Abstract
Recent data suggests that the levels of many synaptic proteins may be tightly controlled by the opposing processes of new translation and protein turnover in neurons. Alterations in this balance or in the levels of such dosage-sensitive proteins that result in altered stoichiometry of protein complexes at developing and remodeling synapses may underlie several human cognitive diseases including Fragile X Syndrome, autism spectrum disorders, Angelman syndrome and non-syndromic mental retardation. While a significant amount is known about the transduction of membrane signals to the translational apparatus through kinase cascades acting on general translation factors, much less is understood about how such signals may influence the activity of mRNA-specific regulators, their mechanisms of action and the specific sets of mRNAs they regulate. New approaches to the unbiased in vivo identification of maps of binding sites for these proteins on mRNA is expected to greatly increase our understanding of this crucial level of regulation in neuronal development and function.
Introduction
Normal human cognition is dependent on the proper wiring of the central nervous system during critical periods in development, as well as the maintenance and plasticity of this network in response to experience and insult throughout life. Communication between neurons allows the formation and fine-tuning of neuronal connections to coordinate cellular activity into circuits. A fundamental unit of communication in neuronal networks is the synapse. Synapses are comprised of a relatively well-defined set of proteins many of which function in multi-protein complexes. As such, they may be present in defined stoichiometric ratios arising in some cases from the coordinated synthesis, packaging and delivery of “units” of these multi-protein complexes to axons or dendrites where new or modified synapses are needed. One example of such a cellular strategy for achieving proper stoichiometry is the set of presynaptic scaffolding proteins including bassoon, piccolo, RIM and munc13 [1, 2]. These proteins are synthesized in the neuronal cell body and transported down the axon of the presynaptic cell in “piccolo transport vesicles” or PTVs. In response to signals enticing formation of a new synapse, one or more of these quanta of synaptic proteins is inserted into the presynaptic membrane [1].
Perhaps due to these stoichiometric constraints, alterations in the functional levels of several synaptic proteins are believed to underlie defects in cognition and behavior in human disease. Autism is one example; haplo-insufficiency of Shank3, neurexins or neuroligins, can cause the disease [3, 4]. Shank3 is a scaffolding protein in the postsynaptic density (or PSD, the assembly of postsynaptic proteins of excitatory glutamatergic synapses), and is believed to be a key organizer of the PSD as a component of defined multi-protein complexes [5, 6]. Similarly, haplo-insufficient mutations in neurexins and neuroligins, pre- and post-synaptic cell adhesion molecules mediating synapse formation and stabilization, have been linked to autism spectrum disorders (ASDs), Tourette’s syndrome, schizophrenia and nonspecific learning disabilities [7].
Variation in protein expression levels can also arise in individuals due to de novo microdeletions and microduplications, giving rise to one or three alleles of a gene rather than the usual two (referred to as copy number variations; CNVs) [8]. CNVs have been shown to be much more common than expected; as many as one in eight births harbors a microdeletion and one in fifty, a microduplication [9]. Several large-scale studies in human copy number variation have examined the relationship of such events with cognitive diseases such as the ASDs and schizophrenia [10]. A fascinating conclusion from these studies is that 50% increases in levels of certain dosage-sensitive synaptic proteins is linked to cognitive diseases as well as the more commonly appreciated 50% decreases arising from loss-of-function mutations. Interestingly, many of the individual synaptic proteins whose dysregulation or mutation is related to the ASDs have now been linked to disease through both under- and overexpression. Again, Shank3 is a good example; duplication of the 22q13 region encompassing the Shank3 gene has been linked to severe impairment of social communication [4]. This and similar examples can explained by the gene balance hypothesis, which posits that deleterious phenotypes can arise from under- or overexpression of the same dosage-sensitive proteins because either can disrupt the stoichiometry of the same complex [11–13]. In sum, this evidence supports the concept that the expression levels of many synaptic proteins are critical to the formation and maintenance of proper synaptic function.
The expression level of many synaptic proteins may be tightly controlled by the balance between translation and turnover. The growing number of developmental cognitive diseases whose underlying cause is a defect in the regulation of either translation or turnover suggests that the equilibrium between these opposing processes is a sensitive point in establishing normal cognition and behavior. The first such disease to be characterized was Fragile X Syndrome, caused by a triplet repeat expansion which silences expression of the Fragile X Mental Retardation protein, FMRP, thought to repress neuronal activity-dependent translation [14]. Subsequently, some cases of autism were found to be caused by mutations in PTEN, TSC2 and NF1, three proteins with a shared function to repress the mammalian target of rapamycin (mTOR) pathway that is important for activity-dependent initiation of new translation [15]. On the side of protein turnover, both Angelman syndrome and X-linked syndromic mental retardation have been linked to defects or alterations in expression of the ubiquitin ligases UBE3A and HUWE1 respectively [16–18] which mark proteins for degradation by the proteasome. Taken together, a compelling argument can be made that elucidating the mechanisms regulating neuronal protein translation and turnover are likely to shed light on fundamental aspects of human cognition and neuronal function. This review is focused primarily on the function and regulation of neuronal translation, and the reader is directed to excellent reviews on synaptic activity-regulated protein turnover for a complementary view of this equilibrium [19, 20].
Mechanisms for translational regulation in neurons
While most cells have the ability to alter translation in response to environmental signals neurons have an additional need for specific mechanisms of translational control because of their architecture (Figure 1). This is in part due to a need for spatial control, exemplified by the ability of local groups of synapses to alter their “strength” in response to local input using mechanisms dependent on new protein synthesis in the processes rather than cell body. There is an additional need for local synthesis to solve temporal control issues, as somatic polyribosomes can be a great distance from the site where new proteins are needed to bring about activity-dependent changes. In order to effect control over protein expression, specific mRNAs must be transported to the neuronal processes, requiring translational repression during localization. Following delivery to dendrites or axons these mRNAs may be maintained in a repressed form until synaptic stimulation triggers the local activation of translation to make specific proteins needed for changes in synaptic strength. Finally, mechanisms must exist to halt the translation of dosage-sensitive genes following a burst of synthesis.
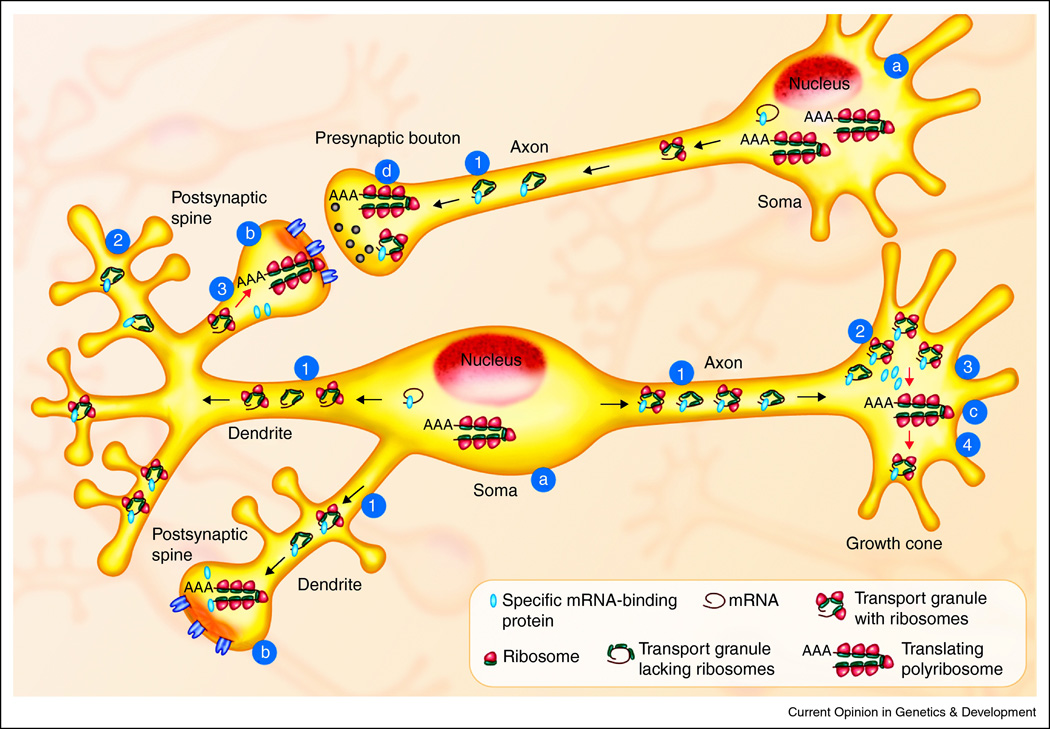
Figure 1. Points of Translational Regulation in Neurons.
Neuronal networks (light blue) have a greater need for spatial and temporal control of translation than many other cell types because of their architecture and need to rapidly alter protein synthesis in response to signals. While the soma was originally believed to be the site of all protein synthesis in the neuron (a) it is now clear that actively translating polyribosomes are present in and near the dendritic spines (the sites of postsynaptic excitatory input, b), in growth cones during development and regeneration after injury (c) and likely on the presynaptic side of synapses as well (d). Localized protein synthesis permits rapid changes in the local proteome at sites distant from the soma but requires delivery of the mRNA templates (black spirals) and synthetic machinery to these sites in the form of transport granules with or without ribosomes (40S and 60S subunits are yellow and blue dots, respectively). The prevailing theory is that specific mRNA-binding proteins (red dots) repress translation during transport (1) and maintain the mRNA in a repressed state until new protein synthesis is needed (2). Mechanisms exist to activate the synthesis of specific proteins in the dendrites and growth cones (3) and finally, specific mechanisms halt their translation as well (4).
Several fundamental pieces of information are needed to understand how activity-dependent translation controls the formation, maintenance and plasticity of synapses in a neuronal network. These include elucidation of the molecular pathways transducing synaptic activity into new protein synthesis, the regulatory factors that stimulate or repress this translation, and the set of activity-dependent plasticity proteins being synthesized.
Translational regulatory factors might be divided into three categories: (1) general translation factors, including the initiation and elongation factors regulated by phosphorylation, (2) sequence-specific RNA binding proteins (RNABPs) and (3) small noncoding RNAs such as miRNAs that regulate translation of specific sets of mRNAs. Two primary pathways for signal transduction from neuronal receptors to these regulatory factors, including the PI3K/Akt/mTOR and MEK/ERK kinase cascades have been extensively reviewed [21–25]. Activation of mTOR affects initiation by phosphorylation of the eIF4E binding proteins (4EBPs) causing their release from eIF4E which increases initiation; activation of the MEK/ERK cascade leads to phosphorylation of eIF4E with the same result. Inhibition of initiation can result from stimuli that cause phosphorylation of eIF2〈 through the activation of eIF2〈 kinases. In addition, inhibition of translation during elongation can be elicited through glutamate receptor activation which leads to eEF2 phosphorylation [26, 27]. Effects on general factors such as eIF4E, 4EBPs, eIF2〈 and eEF2 might be expected to globally alter local translation, although in some cases specificity may be mediated by mechanisms involving competition of specific mRNAs for the translation apparatus [26, 28–31]. Nonetheless, it seems unlikely that general mechanisms of translational control can fully account for the complexity of protein synthesis-dependent synaptic plasticity.
Specific changes in gene expression may also be mediated by mRNA-specific RNABPs or Argonaute(Ago)/microRNA(miRNA) complexes that respond to signaling pathways to fine-tune translation of specific mRNAs or sets of functionally related mRNAs. The identification of relevant RNABPs and the sets of mRNAs that they regulate has been a major hurdle in connecting the compelling evidence for the role of local translation in human cognition and synaptic function with a molecular understanding of its function.
Evaluation of neuronal RNABPs that are implicated in translation regulation underlying neuronal development, activity-dependent synapse formation, stabilization and plasticity should consider several relevant questions: (1) Is there a human disease of cognition or behavior linked to dysfunction of the RNABP? (2) Do bidirectional RNABP level changes in model organisms cause relevant phenotypes such as defects in synaptic plasticity, spine structure, synapse number, or axonal/dendritic morphology? (3) Is the RNABP associated with polyribosomes, stalled initiation complexes, or transport/stress/P-body granules? (4) Is the expression or phosphorylation state of the RNABP activity- or experience-dependent? (5) What is known about how the RNABP regulates translation at the mechanistic level and is this regulation associated with an additional function in regulating stability or localization? This type of analysis suggests several mRNA binding proteins and non-coding RNAs that may serve this function in neurons including FMRP, the CPEBs, pumilio1/2, ZBP1 (and mammalian homologs IGF2BP1–3), caprin1/2, HuB, C and D, Ago/miRNA complexes and the noncoding RNAs BC1 and BC200.
FMRP: an example of an mRNA-specific regulatory protein
Perhaps the most appealing approach to identifying the most relevant mRNA-specific translation factors is to mine the documentation of naturally occurring human mutations in neuronal RNABPs whose function is likely to include translational control and that are causally linked to cognitive and behavioral symptoms. Fragile X Syndrome, characterized by mental retardation, autistic symptoms, and childhood seizures is a model example. In Fragile X patients, a CGG triplet repeat expansion in the 5’UTR of the Fragile X Mental Retardation (FMR1) gene results in loss of expression of the encoded RNA binding protein FMRP [32, 33]. FMRP is recognized to be an RNABP by the presence of three canonical RNA binding domains, two KH-type and a C-terminal RGG box [34] and a role in regulating translation in neurons was suggested by its polyribosome association in brain [35–37]. Loss of polyribosome association due to a point mutation (I304N) in the KH2 RNA binding domain was reported in a severely affected Fragile X Syndrome patient [38, 39] and this mutation was subsequently confirmed to cause a Fragile X phenotype in a knock-in mouse model of the I304N mutation [40]. These observations suggest that loss of the specific function of FMRP in regulating translation in association with polyribosomes underlies Fragile X Syndrome, and has incited a great deal of interest in understanding the mechanisms by which FMRP controls translational and in identifying its mRNA targets.
Loss of function of FMRP has been linked to many defects in synaptic plasticity in a knockout (KO) mouse model of the disease (reviewed by [41–43]) including the finding of enhanced Group 1 metabotropic glutamate receptor-dependent long term depression (mGluR-LTD) in hippocampal CA1 neurons in the Fmr1 KO mouse [44] which led to the “mGluR theory of Fragile X Syndrome”, a compelling explanation for the neurologic and psychiatric aspects of the disease based on known properties of the mGluR signaling pathways [45]. mGluR-LTD is a form of plasticity that requires local postsynaptic protein synthesis in dendrites for its expression [46]. Remarkably, mGluR-LTD loses protein-synthesis dependence in the absence of FMRP [47, 48] suggesting that the plasticity-related proteins needed for LTD expression are already present in excess due to loss of translational repression by FMRP. This hypothesis has been extensively reviewed [14, 41, 49]. FMRP has been implicated in other forms of protein synthesis-dependent long-term plasticity as well, and taken together, the data suggest that FMRP plays a widespread role in regulating synaptic strength in response to activity in the central nervous system by regulating translation (reviewed in [41–43]).
Interest in the function of FMRP was heightened by reports of its rapid synthesis in synaptoneurosomes in response to mGluR activation [50], in hippocampal slices in which mGluR-LTD is induced [51], in mouse brain after behavioral stimulation [52], visual experience [53], or whisker stimulation [54]. The Drosophila homolog, dFMRP, was strongly up-regulated after spaced training which induces a protein synthesis-dependent form of long-term memory [55]. Klann and coworkers demonstrated that FMRP was also rapidly degraded by the proteasome during mGluR-LTD and turnover was necessary for this form of synaptic plasticity [51] suggesting that much of the activity-dependent alteration in FMRP levels might be mediated by a decrease in turnover rate. In light of the importance of proteasome-mediated turnover in plasticity and neurologic disease, further understanding of the fine control of FMRP levels is fundamental to understanding its role in synaptogenesis and plasticity.
An important issue for translational regulation in neurons is to what extent presynaptic regulation of local translation will affect synapse formation and plasticity. A presynaptic function for FMRP has been suggested by localization studies that have found FMRP in axons and growth cones [35, 56, 57] as well as the characterization of axonal growth cone motility [57, 58], elongation [59] and pathfinding [60] defects in both fly and mouse models of Fragile X Syndrome. In addition, there is experimental support for a presynaptic role in synapse formation and the establishment of circuitry [61–64]. Recent studies on the role of the Aplysia FMRP homolog (ApFMRP) in sensory to motor neuron synaptic plasticity supports both a pre- and post-synaptic role for FMRP in regulating protein synthesis in response to synaptic stimulation [65]. A fruitful area for further research is to connect these observations with the set of presynaptic mRNAs whose translation is regulated by FMRP [66].
Historically, two independent studies suggested that FMRP acted to repress translation but were met with some skepticism as neither the FMRP nor the “mRNA” reporters represented endogenous interactions and specificity was lacking [67, 68]. However, several ensuing studies support a role for FMRP as a translational repressor though there is little consensus as to its mechanism of action [59, 69–74]. Two studies addressing mechanism arrived at different conclusions as to whether FMRP inhibits elongation [75] or initiation [76]. Very recent work used in vivo UV-crosslinking and a brain polyribosome-programmed in vitro translation assay designed to preserve endogenous interactions between FMRP and its mRNA targets in neurons. This study found that FMRP interacts along the length of the coding region of target mRNAs and stalls ribosome translocation to repress translation during elongation [66].
Challenges inherent in the identification of sets of regulated mRNAs
While identification of important neuronal regulators of translation (illustrated above by work on FMRP but also including CPEB, pumilio, ZBP, caprin, Hu, Ago/miRNA complexes and the noncoding RNAs BC1 and BC200) through genetic and biochemical approaches has been quite successful, identification of the mRNAs regulated by these binding proteins remains a major hurdle. Previous approaches have included in vitro RNA selection [77, 78] and co-IP followed by either microarray (RIP-Chip, also called “ribonomics”) [79] or by directed PCR for candidate mRNA ligands. In vitro RNA selection studies using fusion proteins with pools of random RNA ligands have been performed in the hope of characterizing high affinity RNA ligands for a given RNABP so that this information can be used to identify in vivo binding sites bioinformatically. While these experiments have been somewhat successful at reproducing in vivo binding motifs [80–82], their bioinformatic identification in vivo still presents a formidable challenge, especially in cases where secondary structure is involved [83, 84] or when high affinity binding requires multimers of a binding motif with variable spacing [85]. Because important cofactors may be missing, the RNA may not fold properly, and binding is influenced by lab buffers the experiment is more similar to a filter binding assay than to the milieu of a cell.
Co-IP of RNABP:RNA complexes is significantly more physiologic, except that once cells are lysed in a particular buffer, and cellular compartments are broken down and contents significantly diluted, the binding of RNABPs to RNA again takes on the character of an equilibrium binding assay, shown experimentally by Joan Steitz and colleagues [86]. In addition, the approach suffers from a failure to identify the sites of RNA binding, and from the need to use relatively gentle IP conditions so as not to lose the RNABP:RNA interaction which precludes the use of very high or low salt buffers, or ionic detergents. This results in a low stringency situation where the RNABP of interest may be IPed in a complex with other RNABPs in addition to the problem of crossreactivity of the antibody with nonspecific proteins under low stringency IP conditions.
Attempts to identify the set of mRNAs whose translation is regulated by FMRP illustrate these issues. FMRP was originally defined as an RNABP by the presence of two KH domains and a C-terminal RGG box [34] and in vitro RNA selection revealed that FMRP binds a G-quadruplex RNA motif through its RGG box [83, 87], and a kissing complex RNA motif through the disease-associated KH2 domain [84]. G-quadruplex forming sequences have been reported in a number of mRNAs, and have been shown to interact with FMRP by in vitro binding assays, but their relevance to in vivo RNA binding remains unclear. Kissing complex RNA structures (kcRNA) cannot be predicted bioinformatically [84] although this motif has been identified structurally in other RNAs [88]. FMRP has been shown to interact with hundreds of neuronal mRNAs by RIP-Chip assays from mouse brain [89] and other attempts to identify its targets have been reviewed [41], however these studies have resulted in little consensus in FMRP target identification.
A significant advance in this area uses UV-crosslinking to introduce a covalent bond between the RNABP and RNA. We have recently combined UV crosslinking of RNABP-RNA complexes in intact cells or tissue with stringent immunoprecipitation (CLIP) to purify RNABPs away from nonspecific cellular RNABPs (Figure 2) [90–92]. Following linker addition the crosslinked RNA can be sequenced by high throughput techniques (HITS-CLIP, also referred to as CLIP-seq), allowing genome-wide assessment of bound RNAs. The advantages of this technique are that 1) UV crosslinking creates a bond between the RNABP and RNA only if the distance between them is within a bond length, in contrast to formaldehyde crosslinking which crosslinks molecules at some distance; 2) because intact tissue is crosslinked the physiologic/endogenous state of RNABP:RNA interactions is preserved; and 3) the covalent crosslink between RNABP and RNA allows the use of very stringent wash conditions to purify the RNABP of interest and remove free RNA. The sum of these, performed correctly, is that the sequenced RNAs represent a snapshot of what the RNABP was bound to in vivo at the time of crosslinking, with very little noise (false positives) in the picture. Because limiting RNAse digestion is used to reduce the size of the RNA “tags” this technique also reveals binding sites. Validation experiments can then be designed to examine altered metabolism (translation, splicing, turnover) of these RNAs in vivo, depending on where the RNA binding sites lie in the transcripts, ideally using animal models lacking the RNABP [93]. Recent studies using HITS-CLIP to identify mRNAs directly bound by FMRP on neuronal polyribosomes have confirmed that approximately 50% of those mRNAs identified by RIP-Chip are directly bound (and therefore likely to be directly regulated) by FMRP and have expanded this set of targets to more than 800 high confidence mRNA targets for FMRP directly bound in vivo [66]. Significantly, a large proportion of these mRNAs are components of either the pre-or postsynaptic proteomes of neurons.
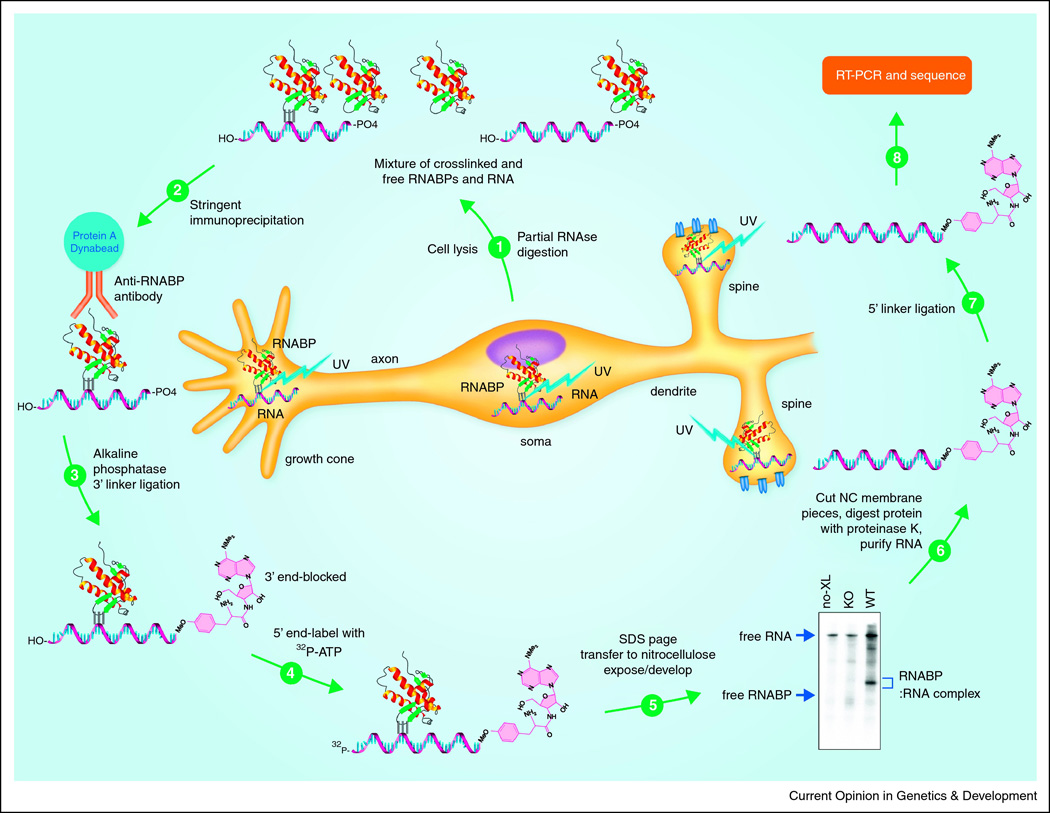
Figure 2. Schematic of the HITS-CLIP protocol to identify the in vivo RNA ligands of a neuronal RNABP.
UV crosslinking at 254 nm (lightning bolts) captures endogenous interactions between RNABPs (colored ribbon structures) interaction with RNA (grey helices) occurring at the time of crosslinking of the tissue. A stylized neuron is shown to illustrate that interactions in the growth cones, dendrites, neuronal soma and other cellular compartments are captured by this technique which creates a covalent bond (triple black lines) between the RNABP and the RNA. 1. Following crosslinking of cells or tissue, cells are lysed and treated with limiting RNAse digestion to reduce the modal size of the RNA “tags” to around 60 nucleotides to permit identification of RNA binding sites with good resolution, and improve purification in subsequent steps. This creates a mixture of RNA fragments, cellular RNABPs and RNABPs crosslinked to their RNA binding sites. The art of CLIP is in the purification of the RNABP of interest from this mixture. 2. The first step in this purification is typically a stringent immunoprecipitation in a buffer that dissociates endogenous RNP complexes. 3. The IP beads are then stringently washed, treated with alkaline phosphatase, and a blocked 3’ RNA linker added. 4. The RNA tags are labeled with T4 polynucleotide kinase and 32P-γATP. 5. A second important purification step is SDS-PAGE gel electrophoresis to separate complexes by size. After transfer to nitrocellulose crosslinked RNABP:32P-RNA complexes can be visualized and success of the experiment evaluated by comparison of a sample (WT) with a control lacking the RNABP of interest (KO) or additional antibodies, both irrelevant (negative control) and directed against other RNABP epitopes on the same RNABP (positive controls). An otherwise identical sample that has not been crosslinked serves as a negative control for contaminating free 32P-RNA. 6. The pieces of nitrocellulose containing complexes of interest, migrating approx. 20 kDa larger than the free RNABP (accounting for the 60 nt RNA tag) are excised, RNABP digested away with proteinase K, and the RNA purified. 7. After ligation of an RNA linker to the 5’end of RNA tags, 8. the tags can be amplified by RT-PCR and products sent for high throughput sequencing (HITS). Resulting sequences can be aligned with the genome or any database of interest, including noncoding RNAs, to identify novel, physiologically relevant maps of RNA binding for a given RNABP. References for a more detailed method for HITSCLIP are given in the text.
Concluding remarks
Application of HITS-CLIP to other specific mRNA-binding proteins implicated in the control of activity-dependent translation in brain, such as CPEB, ZBP, Hu, caprin or pumilio should greatly expand our understanding of the mRNA targets they regulate. Indeed, application to Ago/miRNA complexes in P13 mouse brain has yielded a compelling map of in vivo Ago binding sites on mRNA [92]. Using HITS-CLIP analysis to quantify changes in binding due to activity, or in subcellular fractions such as polyribosomes or purifiable granules, or during development is expected to lead to dramatic advances in our understanding of how these proteins act individually and in concert to control the synthesis of neuronal proteins underlying development and plasticity.
Highlights.
Activity-dependent translation is required for proper synaptic development and plasticity
Specific mRNA-binding proteins including FMRP may regulate such translation in neurons
New methods are being used to identify physiologically relevant mRNA targets of RNABPs
Acknowledgements
I apologize to the many authors whose relevant work could not be cited due to space limitations and thank Robert Darnell, Alicia Darnell and Sarah van Dreische for their critical review of the manuscript during its preparation, as well the past and present members of the Darnell laboratory for their insights. JCD is supported by NICHD grant R01 HD40647.
References and recommended reading
References of special (*) or outstanding (**) interest:
- 1.Shapira M, Zhai RG, Dresbach T, Bresler T, Torres VI, Gundelfinger ED, Ziv NE, Garner CC. Unitary assembly of presynaptic active zones from Piccolo-Bassoon transport vesicles. Neuron. 2003;38:237–252. doi: 10.1016/s0896-6273(03)00207-1. [DOI] [PubMed] [Google Scholar]
- 2.Zhai RG, Vardinon-Friedman H, Cases-Langhoff C, Becker B, Gundelfinger ED, Ziv NE, Garner CC. Assembling the presynaptic active zone: a characterization of an active one precursor vesicle. Neuron. 2001;29:131–143. doi: 10.1016/s0896-6273(01)00185-4. [DOI] [PubMed] [Google Scholar]
- 3.Wilson HL, Wong AC, Shaw SR, Tse WY, Stapleton GA, Phelan MC, Hu S, Marshall J, McDermid HE. Molecular characterisation of the 22q13 deletion syndrome supports the role of haploinsufficiency of SHANK3/PROSAP2 in the major neurological symptoms. J Med Genet. 2003;40:575–584. doi: 10.1136/jmg.40.8.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Durand CM, Betancur C, Boeckers TM, Bockmann J, Chaste P, Fauchereau F, Nygren G, Rastam M, Gillberg IC, Anckarsater H, et al. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat Genet. 2007;39:25–27. doi: 10.1038/ng1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Naisbitt S, Kim E, Tu JC, Xiao B, Sala C, Valtschanoff J, Weinberg RJ, Worley PF, Sheng M. Shank, a novel family of postsynaptic density proteins that binds to the NMDA receptor/PSD-95/GKAP complex and cortactin. Neuron. 1999;23:569–582. doi: 10.1016/s0896-6273(00)80809-0. [DOI] [PubMed] [Google Scholar]
- 6.Boeckers TM, Bockmann J, Kreutz MR, Gundelfinger ED. ProSAP/Shank proteins - a family of higher order organizing molecules of the postsynaptic density with an emerging role in human neurological disease. J Neurochem. 2002;81:903–910. doi: 10.1046/j.1471-4159.2002.00931.x. [DOI] [PubMed] [Google Scholar]
- 7.Sudhof TC. Neuroligins and neurexins link synaptic function to cognitive disease. Nature. 2008;455:903–911. doi: 10.1038/nature07456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang F, Gu W, Hurles ME, Lupski JR. Copy number variation in human health, disease, and evolution. Annu Rev Genomics Hum Genet. 2009;10:451–481. doi: 10.1146/annurev.genom.9.081307.164217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Ommen GJ. Frequency of new copy number variation in humans. Nat Genet. 2005;37:333–334. doi: 10.1038/ng0405-333. [DOI] [PubMed] [Google Scholar]
- 10.Merikangas AK, Corvin AP, Gallagher L. Copy-number variants in neurodevelopmental disorders: promises and challenges. Trends Genet. 2009;25:536–544. doi: 10.1016/j.tig.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 11.Conrad B, Antonarakis SE. Gene duplication: a drive for phenotypic diversity and cause of human disease. Annu Rev Genomics Hum Genet. 2007;8:17–35. doi: 10.1146/annurev.genom.8.021307.110233. [DOI] [PubMed] [Google Scholar]
- 12.Birchler JA, Bhadra U, Bhadra MP, Auger DL. Dosage-dependent gene regulation in multicellular eukaryotes: implications for dosage compensation, aneuploid syndromes, and quantitative traits. Dev Biol. 2001;234:275–288. doi: 10.1006/dbio.2001.0262. [DOI] [PubMed] [Google Scholar]
- 13.Papp B, Pal C, Hurst LD. Dosage sensitivity and the evolution of gene families in yeast. Nature. 2003;424:194–197. doi: 10.1038/nature01771. [DOI] [PubMed] [Google Scholar]
- 14.Bassell GJ, Warren ST. Fragile X syndrome: loss of local mRNA regulation alters synaptic development and function. Neuron. 2008;60:201–214. doi: 10.1016/j.neuron.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kelleher RJr, Bear MF. The autistic neuron: troubled translation? Cell. 2008;135:401–406. doi: 10.1016/j.cell.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 16.Matsuura T, Sutcliffe JS, Fang P, Galjaard RJ, Jiang YH, Benton CS, Rommens JM, Beaudet AL. De novo truncating mutations in E6-AP ubiquitin-protein ligase gene (UBE3A) in Angelman syndrome. Nat Genet. 1997;15:74–77. doi: 10.1038/ng0197-74. [DOI] [PubMed] [Google Scholar]
- 17.Kishino T, Lalande M, Wagstaff J. UBE3A/E6-AP mutations cause Angelman syndrome. Nat Genet. 1997;15:70–73. doi: 10.1038/ng0197-70. [DOI] [PubMed] [Google Scholar]
- 18.Froyen G, Corbett M, Vandewalle J, Jarvela I, Lawrence O, Meldrum C, Bauters M, Govaerts K, Vandeleur L, Van Esch H, et al. Submicroscopic duplications of the hydroxysteroid dehydrogenase HSD17B10 and the E3 ubiquitin ligase HUWE1 are associated with mental retardation. Am J Hum Genet. 2008;82:432–443. doi: 10.1016/j.ajhg.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cajigas IJ, Will T, Schuman EM. Protein homeostasis and synaptic plasticity. EMBO J. 2010;29:2746–2752. doi: 10.1038/emboj.2010.173.. This comprehensive recent review discusses the evidence that both synthesis and turnover of proteins contribute to changes in synaptic plasticity.
- 20.Steward O, Schuman EM. Compartmentalized synthesis and degradation of proteins in neurons. Neuron. 2003;40:347–359. doi: 10.1016/s0896-6273(03)00635-4. [DOI] [PubMed] [Google Scholar]
- 21.Hoeffer CA, Klann E. mTOR signaling: at the crossroads of plasticity, memory and disease. Trends Neurosci. 2010;33:67–75. doi: 10.1016/j.tins.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Richter JD, Klann E. Making synaptic plasticity and memory last: mechanisms of translational regulation. Genes Dev. 2009;23:1–11. doi: 10.1101/gad.1735809. [DOI] [PubMed] [Google Scholar]
- 23.Kelleher RJr, Govindarajan A, Tonegawa S. Translational regulatory mechanisms in persistent forms of synaptic plasticity. Neuron. 2004;44:59–73. doi: 10.1016/j.neuron.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 24.Costa-Mattioli M, Sossin WS, Klann E, Sonenberg N. Translational control of long-lasting synaptic plasticity and memory. Neuron. 2009;61:10–26. doi: 10.1016/j.neuron.2008.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klann E, Dever TE. Biochemical mechanisms for translational regulation in synaptic plasticity. Nat Rev Neurosci. 2004;5:931–942. doi: 10.1038/nrn1557. [DOI] [PubMed] [Google Scholar]
- 26.Scheetz AJ, Nairn AC, Constantine-Paton M. N-methyl-D-aspartate receptor activation and visual activity induce elongation factor-2 phosphorylation in amphibian tecta: a role for N-methyl-D-aspartate receptors in controlling protein synthesis. Proc Natl Acad Sci U S A. 1997;94:14770–14775. doi: 10.1073/pnas.94.26.14770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sutton MA, Taylor AM, Ito HT, Pham A, Schuman EM. Postsynaptic decoding of neural activity: eEF2 as a biochemical sensor coupling miniature synaptic transmission to local protein synthesis. Neuron. 2007;55:648–661. doi: 10.1016/j.neuron.2007.07.030.. This fascinating paper is the culmination of a series of papers by Sutton, Schuman and colleagues in which they demonstrate that NMDAR-dependent mini EPSCs result in a tonic repression of protein synthesis in the postsynaptic compartment. In this latest in the series of papers the authors demonstrate that the mechanism for the observed translational repression involves phosphorylation of the general elongation factor eEF2, presumably stalling ribosomes during the elongation step of translation. This is an important addition to the understanding of general translational control underlying synaptic plasticity, previously demonstrated to primarily affect translation initiation factors.
- 28.Scheetz AJ, Nairn AC, Constantine-Paton M. NMDA receptor-mediated control of protein synthesis at developing synapses. Nat Neurosci. 2000;3:211–216. doi: 10.1038/72915. [DOI] [PubMed] [Google Scholar]
- 29.Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, Ron D. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell. 2000;6:1099–1108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- 30.Vattem KM, Wek RC. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc Natl Acad Sci U S A. 2004;101:11269–11274. doi: 10.1073/pnas.0400541101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Park S, Park JM, Kim S, Kim JA, Shepherd JD, Smith-Hicks CL, Chowdhury S, Kaufmann W, Kuhl D, Ryazanov AG, et al. Elongation factor 2 and fragile X mental retardation protein control the dynamic translation of Arc/Arg3.1 essential for mGluR-LTD. Neuron. 2008;59:70–83. doi: 10.1016/j.neuron.2008.05.023.. This paper finds that the mGluR5-dependent increase in translation of Arc required for mGluR-LTD paradoxically requires phosphorylation of eEF2 which represses general translation. Intriguingly, this pathway is disrupted in the absence of FMRP.
- 32.Verkerk AJ, Pieretti M, Sutcliffe JS, Fu YH, Kuhl DP, Pizzuti A, Reiner O, Richards S, Victoria MF, Zhang FP, et a. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991;65:905–914. doi: 10.1016/0092-8674(91)90397-h. [DOI] [PubMed] [Google Scholar]
- 33.Pieretti M, Zhang FP, Fu YH, Warren ST, Oostra BA, Caskey CT, Nelson DL. Absence of expression of the FMR-1 gene in fragile X syndrome. Cell. 1991;66:817–822. doi: 10.1016/0092-8674(91)90125-i. [DOI] [PubMed] [Google Scholar]
- 34. Siomi H, Siomi MC, Nussbaum RL, Dreyfuss G. The protein product of the fragile X gene, FMR1, has characteristics of an RNA-binding protein. Cell. 1993;74:291–298. doi: 10.1016/0092-8674(93)90420-u.. This paper presented the first evidence that FMRP, whose loss of expression results in Fragile X Syndrome, is an RNA binding protein, and stimulated a great deal of interest in identifying the in vivo RNA targets of FMRP.
- 35.Feng Y, Gutekunst CA, Eberhart DE, Yi H, Warren ST, Hersch SM. Fragile X mental retardation protein: nucleocytoplasmic shuttling and association with somatodendritic ribosomes. J Neurosci. 1997;17:1539–1547. doi: 10.1523/JNEUROSCI.17-05-01539.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stefani G, Fraser CE, Darnell JC, Darnell RB. Fragile X mental retardation protein is associated with translating polyribosomes in neuronal cells. J Neurosci. 2004;24:7272–7276. doi: 10.1523/JNEUROSCI.2306-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khandjian EW, Huot ME, Tremblay S, Davidovic L, Mazroui R, Bardoni B. Biochemical evidence for the association of fragile X mental retardation protein with brain polyribosomal ribonucleoparticles. Proc Natl Acad Sci U S A. 2004;101:13357–13362. doi: 10.1073/pnas.0405398101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. De Boulle K, Verkerk AJ, Reyniers E, Vits L, Hendrickx J, Van Roy B, Van den Bos F, de Graaff E, Oostra BA, Willems PJ. A point mutation in the FMR-1 gene associated with fragile X mental retardation. Nat Genet. 1993;3:31–35. doi: 10.1038/ng0193-31.. This paper describes the only Fragile X patient with a documented point mutation in the gene that leads to the disease. The finding that this mutation involves one of the most highly conserved amino acids in the KH2 RNA binding domain suggested that it was the loss of RNA binding by the FMRP KH2 domain that caused the symptoms of the disease. The mutation was later modeled in a mouse and shown definitively to cause a Fragile X phenotype (ref. [40]) as well as loss of RNA binding and polyribosome association of mutant FMRP in brain.
- 39.Feng Y, Absher D, Eberhart DE, Brown V, Malter HE, Warren ST. FMRP associates with polyribosomes as an mRNP, and the I304N mutation of severe fragile X syndrome abolishes this association. Mol Cell. 1997;1:109–118. doi: 10.1016/s1097-2765(00)80012-x. [DOI] [PubMed] [Google Scholar]
- 40.Zang JB, Nosyreva ED, Spencer CM, Volk LJ, Musunuru K, Zhong R, Stone EF, Yuva-Paylor LA, Huber KM, Paylor R, et al. A mouse model of the human Fragile X syndrome I304N mutation. PLoS Genet. 2009;5:e1000758. doi: 10.1371/journal.pgen.1000758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gatto CL, Broadie K. The fragile X mental retardation protein in circadian rhythmicity and memory consolidation. Mol Neurobiol. 2009;39:107–129. doi: 10.1007/s12035-009-8057-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pfeiffer BE, Huber KM. The state of synapses in fragile X syndrome. Neuroscientist. 2009;15:549–567. doi: 10.1177/1073858409333075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mercaldo V, Descalzi G, Zhuo M. Fragile X mental retardation protein in learning-related synaptic plasticity. Mol Cells. 2009;28:501–507. doi: 10.1007/s10059-009-0193-x. [DOI] [PubMed] [Google Scholar]
- 44. Huber KM, Gallagher SM, Warren ST, Bear MF. Altered synaptic plasticity in a mouse model of fragile X mental retardation. Proc Natl Acad Sci U S A. 2002;99:7746–7750. doi: 10.1073/pnas.122205699.. This paper presents the finding that the Fmr1 knockout mouse model for Fragile X Syndrome has enhanced mGluR-dependent long term depression compared with wild-type littermates. This form of synaptic plasticity is dependent on new local translation for its maintenance, suggesting an important role for FMRP in regulating local protein synthesis to bring about the observed changes in synaptic strength. This paper and the accompanying review [45] discuss the relevance of this phenotype to the symptoms of Fragile X Syndrome.
- 45.Bear MF, Huber KM, Warren ST. The mGluR theory of fragile X mental retardation. Trends Neurosci. 2004;27:370–377. doi: 10.1016/j.tins.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 46.Huber KM, Kayser MS, Bear MF. Role for rapid dendritic protein synthesis in hippocampal mGluR-dependent long-term depression. Science. 2000;288:1254–1257. doi: 10.1126/science.288.5469.1254. [DOI] [PubMed] [Google Scholar]
- 47.Nosyreva ED, Huber KM. Metabotropic receptor-dependent long-term depression persists in the absence of protein synthesis in the mouse model of fragile X syndrome. J Neurophysiol. 2006;95:3291–3295. doi: 10.1152/jn.01316.2005. [DOI] [PubMed] [Google Scholar]
- 48.Zhang J, Hou L, Klann E, Nelson DL. Altered hippocampal synaptic plasticity in the FMR1 gene family knockout mouse models. J Neurophysiol. 2009;101:2572–2580. doi: 10.1152/jn.90558.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dolen G, Bear MF. Role for metabotropic glutamate receptor 5 (mGluR5) in the pathogenesis of fragile X syndrome. J Physiol. 2008;586:1503–1508. doi: 10.1113/jphysiol.2008.150722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weiler IJ, Irwin SA, Klintsova AY, Spencer CM, Brazelton AD, Miyashiro K, Comery TA, Patel B, Eberwine J, Greenough WT. Fragile X mental retardation protein is translated near synapses in response to neurotransmitter activation. Proc Natl Acad Sci U S A. 1997;94:5395–5400. doi: 10.1073/pnas.94.10.5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hou L, Antion MD, Hu D, Spencer CM, Paylor R, Klann E. Dynamic translational and proteasomal regulation of fragile X mental retardation protein controls mGluR-dependent long-term depression. Neuron. 2006;51:441–454. doi: 10.1016/j.neuron.2006.07.005.. This paper found that induction of protein-synthesis dependent mGluR-LTD in hippocampus was accompanied by a transient increase in the translation of FMRP followed by turnover of FMRP mediated by ubiquitination and the proteasome. Both were found to be required for this form of synaptic plasticity. This important discovery suggests that the local level of FMRP at synapses regulates expression of mGluR-LTD.
- 52.Irwin SA, Swain RA, Christmon CA, Chakravarti A, Weiler IJ, Greenough WT. Evidence for altered Fragile-X mental retardation protein expression in response to behavioral stimulation. Neurobiol Learn Mem. 2000;74:87–93. [PubMed] [Google Scholar]
- 53.Gabel LA, Won S, Kawai H, McKinney M, Tartakoff AM, Fallon JR. Visual experience regulates transient expression and dendritic localization of fragile X mental retardation protein. J Neurosci. 2004;24:10579–10583. doi: 10.1523/JNEUROSCI.2185-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Todd PK, Malter JS, Mack KJ. Whisker stimulation-dependent translation of FMRP in the barrel cortex requires activation of type I metabotropic glutamate receptors. Brain Res Mol Brain Res. 2003;110:267–278. doi: 10.1016/s0169-328x(02)00657-5. [DOI] [PubMed] [Google Scholar]
- 55.Bolduc FV, Bell K, Cox H, Broadie KS, Tully T. Excess protein synthesis in Drosophila fragile X mutants impairs long-term memory. Nat Neurosci. 2008;11:1143–1145. doi: 10.1038/nn.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Christie SB, Akins MR, Schwob JE, Fallon JR. The FXG: a presynaptic fragile X granule expressed in a subset of developing brain circuits. J Neurosci. 2009;29:1514–1524. doi: 10.1523/JNEUROSCI.3937-08.2009.. This paper is an excellent example of a well-controlled protein localization study. Christie and colleagues used multiple antibodies against FMRP as a positive control and Fmr1 knockout littermates as a negative control for experiments that convincingly demonstrated FMRP localization to axonal granules as well as the dendrites and soma of neurons.
- 57.Antar LN, Li C, Zhang H, Carroll RC, Bassell GJ. Local functions for FMRP in axon growth cone motility and activity-dependent regulation of filopodia and spine synapses. Mol Cell Neurosci. 2006;32:37–48. doi: 10.1016/j.mcn.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 58.Li C, Bassell GJ, Sasaki Y. Fragile X Mental Retardation Protein is Involved in Protein Synthesis-Dependent Collapse of Growth Cones Induced by Semaphorin-3A. Front Neural Circuits. 2009;3:11. doi: 10.3389/neuro.04.011.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tessier CR, Broadie K. Drosophila fragile X mental retardation protein developmentally regulates activity-dependent axon pruning. Development. 2008;135:1547–1557. doi: 10.1242/dev.015867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Michel CI, Kraft R, Restifo LL. Defective neuronal development in the mushroom bodies of Drosophila fragile X mental retardation 1 mutants. J Neurosci. 2004;24:5798–5809. doi: 10.1523/JNEUROSCI.1102-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang YQ, Bailey AM, Matthies HJ, Renden RB, Smith MA, Speese SD, Rubin GM, Broadie K. Drosophila fragile X-related gene regulates the MAP1B homolog Futsch to control synaptic structure and function. Cell. 2001;107:591–603. doi: 10.1016/s0092-8674(01)00589-x. [DOI] [PubMed] [Google Scholar]
- 62. Hanson JE, Madison DV. Presynaptic FMR1 genotype influences the degree of synaptic connectivity in a mosaic mouse model of fragile X syndrome. J Neurosci. 2007;27:4014–4018. doi: 10.1523/JNEUROSCI.4717-06.2007.. This study uses a clever genetic approach to begin to dissect whether FMRP plays a presynaptic role in synaptic plasticity. Because the FMRP gene is on the X chromosome heterozygous females are mosaic for FMRP expression due to random X inactivation in each neuron. Breeding with a line expressing X-linked GFP yielded female mice in which each neuron could identified as having either normal or absent FMRP expression by GFP fluorescence. Paired recording from pre- and post-synaptic pairs of neurons was then used to demonstrate that synaptic connectivity was dependent on FMRP expression in the presynaptic neuron rather than the postsynaptic neuron, a surprising result in the field.
- 63.Gibson JR, Bartley AF, Hays SA, Huber KM. Imbalance of neocortical excitation and inhibition and altered UP states reflect network hyperexcitability in the mouse model of fragile X syndrome. J Neurophysiol. 2008;100:2615–2626. doi: 10.1152/jn.90752.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bureau I, Shepherd GM, Svoboda K. Circuit and plasticity defects in the developing somatosensory cortex of FMR1 knock-out mice. J Neurosci. 2008;28:5178–5188. doi: 10.1523/JNEUROSCI.1076-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Till SM, Li HL, Miniaci MC, Kandel ER, Choi YB. A presynaptic role for FMRP during protein synthesis-dependent long-term plasticity in Aplysia. Learn Mem. 2011;18:39–48. doi: 10.1101/lm.1958811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Darnell JC, vanDreische SJ, Zhang C, Hung KYS, Mele A, Fraser CE, Stone EF, Chen C, Fak JJ, Chi SW, et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. doi: 10.1016/j.cell.2011.06.013. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li Z, Zhang Y, Ku L, Wilkinson KD, Warren ST, Feng Y. The fragile X mental retardation protein inhibits translation via interacting with mRNA. Nucleic Acids Res. 2001;29:2276–2283. doi: 10.1093/nar/29.11.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Laggerbauer B, Ostareck D, Keidel EM, Ostareck-Lederer A, Fischer U. Evidence that fragile X mental retardation protein is a negative regulator of translation. Hum Mol Genet. 2001;10:329–338. doi: 10.1093/hmg/10.4.329. [DOI] [PubMed] [Google Scholar]
- 69.Zalfa F, Giorgi M, Primerano B, Moro A, Di Penta A, Reis S, Oostra B, Bagni C. The fragile X syndrome protein FMRP associates with BC1 RNA and regulates the translation of specific mRNAs at synapses. Cell. 2003;112:317–327. doi: 10.1016/s0092-8674(03)00079-5. [DOI] [PubMed] [Google Scholar]
- 70. Qin M, Kang J, Burlin TV, Jiang C, Smith CB. Postadolescent changes in regional cerebral protein synthesis: an in vivo study in the FMR1 null mouse. J Neurosci. 2005;25:5087–5095. doi: 10.1523/JNEUROSCI.0093-05.2005.. Using a sensitive technique for measuring protein synthesis in living mice, Smith and colleagues found that Fmr1 knockout mice have significantly higher rates of protein synthesis in several regions of the brain, supporting the hypothesis that FMRP functions as a translational repressor in living brain.
- 71.Sung YJ, Dolzhanskaya N, Nolin SL, Brown T, Currie JR, Denman RB. The fragile X mental retardation protein FMRP binds elongation factor 1A mRNA and negatively regulates its translation in vivo. J Biol Chem. 2003;278:15669–15678. doi: 10.1074/jbc.M211117200. [DOI] [PubMed] [Google Scholar]
- 72.Lu R, Wang H, Liang Z, Ku L, O'donnell WT, Li W, Warren ST, Feng Y. The fragile X protein controls microtubule-associated protein 1B translation and microtubule stability in brain neuron development. Proc Natl Acad Sci U S A. 2004;101:15201–15206. doi: 10.1073/pnas.0404995101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Westmark CJ, Malter JS. FMRP mediates mGluR5-dependent translation of amyloid precursor protein. PLoS Biol. 2007;5:e52. doi: 10.1371/journal.pbio.0050052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Muddashetty RS, Kelic S, Gross C, Xu M, Bassell GJ. Dysregulated metabotropic glutamate receptor-dependent translation of AMPA receptor and postsynaptic density-95 mRNAs at synapses in a mouse model of fragile X syndrome. J Neurosci. 2007;27:5338–5348. doi: 10.1523/JNEUROSCI.0937-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ceman S, O'Donnell WT, Reed M, Patton S, Pohl J, Warren ST. Phosphorylation influences the translation state of FMRP-associated polyribosomes. Hum Mol Genet. 2003;12:3295–3305. doi: 10.1093/hmg/ddg350. [DOI] [PubMed] [Google Scholar]
- 76.Napoli I, Mercaldo V, Boyl PP, Eleuteri B, Zalfa F, De Rubeis S, Di Marino D, Mohr E, Massimi M, Falconi M, et al. The fragile X syndrome protein represses activity-dependent translation through CYFIP1, a new 4E-BP. Cell. 2008;134:1042–1054. doi: 10.1016/j.cell.2008.07.031. [DOI] [PubMed] [Google Scholar]
- 77.Szostak JW. In vitro genetics. Trends Biochem Sci. 1992;17:89–93. doi: 10.1016/0968-0004(92)90242-2. [DOI] [PubMed] [Google Scholar]
- 78.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 79.Tenenbaum SA, Lager PJ, Carson CC, Keene JD. Ribonomics: identifying mRNA subsets in mRNP complexes using antibodies to RNA-binding proteins and genomic arrays. Methods. 2002;26:191–198. doi: 10.1016/S1046-2023(02)00022-1. [DOI] [PubMed] [Google Scholar]
- 80.Buckanovich RJ, Darnell RB. The neuronal RNA binding protein Nova-1 recognizes specific RNA targets in vitro and in vivo. Mol Cell Biol. 1997;17:3194–3201. doi: 10.1128/mcb.17.6.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Levine TD, Gao F, King PH, Andrews LG, Keene JD. Hel-N1: an autoimmune RNA-binding protein with specificity for 3' uridylate-rich untranslated regions of growth factor mRNAs. Mol Cell Biol. 1993;13:3494–3504. doi: 10.1128/mcb.13.6.3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Farina KL, Huttelmaier S, Musunuru K, Darnell RB, Singer RH. Two ZBP1 KH domains facilitate beta-actin mRNA localization, granule formation, and cytoskeletal attachment. J Cell Biol. 2003;160:77–87. doi: 10.1083/jcb.200206003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Darnell JC, Jensen KB, Jin P, Brown V, Warren ST, Darnell RB. Fragile X mental retardation protein targets G Quartet mRNAs important for neuronal function. Cell. 2001;107:489–499. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- 84.Darnell JC, Fraser CE, Mostovetsky O, Stefani G, Jones TA, Eddy SR, Darnell RB. Kissing complex RNAs mediate interaction between the Fragile-X mental retardation protein KH2 domain and brain polyribosomes. Genes Dev. 2005;19:903–918. doi: 10.1101/gad.1276805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dredge BK, Darnell RB. Nova regulates GABA(A) receptor gamma2 alternative splicing via a distal downstream UCAU-rich intronic splicing enhancer. Molecular & Cellular Biology. 2003;23:4687–4700. doi: 10.1128/MCB.23.13.4687-4700.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Mili S, Steitz JA. Evidence for reassociation of RNA-binding proteins after cell lysis: implications for the interpretation of immunoprecipitation analyses. RNA. 2004;10:1692–1694. doi: 10.1261/rna.7151404.. This important paper experimentally demonstrates a major limitation of a commonly used method for identifying the RNAs bound by an RNA binding protein in vivo. Mili and Steitz show that once cell compartments are broken and contents are diluted in a lab buffer the reaction takes on the character of an equilibrium binding assay rather than a true reflection of what was bound in the cell.
- 87.Schaeffer C, Bardoni B, Mandel JL, Ehresmann B, Ehresmann C, Moine H. The fragile X mental retardation protein binds specifically to its mRNA via a purine quartet motif. EMBO J. 2001;20:4803–4813. doi: 10.1093/emboj/20.17.4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.de la Pena M, Dufour D, Gallego J. Three-way RNA junctions with remote tertiary contacts: a recurrent and highly versatile fold. RNA. 2009;15:1949–1964. doi: 10.1261/rna.1889509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Brown V, Jin P, Ceman S, Darnell JC, O'Donnell WT, Tenenbaum SA, Jin X, Feng Y, Wilkinson KD, Keene JD, et al. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in Fragile X Syndrome. Cell. 2001;107:477–487. doi: 10.1016/s0092-8674(01)00568-2. [DOI] [PubMed] [Google Scholar]
- 90. Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488.. This paper was the first to apply high throughput sequencing approaches to the in vivo crosslinking-IP (CLIP) technology to generate a detailed genome-wide maps for physiologic RNA interactions of an RNA binding protein. These compelling findings outlined an attractive approach applicable to other RNABPs in other tissues or organisms.
- 91. Ule J, Stefani G, Mele A, Ruggiu M, Wang X, Taneri B, Gaasterland T, Blencowe BJ, Darnell RB. An RNA map predicting Nova-dependent splicing regulation. Nature. 2006;444:580–586. doi: 10.1038/nature05304.. This paper was unique in applying a combination of mapping in vivo binding of an RNABP on an unbiased genome-wide scale with a functional assay predicting the functional outcome of RNA binding. Specifically, in vivo UV-crosslinking was used to show that the neuron-specific splicing factor Nova bound intronic YCAY-rich sites in a pattern specifying the direction of alternative splicing of an exon.
- 92. Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170.. This paper used HITS-CLIP to reveal a genome-wide map of Ago/miRNA binding sites on mRNAs in brain. The ability to capture real Ago/miR binding sites on mRNA by in vivo UV crosslinking is a marked improvement over bioinformatic methods for predicting such sites. This method for determination of miRNA regulation of mRNA targets can now be applied to a number of other biologic systems and physiologic conditions.
- 93.Licatalosi DD, Darnell RB. RNA processing and its regulation: global insights into biological networks. Nat Rev Genet. 2010;11:75–87. doi: 10.1038/nrg2673. [DOI] [PMC free article] [PubMed] [Google Scholar]