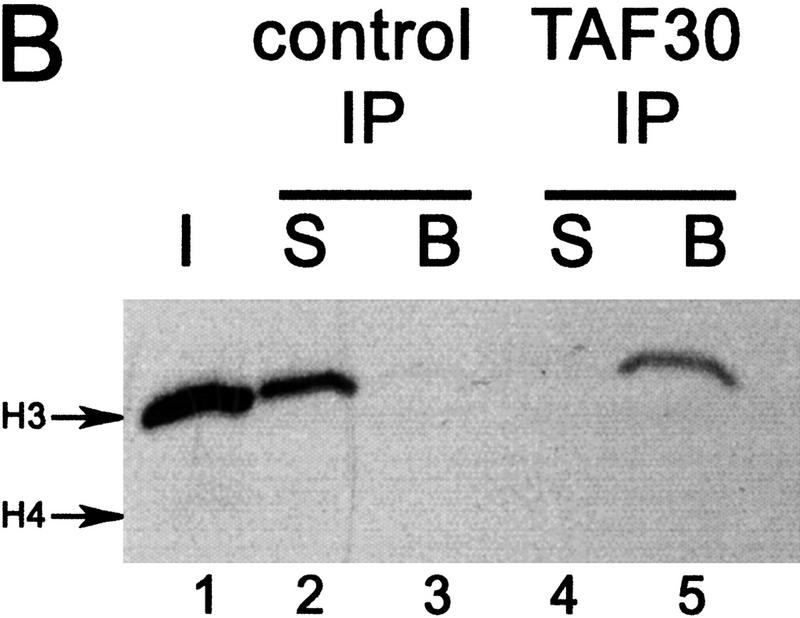
Protein sequencing results suggested that, in addition to Sas3, TAF30 is also a component of NuA3. The SWI/SNF complex, another TAF30-containing complex, coelutes closely with NuA3 (fractions 30–34 for SWI/SNF and 34–36 for NuA3, data not shown and Grant et al. 1997) making it difficult to resolve the two complexes using MonoQ fractions. To overcome this, MonoQ fractions 34–36 of both wild-type and sas3 mutant strains were put over a Superose 6 gel-filtration column to separate NuA3 from the significantly larger SWI/SNF complex. The Superose 6 fractions were assayed for HAT activity and the presence of TAFII30 detected by Western blotting (Fig. 4A). A TAF30 signal was observed coeluting with the NuA3 HAT activity (fractions 27–28) from wild-type strains. However, in a sas3 deletion strain, neither NuA3 HAT activity nor a TAF30 signal was observed. This result likely suggests that the sas3 deletion has compromised the overall integrity of the complex or at the very least affected the biochemical properties of the complex such that it fails to elute as predicted. We have been unable to detect NuA3 in other parts of the fractionation scheme in a sas3 mutant (such as the flowthrough of nickel agarose or MonoQ columns). Immunoprecipitations with antibodies to TAF30 also confirm that this protein is a stable component of the NuA3 complex (Fig. 4B, lanes 4,5). The control antibody used (Fig. 4B, lanes 2,3) is to Ahc1, a component of the ADA complex (Eberharter et al. 1999).
Figure 4.
TAF30 is a component of the NuA3 complex. (A) HAT activity (oligonucleosomes) and TAF30 Western blot analysis of Superose 6 fractions from wild-type and sas3 deletion strains. MonoQ fractions 33–37 from wild-type and sas3 deletion strains were concentrated and put over a Superose 6 sizing column. (Arrows) Fraction in which molecular weight standards elute. (B) Fluorogram of supernatant and beads of immunoprecipitates (IP) of Superose 6 NuA3 fractions using a control Ahc1 antibody (lanes 2,3) or TAF30 antibody (lanes 4,5). I, Input; S, supernatant; B, beads.
Sas3 encodes the catalytic subunit of the NuA3 complex
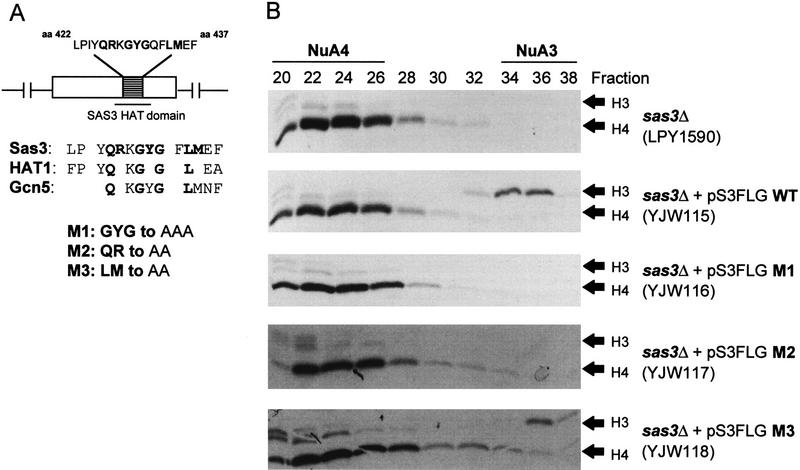
Reifsnyder and co-workers have described Sas3 as a homolog of the yeast silencing protein Sas2. Both proteins share homology with each other and other MYST family proteins (Reifsnyder et al. 1996). In addition, both Sas2 and Sas3 share homology over a small region (HAT domain or acetyl-CoA binding domain) with a larger group of HAT and NAT (N-acetyltransferase) proteins. Although no histone acetyltransferase activity has yet been documented for Sas2, Sas3 has been shown to be able to acetylate histones but not nucleosomes (Takechi and Nakayama 1999). Identification of Sas3 peptides in the purified NuA3 complex suggested that Sas3 might be the catalytic subunit of NuA3. However, because the presence of the NuA3 complex required the presence of Sas3 it was formally possible that while Sas3 played a structural role another subunit may provide the detected HAT activity. To directly link Sas3 as the catalytic subunit of the NuA3 complex, we constructed point mutants in the highly conserved HAT domain (Reifsnyder et al. 1996; Dutnall et al. 1998). A comparison of HAT domain sequences between Sas3 (amino acids 422–437) and other HATs show numerous conserved amino acids (Fig. 5A). One absolutely invariant amino acid between all HATs and NATs is a glycine at amino acid position 429 in Sas3. Other conserved residues include glutamine (amino acid 426) and leucine (amino acid 434). A set of mutants was constructed changing these conserved amino acids and neighboring residues to alanines (Fig. 5A). These mutations were introduced into a sas3 deletion strain on a CEN/ARS plasmid and functional rescue (or lack thereof) of the NuA3 complex was determined. The wild-type SAS3 gene can functionally complement the NuA3 deficiency in a sas3 deletion, but mutant strains harboring either mutation M1 or M2 (strains YJW116, YJW117) could no longer rescue the HAT activity of the NuA3 complex (Fig. 5B, fractions 34–36). Mutation M3 (strain YJW118) was able to complement in a manner similar to wild-type Sas3 suggesting that this mutation did not affect the ability of Sas3 to catalyze the acetylation reaction. The recently determined crystal structure of HAT1 (Dutnall et al. 1998) indicates that the catalytically inactive mutants M1 (glycine, amino acid 429) and M2 (glutamine, amino acid 426) both reside within the acetyl-CoA binding pocket while the innocuous mutation M3 (leucine, amino acid 434) falls within a hydrophobic pocket somewhat distal to the acetyl-CoA binding site.
Figure 5.
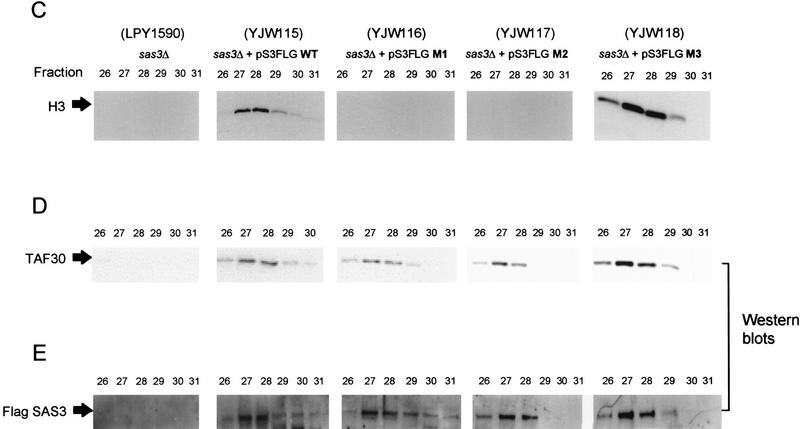
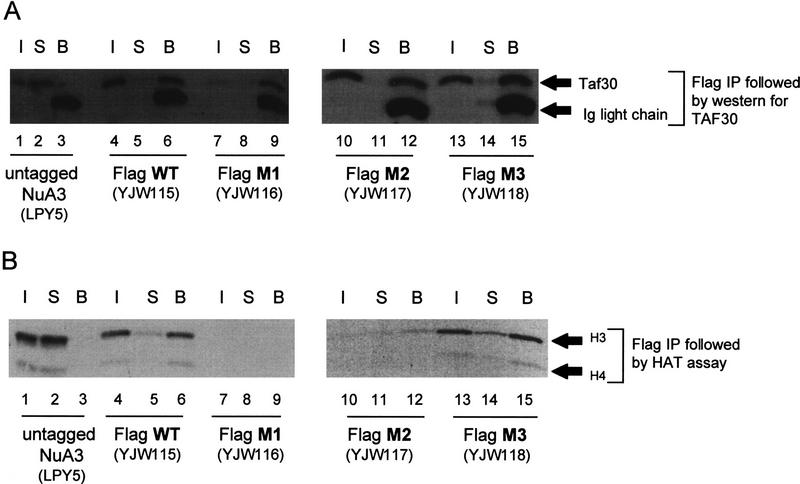
Point mutations in the Sas3 HAT domain compromise NuA3 HAT activity. An sas3 deletion strain was transformed with a CEN/ARS plasmid carrying the wild-type or mutant forms of the SAS3 gene under the control of its own promoter. (A) Region comprising the HAT domain in Sas3 and the related HATs, HAT1 and Gcn5. Sas3 amino acids depicted in bold have been mutated to alanines. (B) Fluorogram using 1 μl of MonoQ fractions prepared from various strains. (C) Fractions 33–37 from the various strains were concentrated and put over a Superose 6 sizing column. Each fraction (2.5 μl) was assayed on oligonucleosomes. Note the catalytically inactive M1 and M2 mutants. (D) Western blot with Superose 6 fractions and antibodies to TAF30. (E) Western blot with Superose 6 fractions and antibodies to Flag-Sas3. Note the coelution of HAT activity [wild-type (WT) and M3] with TAF30 and Sas3. Inactive mutants (M1 and M2) also show a similar coelution of TAF30 and Sas3 suggesting that the mutant complexes are largely intact.
The lack of NuA3 activity with mutants M1 and M2 could be due to a lack of expression of the protein or more importantly, a failure to become incorporated into the NuA3 complex. To address this issue, NuA3 was partially purified from wild-type and mutant strains (YWJ115–YJW118). Fractions from a sizing column (Superose 6) were assayed for HAT activity (Fig. 5C) and the presence of Sas3 and TAF30 monitored by Western blotting (Fig. 5D,E). Peak NuA3 activity elutes from a sizing column typically in fractions 27 and 28. As evidenced by Western blot signals for TAF30 or Sas3 (Flag Western), the size of the complex in either the active (M3) or inactive (M1, M2) mutants remains unchanged relative to that of the wild-type complex.
The successful incorporation of the Sas3 mutants into NuA3 is supported further by immunoprecipitation experiments against the Flag tag on Sas3. Immunoprecipitations with Flag antibodies followed by Western blotting to detect TAF30 show that both wild-type and mutant forms of Sas3 are incorporated into complexes containing TAF30 (Fig. 6A), indicating that both proteins are stable NuA3 components. As expected, the HAT activity was associated with the anti-Flag immunoprecipitated NuA3 complex in the wild-type and M3-containing complex but not in the M1 and M2-containing complexes (Fig. 6B).
Figure 6.
Flag immunoprecipitates demonstrate TAF30 and Sas3 to be stable components of the NuA3 complex. Superose 6 fractions from various strains were immunoprecipitated with Flag antibodies. (A) Supernatant and beads from Flag immunoprecipitates (IP) were run on 10% SDS-PAGE gels, transferred to nitrocellulose and probed with TAF30 antibodies. In all cases (except for untagged NuA3), the Flag antibody co-immunoprecipitates TAF30. Note, lane 7 is slightly underloaded. I, Input; S, supernatant; B, beads. (B) Fluorogram of supernatant and beads of Flag immunoprecipitates using Superose 6 fractions from various strains. Activity is detected in the Flag beads only with WT and M3 fractions. Abbreviations as in A.
The data in Figures 5 and 6 demonstrate that Sas3 proteins bearing point mutations within the HAT domain were successfully incorporated into NuA3 complexes but were unable to acetylate histones. This result confirms that Sas3 is indeed the sole catalytic subunit of the NuA3 HAT complex. Indeed, a similar mutation constructed in the HAT domain of Gcn5 is also catalytically inactive yet maintains the overall structural integrity of the Gcn5-containing SAGA and ADA complexes (Wang et al. 1998). Also similar to Gcn5, a bacterially expressed Sas3 fusion protein has been shown to acetylate histones but lacks the ability to acetylate histones within nucleosomes (Takechi and Nakayama 1999). Thus, one of the functions of the additional proteins within the NuA3 complex is to allow Sas3 to access these nucleosomal histones.
Sas3 interaction with Spt16, a component of the FACT–CP complex
Previously, we have shown that the ADA, NuA4, NuA3, and SAGA complexes can stimulate transcription in an acetyl-CoA-dependent fashion specific to chromatin templates (Steger et al. 1998). The SAGA and NuA4 complexes have been shown to interact with a broad range of acidic transcriptional activators and general transcription factors (Utley et al. 1998; Sterner et al. 1999). These activator–HAT interactions can target these HATs to specific nucleosomes (Utley et al. 1998; Ikeda et al. 1999). The activator bound HATs may in turn stimulate transcription by bridging activators to components of the basal machinery. However, the NuA3 complex, whilst stimulating transcription, failed to demonstrate interactions with any activator proteins tested to date (Utley et al. 1998; Ikeda et al. 1999; and data not shown).
Insight into possible recruitment mechanisms for NuA3 came from a two- hybrid screen using a LexA–Spt16(1–464) fusion protein as bait. One million transformants were screened, and five of the seven that passed the tests for specificity (Park and Sternglanz 1999) were idenitified as fragments of the SAS3 gene (Fig. 7A). No known components of ADA, NuA4, or SAGA were obtained in this screen. Interestingly, all the Sas3 fragments obtained have variable amino termini but contain a common acidic carboxyl terminus. The smallest Sas3 fragment obtained is a 148-amino-acid stretch that encompasses the entire acidic domain and lacks the HAT domain (Fig. 7A). The mammalian homolog of Spt16 is a component of FACT (Orphanides et al. 1999), a heterodimeric complex that facilitates transcription on chromatin templates by helping RNA Pol II overcome nucleosome-mediated blocks to transcriptional elongation (LeRoy et al. 1998; Orphanides et al. 1998). Spt16 is identical to Cdc68 (Malone et al. 1991) and has demonstrated roles in activation and repression of genes as well as in cell cycle regulation (Rowley et al. 1991). In yeast, Cdc68/Spt16 is tightly associated with Pob3 (polymerase one binding) in the CP complex. Furthermore, Spt16 and Pob3 have been shown to be chromatin-associated proteins that co-purify with yeast DNA polymerase α, suggesting that the FACT–CP complex may, in addition, play a role in permitting access or facilitating progress of replication proteins on chromatin templates (Wittmeyer and Formosa 1997).
Figure 7.
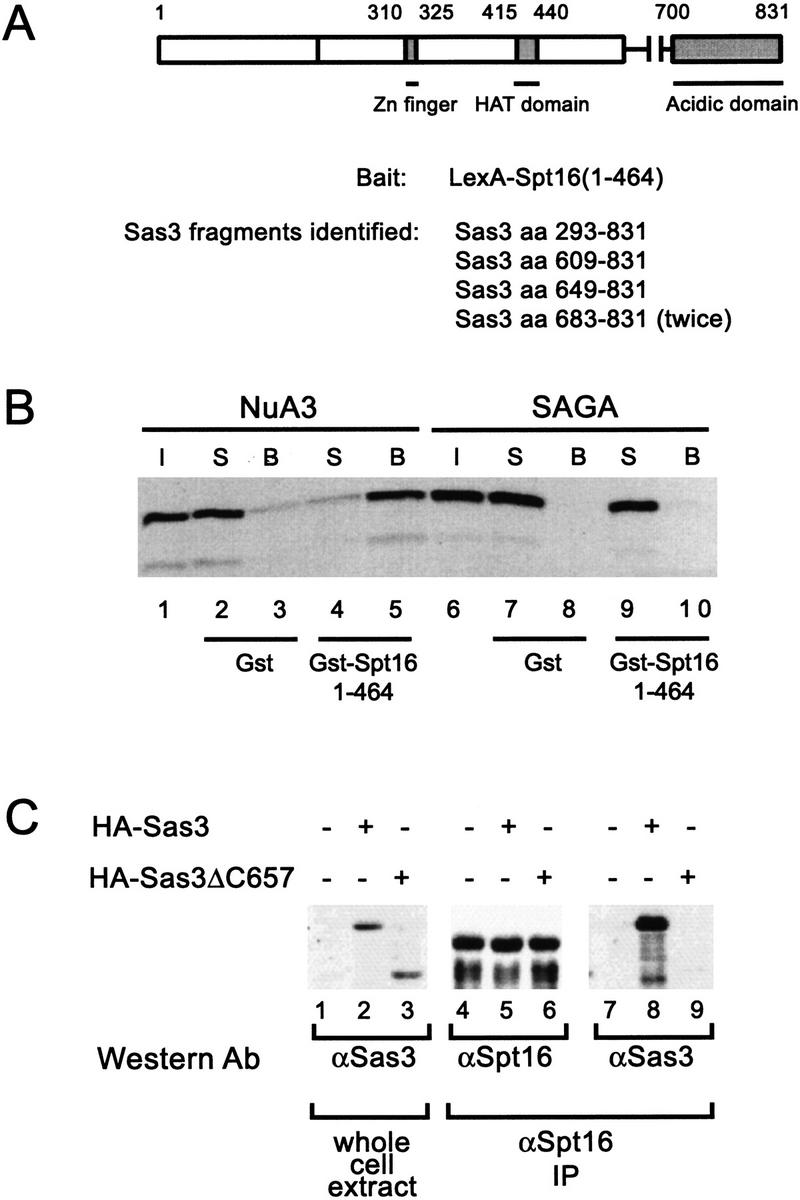
The carboxyl terminus of Sas3 interacts with Spt16, a component of a transcriptional elongation complex. In vivo and in vitro interactions between Sas3 and Spt16. (A) Schematic representation of the Sas3 protein and the Sas3 fragments identified by a two-hybrid screen using a LexA–Spt16 fusion to screen a yeast library. Sas3 fragments identified all share the common acidic carboxy-terminus. (B) Fluorogram of supernatant and beads of NuA3 or SAGA fractions incubated with either Gst or Gst–Spt16. Only NuA3 interacts with Gst–Spt16. I, Input; S, supernatant; B, beads. (C) Co-immunoprecipitation on Sas3 with Spt16. Yeast whole-cell extracts from HA-tagged Sas3 strains were incubated with anti-Spt16 antisera and precipitated with protein A–sepharose beads. Whole-cell extracts and immunoprecipitates were assayed by Western blotting with the antibodies indicated.
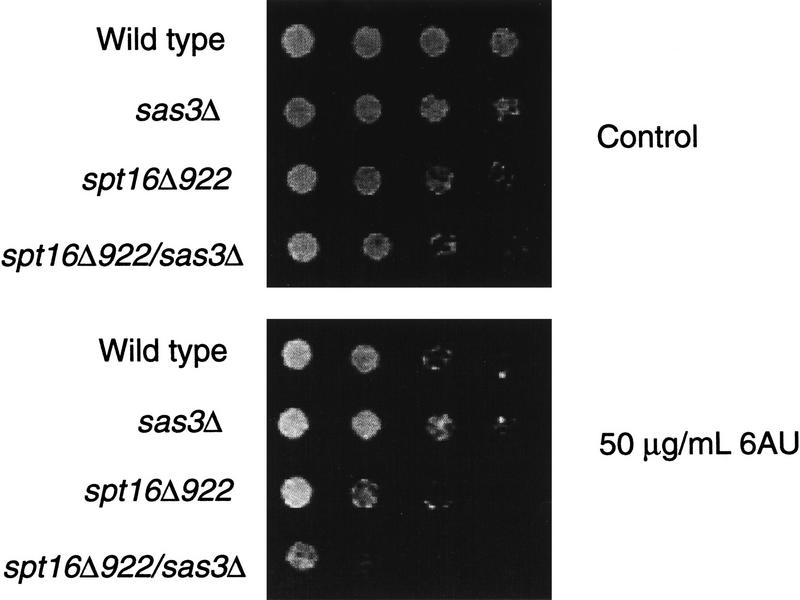
To determine whether the intact NuA3 complex might interact with Spt16 we tested for direct interactions using a Gst–Spt16(1–464N) fusion protein and partially purified NuA3 fractions. As seen in Figure 7B, the NuA3 complex interacted with the amino terminus of Spt16 (Fig. 7B, lanes 4–5) while SAGA failed to interact (Fig. 7B, lanes 9–10). Conversely, identical NuA3 fractions fail to interact with VP16 while SAGA fractions are quantitatively depleted onto Gst–VP16 beads (Utley et al. 1998; data not shown). Gst–Pob3 fusion proteins do not interact with either NuA3 or SAGA (data not shown) further arguing for the specificity of the NuA3–Spt16 interaction. To provide support that a Sas3–Spt16 interaction occurs in vivo, whole-cell extracts were immunoprecipitated with anti-Spt16 polysera and the precipitated material probed for the presence of an HA-tagged Sas3. Figure 7C (lane 8) shows that Sas3 can be co-immunoprecipitated with Spt16 and this association is dependent on the presence of the carboxy-terminal 174 amino acids of Sas3 (lane 9). To provide genetic evidence for an interaction between Spt16 and Sas3, the first 921 nucleotides of the essential SPT16 gene were replaced with a GAL promoter resulting in the production of an amino-terminal truncated Spt16 protein. We find that this spt16 mutant is partially sensitive to the drug 6-azauracil (6AUs; Fig. 8), a phenotype associated with defects in transcription elongation (Exinger and Lacroute 1992). The 6AUs sensitivity of our strain may indicate that the truncated Spt16 protein does not completely complement the function of a wild-type SPT16 gene. The 6AUs phenotype of this strain is greatly enhanced by disruption of the SAS3 gene (Fig. 8), suggesting a further impairment of Spt16 function in this strain. These data suggest that the NuA3 complex might function in concert with FACT–CP to stimulate transcription or replication elongation through nucleosomes by providing a coupled acetyltransferase activity.
Figure 8.
Disruption of SAS3 enhances the 6AU sensitivity of a truncated SPT16 gene. Tenfold serial dilutions of strains with the indicated genotypes were spotted onto synthetic complete media with galactose and with or without (control) 50 μg/ml 6-azauracil. The plates were incubated at 30°C for three days.
Discussion
NuA3 is a Gcn5-independent HAT complex containing Sas3, a MYST family protein
The amino-terminal ends of histones lie outside the limits of core DNA and, therefore, are strategically situated for interactions with chromatin-modifying (HATs) and chromatin-remodeling activities (e.g., SWI/SNF). For example, the amino terminus of histone H2B is involved in the formation of repressive chromatin structures that can be antagonized by SWI/SNF (Recht and Osley 1999). Moreover, the amino terminus of histone H4 has been shown to be required for repression at the silent mating loci, and acetylation of lysine 16 of this histone correlates with derepression (Johnson et al. 1992). Numerous studies over the years have established a strong association between acetylation of the amino termini of histones and transcription (Allfrey et al. 1964; Turner and O'Neill 1995; Workman and Kingston 1998). Acetylation of histone tails has been shown to weaken interactions between the global repressor Tup1 and histone H3 and H4 tails (Edmondson et al. 1996) and promote the binding of activator proteins and basal transcription factors to chromatin templates (Imbalzano et al. 1994; Vettese-Dadey et al. 1996).
We have identified four high-molecular-weight Type A HAT complexes in yeast. Three of these complexes (ADA, NuA3, and SAGA) acetylate primarily histone H3 while NuA4 acetylates primarily histone H4. The 0.4–0.5 MD NuA3 complex is a Gcn5-independent HAT complex, as gcn5 deletions do not affect the activity or integrity of the complex (Fig. 1). Peptide sequencing of proteins from purified NuA3 fractions identified the yeast protein Sas3 as a component of the complex. Sas3 belongs to the MYST family of proteins and shares multiple regions of homology with the related yeast protein Sas2 and the human proteins MOZ and Tip60 (Borrow et al. 1996; Reifsnyder et al. 1996; Yamamoto and Horikoshi 1997). In addition, these proteins share a remarkable degree of similarity with the GNAT family of proteins in the conserved HAT domain (Reifsnyder et al. 1996; Neuwald and Landsman 1997). To date, there is no evidence for HAT activity associated with either Sas2 or MOZ. In fact, recombinant Sas2 has thus far failed to acetylate histones in vitro (S.Tatrov and R. Sternglanz, unpubl.).
Sas2 was identified in a yeast genetic screen conducted to identify enhancers of sir1 epigenetic silencing defects (Reifsnyder et al. 1996). Sas3, in turn, was identified as a homolog of Sas2. Disruption of SAS3 restores silencing to a derepressed HMR locus, suggesting that at this locus, Sas3 functions in transcription activation. In contrast, similar to sas2, sas3 mutants enhance the silencing defects of the HML locus in a sir1 background, albeit to a lesser extent than sas2. This result suggests that, depending on gene context, Sas3 can function as either an activator or repressor of transcription. Similar gene-specific activating and repressive functions have been ascribed to Rap1 depending on the context of the gene examined (Shore 1994). Moreover, this observation is analogous to the observation that transcription of yeast genes is both increased and decreased in a gcn5-deleted strain (Holstege et al. 1998; Spellman et al. 1998).
Sas3 is the acetyltransferase subunit of a multisubunit complex
Our results identify the Sas2-related protein, Sas3, as being the HAT component of the multisubunit NuA3 complex. There are several lines of evidence that lead us to this conclusion. First, a deletion of the entire SAS3 gene specifically abolishes NuA3 activity (Fig. 3A). Second, Sas3 is a bona fide stable component of the HAT complex as demonstrated by immunoprecipitations, thereby, ruling out indirect effects (Fig. 3B, C). Third, point mutations within the HAT domain of Sas3 inactivate the HAT activity of the complex but maintain the overall integrity of the complex. Fourth, a bacterially expressed recombinant form of Sas3 can acetylate histones in vitro (Takechi and Nakayama 1999).
We have also identified TAF30 as a component of the NuA3 complex. TAF30 is an integral member of at least three other yeast transcription-related complexes, TFIID, TFIIF, and SWI/SNF. Coelution and immunoprecipitations (Figs. 4B and 6) demonstrate the stable incorporation of TAF30 into the Sas3-containing HAT complex. TAF30 is quite remarkable in that it is an abundant TAF, present in numerous regulatory complexes, and yet, unlike the other TAFs, is not essential for cell viability. TAF30 appears to be dispensable for basal level transcription or transcription mediated by certain activators (Cairns et al. 1996a). taf30 deletions do not affect the structural integrity of the SWI/SNF complex and a taf30-deficient SWI/SNF complex is intact and catalytically active. Similarly, taf30 deletions result in only modest decreases in NuA3 HAT activity and the size of the complex is unchanged relative to that of wild-type (S. John and J.L. Workman, data not shown). The role of TAF30 in NuA3 and other complexes remains unclear. As has been postulated, it may play a role in facilitating nonessential functions such as promoting binding of these complexes to chromatin components or to the transcriptional machinery (Cairns et al. 1996a).
Sas3–Spt16 interactions link NuA3 to transcription and DNA replication
Previously, we have demonstrated the ability of yeast HATs to stimulate transcription on chromatin templates in an acetyl-CoA-dependent fashion (Steger et al. 1998). Recent studies have shown that direct interactions between chromatin-associated transcriptional activators and HATs target acetylation to proximal nucleosomes leading to gene-specific transcription (Utley et al. 1998).
Studies examining genes regulated by Gcn5 in vivo have demonstrated that hyperacetylated histones are typically localized to defined regions proximal to the promoter (Cosma et al. 1999; Krebs et al. 1999; Kuo et al. 1998). However, earlier studies have shown that broad regions of transcriptionally active chromatin are associated with hyperacetylated histones (Hebbes et al. 1994; O'Neill and Turner 1995). It is possible that distinct HAT complexes are responsible for targeted acetylation of specific genes within a transcriptionally active domain. An intriguing alternative is that there are HAT complexes that function in an elongation-dependent fashion, thereby, spreading hyperacetylation over an entire transcription domain. The association of the NuA3 complex with Spt16 might suggest a role for this HAT in elongation-associated histone acetylation. Interestingly, the FACT complex has been shown to enhance transcription from chromatin templates in a highly purified system in the absence of any added HAT complex (LeRoy et al. 1998; Orphanides et al. 1998). Furthermore, it was also postulated that the mode of action of the FACT complex is to dissociate histones H2A and H2B from nucleosomal templates during transcription (Orphanides et al. 1999). Thus, although FACT does possess some ability on its own to facilitate transcription through chromatin templates, the recruitment of the histone H3-specific HAT complex NuA3 might provide an additional mechanism whereby the FACT complex can modify chromatin structure.
LeRoy et al. (1998) have shown that FACT-dependent transcriptional elongation only requires a remodeled promoter template, and remodeling complexes like SWI/SNF have been implicated in transcriptional elongation (Brown et al. 1996; LeRoy et al. 1998; Wilson et al. 1996). Analysis of the HO gene provides strong evidence for the interplay between remodeling complexes and HATs (Cosma et al. 1999; Krebs et al. 1999). DNA-bound factors at the HO promoter recruit remodeling complexes like SWI/SNF, which in turn facilitates the targeting of HATs to the same promoter. Therefore, the two types of complexes appear to function in sequential and separate steps (Krebs et al. 1999). Thus, multiple combinations of distinct activities may be needed to overcome the chromatin hurdle at different promoters (John and Workman 1998). Interactions between FACT–CP and the replicative DNA polymerase α, Pol1, suggest an additional role for this complex in DNA replication. Indeed, depletion of a related Xenopus complex (DUF) from Xenopus egg extracts has been shown to drastically reduce the DNA and chromatin replication potential of the egg extract (Okuhara et al. 1999). Therefore, complexes like FACT, working in conjunction with HATs, may represent yet another novel combination of activities that allow for efficient transcription and/or replication on chromatin templates. The identification of NuA3-dependent genes and the establishment of in vitro assays using defined components will enable us to address targeting and transcription or replication mechanisms mediated by NuA3–Spt16 interactions.
Material and methods
Yeast strains and manipulations
Yeast strain CY396 (swi2::HIS3, HO–LacZ, SWI2-HA-6HIS::URA3; Peterson et al. 1994) was used for the biochemical purification of the NuA3 complex. Yeast strain PSY316 (MAT αade2-101 his3-200 leu2-3,112 lys2 ura3-52) and its derivative PSY316Δgcn5 have been described previously (Candau et al. 1996). LPY5 (MAT a ade2-1 can 1-100 his 3-11,15 leu2-3,112 trp1-1 ura 3-1) and its derivative LPY1590 (Δsas3) have also been described previously (Reifsnyder et al. 1996). The YJW115-118 strains are all derivatives of LPY1590 (Δsas3) that contain CEN/ARS LEU2 plasmids bearing wild-type or mutant forms of a carboxy terminal Flag-tagged SAS3 gene under the control of its own promoter. All mutations are within the highly conserved HAT domain of Sas3. YJW115 contains the wild-type SAS3 gene; YJW116, YJW117, and YJW118 contain Sas3 HAT mutations M1, M2, and M3, respectively (for details, see Fig. 5A). The YJW119 and 120 strains are also derivatives of LPY1590 that contain CEN/ARS TRP1 plasmids bearing wild-type and carboxy-terminal truncated forms, respectively, of an amino-terminal HA-tagged SAS3 under the control of a GAL promoter. Disruption strains sas3Δ (YJW121), spt16Δ922 (YJW122), and sas3Δspt16Δ922 (YJW123), were derived from FY602 (MAT a, his3Δ200, leu2Δr1, lys2–128Δ, ura3-52, trp1Δ63) as described previously (Longtine et al. 1998).
Standard yeast manipulations were performed as described (Ausubel 1995). Yeast grown in minimal medium were grown in 0.67% yeast nitrogen base without amino acids, 2% dextrose, and supplemented with the appropriate amino acids as needed.
Epitope tagging of Sas3 and generation of mutants
Plasmids expressing Sas3 from its endogenous promoter were created by PCR amplification of the SAS3-coding sequences along with 1 kb of upstream promoter sequence from yeast genomic DNA. The amplified product was ligated into pRS415 (CEN/ARS LEU2) along with a Flag–Cyc terminator cassette that placed the epitope tag at the carboxy terminus of the SAS3 gene. The construct is called pS3FLG. Mutants within the HAT domain of the SAS3 gene (pS3FLGM1-M3) were constructed using the QuikChange site-directed mutagenesis kit (Stratagene) following manufacturer recommended protocols. All mutations were verified by sequencing over the region of change. Plasmids expressing HA-tagged Sas3 under the control of a GAL promoter were prepared using the Univector Plasmid Fusion System as described elsewhere (Liu et al. 1998).
Purification of the NuA complex
Approximately 300 liters of yeast strain CY396 was grown to mid-log phase in a fermentor. Whole-cell extracts from 90 liters of cells were prepared in a scaled-up version of a previously described protocol (Eberharter et al. 1998). The resulting 350 ml of extract was bound to 90 ml of Ni2+–NTA agarose resin (Qiagen) on a rotating wheel for 12 hr at 4°C. Then, the resin was washed in a column with extraction buffer [40 mm HEPES (pH 7.5), 350 mm NaCl, 10% glycerol, 0.1% Tween-20, 2 μg/ml leupeptin, 2 μg/ml pepstatin A, 5 μg/ml aprotinin, and 1 mm PMSF] and bound proteins eluted with three column volumes (∼270 mls) of 300 mm imidazole buffer containing 100 mm NaCl, 10% glycerol, 0.1% Tween-20, and protease inhibitors as described above. The imidazole eluate was loaded onto a 20-ml MonoQ column HR 16/10 (Pharmacia). Bound proteins were eluted in a 25-column-volume gradient from 100 mm to 500 mm NaCl in buffer containing 50 mm Tris (pH 8.0), 10% glycerol, 0.1% Tween-20, 1 mm DTT and protease inhibitors using a Pharmacia FPLC system (LCC-500). Peak NuA3 fractions were identified by liquid HAT assays and fluorography (see methods below) and the active fractions pooled, diluted to 100 mm NaCl, and concentrated over a 1-ml MonoQ HR 5/5 column (Pharmacia) using a similar gradient as described above. NuA3 fractions were dialyzed into 50 mm HEPES (pH 7.5), 100 mm NaCl, 0.1% Tween-20, 1 mm DTT and protease inhibitors (Buffer A) and applied to a Mono S column HR 5/5 (Pharmacia). Bound proteins were eluted in a 10-column-volume linear gradient from 100 mm to 1 mm NaCl. Peak fractions were dialyzed into Buffer A and loaded onto a 1-ml histone–agarose column. Proteins were eluted in a 10-column-volume gradient from 100 mm to 1 m NaCl in Buffer A. NuA3 fractions were pooled, dialyzed, and applied to a 1-ml heparin–Sepharose column, and a similar 10-column-volume gradient was used. Peak fractions were concentrated in a Centricon-30 (Amicon) to ∼400 μl and put over a 24-ml gel-filtration column (Superose 6 HR 10/30; Pharmacia). The Superose 6 calibration in Buffer A is as follows: blue dextran (void volume, fractions 14 and 15), thyroglobulin (669 kD, fraction 22), ferritin (440 kD, fraction 27), aldolase (220 kD, fractions 29 and 30), and ovalbumin (45 kD, fraction 32). Peak Superose 6 fractions (fractions 26–29) were pooled and run on a preparative 10% SDS-PAGE gel. Following Coomassie staining, bands were excised and subject to peptide sequencing (see sequence below). A single peak Superose 6 fraction was diluted to 100 mm NaCl in Buffer A (lacking NaCl) and loaded onto a 240 μl MiniQ PC 3.2/3 column on a SMART system (Pharmacia). Proteins were eluted with a 20-column-volume gradient from 100 mm to 500 mm NaCl and assayed for HAT activity. The volume of all column fractions was 500 μl. The protein profile of the peak MiniQ fraction is shown as a silver stain gel in Figure 2B. The estimated fold purification of the NuA3 complex is ∼100,000.
Peptide sequence obtained for Sas3 is from amino acids 799–813 (KGQEQDENDIESHIR). The TAF30 peptide sequence is from amino acids 181–201 (LNEDDLVGVVQMVDT).
Preparation of extracts from various mutant strains was performed essentially as described previously (Eberharter et al. 1998). NuA3 fractions from these strains are only partially purified (three columns). Briefly, 6 liters of the appropriate strain were grown to mid-log phase, disrupted, and whole-cell extracts bound to nickel–agarose. Following fractionation over a MonoQ column, peak NuA3 fractions were purified further over a Superose 6 sizing column (see above).
HAT assays
HeLa core histones and oligonucleosomes were isolated as described previously (Côté et al. 1995). HAT assays were performed as follows: 1 μg of HeLa core histones or oligonucleosomes was incubated with column fractions and 3H-labeled acetyl-CoA (0.125 μCi) in HAT buffer [50 mm Tris-HCl (pH 8.0)], 50 mm KCl, 5% glycerol, 0.1 mm EDTA, 1 mm DTT, 1 mm PMSF and 10 mm sodium butyrate] at 30°C for 30 min. The reactions were stopped with Laemmli buffer, loaded onto 18% SDS-PAGE gels and electrophoresed at 150 V for 90 min. Gels were stained, destained, and fluorographed with Enhance (Dupont, NEN) according to the manufacturer's recommendations. Typical exposure times were 2–4 days.
Antibodies, Western blots, immunoprecipitations, and GST pulldowns
Standard Western blotting conditions involved electrophoresis of 15 μl of a column fraction on 10% SDS-PAGE gels. Proteins were transferred to nitrocellulose at 100 V for 2 hr and processed for immunoblotting according to standard protocols (Ausubel 1995). In some cases, slower transfers allowed for better signal detection (20 V for 10 hr).
TAF30 antibody is a polyclonal antibody raised to the entire protein (J. Reese, pers. comm.). A monoclonal antibody to the Flag epitope, M2, (Sigma) was used in Western blots at concentrations of 1 μg/ml. All antibodies were incubated with blots for 12 hr at room temperature on a rocking platform. Typical exposure times to visualize Flag signals were 10–15 min.
Immunoprecipitations with TAF30 antibody were performed with 10 μl of polyclonal serum bound to 20 μl of Buffer A-equilibrated protein A–Sepharose beads (Pharmacia). Antibody-bound beads were washed in Buffer A and mixed with 40 μl of pooled Superose 6 column fractions diluted to Buffer A conditions (∼15,000 dpm of HAT activity total). Samples were incubated on a rotating wheel for 2 hr at 4°C. Beads were gently spun down at 2000 rpm for 2 min and then washed three times with 100 μl (five bed volumes) of Buffer A. Beads were resuspended in 20 μl of Buffer A and proportional amounts of the supernatant and beads used in HAT assays or Western blots (as described above). Flag immunoprecipitations were performed with 5 μl of M2 antibody bound to 20 μl of protein A–Sepharose beads and processed as described above for the TAF30 antibody. In some cases, cross-linking the antibody to protein A–Sepharose resin decreased the background and enhanced the clarity of the Western blot signal. Cross-linking was performed with dimethylpimelidate (Sigma) using previously described protocols (Harlow and Lane 1988) or purchased (M2–agarose, Sigma). Preparation and immunoprecipitation of yeast whole-cell extracts was as described previously (Saleh et al. 1997). For the GST-pulldown assay, 20 μl of GST-fusion protein (∼2–3 μg protein) bound to glutathione–sepharose beads equilibrated in Buffer A, was incubated with pooled Superose 6 HAT fractions diluted to Buffer A conditions (∼15,000 dpm total HAT activity in input). Fusion protein and HATs were incubated at 4°C with rotation and then processed in a manner similar to that described for immunoprecipitations (see above).
Two-hybrid screening
Two-hybrid screens were performed exactly as described (Park and Sternglanz 1999). A LexA–Spt16 hybrid containing the amino-terminal 464 amino acids of Spt16 was used to screen a yeast two-hybrid library (James et al. 1996). After screening, only those candidates that were β-galactosidase positive with the original LexA target (and not with other LexA hybrids) were analyzed further (Park and Sternglanz 1999). The library plasmids of relevant clones were isolated and sequenced.
Acknowledgments
We gratefully acknowledge the valuable comments provided by members of the Workman, Simpson, and Reese Laboratories. We thank Lorraine Pillus for providing the LPY family of yeast strains and sharing unpublished information. We are also grateful to Joe Reese for the TAF30 antisera, Tim Formosa for the Spt16 antisera, and Steve Elledge for the Univector Plasmid Fusion System. We thank Song Tan for modeling the HAT domain mutants and Bill Lane (Harvard Microchemistry Facility) for peptide sequencing. In addition, we would like to acknowledge Richard Petrik and Kristina Havas for technical assistance during the course of these experiments. Support for this work was provided by grants from the National Institute of General Medical Sciences (NIGMS) to J.L.W and R.S. S.J. was a postdoctoral associate of the HHMI. L.H. is supported by a post-doctoral fellowship from the Medical Research Council of Canada. P.A.G. is funded by a postdoctoral fellowship from the American Cancer Society. J.L.W. is an Associate Investigator of the HHMI.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL jlw10@psu.edu; FAX (814) 863-0099.
References
- Allfrey VG, Faulkner R, Mirsky AE. Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc Natl Acad Sci. 1964;51:786–794. doi: 10.1073/pnas.51.5.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM. Short protocols in molecular biology: A compendium of methods from Current protocols in molecular biology. NY: John Wiley & Sons; 1995. [Google Scholar]
- Bannister AJ, Kouzarides T. The CBP coactivator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- Borrow J, Stanton VPJ, Andresen JM, Becher RB, Chaganti FG, RS, Civin CI, Disteche C, Dube I, Frischauf AM, Horsman D, Mitelman F, Volinia S, Watmore AE, Housman DE. The translocation t(8;16)(p11;p13) of acute myeloid leukaemia fuses a putative acetyltransferase to the CREB-binding protein. Nat Genet. 1996;14:33–41. doi: 10.1038/ng0996-33. [DOI] [PubMed] [Google Scholar]
- Brewster NK, Johnston GC, Singer RA. Characterization of the CP complex, an abundant dimer of Cdc68 and Pob3 proteins that regulates yeast transcriptional activation and chromatin repression. J Biol Chem. 1998;273:21972–21979. doi: 10.1074/jbc.273.34.21972. [DOI] [PubMed] [Google Scholar]
- Brown SA, Imbalzano AN, Kingston RE. Activator-dependent regulation of transcriptional pausing on nucleosomal templates. Genes & Dev. 1996;10:1479–1490. doi: 10.1101/gad.10.12.1479. [DOI] [PubMed] [Google Scholar]
- Brownell JE, Allis CD. An activity gel assay detects a single, catalytically active histone acetyltransferase subunit in Tetrahymena macronuclei. Proc Natl Acad Sci. 1995;92:6364–6368. doi: 10.1073/pnas.92.14.6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownell JE, Zhou J, Ranalli T, Kobayashi R, Edmondson DG, Roth SY, Allis CD. Tetrahymena histone acetyltransferase A: A homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- Cairns BR, Henry LN, Kornberg RD. TFG3/TAF30/ANC1, a component of the yeast SWI/SNF complex that is similar to the leukemeogenic proteins ENL and AF-9. Mol Cell Biol. 1996a;16:3308–3316. doi: 10.1128/mcb.16.7.3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns BR, Lorch Y, Li Y, Lacomis L, Erdjument-Bromage H, Tempst P, Laurent B, Kornberg RD. RSC, an essential, abundant chromatin-remodeling complex. Cell. 1996b;87:1249–1260. doi: 10.1016/s0092-8674(00)81820-6. [DOI] [PubMed] [Google Scholar]
- Candau R, Moore PA, Wang L, Barlev N, Ying CY, Rosen CA, Berger SL. Identification of human proteins functionally conserved with the yeast putative adaptors ADA2 and GCN5. Mol Cell Biol. 1996;16:593–602. doi: 10.1128/mcb.16.2.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Lin RJ, Schiltz RL, Chakravarti D, Nash A, Nagy L, Privalsky ML, Nakatani Y, Evans RM. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- Clarke AS, Lowell JE, Jacobson SJ, Pillus L. Esa1p is an essential histone acetyltransferase required for cell cycle progression. Mol Cell Biol. 1999;19:2515–2526. doi: 10.1128/mcb.19.4.2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosma MP, Tanaka T, Nasmyth K. Ordered recruitment of transcription and chromatin remodeling factors to cell cycle and developmentally regulated promoters. Cell. 1999;97:299–311. doi: 10.1016/s0092-8674(00)80740-0. [DOI] [PubMed] [Google Scholar]
- Côté J, Utley RT, Workman JL. Basic Analysis of transcription factor binding to nucleosomes. Methods MolGenet. 1995;6:108–129. doi: 10.1016/s0076-6879(96)74024-7. [DOI] [PubMed] [Google Scholar]
- Dutnall RN, Tafrov ST, Sternglanz R, Ramakrishnan V. Structure of the histone acetyltransferase Hat1: A paradigm for the Gcn5-related N-acetyltransferase superfamily. Cell. 1998;94:427–438. doi: 10.1016/s0092-8674(00)81584-6. [DOI] [PubMed] [Google Scholar]
- Eberharter A, John S, Grant PA, Utley RT, Workman JL. Identification and analysis of yeast nucleosomal histone acetyltransferase complexes. Methods. 1998;15:315–321. doi: 10.1006/meth.1998.0635. [DOI] [PubMed] [Google Scholar]
- Eberharter A, Sterner D, Schieltz D, Hassan A, Yates J, III, Berger S, Workman J. The ADA complex is a distinct histone acetyltransferase complex in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:6621–6631. doi: 10.1128/mcb.19.10.6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmondson DG, Smith MM, Roth SY. Repression domain of the yeast global repressor Tup1 interacts directly with histones H3 and H4. Genes & Dev. 1996;10:1247–1259. doi: 10.1101/gad.10.10.1247. [DOI] [PubMed] [Google Scholar]
- Exinger F, Lacroute F. 6-Azauracil inhibition of GTP biosynthesis in Saccharomyces cerevisiae. Curr Genet. 1992;22:9–11. doi: 10.1007/BF00351735. [DOI] [PubMed] [Google Scholar]
- Grant PA, Berger SL. Histone acetyltransferase complexes. Sem Cell Dev Biol. 1999;10:169–177. doi: 10.1006/scdb.1999.0298. [DOI] [PubMed] [Google Scholar]
- Grant PA, Duggan L, Côté J, Roberts SM, Brownell JE, Candau R, Ohba R, Owen-Hughes T, Allis CD, Winston F, Berger SL, Workman JL. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: Characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes & Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- Grant PA, Schieltz D, Pray-Grant MG, Yates JRR, Workman JL. The ATM-related cofactor Tra1 is a component of the purified SAGA complex. Mol Cell. 1998;2:863–867. doi: 10.1016/s1097-2765(00)80300-7. [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane D. Antibodies: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1988. pp. 522–523. [Google Scholar]
- Hebbes TR, Clayton AL, Thorne AW, Crane-Robinson C. Core histone hyperacetylation co-maps with generalized DNase I sensitivity in the chicken β-globin chromosomal domain. EMBO J. 1994;13:1823–1830. doi: 10.1002/j.1460-2075.1994.tb06451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holstege FCP, Jennings EG, Wyrick JJ, Lee TI, Hengarter CJ, Green MR, Golub TR, Lander ES, Young RA. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- Ikeda K, Steger DJ, Eberharter A, Workman JL. Activation domain-specific and general transcription stimulation by native histone acetyltransferase complexes. Mol Cell Biol. 1999;19:855–863. doi: 10.1128/mcb.19.1.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imbalzano AN, Kwon H, Green MR, Kingston RE. Facilitated binding of TATA-binding protein to nucleosomal DNA. Nature. 1994;370:481–485. doi: 10.1038/370481a0. [DOI] [PubMed] [Google Scholar]
- Ito T, Bulger M, Pazin MJ, Kobayashi R, Kadonaga JT. ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell. 1997;90:145–155. doi: 10.1016/s0092-8674(00)80321-9. [DOI] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John S, Workman JL. Just the facts of chromatin transcription. Science. 1998;282:1836–1837. doi: 10.1126/science.282.5395.1836. [DOI] [PubMed] [Google Scholar]
- Johnson LM, Fisher-Adams G, Grunstein M. Identification of a nonbasic domain in the histone H4 N-terminus required for repression of the yeast silent mating loci. EMBO J. 1992;11:2201–2209. doi: 10.1002/j.1460-2075.1992.tb05279.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamine J, Elangovan B, Subramanian T, Coleman D, Chinnadurai G. Identification of a cellular protein that specifically interacts with the essential cysteine region of the HIV-1 Tat transactivator. Virology. 1996;216:356–366. doi: 10.1006/viro.1996.0071. [DOI] [PubMed] [Google Scholar]
- Kimura A, Horikoshi M. Tip60 acetylates six lysines of a specific class in core histones in vitro. Genes Cells. 1998;3:789–800. doi: 10.1046/j.1365-2443.1998.00229.x. [DOI] [PubMed] [Google Scholar]
- Kleff S, Andrulis ED, Anderson CW, Sternglanz R. Identification of a gene encoding a yeast histone H4 acetyltransferase. J Biol Chem. 1995;270:24674–24677. doi: 10.1074/jbc.270.42.24674. [DOI] [PubMed] [Google Scholar]
- Krebs JE, Kuo M-H, Allis CD, Peterson CL. Cell cycle-regulated histone acetylation required for expression of the yeast HO gene. Genes & Dev. 1999;13:1412–1421. doi: 10.1101/gad.13.11.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu TK, Wang Z, Roeder RG. Human TFIIIC relieves chromatin-mediated repression of RNA polymerase III transcription and contains an intrinsic histone acetyltransferase activity. Mol Cell Biol. 1999;19:1605–1615. doi: 10.1128/mcb.19.2.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo M-H, Zhou J, Jambeck P, Churchill MEA, Allis CD. Histone acetyltransferase activity of yeast Gcn5p is required for the activation of target genes in vivo. Genes & Dev. 1998;12:627–639. doi: 10.1101/gad.12.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon H, Imbalzano AN, Khavari P, Kingston RE, Green MR. Nucleosome disruption and enhancement of activator binding by a human SWI/SNF complex. Nature. 1994;370:477–481. doi: 10.1038/370477a0. [DOI] [PubMed] [Google Scholar]
- LeRoy G, Orphanides G, Lane WS, Reinberg D. Requirement of RSF and FACT for transcription of chromatin templates in vitro. Science. 1998;282:1900–1904. doi: 10.1126/science.282.5395.1900. [DOI] [PubMed] [Google Scholar]
- Lin Y, Fletcher CM, Zhou J, Allis CD, Wagner G. Solution structure of the catalytic domain of Gcn5 histone acetyltransferase bound to coenzyme A. Nature. 1999;400:86–89. doi: 10.1038/21922. [DOI] [PubMed] [Google Scholar]
- Liu Q, Li MZ, Leibham D, Cortez D, Elledge SJ. The univector plasmid-fusion system, a method for rapid construction of recombinant DNA without restriction enzymes. Curr Biol. 1998;8:1300–133. doi: 10.1016/s0960-9822(07)00560-x. [DOI] [PubMed] [Google Scholar]
- Loidl P. Histone acetylation: Facts and questions. Chromosoma. 1994;103:441–449. doi: 10.1007/BF00337382. [DOI] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Malone EA, Clark CD, Chiang A, Winston F. Mutations in SPT16/CDC68 suppress cis- and trans-acting mutations that affect promoter functin in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:5710–5717. doi: 10.1128/mcb.11.11.5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizzen CA, Yang X-J, Kokubo T, Brownell JE, Bannister AJ, Owen-Hughes T, Workman JL, Wang L, Berger SL, Kouzarides T, Nakatani Y, Allis CD. The TAFII250 subunit of TFIID has histone acetyltransferase activity. Cell. 1996;87:1261–1270. doi: 10.1016/s0092-8674(00)81821-8. [DOI] [PubMed] [Google Scholar]
- Neuwald AF, Landsman D. Gcn5-related histone N-acetyltransferases belong to a diverse superfamily that includes the yeast Spt10 protein. Trends Biochem Sci. 1997;22:154–155. doi: 10.1016/s0968-0004(97)01034-7. [DOI] [PubMed] [Google Scholar]
- Nightingale KP, Wellinger RE, Sogo JM, Becker PB. Histone acetylation facilitates RNA polymerase II transcription of the Drosophila hsp26 gene in chromatin. EMBO J. 1998;17:2865–2876. doi: 10.1093/emboj/17.10.2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill LP, Turner BM. Histone H4 acetylation distinguishes coding regions of the human genome from heterochromatin in a differentiation-dependent but transcription-independent manner. EMBO J. 1995;14:3946–3957. doi: 10.1002/j.1460-2075.1995.tb00066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuhara K, Ohta K, Seo H, Shioda M, Yamada T, Tanaka Y, Dohmae N, Seyama Y, Shibata T, Murofushi H. A DNA unwinding factor involved in DNA replication in cell-free extracts of Xenopus eggs. Curr Biol. 1999;9:341–350. doi: 10.1016/s0960-9822(99)80160-2. [DOI] [PubMed] [Google Scholar]
- Orphanides G, LeRoy G, Chang CH, Luse DS, Reinberg D. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell. 1998;92:105–116. doi: 10.1016/s0092-8674(00)80903-4. [DOI] [PubMed] [Google Scholar]
- Orphanides G, Wu WH, Lane WS, Hampsey M, Reinberg D. The chromatin-specific transcription elongation factor FACT comprises human SPT16 and SSRP1 proteins. Nature. 1999;400:284–288. doi: 10.1038/22350. [DOI] [PubMed] [Google Scholar]
- Park H, Sternglanz R. Identification and characterization of the genes for two topoisomerase I-interacting proteins from Saccharomyces cerevisiae. Yeast. 1999;15:35–41. doi: 10.1002/(SICI)1097-0061(19990115)15:1<35::AID-YEA340>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Parthun MR, Widom J, Gottschling DE. The major cytoplasmic histone acetyltransferase in yeast: Links to chromatin replication and histone metabolism. Cell. 1996;87:85–94. doi: 10.1016/s0092-8674(00)81325-2. [DOI] [PubMed] [Google Scholar]
- Peterson CL, Dingwall A, Scott MP. Five SWI/SNF gene products are components of a large multisubunit complex required for transcriptional enhancement. Proc Natl Acad Sci. 1994;91:2905–2908. doi: 10.1073/pnas.91.8.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recht J, Osley MA. Mutations in both the structured domain and N-terminus of histone H2B bypass the requirement for Swi-Snf in yeast. EMBO J. 1999;18:229–240. doi: 10.1093/emboj/18.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reifsnyder C, Lowell J, Clarke A, Pillus L. Yeast SAS silencing genes and human genes associated with AML and HIV-1 Tat interactions are homologous with acetyltransferases. Nature Genet. 1996;14:42–49. doi: 10.1038/ng0996-42. [DOI] [PubMed] [Google Scholar]
- Rowley A, Singer RA, Johnston GC. CDC68, a yeast gene that affects regulation of cell proliferation and transcription, encodes a protein with a highly acidic carboxyl terminus. Mol Cell Biol. 1991;11:5718–5726. doi: 10.1128/mcb.11.11.5718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh A, Lang V, Cook R, Brandl CJ. Identification of native complexes containing the yeast coactivator/repressor proteins NGG1/ADA3 and ADA2. J Biol Chem. 1997;272:5571–5578. doi: 10.1074/jbc.272.9.5571. [DOI] [PubMed] [Google Scholar]
- Shore D. RAP1: A protean regulator in yeast. Trends Genet. 1994;10:408–412. doi: 10.1016/0168-9525(94)90058-2. [DOI] [PubMed] [Google Scholar]
- Simpson RT, Thoma F, Brubaker JM. Chromatin reconstituted from tandemly repeated cloned DNA fragments and core histones: A model system for study of higher order structure. Cell. 1985;42:799–808. doi: 10.1016/0092-8674(85)90276-4. [DOI] [PubMed] [Google Scholar]
- Smith ER, Eisen A, Gu W, Sattah M, Pannuti A, Zhou J, Cook RG, Lucchesi JC, Allis CD. ESA1 is a histone acetyltransferase that is essential for growth in yeast. Proc Natl Acad Sci. 1998;95:3561–3565. doi: 10.1073/pnas.95.7.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellman PT, Sherlock G, Zhang MQ, Iyer VR, Anders K, Eisen MB, Brown PO, Botstein D, Futcher B. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer TE, Jenster G, Burcin MM, Allis CD, Zhou J, Mizzen CA, McKenna NJ, Onate SA, Tsai SY, Tsai M-J, O'Malley BW. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature. 1997;389:194–198. doi: 10.1038/38304. [DOI] [PubMed] [Google Scholar]
- Steger DJ, Eberharter A, John S, Grant PA, Workman JL. Purified histone acetyltransferase complexes stimulate HIV-1 transcription from preassembled nucleosomal arrays. Proc Natl Acad Sci. 1998;95:12924–12929. doi: 10.1073/pnas.95.22.12924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner DE, Grant PA, Roberts SM, Duggan L, Belotserkovskaya R, Pacella LA, Winston F, Workman JL, Berger SL. Functional organization of the yeast SAGA complex: Distinct components involved in structural integrity, nucleosome acetylation, and TATA-binding protein interaction. Mol Cell Biol. 1999;19:86–98. doi: 10.1128/mcb.19.1.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes & Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- Takechi S, Nakayama T. Sas3 is a histone acetyltransferase and requires a zinc finger motif. Biochem Biophys Res Commun. 1999;266:405–410. doi: 10.1006/bbrc.1999.1836. [DOI] [PubMed] [Google Scholar]
- Tsukiyama T, Wu C. Purification and properties of an ATP dependent nucleosome remodeling factor. Cell. 1995;83:1011–1020. doi: 10.1016/0092-8674(95)90216-3. [DOI] [PubMed] [Google Scholar]
- Turner BM, O'Neill LP. Histone acetylation in chromatin and chromosomes. Sem Cell Biol. 1995;6:229–236. doi: 10.1006/scel.1995.0031. [DOI] [PubMed] [Google Scholar]
- Utley RT, Ikeda K, Grant PA, Côté J, Steger DJ, Eberharter A, John S, Workman JL. Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature. 1998;394:498–502. doi: 10.1038/28886. [DOI] [PubMed] [Google Scholar]
- Varga–Weisz P, Wilm M, Bonte E, Dumas K, Mann M, Becker PB. Chromatin-remodelling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature. 1997;388:598–602. doi: 10.1038/41587. [DOI] [PubMed] [Google Scholar]
- Vettese-Dadey M, Grant PA, Hebbes TR, Crane-Robinson C, Allis CD, Workman JL. Acetylation of histone H4 plays a primary role in enhancing transcription factor binding to nucleosomal DNA in vitro. EMBO J. 1996;15:2508–2518. [PMC free article] [PubMed] [Google Scholar]
- Wang L, Liu L, Berger SL. Critical residues for histone acetylation by Gcn5, functioning in Ada and SAGA complexes, are also required for transcriptional function in vivo. Genes & Dev. 1998;12:640–653. doi: 10.1101/gad.12.5.640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Côté J, Xue Y, Khavari P, Biggar S, Muchardt C, Kalpana G, Goff S, Yaniv M, Workman JL, Crabtree G. Purification and biochemical heterogeneity of the mammalian SWI–SNF complex. EMBO J. 1996;15:5370–5382. [PMC free article] [PubMed] [Google Scholar]
- Wilson CJ, Chao DM, Imbalzano AN, Schnitzer GR, Kingston RE, Young RA. RNA polymerase II holoenzyme contains SWI/SNF regulators involved in chromatin remodeling. Cell. 1996;84:235–244. doi: 10.1016/s0092-8674(00)80978-2. [DOI] [PubMed] [Google Scholar]
- Winston F, Sudarsanam P. The SAGA of Spt proteins and transcriptional analysis in yeast: Past, present, and future. Cold Spring Harbor Symp Quant Biol. 1998;63:553–561. doi: 10.1101/sqb.1998.63.553. [DOI] [PubMed] [Google Scholar]
- Wittmeyer J, Formosa T. The Saccharomyces cerevisiae DNA polymerase alpha catalytic subunit interacts with Cdc68/Spt16 and with Pob3, a protein similar to an HMG1-like protein. Mol Cell Biol. 1997;17:4178–4190. doi: 10.1128/mcb.17.7.4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman JL, Kingston RE. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu Rev Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- Yamamoto T, Horikoshi M. Novel substrate specificity of the histone acetyltransferase activity of HIV-1-Tat interactive protein Tip60. J Biol Chem. 1997;272:30595–30598. doi: 10.1074/jbc.272.49.30595. [DOI] [PubMed] [Google Scholar]
- Yang X-J, Ogryzko VV, Nishikawa J-I, Howard BH, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]