Abstract
Polycomb group (PcG) proteins regulate gene expression in embryonic and adult stem cells, but the mechanisms responsible for PcG gene targeting and regulation remain largely unknown. Recent evidence shows that EZH2, the enzymatic subunit of Polycomb Repressive Complex 2 (PRC2), is a nuclear phosphoprotein linking cell-cycle-intrinsic or extracellular signals to specific epigenetic signatures.
The establishment and maintenance of embryonic stem cells (ESCs) and generation of somatic cells with highly specialized functions are achieved through an exquisitely regulated control of chromatin modifications that influence gene expression. Critical in this process is the interpretation of developmental signals at the chromatin level. Polycomb group (PcG) proteins are critical regulators of embryonic and adult stem cell differentiation that operate via repression of gene transcription (reviewed in Surface et al., 2010; Sauvageau and Sauvageau, 2010). While their function in transcriptional repression via histone 3 methylation of lysine 27 (H3K27me3) has been extensively demonstrated, the mechanisms by which PcGs are recruited to chromatin in response to external signals remain elusive. Both proteins and noncoding (nc) RNAs have been found to regulate PcG chromatin engagement (Sawarkar and Paro, 2010; Margueron and Reinberg, 2011). A systematic identification of PcG-associated RNAs has revealed that RNAs occupy up to 18% of the sites at which SUZ12 (an essential component of Polycomb Repressive Complex 2 [PRC2]) is known to bind chromatin in ESCs (Zhao et al., 2010), indicating that additional molecular determinants participate in the recognition of the remaining 82% SUZ12-binding sites. These determinants may include CpG islands, PRC2-associated JARID2, and PRC1-related YY1, among others. In addition, beyond the question of how PcG are recruited, the dynamics of PcG targeting to specific genomic loci in response to intrinsic cell-cycle signals or to extrinsic input emanating from the environment remain largely obscure (Simon and Kingston, 2009). Clarifying how specific signals translate into specific PcG targeting is a particularly important goal, as PcG-mediated repression and derepression of genes in ESCs and adult stem cells is regulated in a cell-cycle-specific manner and in response to external factors that direct lineage commitment and differentiation into specialized cell types. By extension, deregulation of these signals might hijack PcGs from their intended targets and subsequently induce gene-expression profiles that may leave a cell susceptible to malignant transformation or developmental defects.
Global proteomic profiling of phosphopeptides has provided evidence that the enzymatic subunit of PRC2, the histone methyltransferase EZH2, is highly phosphorylated (Dephoure et al., 2008; Mayya et al., 2009; Brill et al., 2009; Huttlin et al., 2010). However, the functional relevance of these phosphorylated marks has remained, until recently, unaddressed. A flurry of recent papers has identified kinases that directly phosphorylate EZH2 and that ascribe mechanistic functions to each individual phosphorylation event (Cha et al., 2005; Palacios et al., 2010; Kaneko et al., 2010; Chen et al., 2010; Wei et al., 2011). Collectively, these studies indicate that EZH2 is a nuclear phosphoprotein that assembles and integrates distinct cues and directly converts them into chromatin modifications (i.e., H3K27me3) that control gene expression. While current studies have focused on the involvement of EZH2 phosphorylation in the regulation of gene expression during the cell cycle and tissue regeneration, other well known biological functions of EZH2—including the regulation of ESC maintenance and lineage commitment—may also be influenced via phosphorylation events. Here, we discuss recent discoveries of specific EZH2 residues found to be phosphorylated by the cell-cycle-regulated cyclin-dependent kinases 1 and 2 (CDK1 and CDK2) and the terminal effectors of two intra-cellular signaling pathways elicited by regeneration and other environmental cues—the AKT and p38 alpha kinases.
EZH2 Phosphorylation in Response to Intrinsic, Cell-Cycle-Dependent Signals
Three recent, independent studies have identified EZH2 residues phosphorylated during the cell cycle by CDK1 and CDK2 (Kaneko et al., 2010; Chen et al., 2010; Wei et al., 2011). These kinases are typically activated through association with cyclins A and B, during the cell-cycle progression from S phase to G2/M. Reinberg and colleagues identified an enrichment of EZH2 threonine 345 and 487 (Thr345 and Thr487) phosphorylation by CDK1 during the G2/M phase, using ectopic expression of mouse EZH2 in 293 cells (Kaneko et al., 2010). Based on experiments involving the ectopic expression of phosphomimetic EZH2 mutants, it was concluded that phosphorylation of Thr345 increased the binding of EZH2 to two ncRNAs, HOTAIR and Xist RepA, which in turn mediate PRC2 chromatin recruitment to specific loci, the hox genes and the inactivation center on the X chromosome, respectively. While ncRNA recruitment to these loci might be expected to increase during the G2/M transition, this hypothesis was not examined. Surprisingly, it was estimated that only a small fraction (1%) of nuclear EZH2 appears to be phosphorylated in ESCs during the G2/M phase. This minimal amount of EZH2 Thr345 phosphorylation prompted the authors to suggest a model whereby two distinct pools of PRC2 operate sequentially. According to this model, the Thr345-phosphylated EZH2-containing pool is responsible for initial recognition of target genes and primes them for subsequent recruitment of PCR2 (presumably containing unphosphorylated Thr345 EZH2), which would in turn spread the repression by H3K27me3 (Kaneko et al., 2010). In addition to Thr345, CDK1 was also found to phosphorylate Thr487 and Thr416 EZH2 residues (Kaneko et al., 2010). However, in contrast to the EZH2 Thr345 mutant, binding of the EZH2 Thr487 mutant to ncRNAs was not altered, and thus, the functional significance of Thr487 phosphorylation remains unclear.
The model proposed by Kaneko et al. is consistent with studies from Huang and colleagues, who demonstrate that phosphorylation of Thr350 of human EZH2 (which corresponds to mouse EZH2 Thr345) by CDK1 and CDK2 results in the epigenetic silencing of the hoxA9 and dab2ip genes, in addition to members of the developmental regulatory families hox, fox, and sox (Chen et al., 2010). These authors also showed that a Thr350 phosphomutant EZH2 is impaired in its ability to mediate gene repression and to stimulate cell proliferation and migration, suggesting the potential contribution of EZH2 Thr350 phosphorylation to tumor formation (Chen et al., 2010). Furthermore, this study established a tighter functional correlation between CDK1, CDK2, and EZH2. Specifically, overexpression of CDK1 and CDK2 was found to repress the EZH2 target hoxA9, an effect that was abrogated when EZH2 levels were reduced. In contrast, silencing of CDK1 and CDK2 increased hoxA9 expression.
Intriguingly, EZH2 phosphorylation at Thr350 has been found to regulate expression of 74%–78% of all the EZH2 targets in normal human fibroblasts and the prostate cancer cell line, LNCaP, respectively (Chen et al., 2010). This is an unexpected finding considering that only ~1% of mouse EZH2 Thr345 is phosphorylated in ESCs (Kaneko et al., 2010), as mentioned above. However, cell-type-specific stoichiometries of EZH2 Thr345/350 phosphorylation may account for this discrepancy.
Both Kaneko et al. and Chen et al. found that EZH2 Thr345/350 phosphodefective mutants are still assembled into a PRC2 complex and do not significantly differ in their enzymatic activity from wild-type EZH2. However, recruitment of the EZH2 Thr350 phosphodefective mutant to EZH2-specific targets was impaired. These findings suggest that ncRNAs or proteins interacting with EZH2 phospho-Thr345 may be required to help stabilize PRC2 chromatin binding.
Adding complexity to PRC2 regulation by EZH2 phosphorylation, Wei et al. (2011) reported that Thr487 phosphorylation by CDK1 indirectly inhibits EZH2 methyltransferase activity by disrupting interactions with the other PRC2 components, SUZ12 and EED. However, these results differ from findings reported by Kaneko et al., who observed that an EZH2 Thr487 phosphomimetic could be effectively assembled into a PRC2 complex and maintain its enzymatic activity. Furthermore, in contrast to the findings of Chen et al. (2010), Wei and colleagues reported that inhibiting CDK1 suppressed, rather than promoted, hoxA gene expression, presumably via increased H3K27me3-mediated repression.
A hypothesis that may reconcile the results of these studies posits that a combinatorial pattern of EZH2 phosphorylation controls the spatiotemporal chromatin distribution of PRC2 during G2/M phase progression. That is, according to this model, the sequential phosphorylation of mouse EZH2 Thr345/human EZH2 Thr350 and Thr487 controls dynamic PRC2 association with and dissociation from target genes, as well as its enzymatic activity. More specifically, phosphorylation of mouse Thr345/human Thr350 may precede Thr487 phosphorylation. In this case, the latter modification could be interpreted as the signal that terminates EZH2-mediated gene repression of the G2/M phase, by disassembling the PRC2 complex, thus resuming the expression of genes necessary for initiation of the cell cycle in daughter cells. Conversely, if EZH2 phosphorylation on Thr487 takes place at the early G2/M phase, prior to phosphorylation of mouse Thr345/human Thr350, it may signal termination of EZH2-mediated gene repression during G1/S phase and make PRC2 available for gene repression in G2/M. Alternatively, specific EZH2 phosphorylation might occur within distinct PRC2 pools to regulate independent subsets of genes, possibly in a cell-type-specific manner. For instance, EZH2 phosphorylation on Thr487 appears to mediate derepression of osteogenic genes in mesenchymal stem cells (Wei et al., 2011), which may be a specific feature restricted to the lineage-specific conversion of multipotent cells. Future studies are expected to resolve these apparent discrepancies.
EZH2 Phosphorylation in Response to Extracellular-Regulated Kinases
In adult stem cells, regeneration signals are communicated by converting extracellular signal transduction pathways into epigenetic information in order to control gene expression. The signals that mediate these epigenetic changes may be targeted to the chromatin directly (via histone phosphorylation) or indirectly (via phosphorylation of chromatin-modifying complexes). Important effectors that accomplish this signaling to the chromatin are p38 and AKT, as well as other extracellular signal-activated kinases (Guasconi and Puri, 2009; Chow and Davis 2006).
Several years ago, AKT was shown to have the capacity to suppress EZH2 methyltransferase activity through phosphorylation of serine 21 (Ser21) (Cha et al., 2005), indicating that PRC2 function could be linked with the activation of specific intracellular signals. In this work, the authors demonstrated that AKT-mediated phosphorylation of Ser21 decreased the affinity of EZH2 for histone H3, indicating that the phosphorylation event had an indirect effect on the enzymatic activity of EZH2. However, AKT kinases are downstream of a variety of signals, including mitogenic, differentiation, and metabolic cues. To date, it is not clear what specific cellular event(s) are regulated by EZH2 phosphorylation on Ser21, or which extracellular signals or environmental settings might serve as the upstream trigger(s) for AKT-mediated chromatin modifications. The recent discovery that p38 alpha kinase phosphorylates human EZH2 at Thr372 (corresponding to mouse EZH2 Thr367) in skeletal muscle (satellite) stem cells exposed to TNF—an inflammatory cytokine highly expressed in regenerating muscle—(Palacios et al., 2010) adds stress-activated kinases to the list of EZH2 regulators that do so via direct phosphorylation. EZH2 phosphorylation by p38 alpha promotes PRC2-mediated repression of the skeletal muscle stem cell lineage marker Pax7 during the transition from proliferating satellite cells to differentiated myotubes (Palacios et al., 2010). Thus, this context-dependent example of phosphorylation-mediated repression is restricted to a discrete stage of muscle stem cell differentiation and to a specific subpopulation of differentiating cells in which Pax7 is repressed (Mozzetta et al., 2011). Of note, p38 alpha-directed phosphorylation of EZH2 at human Thr372/mouse Thr367 promotes the interaction between EZH2 and the PRC1-related YY1 protein and correlates with the G0/G1 cell-cycle arrest typical of differentiated muscle cells. Thus, because of the cell-cycle phase in which it occurs, this phosphorylation is predicted to regulate the activity of a distinct PRC2 pool from that subjected to regulation by G2/M-activated CDK1 and CDK2. Furthermore, phosphorylation by TNF-activated p38 alpha is temporally regulated and affects only a residual amount of EZH2, which is typically downregulated in differentiating satellite cells (Caretti et al., 2004; Juan et al., 2009). While these data indicate a cell-type- and stage-specific phosphorylation of EZH2 in response to cues from regenerating muscles, phosphoproteomic analysis of human ESCs has revealed EZH2 phosphorylation at Thr372 (corresponding to mouse Thr367) (Brill et al., 2009), indicating that this phosphorylation event might regulate PRC2 distribution and activity in other cellular contexts.
An interesting implication of the presence of combinatorial phosphorylation patterns is the potential for coordinated, reciprocal relationships between the cell cycle and differentiation machineries, such as repression of differentiation genes by cyclins/CDKs during proliferation, and repression of lineage and cell-cycle genes (Pax7, cyclin A) in response to differentiation cues. It is possible that various cell-cycle-specific patterns of EZH2 phosphorylation may control this switch.
Discussion and Future Directions
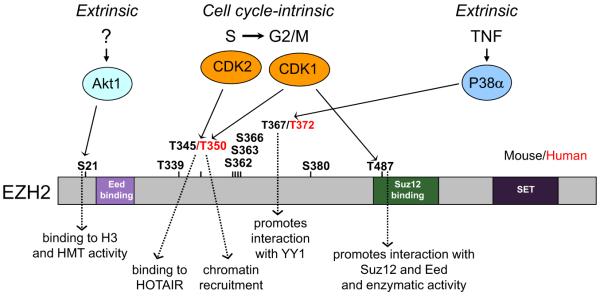
From these studies, a complex scenario emerges where individual EZH2 phosphorylation sites regulate interactions with chromatin, other PcG members, or ncRNAs during the cell cycle and in response to external signals (Figure 1). Similar to other phosphoproteins, it is likely that combinatorial phosphorylation patterns, and perhaps other posttranslational modifications, regulate EZH2 activity, interactions, and nuclear topography to achieve spatiotemporal coordination of gene expression in diverse cell types.
Figure 1.
Illustrative Scheme of Phosphorylation Sites on EZH2, Their Respective Kinases, and Functional Outcomes
The capacity to allow for alternative patterns of combinatorial phosphorylations might enable EZH2 to modulate PcG activity, by dictating the composition of PcG protein/ncRNA complexes that consequently exhibit distinct chromatin affinities and, thus, directly transmit signals to the chromatin. Direct regulation of chromatin structure by EZH2 phosphorylation, in response to signals from the cell-cycle machinery or the environment, opens a number of exciting new avenues of research and testable hypotheses. For instance, EZH2 phosphorylation may be a key event that mediates the effect of media conditioned with specific factors that maintain pluripotency or promote differentiation of human ESCs. Within this context, it will be interesting to determine the effect of inhibiting the activity of distinct pools of PRC2, by virtue of the different pattern of EZH2 phosphorylation in response to specific kinases. Likewise, future studies will likely address the relationship between specific patterns of EZH2 phosphorylation, posttranslation modifications of other components of PRC1 and PRC2 (Niessen et al., 2009), the formation of distinct PRC pools, and their cell type and tissue-specific signaling distribution. Moreover, unraveling the relationship between EZH2 phosphorylation and cell-cycle regulation will have an obvious impact on our understanding of EZH2’s role in the control of proliferation in normal and neoplastic cells.
In defined cellular contexts, simultaneous phosphorylation of target(s) besides PcG by additional kinases might coordinate more complex events, as appears to be the case for MSK-mediated H3S28 phosphorylation, which regulates the accessibility for PRC2-mediated methylation to derepress target genes (Gehani et al., 2010).
EZH2 phosphorylation can be also regarded as an additional cellular control mechanism that limits postmitotic cell responses to external signals. For instance, it may be involved in the resistance of terminally differentiated cells, such as skeletal myotubes (Caretti et al., 2004) and keratinocytes (Ezhkova et al., 2009), to certain environmental cues. In this model, the downregulation of EZH2 that occurs in terminally differentiated cells (Juan et al., 2009) may serve to insulate the chromatin at specific genes, which were repressed by PRC2 in proliferating progenitors from external signals. In other words, the absence of EZH2 would compromise chromatin responsiveness at those loci, which were previously regulated through EZH2 phosphorylation. This scenario may be the case for Pax7, which is dynamically regulated by TNF signaling in activated satellite cells (Palacios et al., 2010) but becomes unresponsive to the same signal upon satellite cell differentiation into myotubes and which coincides with the disappearance of EZH2 (Mozzetta et al., 2011). A mechanism that followed this pattern could allow for the observed cell specificity of signals delivered within a heterogeneous environment, such as the “niche” in which various adult tissue stem cells are regulated.
In addition to containing multiple phosphorylation sites, EZH2 contains several predicted acetylation sites (www.phosida.com). It is tempting to speculate that, analogously to histones, an “EZH2 code” may regulate the dynamics of PcG recruitment by altering protein and RNA interactions, substrate selection, and cell compartmentalization (Su et al., 2005).
As often occurs in the “early days” following important new discoveries, the studies discussed here have begun to shed an imperfect, but nonetheless vivid, light to illuminate how EZH2 phosphorylation has the potential to directly transmit signals generated by the cell-cycle machinery and by extracellular-regulated kinases down to the level of chromatin. Collectively, these recent examples suggest that EZH2 phosphorylation is a key mechanism by which mitogens and growth factors can influence numerous cell processes, such as proliferation, cell lineage determination, and differentiation in stem cells, through directing genome distribution and activity of PcG. We expect that the generation of cells and knockin animals with individual and combinatorial mutations of EZH2 phosphorylation sites will reveal additional intricacies and surprises to the Polycomb world.
Within this regulatory context, it will be important to clarify how conformational changes potentially induced by posttranslational modifications or by interaction with proteins or ncRNAs affect the enzymatic activity of EZH2. This impact may occur physiologically in response to developmental cues but also in pathological conditions, such as cancer, where EZH2 dysregulation has been shown to play an important role in conferring both oncogenic and metastatic potential.
Likewise, it will be important to understand how EZH2 regulation by phosphorylation is integrated within a broader scenario in which other components of the transcriptional machinery (i.e., other chromatin-modifying complexes and transcription factors) and histones themselves are altered by posttranslational modifications in response to specific signals.
ACKNOWLEDGMENTS
P.L.P. is an Associate Telethon Scientist of the Dulbecco Telethon Institute (DTI) and Associate Investigator of Sanford Children’s Health Research Center. This work has been supported by R01 AR053779 from the National Institute of Health/National Institute of Arthritis and Musculoskeletal and SkinDiseases (NIAMS) (PLP) and by the Intramural Program of the National Institute of Arthritis, Musculoskeletal, and Skin Diseases of the National Institutes of Health (V.S.). G.C. is supported by AIRC MFAG 5386. We thank Dr. D. Pasini and Dr. J.G. Ryall for critical reading of the manuscript.
REFERENCES
- Brill LM, Xiong W, Lee KB, Ficarro SB, Crain A, Xu Y, Terskikh A, Snyder EY, Ding S. Cell Stem Cell. 2009;5:204–213. doi: 10.1016/j.stem.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caretti G, Di Padova M, Micales B, Lyons GE, Sartorelli V. Genes Dev. 2004;18:2627–2638. doi: 10.1101/gad.1241904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha TL, Zhou BP, Xia W, Wu Y, Yang CC, Chen CT, Ping B, Otte AP, Hung MC. Science. 2005;310:306–310. doi: 10.1126/science.1118947. [DOI] [PubMed] [Google Scholar]
- Chen S, Bohrer LR, Rai AN, Pan Y, Gan L, Zhou X, Bagchi A, Simon JA, Huang H. Nat. Cell Biol. 2010;12:1108–1114. doi: 10.1038/ncb2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow CW, Davis RJ. Cell. 2006;127:887–890. doi: 10.1016/j.cell.2006.11.015. [DOI] [PubMed] [Google Scholar]
- Dephoure N, Zhou C, Villen J, Beausoleil SA, Bakalarski CE, Elledge SJ, Gygi SP. Proc. Natl. Acad. Sci. USA. 2008;105:10762–10767. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezhkova E, Pasolli HA, Parker JS, Stokes N, Su IH, Hannon G, Tarakhovsky A, Fuchs E. Cell. 2009;136:1122–1135. doi: 10.1016/j.cell.2008.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehani SS, Agrawal-Singh S, Dietrich N, Christophersen NS, Helin K, Hansen K. Mol. Cell. 2010;39:886–900. doi: 10.1016/j.molcel.2010.08.020. [DOI] [PubMed] [Google Scholar]
- Guasconi V, Puri PL. Trends Cell Biol. 2009;19:286–294. doi: 10.1016/j.tcb.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttlin EL, Jedrychowski MP, Elias JE, Goswami T, Rad R, Beausoleil SA, Villén J, Haas W, Sowa ME, Gygi SP. Cell. 2010;143:1174–1189. doi: 10.1016/j.cell.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juan AH, Kumar RM, Marx JG, Young RA, Sartorelli V. Mol. Cell. 2009;36:61–74. doi: 10.1016/j.molcel.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko S, Li G, Son J, Xu CF, Margueron R, Neubert TA, Reinberg D. Genes Dev. 2010;24:2615–2620. doi: 10.1101/gad.1983810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Reinberg D. Nature. 2011;46:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayya V, Lundgren DH, Hwang SI, Rezaul K, Wu L, Eng JK, Rodionov V, Han DK. Sci. Signal. 2009;2:ra46. doi: 10.1126/scisignal.2000007. [DOI] [PubMed] [Google Scholar]
- Mozzetta C, Consalvi S, Saccone V, Forcales SV, Puri PL, Palacios D. Cell Cycle. 2011;10:191–198. doi: 10.4161/cc.10.2.14441. [DOI] [PubMed] [Google Scholar]
- Niessen HE, Demmers JA, Voncken JW. Epigenetics Chromatin. 2009;2:10. doi: 10.1186/1756-8935-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios D, Mozzetta C, Consalvi S, Caretti G, Saccone V, Proserpio V, Marquez VE, Valente S, Mai A, Forcales SV, et al. Cell Stem Cell. 2010;7:455–469. doi: 10.1016/j.stem.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvageau M, Sauvageau G. Cell Stem Cell. 2010;7:299–313. doi: 10.1016/j.stem.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawarkar R, Paro R. Dev. Cell. 2010;19:651–661. doi: 10.1016/j.devcel.2010.10.012. [DOI] [PubMed] [Google Scholar]
- Simon JA, Kingston RE. Nat. Rev. Mol. Cell Biol. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- Su IH, Dobenecker MW, Dickinson E, Oser M, Basavaraj A, Marqueron R, Viale A, Reinberg D, Wulfing C, Tarakhovsky A. Cell. 2005;121:425–436. doi: 10.1016/j.cell.2005.02.029. [DOI] [PubMed] [Google Scholar]
- Surface LE, Thornton SR, Boyer LA. Cell Stem Cell. 2010;7:288–298. doi: 10.1016/j.stem.2010.08.004. [DOI] [PubMed] [Google Scholar]
- Wei Y, Chen YH, Li LY, Lang J, Yeh SP, Shi B, Yang CC, Yang JY, Lin CY, Lai CC, et al. Nat. Cell Biol. 2011;13:87–94. doi: 10.1038/ncb2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Ohsumi TK, Kung JT, Ogawa Y, Grau DJ, Sarma K, Song JJ, Kingston RE, Borowsky M, Lee JT. Mol. Cell. 2010;40:939–953. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]