Abstract
The Ikaros gene encodes multiple protein isoforms that contribute critical functions during the development of lymphocytes and other hematopoietic cell types. The intracellular functions of Ikaros are not known, although recent studies have shown that Ikaros proteins colocalize with inactive genes and centromeric heterochromatin. In this study, Ikaros proteins were found to be components of highly stable complexes. The complexes from an immature T cell line were purified, revealing associated proteins of 70 and 30 kD. The p70 gene, named Helios, encodes two protein isoforms with zinc finger domains exhibiting considerable homology to those within Ikaros proteins. Helios and Ikaros recognize similar DNA sequences and, when overexpressed, Helios associates indiscriminately with the various Ikaros isoforms. Although Ikaros is present in most hematopoietic cells, Helios was found primarily in T cells. The relevance of the Ikaros–Helios interaction in T cells is supported by the quantitative association of Helios with a fraction of the Ikaros. Interestingly, the Ikaros–Helios complexes localize to the centromeric regions of T cell nuclei, similar to the Ikaros localization previously observed in B cells. Unlike the B cell results, however, only a fraction of the Ikaros, presumably the fraction associated with Helios, exhibited centromeric localization in T cells. These results establish immunoaffinity chromatography as a useful method for identifying Ikaros partners and suggest that Helios is a limiting regulatory subunit for Ikaros within centromeric heterochromatin.
Keywords: Ikaros, Helios, T lymphocyte, lymphocyte development, heterochromatin
The molecular mechanisms by which B and T lymphocytes are generated from hematopoietic stem cells have been the subject of intensive investigation. By analysis of the immunoglobulin and T cell receptor (TCR) gene recombination events and the differential expression of lymphocyte-specific genes, much has been learned about the regulation of B and T cell maturation (Clevers et al. 1993; Hagman and Grosschedl 1994; Ernst and Smale 1995; Clevers and Grosschedl 1996; Shortman and Wu 1996; Willerford et al. 1996). In contrast, little is known about the earliest stages of lymphocyte development, including commitment to the lymphocyte lineages and the maturation events that precede gene recombination (Ikuda et al. 1992; Morrison et al. 1995; Orkin 1995; Singh 1996; Georgopoulos et al. 1997).
In recent years, insight into the early regulatory events has been provided by the phenotypes of mice containing homozygous disruptions of genes encoding sequence-specific DNA-binding proteins (Orkin 1995; Clevers and Grosschedl 1996; Singh 1996; Ting et al. 1996; Georgopoulos et al. 1997). PU.1, Ikaros, E2A, EBF, BSAP, and GATA-3 are among the proteins that are critical for early lymphocyte development. Most of these proteins act as typical transcription factors, which bind to regulatory elements within specific target genes and direct gene activation.
Ikaros is unusual among the above DNA-binding proteins as none of its targets has been clearly established and its intracellular functions remain unknown. The Ikaros gene was first identified through an expression library screen for proteins that interact with an enhancer for the TCR CD3δ gene (Georgopoulos et al. 1992). The gene was later found to encode the LyF-1 protein that interacts with a critical control element in the promoter for the lymphocyte-specific terminal transferase (TdT) gene (Lo et al. 1991; Hahm et al. 1994). Primary Ikaros transcripts undergo alternative pre-mRNA splicing to generate several protein isoforms. The isoforms vary within an amino-terminal zinc finger domain that is responsible for sequence-specific DNA binding (Hahm et al. 1994; Molnar and Georgopoulos 1994; Molnar et al. 1996; Sun et al. 1996). The largest Ikaros isoform (isoform VI; Ik-1) contains four zinc fingers near the amino terminus, whereas the smaller isoforms contain fewer or no amino-terminal zinc fingers. All isoforms contain two additional zinc finger motifs at their carboxyl terminus, which do not bind DNA (Hahm et al. 1994), but serve as protein–protein interaction domains (Sun et al. 1996). Given the apparent indiscriminate nature of the protein–protein interactions, a large number of dimeric or multimeric species can be generated. Recently, a protein related to Ikaros was identifed by degenerate PCR with primers complementary to sequences encoding the carboxy-terminal zinc fingers (Morgan et al. 1997). This protein, Aiolos, exhibits considerable homology to Ikaros and interacts with Ikaros through its carboxy-terminal fingers (Morgan et al. 1997).
Ikaros isoforms are expressed in most cells of the hematopoietic lineages, including multipotent stem cells (Georgopoulos et al. 1992; Hahm et al. 1994; Molnar and Georgopoulos 1994; Morgan et al. 1997; Klug et al. 1998). Many cell types express the two largest isoforms (V and VI), but isoform expression patterns vary to some extent in a cell-specific manner. Aiolos is expressed in most of the cell types that express Ikaros, except the earliest hematopoietic progenitors (Morgan et al. 1997). Gene disruption experiments have demonstrated that the Ikaros proteins are critical for multiple hematopoietic events (Georgopoulos et al. 1994; Winandy et al. 1995; Wang et al. 1996). The most striking defect in Ikaros−/− mice is the absence of B cells, natural killer cells, and some T lineage cells, including fetal-derived T cells and some γδ T cells (Wang et al. 1996). Ikaros−/− mice also exhibit a biased distribution and expansion of CD4+ T cells and defects in other hematopoietic lineages (Wang et al. 1996). A second Ikaros gene disruption, which eliminates the DNA-binding domain, but allows expression of smaller isoforms, results in a more severe lymphopoietic defect; these mice contain no B or T lymphocytes or any of the known B or T cell progenitors (Georgopoulos et al. 1994). Heterozygotes of these mice contain normal lymphocyte phenotypes and numbers at birth, but rapidly develop lymphoproliferative disorders (Winandy et al. 1995).
Ikaros-binding sites have been identified in the promoters or enhancers of several genes (Lo et al. 1991; Georgopoulos et al. 1992; Omori and Wall 1993; Wargnier et al. 1995; Babichuk et al. 1996; Haag et al. 1996; Santee and Owen-Schaub 1996; Wang et al. 1996), but none of these genes has been shown to be an authentic Ikaros target. A well-characterized binding site for Ikaros within the TdT promoter is critical for promoter activity in immature lymphocytes (Lo et al. 1991). Ets family proteins also bind this site however, and several findings suggest that the Ets protein Elf-1 is the functional activator of TdT transcription (Ernst et al. 1993, 1996). Ikaros and Elf-1 cannot bind simultaneously to this element (K. Hahm, L. Trinh, P. Ernst, and S.T. Smale, in prep.), suggesting that if Ikaros contributes to TdT promoter activity, it does so as a repressor or competitive inhibitor of the Elf-1 activator.
Recent studies of subnuclear localization in B cells have provided further evidence that Ikaros may not be a simple transcriptional activator, as it was found by immunogold electron microscopy and confocal microscopy to localize to heterochromatin (Brown et al. 1997; Klug et al. 1998). More specifically, by combining fluorescence in situ hybridization with confocal immunofluorescence assays, Ikaros colocalized with centromeric heterochromatin and with inactive genes, which themselves migrate to foci of centromeric heterochromatin (Brown et al. 1997).
The centromeric localization of Ikaros makes an understanding of its precise intracellular functions difficult to establish. The ability of the many Ikaros isoforms to associate with one another indiscriminately into a large number of dimeric or multimeric species adds further complexity, as each species may carry out a distinct function. Finally, the complicated phenotypes of the Ikaros−/− mice suggest that Ikaros proteins may carry out different functions in the different hematopoietic cell lineages, perhaps in association with lineage-restricted partners. As an essential step toward an understanding of these issues, we performed a biochemical analysis of the native Ikaros proteins. This analysis revealed that Ikaros proteins form highly stable complexes that can be purified to near homogeneity by immunoaffinity chromatography, providing a means of isolating Ikaros partners in various cell types. The Ikaros complexes purified from an immature T cell line contain an Ikaros-associated protein, Helios, which possesses a zinc finger structure similar to that found in Ikaros. The restricted expression pattern of Helios, its presence at limiting quantities, its quantitative association with Ikaros, its assembly into a relatively homogeneous complex, and its specific localization to centromeres suggest that it functions as a critical regulator of Ikaros within the centromeric heterochromatin regions of the nucleus.
Results
Expression of Ikaros isoforms by RLm11 and VL3-3M2 T cells
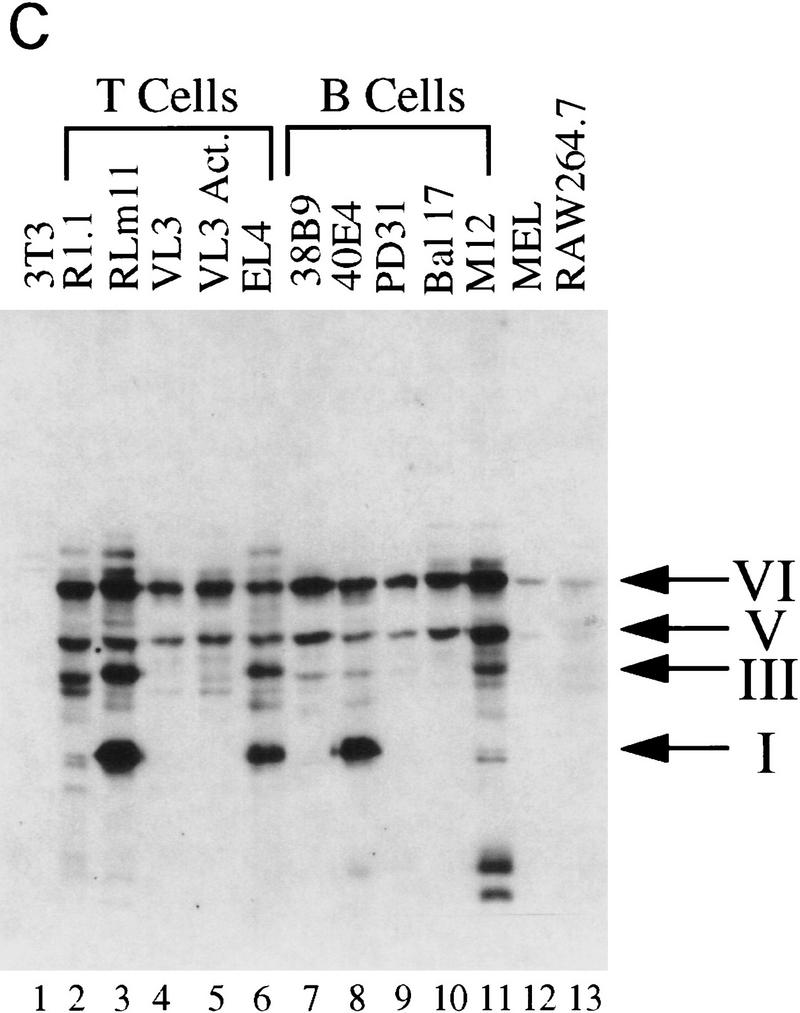
For most of the studies described in this report, a murine radiation-induced thymoma cell line, RLm11, was employed. This cell line produces large quantities of TdT mRNA and protein and has been used for several years to study the transcriptional regulation of the TdT gene, a potential Ikaros target (Lo et al. 1991; Ernst et al. 1993, 1996; Hahm et al. 1994). Immunoblot and RT-PCR analyses have demonstrated that RLm11 cells actively express 4 of the 10 Ikaros isoforms (see Fig. 1, isoforms I, III, V, and VI; Hahm et al. 1994; Molnar and Georgopoulos 1994; Sun et al. 1996). All four of these isoforms have been detected in populations of primary immature lymphocytes (Molnar and Georgopoulos 1994; Molnar et al. 1996; Klug et al. 1998).
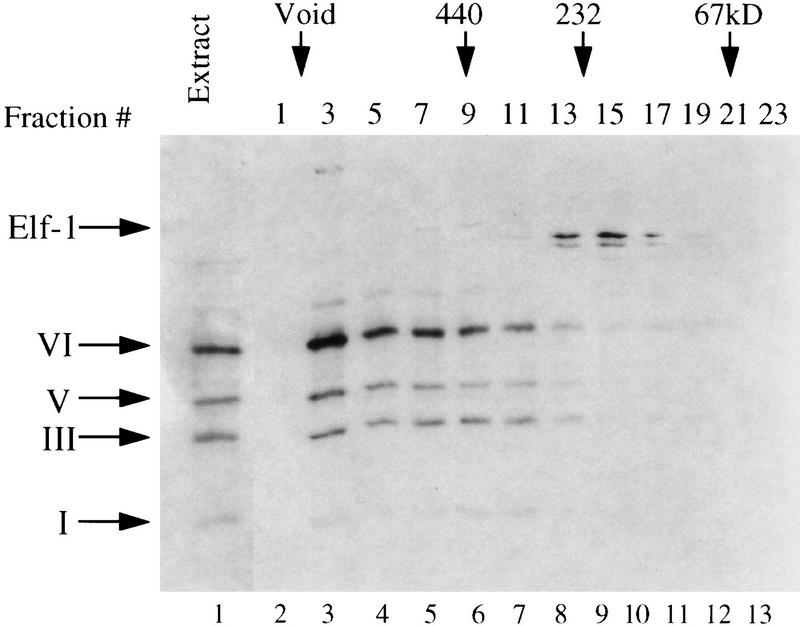
Figure 1.
Coelution of Ikaros isoforms in a broad peak from gel filtration columns. RLm11 nuclear extracts (1 mg) were analyzed by Superdex 200 (Pharmacia) gel filtration chromatography. Five micrograms of nuclear extract (lane 1) and 45 μl of every other column fraction (lanes 2–13) were separated by SDS-PAGE and analyzed by immunoblotting, with antiserum directed against Ikaros and Elf-1. The fractions in which standard molecular mass markers elute are indicated by arrows at the top, and the bands corresponding to Elf-1 and Ikaros isoforms I, III, V, and VI are (left).
For some experiments, another T cell line, VL3-3M2, was used (Groves et al. 1995). This cell line expresses large quantities of the TdT, RAG-1, and RAG-2 mRNAs and exhibits properties of double-positive CD4+CD8+ cells (Groves et al. 1995). VL3-3M2 cells produce primarily two Ikaros isoforms, V and VI (see Fig. 8, below), the two most abundant isoforms detected in primary thymocytes (data not shown; Molnar and Georgopoulos 1994).
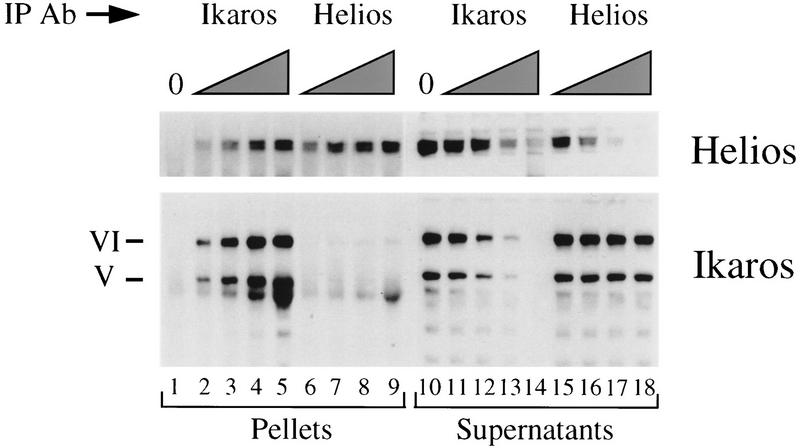
Figure 8.
Quantitative association of Helios with a subset of the Ikaros within VL3-3M2 cells. Quantitative immunoprecipitation experiments were performed with purified IgG directed against the amino-terminal domains of either Ikaros (lanes 2–5,11–14) or Helios (lanes 6–9,15–18). Control immunoprecipitations contained no added antibody (lanes 1,10). The proteins present in the immunoprecipitation pellets (lanes 1–9) and supernatants (lanes 10–18) were analyzed by immunoblot, with the membranes probed with antibodies directed against either Helios (top) or Ikaros (bottom). The two isoforms predominantly expressed in VL3-3M2 cells are indicated to the left of the bottom panel. The amounts of anti-Ikaros or anti-Helios IgGs used in the immunoprecipitations were as follows: 1.5 μg (lanes 2,6,11,,15), 4.5 μg (lanes 3,7,12,16), 15 μg (lanes 4,8,13,,17), and 45 μg (lanes 5,9,14,,18).
Ikaros isoforms coelute in a broad peak from gel fitration columns
Gel filtration chromatography was used to study the properties of the Ikaros proteins within RLm11 nuclear extracts. Immunoblot analysis of column fractions collected from a Superdex 200 FPLC column (Pharmacia) revealed that all four isoforms coelute in a broad peak between the excluded volume (exclusion limit, 1.3 × 106 kD) and a 232-kD molecular mass marker (Fig. 1, lanes 3–8). In contrast, Elf-1, with a calculated molecular mass of 76 kD (Davis and Roussel 1996), eluted in a sharp peak at ∼200 kD (Fig. 1A, lanes 8–10). When using a gel filtration column with a larger exclusion limit, the four Ikaros isoforms again coeluted with each other in a broad peak between 750 and 200 kD (data not shown; see Fig. 9, below). Similar results were obtained in several experiments with independent extract preparations and with column buffers at 0.1 and 0.45 m KCl, both in the presence and absence of NP-40 detergent (data not shown).
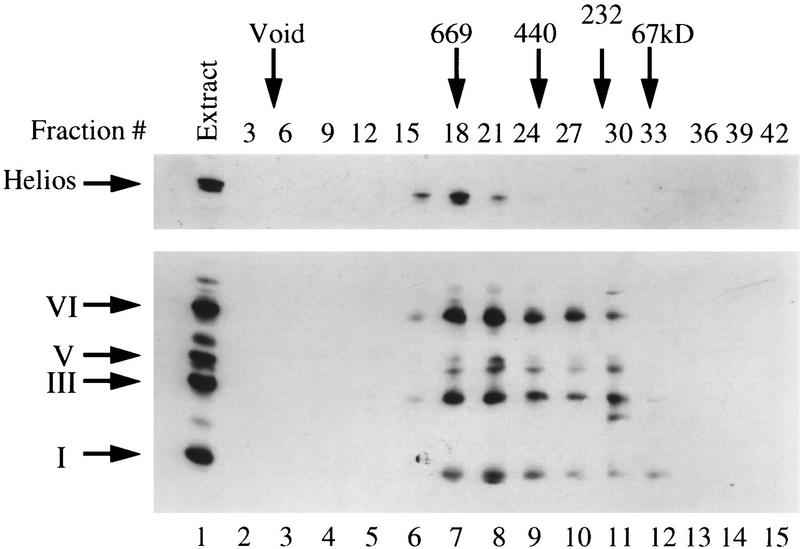
Figure 9.
The Ikaros–Helios complexes appear to be relatively homogeneous when analyzed by gel-filtration chromatography. Proteins in Superose 6 (Pharmacia) gel-filtration column fractions were separated by 10% SDS-PAGE and analyzed by Western blot analysis involving probing of the blot sequentially with anti-Helios serum (top) followed by anti-Ikaros serum (bottom). Four micrograms of RLm11 nuclear extracts was also analyzed (lane 1). The fractions where standard molecular mass markers migrate in Superose 6 (Pharmacia) gel-filtration chromatography are indicated by arrows on the top with their molecular masses. Isoforms I, III, V and VI are indicated.
The above results raise the possibility that Ikaros proteins form an array of multimeric complexes. The results of other biochemical experiments are consistent with the multimer hypothesis, but some results have suggested that the proteins exist as highly stable dimers (A.S. McCarty, K. Hahm, R. Lee, B.S. Cobb, unpubl.). Further experiments will be needed to clarify the precise stoichiometry of the Ikaros complexes, but for the purposes of this study, the relevant findings are: (1) Ikaros proteins appear to exist as stable dimeric or multimer complexes in solution, and (2) the unusually broad elution profile suggests that a given cell contains a large number of distinct complexes. This latter suggestion is consistent with previous studies in which the carboxy-terminal zinc finger domains of Ikaros were found to promote indiscriminate protein–protein interactions between Ikaros isoforms (Sun et al. 1996; K. Hahm, unpubl.).
Purification of Ikaros complexes
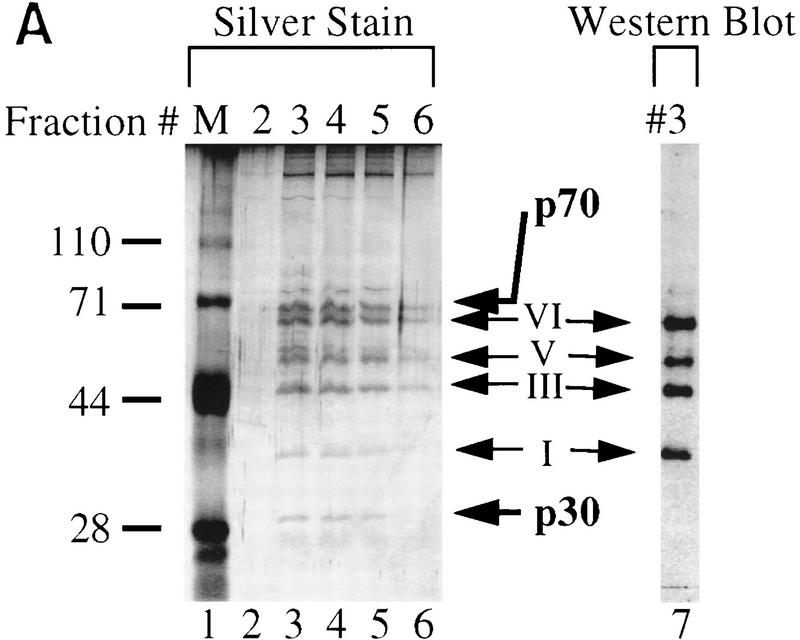
To elucidate the intracellular functions of a protein, a critical objective is to identify other proteins with which it carries out relevant interactions. The biochemical properties summarized above and the complicated phenotypes of the Ikaros−/− mice raise the possibility that the Ikaros isoforms stably associate with proteins that have not been identified. Because of the apparent stability of the complexes, purification by immunoaffinity chromatography was an attractive method for identifying relevant Ikaros-associated proteins. Polyclonal antibodies directed against the carboxy-terminal half of Ikaros were initially used for the purification. RLm11 nuclear extracts in a buffer containing 0.45 m KCl were applied to a protein A–Sepharose column containing covalently linked antibodies (see Materials and Methods). The column was washed extensively with 0.45 and 1 m KCl and, in some experiments, with 0 m KCl to disrupt nonspecific hydrophobic interactions. Tightly bound proteins were eluted with a buffer containing 100 mm trimethyl ethanolamine (pH 11.0). Analysis of the eluted proteins by SDS-PAGE followed by silver staining revealed six bands that were consistently observed at comparable molar amounts (Fig. 2A, lanes 3–6). Four of these bands correspond to Ikaros isoforms I, III, V, and VI, based on immunoblot analysis with Ikaros antibodies (lane 7). The other two bands, which migrate at 30 and 70 kD (p30 and p70, respectively), did not interact with the Ikaros antibodies by immunoblot analysis. These proteins also did not cross-react with three Ikaros antisera raised against other domains of the Ikaros isoforms (data not shown). These results suggest that p30 and p70 bound to the column through specific interactions with Ikaros proteins.
Figure 2.
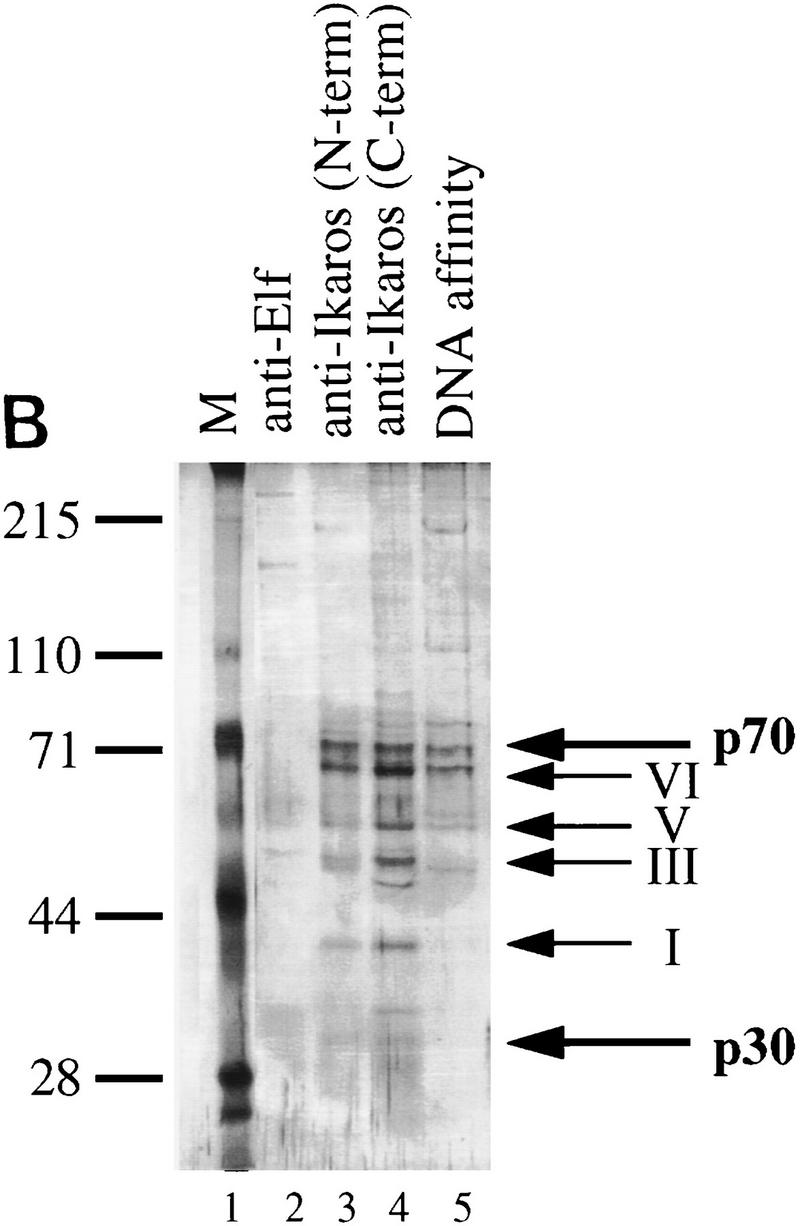
Immunoaffinity purification of Ikaros complexes from RLm11 nuclear extracts. (A) RLm11 nuclear extracts were applied to a protein A–Sepharose column containing covalently linked antibodies directed against the carboxy-terminal half of Ikaros (see Materials and Methods). After washing the resin with buffers containing 0.45 and 1 m KCl, bound proteins were eluted with 100 mm trimethyl ethanolamine (pH 11.0). The trimethyl ethanolamine fractions (numbers 2 through 6, lanes 2–6) were analyzed by SDS-PAGE followed by silver staining. Molecular mass markers are shown in lane 1 and are indicated to the left. Fraction 3 was also analyzed by immunoblotting with anti-Ikaros serum (lane 7). Ikaros isoforms I, III, V, and VI are indicated between lanes 6 and 7. Two proteins, p30 and p70, were detected by silver staining that did not interact with the Ikaros antibodies. (B) RLm11 extracts were applied to protein A–Sepharose columns containing covalently linked antibodies directed against Elf-1 (lane 2), the carboxyl-terminus of Ikaros (lane 3), or the amino-terminus of Ikaros (lane 4). The columns were washed and proteins eluted as described above. Portions of the trimethyl ethanolamine eluates were analyzed by SDS-PAGE followed by silver-staining. Also analyzed were proteins purified by sequence-specific DNA-affinity chromatography with a resin containing covalently linked multimers of the TdT D element (lane 5, see Materials and Methods and Hahm et al. 1994). Molecular mass markers are shown in lane 1 and are indicated at left in kD. Ikaros isoforms I, III, V, and VI, p70, and p30, are indicated at right.
Although denatured p30 and p70 did not interact with the Ikaros antibodies by immunoblot analysis, it remained possible that they copurified with Ikaros because they were recognized in their native conformations by the antibodies. To address this possibility, immunoaffinity chromatography was performed with a resin containing an antibody preparation directed against an amino-terminal domain of Ikaros (Fig. 2B, lanes 3,4). The p70 and p30 proteins bound to this resin as efficiently as to the carboxy-terminal antibody resin. In contrast, an immunoaffinity resin containing an unrelated antibody preparation of similar titer, directed against murine Elf-1, did not yield detectable proteins of 30 and 70 kD (Fig. 2B, lane 2).
As a final control, Ikaros was purified by DNA-affinity chromatography with a resin containing covalently linked multimers of the TdT D element, which comprises an Ikaros-binding site (Lo et al. 1991). The DNA-affinity chromatography procedure was an improved version of the procedure used previously to purify and clone Ikaros (Hahm et al. 1994). In the previous experiments, only a single 65-kD band was observed. This band contained a mixture of Ikaros isoform VI and YY1, which also binds to a sequence within the D element (see Hahm et al. 1994). In the new experiment, which diminished YY1 copurification, Ikaros isoforms III, V, VI, and the p70 protein, were readily detected (Fig. 2B, lane 5). Ikaros isoform I and p30 were less apparent and possibly less abundant, but were detected in some experiments (data not shown).
Isolation of Helios
To isolate the gene encoding p70, the immunoaffinity purification procedure was carried out with large quantities of RLm11 extract (see Materials and Methods). The purified fractions were then concentrated and the proteins separated on a preparative SDS–polyacrylamide gel. After transferring to a PVDF membrane, the p70 band was excised, proteolyzed with endopeptidase C, and sequences of two peptides were obtained (Fernandez et al. 1994). The peptide sequences (see Fig. 3) were unrelated to proteins or genes described in various databases (data not shown).
Figure 3.

Helios A and Helios B contain zinc fingers with homology to those in Ikaros and Aiolos. An amino acid sequence alignment compiled by use of the PILEUP program is shown. Compared are the largest isoforms of the Ikaros family members, Helios B, Ikaros isoform VI (Ik-1), and Aiolos. Lowercase lettering in the Helios B sequence designates the sole difference between Helios B and Helios A. Amino acid sequences obtained by microsequencing of purified Helios protein are indicated by a line above the Helios sequence. Shaded boxes in blue emphasize the highly conserved zinc finger motifs. Yellow bars indicate the conserved cysteines and histidines in the zinc fingers. The red box indicates the unusual cysteine in the apparent C2HC finger. Overall sequence similarities are as follows: Helios B–Ikaros, 55%; Ikaros–Aiolos, 53%; Helios B–Aiolos, 50%. The four amino-terminal zinc fingers of Helios share 94% identity with the Ikaros fingers and the Aiolos fingers share 86% identity with the Ikaros fingers. The two carboxy-terminal zinc fingers of Helios share 85% identity with the corresponding Ikaros fingers, with 80% identity between the Aiolos and Ikaros fingers. The Helios cDNA and amino acid sequences have been deposited in the GenBank/Swiss Prot databases (accession nos. AF044257 and P81183, respectively).
Degenerate oligonucleotides were used to isolate a 216-bp fragment of the p70 gene by RT-PCR from RLm11 mRNA. A full-length cDNA was then isolated from a library prepared from newborn mouse thymus mRNA (Stratagene). Subsequent studies revealed two distinct cDNA products, p70A and p70B, that presumably are generated by alternative pre-mRNA splicing. RT-PCR analysis of RLm11 mRNA revealed that the two RNA products are present at comparable amounts (data not shown).
DNA sequencing of the p70A and p70B cDNAs revealed 1500- and 1578-bp ORFs, respectively, encoding proteins of 55 and 58 kD (Fig. 3). The proteins contain domains with considerable homology to domains within Ikaros. Because of this homology, we named the p70 gene Helios. The small sizes of the Helios proteins relative to their apparent molecular masses based on SDS-PAGE are consistent with the properties of the Ikaros proteins; the calculated molecular mass of Ikaros isoform VI, for example, is 57 kD, whereas its apparent molecular mass by SDS-PAGE is 65 kD. The most striking homology between Helios and Ikaros is within the amino- and carboxy-terminal zinc finger domains. Helios A (which lacks the lower-case amino acids in Fig. 3) contains two C2H2 zinc fingers (yellow) and one C2HC zinc finger (red) near its amino terminus, similar to Ikaros isoform V. Helios B (which contains the lower-case amino acids in Fig. 3) contains three C2H2 zinc fingers and one C2HC zinc finger near its amino terminus, similar to Ikaros isoform VI. Both Helios isoforms, like all of the Ikaros isoforms, contain two carboxy-terminal zinc finger motifs (yellow). The Helios and Ikaros zinc finger domains are highly homologous at the amino acid level. Surrounding the two zinc finger domains, a few short stretches of identity and similarity between Helios and Ikaros are apparent, but most of the Helios sequence exhibits little homology to Ikaros.
Recently, another protein with zinc finger domains homologous to Ikaros, called Aiolos, was isolated from a cDNA library by PCR with degenerate primers directed against the carboxy-terminal zinc-finger domain of Ikaros (Morgan et al. 1997). The Ikaros, Aiolos, and Helios zinc finger domains are highly homologous to each other (Fig. 3). The stretches of Ikaros–Helios homology surrounding the zinc finger domains are also homologous to Aiolos (e.g., amino acids 282–295 and 312–362).
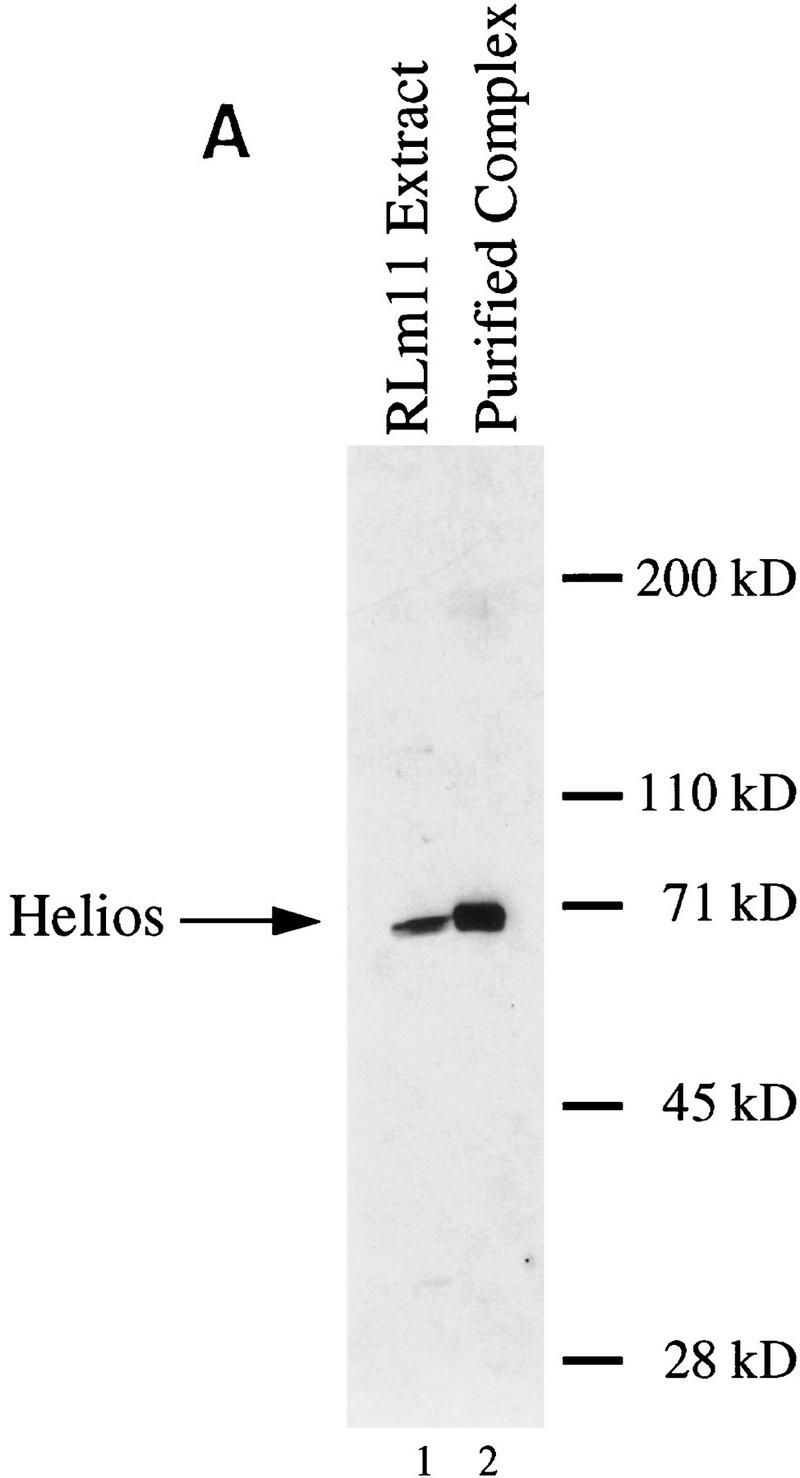
The isolated Helios gene appears to encode the protein purified by immunoaffinity chromatography on the basis of two criteria: First, both peptide sequences obtained from the purified protein are encoded by the Helios gene (Fig. 3, overlined). Second, immunoblot analysis of the purified protein with antibodies generated against the amino-terminal 109 amino acids of Helios, reacted strongly with a protein of the expected size (Fig. 4A).
Figure 4.
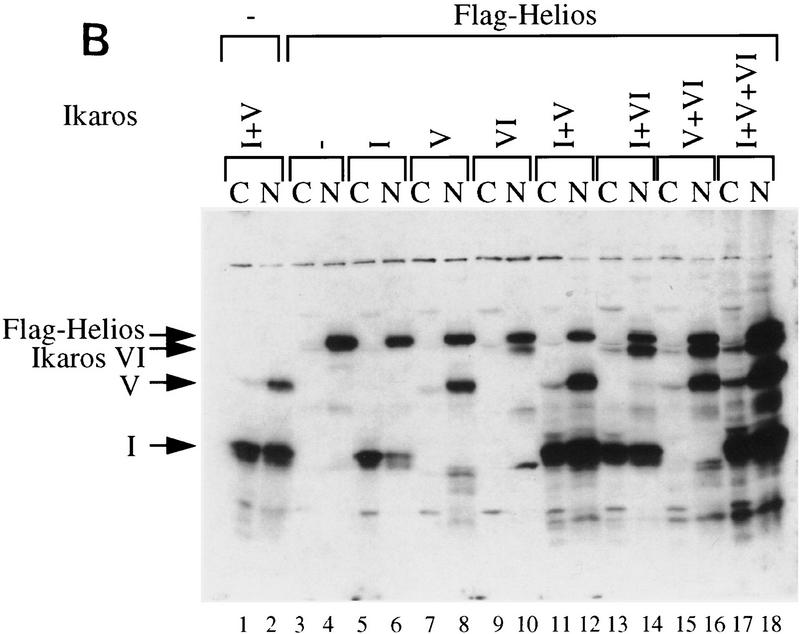
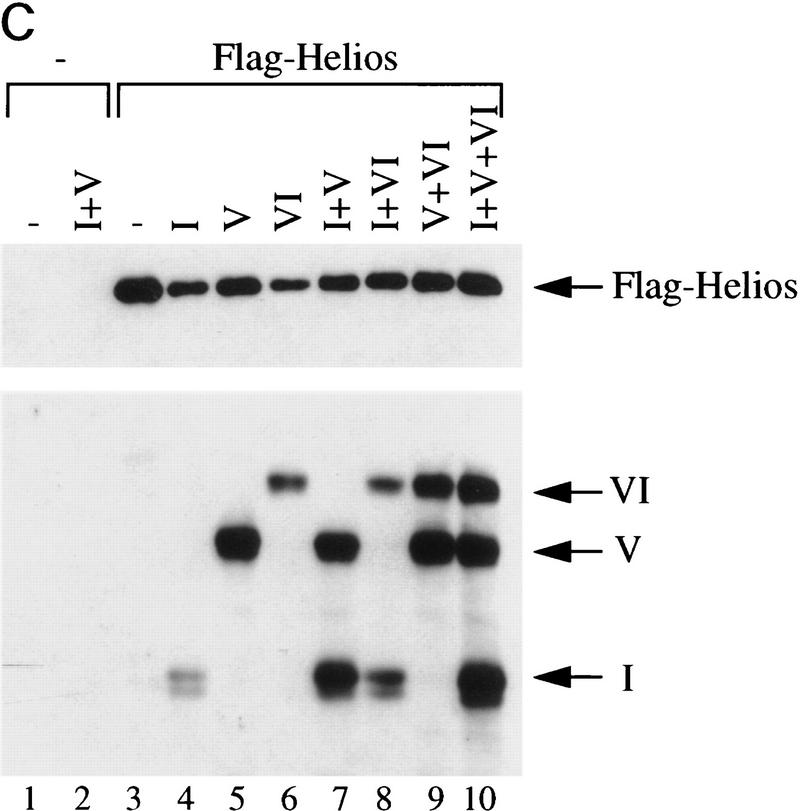
Indiscriminate interactions between Helios A and Ikaros isoforms in 293T cells. (A) The Helios protein within an RLm11 extract (lane 1) and the immunoaffinity purified Ikaros complex (lane 2) can be detected by immunoblot analysis with antisera directed against recombinant Helios (amino acids 1–109). Molecular mass markers are indicated at right, and the location of the Helios band is indicated at left. (B) 293T cells were transfected with 10 or 15 μg of expression plasmids for various Ikaros isoforms (lanes 1–18; specific isoforms indicated above each lane), in the absence (lanes 1,2) or presence (lanes 3–18) of an expression plasmid for FLAG-tagged Helios A (5 μg). Cytoplasmic (odd-numbered lanes) and nuclear (even numbered lanes) extracts from the transfected cells were analyzed by immunoblotting with antibodies directed against both Helios and Ikaros. The bands corresponding to FLAG-tagged Helios A and Ikaros isoforms I, V, and VI are indicated to the left. (C) Interactions between Helios A and Ikaros isoforms were assessed by immunoprecipitation from the nuclear extracts shown in part B with a monoclonal antibody directed against the FLAG epitope. Proteins within the immunoprecipitated pellet were analyzed by immunoblotting with antisera directed against Helios (top) or Ikaros (bottom). The extacts used for immunoprecipitation contained (lanes 3–10) or lacked (lanes 1,2) the FLAG–Helios A protein and zero, one, two, or three Ikaros isoforms (isoforms indicated above each lane). The locations of the bands corresponding to FLAG–Helios A and Ikaros isoforms I, V, and VI are indicated to the right.
Specific, indiscriminate protein–protein interactions between Helios and Ikaros isoforms
The above data suggest that Helios copurifies with Ikaros by immunoaffinity chromatography because the two proteins are tightly associated with each other in RLm11 cells. To confirm that recombinant Helios can interact with recombinant Ikaros isoforms, an expression vector for Helios A containing a FLAG epitope tag was cotransfected into 293T cells along with expression vectors for Ikaros isoforms I, V, and/or VI. Interactions were monitored by immunoprecipitation with anti-FLAG antibodies, followed by immunoblot analysis with either Ikaros or Helios antibodies. Figure 4B shows the Helios A and Ikaros proteins in 293T cytoplasmic and nuclear extracts prior to immunoprecipitation. (Ikaros and Helios antibodies were added to the immunoblot to visualize the products of both genes, but the relative amounts of the Ikaros and Helios gene products cannot be determined from these data.) Epitope-tagged Helios A localized to the nucleus and migrated slightly slower than Ikaros isoform VI (e.g., Fig. 4B, lane 14), agreeing with the migration pattern of Helios observed in the purified complexes. Immunoprecipitation with FLAG antibodies (Fig. 4C) revealed that FLAG–Helios A interacts with all three Ikaros isoforms with similar efficiencies (on the basis of a comparison on the relative protein amounts before and after immunoprecipitation). The carboxy-terminal zinc fingers are likely to be responsible for the interactions, given the strong homology between the Ikaros and Helios domains.
DNA-binding-site specificity of Helios A
The above results suggest that intracellular complexes exist containing a combination of Helios and Ikaros proteins. Helios complexes may also exist without Ikaros components, and Ikaros complexes may exist without Helios components. Perhaps, the various complexes carry out different functions by recognizing different DNA sequences. To determine the DNA sequence specificity of Helios A, a binding-site selection–PCR ampification strategy was employed with a fusion protein containing the Helios A DNA-binding domain and GST (Zweidler-McKay et al. 1996; see Materials and Methods).
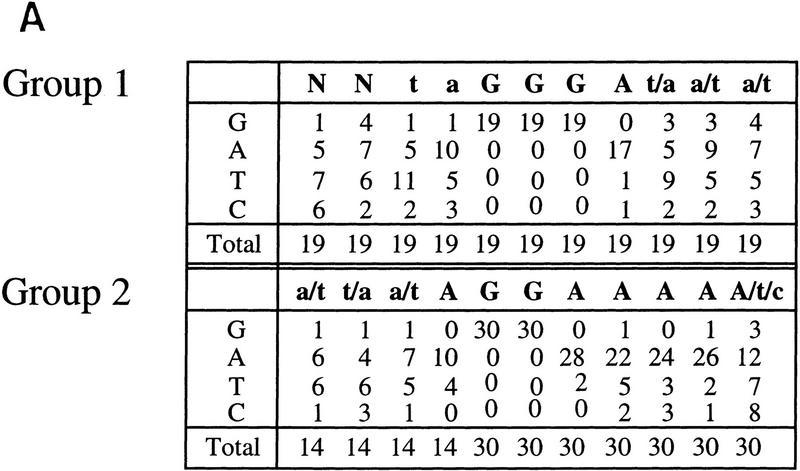
The selected clones revealed potential recognition sequences that can be divided into two groups (Fig. 5A). The first group, containing 19 sequences, was characterized by the core sequence GGGA. The second group, containing 30 sequences, was characterized by the core sequence GGAAAA. A simple interpretation of these data is that the core sequence GGA is generally sufficient for high-affinity DNA binding if it is flanked by a guanine at the 5′ end or by three adenines at the 3′ end. The first group is very similar to a previously described consensus sequence for Ikaros isoform V (Molnar and Georgopoulos 1994), which is the most homologous to Helios A. The second group did not match the Ikaros consensus sequence.
Figure 5.
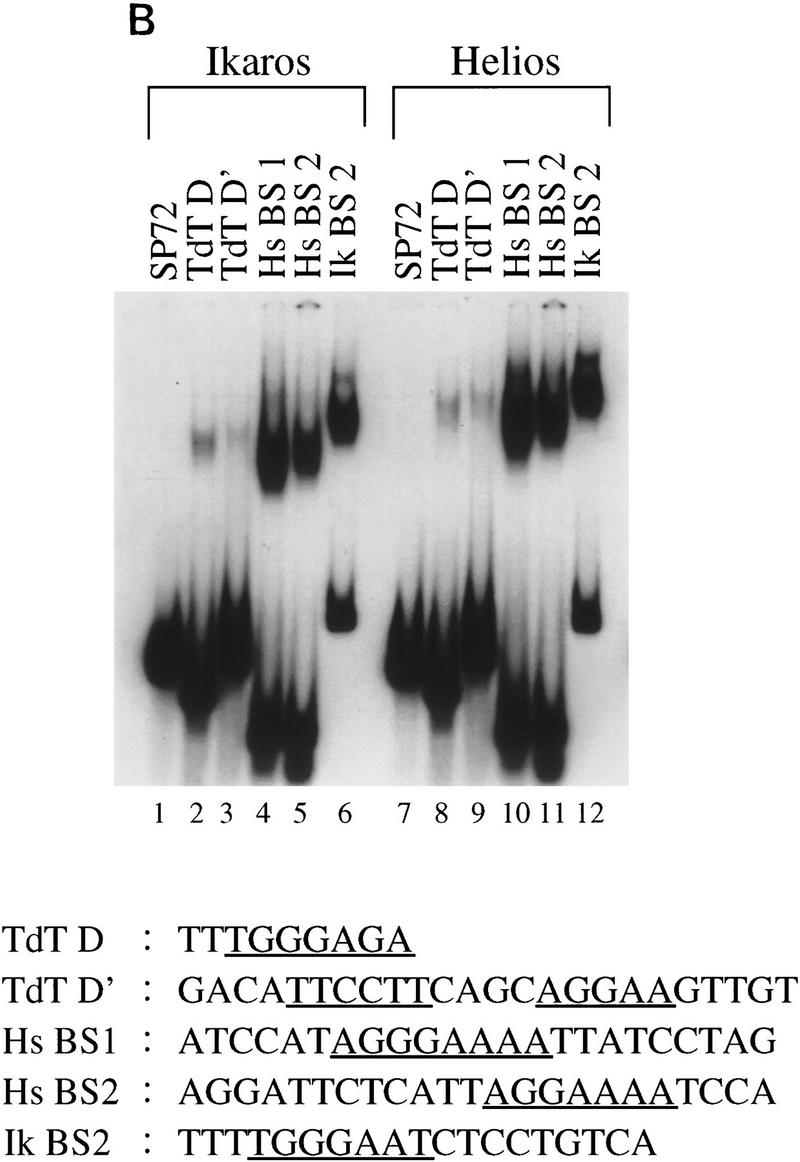
Helios A and Ikaros isoform V bind to similar DNA sequences. (A) High-affinity Helios A-binding sites were selected by a GST pull-down assay (Zweidler-McKay et al. 1996). DNA fragments selected by the GST–Helios A fusion protein were sequenced and tabulated. The selected sequences can be divided into two groups, both containing a core sequence of GGA. Group 1 was represented by 19 sequences and Group 2, by 30 sequences. The number of fragments containing each nucleotide at a given position following alignment are indicated for each group. Only 14 sequences are shown for the first four positions of the Group 2 sequence because the remaining 16 were derived from fragments in which these four positions were contained within the invariant primer-binding sites flanking the variable sequences. (B) Gel mobility shift assays were performed with recombinant GST–Ikaros isoform V (lanes 1–6) and recombinant Helios A (lanes 7–12) with probes containing the five sequences shown at the bottom (lanes 2–6, 8–12) and a negative control probe (lanes 1,7).The specific probes were derived from the pSP72 (Promega) multiple cloning site region (lanes 1,7), the TdT D (lanes 2,8) and D′ (lanes 3,9) elements, the Hs BS1 (i.e., Helios binding site 1; lanes 4,10) and Hs BS2 (lanes 5,11) sequences selected in this study, and the IkBS2 (lanes 6,12) sequence selected previously (Molnar and Georgopoulos 1994).
To determine whether the two groups of Helios A consensus sequences provide a functional distinction between Ikaros and Helios, gel mobility shift assays were performed with a probe representative of the first group (Fig. 5B, lanes 6,12; Ik BS2; Molnar and Georgopoulos 1994), a probe representative of the second group (lanes 5,11; Hs BS2), and a probe containing a sequence that conforms to both groups (lanes 4,10; Hs BS1). A negative control was also included (lanes 1,7), along with probes containing two TdT promoter elements that are known to bind Ikaros (lanes 2,3,8,9). The results demonstrate that Ikaros isoform V and Helios A possess very similar sequence specificities, as the relative complex abundances observed with each probe were very similar for the two proteins. Thus, if Ikaros–Helios complexes carry out different functions from either Ikaros–Ikaros or Helios–Helios complexes, they are likely to do so as a result of other functional differences between Helios and Ikaros.
T cell-restricted expression pattern of Helios
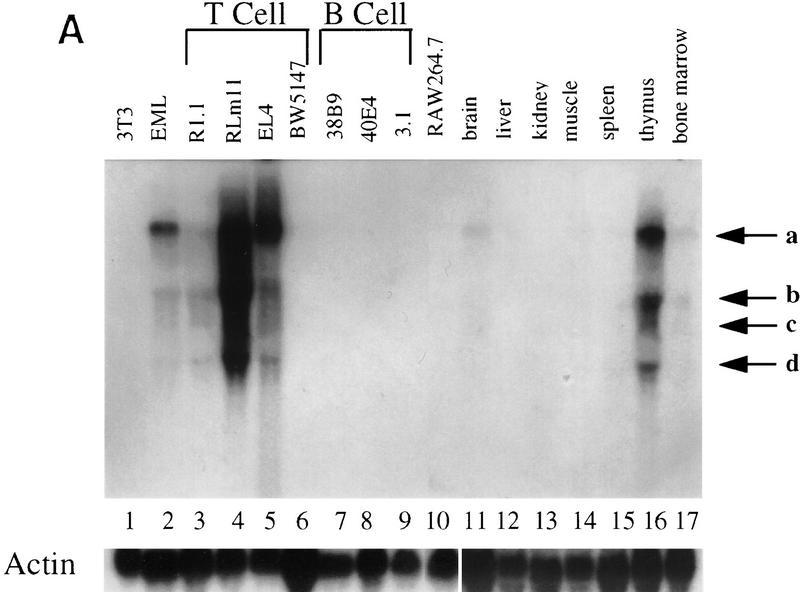
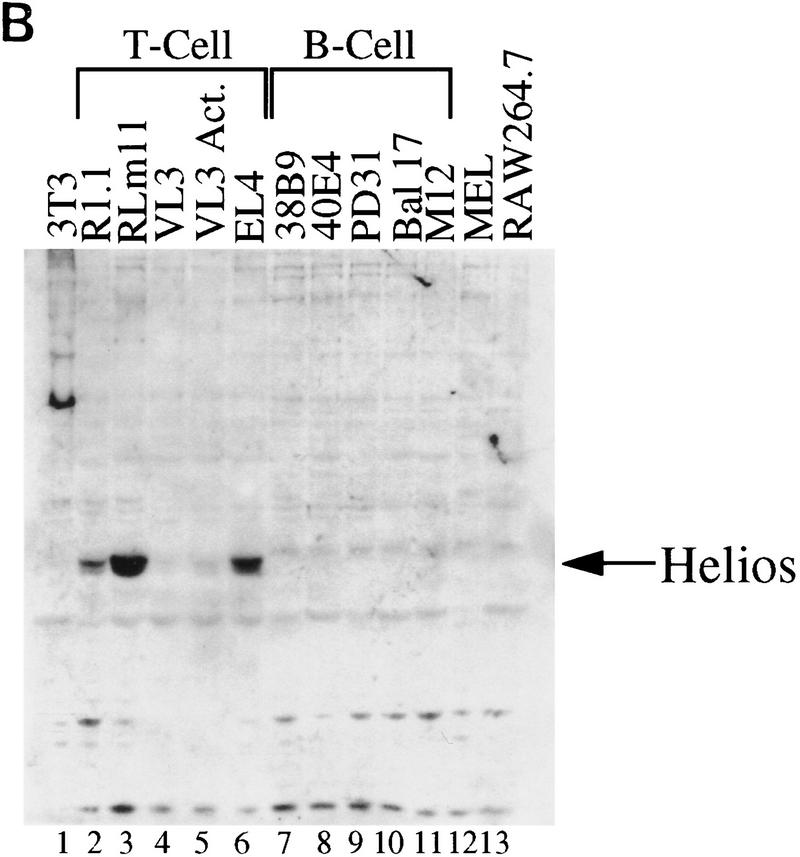
To determine the expression pattern of Helios, Northern blots were carried out with mRNA from transformed cell lines and murine tissues. Surprisingly, Helios mRNA was largely restricted to the T cell lineage. Four major transcripts were readily detected in three of four T cell lines tested (Fig. 6A, lanes 3–6) and in a cell line containing early progenitors of multiple hematopoietic lineages (lane 2; Tsai et al. 1994). No expression was observed in B lineage cell lines (lanes 7–9) or in fibroblast (lane 1) or macrophage (lane 10) lines. In contrast, the Ikaros gene was expressed at high levels in all three B cell lines and at lower levels in the macrophage line (data not shown). Consistent with the expression pattern in the cell lines, abundant Helios transcripts were detected in the thymus (lane 16), with very little expression in the bone marrow (lane 17) and brain (lane 11), and no detectable expression in spleen, liver, kidney, or muscle (lanes 12–15). By contrast, Ikaros transcripts were readily detected in both the spleen and thymus (Georgopoulos et al. 1992; data not shown). The reason for the existence of transcripts of different sizes has not been explored, but all four transcripts were detected when the blots were probed with gene fragments encoding amino acids 221–292 (Fig. 6A) or amino acids 1–109 (data not shown).
Figure 6.
Helios expression is largely restricted in T cells. (A) Northern blot analysis was used to measure Helios mRNA expression in murine hematopoietic cell lines and tissues. Cell lines and tissues analyzed are indicated at the top. Twenty micrograms of total RNAs was analyzed in each lane. Nitrocellulose membranes were hybridized sequentially with a Helios probe (fragment encoding amino acids 221–292, top) and a human β-actin probe (500 bp, bottom). Positions of the 28S and 18S rRNAs are indicated. Actin transcripts and four major transcripts detected by Helios probe are indicated. (B) Helios protein in murine cell lines was analyzed by immunoblotting. Cell lines analyzed are indicated at the top. Twenty micrograms of protein from nuclear extracts estimated by Coomassie Assay (Pierce) were loaded in each lane. (p70) Protein migrating close to 70 kD and detected with anti-Helios serum. (C) The nuclear extracts analyzed in B were probes with antibodies directed against the carboxy-terminal half of Ikaros.
Immunoblot experiments provided evidence that the Helios protein is restricted to the T cell lineage (Fig. 6B). The expected 70-kD band was observed in four different T cell lines (Fig. 6B, lanes 2–6), but not in 5 cell lines of the B lineage (lanes 7–11) or in erythroid (lane 12), macrophage (lane 13), or fibroblast (lane 14) lines. The T cell-restricted expression pattern of Helios is in striking contrast to the expression pattern of the Ikaros proteins, which were present in all of the hematopoietic cell lines (Fig. 6C).
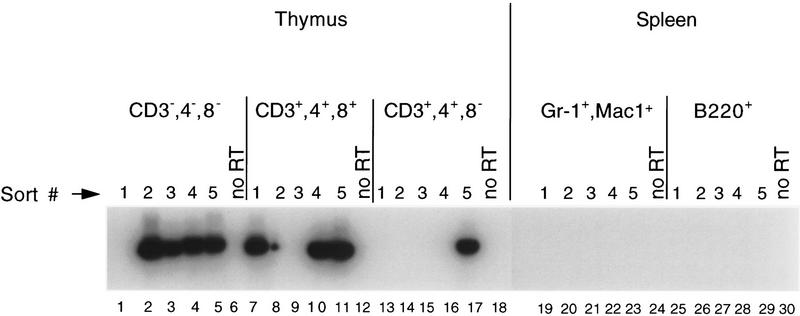
To examine Helios expression during T cell development, primary thymocytes were sorted by FACS into triple negative (CD3−4−8−), double-positive (CD3+4+8+), and CD4+ single-positive (CD3+4+8−) populations (see Materials and Methods and Klug et al. 1998). Five different samples of 20 cells each were analyzed by RT–PCR for each population. PCR products were efficiently amplified in four of five samples of triple-negative cells (Fig. 7, lanes 1–5), three of five samples of double-positive cells (lanes 7–11), and only one of five samples of single-positive cells (lanes 13–17). DNA sequencing of the PCR products confirmed that they contain Helios sequences. The sequences also revealed that the small size difference between the products in lanes 2 and 3 versus 4 and 5 was attributable to the amplification of the Helios A sequence in lanes 2 and 3 and the Helios B sequence in lanes 4 and 5 (data not shown). No PCR products were amplified from primary splenic neutrophils (Gr-1+Mac1+, lanes 19–23) or splenic B cells (B220+, lanes 25–29). These results confirm the T cell-restricted expression of Helios and suggest that it might be most abundant in immature cells of the T lineage.
Figure 7.
Helios expression in thymocyte and splenic cell subsets. Twenty cells of the indicated cell-surface phenotypes were sorted by FACS for analysis of Helios expression during thymocyte maturation and in non-T-lineage cell subsets. Primers for nested PCR amplified across sequences that encode the four amino-terminal zinc fingers of Helios. The PCR products observed with the various samples exhibit slightly different migrations, which represent the amplification of both the Helios A and Helios B products with predicted sizes of 712 and 790 bp, respectively.
Quantitative association of Helios with Ikaros
The stability of the Ikaros–Helios interaction provides strong evidence that the interaction is relevant. To determine the percentage of Helios that is stably associated with Ikaros and, conversely, the percentage of Ikaros that is stably associated with Helios, quantitative immunoprecipitation experiments were performed with VL3-3M2 (Fig. 8) and RLm11 (data not shown) cell extracts.
The immunoprecipitation reactions were carried out with increasing amounts of either Ikaros antibodies (lanes 2–5, 11–14) or Helios antibodies (lanes 6–9, 15–18). The amounts of Ikaros and Helios within the immunoprecipitation pellets (lanes 1–9) and supernatants (lanes 10–18) were determined by immunoblot analysis. Ikaros antibodies were capable of depleting almost all of the Helios from the VL3-3M2 extracts, as determined by the depletion of almost all of the Helios from the immunoprecipitation supernatants (lanes 10–14, top). This result suggests that virtually all of the Helios within the cell is associated with Ikaros isoforms. In contrast, Helios antibodies depleted only a small fraction of the Ikaros (lanes 15–18, bottom), despite the ability of these antibodies to deplete all of the Helios (lanes 15–18, top). This result suggests that only a fraction of the Ikaros is associated with Helios, and therefore that Ikaros is in considerable excess. Similar results were obtained with RLm11 extracts (data not shown). The quantitative association of Helios with Ikaros provides strong support for the functional relevance of the Ikaros–Helios interaction.
Helios and Ikaros exist as a relatively homogeneous complex
The overexpression experiments in 293T cells suggest that the four Ikaros isoforms and two Helios isoforms present in RLm11 cells are capable of forming stable complexes with each other in an indiscriminate manner. With six different proteins interacting through highly homologous carboxy-terminal zinc finger domains, 21 different dimers could be produced. If these dimers associate into multimers, as suggested by some of the data, a much larger number of species is possible. Each species might carry out a distinct function within the cell, or many of the complexes might carry out redundant functions. Alternatively, the interactions between the endogenous proteins in RLm11 cells might not be as indiscriminate as suggested by the 293T experiments. The relative elution profiles of Ikaros and Helios from a gel filtration column support this latter hypothesis (Fig. 9). As shown above (Fig. 1), Ikaros isoforms elute from gel filtration columns in a broad peak and at large molecular masses. It is not known whether this elution profile accurately reflects the sizes of the complexes, but the broad peak suggests that the complexes are quite heterogeneous. Surprisingly, Helios eluted as a sharp peak from a Superose 6 gel filtration column, coeluting with the largest of the Ikaros complexes (Fig. 9, lanes 6–8). This sharp elution profile suggests that Helios is not assembled into the same heterogeneous array of complexes as Ikaros, but rather is assembled into one discrete complex, or at least a much more homogeneous set of complexes than Ikaros.
The gel filtration result raises the possibility that Helios is a limiting regulatory molecule that dictates the function of Ikaros isoforms. The apparently homogeneous complex containing Helios and Ikaros might carry out a specific function. The excess Ikaros that is not associated with Helios might assemble into specific complexes with other Ikaros-related proteins, such as the p30 protein (Fig. 2A). Indeed, a partial peptide sequence of p30 has revealed that it is another member of the Ikaros family (B.S. Cobb, unpubl.). Some of the excess Ikaros might instead exist as partially formed complexes that lack subunits essential for function.
Selective association of complexes containing Helios with centromeric regions of T cell nuclei
A recent study revealed that Ikaros might not be a typical transcriptional activator, as it was found in B cells to localize primarily to centromeric heterochromatin (Brown et al. 1997). This study also revealed that a variety of inactive genes colocalize with Ikaros to the centromeric regions, leading to the hypothesis that Ikaros might play a role in recruiting genes to centromeric foci that are destined for inactivation. Consistent with the hypothesis that both Helios and Ikaros are not typical transcriptional activators, we were unable to detect activation of reporter constructs containing multiple high-affinity Ikaros/Helios binding sites when various Ikaros and/or Helios isoforms were overexpressed (data not shown). In addition, none of these proteins appears to function as a simple transcriptional repressor in either transient or stable transfection experiments (data not shown).
Confocal microscopy was employed to examine whether the subnuclear localization of Helios and Ikaros in T cells is similar to the centromeric localization of Ikaros observed in B cells (Brown et al. 1997). For these experiments, VL3-3M2 and primary activated lymph node T cells were used. Surprisingly, with both T cell sources, Ikaros was distributed throughout the nucleoplasm and did not exhibit the predominant centromeric localization that had been observed in B cells. These results are apparent in Figure 10 (g,h,i), which shows a single optical section of a representative lymph node T cell stained with an Ikaros antibody (h) and with a fluorescent DNA probe for gamma satellite repeats (g), which are found primarily at centromeric regions of chromosomes (see Materials and Methods and Brown et al. 1997). The costained image (i) reveals patches of Ikaros staining at the edges of the centromeric foci (yellow), but most of the Ikaros was distributed throughout the nucleoplasm (green). Similar results were observed in VL3-3M2 cells (data not shown). These results are in striking contrast to the results observed in B cells (j,k,l), in which the Ikaros (j) and gamma-satellite (k) staining patterns are very similar to each other, and closely coincide in the costained image (l).
Figure 10.
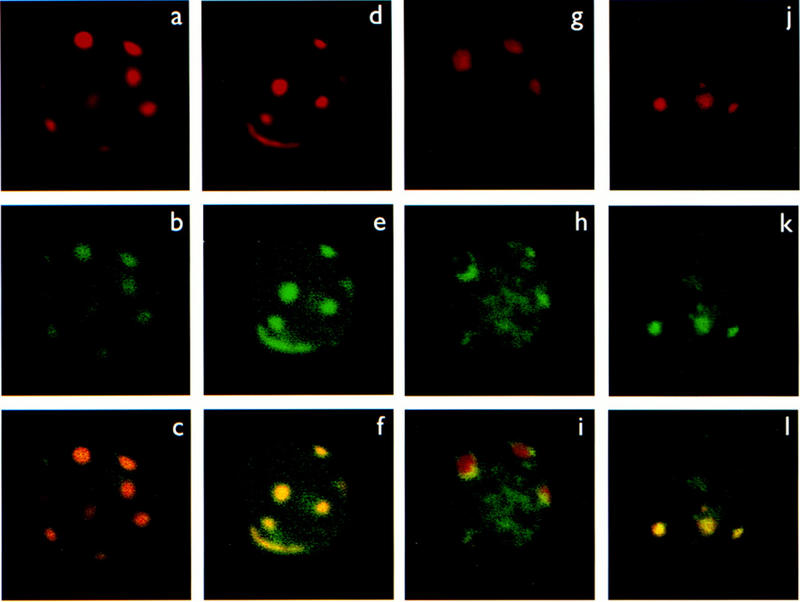
Distribution of Helios and Ikaros proteins within the nuclei of T and B lymphocytes. Confocal images are shown of single optical sections through the nucleus of representative individual concavalin-A stimulated lymph node T cells (a,b,c,g,h,i), activated VL3-3M2 T cells (d,e,f), and B3 pre-B cells (j,k,l). The cells were labeled simultaneously with a probe for gamma satellite sequences (red) and specific antisera for Helios (a–f) or Ikaros (g–l) (shown in green). The red and green components of costained nuclei (bottom, c,f,i,l) are shown separately in the top (gamma satellite-red a,d,g,j) and middle (p70-green b,e, Ikaros-green h,k) rows.
Interestingly, in both the lymph node T cells (a,b,c) and the VL3-3M2 T cells (d,e,f), Helios colocalized with the gamma satellites at the centromeric foci, similar to the predominant localization of Ikaros in B cells. The colocalization is apparent by comparison of the gamma-satellite staining pattern (a,d) with the Helios staining pattern (b,e) and by examination of the costained images (b,e), in which colocalization is evident by the orange and yellow colors. The yellow color observed in costained VL3-3M2 cells indicates that the intensity of Helios staining is greater than in the lymph node T cells, which yield an orange color. It is worth noting, however, that the intensity of Helios staining in both of these cell types was much weaker than the intensity of Ikaros staining. (The images in Fig. 10 do not reflect the lower abundance of Helios because different settings were used for the Helios images to enhance detection.) These results are consistent with those in Figure 8, which demonstrate that Helios is much less abundant than Ikaros in T cells. Because the data in Figure 8 demonstrate that Helios is quantitatively associated with a subset of the Ikaros, the most likely interpretation of the confocal results is that the Ikaros–Helios complexes are predominantly localized to the centromeric foci. In contrast, the excess Ikaros that is not associated with Helios appears to be distributed more broadly throughout the nucleoplasm.
Although not easily apparent from the images shown in Figure 10, the similar staining patterns of Helios and gamma satellite repeats may not represent true colocalization (i.e., identity of spatial location). Instead, the Helios staining appears to extend slightly beyond the gamma satellite staining. Preliminary studies of deconvolved images are consistent with this observation (K.E. Brown and A.G. Fisher, unpubl.), suggesting that Helios and gamma satellites are interlaced rather than spatially identical.
Discussion
Gene disruption experiments have shown that Ikaros isoforms carry out critical functions during the development of B and T lymphocytes, as well as other hematopoietic cell types (Georgopoulos et al. 1994; Winandy et al. 1995; Wang et al. 1996). Nevertheless, in light of the centromeric heterochromatin localization that has been observed (Brown et al. 1997; Klug et al. 1998), the precise intracellular functions of Ikaros almost certainly will be difficult to elucidate. The finding that Ikaros protein complexes are sufficiently stable to allow their purification by immunoaffinity chromatography provides a means of identifying relevant Ikaros partners within any given cell type or cell line, information which will be essential for fully understanding Ikaros functions. Our analysis of the complexes within the RLm11 T cell line led to the identification of a new member of the Ikaros family, Helios. The relevance of the Helios–Ikaros interaction was demonstrated by the quantitative association in the two cell lines examined. The specific functions of Helios and of the Helios–Ikaros complex remain unknown, but the colocalization of Helios–Ikaros complexes with gamma satellites suggests that Helios might be a limiting regulatory factor that recruits a subset of the Ikaros within a T cell to centromeric foci.
The function of the Helios–Ikaros complexes at the centromeric regions, and the function of the excess Ikaros distributed throughout the nucleus, remains to be elucidated. One hypothesis is that Helios–Ikaros complexes bind to DNA sequence elements within the promoters, enhancers, or silencers of genes that are destined for inactivation. Binding of Helios–Ikaros might result in the recruitment of those genes to the centromeric foci, where they might be assembled into inactive heterochromatin structures. A more extensive discussion of the possible functions for these proteins at centromeric regions, and a discussion of related studies of Drosophila heterochromatin regulation and position effect variegation, can be found in Brown et al. (1997).
The reason for the broad distribution of Ikaros in T cell nuclei, relative to its predominant centromeric localization in B cells, also remains unknown. Perhaps, B cells contain a more abundant partner for Ikaros that recruits a larger fraction of the Ikaros pool to the centromeric foci. Alternatively, Ikaros may be capable of localizing to the centromeric regions in B cells in the absence of a partner. The large pool of Ikaros in T cells that is not localized to the centromeres may simply be excess protein that carries out no specific functions. Alternatively, this pool may carry out one or more critical functions related to the activation or inactivation of specific genes, perhaps acting as a more typical activator or repressor in combination with other transcription factors, coactivators, or corepressors. Although our discussion of Ikaros and Helios has focused on their possible roles in transcriptional regulation, it is important to note that no compelling evidence has been published demonstrating that Ikaros or Helios are actually involved, either directly or indirectly, in transcription. Alternative functions that must be considered are involvement in nuclear structure, DNA synthesis, or mitosis.
The discovery of a T cell-restricted member of the Ikaros family leads to models that might explain some of the phenotypes observed in mice containing Ikaros gene disruptions. Ikaros−/− mice lack all cells of the B lineages, but exhibit less severe defects in some of the T cell lineages (see introductory section; Wang et al. 1996) even though Ikaros normally appears to be expressed in all B and T cells. Perhaps complete disruption of the Ikaros gene has relatively modest effects on T cell development because Helios compensates for the absence of Ikaros. This hypothesis is supported by the finding that Helios and Ikaros recognize similar DNA sequences. A more severe T cell defect was observed in mice containing a specific disruption of the zinc finger DNA-binding domains of Ikaros; these mice, which retain the capacity to produce smaller Ikaros proteins containing the carboxy-terminal protein–protein interaction domain, do not produce any progenitor or mature T cells (Georgopoulos et al. 1994). The severity of this mutation within the T cell lineage might be due to the fact that the small Ikaros proteins act in a dominant negative manner, sequestering Helios and preventing it from compensating for the loss of Ikaros.
Consistent with the hypothesis that Ikaros and Helios might not simply be transcriptional activators, we have been unable to detect transcriptional activation functions for either protein by standard transient and stable transfection assays (B.S. Cobb and K. Hahm, unpubl.). Ikaros proteins contain a domain that functions as a strong transactivation domain when fused to a GAL4 DNA-binding domain (Sun et al. 1996; K. Hahm, L. Trinh, P. Ernst, and S.T. Smale, in prep.). Transactivation of reporter plasmids has also been reported with the full-length protein (Molnar and Georgopoulos 1994). In our hands, however, we have been unable to detect transactivation with either full-length Ikaros or Helios, or a combination of the two, despite efficient expression of the proteins and despite the fact that extracts from those cells contain the expected DNA-binding activities (B.S. Cobb and A.S. McCarty, unpubl.). Because these results are negative, they can only be substantiated by the demonstration that Ikaros and Helios carry out a different function. It remains possible that Ikaros functions as a activator of some genes through combinatorial interactions, yet is involved in heterochromatin formation on other genes at the centromeric foci. Such a function would be similar to that proposed for the Drosophila Hunchback protein, which contains zinc finger domains that are highly related to those in Ikaros and Helios. Hunchback acts as a simple activator during embryogenesis and also has been proposed to establish silencing complexes in Drosophila by recruiting Polycomb-group proteins (Zhang and Bienz 1992; Poux et al. 1996).
The stoichiometry and structure of the complexes observed by gel filtration chromatography and other techniques remain unknown. Some techniques strongly suggest that Ikaros complexes are composed of highly stable dimers. Other techniques suggest that the complexes are multimeric (see Results). Additional experiments will be needed to clarify these results and, if multimeric complexes are indeed present in cell extracts, to determine whether these complexes are the functional species in vivo. On the basis of all the data that has been obtained, our working model is that the complexes contain highly stable dimers that associate into multimers.
The homogeneous nature of the Ikaros complexes that contain Helios, as judged by gel filtration chromatography, is particularly intriguing. Because Helios appears to interact indiscriminately with the various Ikaros isoforms following overexpression in 293T cells, it is not clear why Helios would elute in a sharper peak than Ikaros. Apparently, within RLm11 cells, the interactions are not as indiscriminate as predicted. Perhaps, the smaller Ikaros complexes observed by gel filtration are partially assembled complexes, with Helios the final component added to the complex. Whatever the reason for the homogeneity of the Helios complexes, this distinction from Ikaros is likely to rely on domains other than the zinc finger domains, since the zinc fingers are extremely well conserved.
Materials and methods
Plasmid DNAs
Mammalian expression plasmids for Ikaros isoforms I, III, V, and VI were prepared in the pcDNA1neo vector (InVitrogen). cDNAs for these isoforms in the pSP72 vector (see Hahm et al. 1994) were excised with BglII and XhoI and inserted into pcDNA1neo cleaved with BamHI and XhoI. Plasmids for expression in Escherichia coli of fusion proteins between Helios A and GST were prepared for antibody preparation and DNA-binding studies. Fragments encoding Helios A amino acids 1–109 (for antibody preparation) and 1–292 (for DNA-binding studies) were generated by PCR from the full-length Helios A cDNA with a primer spanning the amino-terminal coding region, 5′-GATAGATCTATGGAAACAGACGCAATTGA-3′, and the reverse primers 5′-GATGAATTCGTCCATCATATGAGACTGCATCAG-3′ or 5′-GATGAATTCGCCTTGAAGGTCCTGGACTTT-3′, respectively. The 327- and 876-bp PCR products, respectively, were cleaved with BglII and EcoRI and inserted into pGEX 2T (Pharmacia) cleaved with BamHI and EcoRI.
The mammalian expression plasmid for FLAG-tagged Helios A was prepared by amplifying the protein-coding sequence with the following PCR primers: 5′-GATAGATCTATGGAAACAGACGCAATTGA-3′ and 5′-GATGAATTCCTAGTGGAATGTGTGCTCCCC-3′. The PCR product was cleaved with BglII and EcoRI and inserted into pSP72 cleaved with the same enzymes. The following FLAG-encoding oligonucleotide and its complement were then annealed, cleaved with BglII and BamHI, and inserted into the BglII-cleaved plasmid: 5′-GATAG A T C T A C C A T G G A C T A C A A G G A C G A CGATGACAAGGGATCCGAT-3′. Finally, the entire coding sequence was transferred to the pcDNA3 expression vector (InVitrogen) following cleavage of the pSP72 plasmid with BglII and XhoI and the vector with BamHI and XhoI.
Cell culture and transient transfections
RLm11, VL3-3M2, and other cell lines were maintained as described previously (Groves et al. 1995; Ernst et al. 1996). Transient transfections of 293T cells were performed by a calcium phosphate coprecipitation method (Ausubel et al. 1989) with the amounts of plasmid DNA indicated in the figure legends.
Antibodies
Ikaros antisera were generated against GST-fusion proteins containing amino-terminal amino acids 1–80 of isoform I and carboxy-terminal amino acids 54–286 of isoform I. Helios antisera were generated against a GST fusion protein containing amino-terminal amino acids 1–109. These fusion proteins were purified by glutathione–Sepharose chromatography and used to immunize rabbits as described previously (Hahm et al. 1994). The Elf-1 antiserum was described previously (Ernst et al. 1996). IgGs from preimmune and immune sera were purified by protein A–Sepharose (Pharmacia) chromatography as described by Harlow and Lane (1988).
Gel filtration and DNA-affinity chromatography
Gel filtration chromatography was performed with prepacked Superdex 200 and Superose 6 FPLC columns (Pharmacia). Approximately 1 mg of RLm11 nuclear extract in a volume of 500 μl was applied to the column in HGED.45 buffer (20 mm HEPES at pH 7.9, 20% glycerol, 0.2 mm EDTA, 1 mm dithiothreitol, 0.1 mm PMSF, and 0.45 m KCl) containing 0.015% NP-40. Some experiments were performed with 0.15 m KCl and without NP-40. Fractions (500 μl) were collected and 45 μl of each was analyzed by immunoblot. Molecular size markers were thyroglobulin (669 kD), ferritin (440 kD), catalase (232 kD), and albumin (67 kD) (Pharmacia).
Sequence-specific DNA-affinity chromatography was performed with 0.5 ml of resin containing covalently linked multimers of the TdT D sequence as described previously (Hahm et al. 1994).
Purification of Ikaros complexes and peptide sequencing
Immunoaffinity columns were prepared by covalent coupling of antibodies to protein A–Sepharose (Pharmacia) by the method described in Harlow and Lane (1988). Ikaros complexes were purified by the following method. RLm11 nuclear extracts (200 mg) in buffer D (20 mm HEPES at pH 7.9, 20% glycerol, 0.2 mm EDTA, 1 mm dithiothreitol, 0.1 mm PMSF, 0.42 m KCl) were applied to a precolumn containing 500 μl of unmodified protein A–Sepharose, with the eluate flowing directly onto the bed of a 4-ml immunoaffinity column. The extract was passed through both columns three times. The immunoaffinity column was then washed with 10 column volumes of HGED buffer (20 mm HEPES at pH 7.9, 20% glycerol, 1 mm EDTA, 1 mm DTT, 0.1 mm PMSF) containing 0.45 m KCl and 1 m KCl. In some experiments, the column was also washed with HGED buffer containing no KCl. Bound proteins were eluted in 1-ml fractions into tubes containing 20 μl of 2 m Tris-HCl (pH 6.8) with 100 mm trimethyl ethanolamine (pH 11.0). Fractions were analyzed by SDS-PAGE followed by silver staining.
To isolate the 70-kD protein for microsequencing, appropriate fractions from the pH 11.0 elution were pooled. After three runs of a 5-ml affinity column, ∼10 μg of the 70-kD protein was obtained. The pooled proteins were precipitated by 20% trichloroacetic acid (Fisher), separated by SDS-PAGE, transferred to PVDF membrane (Biorad), and stained with Ponceau S as described previously (Hahm et al. 1994). The 70-kD band was excised and subjected to endopetidase C digestion followed by peptide sequencing as described (Fernandez et al. 1994).
Isolation of Helios cDNAs
To isolate the Helios gene, first strand cDNA was generated from RLm11 mRNA and used for PCR with the following degenerate primer pairs designed from the two peptide sequences; 5′-GATGAATTCCA(A/G)GA(A/G)CC(A/C/G/T)AT(T/C)A TGGA(C/T)AA(C/T)AA-3′, 5′-GATGAATTCTT(C/T)TC(A/G)TA(A/C/T)GT(C/T)AG(A/G)TTCAT-3′. A 216-bp PCR product was isolated and inserted into pSP72 (Promega) digested with BglII and EcoRI. The sequence of the insert was determined and found to contain codons encoding the amino acids within the two original peptides that were not included in the PCR primers, confirming that the fragment was derived from the correct gene. To isolate a full-length cDNA, the 216-bp fragment was radiolabeled and used to screen a newborn thymus cDNA library (Stratagene) as previously described (Hahm et al. 1994). The resulting full-length cDNA encodes the Helios A protein. The Helios B cDNA was isolated by RT-PCR with the following primers flanking the amino-terminal zinc finger domain.
Plasmids encoding Helios A–GST fusion proteins were introduced into the SCS-1 strain of E. coli (Stratagene). Fusion proteins were then induced, purified and stored at −80°C as described previously (Hahm et al. 1994).
Immunoprecipitation and immunoblot analyses
293T cell immunoprecipitation assays were performed from cytoplasmic or nuclear extracts with the following method. First, cytoplasmic and nuclear extracts were prepared from four 100-mm plates of transfected cells by Dounce homogenization as described by Lo et al. (1991). Nuclear extracts (400 μg) were incubated with 3–6 μg of FLAG M2 monoclonal antibody (Kodak IBI) for 2–4 hr at 4°C, followed by centrifugation for 10 min. Supernatants were transferred to a new tube and mixed with 40 μl of protein A–Sepharose (Pharmacia). The slurry was incubated for 1 hr at 4°C. After brief centrifugation, pellets were washed 5 times with buffer containing 10 mm HEPES (pH 7.9), 0.45 m KCl, 1 mm EDTA, 0.015% NP-40, 10% glycerin, and 1 mm dithiothreitol. The washed pellets were analyzed by SDS-PAGE followed by immunoblot as described previously (Hahm et al. 1994).
The quantitative immunoprecipitations were performed with 100 μl of RLm11 (500 μg) nuclear extract or 200 μl of VL3-3M2 (1.2 mg) nuclear extract. The extract was mixed with 167 μl of a buffer containing 100 mm NaCl, 20 mm Tris (pH 8), and 0.5% NP-40. To this mixture was added the indicated amount of IgG (against the amino terminus of either Ikaros or Helios), which was diluted to 33 μl with PBS. Binding proceeded for 1 hr on ice after which 100 μl of a 50% slurry of protein A–Sepharose was added. The mixture was incubated for an additional hour at 4°C on a rocker. The resin was pelleted by brief centrifugation and the supernatants were transferred to a new tube. The pellets were washed four times with the above buffer. The immune complexes and a constant proportion of the supernatants were analyzed by SDS-PAGE, followed by immunoblot analysis with antisera against the carboxyl terminus of Ikaros or the amino-terminus of Helios.
Binding-site selection analysis and gel mobility shift assays
Binding-site selection assays were performed as described by Zweidler-McKay et al. (1996). The double-stranded oligonucleotide containing random sequences was generated from the following 66-nucleotide fragment: 5′-GGTAGAATTCAACTGCCATCTAGGNNNNNNNNNNNNNNNNNNACACCGAG TCCAGTGGATCCTACG-3′. The complementary strand was generated by annealing the following primer and extending with the E. coli DNA polymerase Klenow fragment: 5′-CGTAGGATCCACTGGACTCGGTG-3′. The DNA fragments containing random sequences were incubated with the recombinant GST–Helios A fusion protein. Bound DNA molecules were separated from unbouind molecules by incubation with GST–Sepharose, followed by centrifugation (Zweidler-McKay et al. 1996). Bound DNA molecules were eluted and amplified by PCR with the above 23-mer and the following reverse primer: 5′-GGTAGAATTCAACTGCCA-3′. After four binding cycles, the final PCR products were digested with EcoRI and HindIII and inserted into pSP72 (Promega). Thirty-two clones were analyzed by sequencing.
For gel mobility shift analysis, the IkBS1 probe was prepared from a plasmid containing the following oligonucleotide and its complement inserted into the BamHI site of pSP72: 5′-GATCTTCAGCTTTTGGGAATCTCCTGTCAG-3′. The Hs BS1 and Hs BS2 probes were prepared from plasmids containing the following oligonucleotides and their complements inserted into the HindIII and EcoRI sites of pSP72: Hs BS1, 5′-CGTGTATCCATAGGGAAAATTATCCTAGAT-3′; Hs BS2, 5′-GATCTCGTGTGATTTTCCTAATGAGAATCCTAGATG-3′. The TdT D and D′ probes were described previously (Hahm et al. 1994). Radiolabeled probes were prepared as described previously (Ernst et al. 1993) following cleavage with HindIII and EcoRI. The SP72 control probe was cleaved with BglII and XhoI. Binding reactions were performed as described by Lo et al. (1991). Samples were analyzed as described in Ernst et al. (1993).
Northern blot analysis
RNAs from cell lines and tissues were prepared by a guanidine thiocyanate centrifugation method (Ausubel et al. 1989). Tissues used to generate RNAs were isolated from 1-month-old Balb/c mice. Northern blot probes were prepared from a 216-bp p70 fragment encoding amino acids 221–292, and were labeled using the Prime-It Kit (Stratagene).
RT–PCR of Helios in thymocyte subsets and in splenic myeloid and B cell populations
Thymocyte subsets (CD-3−CD-4−CD-8−, CD-3+CD-4+CD-8+, and CD-3+CD-4+CD-8−) were sorted by use of the following combination of fluorochromes: CD-3PE, CD-4FITC, and CD-8APC. Splenic myeloid cells were sorted as Gr-1HIMac-1+ and splenic B cells were isolated as B220+ cells. Each cell population was sorted once and then clone-sorted directly into 0.2-ml tubes containing 20 μl of RT lysis buffer [5× first strand buffer (Gibco-BRL), 10 mm DTT, 2% Triton X-100, 0.01% BSA, 0.2 mm spermidine, 0.4 units of RNasin (Promega), 100 ng of RT primer, 0.5 mm each dNTP]. Reactions were initiated by adding 1 μl of MMLV reverse transcriptase. Reactions were incubated for 75 min at 37°C. About 10%–15% of the cDNA reaction was used as a template for 35 cycles of PCR with the outside primers listed below (PCR conditions: 94°C for 30 sec, 55°C for 30 sec, and 72°C for 30 sec). Five percent of the first PCR reaction was used as template for a second round of 35 cycles with the inside primer set and the same PCR conditions described above. Products were resolved on a 1.5% agarose gel and then blotted for Southern analysis with an oligonucleotide (Helios probe) complementary to an exon located just upstream of the first zinc finger exon in Helios. Control reactions received sorted cells but no reverse transcriptase. At least four large introns occur between the exon sequences being amplified by the inside primer set.
The following oligonucleotides used for the RT-PCR and Southern blots are written in a 5′ to 3′ orientation: RT primer, GCATTGTTGATGGCTTGGTC; outside primers, GGGGAACA CGCCAATATGGC (5′ end) and GCATCAGCTCAGCCTCCTTC (3′ end); inside primers, CCAATGGACAGCACGCCTCG (5′ end) and ATATCTGGGTAGCTGAATCGC (3′ end); p70 probe, CCCTGAGCCGTGAGGATGAG.
Confocal microscopy
Lymph node T cells were prepared as follows: Lymph nodes were removed from a Balb/c mouse and minced in sterile medium to yield a single cell suspension. After washing, cells were resuspended at 2.5 × 106/ml and cultured at 37°C in AM DM medium containing 10% FCS, antibiotics, concanavalin A (5 μg/ml) and IL-2 (20 U/ml). After 36 hr, live cells were enriched by ficoll separation and cultured for a further 24 hr in 1 m DM containing 10% FCS and IL-2 (20 U/ml) at 37°C. VL3-3M2 cells were activated with PMA (10 ng/ml) and ionomycin (250 ng/ml) as described previously (Groves et al. 1995). B3 cells, the gamma satellite probe, and the immunofish protocol were described previously (Brown et al. 1997).
Acknowledgments
K.H. and R.L. were supported by U.S. Public Health Service (U.S. PHS) Training Grants GM-07104 and GM-08042, respectively. C.A.K. was a fellow of the Irvington Institute, I.L.W. was supported by grants from the National Cancer Institute (CA42551) and from SyStemix/Sandoz, A.G.F. was supported by the Medical Research Council, UK, and S.T.S. was supported by U.S. PHS grant DK43726. S.T.S. is an Associate Investigator with the Howard Hughes Medical Institute.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL steves@hhmi.ucla.edu; FAX (310) 206-8623.
References
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current protocols in molecular biology. New York, NY: John Wiley; 1989. [Google Scholar]
- Babichuk CK, Duggan BL, Bleackley RC. In vivo regulation of murine granzyme B gene transcription in activated primary T cells. J Biol Chem. 1996;271:16485–16493. doi: 10.1074/jbc.271.28.16485. [DOI] [PubMed] [Google Scholar]
- Brown KE, Guest SS, Smale ST, Hahm K, Merkenschlager M, Fisher AG. Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell. 1997;91:845–854. doi: 10.1016/s0092-8674(00)80472-9. [DOI] [PubMed] [Google Scholar]
- Clevers HC, Grosschedl R. Transcriptional control of lymphoid development: Lessons from gene targeting. Immunol Today. 1996;17:336–343. doi: 10.1016/0167-5699(96)10019-0. [DOI] [PubMed] [Google Scholar]
- Clevers HC, Oosterwegel MA, Georgopoulos K. Transcription factors in early T-cell development. Immunol Today. 1993;14:591–596. doi: 10.1016/0167-5699(93)90198-T. [DOI] [PubMed] [Google Scholar]
- Davis JN, Roussel MF. Cloning and expression of the murine Elf-1 cDNA. Gene. 1996;171:265–269. doi: 10.1016/0378-1119(96)00013-3. [DOI] [PubMed] [Google Scholar]
- Ernst P, Smale ST. Combinatorial regulation of transcription II: The immunoglobulin μ heavy chain gene. Immunity. 1995;2:427–438. doi: 10.1016/1074-7613(95)90024-1. [DOI] [PubMed] [Google Scholar]
- Ernst P, Hahm K, Smale ST. Both LyF-1 and an Ets protein interact with a critical promoter element in the murine terminal transferase gene. Mol Cell Biol. 1993;13:2982–2992. doi: 10.1128/mcb.13.5.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst P, Hahm K, Trinh L, Davis JN, Roussel MF, Turck CW, Smale ST. A potential role for Elf-1 in terminal transferase gene regulation. Mol Cell Biol. 1996;16:6121–6131. doi: 10.1128/mcb.16.11.6121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez J, Andrews L, Mische SM. An improved procedure for enzymatic digestion of polyvinylidene difluoride-bound proteins for internal sequence analysis. Annal Biochem. 1994;218:112. doi: 10.1006/abio.1994.1148. [DOI] [PubMed] [Google Scholar]
- Georgopoulos K. Transcription factors required for lymphoid lineage commitment. Curr Opin Immun. 1997;9:222–227. doi: 10.1016/s0952-7915(97)80139-2. [DOI] [PubMed] [Google Scholar]
- Georgopoulos K, Moore DD, Derfler B. Ikaros, an early lymphoid-specific transcription factor and a putative mediator for T cell commitment. Science. 1992;258:808–812. doi: 10.1126/science.1439790. [DOI] [PubMed] [Google Scholar]
- Georgopoulos K, Bigby M, Wang J-H, Molnar A, Wu P, Winandy S, Sharpe A. The Ikaros gene is required for the development of all lymphoid lineages. Cell. 1994;79:143–156. doi: 10.1016/0092-8674(94)90407-3. [DOI] [PubMed] [Google Scholar]
- Georgopoulos K, Winandy S, Avitahl N. The role of the Ikaros gene in lymphocyte development and homeostasis. Annu Rev Immunol. 1997;15:155–176. doi: 10.1146/annurev.immunol.15.1.155. [DOI] [PubMed] [Google Scholar]
- Groves T, Katis P, Madden Z, Manickam K, Ramsden D, Wu G, Guidos CJ. In vitro maturation of clonal CD4+CD8+ cell lines in response to TCR engagement. J Immunol. 1995;154:5011–5022. [PubMed] [Google Scholar]
- Haag FA, Kuhlenbaumer G, Koch-Nolte F, Wingender E, Thiele HG. Structure of the gene encoding the rat T cell ecto-ADP-ribosyltransferase RT6. J Immunol. 1996;157:2022–2030. [PubMed] [Google Scholar]
- Hagman J, Grosschedl R. Regulation of gene expression at early stages of B-cell differentiation. Curr Opin Immunol. 1994;6:222–230. doi: 10.1016/0952-7915(94)90095-7. [DOI] [PubMed] [Google Scholar]
- Hahm K, Ernst P, Lo K, Kim G, Turck C, Smale ST. The lymphoid transcription factor LyF-1 is encoded by a specific, alternatively spliced mRNA derived from the Ikaros gene. Mol Cell Biol. 1994;14:7111–7123. doi: 10.1128/mcb.14.11.7111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow E, Lane D. Antibodies: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- Ikuda K, Uchida N, Friedman J, Weissman IL. Lymphocyte development from stem cells. Annu Rev Immunol. 1992;10:759–783. doi: 10.1146/annurev.iy.10.040192.003551. [DOI] [PubMed] [Google Scholar]
- Klug CA, Morrision SJ, Masek M, Hahm K, Smale ST, Weissman IL. Hematopoietic stem cells and lymphoid progenitors express different Ikaros isoforms and Ikaros is localized to heterochromatin in immature lymphocytes. Proc Natl Acad Sci. 1998;95:657–662. doi: 10.1073/pnas.95.2.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo K, Landau NR, Smale ST. LyF-1, a transcriptional regulator that interacts with a novel class of promoters for lymphocyte-specific genes. Mol Cell Biol. 1991;11:5229–5243. doi: 10.1128/mcb.11.10.5229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar A, Georgopoulos K. The Ikaros gene encodes a family of functionally diverse zinc finger DNA-binding proteins. Mol Cell Biol. 1994;14:8292–8303. doi: 10.1128/mcb.14.12.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar A, Wu P, Largespada DA, Vortkamp A, Scherer S, Copeland NG, Jenkins NA, Bruns G, Georgopoulos K. The Ikaros gene encodes a family of lymphocyte-restricted zinc finger DNA binding proteins, highly conserved in human and mouse. J Immunol. 1996;156:585–592. [PubMed] [Google Scholar]
- Morgan B, Sun L, Avitahl N, Andrikopoulos K, Ikeda T, Gonzales E, Wu P, Neben S, Georgopoulos K. Aiolos, a lymphoid restricted transcription factor that interacts with Ikaros to regulate lymphocyte differentiation. EMBO J. 1997;16:2004–2013. doi: 10.1093/emboj/16.8.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison SJ, Uchida N, Weissman IL. The biology of hematopoietic stem cells. Annu Rev Cell Devel Biol. 1995;11:35–71. doi: 10.1146/annurev.cb.11.110195.000343. [DOI] [PubMed] [Google Scholar]
- Omori SA, Wall R. Multiple motifs regulate the B-cell-specific promoter of the B29 gene. Proc Natl Acad Sci. 1993;90:11723–11727. doi: 10.1073/pnas.90.24.11723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin SH. Hematopoiesis: How does it happen? Curr Opin Cell Biol. 1995;7:870–877. doi: 10.1016/0955-0674(95)80072-7. [DOI] [PubMed] [Google Scholar]
- Poux S, Kostic C, Pirrotta V. Hunchback-independent silencing of the late UBX enhancers by a polycomb group response element. EMBO J. 1996;15:4713–4722. [PMC free article] [PubMed] [Google Scholar]
- Santee SM, Owen-Schaub LB. Human tumor necrosis factor receptor p75/80 (CD120b) gene structure and promoter characterization. J Biol Chem. 1996;271:21151–21159. doi: 10.1074/jbc.271.35.21151. [DOI] [PubMed] [Google Scholar]
- Shortman K, Wu L. Early T lymphocyte progenitors. Annu Rev Immunol. 1996;14:29–47. doi: 10.1146/annurev.immunol.14.1.29. [DOI] [PubMed] [Google Scholar]
- Singh H. Gene targeting reveals a hierarchy of transcription factors regulating specification of lymphoid cell fates. Curr Opin Immunol. 1996;8:160–165. doi: 10.1016/s0952-7915(96)80053-7. [DOI] [PubMed] [Google Scholar]
- Sun L, Liu A, Georgopoulos K. Zinc finger-mediated protein interactions modulate Ikaros activity, a molecular control of lymphocyte development. EMBO J. 1996;15:5358–5369. [PMC free article] [PubMed] [Google Scholar]
- Ting CN, Olson MC, Barton KP, Leiden JM. Transcription factor GATA-3 is required for development of the T-cell lineage. Nature. 1996;384:474–478. doi: 10.1038/384474a0. [DOI] [PubMed] [Google Scholar]
- Tsai S, Bartelmez S, Sitnicka E, Collins S. Lymphohematopoietic progenitors immortalized by a retroviral vector harboring a dominant-negative retinoic acid receptor can recapitulate lymphoid, myeloid, and erythroid development. Genes & Dev. 1994;8:2831–2841. doi: 10.1101/gad.8.23.2831. [DOI] [PubMed] [Google Scholar]
- Wang J, Walker H, Lin Q, Jenkins N, Copeland NG, Watanabe T, Burrows PD, Cooper MD. The mouse BP-1 gene: Structure, chromosomal localization, and regulation of expression by type I interferons and interleukin-7. Genomics. 1996;33:167–176. doi: 10.1006/geno.1996.0180. [DOI] [PubMed] [Google Scholar]
- Wang J-H, Nichogiannopoulou A, Wu L, Sun L, Sharpe AH, Bigby M, Georgopoulos K. Selective defects in the development of the fetal and adult lymphoid system in mice with an Ikaros null mutation. Immunity. 1996;5:537–549. doi: 10.1016/s1074-7613(00)80269-1. [DOI] [PubMed] [Google Scholar]
- Wargnier A, Legros-Maida S, Bosselut R, Bourge JF, Lafaurie C, Ghysdael CJ, Sasportes M, Paul P. Identification of human granzyme B promoter regulatory elements interacting with activated T-cell-specific proteins: Implication of Ikaros and CBF binding sites in promoter activation. Proc Natl Acad Sci. 1995;92:6930–6934. doi: 10.1073/pnas.92.15.6930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willerford DM, Swat W, Alt FW. Developmental regulation of V(D)J recombination and lymphocyte differentiation. Curr Opin Genet Dev. 1996;6:603–609. doi: 10.1016/s0959-437x(96)80090-6. [DOI] [PubMed] [Google Scholar]
- Winandy S, Wu P, Georgopoulos K. A dominant mutation in the Ikaros gene leads to rapid development of leukemia and lymphoma. Cell. 1995;83:289–299. doi: 10.1016/0092-8674(95)90170-1. [DOI] [PubMed] [Google Scholar]
- Zhang CC, Bienz M. Segmental determination in Drosophila conferred by hunchback (hb) a repressor of the homeotic gen Ultrabithorax (Ubx) Proc Natl Acad Sci. 1992;89:7511–7515. doi: 10.1073/pnas.89.16.7511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zweidler-McKay PA, Grimes HL, Flubacher MM, Tsichlis PN. Gfi-1 encodes a nuclear zinc finger protein that binds DNA and functions as a transcription repressor. Mol Cell Biol. 1996;16:4024–4034. doi: 10.1128/mcb.16.8.4024. [DOI] [PMC free article] [PubMed] [Google Scholar]