Abstract
Findings
At first 32 housekeeping genes were analyzed in six randomly chosen meningiomas, brain and dura mater using geNorm, NormFinder, Bestkeeper-1 software and the comparative ΔCt method. Reference genes were ranked according to an integration tool for analyzing reference genes expression based on those four algorithms. Eight highest ranked reference genes (CASC3, EIF2B1, IPO8, MRPL19, PGK1, POP4, PPIA, and RPL37A) plus GAPDH and ACTB were then analyzed in 35 meningiomas, arachnoidea, dura mater and normal brain. NormFinder and Bestkeeper-1 identified RPL37A as the most stable expressed gene in meningiomas and their normal control tissue. NormFinder also determined the best combination of genes: RPL37A and EIF2B1. Commonly used reference genes GAPDH and ACTB were considered least stable genes. The critical influence of reference genes on qPCR data analysis is shown for VEGFA transcription patterns.
Background
In meningiomas quantitative real-time reverse transcription-polymerase chain reaction (qPCR) is most frequently used for accurate determination of gene expression using various reference genes. Although meningiomas are a heterogeneous group of tissue, no data have been reported to validate reference genes for meningiomas and their control tissues.
Conclusions
RPL37A is the optimal single reference gene for normalization of gene expression in meningiomas and their control tissues, although the use of the combination of RPL37A and EIF2B1 would provide more stable results.
Background
Meningiomas are the most frequent intracranial tumours. They originate from the arachnoidal cap cells of the meningeal coverings of the spinal cord and brain, constituting for approximatively 13 to 26% of all intracranial pathologies [1,2]. The conventional strategy for meningiomas is surgery [3,4]. However, some meningiomas recur as resection might be sub totally due to their delicate location at skull-based structures. The definition of malignant potential is beset by the frequent discordance between histology and biology [5,6]. Meningiomas are categorized in three WHO grades, in which there are several subtypes differentiated by their histological features.
Real-time quantitative reverse transcription polymerase chain reaction (RT-qPCR) is a sensitive and reliable method for quantifying gene expression. Most frequently the relative quantification method is used, which requires the use of an internal control gene for normalization. Reference genes are mostly genes, which are involved in basic metabolism and maintenance of the cell. An ideal reference gene should be expressed at a constant level in all examined tissues and cells, and should not be influenced by experimental conditions. However several studies have shown, that genes used as reference gene display significantly different gene expression levels [7-9].
Established housekeeping genes in meningioma RT-qPCR experiments are genes such as glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and β-Actin (ACTB) [10-18] as well as ribosomal RNA (18S rRNA) and TATA binding box protein (TBP) [19-21]. As the application of these various housekeeping genes shows, there are no reports that candidate reference genes in meningiomas were validated. Due to the heterogeneity of meningioma tissue and the necessity to compare meningiomas and their control tissue reliably, the selection of an appropriate reference gene with stable gene expression throughout the various tissues is essential for further use of RT-qPCR in meningioma research.
In this study, we investigate the gene expression levels of 32 commonly used housekeeping genes in meningiomas and their control tissues arachnoidea, dura and normal brain. The RT-qPCR results were analyzed with four different algorithms, to select eight suitable reference genes. Those genes plus GAPDH and ACTB were compared in an increased number of meningiomas and control tissues. These RT-qPCR results were further analyzed with two different algorithms: NormFinder and Bestkeeper-1.
Methods
Tumour Specimens and Cell Culture
Meningioma surgical specimens as well as arachnoidea and dura mater were obtained from the Neurosurgical Department in accordance to regulations of the Ethic Committee of the University of Tuebingen. Primary cultures were obtained from tumour tissue samples within 30 minutes of surgical removal. Samples were first washed in phosphate-buffered saline (PBS), reduced and mashed through a filter and placed in Dulbecco's modified Eagle's medium (DMEM) with fetal bovine serum (FBS), 2 mmol/L L-glutamine and 0,1% 10 mg/ml Gentamicin (Invitrogen, Grand Island, NY). Cells were plated in 25-mm2 tissue culture flasks and incubated at 37°C in a humidified atmosphere of 5% CO2. Medium was changed every 3 to 4 days and cultures were split using 600 μl Accutase (PAA, Pasching; Austria). Viable cells were stored in liquid nitrogen in 90% medium/10% dimethyl sulfoxide.
RNA isolation and reverse transcription
Meningioma total ribonucleic acid (RNA) was isolated directly from primary cell cultures before splitting and RNA of Arachnoidea and dura was isolated from fresh tissue using PARIS® kit (Ambion, Inc., Austin, TX), according to the manufacturer's protocol. RNA was treated with DNA-free™ (Ambion, Inc., Austin, TX) to remove residual genomic DNA. The concentration of the isolated RNA and the 260/280 absorbance ratio was measured in triplicates with Eppendorf Biophotometer (Eppendorf, Hamburg, Germany). The integrity of RNA samples was confirmed by electrophoresis on a 2% Sybr Green agarose gel (Invitrogen Inc., Carlsbad, CA). The criterion to include RNA samples was 260/280 ~ 2 (1.9 to 2.2) and 28S/18S ratio ≥ 1.7. The probes were stored at - 80°C until use. For normal brain FirstChoice® Human Brain Reference RNA (Ambion, Inc., Austin, TX) was used, which pools RNA from different donors and several brain regions. RNA samples were DNase treated using DNAfree kit (Ambion Inc., Austin, TX). Total RNA (1 μg) was reverse-transcribed to cDNA using des High Capacity RNA-to-cDNA Kits (Applied Biosystems, Foster City, CA) in a total volume of 20 μl, according to the manufacturer's protocol.
Primer selection
For investigation to identify the most stable reference gene that could be used for normalization in RT-qPCR studies in meningiomas TaqMan® Express Plate Human Endogenous Control Plates (Applied Biosystems, Foster City, CA) were used, which contained 32 different genes plated in triplicates (Table 1). This collection of genes has been selected from literature searches and/or whole genome microarray tests carried out on numerous human tissues. They have been shown to be expressed constitutively and at moderate abundance across most test samples. All primers further evaluated spanned an exon junction to minimize inaccuracies due to genomic DNA contamination in RNA samples except the primer for S18. Additional information on assay optimisation and validation such as primer sequence for each TaqMan® Assay are available from Applied Biosystems. Priming conditions, primer concentration and annealing temperature was identical in all used TaqMan® Gene Expression Assays. TaqMan®-based detection was chosen, because this detection method detects only specific amplification products, whereas SYBR®-Green based detection detects all amplified double-stranded DNA, including nonspecific double-stranded DNA sequences, which may generate false positive signals. Melt curves were not assessed because they are only suitable for SYBR®-Green based detection.
Table 1.
Candidate reference genes evaluated in this study
| Gene Symbol | Gene Name | Genbank Acession No. | TaqMan®Assay ID | Amplicon length |
|---|---|---|---|---|
| 18S | Eukaryotic 18S rRNA | X03205.1 (mRNA) | Hs99999901_s1 | 187 |
| ABL1 | v-abl Abelson murine leukemia oncogene homolog 1 | NM_005157.3 + NM_007313.2 | Hs00245445_m1 | 91 |
| ACTB | Actin, Beta, cytoplasmic | NM_001101.3 | Hs99999903_m1 | 171 |
| B2M | Beta-2-microglobulin | NM_004048.2 | Hs99999907_m1 | 75 |
| CASC3 | cancer susceptibility candidate 3 | NM_007359.4 | Hs00201226_m1 | 67 |
| CDKN1A | cyclin-dependent kinase inhibitor 1A (p21, Cip1) | NM_078467.1 + NM_000389.3 | Hs00355782_m1 | 66 |
| CDKN1B | cyclin-dependent kinase inhibitor 1B (p27, Kip1) | NM_004064.3 | Hs00153277_m1 | 71 |
| EIF2B1 | eukaryotic translation initiation factor 2B, subunit 1 alpha, 26 kDa | NM_001414.3 | Hs00426752_m1 | 75 |
| ELF1 | E74-like factor 1/ets domain transcription factor) | NM_172373.3 + NM_001145353.1 | Hs00152844_m1 | 76 |
| GADD45A | growth arrest and DNA-damage-inducible, alpha | NM_001924.2 | Hs00169255_m1 | 123 |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | NM_002046.3 | Hs99999905_m1 | 122 |
| GUSB | Glucuronidase, Beta | NM_000181.3 | Hs99999908_m1 | 81 |
| HMBS | Hydromethylbilane synthase | NM_000190.3 | Hs00609297_m1 | 64 |
| HPRT1 | Hypoxanthine guanine phospho- ribosyl transferase 1 | NM_000194.2 | Hs99999909_m1 | 100 |
| IPO8 | Improtin 8 | NM_006390.2 | Hs00183553_m1 | 71 |
| MRPL19 | mitochondrial ribosomal protein L19 | NM_014763.3 | Hs00608519_m1 | 72 |
| MT-ATP6 | mitochondrially encoded ATP synthase 6 | NC_001807.ATP6.0 | Hs02596862_g1 | 150 |
| PES1 | pescadillo homolog 1, containing BRCT domain (zebrafish) | NM_014303.2 | Hs00362795_g1 | 56 |
| PGK1 | Phosphoglycerate kinase 1 | NM_000291.3 | Hs99999906_m1 | 75 |
| POLR2A | Polymerase (RNA) II (DNA directed) polypeptide A, 220 kDa | NM_000937.3 | Hs00172181_m1 | 61 |
| POP4 | processing of precursor 4, ribonuclease P/MRP subunit | NM_006627.2 | Hs00198357_m1 | 68 |
| PPIA | Peptidylprolyl Isomerase A | NM_021130.3 | Hs99999904_m1 | 98 |
| PSMC4 | proteasome (prosome, macropain) 26S subunit, ATPase, 4 | NM_153001.1 + NM_006503.2 | Hs00197826_m1 | 83 |
| PUM1 | pumilio homolog 1 (Drosophila) | NM_001020658.1 + NM_014676.2 | Hs00206469_m1 | 89 |
| RPL30 | ribosomal protein L30 | NM_000989.2 | Hs00265497_m1 | 149 |
| RPL37A | ribosomal protein L37A | NM_000998.4 | Hs01102345_m1 | 125 |
| RPLP0 | Ribosomal protein, large, P0 | NM_053275.3 + NM_001002.3 | Hs99999902_m1 | 105 |
| RPS17 | ribosomal protein S17 | NM_001021.3 | Hs00734303_g1 | 93 |
| TBP | TATA binding box protein | NM_008907 | Hs99999910_m1 | 127 |
| TFRC | Transferrin receptor | NM_001128148.1 + NM_003234.2 | Hs99999911_m1 | 105 |
| UBC | Ubiquitin C | NM_021009.4 | Hs00824723_m1 | 71 |
| YWHAZ | Tyrosine 3-monooxygenase | NM_003406.3 | Hs00237047_m1 | 70 |
Thickly printed reference genes were further evaluated. TaqMan® Assay ID ending with "_m" indicates an assay whose probe spans an exon junction and will not detect genomic DNA. "_g" indicates an assay that may detect genomic DNA. The assay primers and probe may also be within a single exon. "_s" indicates an assay whose primers and probes are designed within a single exon, such assays will, by definition detect genomic DNA. Additional information for each TaqMan® Assay is available from Applied Biosystems.
For further evaluation single TaqMan® Gene Expression Assays for ACTB, CASC3, EIF2B1, GAPDH, IPO8, MRPL19, PGK1, POP4, PPIA, RPL37A (Applied Biosystems, Foster City, CA) were used, which were identical with the assays used in TaqMan® Express Plate Human Endogenous Control Plates.
Real-time PCR
TaqMan® real-time PCR was run in triplicates in 48-well reaction plates with a StepOne™ (Applied Biosystems, Foster City, CA). Real-time PCR reaction was performed with 1 μl cDNA (5 ng/μl) in 20 μl reaction mix containing 10 μl TaqMan® Gene Expression Master Mix (Applied Biosystems, Foster City, CA) and 1 μl TaqMan® Gene Expression Assays (Applied Biosystems, Foster City, CA). The cycling conditions were as follows: initial holding period at 95°C for 10 min, followed by a two-step PCR program consisting of 95°C for 15 s and 60°C for 1 min for 40 cycles. Reverse transcriptase negative controls and "no template controls" (without cDNA in PCR) were included. Data were collected and quantitatively analyzed using StepOne™ Software v2.1. Relative quantitation analysis of gene expression data for VEGFA analysis was conducted according to the 2-ΔΔCt method [22].
For PCR efficiency a 5-fold dilution series was created from a random pool of cDNA from our sample group ranging from 50 ng to 0.08 ng. PCR were performed as described above in triplicate. The PCR efficiency and correlation coefficients (R2) of each TaqMan® Gene Expression Assay were generated using the slops of the standard curves. The efficiencies were calculated by the formula: efficiency (%) = (10(-1/slope) -1) * 100. All assays displayed efficiencies between 93.2% and 100.2% (Table 2).
Table 2.
Efficiency data for evaluated genes
| Gene symbol | Slope | R2 | Efficiency (100%) |
|---|---|---|---|
| ACTB | - 3.420 | 1.000 | 96.1 |
| CASC3 | - 3.442 | 0.999 | 95.2 |
| EIF2B1 | - 3.434 | 0.997 | 95.5 |
| GAPDH | - 3.430 | 1.000 | 95.7 |
| IPO8 | - 3.390 | 1.00 | 97.3 |
| MRPL19 | - 3.364 | 0.999 | 98.3 |
| PGK1 | - 3.266 | 0.994 | 100.2 |
| POP4 | - 3.410 | 0.999 | 96.4 |
| PPIA | - 3.497 | 1.00 | 93.2 |
| RPL37A | - 3.406 | 1.000 | 96.6 |
Statistical analysis
To compare the stability of candidate reference genes, four validation software programs were used according to their original publication: geNorm http://medgen.ugent.be/~jvdesomp/genorm[23], NormFinder http://www.mdl.dk/publicationsnormfinder.htm[24], BestKeeper-1 http://www.gene-quantification.de/bestkeeper.html[25] and the comparative delta Ct method [26]. For geNorm and NormFinder the raw Ct values were transformed to quantities by using the delta Ct method [27]. The highest relative quantities for each gene were set to 1. Bestkeeper-1 and the comparative delta Ct method use raw Ct values. To evaluate the results from the four algorithms an integration tool for analyzing reference genes expression was used http://www.leonxie.com/referencegene.php. First according to the reference genes ranking by every algorithm from the most stable gene to the least stable gene, a series of continuous integers starting from 1 as weight to each reference gene is assigned. The geomean of each gene weights across the four algorithms is calculated and then these reference genes are re-ranked. The gene with the less geomean is viewed as more stable reference gene. Input data is value data from Real-Time qRT-PCR. Statistical analysis was performed with GraphPad Prism V5.03 (GraphPad Software, La Jolla, USA). Normality was assessed according to D'Agostino-Pearson tests with alpha = 0.05. For evaluation of statistical equivalence a confidence-interval version of the Two One-Sided Tests (TOST) procedure of Schuirmann was used [28]. The groups are considered equivalent at a 5% significance level if their difference has a 90% confidence interval that lies entirely inside the upper and lower equivalence limits. Therefore we considered ± δ = ± 1.5 to be reasonable limits of equivalence.
Results
Expression levels of 32 reference genes in meningioma and normal tissue
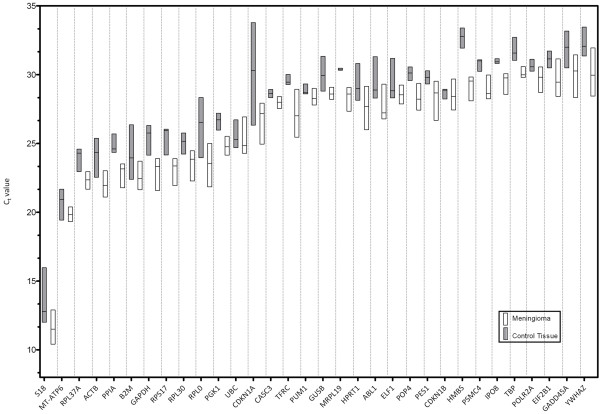
To select suitable reference genes TaqMan® human endogenous control plates (Applied Biosystems, Foster City, CA, USA) were used containing 32 known housekeeping genes (Table 1). Four different meningiomas, the malignant meningioma cell line IOMM-Lee, normal brain, cerebral meninges and dura mater were analyzed regarding the gene expression levels of those housekeeping genes. The mean Ct values displayed a wide range of expression levels between 10.41 and 33.78 as shown in Figure 1. The most abundant transcript was S18 with median Ct value of 11.50 in meningiomas and a mean Ct value of 13.59 in normal tissue. In meningioma the lowest expressed genes were YHWAZ with median Ct value of 30.27 and TBP with 30.08. In normal tissue HMBS had the lowest expression with a median Ct value of 32.70. Tumour tissue and normal tissue group of five candidate reference genes (CASC3, CDKN1B, POLR2A, PUM1 and UBC) were statistically equivalent to within ± 1.5.
Figure 1.
Expression levels of 32 candidate reference genes. Expression levels of 32 reference genes in four meningiomas, the malignant meningioma cell line IOMM-Lee, normal brain, cerebral meninges and dura mater. Real-Time PCR cycle threshold numbers are shown (Ct value). Box plots represent maximum and minimum values with median. Lower Ct values indicate higher gene expression.
Expression stability of 32 candidate reference genes in meningioma and brain
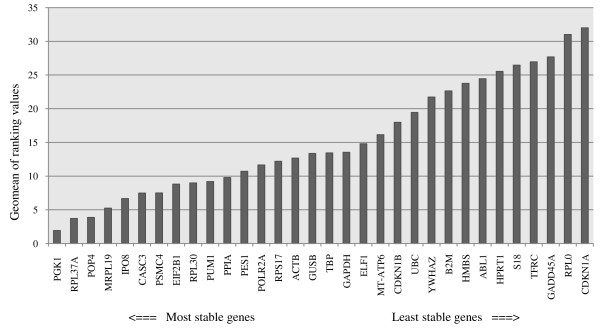
All 32 housekeeping genes were analyzed using four different algorithms, geNorm NormFinder, Bestkeeper-2 and the comparative delta Ct method. An integration tool calculated the geomean of each gene across the four algorithms and ranked the reference genes according to their comprehensive gene stability (Figure 2). The three most stable reference genes were PGK1 > RPL37A > POP4. The least stable reference genes were CDKN1A > RPL0 > GADD45A. Three of four used algorithms ranked PGK1 highest, only Bestkeeper-1 ranked CASC3 highest and PGK1 only in fourteenth place. For further analysis eight of the highest ranked expression genes were chosen: PGK1, RPL37A, POP4, MRPL19, IPO8 and CASC3. Additionally the most used reference genes in meningioma qPCR experiments ACTB and GAPDH were also chosen although being only ranked in fifteenth respectively eighteens place and being considered inconsistent with a standard deviation (SD) higher than 1 by Bestkeeper-1. Three reference genes (CDKN1B, UBC and POLR2A) with equivalent tumour and normal tissue group were ranked low from position 19 to 22 respectively 13. CASC3 was the only one included for further investigation with statistically equal groups.
Figure 2.
Comprehensive gene stability of 32 candidate reference genes in meningiomas and control tissue. Geomean of ranking values (y-axis) of 32 reference genes with their ranking from least to most stable gene expression (x-axis). Lower geomean values indicate more stable gene expression.
Expression stability of eight reference genes plus GAPDH and ACTB in meningiomas, arachnoidea, dura and normal brain
To validate the expression stability of CASC3, EIF2B1, IPO8, MRPL19, PGK1, POP4, PPIA, RPL37A plus GAPDH and ACTB, thirty-four randomly chosen primary cultured meningiomas, the meningioma cell line IOMM-Lee, two arachnoidea, six dura mater, one cerebral meninges and two pooled normal brain samples were screened for these reference genes. For analysis two different algorithms were chosen: NormFinder and Bestkeeper-1. NormFinder has a model-based approach whereas Bestkeeper-1 employs a pair-wise correlation analysis. NormFinder also estimates the variation between subgroups such as normal and cancer tissue.
Both algorithms identified RPL37A as the most stable gene in meningiomas and normal control tissue with an average expression stability value (M) value of 0.54 (Bestkeeper-1) respectively 0.12 (NormFinder). NormFinder not only determines the most stable gene, but also the best combination of two genes, which are RPL73A and EIF2B1 with a stability value of 0.088. The remaining ranking differed significantly for Bestkeeper-1 and NormFinder (Table 3). Bestkeeper-1 considered ACTB inconsistent with SD = 1.00 in meningiomas and their control tissue, whereas PGK1 was inconsistent in meningiomas (SD = 1.04). Both algorithms determined EIF2B1 and RPL37A as the two most stable genes in normal tissue (Table 4). In contrast there were significant differences between the ranking of Bestkeeper-1 and NormFinder in meningiomas (Table 5). Bestkeeper-1 ranked CASC3 and RPL37A highest. NormFinder identified MRPL19 and POP4 as the two most suitable genes, ranking CASC3 and RPL37A in eighth respectively ninth position.
Table 3.
Ranking of ten candidate reference genes in meningiomas and their control tissue based on average expression stability value as calculated by Bestkeeper-1 and NormFinder.
| Rank | Bestkeeper-1 | NormFinder | ||
|---|---|---|---|---|
| Gene name | Stability value | Gene name | Stability value | |
| 1 | RPL37A | 0.54 | RPL37A | 0.118 |
| 2 | CASC3 | 0.54 | EIF2B1 | 0.122 |
| 3 | MRPL19 | 0.65 | POP4 | 0.155 |
| 4 | IPO8 | 0.66 | MRPL19 | 0.163 |
| 5 | POP4 | 0.70 | PGK1 | 0.169 |
| 6 | PPIA | 0.73 | PPIA | 0.200 |
| 7 | EIF2B1 | 0.74 | GAPDH | 0.286 |
| 8 | PGK1 | 0.76 | ACTB | 0.287 |
| 9 | GAPDH | 0.83 | CASC3 | 0.289 |
| 10 | ACTB | 1.00 | IPO8 | 0.380 |
Table 4.
Ranking of ten candidate reference genes in normal control tissue based on average expression stability value as calculated by Bestkeeper-1 and NormFinder.
| Rank | Bestkeeper-1 | NormFinder | ||
|---|---|---|---|---|
| Gene name | Stability value | Gene name | Stability value | |
| 1 | EIF2B1 | 0.45 | EIF2B1 | 0.306 |
| 2 | RPL37A | 0.56 | RPL37A | 0.318 |
| 3 | PPIA | 0.65 | MRPL19 | 0.329 |
| 4 | MRPL19 | 0.69 | PPIA | 0.404 |
| 5 | ACTB | 0.75 | GAPDH | 0.415 |
| 6 | POP4 | 0.77 | POP4 | 0.419 |
| 7 | IPO8 | 0.82 | CASC3 | 0.462 |
| 8 | GAPDH | 0.82 | ACTB | 0.542 |
| 9 | CASC3 | 0.85 | PGK1 | 0.590 |
| 10 | PGK1 | 1.04 | IPO8 | 0.621 |
Table 5.
Ranking of ten candidate reference genes in meningiomas based on average expression stability value as calculated by Bestkeeper-1 and NormFinder.
| Rank | Bestkeeper-1 | NormFinder | ||
|---|---|---|---|---|
| Gene name | Stability value | Gene name | Stability value | |
| 1 | CASC3 | 0.43 | MRPL19 | 0.197 |
| 2 | RPL37A | 0.45 | POP4 | 0.269 |
| 3 | MRPL19 | 0.59 | IPO8 | 0.277 |
| 4 | IPO8 | 0.62 | PPIA | 0.312 |
| 5 | PPIA | 0.63 | PGK1 | 0.332 |
| 6 | POP4 | 0.63 | EIF2B1 | 0.355 |
| 7 | PGK1 | 0.67 | GAPDH | 0.386 |
| 8 | EIF2B1 | 0.70 | CASC3 | 0.388 |
| 9 | GAPDH | 0.75 | RPL37A | 0.447 |
| 10 | ACTB | 0.95 | ACTB | 0.521 |
TOST procedure showed statistical equivalence between normal tissue and meningiomas (± δ = ± 1.5) for three reference genes: CASC3 (+0.87), IPO8 (+0.57) and POP4 (+1.36). Those three genes were not normally distributed in meningiomas (CASC3 (P-value = 0.002), IPO8 (P-value < 0.0001) and POP4 (P-value = 0.0005). After inclusion of the normal tissue group IPO8 and POP4 remained not normally distributed.
Contribution of reference genes on expression levels of target genes
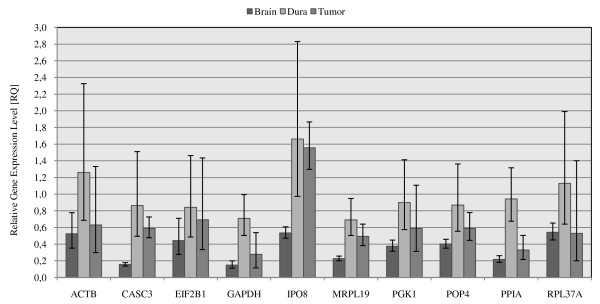
The selection of a reference gene for normalisation of qPCR can have a distinct influence on the expression profile of target genes [29]. To show the influence of different reference genes on the determination of gene expression levels, VEGFA expression levels in meningiomas and their control tissues were sequentially normalized with the analyzed ten reference genes. The Ct values for VEGFA were between 29 and 31. The expression level of VEGFA was normalized to each single reference gene as shown in Figure 3. The relative gene expression level (RQ) of VEGFA was calculated relative to the arachnoidea group (RQArachnoidea = 1). Subsequent bars represented the different expression levels of VEGFA in normal brain, dura and meningiomas normalized by different reference genes. Most reference genes maintained the ratio between brain, dura and meningioma except IPO8 and CASC3. Normalization with IPO8 or CASC3 showed significantly increased ratio for brain to dura and brain to tumour.
Figure 3.
Contribution of reference gene selection to determination of target gene expression levels. Data is expressed as relative gene expression levels (RQ) to arachnoidea (RQarachnoidea = 1). RQmin. and RQmax. are calculated with a confidence level of 95%.
Discussion
The requirement for distinct and reproducible results from quantitative gene expression analysis is accurate data normalization [23,24,29,30]. The application of an inappropriate reference gene can lead to false experimental conclusions [31-33]. Therefore one or more reference need to be chosen dependent on used tissue and experimental conditions.
To our knowledge, this is the first systematic analysis of average expression stability of reference genes in meningiomas for data normalisation in qPCR experiments. To evaluate the average expression stability four analysis software programs (geNorm, NormFinder, Bestkeeper-1 and the comparative delta Ct method) based on different algorithms were used. So far various reference genes (GAPDH, ACTB, S18, TBP) were used in qPCR experiments in meningiomas [11-21,34], although GAPDH was mainly used for normalizations. This study demonstrates that none of these reference genes were ranked under the ten most stable genes of 32 analyzed reference genes. However GAPDH and ACTB as the most used reference genes in meningioma qPCR experiments were further analyzed. After reducing the number of reference genes and increasing the number of samples both reference genes were considered one of the least stable genes. Bestkeeper-1 considered ACTB unsuitable as reference gene in meningiomas and their control tissues.
Because there is so few data available for gene expression of reference genes in meningiomas a large number of reference genes were screened. Using four randomly chosen meningiomas, the malignant meningioma cell line IOMM-Lee, pooled normal brain, cerebral meninges and dura mater was sufficient to determine expression levels of all reference genes as shown in Table 1. Because the four algorithms use different approaches for their rankings of the 32 reference genes, the ranking differed significantly making a selection of genes for further investigation difficult. Using the integration tool which weighs the ranking of each algorithm made the selection easier and more comprehensible. The six most stable reference genes according to the integration tool (PGK1, RPL37A, POP4, MRPL19, IPO8 and CASC3) were chosen for further analysis. Additionally PPIA and EIF2B1 were selected. PPIA was the highest ranked gene, which displayed high expression levels. EIF2B1 was the most stable gene with low expression levels. Because RPL30 is potentially co-regulated with RPL37A, it was not chosen, so the outcome of the result would not be affected.
For a more detailed analysis the remaining ten reference genes were analyzed using an increased number of samples (ntotal = 46 with nnormal = 11 and nmeningioma = 35) but a decreased number of software (NormFinder and Bestkeeper-1). NormFinder was chosen because of the model-based approach and the additional estimation of variation between normal and cancer tissue. In contrast Bestkeeper-1 employs a pair-wise correlation analysis and uses raw Ct values whereas NormFinder uses transformed quantities. Also Bestkeeper-1 directly includes qPCR efficiency.
Both algorithms considered RPL37A as the most suitable reference gene for normalization in qPCR in meningiomas and their control tissue. The following ranking differed significantly especially for CASC3, IPO8 and EIF2B1. Bestkeeper-1 considered CASC3 as the most stable genes in meningiomas, but ranked CASC3 only in ninth place for normal control tissue. This led to a second place in the combined ranking due to the higher number of tumour samples. In contrast NormFinder ranked EIF2B1 highest for normal control tissue and only in sixth place in meningiomas. Because NormFinder weighs the two subgroups, normal tissue versus meningiomas, the ranking of the control tissue has more influence on the combined ranking. This is also demonstrated with IPO8 and conversely with RPL37A. NormFinder ranks RPL37A in meningiomas only in ninth place and in normal control tissue in second place. But after including the variation between those subgroups NormFinder displays RPL37A as the most stable gene for both subgroups.
Considering the results of the normalization of VEGFA against every single reference genes with significantly altered results for CASC3 and IPO8, NormFinder displays a more accurate ranking for meningiomas and their control tissue.
Some researchers recommend the use of multiple reference genes for calculating a normalization factor [23]. NormFinder also determines the best combination of two genes, when subgroups are included. For meningiomas and their normal control tissue the combination is RPL37A and EIF2B1.
Conclusions
In conclusion, the results from the current study demonstrate that RPL37A is the most appropriate single reference gene for the normalization process of gene profiling studies in meningiomas and their normal control tissue arachnoidea, dura mater and normal brain. If a combination of reference genes is applicable RPL37A and EIF2B1 are most suitable. Additionally results from the current study indicate that widely used GAPDH and ACTB are both inappropriate reference genes for meningiomas.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
CP carried out cell cultivation and the Real-Time PCR studies, performed the statistical analysis and drafted the manuscript. MS supervised the study and reviewed the final version of the manuscript. FR conceived of the study, and participated in its design and coordination and helped to draft the manuscript. All authors read and approved the final manuscript.
Contributor Information
Christina Pfister, Email: christina.pfister@med.uni-tuebingen.de.
Marcos S Tatabiga, Email: marcos.tatagiba@med.uni-tuebingen.de.
Florian Roser, Email: f.roser@gmx.de.
Acknowledgements
We are grateful to Anita Lal (UCSF, USA) for kindly providing the IOMM-Lee cell line.
References
- Kleihues P, Burger PC, Scheithauer B. The new WHO classification of brain tumors. Brain Pathol. 1993;3:255–268. doi: 10.1111/j.1750-3639.1993.tb00752.x. [DOI] [PubMed] [Google Scholar]
- Riemenschneider MJ, Perry A, Reifenberger G. Histological classification and molecular genetics of meningiomas. Lancet Neurol. 2006;5(12):1045–1054. doi: 10.1016/S1474-4422(06)70625-1. [DOI] [PubMed] [Google Scholar]
- McMullen KP, Stieber VW. Meningioma: current treatment options and future directions. CurrTreatOptionsOncol. 2004;5(6):499–509. doi: 10.1007/s11864-004-0038-y. [DOI] [PubMed] [Google Scholar]
- Whittle IR, Smith C, Navoo P, Collie D. Meningiomas. Lancet. 2004;363(9420):1535–1543. doi: 10.1016/S0140-6736(04)16153-9. [DOI] [PubMed] [Google Scholar]
- Mahmood A, Caccamo DV, Tomecek FJ, Malik GM. Atypical and malignant meningiomas: a clinicopathological review. Neurosurgery. 1993;33(6):955–963. doi: 10.1227/00006123-199312000-00001. [DOI] [PubMed] [Google Scholar]
- Schittenhelm J, Mittelbronn M, Roser F, Tatagiba M, Mawrin C, Bornemann A. Patterns of SPARC expression and basement membrane intactness at the tumour-brain border of invasive meningiomas. Neuropathol Appl Neurobiol. 2006;32(5):525–531. doi: 10.1111/j.1365-2990.2006.00761.x. [DOI] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 2005;139(1):5–17. doi: 10.1104/pp.105.063743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD, Zakrajsek BA. Effect of experimental treatment on housekeeping gene expression: validation by real-time, quantitative RT-PCR. J Biochem Biophys Methods. 2000;46(1-2):69–81. doi: 10.1016/S0165-022X(00)00129-9. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Higgins PJ, Crawford DR. Control selection for RNA quantitation. Biotechniques. 2000;29(2):332–337. doi: 10.2144/00292rv02. [DOI] [PubMed] [Google Scholar]
- Buccoliero AM, Castiglione F, Degl'Innocenti DR, Arganini L, Taddei A, Ammannati F, Mennonna P, Taddei GL. Cyclooxygenase-2 (COX-2) overexpression in meningiomas: real time PCR and immunohistochemistry. Appl Immunohistochem Mol Morphol. 2007;15(2):187–192. doi: 10.1097/01.pai.0000201807.58801.fc. [DOI] [PubMed] [Google Scholar]
- Buccoliero AM, Castiglione F, Degl'Innocenti DR, Gheri CF, Garbini F, Taddei A, Ammannati F, Mennonna P, Taddei GL. NF2 gene expression in sporadic meningiomas: relation to grades or histotypes real time-pCR study. Neuropathology. 2007;27(1):36–42. doi: 10.1111/j.1440-1789.2006.00737.x. [DOI] [PubMed] [Google Scholar]
- Huang H, Held-Feindt J, Buhl R, Mehdorn HM, Mentlein R. Expression of VEGF and its receptors in different brain tumors. Neurol Res. 2005;27(4):371–377. doi: 10.1179/016164105X39833. [DOI] [PubMed] [Google Scholar]
- Miracco C, Cosci E, Oliveri G, Luzi P, Pacenti L, Monciatti I, Mannucci S, De Nisi MC, Toscano M, Malagnino V. et al. Protein and mRNA expression of autophagy gene Beclin 1 in human brain tumours. Int J Oncol. 2007;30(2):429–436. [PubMed] [Google Scholar]
- Paek SH, Kim DG, Park CK, Phi JH, Kim YY, Im SY, Kim JE, Park SH, Jung HW. The role of matrix metalloproteinases and tissue inhibitors of matrix metalloproteinase in microcystic meningiomas. Oncol Rep. 2006;16(1):49–56. [PubMed] [Google Scholar]
- Puri S, Joshi BH, Sarkar C, Mahapatra AK, Hussain E, Sinha S. Expression and structure of interleukin 4 receptors in primary meningeal tumors. Cancer. 2005;103(10):2132–2142. doi: 10.1002/cncr.21008. [DOI] [PubMed] [Google Scholar]
- Rollison DE, Utaipat U, Ryschkewitsch C, Hou J, Goldthwaite P, Daniel R, Helzlsouer KJ, Burger PC, Shah KV, Major EO. Investigation of human brain tumors for the presence of polyomavirus genome sequences by two independent laboratories. Int J Cancer. 2005;113(5):769–774. doi: 10.1002/ijc.20641. [DOI] [PubMed] [Google Scholar]
- Uesaka T, Shono T, Suzuki SO, Nakamizo A, Niiro H, Mizoguchi M, Iwaki T, Sasaki T. Expression of VEGF and its receptor genes in intracranial schwannomas. J Neurooncol. 2007;83(3):259–266. doi: 10.1007/s11060-007-9336-0. [DOI] [PubMed] [Google Scholar]
- Yang Y, Shao N, Luo G, Li L, Nilsson-Ehle P, Xu N. Relationship between PTEN gene expression and differentiation of human glioma. Scand J Clin Lab Invest. 2006;66(6):469–475. doi: 10.1080/00365510600763285. [DOI] [PubMed] [Google Scholar]
- Andersson U, Guo D, Malmer B, Bergenheim AT, Brannstrom T, Hedman H, Henriksson R. Epidermal growth factor receptor family (EGFR, ErbB2-4) in gliomas and meningiomas. Acta Neuropathol. 2004;108(2):135–142. doi: 10.1007/s00401-004-0875-6. [DOI] [PubMed] [Google Scholar]
- Denizot Y, De Armas R, Durand K, Robert S, Moreau JJ, Caire F, Weinbreck N, Labrousse F. Analysis of several PLA2 mRNA in human meningiomas. Mediators Inflamm. 2009;2009:689430. doi: 10.1155/2009/689430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurendeau I, Ferrer M, Garrido D, D'Haene N, Ciavarelli P, Basso A, Vidaud M, Bieche I, Salmon I, Szijan I. Gene expression profiling of ErbB receptors and ligands in human meningiomas. Cancer Invest. 2009;27(6):691–698. doi: 10.1080/07357900802709175. [DOI] [PubMed] [Google Scholar]
- Erickson HS, Albert PS, Gillespie JW, Wallis BS, Rodriguez-Canales J, Linehan WM, Gonzalez S, Velasco A, Chuaqui RF, Emmert-Buck MR. Assessment of normalization strategies for quantitative RT-PCR using microdissected tissue samples. Lab Invest. 2007;87(9):951–962. doi: 10.1038/labinvest.3700659. [DOI] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3(7):RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen CL, Jensen JL, Orntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64(15):5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper--Excel-based tool using pair-wise correlations. Biotechnol Lett. 2004;26(6):509–515. doi: 10.1023/b:bile.0000019559.84305.47. [DOI] [PubMed] [Google Scholar]
- Nicholas Silver SB, Jiang Jie, Thein Swee Lay. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Molecular Biology. 2006;7:33. doi: 10.1186/1471-2199-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Schuirmann DJ. A comparison of the two one-sided tests procedure and the power approach for assessing the equivalence of average bioavailability. J Pharmacokinet Biopharm. 1987;15(6):657–680. doi: 10.1007/BF01068419. [DOI] [PubMed] [Google Scholar]
- Nolan T, Hands RE, Bustin SA. Quantification of mRNA using real-time RT-PCR. Nat Protoc. 2006;1(3):1559–1582. doi: 10.1038/nprot.2006.236. [DOI] [PubMed] [Google Scholar]
- Gutierrez L, Mauriat M, Pelloux J, Bellini C, Van Wuytswinkel O. Towards a systematic validation of references in real-time rt-PCR. Plant Cell. 2008;20(7):1734–1735. doi: 10.1105/tpc.108.059774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin SA, Nolan T. Pitfalls of quantitative real-time reverse-transcription polymerase chain reaction. J Biomol Tech. 2004;15(3):155–166. [PMC free article] [PubMed] [Google Scholar]
- Dheda K, Huggett JF, Chang JS, Kim LU, Bustin SA, Johnson MA, Rook GA, Zumla A. The implications of using an inappropriate reference gene for real-time reverse transcription PCR data normalization. Anal Biochem. 2005;344(1):141–143. doi: 10.1016/j.ab.2005.05.022. [DOI] [PubMed] [Google Scholar]
- Huggett J, Dheda K, Bustin S, Zumla A. Real-time RT-PCR normalisation; strategies and considerations. Genes Immun. 2005;6(4):279–284. doi: 10.1038/sj.gene.6364190. [DOI] [PubMed] [Google Scholar]
- Laurendeau I, Ferrer M, Garrido D, D'Haene N, Ciavarelli P, Basso A, Vidaud M, Bieche I, Salmon I, Szijan I. Gene expression profiling of the hedgehog signaling pathway in human meningiomas. Mol Med. 2010;16(7-8):262–270. doi: 10.2119/molmed.2010.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]