Abstract
Eukaryotic organisms from yeast to human contain a multiprotein complex that includes Rpd3 histone deacetylase and Sin3 corepressor. The Sin3–Rpd3 complex, when recruited to promoters by specific DNA-binding proteins, can direct transcriptional repression of specific classes of target genes. It has been proposed that the histone deacetylase activity of Rpd3 is important for repression, but direct evidence is lacking. Here, we describe four Rpd3 derivatives with mutations in evolutionarily invariant histidine residues in a putative deacetylation motif. These Rpd3 mutants lack detectable histone deacetylase activity in vitro, but interact normally with Sin3 in vivo. In yeast cells, these catalytically inactive mutants are defective for transcriptional repression. They retain some residual Rpd3 function in vivo, however, suggesting that repression by the Sin3–Rpd3 complex may not be attributable exclusively to its intrinsic histone deacetylase activity. Finally, we show that a human Rpd3 homolog can interact with yeast Sin3 and repress transcription when artificially recruited to a promoter. These results suggest that the histone deacetylase activity of Rpd3 is important, but perhaps not absolutely required, for transcriptional repression in vivo.
Keywords: Chromatin, transcriptional repression, histone deacetylation, gene regulation
In recent years, it has become increasingly apparent that histone acetylases and deacetylases play an important role in transcriptional regulation (Pazin and Kadonaga 1997; Pennisi 1997). Proteins that are components of the basic Pol II transcription machinery or are involved in transcriptional activation possess intrinsic histone acetylase activity. These proteins include Gcn5 (Brownell et al. 1996), P/CAF (Yang et al. 1996b), p300/CBP (Bannister and Kouzarides 1996; Ogryzko et al. 1996), and the TAF130/250 subunit of the TFIID complex (Mizzen et al. 1996). Conversely, proteins that play a role in transcriptional repression, such as Sin3, Mad, YY1, SMRT, and NCoR, are associated with the histone deacetylase Rpd3/HDAC1 (Yang et al. 1996a; Alland et al. 1997; Hassig et al. 1997; Heinzel et al. 1997; Kadosh and Struhl 1997; Laherty et al. 1997; Nagy et al. 1997).
The histone acetylases and deacetylases mentioned above have been defined by virtue of their enzymatic activities in vitro. These enzymes have been assayed on a variety of substrates including peptides corresponding to the amino-terminal histone tails, isolated histones, or nucleosomes. The enzymes display distinct specificities in terms of the individual lysine residues and particular histones that are affected, and they differ in their ability to act on nucleosomal or free histones. A critical and generally unresolved issue is whether these enzymes can acetylate or deacetylate nonhistone proteins, and whether histones are physiological substrates. However, yeast Rpd3 and Hda1 deacetylate histones under physiological conditions, because rpd3 or hda1 mutant strains show increased acetylation at lysines 5 and 12 of histone H4 (Rundlett et al. 1996).
In yeast, Sin3 corepressor and Rpd3 histone deacetylase negatively regulate target genes involved in diverse processes such as meiosis, cell-type specificity, potassium transport, phosphate metabolism, methionine biosynthesis, and phospholipid metabolism (Vidal and Gaber 1991; Vidal et al. 1991; McKenzie et al. 1993; Stillman et al. 1994; Jackson and Lopes 1996). Sin3 and Rpd3 are tightly associated in a complex that is distinct from TFIID and Pol II holoenzyme (Kadosh and Struhl 1997; Kasten et al. 1997). The Sin3–Rpd3 complex can be recruited to the promoters of target genes by Ume6 (Kadosh and Struhl 1997), a repressor that specifically binds an upstream repression sequence (URS1) present in a wide variety of yeast promoters (Strich et al. 1994). Artificial recruitment of either Sin3 (Wang and Stillman 1993) or Rpd3 (Kadosh and Struhl 1997) to a heterologous promoter by the LexA DNA-binding domain results in transcriptional repression. Interestingly, however, LexA–Sin3 repression requires Rpd3 whereas LexA–Rpd3 can direct at least some repression in the absence of Sin3 (Kadosh and Struhl 1997). This result implies that Rpd3 accounts for the majority of repression activity in the complex.
Several lines of evidence have suggested that targeted histone deacetylation is the mechanism for transcriptional repression in vivo. First, Ume6- and URS1-mediated repression in yeast is significantly reduced in an Rpd3 deletion strain (Kadosh and Struhl 1997). These experiments are limited, however, in that they show only that the Rpd3 protein, rather than the histone deacetylase activity per se, is necessary for repression. Indeed, these observations do not exclude the possibility that Rpd3 possesses some other function, distinct from histone deacetylase activity, that accounts for its ability to mediate transcriptional repression. Second, potent histone deacetylase inhibitors, such as trapoxin and trichostatin A, significantly reduce or abolish repression mediated by Mad and SMRT in mammalian cells (Hassig et al. 1997; Laherty et al. 1997; Nagy et al. 1997). Little is known about the specificity of these histone deacetylase inhibitors, however, and it is quite possible that these drugs may inhibit other cellular deacetylases in addition to HDAC1. Furthermore, trichostatin A affects a variety of cellular processes, including apoptosis, differentiation, and DNA synthesis (Almouchi et al. 1994; Kimura et al. 1994; Medina et al. 1997), and trapoxin is an anti-tumor agent (Itazaki et al. 1990). Thus, these experiments cannot determine whether the histone deacetylase activity of the Sin3–Rpd3/HDAC1 complex per se is required for transcriptional repression in vivo.
Here, we directly examine the relationship between histone deacetylase activity and transcriptional repression. Specifically, we have generated point mutations in Rpd3 that abolish histone deacetylase activity in vitro, and analyze their transcriptional repression properties in vivo. Our results indicate that Rpd3 histone deacetylase activity is important, but perhaps not absolutely required, for transcriptional repression.
Results
Identification of a putative deacetylation motif
Rpd3 homologs are found in humans, mice, frogs, flies, worms, and plants. In Saccharomyces cerevisiae, there are four Rpd3-like proteins in addition to Rpd3 itself. The most highly conserved region among all of these proteins is a 60- to 70-amino-acid stretch (Fig. 1A,B). Interestingly, this highly conserved region strongly resembles sequences in acetylpolyamine amidohydrolase (aphA) (Sakurada et al. 1996) and in acuC, a protein involved in acetate utilization (Fig. 1C; Grundy et al. 1993). Of particular relevance, aphA from Mycoplana ramosa is a deacetylase that functions on a variety of acetylated polyamine substrates (Sakurada et al. 1996). This motif also is found in a number of prokaryotic and eukaryotic proteins whose functions are presently unknown. Thus, we suggest that this highly conserved region encodes a putative deacetylation motif.
Figure 1.

Evolutionarily invariant histidine residues lie in a putative deacetylation motif. Comparison of the region between amino acids 139 and 200 in yeast Rpd3 (yRpd3) with sequences from the following proteins. (A) Rpd3-related proteins (GenBank accession nos. indicated): human HDAC1 (Taunton et al. 1996); HDAC2 (Yang et al. 1996a); HDAC3, U66914: mouse mHD1 (Bartl et al. 1997); mRpd3 (Yang et al. 1996): Xenopus laevis xRpd3, X78454: Drosophila melanogaster dRpd3 (De Rubertis et al. 1996): Arabidopsis thaliana aRpd3, AF014824: Caenorhabditis elegans Ce-HDA1, Z81486; Ce-HDA2, Z46676; Ce-HDA5, AF026202. (B) Rpd3-related proteins in S. cerevisiae (Hda1, Hos1, Hos2, Hos3) (Rundlett et al. 1996). (C) Acetate utilization protein (acuC) in Bacillus subtilis (Bs) (Grundy et al. 1993), C. elegans (Ce; U61954), and Staphylococcus xylosus (Sx) (Egeter and Bruckner 1996) and from acetylpolyamine amidohydrolase (aphA) in Methanococcus jannaschii (Mj; U67502), M. ramosa (Mr) (Sakurada et al. 1996) and Synechocystis sp. (Ss; D90900). A second region in the amino terminus of acuC (Ce), bearing similarity to yRpd3139–200, is not shown. Sequences were aligned by use of the PIMA program (Human Genome Center, Baylor College of Medicine, Houston, TX). With respect to yeast Rpd3, regions of similarity are shaded and regions of identity are boxed (with the SeqVu program, Garvan Institute of Medical Research, Sydney, Australia). The four highly conserved histidine residues (corresponding to H150, H151, H188, and H189 in yeast Rpd3) are indicated in bold.
Generation of Rpd3 mutants defective for histone deacetylase activity
To investigate the role of histone deacetylase activity in transcriptional repression by Rpd3, we wished to obtain catalytically inactive derivatives, preferably those affecting residues involved in the active site. Although there is virtually no information about the enzymological mechanism of histone deacetylases, we followed up the suggestion of Christopher Walsh (Harvard Medical School, Boston, MA) that histidine residues might be directly involved in the histone deacetylation reaction. Within the putative deacetylation motif, there are four histidine residues that are organized in two pairs separated by 37 residues. Thus, we generated three different alanine substitutions (H150A, H151A, H188A) as well as a double substitution (H150A; H151A).
These mutants, as well as wild-type Rpd3, were tagged with the FLAG epitope at the carboxyl terminus and expressed in an Rpd3 deletion strain. FLAG-tagged Rpd3 derivatives were immunoprecipitated from extracts of these strains and histone deacetylase activity was measured directly from the immunoprecipitates. As expected, wild-type Rpd3-F shows a significant level of histone deacetylase activity by use of, as substrate, an acetylated peptide corresponding to the amino terminus of histone H4 (Fig. 2A). In contrast, immunoprecipitates from extracts of strains expressing any of the four mutants, or non-FLAG-tagged Rpd3, show the same background level as observed when no protein is added to the assay. Thus, all four mutants appear to be completely defective for histone deacetylase activity. Because of inherent limitations of the assay, we cannot exclude the possibility that the mutants retain a small, residual amount of histone deacetylase activity (<3%). All of the mutants are expressed (Fig. 2B) and immunoprecipitated (data not shown) at levels equivalent to that of wild-type Rpd3-F.
Figure 2.
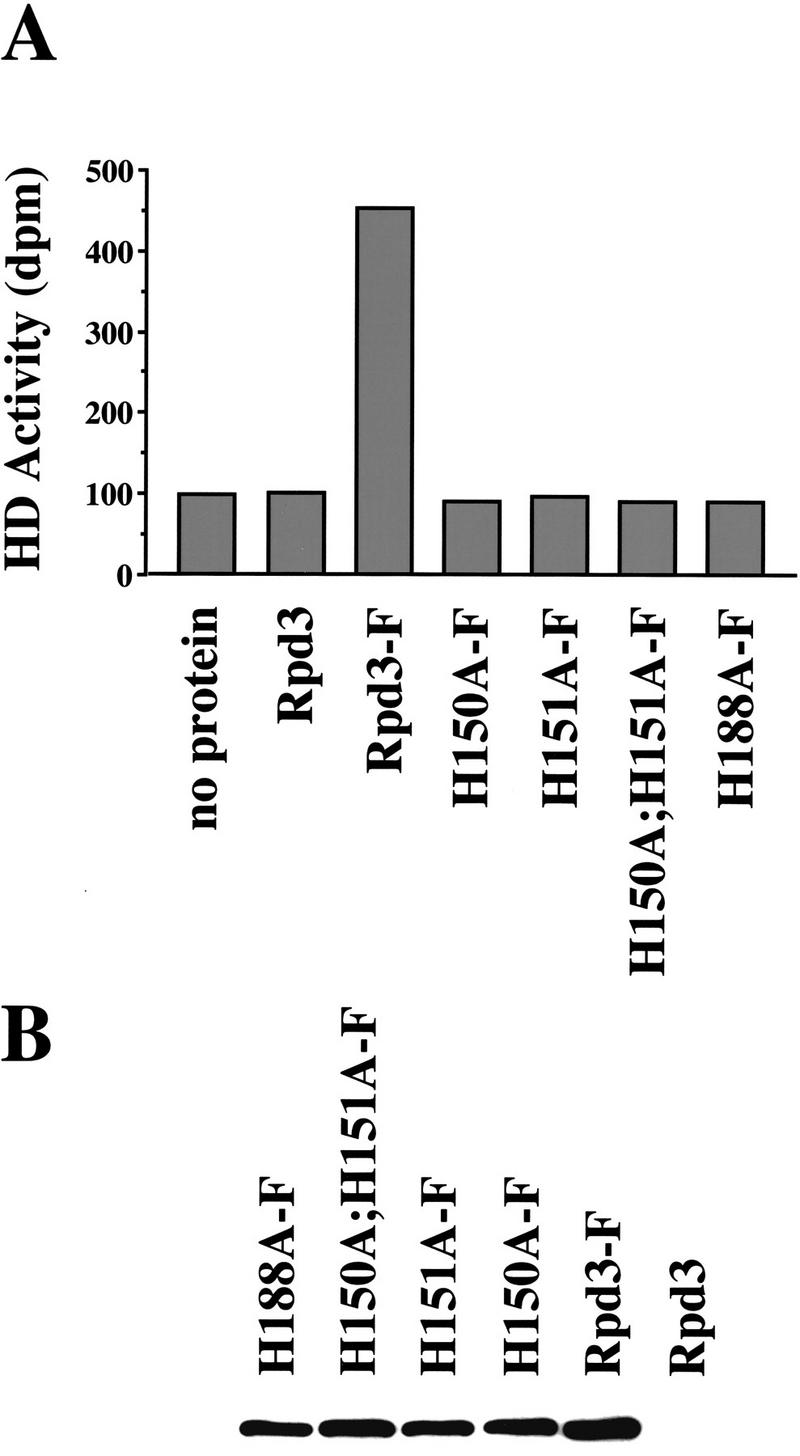
Rpd3 mutants are defective for histone deacetylase activity in vitro. (A) Whole-cell extracts were prepared from Rpd3 deletion strains expressing the indicated FLAG-tagged (F) Rpd3 derivatives. These derivatives were immunoprecipitated with anti-FLAG-conjugated agarose beads and histone deacetylase activity was assayed directly from the immunoprecipitates by use of an acetylated peptide corresponding to the amino-terminal tail of histone H4 as substrate. Control assays, with either no protein or extract from a Rpd3 deletion strain expressing non-FLAG-tagged Rpd3, are shown at left. Histone deacetylase (HD) activity is given as dpm of 3H-labeled acetic acid released. (B) Western analysis was performed by use of the protein extracts described in A. The immunoblot was probed with anti-FLAG antibodies.
Rpd3 catalytic mutants are defective for transcriptional repression
We first tested the ability of the histone deacetylase mutants to complement the rpd3 transcriptional defect in vivo. The Ume6 DNA-binding protein directs transcriptional repression activity in an Rpd3-dependent manner (Kadosh and Struhl 1997). Using the minimal repression domain of Ume6 fused to the LexA DNA-binding domain, we examined repression under various conditions (Fig. 3A). As reported previously, LexA–Ume6508–594 strongly represses transcription in a wild-type strain (11-fold), and this repression is nearly abolished (to 1.3-fold) in an Rpd3 deletion strain. When this strain is transformed with a plasmid expressing wild-type Rpd3, the transcriptional defect is fully complemented. In contrast, when any of the four histone deacetylase mutants are expressed in the Δrpd3 strain, repression by LexA–Ume6508–594 is severely reduced (1.2- to 2.9-fold depending on the derivative). Thus, these Rpd3 derivatives poorly complement the rpd3 phenotype, and the H188A mutant shows so little repression activity that it can not be distinguished from a null allele.
Figure 3.
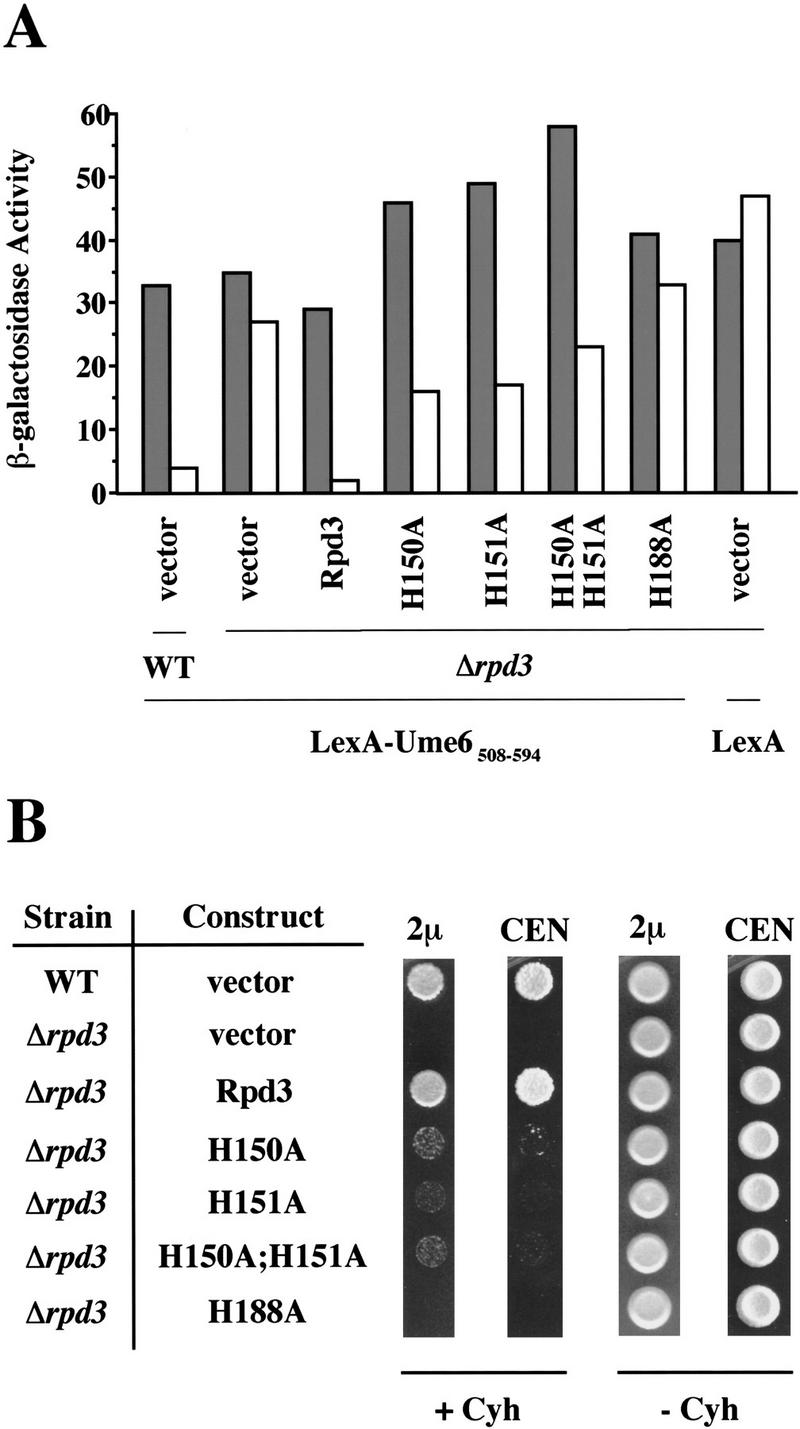
Rpd3 mutants poorly complement rpd3 transcriptional and functional defects in vivo. (A) β-Galactosidase activities of wild-type (WT) or rpd3 deletion strains expressing LexA–Ume6508–594, or LexA, and the indicated Rpd3 derivatives or vector only. Fold repression represents the ratio of β-galactosidase activities in strains containing plasmids that either lack (shaded bars) or contain (open bars) four LexA operators upstream of the CYC1 promoter. Values are normalized to A600 and represent the average of at least seven independent transformants; they are accurate to ±10%. (B) Cells (104) of wild-type (WT) or rpd3 deletion strains expressing the indicated Rpd3 derivatives (or vector only), on high-copy (2μ) or low copy (CEN) plasmids, were grown in the presence (+ Cyh) or absence (− Cyh) of 0.2 μg/ml of cycloheximide for 5 days at 30°C.
The mutants were also tested for their ability to complement the cycloheximide-hypersensitivity phenotype of an Rpd3 deletion strain (Fig. 3B). The H150A, H151A, and H150A;H151A mutants all poorly complement this phenotype, but clearly display a low level of Rpd3 function. Complementation by these mutant proteins is slightly improved when they are overexpressed on multicopy plasmids. H188A fails to complement the cycloheximide hypersensitivity defect, and the phenotype is indistinguishable from that of a strain totally lacking Rpd3. This pattern of growth phenotypes is very similar to that observed with the Ume6 repression assay (Fig. 3A), suggesting that the cycloheximide hypersensitivity phenotype is caused by a transcriptional repression defect.
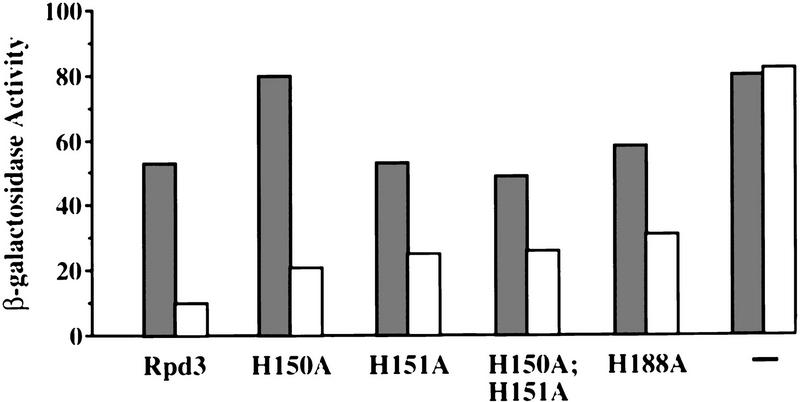
Finally, the Rpd3 mutants were tested for their ability to direct transcriptional repression when artificially recruited to a heterologous promoter by a DNA-binding domain (Fig. 4). As expected (Kadosh and Struhl 1997), a LexA–Rpd3 (wild-type) fusion protein represses transcription from a promoter containing LexA operators by a factor of 6 (Kadosh and Struhl 1997). When placed in the context of a LexA fusion protein, all four Rpd3 derivatives are partially defective for repression activity (two- to threefold repression). This result indicates that the mutants are defective for a step in the repression mechanism following promoter recruitment. It is important to note that the LexA–Rpd3 mutant derivatives should artificially recruit the Sin3–Rpd3 complex (see below), and that the Sin3–Rpd3 complex can partially repress transcription in the absence of Rpd3 (Kadosh and Struhl 1997). Thus, it is likely that the partial repression conferred by LexA–Rpd3 mutant derivatives is mediated primarily by another component(s) of the Sin3–Rpd3 complex rather than by Rpd3 itself.
Figure 4.
Rpd3 mutants are partially defective for repression function when artificially recruited to a heterologous promoter. β-Galactosidase activities of rpd3 deletion strains expressing the indicated Rpd3 derivatives fused to the LexA DNA-binding domain. (−) The LexA only control. Fold repression represents the ratio of β-galactosidase activities in strains containing plasmids that either lack (shaded bars) or contain (open bars) four LexA operators upstream of the CYC1 promoter. Values are normalized to A600 and represent the average of at least seven independent transformants; they are accurate to ±10%.
The H150A and H151A derivatives exhibit dominant-negative effects in vivo
Next, we sought to determine whether overexpression of the Rpd3 mutants could disrupt wild-type Rpd3 function in vivo. All of the mutants, as well as wild-type Rpd3, were expressed on a high-copy plasmid, under control of the ADH1 promoter, in a wild-type strain (Fig. 5A). Under these conditions, repression by LexA–Ume6508–594 is unaffected when wild-type Rpd3 is overexpressed. In contrast, repression is significantly disrupted when the H150A mutant is overexpressed and mildly reduced when H151A or H150A;H151A are overexpressed. No significant change in LexA–Ume6508–594 repression activity is observed upon overexpression of the H188A mutant.
Figure 5.
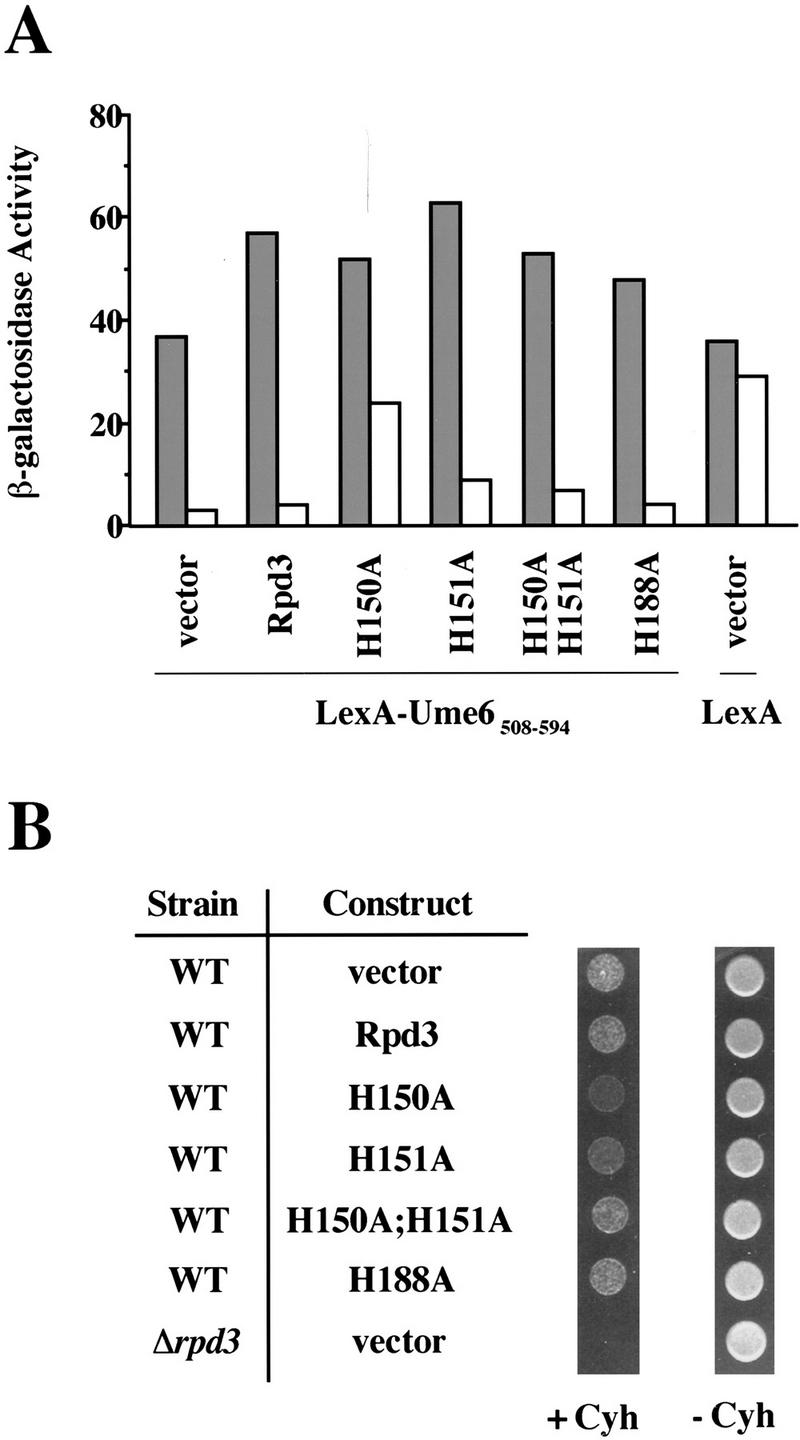
Dominant-negative phenotypes. (A) β-Galactosidase activities of wild-type strains expressing LexA–Ume6508–594, or LexA, and the indicated Rpd3 derivatives (or vector only) on high-copy plasmids, under control of the ADH1 promoter. Fold repression represents the ratio of β-galactosidase activities in strains containing plasmids that either lack (shaded bars) or contain (open bars) four LexA operators upstream of the CYC1 promoter. Values are normalized to A600 and represent the average of at least seven independent transformants; they are accurate to ±10%. (B) Cells (104) of wild-type (WT) or rpd3 deletion strains expressing the indicated Rpd3 derivatives on a high-copy plasmid under control of the ADH1 promoter, or vector only, were grown in the presence (+ Cyh) or absence (− Cyh) of 0.2 μg/ml of cycloheximide for 5 days at 30°C.
In a separate experiment, we determined whether overexpression of the Rpd3 mutants would confer a dominant-negative phenotype as assayed by cycloheximide hypersensitivity (Fig. 5B). A wild-type strain overexpressing the H150A mutant is clearly more sensitive to cycloheximide than a strain overexpressing wild-type Rpd3 (or a control strain containing the vector only). Overexpression of the H151A mutant confers mild hypersensitivity to cycloheximide, whereas overexpression of the H150A;H151A and H188A derivatives has no effect. Again, the LexA–Ume6508–594 repression properties of the various derivatives are in good accord with the cycloheximide-hypersensitivity phenotypes. By both assays, H150A and H151A mutants, respectively, show strong and modest dominant-negative effects when overexpressed in a wild-type strain.
Rpd3 catalytic mutants are not defective for interaction with Sin3
One possible explanation for the mutant phenotypes described above is that these residues are important for interaction with Sin3, and that Sin3 (or some other component of the complex) is responsible for the observed effects. To address this question, cell-free extracts were prepared from an Rpd3 deletion strain expressing an HA-epitope-tagged version of Sin3 in addition to either wild-type Rpd3 or each of the four mutants. Extract from a control strain, expressing nontagged Sin3 and wild-type Rpd3, was also prepared. By use of antibodies to the HA-1 epitope, Rpd3 specifically coimmunoprecipitates with the tagged Sin3 derivative (Fig. 6, cf. lanes 7 and 8), as previously reported (Kadosh and Struhl 1997). All of the Rpd3 mutants coimmunoprecipitate with Sin3–HA at levels comparable with that of wild-type Rpd3 (Fig. 6, cf. lanes 8–12). The physical association between Sin3–HA and the Rpd3 derivatives is strong, as the immunoprecipitates were washed under relatively stringent conditions for protein–protein interactions (500 mm potassium acetate, 1% NP-40). These results indicate that the Rpd3 histone deacetylase mutants are not defective for physically associating with Sin3.
Figure 6.
Rpd3 mutants are not defective for interaction with Sin3. Extracts from rpd3 deletion strains expressing Sin3 (S) (lane 1) or HA-tagged Sin3 (SH) (lan es 2–6) and the indicated Rpd3 derivatives were immunoprecipitated with antibodies to the HA-1 epitope (lanes 7–12). Immunoprecipitates were washed with 500 mm of potassium acetate, 1% NP-40, and the resulting samples were immunoblotted and probed with antibodies to Rpd3.
Artificial recruitment of HDAC1 results in transcriptional repression in yeast
The human Rpd3 homolog, HDAC1, possesses histone deacetyalse activity, and it can be recruited to target promoters by promoter-specific DNA-binding proteins (Taunton et al. 1996; Hassig et al. 1997; Laherty et al. 1997; Nagy et al. 1997). To determine whether HDAC1 carries out a similar function in yeast, we fused this protein to the LexA DNA-binding domain. As shown in Figure 7A, LexA–HDAC1, when artificially recruited to a heterologous promoter containing LexA operators, functions as a weak (two-fold) repressor in yeast, compared with LexA–Rpd3 (six-fold). LexA–HDAC1, like LexA–Rpd3, coimmunoprecipitates with the HA-tagged yeast Sin3 derivative (Fig. 7B, cf. lanes 1 and 2). A FLAG-tagged HDAC1 derivative (HDAC1-F), however, unlike Rpd3-F, fails to complement the rpd3 cycloheximide-hypersensitivity defect (data not shown). This lack of complementation might be explained by a defect in Sin3–Rpd3 complex formation and/or interaction with yeast DNA-binding proteins, but other explanations are possible. Nevertheless, the ability of Rpd3 to physically associate with Sin3 and to direct transcriptional repression when artificially recruited to a promoter, is functionally conserved from yeast to humans.
Figure 7.
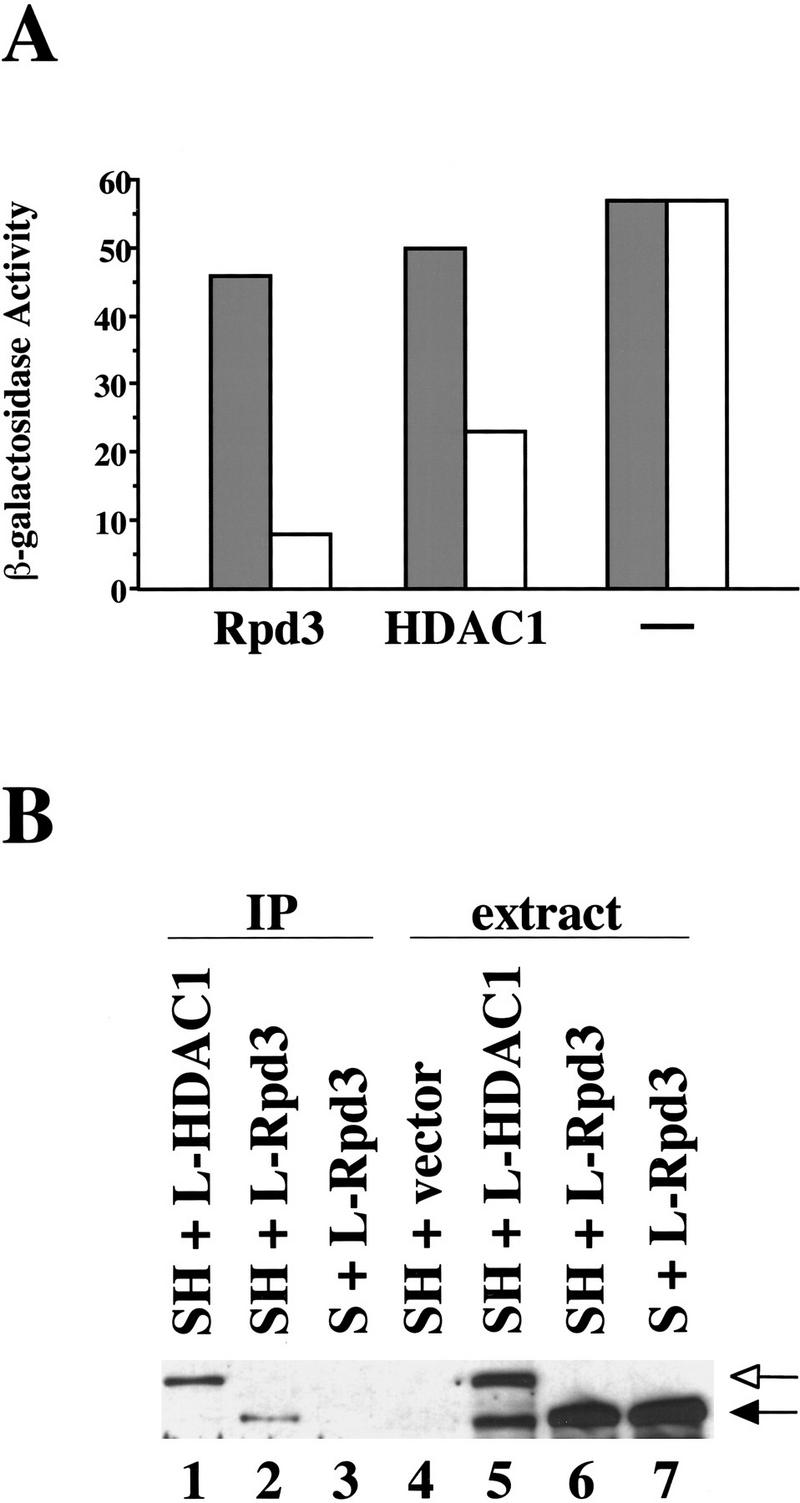
Artificial recruitment of HDAC1 to a heterologous promoter represses transcription in yeast. (A) β-Galactosidase activities of rpd3 deletion strains expressing either Rpd3 or HDAC1 fused to the LexA DNA-binding domain. (−) The LexA only control. Fold repression represents the ratio of β-galactosidase activities in strains containing plasmids that either lack (shaded bars) or contain (open bars) four LexA operators upstream of the CYC1 promoter. Values are normalized to A600 and represent the average of at least seven independent transformants; they are accurate to ±10%. (B) Extracts from rpd3 deletion strains expressing Sin3 (S, lane 7) or HA-tagged Sin3 (SH, lanes 4–6) and LexA–Rpd3 (L-Rpd3, lanes 6,7) or LexA–HDAC1 (L-HDAC1, lane 5) were immunoprecipitated with antibodies to the HA-1 epitope (lanes 1–3). Immunoprecipitates were washed with 125 mm of potassium acetate, 1% NP-40. The resulting samples were probed with antibodies to LexA. The positions of LexA–Rpd3 and LexA–HDAC1 are indicated by solid and open arrows, respectively. The lower band in lane 5 corresponds to a degradation product of LexA–HDAC1.
Discussion
Invariant histidine residues in a putative deacetylation motif are required for Rpd3 histone deacetylase activity
In this paper we have defined a putative deacetylation motif in Rpd3 by two criteria. First, this motif represents the most highly conserved region among Rpd3-related proteins in yeast and other eukaryotic species. Moreover, this motif is found in aphA, which can catalyze the deacetylation of a variety of acetylpolyamines, and in acuC, a protein that is important for acetate utilization. Second, three histidine residues located within the motif appear to be essential for histone deacetylase activity in vitro. We speculate that the motif plays a role in the deacetylation reaction per se, whereas other parts of the protein (which are not conserved between Rpd3- and aphA-related proteins) are involved in substrate specificity.
Histidines are found in the active sites of a variety of enzymes, and the presence of these histidines within a putative deacetylation motif prompts us to hypothesize that they may play a direct role in catalysis. For example, a proton from the imidizole ring of histidine might be transferred to the target lysine residue on the amino-terminal tail of histone H4, thus displacing the acetyl group. Alternatively, the histidines might be important for stabilizing an active site intermediate. A third possibility is that these residues play a role in coordinating the binding of a metal ion(s), such as zinc, that may be important for catalysis. In this regard, acetylpolyamine amidohydrolase contains one zinc atom per subunit (Sakurada et al. 1996). It is unclear whether the positioning of four conserved histidines as two pairs spaced 37 residues apart, has a bearing on their function. At least three of these histidine residues, however, do not play a redundant function with regard to histone deacetylase activity.
Histone deacetylase activity is important for transcriptional repression in vivo
Three independent lines of evidence indicate that histone deacetylase activity directly affects transcriptional repression in vivo. First, Rpd3 mutants defective for deacetylase activity poorly complement, or fail to complement, the defects in LexA–Ume6508–594 repression and cycloheximide hypersensitivity that are observed in rpd3 deletion strains. Second, LexA fusions to the Rpd3 mutants are partially defective for repression activity when recruited to a heterologous promoter. Third, overexpression of two mutants (H150A and H151A) causes a dominant-negative phenotype with respect to Ume6-mediated repression and cycloheximide hypersensitivity.
Although all four mutants appear to be completely defective for histone deacetylase activity in vitro, their transcriptional phenotypes vary to some extent. In particular, the H188A derivative fails to complement the cycloheximide hypersensitivity phenotype of an rpd3 null strain, whereas the other three mutants partially complement. One possible explanation is that H150A, H151A, and H150A;H151A retain a residual amount of histone deacetylase activity (undetectable by our assay) and that H188A completely lacks the activity. We disfavor this possibility because the H150A;H151A double mutant is likely to have considerably less enzymatic activity than the corresponding single mutants, yet there is not much phenotypic difference in vivo. Alternatively, the H188 residue may play an additional role, independent from its role in deacetylase activity, that is required for full repression function.
The Rpd3 derivatives also vary in their ability to act as dominant-negative mutants. Perhaps the H150A (and to a lesser extent the H151A) mutant exerts a dominant-negative effect because it acquires some structural feature that allows it to better compete with wild-type Rpd3 for integration in the Sin3–Rpd3 complex. Alternatively, H188A may be slightly defective for Sin3–Rpd3 complex formation, compared with H150A or wild-type Rpd3. Such a defect would not be detected except in situations where different Rpd3 derivatives are competing for a limited amount of Sin3 and associated proteins. Interestingly, the double mutant (H150A;H151A) shows a negligable dominant-negative phenotype, suggesting that whatever competitive advantage H150A confers, it is negated by the H151A mutation (i.e., H151A is epistatic to H150A with respect to this phenotype).
Evidence that histone deacetylase activity is not sufficient to account for the full repression function of the Sin3–Rpd3 complex
It is of interest that the Rpd3 catalytic mutants appear to be partially, rather than fully, defective for the transcriptional repression function of Rpd3 in vivo (Fig. 3). With the exception of H188A, all the Rpd3 derivatives show residual repression by LexA–Ume6508–594, and they confer cycloheximide–hypersensitivity phenotypes that are distinct from that of the rpd3 deletion strain. Such partial effects in vivo could be caused by incomplete inactivation of the catalytic activity, and in this view, the H188A derivative could be interpreted as having totally abolished histone deacetylase activity. Alternatively, Rpd3 could perform a distinct repression function that does not rely on its intrinsic histone deacetylase activity; such a secondary function could involve interactions with Sin3, other components of the Sin3–Rpd3 complex, or other Rpd3-related histone deacetylases. Even if such a secondary function exists, however, it would make a minor contribution to overall repression by Rpd3.
All four of the Rpd3 derivatives retain some ability to repress transcription when artificially recruited to a heterologous promoter (Fig. 4). One possibility is that such repression represents an artifactual function of Rpd3 that is only observed when it is artificially recruited to promoters in a manner independent of other components of the Sin3–Rpd3 complex. A more likely explanation is that tethering of Rpd3 via the LexA DNA-binding domain results in recruitment of the intact Sin3–Rpd3 complex, and that this complex performs an Rpd3-independent repression function. This hypothesis is supported by the findings that deletion of the HDAC1-interaction domain of mSin3 does not abolish mSin3-mediated repression in mammalian systems (Laherty et al. 1997), and that URS1- and Ume6-mediated repression in yeast is only partially reduced in an Rpd3 deletion strain (Kadosh and Struhl 1997). Thus, it is likely that the partial repression conferred by LexA–Rpd3 mutant derivatives is mediated primarily by another component(s) of the Sin3–Rpd3 complex rather than by Rpd3 itself.
In summary, our results demonstrate that the intrinsic histone deacetylase activity of Rpd3 is important for transcriptional repression in vivo. Our results provide further evidence, however, that histone deacetylation is not exclusively responsible for transcriptional repression by the Sin3–Rpd3 complex, and they suggest the possibility that Rpd3 has a secondary function that is distinct from its catalytic activity.
Materials and methods
Strains and DNAs
The FT5 (wild-type) and FT5 Δrpd3::HIS3 strains used in this work have been described previously (Tzamarias and Struhl 1994; Kadosh and Struhl 1997). For phenotypic assays, cells were grown on plates containing glucose-minimal medium with casamino acids in the presence or absence of cycloheximide (0.2μg/ml). Plasmids expressing the Rpd3 mutants were generated by substituting mutant PCR fragments spanning the region between the MluI and HindIII (for H150A, H151A, and H150A;H151A) or NcoI (for H188A) sites of Rpd3 for the corresponding region of YEplac112–Rpd3 (Kadosh and Struhl 1997). All PCR inserts were sequenced in their entirety to confirm that no other mutations were present. For some assays, it was necessary to place the wild-type and mutant versions of Rpd3 on a LEU2-marked plasmid. Rpd3 derivatives were cloned between the BamHI and SacI sites of YEplac181 and YCplac22 by three-piece ligation. LexA fusions of H150A, H150A;H151A, and H188A were constructed by digesting plasmids containing these mutants with NdeI and BglII, and substituting the relevant fragment for the corresponding region of YEplac112–Rpd3–LexA (Kadosh and Struhl 1997). The LexA fusion to H151A was generated by cloning a 1.1-kb BstBI–SacI fragment containing LexA1–202 and the CYC8 terminator from YEplac112–Rpd3–LexA into YEplac112–H150A. FLAG-tagged wild-type and mutant Rpd3 derivatives were constructed as follows: two oligonucleotides (GGACTAACAGGTACCGACTACAAGGACGACGATGACAAGTGAACGCGTGGTACCTTGATCTC and GAGATCAAGGTACCACGCGTTCACTTGTCATCGTCGTCCTTGTAGTCGGTACCGTTAGTCC) were annealed and digested with KpnI, and the resulting fragment was cloned into the KpnI site at the carboxyl terminus of Rpd3 in YEplac112–Rpd3 to generate YEplac112–Rpd3-F. A BstBI–SacI fragment containing the FLAG epitope was excised from YEplac112-Rpd3-F and cloned into each of the YEplac112 mutant constructs (to generate carboxy-terminally FLAG-tagged mutants). The HDAC1–LexA fusion protein was generated by ligating a BamHI–SalI fragment containing HDAC1 (Taunton et al. 1996) with a SalI–EcoRI PCR fragment containing LexA1–202 into a p424 vector (2μ, TRP1) (Mumberg et al. 1995) digested with BamHI and EcoRI. Plasmids expressing LexA–Ume6508–594 or LexA only have been described previously (Kadosh and Struhl 1997).
Histone deacetylase assays
Whole-cell protein extracts were prepared from rpd3 strains expressing FLAG-tagged Rpd3 derivatives as described previously (Kadosh and Struhl 1997). For each immunoprecipitation, 200 μg of extract was precleared by incubation for 1 hr at 4°C with 25 μl of agarose beads in buffer F (20 mm HEPES at pH 7.6, 1 mm EDTA, 150 mm NaCl, 20% glycerol). The extracts were incubated overnight at 4°C with 12.5 μl of FLAG-coupled agarose beads (IBI) in buffer F. The beads were then washed five times with 1 ml of buffer F and resuspended in 200 μl of histone deacetylase buffer (20 mm Tris at pH 7.65, 150 mm NaCl, 10% glycerol). As substrate for the histone deacetylase assays, a peptide corresponding to the first 24 amino acids of yeast histone H4 was synthesized and HPLC-purified. The peptide was chemically acetylated with 3H-labeled sodium acetate (2.5 Ci/mmole), in the presence of PyBOP reagent and triethylamine, and eluted from a sephadex LH-20 column with 10% MeOH. Two microliters of 3H-labeled peptide (∼34,000 DPM) was added to each 200-μl assay and the reaction was allowed to proceed at 30°C for 1 to 3 hr. Reactions were quenched by the addition of 50 μl of 1 m HCl/0.16 m acetic acid; released 3H-labeled acetic acid was extracted with 600 μl of ethyl acetate and quantified with a scintillation counter (Taunton et al. 1996).
Transcriptional repression assays
The lacZ reporter constructs used in LexA-dependent repression assays contain either four (JK1621) or no (pLGΔ312S) LexA operator sites upstream of the intact CYC1 promoter (Keleher et al. 1992). β-Galactosidase assays were performed as described previously (Kadosh and Struhl 1997). Values are normalized to A600 and represent the average of at least seven independent transformants; they are accurate to ±10%.
Immunoprecipitation experiments
The preparation of total cell protein extracts and immunoprecipitations were carried out essentially as described previously (Kadosh and Struhl 1997). For Sin3–Rpd3 interaction experiments, 300 μg of extract was used per immunoprecipitation, and the immunoprecipitates were washed in buffer containing 0.5 m KAc (high stringency). For the Sin3–HDAC1–LexA interaction experiment, 250 μg of extract was used, and the immunoprecipitates were washed in buffer containing 0.125 m KAc (moderate stringency). Immunoblots were probed with mouse antibody to FLAG (1:360) or with rabbit polyclonal antibodies to Rpd3 (1:2000) or LexA (1:2000) followed by secondary HRP-conjugated anti-mouse or anti-rabbit IgG.
Acknowledgments
We are especially grateful to Chris Walsh for discussions of potential enzymatic mechanisms for histone deacetylase and the suggestion that we mutate histidine residues. We thank Roger Brent, Michael Grunstein, and Chris Hassig for plasmids and antibodies, Charles Dahl for synthesis and purification of the yeast histone H4 peptide substrate for histone deacetylase assays, Chris Hassig for communicating results prior to publication, and Peter Belshaw and members of the Struhl lab for useful advice and criticism during the course of the experiments. This work was supported by research grant GM 53720 to K.S. from the National Institutes of Health.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL kevin@hms.harvard.edu;FAX (617)-432-2529.
References
- Alland L, Muhle R, Hou H, Potes J, Chin L, Schreiber-Agus N, DePinho RA. Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature. 1997;387:49–55. doi: 10.1038/387049a0. [DOI] [PubMed] [Google Scholar]
- Almouchi G, Khochbin S, Dimitrov S, Wolffe AP. Histone acetylation influences both gene expression and development of Xenopus laevis. Dev Biol. 1994;165:654–659. doi: 10.1006/dbio.1994.1283. [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- Bartl S, Taplick T, Lagger G, Khier H, Kuchler K, Seiser C. Identification of mouse histone deacetylase 1 as a growth factor-inducible gene. Mol Cell Biol. 1997;17:5033–5043. doi: 10.1128/mcb.17.9.5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownell JE, Zhou J, Ranalli T, Kobayashi R, Edmondson DG, Roth SY, Allis CD. Tetrahymena histone acetyltransferase A: A homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- De Rubertis F, Kadosh D, Henchoz S, Pauli D, Reuter G, Struhl K, Spierer P. The histone deacetylase RPD3 counteracts genomic silencing in Drosophila and yeast. Nature. 1996;384:589–591. doi: 10.1038/384589a0. [DOI] [PubMed] [Google Scholar]
- Egeter O, Bruckner R. Catabolite repression mediated by the catabolite control protein CcpA in Staphylococcus xylosus. Mol Microbiol. 1996;21:739–749. doi: 10.1046/j.1365-2958.1996.301398.x. [DOI] [PubMed] [Google Scholar]
- Grundy FJ, Waters DA, Takova TY, Henkin TM. Identification of genes involved in utilization of acetate and acetoin in Bacillus subtilis. Mol Microbiol. 1993;10:259–271. doi: 10.1111/j.1365-2958.1993.tb01952.x. [DOI] [PubMed] [Google Scholar]
- Hassig CA, Fleischer TC, Billin AN, Schreiber SL, Ayer DE. Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell. 1997;89:341–347. doi: 10.1016/s0092-8674(00)80214-7. [DOI] [PubMed] [Google Scholar]
- Heinzel T, Lavinsky RM, Mullen T-M, Soderstrom M, Laherty CD, Torchia J, Yang W-M, Brard G, Ngo SD, Davie JR, et al. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature. 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- Itazaki H, Nagashima K, Sugita K, Yoshida H, Kawamura Y, Yasuda Y, Matsumoto K, Ishii K, Uotani N, Nakai H, et al. Isolation and structural elucidation of new cyclotetrapeptides, trapoxins A and B, having detransformation activities as antitumor agents. J Antibiot. 1990;43:1524–1532. doi: 10.7164/antibiotics.43.1524. [DOI] [PubMed] [Google Scholar]
- Jackson JC, Lopes JM. The yeast UME6 gene is required for both negative and positive transcriptional regulation of phospholipid biosynthetic gene expression. Nucleic Acids Res. 1996;24:1322–1329. doi: 10.1093/nar/24.7.1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadosh D, Struhl K. Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell. 1997;89:365–371. doi: 10.1016/s0092-8674(00)80217-2. [DOI] [PubMed] [Google Scholar]
- Kasten MM, Dorland S, Stillman DJ. A large protein complex containing the yeast Sin3p and Rpd3p transcriptional regulators. Mol Cell Biol. 1997;17:4852–4858. doi: 10.1128/mcb.17.8.4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keleher CA, Redd MJ, Schultz J, Carlson M, Johnson AD. Ssn6-Tup1 is a general repressor of transcription in yeast. Cell. 1992;68:709–719. doi: 10.1016/0092-8674(92)90146-4. [DOI] [PubMed] [Google Scholar]
- Kimura M, Kimura I, Okabe M, Beppu T. Dual modes of action of platelet derived growth factor and its inhibition by trichostatin A for DNA synthesis in primary cultured smooth muscle cells of rat aorta. Biol Pharmacol Bull. 1994;17:399–402. doi: 10.1248/bpb.17.399. [DOI] [PubMed] [Google Scholar]
- Laherty C, Yang W-M, Sun J-M, Davie JR, Seto E, Eisenman RN. Histone deacetylases associated with the mSin3 corepressor mediate Mad:Max transcriptional repression. Cell. 1997;89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- McKenzie EA, Kent NA, Dowell SJ, Moreno F, Bird LE, Mellor J. The centromere and promoter factor 1, CPF1, of Saccharomyces cerevisiae modulates gene activity through a family of factors including SPT21, RPD1 (SIN3), RPD3 and CCR4. Mol & Gen Genet. 1993;240:374–386. doi: 10.1007/BF00280389. [DOI] [PubMed] [Google Scholar]
- Medina V, Edmonds B, Young GP, James R, Appleton S, Zalewski PD. Induction of caspase-3 protease activity and apoptosis by butyrate and trichostatin A (inhibitors of histone deacetylase): Dependence on protein synthesis and synergy with a mitochondrial/cytochrome c-dependent pathway. Cancer Res. 1997;57:3697–3707. [PubMed] [Google Scholar]
- Mizzen CA, Yang X-Y, Kokubo T, Brownell JE, Bannister AJ, Owen-Hughes T, Workman J, Wang L, Berger SL, Kouzarides T, et al. The TAFII250 subunit of TFIID has histone acetyltransferase activity. Cell. 1996;87:1261–1270. doi: 10.1016/s0092-8674(00)81821-8. [DOI] [PubMed] [Google Scholar]
- Mumberg D, Muller R, Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- Nagy L, Kao H-Y, Chakravarti D, Lin RJ, Hassig CA, Ayer DE, Schreiber SL, Evans RM. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- Pazin MJ, Kadonaga JT. What’s up and down with histone deacetylation and transcription? Cell. 1997;89:325–328. doi: 10.1016/s0092-8674(00)80211-1. [DOI] [PubMed] [Google Scholar]
- Pennisi E. Opening the way to gene activity. Science. 1997;275:155–157. doi: 10.1126/science.275.5297.155. [DOI] [PubMed] [Google Scholar]
- Rundlett SE, Carmen AA, Kobayashi R, Bavykin S, Turner BM, Grunstein M. HDA1 and RPD3 are members of distinct yeast histone deacetylase complexes. Proc Natl Acad Sci. 1996;93:14503–14508. doi: 10.1073/pnas.93.25.14503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurada K, Ohta T, Fujishiro K, Hasegawa M, Aisaka K. Acetylpolyamine amidohydrolase from Mycoplana ramosa: Gene cloning and characterization of the metal-substituted enzyme. J Bacteriol. 1996;178:5781–5786. doi: 10.1128/jb.178.19.5781-5786.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillman DJ, Dorland S, Yu Y. Epistasis analysis of suppressor mutations that allow HO expression in the absence of the yeast SWI5 transcriptional activator. Genetics. 1994;136:781–788. doi: 10.1093/genetics/136.3.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strich R, Surosky RT, Steber C, Dubois E, Messenguy F, Esposito RE. UME6 is a key regulator of nitrogen repression and meiotic development. Genes & Dev. 1994;8:796–810. doi: 10.1101/gad.8.7.796. [DOI] [PubMed] [Google Scholar]
- Taunton J, Hassig CA, Schreiber SL. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- Tzamarias D, Struhl K. Functional dissection of the yeast Cyc8-Tup1 transcriptional corepressor complex. Nature. 1994;369:758–761. doi: 10.1038/369758a0. [DOI] [PubMed] [Google Scholar]
- Vidal M, Gaber RF. RPD3 encodes a second factor required to achieve maximum positive and negative transcriptional states in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:6317–6327. doi: 10.1128/mcb.11.12.6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal M, Strich R, Esposito RE, Gaber RF. RPD1 (SIN3/UME4) is required for maximal activation and repression of diverse yeast genes. Mol Cell Biol. 1991;11:6306–6316. doi: 10.1128/mcb.11.12.6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Stillman DJ. Transcriptional repression in Saccharomyces cerevisiae by a SIN3-LexA fusion protein. Mol Cell Biol. 1993;13:1805–1814. doi: 10.1128/mcb.13.3.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W-M, Inouye C, Zeng Y, Bearss D, Seto E. Transcriptional repression by YY1 is mediated by interaction with the mammalian homolog of the yeast global regulator RPD3. Proc Natl Acad Sci. 1996a;93:12845–12850. doi: 10.1073/pnas.93.23.12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X-J, Ogryzko VV, Nishikawa J, Howard BH, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996b;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]