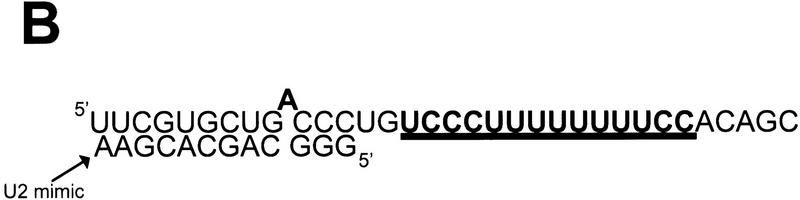Abstract
During the early events of pre-mRNA splicing, intronic cis-acting sequences are recognized and interact through a network of RNA–RNA, RNA–protein, and protein–protein contacts. Recently, we identified a branchpoint sequence binding protein in yeast (BBP). The mammalian ortholog (mBBP/SF1) also binds specifically to branchpoint sequences and interacts with the well studied mammalian splicing factor U2AF65, which binds to the adjacent polypyrimidine (PY) tract. In this paper we demonstrate that the mBBP/SF1-U2AF65 interaction promotes cooperative binding to a branchpoint sequence-polypyrimidine tract-containing RNA, and we suggest that this cooperative RNA binding contributes to initial recognition of the branchpoint sequence (BPS) during pre-mRNA splicing. We also demonstrate the essential nature of the third RBD of U2AF65 for the interaction between the two proteins, both in the presence and absence of RNA.
Keywords: BPS, PYtract, protein–protein interaction, RNA
Pre-mRNA splicing is performed by the spliceosome, a large complex consisting of several small nuclear ribonucleoproteins and many non-snRNP proteins. Spliceosome assembly proceeds through several intermediate complexes, beginning with the E complex, or commitment complex, proceeding to A, B, and, finally, to the C complex, within which chemistry (phosphodiester bond cleavage and formation) takes place. Mammalian splicing substrates contain an intronic 5′ splice site, branchpoint sequence (BPS), polypyrimidine (PY) tract, and 3′ splice site. The 2′-OH of the branchpoint adenosine within the BPS is the nucleophile for attack of the 5′ splice site, which forms a lariat intermediate and a free 5′ exon; in the second step, the 3′ OH of the liberated 5′ exon attacks the 3′ splice site, resulting in the mRNA and lariat intron products (for review, see Moore et al. 1993; Madhani and Guthrie 1994; Kramer 1996).
During the initial steps of in vitro splicing, the 5′ and 3′ ends of introns are recognized by the U1 snRNP and other splicing factors, respectively. This corresponds to formation of the commitment complex in yeast and the E complex in mammals (Moore et al. 1993). There are modest sequence requirement differences between the two systems, especially on the 3′ side of the intron (BPS, PY tract, 3′ splice site). In yeast, the BPS is the almost invariant UACUAAC (Rymond and Rosbash 1992), whereas in mammals, the BPS is degenerate with a consensus sequence of YNCURAY (Keller and Noon 1984; Green 1986). In mammals, a PY tract is highly conserved and an essential 3′ side element (Reed and Maniatis 1985; Ruskin and Green 1985). This is much less conserved and important in yeast introns, although a uridine tract will modestly enhance yeast splicing (Patterson and Guthrie 1991). The splicing components involved in E complex or commitment complex formation include: (1) U1 snRNP base pairing to the 5′ splice site in both systems (Mount et al. 1983; Zhuang and Weiner 1986; Séraphin et al. 1988; Siliciano and Guthrie 1988); (2) U2AF65 binding the PY tract (mammalian) and Mud2p, which cross-links to the 3′ side of yeast introns (Zamore et al. 1992; Abovich et al. 1994; data not shown); (3) SR proteins, which promote U1 snRNP and U2AF65 binding in E complex formation in mammals (Kohtz et al. 1994; Staknis and Reed 1994); (4) the recently identified yeast and mammalian branchpoint sequence binding proteins (BBP and mBBP/SF1, respectively), which recognize the BPS (Abovich and Rosbash 1997; Berglund et al. 1997). mBBP is also known as splicing factor one (SF1), which was initially identified as an essential factor for A complex formation (Kramer 1992; Arning et al. 1996). In both yeast and mammals, a network of interactions between U1 snRNP at the 5′ side and proteins at the 3′ side bridges the two ends of the intron and facilitate intron definition and removal (Reed 1996; Abovich and Rosbash 1997).
The organization of mammalian intron 3′ side sequence elements (from 5′ to 3′) is BPS–PY tract–3′ splice site (AG), and the distance from the branchpoint adenosine to the 3′ splice site is normally between 20 and 40 nucleotides (Reed 1989; Smith and Nadal-Ginard 1989). Thus, the BPS and PY tract are usually adjacent. Because we had shown previously that U2AF65 and mBBP can interact (Abovich and Rosbash 1997), we suspected that this might contribute to intron recognition. A cooperative binding interaction would facilitate BPS selection despite low sequence information content within the degenerate mammalian BPS. Although a similar cooperative interaction may occur between BBP and Mud2p in yeast, this is more difficult to test; the RNA binding properties of Mud2p are not known, and recombinant Mud2p is not available.
For the mammalian proteins, the binding affinity of U2AF65 for the PY tract has been determined, and all three RNA-binding domains (RBDs) affect the strength of the interaction between U2AF65 and the PY tract (Zamore et al. 1992). U2AF65 also has an amino-terminal RS domain, rich in arginines and serines. RS domains are generally involved in protein-protein interactions within the spliceosome, mediated in part through serine phosphorylation (Fu 1995). However, the RS domain of U2AF65 is unusual, as it interacts with RNA and may not mediate protein-protein interactions: U2AF65-RS has been cross-linked to the BPS and facilitates the annealing of the snRNA in U2 to the BPS in biochemical experiments (Valcárcel et al. 1996).
We have shown more recently that the other mammalian 3′ side factor, mBBP, specifically recognizes the yeast BPS (UACUAAC), which is also the preferred BPS in mammals (Zhuang et al. 1989). The putative RNA binding region of mBBP contains a KH and a Zn knuckle domain and is sufficient for specific RNA binding (Berglund et al. 1997). Although mBBP recognizes the BPS, its affinity and specificity are less impressive than those of its yeast ortholog BBP, which recognizes all seven positions within the yeast BPS. In contrast, mBBP binding is only affected by point mutations at the branchpoint adenosine and the conserved uridine two nucleotides upstream; these are the two most conserved nucleotides in the mammalian branchpoint consensus sequence, YNCURAY (Keller and Noon 1984). This poorly conserved, short sequence is probably insufficient to specify a mBBP–BPS interaction. We speculated that if U2AF65 were bound to the PY tract, protein–protein contacts with mBBP would facilitate recognition of the BPS (Berglund et al. 1997).
Here, we use recombinant mBBP and U2AF65 to demonstrate a cooperative interaction during binding to a RNA substrate containing a BPS and PY tract. The formation of this ternary complex (mBBP, U2AF65, and RNA substrate) appears to be aided by the presence of the branchpoint adenosine within the BPS as well as by a contiguous PY tract. We also demonstrate the importance of the third RBD of U2AF65, for the physical interaction between U2AF65 and mBBP both in the presence and absence of RNA. The results suggest that cooperative RNA binding by these two proteins constitutes initial recognition of the BPS region and may even contribute to branchpoint selection.
Results
Cooperative interaction between mBBP and U2AF65
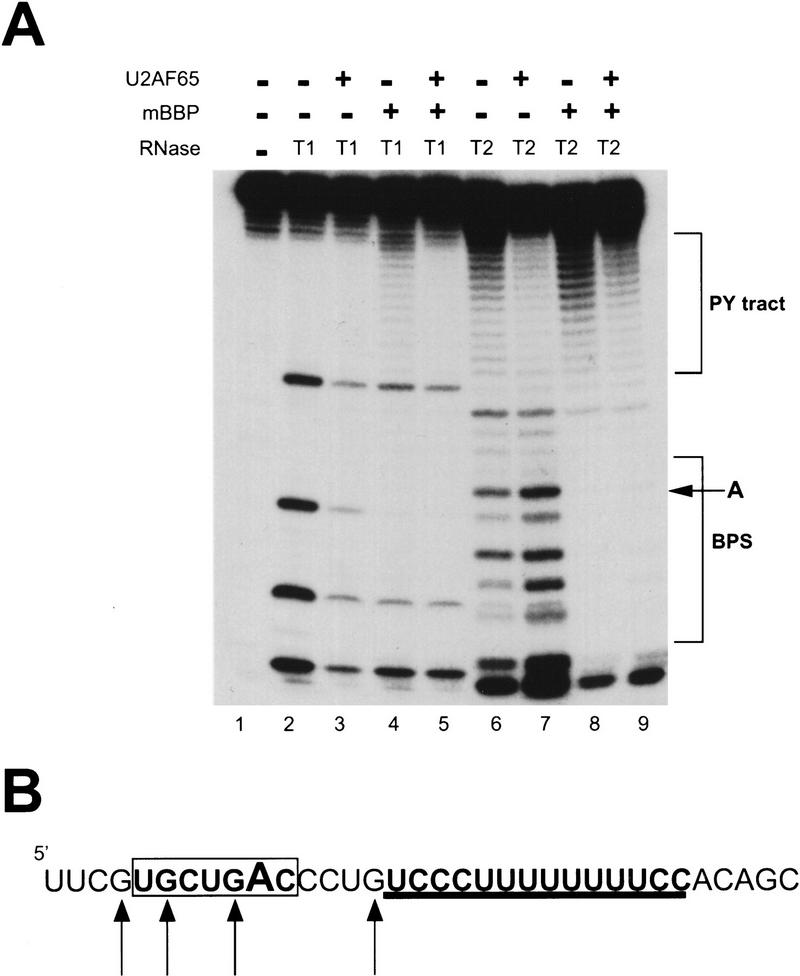
The nucleotides important for specific recognition by mBBP and U2AF65 have been identified within their respective RNA elements (Zamore et al. 1992; Singh et al. 1995; Berglund et al. 1997). However, footprinting of U2AF65 to RNA has only been assayed with CMCT, which does not examine protection of adenosine, cytosine, or guanosine (Singh et al. 1995). Additional experiments might determine whether either protein interacts with RNA on either side of its recognition site. This might influence cooperative binding by the two proteins. To this end, footprinting experiments were performed with purified full-length recombinant U2AF65, mBBP containing amino acids 1–361 (Fig. 1), and a 34-nucleotide RNA substrate derived from the 3′ side of adenovirus major late pre-mRNA. In this commonly employed pre-mRNA substrate, the BPS and PY tracts are separated by only four nucleotides (Fig. 2B). If either protein extends past its specific site, this might influence the interaction between the two proteins.
Figure 1.

Commassie stained SDS–polyacrylamide gel of purified U2AF65, U2AF65-3, and mBBP. Three micrograms of protein was loaded in each lane. Protein markers are from GIBCO.
Figure 2.
Double footprint of mBBP and U2AF65. (A) Purified mBBP, U2AF65, or both proteins together were incubated with 5′-end labeled RNA substrate in the presence of either RNase T1 (lanes 2–5) or RNase T2 (lanes 6–9), and then run in a denaturing polyacrylamide gel as described in Materials and methods. (Lanes 4,8) The concentration of mBBP used was 11 μm; (lanes 3,7) concentration of U2AF65 was 2.8 μm; (lanes 5,9) mBBP was at a concentration of 5.5 μm and U2AF65 at 1.4 μm. The PY tract and BPS are marked by brackets, and the position of the branchpoint adenosine is marked by an arrow. (B) Sequence of the 34-nucleotide RNA substrate derived from the Adenovirus major late pre-mRNA substrate. The BPS is in bold and boxed, and the branchpoint adenosine is in large text. The PY tract is in bold and underlined. Arrows represent guanosines cleaved by RNase T1 (lane 2).
U2AF65 protects the PY tract but not the BPS (Fig. 2A, lane 7), and mBBP protects only the BPS and not the PY tract (Fig. 2A, lane 8). The two proteins, therefore, sit side-by-side, with no more than a 1- or 2-nucleotide space in between. The protection pattern for mBBP is reminiscent of that observed for the yeast ortholog (BBP); both proteins protect the 7 nucleotides of the BPS and only 2 nucleotides on either side (Berglund et al. 1997). With U2AF65 as well as mBBP, the protection spans both the BPS and PY tract (Fig. 2A, lane 9). In this case, the protection of the PY tract is weaker (lane 9), probably the result of a low level of contaminating RNases in the mBBP preparation. Evidence supporting this interpretation is in lane 4: RNase T1 should have only guanosine cleavages, but addition of mBBP induces some cleavage of the PY tract.
RNase T1 was assayed primarily for mapping purposes, because the 34 nucleotide RNA contains only five guanosines, two of which are in the BPS (Fig. 2B). The four visible guanosines are all partially protected by U2AF65, possibly because of steric effects on RNase T1 activity or to additional U2AF65 molecules that bind nonspecifically to the BPS (Fig. 2B, lane 3). mBBP also protects the four guanosines; protection of three is similar to U2AF65, but mBBP completely protects the guanosine next to the branchpoint adenosine (Fig. 2A, lanes 4,5). This suggests that the complete protection of this one guanosine is the result of a strong mBBP interaction at this position, and the partial protection of the other guanosines is caused by weaker interactions or to steric inhibition of RNase T1. The complete protection of this particular guanosine recalls the fact that the branchpoint adenosine and adjacent uridine, the nucleotides to either side of this guanosine, have the strongest effect on mBBP binding (Berglund et al. 1997).
To verify the mBBP footprinting and the sequence-specific interactions of mBBP at the BPS, we used a competition assay involving a 12-nucleotide RNA that base-pairs to the BPS as well as to a few nucleotides upstream and downstream of the BPS. The oligo was designed to encourage a bulged branchpoint adenosine, as has been shown for the U2 snRNP–BPS interaction (Query et al. 1994). This antisense molecule is, therefore, a simplified mimic of U2 snRNP (Fig. 3B, U2 mimic).
Figure 3.
The binding of a 12-nucleotide RNA to the BPS competes with mBBP binding. (A) (Lane 1) radiolabeled RNA alone; (lane 2–11) either the 12-nucleotide RNA (U2 mimic), mBBP, U2AF65, or a combination as shown above the autoradiograph. The different complexes are labeled by arrows (right). (B) Sequences of the two RNA oligoribonucleotides. The branchpoint adenosine is shown in bold and bulged out, and the PY tract is in bold and underlined.
Base-pairing of the U2 mimic to the BPS blocks mBBP binding (Fig. 3A, lanes 4,5). Only at a low concentration of U2 mimic is mBBP binding detectable (lane 6). This suggests that mBBP and U2 snRNP binding are mutually exclusive. In contrast, U2 mimic base-pairing has only a modest effect on U2AF65 binding, and slightly decreases the mobility of the complex (Fig. 3A, lanes 9,10). The data indicate that U2AF65 can form a ternary complex with the 34-mer and U2 mimic (Valcárcel et al. 1996). Because mBBP is present in E complex (Abovich and Rosbash 1997), mBBP probably binds to the BPS and is then replaced by U2 snRNP during or prior to A complex formation.
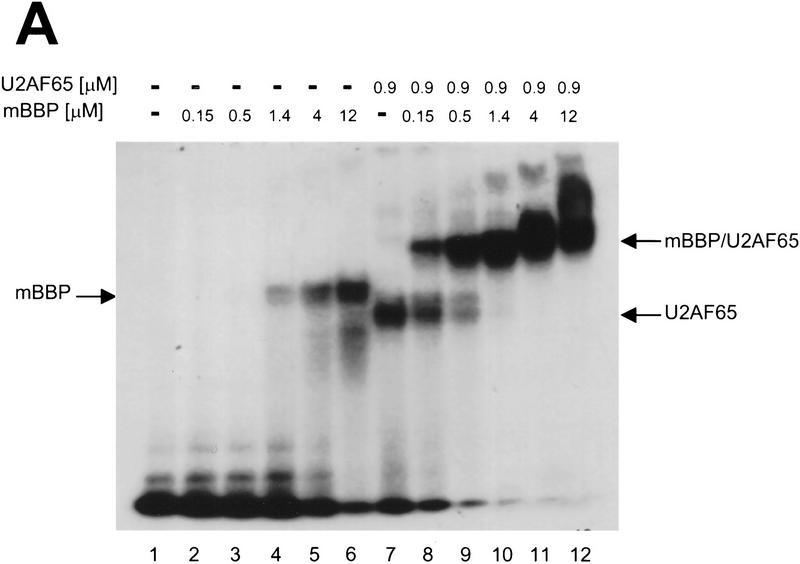
We used the same RNA substrate (Fig. 2B) and purified proteins to establish assays using native polyacrylamide gel electrophoresis. Purified mBBP, U2AF65, or both together were incubated in the presence of radiolabeled RNA, and complexes were then separated on a 6% 0.5× TBE gel (Materials and Methods). The different complexes are easily distinguished (Fig. 4A): lane 7, U2AF65–RNA; lane 5, mBBP–RNA; lane 10, mBBP–U2AF65–RNA. The KDs are ∼1 μm for U2AF65 and 6 μm for mBBP. For both binary complexes, binding appears weakly cooperative. This is apparent in previous studies with U2AF65 (Zamore et al. 1992; Lee et al. 1993) and has been generally more discussed for proteins with multiple RNA binding domains (Birney et al. 1993).
Figure 4.
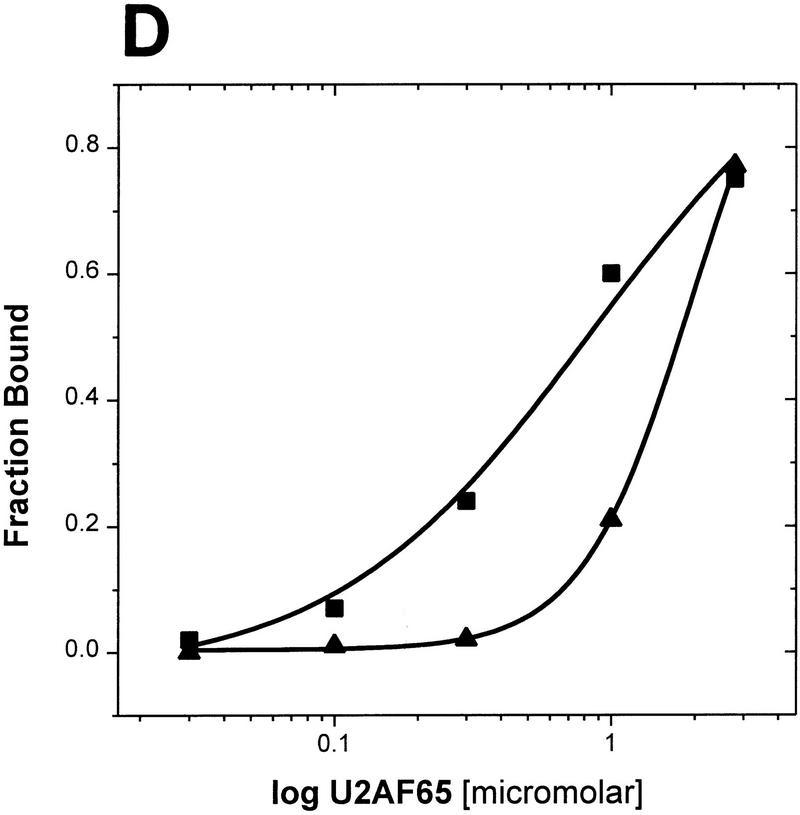
Cooperative binding of mBBP and U2AF65. (A) In a gel-shift assay using a radiolabed 34-nucleotide RNA substrate derived from the Adenovirus major late pre-mRNA and purified proteins mBBP and U2AF65, we assayed for cooperative binding between mBBP and U2AF65 (Materials and Methods). (Lanes 2–6) Increasing concentrations (top) of mBBP; the complex is marked (left) by an arrow. (Lane 7) U2AF65 plus RNA; the complex is again marked by an arrow (right). (Lanes 8–12) U2AF65 at the same concentration as lane 7 plus mBBP at increasing concentrations, the same as those in lanes 2–6. The ternary complex of mBBP/U2AF65/RNA is marked by an arrow (right). (B) Graphical representation of the data shown in A (□) mBBP; (▵) mBBP + U2AF65. (C) The same experiment as in A except that U2AF65 concentration is varied (at top); the concentration of mBBP is held constant at 9 μm. The different complexes are marked at left and right. (D) Graphical representation of the data shown in C (▴) U2AF65; (▪) U2AF65 + mBBP. These experiments were repeated multiple times under multiple conditions with approximately the same 20-fold and 5-fold effects in cooperativity observed.
The ternary complex of U2AF65, mBBP, and RNA is clearly distinguishable from the binary complexes. Cooperativity between U2AF65 and mBBP was defined as the ability of one protein to increase the affinity of the other for RNA. A comparison of lanes 2–6 (mBBP) with lanes 8–12 (mBBP plus U2AF65) indicates cooperative RNA binding by the two proteins. In the presence of U2AF65, mBBP forms a complex at 0.57 μm (lane 9) but fails to bind at this same concentration without U2AF65 (lane 3). Quantification reveals a 20-fold increase in mBBP’s RNA affinity by U2AF65 (Fig. 4B). The effect is based on the difference in apparent Kds (Materials and Methods): 6 μm in the presence of RNA alone and 0.3 μm with U2AF65. The same effect was observed at different U2AF65 concentrations (see legend to Fig. 4; data not shown), but the presence of multiple complexes at saturating U2AF65 concentrations made identification and interpretation of the various complexes difficult.
The experiment was also performed in the opposite way, namely, mBBP concentration was held constant and U2AF65 concentration was varied. With this protocol, cooperativity was also observed, but the effect was less strong: 5-fold compared with 20-fold (Fig. 4C,D). On the basis of thermodynamics, cooperativity should be independent of which protein concentration is held constant and which is varied. The difference could be the result of a failure to achieve equilibrium under the experimental conditions used. For all cooperative binding assays, incubations were for 1 hr at room temperature before separation by native gel electrophoresis. In any case, both protocols indicate that mBBP and U2AF65 bind in a cooperative manner to an RNA substrate containing a BPS and PY tract.
The effect of RNA mutations on the cooperative interaction between U2AF65 and mBBP
To determine the contribution of the substrate RNA elements to the cooperative interaction, we assayed mutations within the BPS and PY tract. A mutation changing the branchpoint adenosine to a cytidine was made (indicated by an * above the change); this decreased the mBBP binding affinity and also changed the complex mobility (cf. Figs. 5A, lanes 5 and 6, and 4A). The decreased affinity agrees with our previous work demonstrating that a mutant branchpoint adenosine within the context of a yeast BPS (UACUAAC) decreased mBBP binding (Berglund et al. 1997). The mobility change could be caused by the binding of multiple mBBP molecules or to mBBP binding to a different region of the RNA substrate. In the presence of U2AF65 (Fig. 5A, lanes 8–12), mBBP may still bind weakly to the mutant BPS, on the basis of a similar migration of the ternary complex (Fig. 5A, lanes 10–12, indicated by an arrow) to the mobility of the ternary complex with wild-type RNA (Fig. 4A). The amount of ternary complex is greatly reduced by mutating the branchpoint adenosine. Because of the weak and aberrant binding of mBBP, it is difficult to determine the extent to which cooperative binding to the mutant substrate is reduced. It is clear, however, that U2AF65 is not able to easily recruit mBBP in the absence of a consensus BPS.
Figure 5.
Mutation of the branchpoint adenosine reduces formation of the ternary complex (mBBP, U2AF65, and RNA substrate). (A) A 34-nucleotide RNA substrate with the branchpoint adenosine mutated gel-shift experiments was used to perform under the same conditions as those in Fig. 4A. The mBBP/RNA complex has a different migration pattern as marked by an arrow to the side of the autoradiograph. (B) Sequence of the mutated 34-nucleotide RNA substrate used in this experiment. The branchpoint adenosine was changed to cytidine and is marked by an asterisk (*).
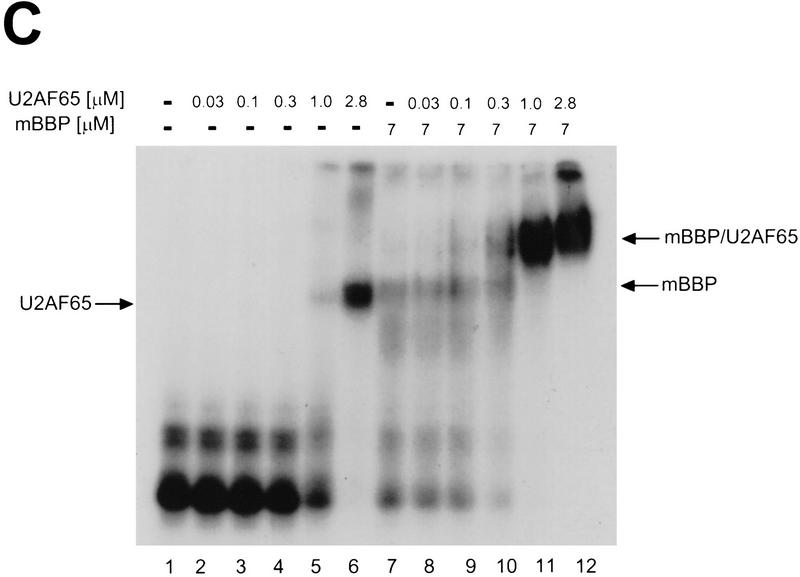
To determine the effect of mutations in the PY tract, we changed two central uridines to guanosines [**; (Fig. 6B)]. In this experiment, mBBP concentration was constant and the concentration of U2AF65 was varied (Fig. 6A). As expected, the PY tract mutation reduces U2AF65 binding (lanes 2–6) compared with the wild-type RNA substrate (Fig. 4C). With this substrate, however, U2AF65 binding is still enhanced in the presence of mBBP (Fig. 6A). This is based on the appearance of the ternary complex at a lower concentration of U2AF65 compared with U2AF65 binding without mBBP (Fig. 6A, cf. lanes 5 and 11). Under these conditions (constant mBBP and variable U2AF65), the cooperative binding interaction to the wild-type RNA is fivefold, and these mutations within the PY tract reduce it (cf. Figs. 4C and 6A). Thus, mutations in both the BPS or PY tract reduce formation of a proper ternary complex.
Figure 6.
Mutating the PY tract reduces cooperative formation of the ternary complex (mBBP, U2AF65, and RNA substrate). (A) This is the same experiment as in Fig. 4C except the PY tract within the 34-nucleotide RNA substrate has been mutated. The different complexes are indicated to left and right. (B) Sequence showing the double mutation of the PY tract. Two uridines in the middle were changed to guanosine, as indicated by asterisks (**).
Domains within U2AF65 and mBBP important for cooperativity
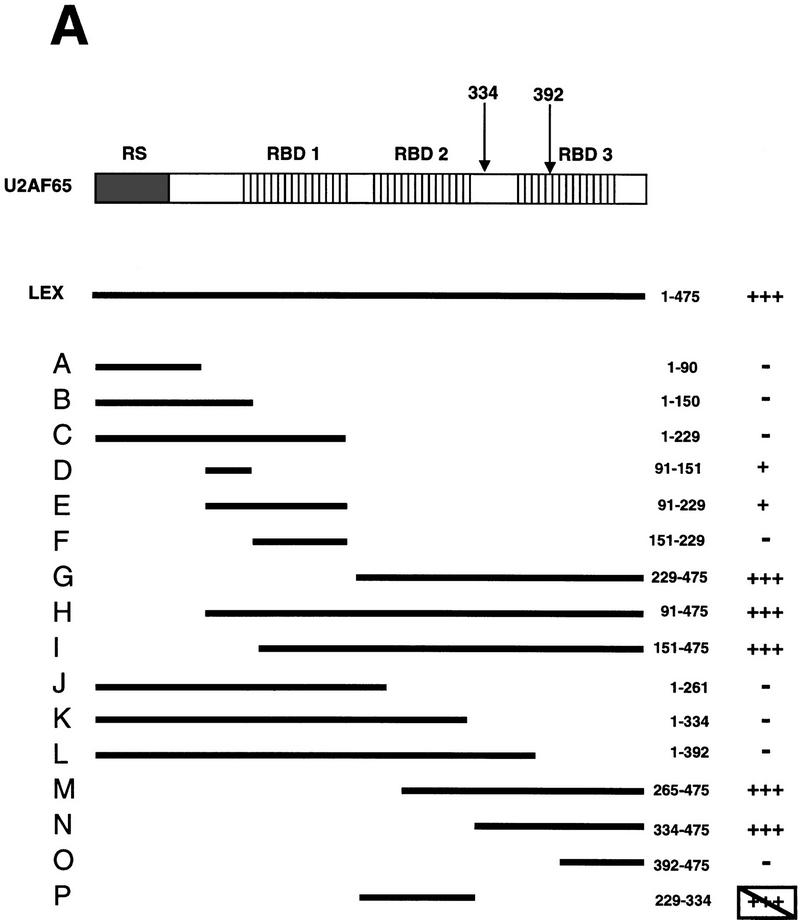
All four domains of U2AF65 have been shown to be involved in RNA interactions (Fig 7A; schematic representation): All three RBDs contribute to binding with a long PY tract, and the RS domain cross-links to the BPS and acts as a chaperone for U2 binding to the BPS (Zamore et al. 1992; Gaur et al. 1995; Valcárcel et al. 1996). To determine which domain(s) contribute to the mBBP interaction, we used yeast two-hybrid and GST-precipitation assays.
Figure 7.
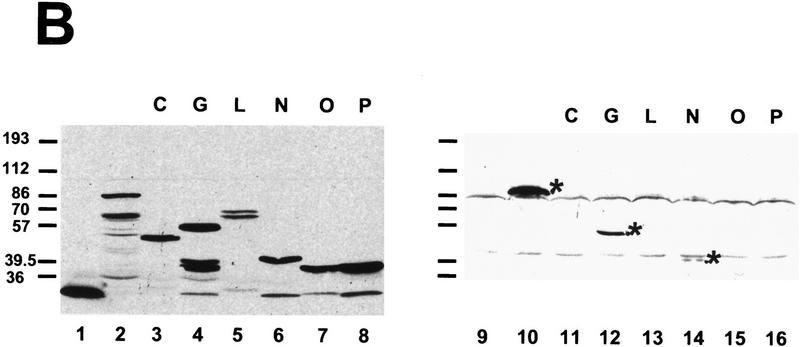
mBBP interacts with the carboxy-terminal RBD of U2AF65. (A) Yeast two-hybrid interactions. Yeast cells carrying the B42–mBBP fusion and the indicated LexA–U2AF65 fusions were obtained and tested for B-galactosidase production as described in Materials and Methods. A schematic representation of U2AF65 depicting the amino-terminal RS domain and the three RNA-binding domains RBD 1–3. The bars indicate the U2AF65 region present in each fusion and the numbers correspond to the U2AF65 amino acids fused to LexA. Plus (+) and minus (−) signs indicate β-galactosidase activity. The box around the three pluses of the P clone indicates that this fusion activates transcription independently of B42–mBBP expression. (B) LexA–U2AF65 fusions interacting with GST–mBBP. (Lane 1) Whole cell extracts were prepared from yeast cells carrying LexA alone, (lane 2) LexA–U2AF65, or (lanes 3–8) the indicated LexA–U2AF65 fusions, and 15 μl was incubated with the GST–mBBP fusion protein bound to glutathione agarose beads as described in Materials and Methods. The bound proteins were eluted in SDS sample buffer and separated in 8% polyacrylamide–SDS gels. After transfer to nitrocellulose the LexA fusion proteins were visualized with anti-LexA antibody. (Left panel) corresponds to 10 μl of the extracts directly loaded on the gel, and (right) proteins after elution from the GST–mBBP beads.
Figure 7A summarizes the yeast two-hybrid results. All clones containing the complete third RBD manifested an interaction with mBBP similar to that observed with full-length U2AF65. Only clone P (Fig. 7A) is inconsistent with this conclusion. Clone P is a false-positive, because in the absence of mBBP (glucose is added to repress mBBP expression) the same strong activation is seen (data not shown). To confirm the two-hybrid results, we made yeast extracts containing several of the different fusion proteins and used GST–mBBP to precipitate these LexA–U2AF65 chimeric proteins. Full-length U2AF65, clones containing the third RBD and the third RBD alone, interacted with GST–mBBP (Fig. 7B, lanes 10,12,14). Clone P (lane 16) was not recovered, consistent with the false-positive interpretation. The two-hybrid and precipitation results indicate that the third RBD of U2AF65 is the relevant domain for interaction with mBBP.
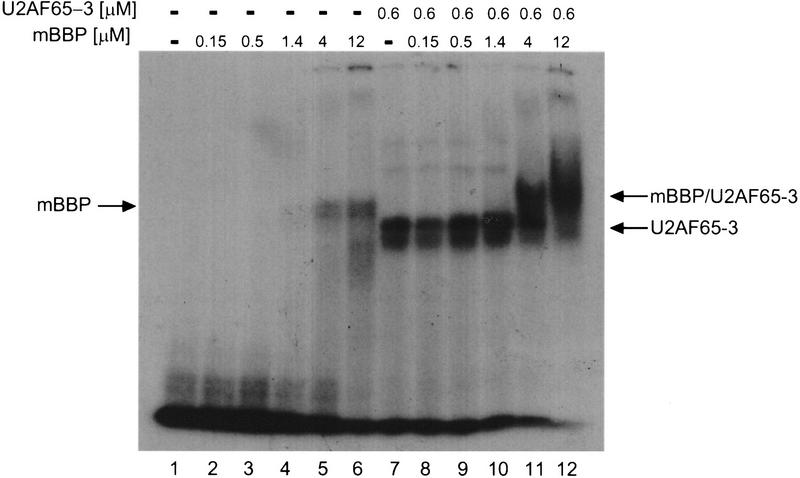
U2AF65 missing the third RBD (U2AF65-3) was then expressed, purified (Fig. 1; Materials and Methods) and used in the gel-shift assay to determine the relevance of the third RBD to the cooperative RNA-binding interaction of U2AF65 and mBBP. As previously reported (Zamore et al. 1992), U2AF65-3 binds to the 34 nucleotide RNA with a similar affinity to that of full-length U2AF65. This presumably reflects the rather short nature of the PY tract of this substrate. In contrast to U2AF65, however, U2AF65-3 cannot promote cooperative binding of mBBP to RNA (Fig. 8). A ternary complex is still formed (lanes 11,12) but only at concentrations where mBBP alone is also able to bind (cf. lanes 5,6 and 11,12). Therefore, removal of the third RBD of U2AF65 eliminates a detectable physical interaction between U2AF65 and mBBP, both in the absence and presence of RNA.
Figure 8.
The third RBD of U2AF65 is necessary for the cooperative interaction between mBBP and U2AF65. Under the same conditions as those for the experiment shown in Fig. 4A (Materials and Methods), we looked for a cooperative interaction between mBBP and a U2AF65 protein missing the third RBD (U2AF65-3). The three different complexes are marked by arrows at left and right.
We had shown previously that the region of mBBP containing the KH domain and Zn knuckle is sufficient for specific binding to the BPS (Berglund et al. 1997). To determine if this same region is sufficient for the cooperative interaction with U2AF65, we used a protein containing amino acids 135–308 from mBBP, termed mBBP(181), in the cooperativity assay. mBBP(181) binds ∼10-fold weaker to this RNA substrate compared with the mBBP (1–361) used previously, suggesting that the amino-terminal portion of mBBP contributes to RNA binding affinity. Importantly, a ternary complex with U2AF65 is visible, but there is no detectable cooperative interaction between the two proteins (data not shown). Taken together with other data, the result provisionally assigns the relevant interacting region of mBBP to the amino-terminal end of the protein (see Discussion).
Discussion
The footprinting data demonstrate that mBBP and U2AF65 can simultaneously interact with this model RNA substrate in a side-by-side manner (Fig. 2). On the basis of experiments in splicing extracts (Abovich and Rosbash 1997), these interactions probably occur during or prior to E complex formation. As this step precedes U2 snRNP addition to the prespliceosome (Michaud and Reed 1991), mBBP binding probably reflects the initial recognition of the BPS.
Previous results indicated that mBBP, as well as BBP, might interact with the bases of the branchpoint region, and the mutually exclusive binding of mBBP or the U2 mimic (Fig. 3) is consistent with this interpretation. This indicates that U2 snRNP addition and formation of the U2 snRNA–BPS duplex destabilizes mBBP, and there is some indication that this protein may no longer be present in the U2 snRNP-containing A complex (R. Reed, pers. comm.). In contrast, U2AF65 is an A complex component as well as an E complex component. Consistent with this role, U2AF65 can form a ternary complex with the U2 mimic and the 34 nucleotide RNA substrate (Fig. 3). Formation of this complex is consistent with the previous suggestion that the U2AF65RS domain contributes to BPS–U2snRNA base-pairing (Valcárcel et al. 1996). The third RBD of U2AF65 interacts with the SAP155 component of U2 snRNP as well as with mBBP (O. Gozani, J. Potashkin, and R. Reed, in prep.). Although it is not known that precisely the same subregion interacts with both proteins, it is tempting to speculate that during this subsequent role of U2AF65 in U2 snRNP recruitment a swap of protein–protein interactions also helps displace mBBP from the spliceosome (Fig. 9).
Figure 9.
A model representing the cooperative interaction between mBBP and U2AF65 at the 3′ end of mammalian introns and the subsequent replacement of mBBP by U2 snRNP. The RNA is represented by the thick black line. The important domains within mBBP (amino terminus) and U2AF65 (third RBD) for the cooperative interaction are shown interacting with one another. The KH domain and Zn knuckles are shown binding the BPS.
The double footprint (Fig. 2) shows that the two proteins can bind simultaneously to their adjacent sites. Taken together with protein–protein interaction studies (Abovich and Rosbash 1997), the results suggested that RNA binding might be cooperative, which is the case (Fig. 4). It is not yet known whether this is the result of a change in off-rate or on-rate, nor is it known for the proposed effect of U2AF65 on U2snRNA–BPS annealing (Valcárcel et al. 1996). But even without a proper biophysical explanation, the mBBP–U2AF65 cooperativity suggests features of mammalian BPS selection and more generally sheds light on intron recognition. Cooperative binding increases the specificity of each protein for its respective site and links the BPS and PY tract together as a large recognition site on the 3′ side of mammalian introns. This notion helps explain how the highly degenerate mammalian BPS is recognized by mBBP: Its binding site is both RNA and protein, the BPS and the adjacent U2AF65. Although the PY binding site of U2AF65 may have more sequence information, mBBP and the BPS may also be viewed as part of the U2AF65 binding site. As mutations in the RNA substrate have demonstrable effects (Figs. 5 and 6), proper RNA–protein interactions also contribute to the wild-type cooperative interaction. The aberrant mobility of the mBBP-branchpoint mutant complex (Fig. 5) might indicate mispositioning of the protein on this substrate, or it might reflect a missing RNA-induced conformational change necessary for the cooperative protein–protein interaction.
We observed a substantial decrease in the amount of ternary complex (mBBP, U2AF65, and the RNA substrate) formed in the presence of mutations in either the BPS or PY tract (Figs. 5 and 6). Although interpretation of the BPS mutation is less certain, the PY tract mutation has a clear effect on cooperativity. Mutations in either the BPS, PY tract, or the presence of a nonconserved BPS have been shown previously to reduce splicing efficiency (Hartmuth and Barta 1988; Reed and Maniatis 1988; Reed 1989). Also, formation of the early splicing complex A has been shown to be effected by mutations in either the BPS or PY (Jamison and Garcia-Blanco 1992; Query et al. 1996). Possibly more relevant is the strong effect of branchpoint mutations, particularly the branchpoint adenosine, on complex formation. These studies used partial RNA substrates missing a 5′ exon and a 5′ splice site, or very short substrates similar to the ones used in our studies (Query et al. 1996, 1997). Taken together, all of these results suggest that the decreases we observed in ternary complex formation and cooperativity may correlate with decreases in complex formation and splicing efficiency.
In addition to contributing to branchpoint recognition, the mBBP–U2AF65 interaction may also contribute to formation or stabilization of E complex. Additional interactions with mBBP or U2AF65 have been postulated or are easy to imagine during this subsequent step of spliceosome assembly: (1) interactions between mBBP/U2AF65 and U1 snRNP (Abovich and Rosbash 1997; Fromont-Racine et al. 1997); (2) a cooperative interaction between splicing factors that bind enhancers and the mBBP/U2AF65 complex (Reed 1996); (3) interactions between U2AF65 and U2AF35 (Zamore and Green 1989); (4) interactions between U2AF35 and U1 snRNP (Wu and Maniatis 1993).
The interaction between mBBP and U2AF65 is reminiscent of previously observed cooperativity between λ repressor molecules and operator DNA (Johnson et al. 1981; Ptashne 1984, 1992). The protein–protein interaction between repressor molecules is relatively weak, only 1–2 kcal/mole (Johnson et al. 1979; Ackers et al. 1982). Assuming the cooperativity observed between U2AF65 and mBBP is relevant to the interaction energy between U2AF65 and mBBP, a 20-fold effect is equal to 1.6 kcal/mol, within the same range as that seen for the λ repressor. The KDs for RNA binding are greater than those between repressor and DNA, however, indicating that the protein–protein interaction might make a greater relative contribution to the protein–RNA interactions. Alternatively, our in vitro experiments might underestimate the effective affinities of these two proteins for many RNA targets. For example, RNA binding of U2AF65 is improved by lengthening the PY tract (Zamore et al. 1992). Although nothing similar has been achieved for mBBP, additional protein–protein interactions within the E complex almost certainly make in vivo binding stronger than what is observed in vitro with recombinant proteins. Finally, relatively weak binding of mBBP may be desirable, as it is probably replaced by U2 snRNP during later spliceosomal assembly steps; a strong mBBP–BPS interaction might be rate-limiting and inhibit U2 snRNP addition.
A similar cooperative interaction probably occurs during yeast intron recognition. BBP, the yeast ortholog of mBBP, binds the yeast BPS (Berglund et al. 1997). Mud2p, the possible yeast ortholog of U2AF65, interacts with BBP (Abovich and Rosbash 1997). Mud2p cross-links to pre-mRNA and may interact with the weakly conserved yeast PY tracts (Abovich et al. 1994). The third RBD of U2AF65 is the region of conservation between U2AF65 and Mud2p (Abovich et al. 1994). This is the region of U2AF65 that interacts with mBBP (Fig. 7), and it is also the region of Mud2p that interacts with yeast BBP (J.-C. Rain, Z. Rafi, Z. Rhani, P. Legrain, and A. Kramer, in prep.). Yeast BBP and mBBP also share a region necessary for the interaction with their respective partners, Mud2p and U2AF65. This is the amino-terminal portion of BBP, upstream of the KH domain (J.-C. Rain, Z. Rafi, Z. Rhani, P. Legrain, and A. Kramer, in prep.), which is highly conserved between the two proteins (Arning et al. 1996). The conservation also suggests that this region contributes in a similar manner in both systems to intron recognition.
In most mammalian introns, the BPS and PY tract are close together (Reed 1989). It has been suggested that the proximity of the BPS to the PY tract allows U2AF65 to aid in U2 snRNP addition to the BPS (Zamore et al. 1992), and more recent experiments support this hypothesis (Valcárcel et al. 1996). On the basis of the results reported here, we suggest that this proximity is also important for the earlier cooperative interaction between mBBP and U2AF65. The footprinting experiment (Fig. 2) indicates that a 4-nucleotide separation does not inhibit binding to the two sites. But it will be interesting to determine whether longer or shorter spacing affects the cooperative formation of this ternary complex, because separating the BPS and PY tract have been shown to have a deleterious effect on both A complex formation and lariat formation (Reed 1989; O. Gozani and R. Reed, pers. comm.). A longer PY tract, which has been shown to bind U2AF65 more tightly (Zamore et al. 1992), might even change U2AF65 conformation and thereby affect its interaction with mBBP.
Materials and methods
Cloning and protein purification
Plasmids for the production of proteins in Escherichia coli were constructed in the pGEX-6p-1 vector (Pharmacia) by standard PCR amplification with oligonucleotides that introduced restriction sites for cloning: pGEX6P–U2AF65 contains the complete coding region of U2AF65 (amino acids 1–476) flanked by BamHI and EcoRI sites; pGEX6P–U2AF65Δ3rd RBD contains amino acids 1–364; pGEX6P–mBBP/SF1 contains amino acids 1–361 of mBBP flanked by BamHI and SalI sites.
All three protein contructs (U2AF65, U2AF65-3, and mBBP) were transformed into BL21 cells (Novagen). Cells were resuspended in 50 mm Tris (pH 7.5), 200 mm NaCl, 1 mm EDTA, and 1 mm DTT. Cells were sonicated, spun at 17,000g for 30 min, and bound to glutathione–Sepharose. Following the protocol from Pharmacia, the protein of interest (U2AF65, etc.) was cleaved from the matrix by use of Precision protease (Pharmacia) and dialyzed overnight against 25 mm Tris (pH 7.5), 25 mm NaCl, 1 mm EDTA and 1 mm DTT. The two U2AF65 proteins were bound to heparin–Sepharose (Pharmacia) and eluted with a salt gradient of 25 mm–1 m NaCl. Peak fractions were collected and concentrated by use of an Amicon ultrafiltration cell and then dialyzed against 25 mm Tris (pH 7.5), 50 mm NaCl, 1 mm DTT and 15% glycerol. Everything was done exactly the same for mBBP except the column matrix used was CM–Sepharose (Pharmacia).
RNA substrates
All RNA oligonucleotides were made with Perseptive RNA amidites on an Expedite 8909 Oligonucleotide Synthesizer. RNAs were kinased with [γ-32P]ATP, gel purified, and further purified on a Bio-Rad P6 spin column.
Footprinting assay
Either mBBP, U2AF65, or both were incubated with radiolabeled RNA in binding buffer [25 mm Tris (pH 7.5), 25 mm NaCl, 1 mm EDTA], 20 mg/ml tRNA, and RNasin (Promega) at final concentration of 1 U/ml. After 20 min incubation, either RNase T1 (Ambion, at a final concentration of 0.4 U/ml) or RNase T2 (GIBCO, at a final concentration of 0.05 U/ml) was added. This mixture was incubated at RT for 5 min, and the reaction was then quenched with phenol/chloroform. After ethanol precipitation, products were separated on a 20% denaturing polyacrylamide gel.
Gel-shift assay
Proteins were incubated with radiolabeled RNA in binding buffer plus 0.5 mg/ml tRNA for 60 min at room temperature. Separation of the RNA and the different complexes was done in 0.5× TBE 6% native polyacrylamide gels. Running time was ∼5 hr at 100 V in the cold room. Radiolabeled RNA was at a final concentration of ∼0.1 nm. The Kd of mBBP alone was obtained by plotting fraction bound (mBBP—RNA complex) versus unbound (free RNA) by use of the program Microcal Origin (Microcal Software Inc.). In the case of the ternary complex, fraction bound was the ternary complex only and unbound was both free RNA and the U2AF65–RNA complex.
The competition assay in which a U2 mimic (12 oligoribonucleotide as shown in Fig. 3) was added to compete mBBP binding was done in a similar manner as above, except that cold U2 mimic was annealed to the radiolabeled 34 oligoribonucleotide at 65°C for 5 min and then placed at room temperature (this was done in binding buffer plus 200 mm NaCl). After 10 min, proteins were added. A 7.5% 0.5× TBE gel was used instead of a 6% gel.
Two-hybrid and GST precipitations
The LexA–U2AF65 deletions A–P were a gift from Or Gozani and Robin Reed (Harvard Medical School, Boston, MA). The B42–mBBP/SF1 fusion has been described previously (Abovich and Rosbash 1997). Yeast two-hybrid assays were performed in diploids obtained by mating strain EGY48–psH18 transformed with each of the LexA–U2AF65 fusions to strain RFY206–psH18 carrying the B42–mBBP/SF1 fusion. After selection of the diploids in His−, Ura−, Trp− selective plates, they were replica-plated to selective indicator plates containing X-gal and either galactose–raffinose or glucose as carbon source.
GST–mBBP/SF1 precipitation of LexA–U2AF65 fusion proteins was performed as previously described (Abovich and Rosbash 1997), except that yeast miniextracts were prepared as described (Abovich et al. 1990). The LexA–U2AF65 fusions were visualized with anti-LexA antibody, a generous gift from Roger Brent (Harvard Medical School, Boston, MA).
Acknowledgments
We thank Or Gozani and Robin Reed for the generous gift of the LexA–U2AF65 fusions, and communication of results before publication. We also appreciate Pierre Legrain and Angela Kramer communicating results before publication. We are grateful to Melissa Moore and members of the Rosbash laboratory for helpful discussions and critical reading of the manuscript. Our thanks also go to Ed Dougherty and Lise-Anne Monaghan for help with figures and secretarial assistance. Supported by National Institutes of Health grant (GM-23549) to M.R.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL rosbash@binah.cc.brandeis.edu; FAX (781) 736-3164.
References
- Abovich N, Rosbash M. Cross-intron bridging interactions in the yeast commitment complex are conserved in mammals. Cell. 1997;89:403–412. doi: 10.1016/s0092-8674(00)80221-4. [DOI] [PubMed] [Google Scholar]
- Abovich N, Legrain P, Rosbash M. The yeast PRP6 gene encodes a U4/U6 small nuclear ribonucleoprotein particle (snRNP) protein, and the PRP9 gene encodes a protein required for U2 snRNP binding. Mol Cell Biol. 1990;10:6417–6425. doi: 10.1128/mcb.10.12.6417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abovich N, Liao XC, Rosbash M. The yeast MUD2 protein: An interaction with PRP11 defines a bridge between commitment complexes and U2 snRNP addition. Genes & Dev. 1994;8:843–854. doi: 10.1101/gad.8.7.843. [DOI] [PubMed] [Google Scholar]
- Ackers GK, Shea MA, Johnson AD. Quantitative model for gene regulation by lamba phage repressor. Proc Natl Acad Sci. 1982;79:1129–1133. doi: 10.1073/pnas.79.4.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arning S, Gruter P, Bilbe G, Kramer A. Mammalian splicing factor SF1 is encoded by variant cDNAs and binds to RNA. RNA. 1996;2:794–810. [PMC free article] [PubMed] [Google Scholar]
- Berglund JA, Chua K, Abovich N, Reed R, Rosbash M. The splicing factor BBP interacts specifically with the pre-mRNA branchpoint sequence UACUAAC. Cell. 1997;89:781–787. doi: 10.1016/s0092-8674(00)80261-5. [DOI] [PubMed] [Google Scholar]
- Birney E, Kumar S, Krainer AR. Analysis of the RNA-recognition motif and RS RGG domains: Conservation in metazoan pre-mRNA splicing factors. Nucleic Acids Res. 1993;21:5803–5816. doi: 10.1093/nar/21.25.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromont-Racine M, Rain J-C, Legrain P. Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nature Genet. 1997;16:277–282. doi: 10.1038/ng0797-277. [DOI] [PubMed] [Google Scholar]
- Fu X-D. The superfamily of arginine/serine-rich splicing factors. RNA. 1995;1:663–680. [PMC free article] [PubMed] [Google Scholar]
- Gaur R, Valcarcel J, Green M. Sequential recognition of the pre-mRNA branch point by U2AF65 and a novel spliceosome-associated 28-kDa protein. RNA. 1995;1:407–417. [PMC free article] [PubMed] [Google Scholar]
- Green MR. Pre-mRNA splicing. Annu Rev Genet. 1986;20:671–708. doi: 10.1146/annurev.ge.20.120186.003323. [DOI] [PubMed] [Google Scholar]
- Hartmuth K, Barta A. Unusual branch point selection in processing of human growth hormone pre-mRNA. Mol Cell Biol. 1988;8:2011–2020. doi: 10.1128/mcb.8.5.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamison SF, Garcia-Blanco MA. An ATP independent U2 small nuclear ribonucleoprotein particle/precursor mRNA complex requires both splice sites and the polypyrimidine tract. Proc Natl Acad Sci. 1992;89:5482–5486. doi: 10.1073/pnas.89.12.5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson AD, Meyer BJ, Ptashne M. Interactions between DNA-bound repressors govern regulation by the lamba phage repressor. Proc Natl Acad Sci. 1979;76:5061–5065. doi: 10.1073/pnas.76.10.5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson AD, Poteete AR, Lauer G, Sauer RT, Ackers GK, Ptashne M. lambda repressor and cro—components of an efficient molecular switch. Nature. 1981;294:217–223. doi: 10.1038/294217a0. [DOI] [PubMed] [Google Scholar]
- Keller EB, Noon WA. Intron splicing: A conserved internal signal in introns of animal pre-mRNAs. Proc Natl Acad Sci. 1984;81:7417. doi: 10.1073/pnas.81.23.7417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohtz JD, Jamison SF, Will CL, Zuo P, Lührmann R, Garcia-Blanco MA, Manley JL. Protein-protein interactions and 5′-splice-site recognition in mammalian mRNA precursors. Nature. 1994;368:119–124. doi: 10.1038/368119a0. [DOI] [PubMed] [Google Scholar]
- Kramer A. Purification of splicing factor SF1, a heat-stable protein that functions in the assembly of a presplicing complex. Mol Cell Biol. 1992;12:4545–4552. doi: 10.1128/mcb.12.10.4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu Rev Biochem. 1996;65:367–409. doi: 10.1146/annurev.bi.65.070196.002055. [DOI] [PubMed] [Google Scholar]
- Lee C-G, Zamore PD, Green MR, Hurwitz J. RNA annealing activity is intrinsically associated with U2AF. J Biol Chem. 1993;268:13472–13478. [PubMed] [Google Scholar]
- Madhani HD, Guthrie C. Dynamic RNA-RNA interactions in the spliceosome. Annu Rev Genet. 1994;28:1–26. doi: 10.1146/annurev.ge.28.120194.000245. [DOI] [PubMed] [Google Scholar]
- Michaud S, Reed R. An ATP-independent complex commits pre-mRNA to the mammalian spliceosome assembly pathway. Genes & Dev. 1991;5:2534–2546. doi: 10.1101/gad.5.12b.2534. [DOI] [PubMed] [Google Scholar]
- Moore MJ, Query CC, Sharp PA. Splicing of precursors to mRNAs by the spliceosome. In: Gesteland RF, Atkins JF, editors. The RNA world. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1993. pp. 303–357. [Google Scholar]
- Mount SM, Pettersson I, Hinterberger M, Karmas A, Steitz JA. The U1 small nuclear RNA-protein complex selectively binds a 5′ splice site in vitro. Cell. 1983;33:509–518. doi: 10.1016/0092-8674(83)90432-4. [DOI] [PubMed] [Google Scholar]
- Patterson B, Guthrie C. A U-rich tract enhances usage of an alternative 3′ splice site in yeast. Cell. 1991;64:181–187. doi: 10.1016/0092-8674(91)90219-o. [DOI] [PubMed] [Google Scholar]
- Ptashne, M. 1984. Repressors. Trends Biochem. Sci. 142–145.
- ————— . A genetic switch. 2nd ed. Cambridge, MA: Cell Press; 1992. [Google Scholar]
- Query CC, Moore MJ, Sharp PA. Branch nucleophile selection in pre-mRNA splicing: Evidence for the bulged duplex model. Genes & Dev. 1994;8:587–597. doi: 10.1101/gad.8.5.587. [DOI] [PubMed] [Google Scholar]
- Query CC, Strobel SA, Sharp PA. Three recognition events at the branch-site adenine. EMBO J. 1996;15:1392–1402. [PMC free article] [PubMed] [Google Scholar]
- Reed R. The organization of 3′ splice-site sequences in mammalian introns. Genes & Dev. 1989;3:2113–2123. doi: 10.1101/gad.3.12b.2113. [DOI] [PubMed] [Google Scholar]
- ————— Initial splice-site recognition and pairing during pre-mRNA splicing. Curr Opin Genet Dev. 1996;6:215–220. doi: 10.1016/s0959-437x(96)80053-0. [DOI] [PubMed] [Google Scholar]
- Reed R, Maniatis T. Intron sequences involved in lariat formation during pre-mRNA splicing. Cell. 1985;41:95–105. doi: 10.1016/0092-8674(85)90064-9. [DOI] [PubMed] [Google Scholar]
- ————— The role of mammalian branchpoint sequences in pre-mRNA splicing. Genes & Dev. 1988;2:1268–1276. doi: 10.1101/gad.2.10.1268. [DOI] [PubMed] [Google Scholar]
- Ruskin B, Green MR. Role of the 3′ splice site consensus sequence in mammalian pre-mRNA splicing. Nature. 1985;317:732–734. doi: 10.1038/317732a0. [DOI] [PubMed] [Google Scholar]
- Rymond BC, Rosbash M. Yeast pre-mRNA splicing. In: Jones EW, Pringle JR, Broach JR, editors. The molecular and cellular biology of the yeast Saccharomyces: Gene expression. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1992. pp. 143–192. [Google Scholar]
- Séraphin B, Kretzner L, Rosbash M. A U1 snRNA: Pre-mRNA base pairing interaction is required early in yeast spliceosome assembly but does not uniquely define the 5′ cleavage site. EMBO J. 1988;7:2533–2538. doi: 10.1002/j.1460-2075.1988.tb03101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siliciano PG, Guthrie C. 5′ splice site selection in yeast: Genetic alterations in base-pairing with U1 reveal additional requirements. Genes & Dev. 1988;2:1258–1267. doi: 10.1101/gad.2.10.1258. [DOI] [PubMed] [Google Scholar]
- Singh R, Valcarcel J, Green MR. Distinct binding specificities and functions of higher eukaryotic polypyrimidine-tract binding proteins. Science. 1995;268:1173–1176. doi: 10.1126/science.7761834. [DOI] [PubMed] [Google Scholar]
- Smith CWJ, Nadal-Ginard BN. Mutualy exclusive splicing of α-tropomyosin exons enforced by an unusual lariat branch point location: Implications for constitutive splicing. Cell. 1989;56:749–758. doi: 10.1016/0092-8674(89)90678-8. [DOI] [PubMed] [Google Scholar]
- Staknis D, Reed R. SR proteins promote the first specific recognition of pre-mRNA and are present together with U1 snRNP in a general splicing enhancer complex. Mol Cell Biol. 1994;14:7670–7682. doi: 10.1128/mcb.14.11.7670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valcárcel J, Gaur RK, Singh R, Green MR. Interaction of U2AF65 RS region with pre-mRNA of branch point and promotion base pairing with U2 snRNA. Science. 1996;273:1706–1709. doi: 10.1126/science.273.5282.1706. [DOI] [PubMed] [Google Scholar]
- Wu JY, Maniatis T. Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell. 1993;75:1061–1070. doi: 10.1016/0092-8674(93)90316-i. [DOI] [PubMed] [Google Scholar]
- Zamore PD, Green MR. Identification, purification, and biochemical characterization of U2 small nuclear ribonucleoprotein auxiliary factor. Proc Natl Acad Sci. 1989;86:9243–9247. doi: 10.1073/pnas.86.23.9243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore PD, Patton JG, Green MR. Cloning and domain structure of the mammalian splicing factor U2AF. Nature. 1992;355:609–614. doi: 10.1038/355609a0. [DOI] [PubMed] [Google Scholar]
- Zhuang Y, Weiner AM. A compensatory base change in U1 snRNA suppresses a 5′ splice site mutation. Cell. 1986;46:827–835. doi: 10.1016/0092-8674(86)90064-4. [DOI] [PubMed] [Google Scholar]
- Zhuang Y, Goldstein M, Weiner AM. UACUAAC is the preferred branch site for mammalian mRNA splicing. Proc Natl Acad Sci. 1989;86:2752–2756. doi: 10.1073/pnas.86.8.2752. [DOI] [PMC free article] [PubMed] [Google Scholar]