Summary
Type I Interferons (IFNs) are important cytokines for innate immunity against viruses and cancer. Sixteen human IFN variants signal through the same cell surface receptors, IFNAR1 and IFNAR2, yet they can evoke markedly different physiological effects. The crystal structures of two human type I IFN ternary signaling complexes containing IFNα2 and IFNω reveal recognition modes and heterotrimeric architectures that are unique amongst the cytokine receptor superfamily, but conserved between different type I IFNs. Receptor-ligand cross-reactivity is enabled by conserved receptor-ligand "anchor-points" interspersed amongst ligand-specific interactions that ‘tune’ the relative IFN binding affinities, in an apparent extracellular ‘ligand proofreading’ mechanism that modulates biological activity. Functional differences between IFNs are linked to their respective receptor recognition chemistries, in concert with a ligand-induced conformational change in IFNAR1, that collectively control signal initiation and complex stability, ultimately regulating differential STAT phosphorylation profiles, receptor internalization rates, and downstream gene expression patterns.
Introduction
IFNs were the first cytokines discovered more than half a century ago as agents that interfered with viral infection (Borden et al., 2007; Isaacs and Lindenmann, 1957). IFNs have been established as pleiotropic, multifunctional proteins in the early immune response, exhibiting antiproliferative effects on cells, in addition to their strong immunomodulatory and antiviral activities. Due to their potency and diverse biological activities, IFNs are used for the treatment of several human diseases, including hepatitis C, multiple sclerosis and certain types of cancer (Borden et al., 2007). Based on the receptor system that mediates their effects, IFNs are grouped into type I, type II and type III IFNs (Uze et al., 2007). The type I IFNs act on, and are produced by almost every nucleated cell, and comprise 16 members with approximately 20–60% sequence identity: IFNβ, IFNε, IFNκ, IFNω and 12 subtypes of IFNα. IFNα, IFNβ and IFNω are produced by cells exposed to viruses or double-stranded RNA (Garcia-Sastre and Biron, 2006) and have been shown to possess antitumor activity (Horton et al., 1999; Pestka et al., 2004) as well as protect cells against parasites and bacterial pathogens (Bogdan, 2000). Although similar in their spectrum of activities, IFNβ, IFNω and IFNα subtypes can vary significantly in their potency against different viruses, their antiproliferative activity and their ability to activate cells of the immune system. The mechanism mediating this differential activity and signaling through a common receptor remains controversial (van Boxel-Dezaire et al., 2006).
Despite their differential activities and broad range of potencies, all 16 human type I IFNs initiate signaling by binding to the same receptor composed of two subunits called IFNAR1 and IFNAR2. Together with the IL-10 family receptors, the IL-20 receptor, IL-22R, IL-22BP, IFNLR1, tissue factor and IFNGR, IFNAR1 and IFNAR2 form the class II helical cytokine receptor family (Pestka et al., 2004; Walter, 2004; Zdanov). In common with other class II helical cytokine receptors, the extracellular domain (ECD) of IFNAR2, whose NMR structure has been characterized (Chill et al., 2003), consists of two fibronectin III (FNIII)-like domains (D1 and D2). The ECD of IFNAR1, however, is unique, comprising a tandem array of four FNIII subdomains, designated SD1 to SD4, which arose from gene duplication of the typical two-domain structure (Gaboriaud et al., 1990).
The intracellular domains (ICDs) of IFNAR1 and IFNAR2 are associated with the Janus kinases (Jaks) Tyk2 and Jak1, respectively (Schindler and Plumlee, 2008; van Boxel-Dezaire et al., 2006). Upon ligand binding by the IFNAR chains and formation of the extracellular signaling complex, these tyrosine kinases initiate a phosphorylation cascade principally mediated by STAT (signal transducer and activator of transcription) activation (Schindler and Plumlee, 2008). Other important signaling pathways activated by type I IFNs include the phosphatidylinositol 3-kinase pathway and the MAP kinase pathway. Studies of the overlapping, yet differential cellular responses elicited by different members of the type I IFNs (Uze et al., 2007) have suggested that the dynamics of ligand interaction with the receptor subunits plays a key role for regulating cellular response patterns (Jaitin et al., 2006; Jaks et al., 2007; Kalie et al., 2007).
There are currently no crystal structures of type I IFN receptor complexes, nor any complete receptor signaling complex in the class II helical cytokine family where structures of binary complexes of ligands (IFN-γ, IL-10, IL-22, IFN-λ) with their high-affinity receptor subunits are known (Bleicher et al., 2008; Jones et al., 2008; Josephson et al., 2001; Miknis et al., 2010; Walter et al., 1995). Here we present structural and functional data that sheds light on how type I IFNs engage their receptor chains, how the receptor system is able to recognize the large number of different ligands, and how the different chemistries of ligand interaction ultimately dictate the stabilities of the receptor complexes, and therefore exert primary control on differential signaling.
Results
Type I IFNs exhibit distinct signaling and functional activities
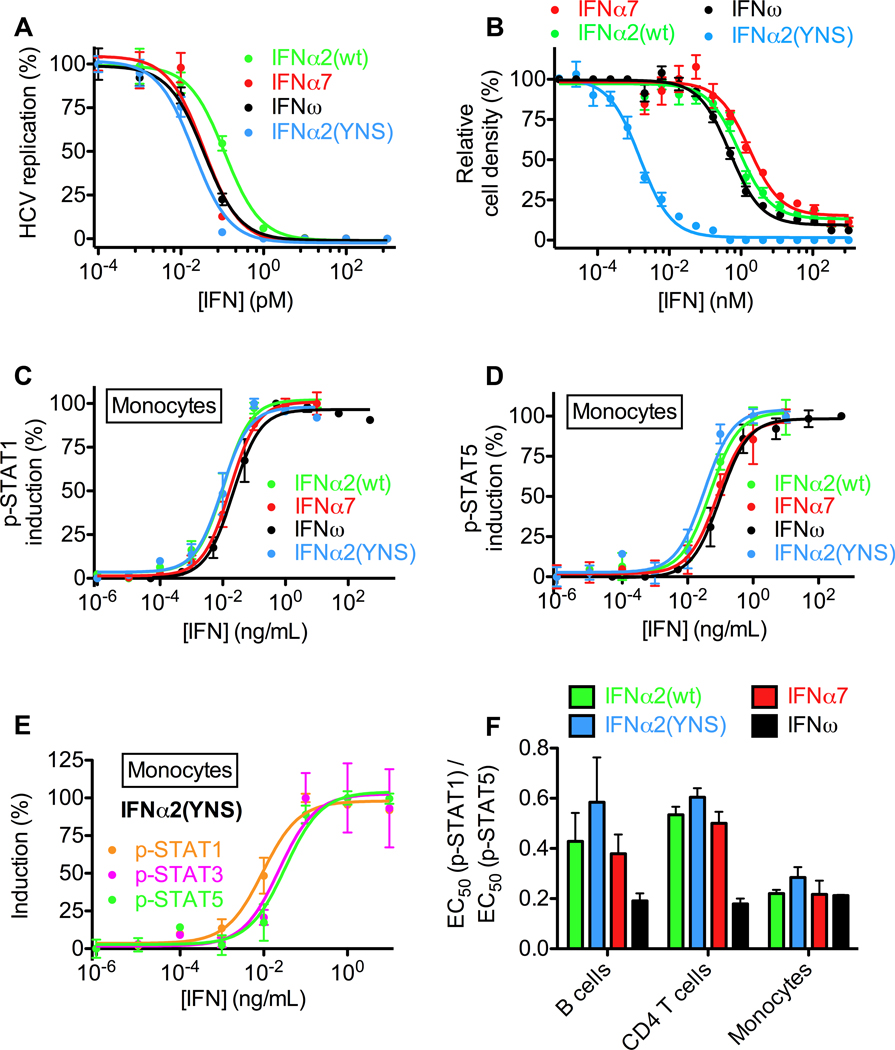
We studied IFNs that differed significantly in their biological activities: IFNω, IFNα2, and a mutant of IFNα2, IFNα2(YNS), that was engineered to have higher affinity for IFNAR1 in order to improve its anti-tumor efficacy (Kalie et al., 2007). We tested their relative antiviral and antiproliferative potencies, as well as another type I IFN, IFNα7 (Figures 1A, 1B, and S5). The EC50 values in a hepatitis C replication assay showed a 2- to 6-fold differences between the IFNs (IFNα7: 36 fM, IFNω: 37 fM, IFNα2(YNS): 20 fM, IFNα2(wt): 116 fM) (Figure 1A), while the antiproliferative activities on WISH cells differed by more than 1000-fold (EC50 values: IFNα7: 1700 pM, IFNω: 490 pM, IFNα2(YNS): 1.5 pM, IFNα2(wt): 890 pM) (Figure 1B).
Figure 1. Differential activities and potencies of type I IFNs.
(A) Antiviral dose-response curves of human hepatoma (Huh7.5) cells transfected with genomic hepatitis C virus (HCV) RNA and treated with IFNα2(wt), IFNα2(YNS), IFNω or IFNα7. (B) Antiproliferative dose-response curves of human amniotic epithelial (WISH) cells treated with IFNα2(wt), IFNα2(YNS), IFNω or IFNα7. (C) Dose-response curves for STAT1 phosphorylation in monocytes from human whole blood, as determined by Phospho-Flow analysis. (D) Dose-response curves for STAT5 phosphorylation in monocytes from human whole blood, as determined by Phospho-Flow cytometry analysis. (E) IFNα2(YNS) more potently induces pSTAT1 than pSTAT3 or pSTAT5 in human primary monocytes. (F) Differential signaling properties of IFNα2(wt), IFNα2(YNS), IFNω and IFNα7 as evidenced by different ratios of pSTAT1 to pSTAT5 EC50 values in different cell types from human whole blood. See also Figure S1.
We also compared the intracellular signaling activities of these IFNs by measuring phosphorylation of STATs in primary cells in whole blood from human donors using Phospho-Flow cytometry coupled with fluorescent cell barcoding (Krutzik and Nolan, 2006). This approach enabled us to measure IFN responses on endogenous IFN receptors of multiple cell subsets (B cells, monocytes, CD8 and CD4 T cells) simultaneously without cell separation (Figures 1C–F, S1, and S7). Although the potency of the different IFNs in inducing phosphorylation of STAT1 and STAT5 in monocytes is similar (Figures 1C, 1D), IFNα2(YNS) has a lower EC50 for pSTAT1 induction versus pSTAT3 and pSTAT5 (Figure 1E). Comparing the ratios of EC50(pSTAT1) versus EC50(pSTAT5), reveals that the different IFNs exhibit significant variability in different cell subsets (Figure 1F), with IFNα2(YNS) displaying the highest ratio of pSTAT1:pSTAT5 EC50 values in B cells, CD4 T cells and monocytes, whereas IFNω produced the lowest ratio in all three cell subsets. Collectively, the cellular and signaling results highlight the puzzling properties of differential signaling through the common IFNAR1/IFNAR2 heterodimeric receptor.
The architecture of the IFN ternary signaling complex
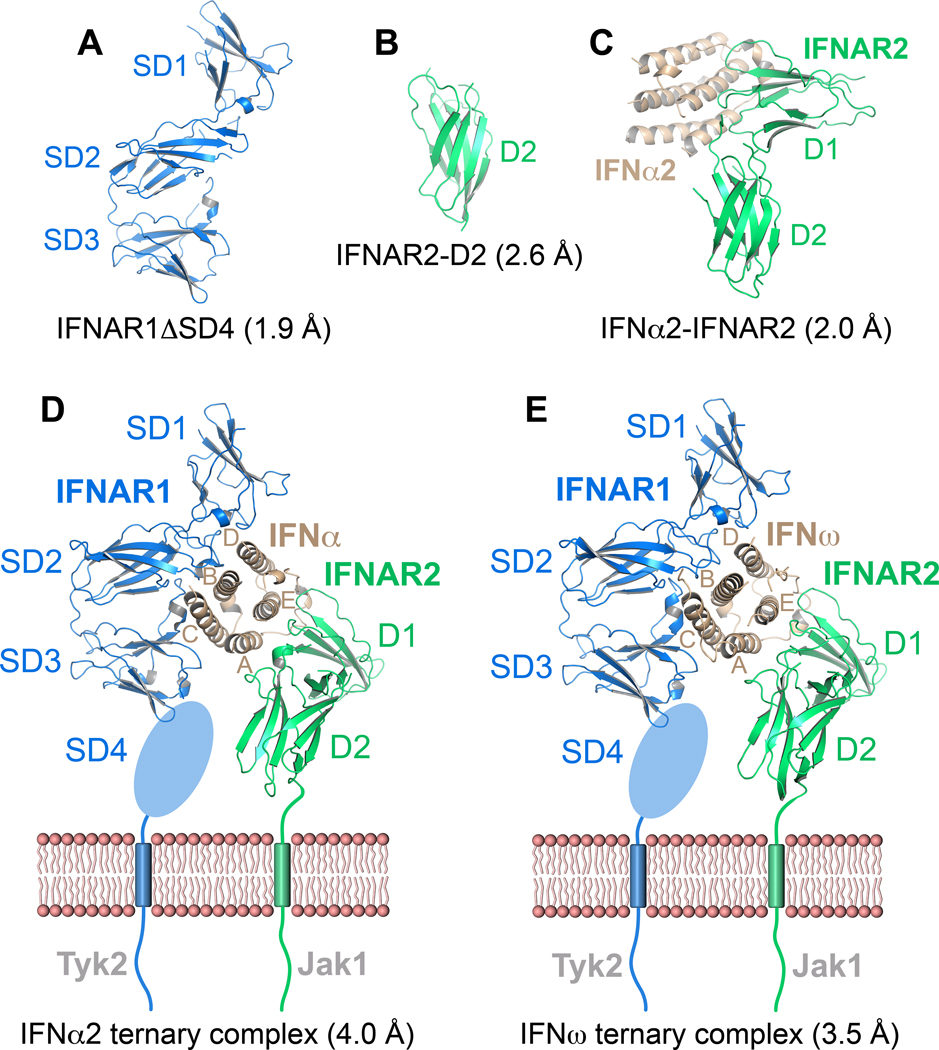
We determined crystal structures of a range of individual IFN receptor components, a sub-complex and ternary complexes at various resolutions (Figure 2 and Table S1): 1- the structure of unliganded IFNAR1 comprising SD1 through SD3 (IFNAR1ΔSD4) at 1.9Å (Figure 2A); 2- the IFNAR2-D2 domain at 2.6Å (Figure 2B); 3- the binary complex between IFNAR2 and IFNα2(HEQ) at 2.0Å (Figure 2C) (HEQ contains three engineered mutations to Ala in its IFNAR1 binding site, but only wild-type IFNα2 residues are in the IFNAR2 binding site (Jaitin et al., 2006)); 4- the ternary ligand-receptor complex of IFNα2(YNS) (hereafter also referred to as IFNα2) at 4.0Å (Figure 2D); 5- the ternary ligand-receptor complex of wild-type IFNω at 3.5Å (Figure 2E). High-resolution structures of sub-components determined here, and previously (IFNα2, and IFNAR2-D1 domain) (Quadt-Akabayov et al., 2006; Radhakrishnan et al., 1996), were used to solve the ternary complexes. Despite their lower resolution, the electron density maps of the ternary complexes (Figure S2) allowed refinement of almost all amino acids, and clear visualization of conformational changes between the free and bound states (details in Supplemental Information). The SD4 of IFNAR1 has been shown to be unnecessary for IFN binding (Lamken et al., 2005) and, consistent with electron-microscopic studies (Li et al., 2008), it was not visible in the electron density maps. All structures can be viewed interactively at (http://proteopedia.org/w/Journal:Cell:1).
Figure 2. Crystal structures of type I IFN receptor components and ligand-receptor complexes.
Ribbon representations and designated resolutions of (A) IFNAR1ΔSD4. (B) IFNAR2-D2. (C) The IFNα2(HEQ)-IFNAR2 binary complex (IFNα2(HEQ) brown, IFNAR2 green). (D) Ternary complex of IFNAR1 (blue), IFNAR2 (green) and IFNα2(YNS). (E) Ternary complex of IFNAR1 (blue), IFNAR2 (green) and IFNω. The membrane-proximal SD4 domain of IFNAR1 is depicted as an oval. See also Table S1 and Figure S2.
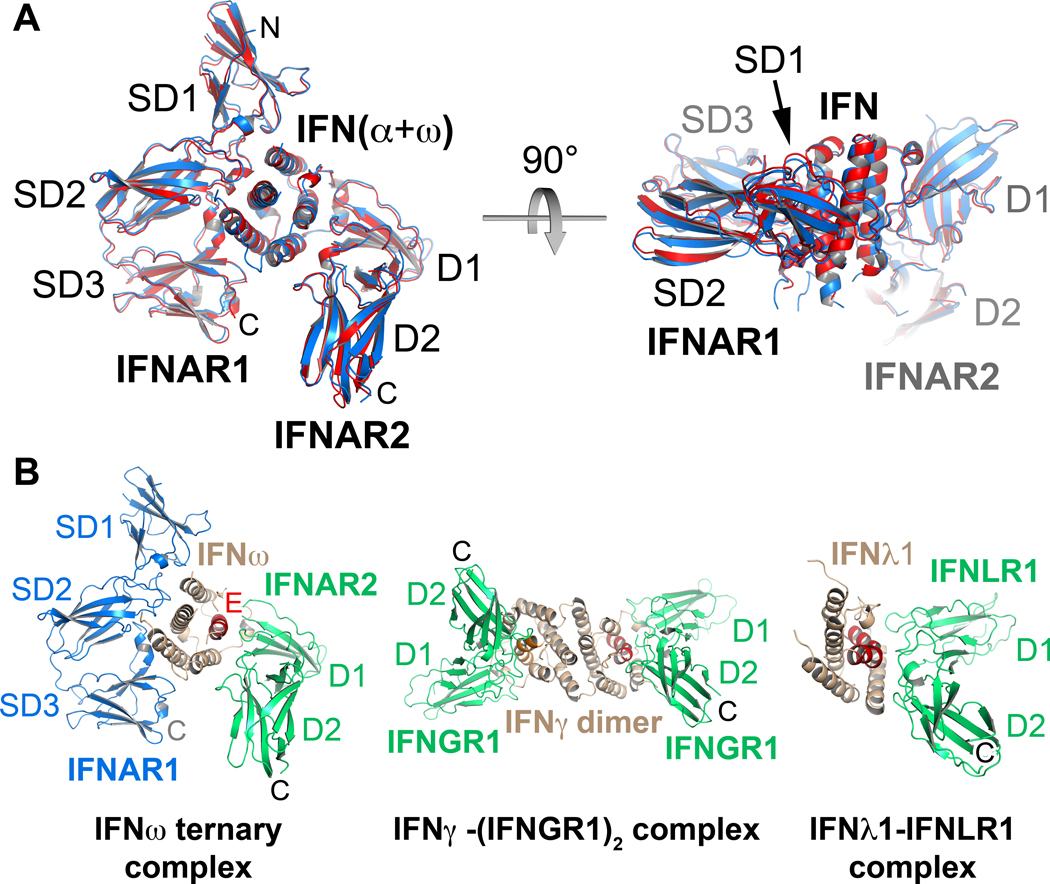
IFNAR1 and IFNAR2 bind on opposing sides of the IFN ligands in a nearly orthogonal architecture that has not been seen previously in crystal structures of cytokine-receptor complexes (Figures 2D, 2E). Both the IFNα2 and IFNω complexes exhibit almost identical overall receptor-ligand docking modes when the two ternary complexes are superimposed (RMSD of Cα = 0.9 Å) (Figure 3A). The IFNAR1-IFN docking mode seen here is unusual and so far without precedent among cytokine-receptor interactions. The IFNAR1-IFN interface is formed by residues of the SD1, SD2 and SD3 sub-domains of IFNAR1 and by helices B, C and D of the IFN molecule (Figures 2D, 2E), burying a total surface area of 2197 Å2 (IFNω ternary complex). The IFN ligand primarily binds to IFNAR1 at the level of the hinge between the SD2 and SD3 domains, with the SD1 domain ‘capping’ the top of the IFN molecule. In prior cytokine receptor complexes of both the Type I (e.g. human Growth Hormone, Interleukin-2, Erythropoietin, etc.) and Type II (e.g. IFNγ, IL-10, etc.) systems, the principal interaction mode is between the cytokine and the loops projecting from the ‘elbow’ formed between two bent Fibronectin-III (FNIII) domains (Figure 3B) (Walter, 2004; Wang et al., 2009). In the case of IFNAR1, the SD2–SD3 tandem FNIII domains appear to be oriented in the opposite direction, such that the loops at the extreme top and bottom ends of the FNIII domains form the major contacts with the IFN ligands in a manner reminiscent of pinchers, while the ‘elbow’ loops that normally bind to cytokines face outward into solvent. The SD1–SD2 tandem FNIII module engages the ligands in a manner that is more representative of a canonical cytokine-binding mode where the elbow contacts the ligand. As the SD1–SD2 and SD3–SD4 modules of IFNAR1 most likely arose by gene duplication, the relative orientation of the domains within the modules is thought to be similar, allowing us to model a position for SD4 (Figure S3A). However, its flexibility implies there is inter-domain variability in its position on a cell surface.
Figure 3. Similar architectures of type I IFN complexes are distinct from type II and type III IFN receptor complexes.
(A) The IFN molecules of the IFNω and IFNα2(YNS) ternary complexes were superimposed and are shown in side view and top view. The RMSD for the overall superposition of both structures is 0.9 Å. (B) The IFNω ternary complex is shown side-by-side with the IFNγ-(IFNGR1)2 complex (PDB accession code: 1FG9), and the IFNλ-IFNLR1 complex (PDB accession code: 3OG6). (N, C: amino- and carboxy-termini. SD1–SD4: subdomains of IFNAR1; D1, D2: N- and C-terminal domain of IFNAR2. See also Figure S3B.
On the opposing side of the ligand, both IFNAR2-IFN interfaces are formed between parts of helices A and E and the A–B loop of the ligand and the IFNAR2-D1 domain and the loop between strands 13 and 14 in the D2 domain, burying ~1841 Å2 (IFNα2 binary complex) of surface area (Figures 2D, 2E, and 4). On IFNAR2, the IFN ligand does not bind at the apex of the elbow region between the D1 and D2 domains of IFNAR2 as seen in most type I and II cytokine-receptor complexes (Figure 3B), but rather almost all of the contact is with the receptor D1 domain. In the ternary complexes, the long axis of the IFN helical bundle is oriented perpendicularly to IFNAR1, but almost parallel to the beta sheets of the IFNAR2 D1 domain. The overall docking position of the ligands bound to IFNAR2 has global similarities to the manner in which the IFNγ dimer engages IFNGR1 (Walter et al., 1995), and also to the IFNλ-IFNLR1 complex (Figure 3B) (Miknis et al., 2010). However there are large differences in the relative receptor-IFN binding orientations between them that clearly distinguish their recognition modes (Figure S3B). The rigid body ligand-binding topology to IFNAR2 is approximately similar to a docking model derived using constraints from NMR and mutagenesis (Nudelman et al., 2010).
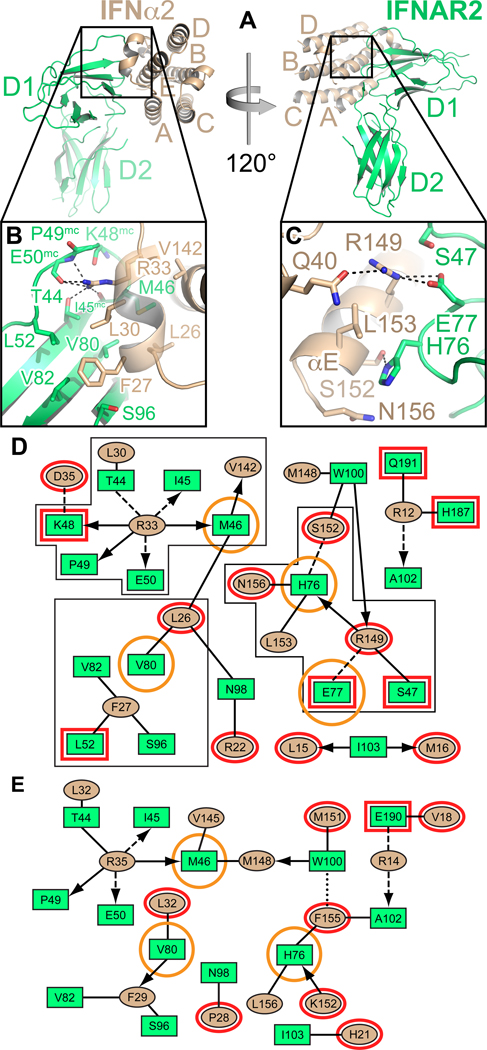
Figure 4. Specificity and cross-reactivity determinants between IFN-IFNAR2.
(A) Two different views of the IFNα2-IFNAR2 binary complex. Helices of IFNα2 are labeled A–E. (B) Hotspot residues Leu30α2 and Arg33α2 of IFNα2 and their environment in the interface with IFNAR2. Hydrogen bonds are shown as dashed lines. (C) Close-up view of Arg149IFN, Glu77R2 and His76R2 and their environment. Hydrogen bonds and salt bridges are depicted as dashed lines. (D) 2D interaction map of the IFNα2-IFNAR2 interface. Amino acids are depicted as nodes in the interaction maps (rectangles: IFNAR2; ellipses: IFN). Interactions between side chains are represented by lines, interactions between side chains and backbone are depicted as arrows pointing toward the backbone. Van-der-Waals interactions and hydrophobic contacts are shown as solid lines, H-bonds or electrostatic interactions as dashed lines and aromatic interactions as dotted lines. Residues shown in panels B and C are bordered with a black line. Structural differences between the IFNα-IFNAR2 and the IFNω-IFNAR2 interfaces are highlighted in red. IFNAR2 residues that, when mutated, differentially affect IFNα and IFNω binding, are encircled in orange. (E) 2D interaction map of the IFNω-IFNAR2 interface. See also Figure S2 and Table S2.
Mechanism of IFN cross-reactivity versus discrimination by IFNAR2
We compared the ligand-IFNAR2 interfaces from the binary IFNα2-IFNAR2 complex (2.0 Å resolution) (Figures 4A–D and S2), and the IFNω ternary complex (3.5 Å resolution) (Figures 4E and S2). We elucidated interactions that are conserved across type I IFNs (i.e. “anchor points”), versus those that would be ligand-specific (Figures 4 and 6). We also assembled previous alanine scanning data (Kalie et al., 2007; Piehler et al., 2000; Roisman et al., 2005; Roisman et al., 2001), together with new site-directed mutations prompted by the structures, in order to reconcile the structures with comprehensive energetic maps of the interfaces (Table S2). Overall, most of the residues involved in the IFNα2-IFNAR2 interaction are also found in the IFNω-IFNAR2 interface of the IFNω ternary complex (Figures 4D, 4E), highlighting that the basis of IFN cross-reactivity is through conservation of interactions rather than through highly divergent binding solutions. For clarity, in the two-dimensional contact maps of Figures 4 and 5, ligand-specific receptor contacts are circled in red, while those with divergent mutational consequences are circled in orange (Table S2). (NOTE: due to non-identical sequence lengths (Figure 6G), the numbering of analogous IFNα2 and IFNω residues will often differ by one to three residues throughout the paper). For example, Arg33α2 (i.e. Arg35 in IFNω), which is conserved in IFNα, IFNω, IFNβ and IFNε (asparagine in IFNκ), appears to be the single most important residue for the interaction of both IFN ligands with IFNAR2 (Table S2, Figures 4B and 4D). It forms an extensive hydrogen-bonding network with the main chain carbonyl oxygen atoms of Ile45R2 and Glu50R2 and the side chain of Thr44R2. Replacing Arg33α2 in IFNα2 by alanine destabilizes binding more than any other mutation in IFNα2 (Table S2). Two hydrophobic interaction clusters are present in the IFNα-IFNAR2 interface: the first one is formed between Leu15α2 and Met16α2 of the IFN molecule and Trp100R2 and Ile103R2 of IFNAR2; the second one comprises Leu26α2, Phe27α2, Leu30α2 and Val142α2 of the ligand and Met46R2, Leu52R2, Val80R2 and Thr44R2 of the receptor. Of these, Trp100R2, Ile103R2, Met46R2, Val80R2, Thr44R2 and the ligand residues corresponding to Met148α2, Phe27α2, Leu30α2 and Val142α2 are also involved in the IFNω-IFNAR2 interface. Substituting Met148α2 in IFNα2, or Ile103R2 of IFNAR2 results in 10–30-fold decreases in binding to the receptor and both IFN ligands, respectively. As another example, Leu30α2 is conserved in all human IFNs, and equates to Leu32ω. Both are involved in similar hydrophobic clusters in IFNAR2 interactions that are also energetically similar (Table S2). Thus, these are energetically critical, shared anchor points mediating IFN cross-reactivity.
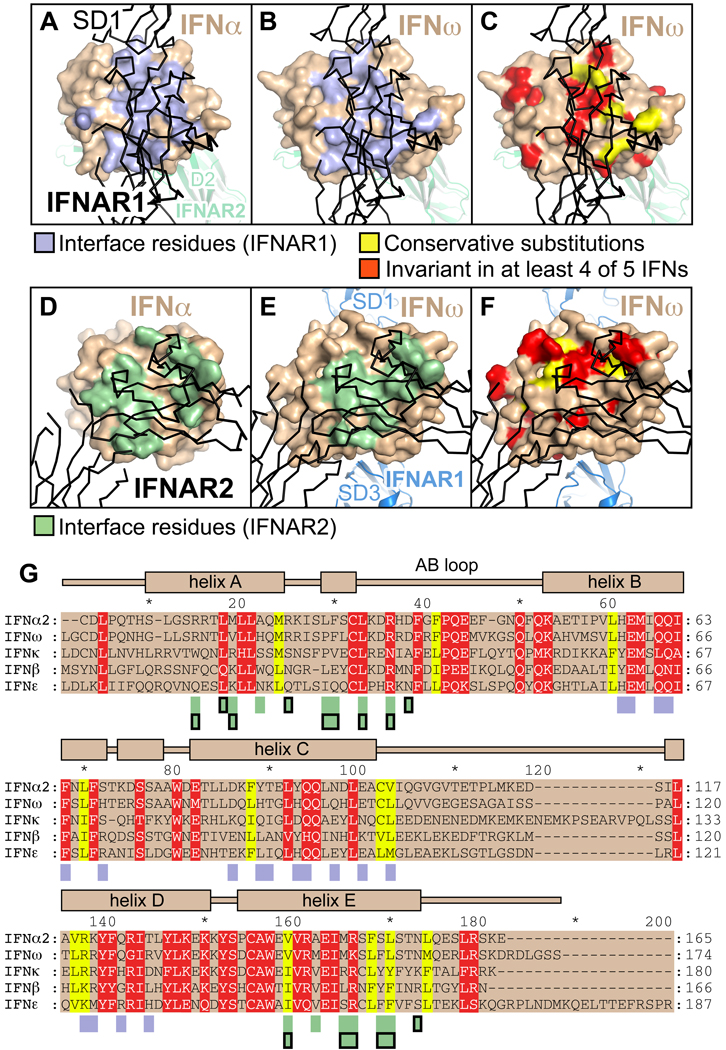
Figure 6. Conservation of residues in the ligand-receptor interfaces.
(A–F) Residues on the surface of IFNω and IFNα involved in the interaction with IFNAR1 (panel A: IFNα ternary complex, panel B: IFNω ternary complex) and IFNAR2 (panel D: IFNα binary complex, panel E: IFNω ternary complex) are colored lightblue and green, respectively. Surface residues on IFNω conserved between IFNs are shown in panels C and F. Physico-chemically conserved amino acids are colored yellow, residues that are invariant in at least four of five IFNs (IFNα2, IFNβ, IFNε, IFNκ and IFNω) are shown in red. (G) Sequence alignment of human IFNs. Conserved and invariant residues are colored as in panels C and F. Interacting residues are denoted by rectangles below the alignment, colored according to panels A, B, D and E. Rectangles outlined in black mark interacting residues in the IFNα2 binary complex. The secondary structural elements of IFNω are depicted on top of the alignment.
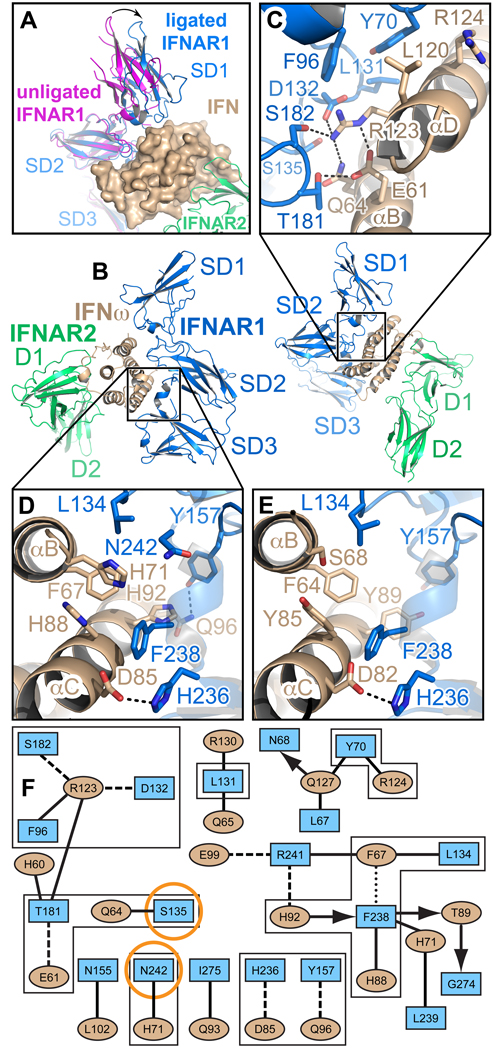
Figure 5. Ligand-induced domain movement in IFNAR1 and the IFN-IFNAR1 interfaces.
(A) Domain movement in IFNAR1 upon IFN binding. Unliganded IFNAR1ΔSD4 (magenta) was superimposed onto subdomains 2 (SD2) and 3 (SD3) of IFNAR1 (blue) in the IFNω ternary complex. The difference in the position of the SD1 domain is depicted as an arrow. The ligand, IFNω, is shown with its molecular surface. See also Figure S3, and http://proteopedia.org/w/Journal:Cell:1. (B) Two different views of the IFNω ternary complex. SD1–SD3: subdomains of IFNAR1; D1 and D2: subdomains of IFNAR2. (C) Environment of the hotspot residues Tyr70R1 and Arg123ω in the IFNAR1-IFNω interface. Dashed lines symbolize hydrogen bonds and salt bridges. (D) Hydrophobic and aromatic interactions between Leu134R1 and the hotspot residue Phe238R1 in IFNAR1 and Phe67 in IFNω. (E) The same region as in (D) in the IFNα2-IFNAR1 interface. Hydrogen bonds in the close-up views are depicted as dashed lines. (F) Interaction map of the IFNω-IFNAR1 interface in the IFNω ternary complex. Amino acids are depicted as nodes in the interaction map (rectangles: IFNAR1; ellipses: IFNω) as used in Figure 3. Residues shown in panels C, D and E are bordered with a black line. IFNAR1 residues that, when mutated, differentially affect IFNα and IFNω binding, are encircled in orange.
In contrast, the mechanism of ligand discrimination appears to derive in large part from differential energetics of shared contact positions among the different IFNs. A major ligand-specific difference between the IFNα and IFNω interface is related to Arg149α2 in IFNα2, and the analogous Lys152IFN in IFNω, and their respective interaction chemistries with Glu77R2. In the IFNα2/IFNAR2 interface these two residues (R149Aα2 and Glu77R2) stabilize the interaction by forming a salt bridge (Figures 4C, 4D) that is worth about 1.9 kcal/mol in free energy (Figure S4). Substituting Arg149α2 by alanine reduces the affinity between IFNα2 and IFNAR2 by two orders of magnitude (Table S2). Arginine at position 149 is the consensus in all type I IFNs except IFNω, where it is replaced by Lys152ω, that forms an intra-molecular salt bridge with Glu149ω, and is within close proximity, but is not directly contacting Glu77R2 of the receptor. The differential contribution of Glu77R2 to the two interfaces is reflected in the observation that its mutation to alanine differentially affects IFNα and IFNω binding: The IFNAR2(E77A) mutant binds IFNα2 with 60-fold lower affinity, while the affinity toward IFNω is reduced only 10-fold (Table S2, highlighted in orange). To mimic the connectivity of the IFNα2 binding interface, we made the Lys152Argω swapping mutation (in addition to K152A), which increases omega binding by 5-fold (Table S2). To establish the connectivity of Arg at position 152 in IFNω with Glu77R2, we performed double mutant cycle analyses between E77R2 and K152Aω and K152Rω (Figure S4). Lysine152ω binds Glu77R2 with a ΔΔGint of 1.3 kcal/mol, while Arg in position 152 binds with a ΔΔGint of 2.2 kcal/mol, clearly establishing that, indeed, the consensus arginine at this position is able to form a significantly stronger interaction, supporting the respective structural organization observed for the two different complexes. That the K152R substitution increases binding in IFNω indicates that this position is a critical modular hotspot for ligand discrimination and signaling (discussed below).
There are additional examples of IFN sequence differences, observed as receptor contacts in the structures, playing a role in ligand subtype discrimination. Leu26α2 in IFNα2 equates to Pro28ω in IFNω (Figures 4D, 4E). The IFNω mutation P28Aω had no effect on receptor binding, while swapping Pro28ω with Leu26α2 (i.e. P28Lω) reduced binding 6-fold in IFNω. Thus, these residues have evolved distinct energetic values by substituting side chains. Another notable IFNAR2 contacting residue that differs between alpha and omega (Table S2), is Ala145α2 that is Met148 in IFNω. In alpha IFNs, alanine is the consensus residue in position 145 (148 in ω). Yet, M148Aω reduces binding by ~2.5-fold. The complementary mutation of Ala145 to Met in IFNα2 reduces binding by ~6-fold. This shows a distinct IFN-specific residue preference, and that this position is not simply degenerate for apolar side chains. The subtle apolar volume differences are keenly sensed in ligand discrimination. On the receptor side, Val80R2 differentially affects IFNω versus IFNα2 binding (Figures 4B, 4D and 4E, and Table S2, highlighted in orange). Two other residues in IFNAR2, His76R2 and Met46R2, also contribute to ligand discrimination (Table S2, highlighted in orange).
IFNAR1 forms a diffuse and broad interface with the IFN ligand
The IFNω and IFNα2(YNS) complexes are essentially identical in their binding footprints to IFNAR1 (Figures 3A, and 5). We focus on the higher resolution IFNω ternary complex for a detailed description of the IFN-IFNAR1 interfaces (Figure S2). When the unliganded IFNAR1ΔSD4 structure and IFNAR1 of the ternary complexes are superimposed, it is apparent that the N-terminal SD1 domain and the SD2–SD3 portion of IFNAR1 move relative to each other upon IFN binding (Figures 5A, and S3C–D), allowing all three subdomains of IFNAR1 to contact the ligand. With the SD2–SD3 domains overlaid and fixed, the conformational change upon complex formation is a quasi-rigid body movement of the SD1 domain by about 10 Å down toward the ligand, bringing Asn68R1, Tyr70R1 and Phe96R1 in contact with helix D of the IFN ligand (Figures 5A and 5C). The aromatic rings of Tyr70R1 and Phe96R1, together with the side chain of Leu131R1 in a loop of the SD2 domain, form a hydrophobic patch that packs against the ligand (Figures 5C, 5F). Arg123ω on helix D of IFNω forms a critical lynchpin for the SD1–SD2 interaction: it hydrogen bonds to Ser182R1 in the SD2 domain, contacts Thr181R1 and Phe96R1, and is engaged in a salt bridge with Asp132R1 (Figures 5C, 5F). Arg123ω is intramolecularly stabilized by Glu61ω that also forms a hydrogen bond with Thr181R1. In addition, the IFNω-IFNAR1 interface is characterized by van-der-Waals and hydrophobic interactions between Leu134R1 (SD2), Phe238R1 (SD3) and Phe67ω in helix B (Figures 5D, 5F). The interactions between Leu134R1, Phe238R1 and the phenylalanine in the ligand (Phe64α2 in IFNα) are conserved in the IFNα-IFNAR1 interface (Figure 4E). Moreover, the contact between His236R1 and an aspartate in the ligand is common to the interfaces of both IFNs (Figures 5D, 5E).
It has been demonstrated that the three N-terminal FNIII domains of IFNAR1 (SD1–SD3) are necessary and sufficient for ligand binding (Lamken et al., 2005). In particular, the SD1 segment spanning residues 62–70 is crucial for ligand binding and biological activity, with Val69R1 and Tyr70R1 as key residues (Cajean-Feroldi et al., 2004). Their role is revealed by our ternary complex structures: Tyr70R1 directly contacts the ligand (Figures 5C, 5F), whereas the preceding Val69R1 stabilizes the S3–S4 loop. Tyr70R1 and F238R1 are the only hotspot residues in the ligand-binding site of IFNAR1 (Table S2), highlighting its comparatively energetically flat binding surface compared to IFNAR2. Substituting these residues by alanine reduces the affinity to all tested IFN ligands by more than 10-fold (Table S2).
Most interactions of the IFNω-IFNAR1 interface are conserved in the IFNα2-IFNAR1 interface (Figures 5D, 5E, and 6C, 6G). Differences include an aromatic interaction between Tyr157R1 of the receptor and Tyr89α2 of IFNα2. In IFNω, Tyr157R1 hydrogen bonds to Gln96ω (Figures 5D, 5E). Furthermore, Tyr85α2 and Tyr89α2 participate in the hydrophobic interaction with Leu134R1 and Phe238R1 of IFNAR1 and Phe64α2 of the ligand (Figure 5E); His71ω, which is contacting Asn242R1 in the IFNω complex, is replaced with a serine in IFNα2. The different chemical environments of Asn242R1 in the two complexes might contribute to ligand specificity (Table S2, highlighted in orange).
IFNs are discriminated through ligand-specific substitutions
In order to analyze the cross-reactivity of the type I IFN receptor, we mapped the interface contact residues and the residues conserved between IFNα2, IFNω, IFNβ, IFNε and IFNκ onto the surface of the IFN molecules in the ternary complexes (Figure 6). A comparison of the maps of contact residues with the degree of sequence conservation reveals that IFNAR1 and IFNAR2 cross-react with different IFNs by using a few conserved residues on their ligands as "anchor points" against a background of less- or non-conserved amino acids (Figures 6C, 6F, and 6G). As our mutational analysis has shown, ligand discrimination occurs primarily through distinct energetics of common contacts, but also through small numbers of IFN subtype- or sequence-specific, contacts. The invariant and conserved ligand residues comprise Leu32ω, Arg35ω, Val145ω, Arg147ω, Glu149ω, Lys152ω, Leu156ω in the IFNAR2 interface and His60ω, Glu61ω, Gln64ω, Gln65ω, Phe67ω, Gln93ω, Gln96ω, Leu102ω, Leu120ω and Arg123ω in the IFNAR1 interface (residues and numbering referring to IFNω). Of the invariant and conserved IFN residues that form direct contacts with the receptor chains, Leu32ω, Arg35ω, Leu156ω, His60ω, Glu61ω, Gln64ω, Phe67ω and Arg123ω influence the energetics of the ligand-receptor interaction, indicated by mutational studies of the corresponding residues in IFNα2 (Leu30α2, Arg33α2, Leu153α2, His57α2, Glu58α2, Gln61α2, Phe64α2 and Arg120α2, see Table S2).
Probing differential IFN signaling with structure-based mutational analysis
We analyzed two types of IFN mutations for their effects on signaling and function. We chose residues that differ in identity between IFNα2 and IFNω and either directly make energetically important receptor contacts in the structures, or are in close proximity to residues that do. The first group includes Ala mutations of these residues (L26Aα2/P28Aω, L30Aα2/L32Aω, A145Gα2/M148Aω, R149Aα2/K152Aω), as well as swaps of energetically important ‘sister’ residues that are in corresponding positions (P28Lω, K152Rω). A second group of mutations was designed to change the binding affinity to both receptors simultaneously. These were based on the IFNα2(YNS) variant (increases binding to IFNAR1 by ~60-fold), and include YNS/M148A and YNS/L153A, which reduce binding to IFNAR2 by 30- and 10-fold, respectively (Table S2). Proteins harboring both mutations will have an altered balance between their affinity to IFNAR1 (higher affinity) and IFNAR2 (lower affinity). The mutant binding affinities are shown in Table S2. This extended set of mutations was then used to assess a range of functional consequences of IFN binding to the IFNAR1/IFNAR2 receptors: 1- antiviral (AV) and antiproliferative (AP) activity (Figures 7 and S5), 2- pSTAT activation with respect to EC50s and their relative ratios in different cell types from whole blood (Figures 8A, 8B, and S7), 3- gene expression (PKR, CXCL11, and TRAIL) in WISH cells (Figures 8A and S6), and 4- receptor internalization (Figure 8C).
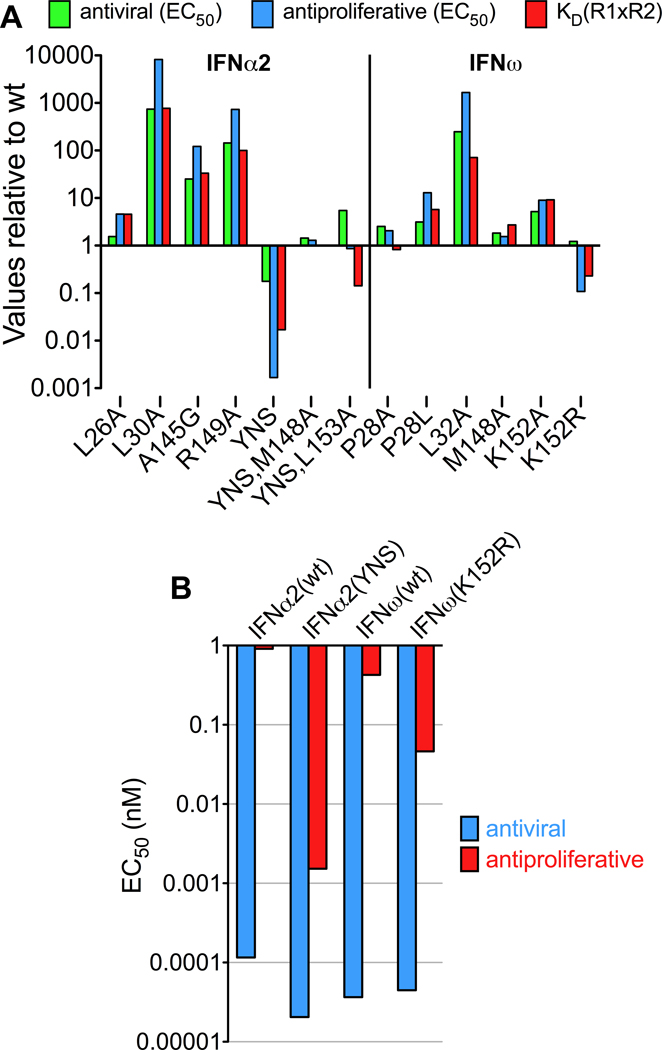
Figure 7. Correlation between complex stability and functional activity.
(A) Antiviral and antiproliferative activity of IFNα2 and IFNω mutants relative to IFNα2(wt) and IFNω(wt), respectively. As a measure of complex stability, the product of the affinities toward IFNAR1 and IFNAR2 was calculated and divided by the value of the respective wildtype protein. (B) Direct comparison of the antiviral and antiproliferative activity (EC50 values) of the high-affinity mutants IFNα2(YNS) and IFNω(K152R) and the corresponding wildtype proteins. See also Figure S5.
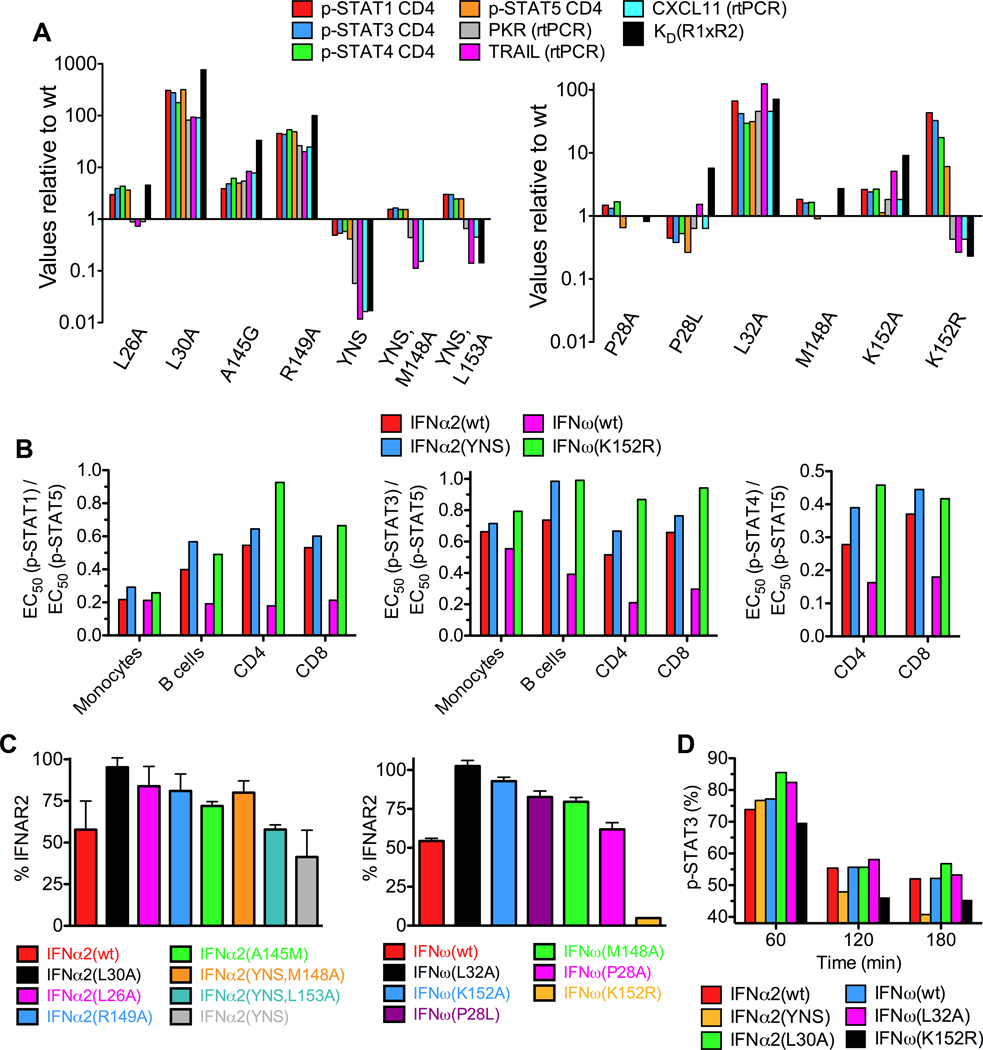
Figure 8. Relationship between STAT phosphorylation, gene expression, and receptor downregulation to IFN mutant binding affinities.
(A) Complex stabilities and induction of STAT phosphorylation in CD4 T cells and gene expression [Protein kinase R (PKR), Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL), chemokine CXCL11] by IFNα2 and IFNω mutants relative to wildtype IFNα2 and IFNω, respectively. (B) Different EC50(p-STAT) ratios for IFNα2(YNS), IFNω(K152R), IFNα2(wt) and IFNω(wt) in different cell subsets of whole blood from human donors. IFNα2(YNS) and IFNω(K152R) show the same trend of ratio deviations from the wildtype proteins. (C) Expression levels of IFNAR2 on the surface of B cell lymphoma (Ramos) cells 5 min after stimulation with IFNα2 and IFNω proteins. (D) Time course of decrease of p-STAT3 activation induced by different IFNα and IFNω proteins. See also Figures S6 and S7.
Antiviral and antiproliferative activity
Structure-based mutations that result in loss of binding affinity also lead to a decrease in potency for both AV and AP activities, and consequently reduce the functional distinction between IFNs (Figures 7 and S5). These data are in general accord with the stability model - antiviral activity is less affected by a loss in binding affinity than antiproliferative activity. Strikingly, mutations increasing binding affinity, as the ones observed for the mutants IFNα2(YNS) and IFNω(K152R), result in a significant increase of the AP activity, but not AV activity (Figure 7B). Indeed, the IFNω(K152R) gain-of-function mutant shows that a single substitution of the corresponding α2 residue results in IFNω behaving more like IFNα2(YNS). This supports the model that IFN-specific polymorphisms engaged in receptor contacts that energetically mediate complex stability play a major role in modulating IFN-specific functional activities (Figure 7B). These results also imply that intermediate affinities are sufficient to induce a maximal AV response, while much higher affinities are required to reach the maximal AP potency – in this respect the AP response is more “tunable” (Levin et al., 2011).
p-STAT activation
Phosphorylation levels of STAT1, 3, 4 and 5 were measured in whole blood samples from two human donors using Phospho-Flow cytometry and fluorescent cell barcoding (Figures 8A, 8B, and S7). Similar to the AV activity, the EC50 values obtained for p-STAT activation proportionally increased for weaker binding mutants, while for the high-affinity mutants (IFNα2(YNS) and IFNω(K152R)) the same trend was not observed. Indeed, although IFNα2(YNS) induced slightly stronger STAT activation than IFNα2(wt), these differences were very small relative to their very substantial differences in affinity. Similarly, with the IFNω(K152R) mutant, despite its higher binding affinity, this mutant is somewhat less potent in pSTAT activation than IFNω(wt). These results are in accord with the current model that AV activity is well correlated with early p-STAT activation, and AV activity is nearly maximal even for weak binders. In contrast, the extent of STAT activation is not sufficient to explain the potency of the AP response.
p-STAT ratios
Variable ratios of STAT activation in cell subsets is a striking example of differential signaling through a common receptor by different IFN sub-types (Figure 1F). We analyzed the p-STAT activation ratios induced by the high affinity IFNα2(YNS) and IFNω(K152R) mutants, as well as the wildtype IFNs, IFNα2 and IFNω, in a mixed population of immune cells, i.e. whole blood samples from humans (Figures 8B and S7). Both mutants followed the same trend in deviations of p-STAT ratios relative to the wildtype IFNs, supporting the idea that by substituting a critical contact residue from one IFN into another, we have narrowed their functional distinction. Since the principal effects of these substitutions are on affinity, this further suggests that the stability of the complex is the key determinant for functional distinction of IFNs, and also highlights the utility of this metric as a readout of differential signaling activities by IFNs.
Receptor internalization
We hypothesized that rapid receptor downregulation could be responsible for the non-proportional p-STAT activation observed for IFN mutants with increased binding affinity. Increased IFNAR2 downregulation by an IFNα2 mutant with increased binding affinity towards IFNAR1 has been observed (Jaitin et al., 2006; Marijanovic et al., 2007). Here we show that mutants exhibiting higher binding affinity for IFNAR2 than wt induced a stronger IFNAR2 downregulation (Figure 8C), and faster decrease in p-STAT activation (Figure 8D). The IFNα2(YNS) reduced IFNAR2 by 60%, while the K152Rω mutant almost completely eliminated surface IFNAR2, as opposed to wildtype IFNα2 or IFNω that only reduced the surface IFNAR2 by ~50% (Figure 8C). Thus, increased binding affinities increase the propensity for receptor endocytosis, which leads to faster termination of signaling. The substantially stronger IFNAR2 downregulation exhibited by K152Rω may explain the surprising increase in EC50 of p-STATs seen for this mutant (Figure 8A) due to more rapid (Figure 8C) receptor inactivation in the endosome.
Gene expression
We asked how receptor-binding affinity regulates the IFN-induced gene expression program. By rtPCR, we measured the levels of PKR, CXCL11 and TRAIL induction following 8 h of treatment with the different IFN mutants (Figures 8A and S6 – note P28Aω and M148Aω were not included in the gene expression analysis). We found a uniform correlation between receptor binding affinity and gene expression levels. That is, mutants with reduced affinity induced lower levels of PKR, CXCL11 and TRAIL, while mutants with higher affinity induced higher levels of these genes. These data also indicate that the extent of STAT activation as measured by tyrosine phosphorylation does not fully explain the level of gene expression and AP response induced by the different IFN mutants, as in the case of the YNS and K152Rω mutants. While YNS is only marginally more potent activating STATs than IFNα2(wt), it is significantly more potent in inducing TRAIL, PKR and CXCL11 than IFNα2(wt). Interestingly, while the EC50 for PKR gene expression activation is ~50-fold lower for most IFNs than the one measured for TRAIL or CXCL11, these ratios are significantly smaller (~15-fold) for the three YNSα2 variants and for the K152Rω mutant (Figure S6), suggesting that tighter binding IFNs lose some of this differential gene activation, perhaps by sacrificing ‘tunability’ for affinity.
Discussion
Type I interferons were discovered over 50 years ago as antiviral agents. Subsequent research has shown that the many IFN sub-types show differential activities through common receptor chains. Our studies show that the overall architectures of receptor binding to both IFNα2 and IFNω are nearly identical (Figure 3A), and that the answer to how different IFNs are capable of inducing differential functional effects appears to result from ligand discrimination through distinct receptor binding chemistries, which dictate the respective stabilities of the receptor-ligand interactions. The distinct binding chemistries are achieved primarily by differential energetics of shared anchor points, and to a lesser extent by key amino acid substitutions between IFNs. These ligand-specific differences in the extracellular complex stabilities manifest as perturbations in downstream signaling cascades, in both linear and non-linear fashions. Mechanistically, different complex stability kinetics could control the relative Jak/Tyk activity towards intracellular substrates of greater or lesser accessibility, which would, in turn lead to distinct downstream effector activation profiles and ultimately impact induction of IFN-responsive genes. In this respect, recognition-mediated tuning of differential signaling by the Type I IFN receptor system is quite unique for a transmembrane receptor, but has parallels to the antigen ‘proofreading’ ability of the T cell receptor to differentially respond to self and foreign peptide-MHC molecules presenting subtly different peptide recognition chemistries.
In the context of prior cytokine receptor structures, IFNAR1 is particularly striking, with participation of three subdomains and a conformational change upon IFN binding (Figures 5A and S3). That this is a bona-fide ligand-induced conformational change is corroborated by the importance of the SD1 domain for ligand binding, and by FRET measurements suggesting conformational changes in the ectodomain of IFNAR1 upon IFN binding (Strunk et al., 2008). The conformational change in IFNAR1 is required to form the full spectrum of interactions with the ligand and to allow the formation of a ternary complex that is stable enough to facilitate trans-phosphorylation between Jak1 and Tyk2. Thus, ligand binding to IFNAR1 will be accompanied by an energetic cost associated with the structural rearrangements required to bring a key hotspot residue into contact, and could play a role in ‘tuning’ responsiveness to different IFN ligands. We suggest that the required conformational change contributes to the reduced binding affinity of IFNAR1 and may result in tighter control of IFN signaling.
In addition to the conformational change, the role of IFNAR1 in ligand responsiveness is also unique compared to IFNAR2. IFNAR1 is not optimized for high binding affinity, but rather for functional plasticity. That is, in contrast to the interaction with IFNAR2, binding energy is distributed over a large number of amino acid contacts with relatively low individual contributions and with much lower cooperativity, altogether resulting in lower affinity. For early STAT activation, which is required for the antiviral cellular response, transient ligand interaction with IFNAR1 appears to be advantageous (Moraga et al., 2009). High stability of the ternary complex seems to be more important for a subset of IFN activities requiring prolonged activation of IFN signaling pathways (Coelho et al., 2005; Jaitin et al., 2006). The relatively large binding interface of IFNAR1 for IFN involving 3 FNIII-like domains provides a versatile means for fine-tuning the binding affinity towards different IFNs and tailoring differential response patterns.
The molecular basis of IFNAR cross-reactivity is unique compared with other shared receptor systems, such as gp130 and common gamma chain (γc), and this likely reflects the fact that the IFN interaction chemistry controls signal initiation. Gp130 engages different cytokines through entirely distinct binding surfaces that do not appear to share anchor points, whereas γc engages in degenerate binding largely through shape complementarity (Wang et al., 2009). What sets the IFNAR system apart is that the IFNAR1/2 heterodimer recognizes and transduces the signal for all 16 IFN sub-types, whereas in the other shared cytokine receptors, signal specificity is determined by different ligand-specific co-receptors hetero-dimerizing with the shared receptor. In this way, the recognition chemistry of gp130 and γc are not important arbiters of signaling specificity.
With regards to function, our mutational and substitution experiments suggest a model whereby ablating or swapping key IFN-specific residues that engage in receptor interactions narrows the functional distinction between IFNs. Importantly, however, the mutational analysis also shows that the local environment of these contacts plays an important role in determining their energetic values in the respective IFN complexes. Mutation of individual positions has complicated energetic consequence. Therefore ligand-specific residues are not “plug-and-play” in a manner that easily allows one to recapitulate IFN sub-type behavior by point mutagenesis. This is to be expected given that the functional distinction of IFN ligands arose, in part, through co-evolution of broad receptor-ligand interaction surfaces over hundreds of millions of years. A surprising exception to this was the K152R gain of function mutation in IFNω, which, clearly, is a highly modular contact point.
Ligand-specific differences in the stabilities of the complexes are also reflected in variances in the kinetics of receptor downregulation, which terminates signaling. Our studies revealed that increased binding affinities towards IFNAR1 (IFNα2(YNS) mutant) or IFNAR2 (IFNω(K152R) mutant) strongly enhance receptor downregulation, which very likely explains a much more rapid decline in p-STAT activation compared to IFNα2(wt) and IFNω(wt). Increased IFNAR2 downregulation by the higher affinity IFNβ, compared to IFNα2, has been previously suggested to be responsible for differential cellular responses (Jaitin et al., 2006; Kalie et al., 2007). Here, we have designed an IFN mutant with increased binding affinity towards IFNAR2, which surprisingly induces even stronger downregulation of IFNAR2. Increased IFNAR2 downregulation could explain why the substantially increased binding affinity of these IFN mutants is not accompanied by a significant increase in their AV potency, because it is very likely responsible for a rapid decrease in p-STAT levels, as seen after stimulation with IFNα2(YNS) and IFNω(K152R).
In contrast to AV activity, which requires only very low doses of IFN to reach saturation, AP activity benefits from an increased binding affinity (Kalie et al., 2008). Cells need to sense very low levels of IFN and act very fast in order to clear viral infections in their initial stages. On the other hand, antiproliferative activity, which is often linked with apoptosis and tissue damage, needs to be under tighter control to prevent unnecessary damage. These activities will therefore be more tunable over a broad range to changes in the kinetics and strength of the downstream signaling. IFNs, by forming a gradient of complex stabilities will induce specific profiles of signal activation that will lead to diverse antiproliferative potencies. Taken together, differential IFN signaling activities are mediated by both non-linear signaling and non-linear receptor desensitization mechanisms. This type of “ligand proofreading” provides a mechanistic model, now together with a structural framework, for how a common receptor can respond in a graded fashion to different ligands.
Experimental procedures
Transient Hepatitis C virus (HCV) replication assay
The transient HCV replication assay was performed using Huh7.5 cells and a Luciferase reporter system as previously described (Cho et al., 2010). Additional details for this and subsequent experimental procedures can be found in Supplementary Information.
Antiproliferative activity assay
Antiproliferative assays were performed using WISH cells as described in (Moraga et al., 2009).
Protein expression, purification and complex formation
The following proteins used in this study were expressed as C-terminally His-tagged constructs from baculovirus using the pAcGp67A vector: Human IFNω (including all IFNω mutants), IFNα7, IFNα2(HEQ), IFNAR1ΔSD4 (amino acids 3–305), full-length IFNAR1 ectodomain, IFNAR2 (amino acids 10–205), IFNAR2-D2 (amino acids 104–205). IFNAR2 used in the binary complex was secreted by Hi-5 cells in the presence of Tunicamycin at a concentration of 0.5 mg/L. IFNAR2 (amino acids 7–205) used for IFNω(N80Q) ternary complex formation was expressed using the BacMam expression vector pVL-AD6-L (Dukkipati et al., 2008) from suspended HEK293 GnTI− cells grown in Pro293 medium, and was deglycosylated with endoglycosidase Hf. Human IFNα2 and all IFNα2 mutants, except HEQ, were expressed in E.coli according to published methods (Kalie et al., 2007). Prior to crystallization, all proteins were treated with 3C protease/TEV protease and/or carboxypeptidase A and B to remove C-terminal His-tags. Selenomethionine(SeMet)-labeled proteins from baculovirus were prepared according to a protocol published earlier (Dong et al., 2009).
The IFNω(N80Q) ternary complex was formed by mixing IFNAR1 and IFNω(N80Q) from insect cells with IFNAR2 expressed in HEK293 cells. The IFNα2(YNS) ternary complex was formed by mixing IFNAR1 and IFNAR2 from insect cells, and IFNα2(YNS) expressed in E.coli. The complexes were formed by mixing individually purified components in approximately stoichiometric ratios; the complexes were purified by gel filtration.
Crystallization and X-ray data collection
All crystallization experiments were carried out using hanging-drop vapor diffusion at 20°C. Individual crystallization conditions can be found in Supplementary Information.
Data sets on frozen crystals were collected at beamlines 9.1 (SeMet-IFNAR1ΔSD4), 9.2 (osmium-derivatized IFNAR1ΔSD4; SeMet-IFNAR2-D2) and 11.1 (native data set of IFNAR1ΔSD4) of the Stanford Synchrotron Radiation Lightsource (SSRL), and at beamlines 8.2.1 (IFNα2(YNS) ternary complex) and 8.2.2 (IFNα2(HEQ)-IFNAR2 binary complex; IFNω(N80Q) ternary complex) of the Advanced Light Source (ALS), Berkeley. All data were indexed, integrated and scaled with the XDS package (Kabsch, 1993).
Structure determination and refinement
Phases for IFNAR1ΔSD4 were obtained by single isomorphous replacement with anomalous scattering (SIRAS) in the program autoSHARP (Vonrhein et al., 2007) using the osmium derivative and the native data set.
The structure of SeMet-labeled IFNAR2-D2 domain was determined by single-wavelength anomalous diffraction (SAD) using autoSHARP.
The IFNα2(HEQ)-IFNAR2 binary complex, IFNα2(YNS) ternary complex and IFNω(N80Q) ternary complex were all solved by molecular replacement using the program Phaser (McCoy et al., 2007). All X-ray structures described were refined with Phenix (Adams et al., 2010). Molecular graphics images were prepared using PyMOL (Schrödinger).
Affinity measurements
All binding data of IFNs and the ECD of IFNAR2 were determined by surface plasmon resonance on a ProteOn XPR36 machine (BIO-RAD) using purified proteins. Binding of IFNα2(YNS) and IFNω to immobilized IFNAR1 was probed by simultaneous total internal reflection fluorescence spectroscopy (TIRFS) and reflectance interference (RIF) detection.
Phospho-Flow analysis of intracellular signaling
Analysis of intracellular signaling in whole blood was performed as previously described (Krutzik and Nolan, 2006). Briefly, whole blood samples from two donors were warmed to 37°C and stimulated with increasing concentrations of the appropriate cytokine for 30 min. After samples were fixed and lysed, samples were fluorescently barcoded with DyLight 800 and Pacific Orange dyes as previously described. After barcoding and combining, samples were stained for one hour with CD3 PE, CD4 Pacific Blue, CD20 PerCP-Cy5.5, CD33 PE-Cy7 and a combination of p-STAT1 Ax647 and p-STAT3 Ax488, or p-STAT5 Ax647 and p-STAT4 Ax488. Analysis was performed on a Becton Dickinson LSRII equipped with 405, 488, and 633 nm lasers. Data analysis was performed in Cytobank software. Log median fluorescence intensity values were plotted against cytokine concentration to yield dose-response curves.
Analysis of IFNAR2 downregulation
Downregulation experiments were performed using Ramos cells stimulated with IFN proteins for 5 minutes, followed by anti-IFNAR2 mAb staining as described in (Jaitin et al., 2006; Marijanovic et al., 2007).
Analysis of STAT phosphorylation kinetics
Ramos cells were stimulated with 10 nM of IFN mutants for the indicated times according to the protocol in (Marijanovic et al., 2007). Samples were analyzed by Phospho-Flow cytometry.
Quantitative PCR
Selected human IFN-stimulated gene expression levels were measured with the ABI Prism 7300 Real-Time PCR System using previously described methods (Kalie et al., 2007).
Supplementary Material
Acknowledgments
We thank Natalia Goriatcheva for expert technical assistance, the staff at SSRL and ALS for their assistance and David Canner for preparing the Proteopedia pages. K.C.G. is an Investigator of the Howard Hughes Medical Institute. This work was also supported by NIH-RO1-AI51321 (K.C.G.) and NIH-RO1-AI087917 (J.S.G.), C.T. is supported by a long-term postdoctoral fellowship of the International Human Frontier Science Program Organization. G.S. and J.P. are supported by the European Community's FP7/2007–2013 under GA no. 223608 (IFNaction).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession numbers
Coordinates and structure factors have been deposited within the Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank (PDB) under accession codes 3S98 (IFNAR1ΔSD4), 3S8W (IFNAR2-D2), 3S9D (binary IFNα-IFNAR2 complex), 3SE4 (ternary IFNω complex) and 3SE3 (ternary IFNα complex).
Supplemental Information
Supplemental Information includes Extended Experimental Procedures, seven figures, and two tables.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleicher L, de Moura PR, Watanabe L, Colau D, Dumoutier L, Renauld JC, Polikarpov I. Crystal structure of the IL-22/IL-22R1 complex and its implications for the IL-22 signaling mechanism. FEBS Lett. 2008;582:2985–2992. doi: 10.1016/j.febslet.2008.07.046. [DOI] [PubMed] [Google Scholar]
- Bogdan C. The function of type I interferons in antimicrobial immunity. Curr Opin Immunol. 2000;12:419–424. doi: 10.1016/s0952-7915(00)00111-4. [DOI] [PubMed] [Google Scholar]
- Borden EC, Sen GC, Uze G, Silverman RH, Ransohoff RM, Foster GR, Stark GR. Interferons at age 50: past, current and future impact on biomedicine. Nat Rev Drug Discov. 2007;6:975–990. doi: 10.1038/nrd2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cajean-Feroldi C, Nosal F, Nardeux PC, Gallet X, Guymarho J, Baychelier F, Sempe P, Tovey MG, Escary JL, Eid P. Identification of residues of the IFNAR1 chain of the type I human interferon receptor critical for ligand binding and biological activity. Biochemistry. 2004;43:12498–12512. doi: 10.1021/bi049111r. [DOI] [PubMed] [Google Scholar]
- Chill JH, Quadt SR, Levy R, Schreiber G, Anglister J. The human type I interferon receptor: NMR structure reveals the molecular basis of ligand binding. Structure. 2003;11:791–802. doi: 10.1016/s0969-2126(03)00120-5. [DOI] [PubMed] [Google Scholar]
- Cho NJ, Dvory-Sobol H, Lee C, Cho SJ, Bryson P, Masek M, Elazar M, Frank CW, Glenn JS. Identification of a class of HCV inhibitors directed against the nonstructural protein NS4B. Sci Transl Med. 2010;2 doi: 10.1126/scitranslmed.3000331. 15ra16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho LF, Magno de Freitas Almeida G, Mennechet FJ, Blangy A, Uze G. Interferon-alpha and -beta differentially regulate osteoclastogenesis: role of differential induction of chemokine CXCL11 expression. Proc Natl Acad Sci U S A. 2005;102:11917–11922. doi: 10.1073/pnas.0502188102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong G, Wearsch PA, Peaper DR, Cresswell P, Reinisch KM. Insights into MHC class I peptide loading from the structure of the tapasin-ERp57 thiol oxidoreductase heterodimer. Immunity. 2009;30:21–32. doi: 10.1016/j.immuni.2008.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dukkipati A, Park HH, Waghray D, Fischer S, Garcia KC. BacMam system for high-level expression of recombinant soluble and membrane glycoproteins for structural studies. Protein Expr Purif. 2008;62:160–170. doi: 10.1016/j.pep.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaboriaud C, Uze G, Lutfalla G, Mogensen K. Hydrophobic cluster analysis reveals duplication in the external structure of human alpha-interferon receptor and homology with gamma-interferon receptor external domain. FEBS Lett. 1990;269:1–3. doi: 10.1016/0014-5793(90)81103-u. [DOI] [PubMed] [Google Scholar]
- Garcia-Sastre A, Biron CA. Type 1 interferons and the virus-host relationship: a lesson in detente. Science. 2006;312:879–882. doi: 10.1126/science.1125676. [DOI] [PubMed] [Google Scholar]
- Horton HM, Hernandez P, Parker SE, Barnhart KM. Antitumor effects of interferon-omega: in vivo therapy of human tumor xenografts in nude mice. Cancer Res. 1999;59:4064–4068. [PubMed] [Google Scholar]
- Isaacs A, Lindenmann J. Virus interference. I. The interferon. Proc R Soc Lond B Biol Sci. 1957;147:258–267. [PubMed] [Google Scholar]
- Jaitin DA, Roisman LC, Jaks E, Gavutis M, Piehler J, Van der Heyden J, Uze G, Schreiber G. Inquiring into the differential action of interferons (IFNs): an IFN-alpha2 mutant with enhanced affinity to IFNAR1 is functionally similar to IFN-beta. Mol Cell Biol. 2006;26:1888–1897. doi: 10.1128/MCB.26.5.1888-1897.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaks E, Gavutis M, Uze G, Martal J, Piehler J. Differential receptor subunit affinities of type I interferons govern differential signal activation. Journal of molecular biology. 2007;366:525–539. doi: 10.1016/j.jmb.2006.11.053. [DOI] [PubMed] [Google Scholar]
- Jones BC, Logsdon NJ, Walter MR. Structure of IL-22 bound to its high-affinity IL-22R1 chain. Structure. 2008;16:1333–1344. doi: 10.1016/j.str.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josephson K, Logsdon NJ, Walter MR. Crystal structure of the IL-10/IL-10R1 complex reveals a shared receptor binding site. Immunity. 2001;15:35–46. doi: 10.1016/s1074-7613(01)00169-8. [DOI] [PubMed] [Google Scholar]
- Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. Journal of Applied Crystallography. 1993;26:795–800. [Google Scholar]
- Kalie E, Jaitin DA, Abramovich R, Schreiber G. An interferon alpha2 mutant optimized by phage display for IFNAR1 binding confers specifically enhanced antitumor activities. J Biol Chem. 2007;282:11602–11611. doi: 10.1074/jbc.M610115200. [DOI] [PubMed] [Google Scholar]
- Kalie E, Jaitin DA, Podoplelova Y, Piehler J, Schreiber G. The stability of the ternary interferon-receptor complex rather than the affinity to the individual subunits dictates differential biological activities. J Biol Chem. 2008;283:32925–32936. doi: 10.1074/jbc.M806019200. [DOI] [PubMed] [Google Scholar]
- Krutzik PO, Nolan GP. Fluorescent cell barcoding in flow cytometry allows high-throughput drug screening and signaling profiling. Nat Methods. 2006;3:361–368. doi: 10.1038/nmeth872. [DOI] [PubMed] [Google Scholar]
- Lamken P, Gavutis M, Peters I, Van der Heyden J, Uze G, Piehler J. Functional cartography of the ectodomain of the type I interferon receptor subunit ifnar1. J Mol Biol. 2005;350:476–488. doi: 10.1016/j.jmb.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Levin D, Harari D, Schreiber G. Stochastic receptor expression determines cell fate upon interferon treatment. Mol Cell Biol. 2011 doi: 10.1128/MCB.05251-11. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Strunk JJ, Lamken P, Piehler J, Walz T. The EM structure of a type I interferon-receptor complex reveals a novel mechanism for cytokine signaling. J Mol Biol. 2008;377:715–724. doi: 10.1016/j.jmb.2007.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marijanovic Z, Ragimbeau J, van der Heyden J, Uze G, Pellegrini S. Comparable potency of IFNalpha2 and IFNbeta on immediate JAK/STAT activation but differential down-regulation of IFNAR2. Biochem J. 2007;407:141–151. doi: 10.1042/BJ20070605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miknis ZJ, Magracheva E, Li W, Zdanov A, Kotenko SV, Wlodawer A. Crystal Structure of Human Interferon-lambda1 in Complex with Its High-Affinity Receptor Interferon-lambdaR1. J Mol Biol. 2010 doi: 10.1016/j.jmb.2010.09.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraga I, Harari D, Schreiber G, Uze G, Pellegrini S. Receptor density is key to the alpha2/beta interferon differential activities. Mol Cell Biol. 2009;29:4778–4787. doi: 10.1128/MCB.01808-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nudelman I, Akabayov SR, Schnur E, Biron Z, Levy R, Xu Y, Yang D, Anglister J. Intermolecular interactions in a 44 kDa interferon-receptor complex detected by asymmetric reverse-protonation and two-dimensional NOESY. Biochemistry. 2010;49:5117–5133. doi: 10.1021/bi100041f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestka S, Krause CD, Walter MR. Interferons, interferon-like cytokines, and their receptors. Immunol Rev. 2004;202:8–32. doi: 10.1111/j.0105-2896.2004.00204.x. [DOI] [PubMed] [Google Scholar]
- Piehler J, Roisman LC, Schreiber G. New structural and functional aspects of the type I interferon-receptor interaction revealed by comprehensive mutational analysis of the binding interface. J Biol Chem. 2000;275:40425–40433. doi: 10.1074/jbc.M006854200. [DOI] [PubMed] [Google Scholar]
- Quadt-Akabayov SR, Chill JH, Levy R, Kessler N, Anglister J. Determination of the human type I interferon receptor binding site on human interferon-alpha2 by cross saturation and an NMR-based model of the complex. Protein Sci. 2006;15:2656–2668. doi: 10.1110/ps.062283006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan R, Walter LJ, Hruza A, Reichert P, Trotta PP, Nagabhushan TL, Walter MR. Zinc mediated dimer of human interferon-alpha 2b revealed by X-ray crystallography. Structure. 1996;4:1453–1463. doi: 10.1016/s0969-2126(96)00152-9. [DOI] [PubMed] [Google Scholar]
- Roisman LC, Jaitin DA, Baker DP, Schreiber G. Mutational analysis of the IFNAR1 binding site on IFNalpha2 reveals the architecture of a weak ligand-receptor binding-site. J Mol Biol. 2005;353:271–281. doi: 10.1016/j.jmb.2005.08.042. [DOI] [PubMed] [Google Scholar]
- Roisman LC, Piehler J, Trosset JY, Scheraga HA, Schreiber G. Structure of the interferon-receptor complex determined by distance constraints from double-mutant cycles and flexible docking. Proc Natl Acad Sci U S A. 2001;98:13231–13236. doi: 10.1073/pnas.221290398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler C, Plumlee C. Interferons pen the JAK-STAT pathway. Semin Cell Dev Biol. 2008;19:311–318. doi: 10.1016/j.semcdb.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunk JJ, Gregor I, Becker Y, Li Z, Gavutis M, Jaks E, Lamken P, Walz T, Enderlein J, Piehler J. Ligand binding induces a conformational change in ifnar1 that is propagated to its membrane-proximal domain. J Mol Biol. 2008;377:725–739. doi: 10.1016/j.jmb.2008.01.017. [DOI] [PubMed] [Google Scholar]
- Uze G, Schreiber G, Piehler J, Pellegrini S. The receptor of the type I interferon family. Curr Top Microbiol Immunol. 2007;316:71–95. doi: 10.1007/978-3-540-71329-6_5. [DOI] [PubMed] [Google Scholar]
- van Boxel-Dezaire AH, Rani MR, Stark GR. Complex modulation of cell type-specific signaling in response to type I interferons. Immunity. 2006;25:361–372. doi: 10.1016/j.immuni.2006.08.014. [DOI] [PubMed] [Google Scholar]
- Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated structure solution with autoSHARP. Methods Mol Biol. 2007;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- Walter MR. Structural analysis of IL-10 and Type I interferon family members and their complexes with receptor. Adv Protein Chem. 2004;68:171–223. doi: 10.1016/S0065-3233(04)68006-5. [DOI] [PubMed] [Google Scholar]
- Walter MR, Windsor WT, Nagabhushan TL, Lundell DJ, Lunn CA, Zauodny PJ, Narula SK. Crystal structure of a complex between interferon-gamma and its soluble high-affinity receptor. Nature. 1995;376:230–235. doi: 10.1038/376230a0. [DOI] [PubMed] [Google Scholar]
- Wang X, Lupardus P, Laporte SL, Garcia KC. Structural biology of shared cytokine receptors. Annu Rev Immunol. 2009;27:29–60. doi: 10.1146/annurev.immunol.24.021605.090616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdanov A. Structural analysis of cytokines comprising the IL-10 family. Cytokine Growth Factor Rev. 21:325–330. doi: 10.1016/j.cytogfr.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.