Abstract
Small RNA molecules (sRNAs) are key mediators of virulence and stress inducible gene expressions in some pathogens. In this work we identify sRNAs in the Gram positive opportunistic pathogen Enterococcus faecalis. We characterized 11 sRNAs by tiling microarray analysis, 5′ and 3′ RACE-PCR, and Northern blot analysis. Six sRNAs were specifically expressed at exponential phase, two sRNAs were observed at stationary phase, and three were detected during both phases. Searches of putative functions revealed that three of them (EFA0080_EFA0081 and EFB0062_EFB0063 on pTF1 and pTF2 plasmids, respectively, and EF0408_EF04092 located on the chromosome) are similar to antisense RNA involved in plasmid addiction modules. Moreover, EF1097_EF1098 shares strong homologies with tmRNA (bi-functional RNA acting as both a tRNA and an mRNA) and EF2205_EF2206 appears homologous to 4.5S RNA member of the Signal Recognition Particle (SRP) ribonucleoprotein complex. In addition, proteomic analysis of the ΔEF3314_EF3315 sRNA mutant suggests that it may be involved in the turnover of some abundant proteins. The expression patterns of these transcripts were evaluated by tiling array hybridizations performed with samples from cells grown under eleven different conditions some of which may be encountered during infection. Finally, distribution of these sRNAs among genome sequences of 54 E. faecalis strains was assessed. This is the first experimental genome-wide identification of sRNAs in E. faecalis and provides impetus to the understanding of gene regulation in this important human pathogen.
Introduction
Some RNA molecules such as riboswitches, transfer-messenger RNA (tmRNA) and small non-cording RNAs (sRNAs) act usually as post-transcriptional regulators in bacteria [1]. sRNAs have become increasingly recognized as an emerging class of gene expression regulators for cellular processes, stress response and virulence genes and their transcription is tightly regulated and induced by distinct environmental conditions [2]. Bacterial sRNAs found on chromosomes are typically 50–400 nucleotides in length and frequently encoded in intergenic regions (IGRs). They may bind to the imperfect complementary sequence of the ribosome binding region of the target mRNA, which is often encoded at separate loci, thus inhibiting 30S ribosomal subunit association and translational initiation [1], [3]. In some Gram positive and Gram negative species such as Escherichia coli [4] and Listeria monocytogenes [5], the formation of sRNA-mRNA duplex requires the RNA chaperon protein Hfq [6], [7] leading to an increase of mRNA degradation by ribonucleases such as RNase E and RNase III [2]. Some sRNAs located in plasmids and phages act as antisense RNAs on cis-encoded mRNAs and mainly control replication initiation, conjugation efficiency and transposition [8], [9]. In addition, plasmid-encoded sRNAs, called hak/sok system of E. coli plasmid R1 [10] and par system of Enterococcus faecalis pAD1 [11], stabilize their host plasmids by programming for death any cell that loses the plasmid [9], [12].
In recent years, several bioinformatic approaches have been performed to identify putative sRNAs in bacterial genomes including E. coli, L. monocytogenes, Bacillus subtilis and Pseudomonas aeruginosa, and identified more than 200 sRNAs [13]. Recently, Livny et al. predicted in silico over 45,000 sRNA candidates from 932 bacterial genomes [14]. In parallel, different experimental strategies including cDNA sequencing, shotgun cloning and isolation from RNA-protein complex have been performed and sometimes lead to the discovery of new transcripts [15], [16]. Tiling microarrays are powerful approaches to identify sRNAs on a genome-wide scale. Thus large numbers of sRNA candidates have been found in Caulobacter crescentus, Streptococcus pyogenes, S. pneumoniae, and L. monocytogenes genomes [17], [18], [19], [20].
E. faecalis is a human commensal Gram-positive bacteria as well as one of the leading causes of hospital acquired infections in United States and Europe [21]. The first whole genome sequence of E. faecalis V583 strain (the first vancomycin resistant enterococci identified in U.S.A.) was determined in 2003 and 53 more sequences are now publically available [22]. In silico study performed by Livny et al. led to the prediction and annotation of 17 putative sRNA-encoding loci in E. faecalis [14]. Surprisingly, in comparison with E. coli and B. subtilis, the number of predicted sRNAs in V583 is roughly 10-fold lower, suggesting that this number is likely under-estimated. Recently, 45 sRNAs and 10 putative mRNAs have been identified in E. faecalis using in silico prediction combined with “5′tag-RACE” [23].
In this work, we developed custom-made tiling microarrays containing only IGRs of E. faecalis V583 chromosome and plasmids, and first performed hybridization with RNA extracted from exponential and stationary-phase cells. Fifty-three statistically significant positive signals were detected and the 12 putative sRNAs most highly expressed were selected for further characterization. Transcription of these candidates under several stress conditions was then analyzed.
Materials and Methods
Bacterial strain and growth conditions
All experiments were performed with E. faecalis V583 strain [24]. For our first tiling array assays, cells were grown at 37°C in M17 0.5% glucose medium and collected at exponential phase (OD600 = 0.5) and at 24 h stationary phase. Growth in BHI medium with or without aeration was tested. Cells were collected at exponential phase (OD600 = 0.5), onset of starvation (OD600 = 2) and late stationary phase (24 h). For experiments under stress conditions, bacterial cells were grown to OD600 = 0.3 in M17 medium and H2O2 (2 mM), lactic acid (pH 5.5), or bile salts (BS) (0.08%), were added before an additional 30 min incubation at 37°C. For the growth in urine and serum, E. faecalis was inoculated into human urine or horse serum (Eurobio, Courtaboeuf, Fr) during overnight. Cells were then pelleted and resuspended into fresh urine or serum for 3 hours at 37°C. Urine collected from four healthy volunteers was pooled, centrifuged and sterilized by filtration (0.22 µm-pore sizes). Written consent from all participants involved in our study was obtained. French CPP (Comité de Protection de Personnes) exempted this study from review because volunteers were informed of the goal of this study, no health information was collected and no biological analysis was performed on these samples.
RNA extraction and tiling microarray hybridization
Total RNA was extracted using Trizol reagent (Invitrogen, Carlsbad, CA) as described by Toledo-Arana et al. [20], with the following modifications. Bacterial cells were resuspended into 200 µl of “max bacterial enhancement reagent” (Invitrogen) and transferred into micro tubes containing glass beads and 400 µl acid phenol (Ambion, Austin, TX). Bacteria were mechanically lysed using Mixer Mill 200 (30/s, 30 min, Retsch, Haan, Germany). After centrifugation for 10 min at 14,000 g at 4°C, aqueous phase was transferred to 2 ml tubes containing 1 ml Trizol reagent, mixed and incubated for 5 min at room temperature (RT). 200 µl chloroform was added, mixed gently and incubated for 3 min at RT. Tubes were centrifuged for 15 min at 12,000 g at 4°C and aqueous phase was transferred into 2 ml tubes containing 200 µl chloroform, mixed gently and centrifuged again. RNAs contained in the aqueous phase were precipitated by addition of 500 µl isopropanol and incubated for 10 min at RT. After centrifugation, RNA pellets were washed with 75% ethanol and dried at RT. Purified RNA pellets were resuspended in DEPC-treated pure water.
To enhance detection sensitivity by enriching of sRNAs and removing non-sRNA, 10 µg RNA were fractionated using flashPAGE Fractionator (Applied Biosystems, Foster City, CA). Fractionated RNA was labelled using mirVana labelling kit (Applied Biosystems) and then hybridized onto the tiling array. 1745 “big intergenic regions (IGR)” (more than 49 nt) and 1070 “small IGR” (from 1 to 49 nt) have been deduced from E. faecalis V583 genome sequence. 50 nt long probes with an overlap of 15 nt were loaded on our IGR custom-made tiling arrays. rRNA and tRNA probes were used as positive control showing signal intensity of hybridization at least 10 fold the threshold level. Since the values of intensity observed in apparent untranslated regions were between 1000 and 2000, 2000 was used as threshold. For each experiment (one sample per growth condition) two chips were used; one corresponding to the forward, and one to the reverse strand. Production, hybridization and data collecting were carried out by Febit biomed GmbH Company (Heidelberg, Germany). The detection was carried out using streptavidin phycoerythrin at different exposure times. Data analyses and visualization were performed by Genedata Phylosopher Business Group (Basel, Switzerland). We have deposed the raw data at GEO/ArrayExpress under accession number GSE28741, we can confirm all details are MIAME compliant.
5′ and 3′ rapid amplification of cDNA ends (RACE) analysis
For these analysis, new RNA samples were prepared as described above. 5′ RACE was performed using 2nd Generation 5′/3′ RACE kit (Roche, Mannheim, Germany) according to the manufacturer's instructions. For polymerase chain reactions (PCR), we used Go Taq polymerase and its buffer (Promega, Madison, WI). The primers used for cDNA synthesis, and for the PCR reactions are listed in Table S1.
For 3′ RACE experiments, total RNAs were treated with poly(A) polymerase (Epicentre, Madison, WI) for 15 min at 37°C. After 3′ end RNA poly(A) tailing, cDNA was synthesized with QuantiTect Reverse Transcription kit (Qiagen, West Sussex, UK) and oligo(dT)-anchor primer supplied in 5′/3′ RACE kit. cDNA products were directly used as templates for PCR performed with the gene-specific primers (Table S1) and the respective PCR anchor primer. After sequencing, 5′ and 3′ ends sequences were determined.
Northern blotting
Northern blots were performed according to standard procedures [25]. Five µg of total RNA were separated on 1.2% formaldehyde agarose gel and transferred to Hybond N+ membrane (Amersham, UK). 0.1–1 kb RNA Marker (Sigma, USA) was used to estimate the sizes of RNA bands. DNA oligonucleotides probes (Table S1) were labeled with α32P-ATP using Terminal Deoxnucleotidyl Transferase Recombinant enzyme (Promega) as recommended by the manufactured protocol. Membranes were prehybridized for 1 h in hybridization buffer (0.25 M NaH2PO4, 0.25 M Na2HPO4, 5% SDS) at 45°C, followed by addition of labelled probes and overnight hybridization at 45°C. Membranes were washed with washing buffer (3×SSC buffer, 0.2% SDS) for 5 min at RT and were then exposed to storage phosphor screen (Packard Instrument Company, Mariden, CT) for 3 h.
In silico analysis
Rho-independent terminators were predicted with TransTerm (http://nbc11.biologie.uni-kl.de/framed/left/menu/auto/right/transterm/) [26]. Blast searches between E. faecalis strains were carried out using a species-level BLAST database (http://www.ncbi.nlm.nih.gov/). The Rfam database was employed to determine putative functions of sRNAs (http://rfam.sanger.ac.uk) [27]. In order to predict target genes for the identified sRNAs sRNATarget (http://ccb.bmi.ac.cn/srnatarget/) [28] and IntaRNA (http://rna.informatik.uni-freiburg.de:8080/IntaRNA.jsp) [29] servers were used.
Construction of EF3314_EF3315 sRNA deletion mutant
For the deletion assay, a DNA fragment containing ligated upstream (869 bp) and downstream (839 bp) sequences of the EF3314_EF3315 sRNA, was cloned into plasmid pMAD [30] (see Table S1 for primers used). 1 µg of recombinant plasmid was finally used to transform competent cells. After electroporation, 300 µl of cell suspension was plated onto GM17 agar containing 50 µg ml−1 of erythromycin and X-Gal (100 µg ml−1). Plates were incubated for 48 hours at 30°C. A few dark blue colonies were obtained and analysed for presence of the plasmid by PCR using primers madR and madF (Table S1). Some blue colonies were then cultured twice in GM17 liquid medium with erythromycin (50 µg ml−1) at 45°C over-night. In the next step, the cultures were used to inoculate (0.05% v/v) GM17 liquid medium without antibiotic. The tubes were incubated for 6 hours at 30°C followed by incubation at 45°C over-night. This step was repeated 2 to 3 times. Serial dilutions of the culture were plated on GM17 agar containing 100 µg ml−1 of X-Gal and incubated for 48 hours at 45°C. White colonies were then isolated on GM17 agar with or without erythromycin. Antibiotic sensitive clones were analysed by PCR on the presence of a deleted sRNA.
Two-dimensional protein gel electrophoresis and protein identification
Protein samples from wild type and ΔEF3314_EF3315 mutant cells harvested in exponential growth phase were performed as described by Giard et al. [31]. First dimensional electrophoresis was carried out using 17 cm ReadyStrip™ IPG Strips (pH 4–7) and Protean®IEF Cell apparatus (Bio-Rad Laboratories, Richmond, CA, USA) as recommended by the manufacturer. Second dimensions were performed in 14% polyacrylamide gels without stacking gel using the Millipore Investigator™ 2-D electrophoresis system (Millipore, Bedford, MA, USA) as described by Giard et al. [31]. 2-D gels were then stained using Coomassie Blue. Spots of interest were excised from the gel, and peptides were digested by trypsin as described by Budin-Verneuil et al. [32]. An electrospray ion trap spectrometer (LCQ DecaXP, ThermoFinnigan, San Jose, CA, USA) coupled on line with HPLC was used for peptides analysis. Mass spectrometry were acquired in a mode that alternated a full MS scan (mass range: 400–1600) and a collision induced dissociation tandem mass spectrometry (MS/MS) of the most abundant ion. Data were analysed using the sequest algorithm incorporated with the ThermoFinnigan BioWorks software.
Results and Discussion
Tiling microarray-based identification of E. faecalis sRNAs
Tiling microarray has become a comprehensive approach to sRNA discovery. Identification of sRNA candidates transcribed by E. faecalis V583 was undertaken with two samples of cells harvested in mid-log growth phase and stationary phase after 24 h of incubation at 37°C in M17 glucose media. Analysis of IGRs tiling microarray data revealed 53 regions with intensity values of hybridization five fold higher than signals from apparent untranslated regions. Importantly, only one (see below) of these putative sRNAs identified by microarray was also predicted by bioinformatic approach as performed by Livny et al. [14]. This low overlap between microarray and in silico analysis is consistent with that observed in other bacteria [18]. These data show that computational and experimental methods are two complementary ways to identify sRNAs. As carried out for identification of sRNAs from S. pneumoniae using tiling arrays, we choose a stringent intensity cutoff to avoid false positives for identifying short length RNA [19]. Using a threshold of intensity of ten fold the background level led to the identification of 12 putative sRNAs (Table 1). No experimental evidence (neither sequence from RACE-PCR nor signal on Northern blot) was obtained for one of them (EF0940_EF0941). Since the IGR between EF0940 and EF0941 is only 51 bp in length, the corresponding probe putatively hybridized with the transcription product of EF0941. Thus, the candidate has been excluded from our study. The 11 other candidates that hybridized in specific intergenic regions were selected for further detailed characterization.
Table 1. sRNAs in E. faecalis V583 detected by tiling microarray.
| Intergenic Region | Left gene | sncRNA strand | Right gene | Size | Flanking genes | Expression valuea | Expression ration(Expo/Stat) | |||||
| start | stop | (nt) | Expo | Stat | ||||||||
| sRNAs expressed at exponential phase | ||||||||||||
| A. | EF3314_EF3315 | ← | ← | ← | 3201675 | 3201582b | 94 | EF3314:cell wall surface anchor family protein | 65025.9 | 1249.6 | 52 | |
| 3201535b | 141 | EF3315:triphosphoribosyl-dephospho-CoA synthase | ||||||||||
| B. | EF0820_EF0822 | ← | ← | → | 784383 | 784014 | 370 | EF0820:rplY; 50S ribosomal protein L25/general stress protein Ctc | 37086.5 | 1376.8 | 26.9 | |
| EF0822:HAD (haloacid dehalogenase) superfamily hydrolase | ||||||||||||
| C. | EFA0080_EFA0081 | → | ← | → | 63478c | 63423b | 99 | EFA0080:UvrC family transcriptional regulator | RNAI | 37537.9 | 3062.9 | 12.3 |
| EF0081:hypothetical protein | ||||||||||||
| D. | EF1368_EF1369 | ← | → | ← | 1345556 | 1346183 | 628 | EF1368:hypothetical protein | 35465.0 | 3058.9 | 11.6 | |
| EF1369:Cro/Cl family transcriptional regulator | ||||||||||||
| EF1370:drug resistance transporter, EmrB/QacA family protein | ||||||||||||
| E. | EF0408_EF0409 | → | → | ← | 381297 | 381708 | 412 | EF0408:PTS (phosphotransferase system) system, IIA component | RNAI | 47418.0 | 11648.3 | 4.1 |
| EF0409:hypothetical protein | ||||||||||||
| F. | EF0605_EF0606 | ← | → | ← | 569151 | 569329 | 179 | EF0605:hypothetical protein | 41977.3 | 11288.0 | 3.7 | |
| EF0606:Dps (DNA-binding protein from starved cells) family protein | ||||||||||||
| sRNAs expressed at stationary phase | ||||||||||||
| G. | EF1097_EF1098* | → | ← | ← | 1067257 | 1066894 | 364 | EF1097:hypothetical protein | tmRNA | 3390.8 | 63399.5 | 0.05 |
| EF1098:hypothetical protein | ||||||||||||
| H. | EF0869_EF0871 | ← | ← | → | 829525 | 829052 | 474 | EF0869:Cro/Cl family transcriptional regulator | 2655.4 | 47286.9 | 0.06 | |
| EF0871:cation transpoter E1–E2 family ATPase | ||||||||||||
| I. | EF0136_EF0137 | → | ← | → | 137278 | 137066d | >213 | EF0136:hypothetical protein | 1755.7 | 28560.7 | 0.06 | |
| EF0137:nucleotidyl transferase domain-containing protein | ||||||||||||
| sRNAs expressed at exponential and stationary phase | ||||||||||||
| J. | EFB0062_EFB0063 | → | ← | → | 55834 | 55623b | 212 | EFB0062:UvrC family transcriptional regulator | RNAI | 49218.4 | 52343.1 | 0.94 |
| EFB0063:replication control protein PrgN | ||||||||||||
| K. | EF2205_EF2206 | → | ← | ← | 2119382 | 2119296 | 87 | EF2205:hypothetical protein | 4.5S | 41604.0 | 55672.1 | 0.75 |
| EF2206:cytidine/deoxycytidylate deaminase family protein | ||||||||||||
: Intensity of hybridization from the intergenic probe showing the highest signal in exponential or stationary phase.
: Computer prediction of the putative 3′ end (using TransTerm software).
: 5′ end corresponding to the 5′ end of probe.
: 3′end corresponding to the 3′ end of probe.
Experimental validation of 11 sRNAs in E. faecalis
One of the main goals of this study was to determine the sequence and the expression pattern of the 11 selected sRNA candidates. First, using a new RNA preparation, we performed Northern blot analysis to confirm the transcription of these RNAs during exponential growth phase and stationary phase and to determine the approximate size of each candidate. We observed a transcript for 10 out of the 11 candidates tested. Six of them (EF3314_EF3315, EF0820_EF0821, EFA0080_EFA0081, EF1368_EF1369, EF0408_EF0409 and EF0605_EF0606) were specifically expressed during exponential phase (Figure 1A–F); 1 sRNA (EF0869_EF0870) was specifically expressed after 24 h of starvation (Figure 1H); and 3 (EF1097_EF1098, EFB0062_EFB0063 and EF2205_EF2206) were detected in comparable amounts in both phases (Figure 1G, J, K). These expression patterns were in good agreement with the results of tiling microarray except for EF1097_EF1098 which was much more expressed in stationary phase than under growing conditions on our chips. For unexplained reasons, no signal has been detected for EF0136_EF0137 (Figure 1I) by Northern blot analysis under our experimental conditions.
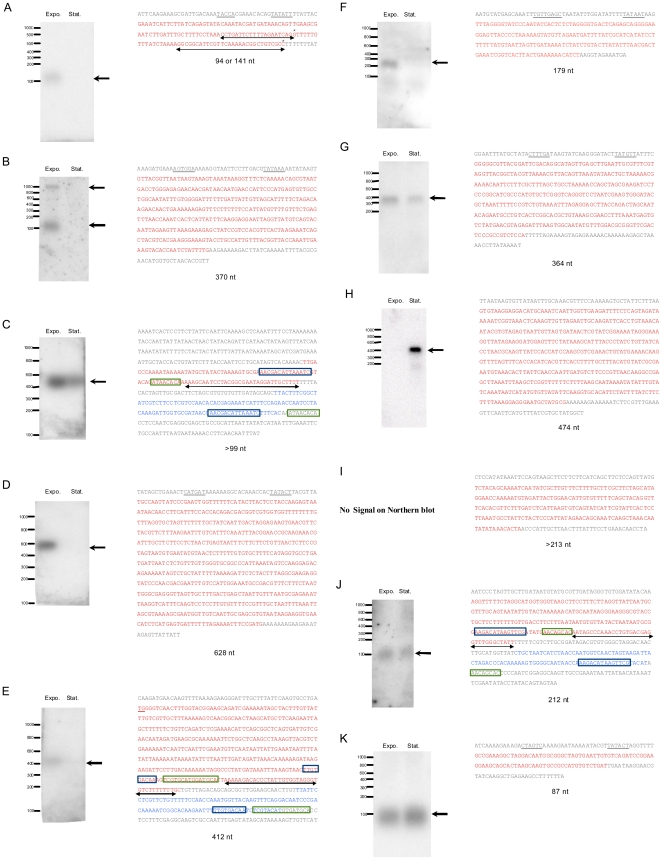
Figure 1. Northern blots and sequences of sRNAs (A: EF3314_EF335, B: EF0820_EF0821, C: EFA0080_EFA0081, D: EF1368_EF1369, E: EF0408_EF0409, F: EF0605_EF0606, G: EF1097_EF1098, H: EF0869_EF0870, I: EF0136_EF0137, J: EFB0062_EFB0063 and K: EF2205_EF2206).
RNA was isolated from cells at exponential (Expo) and stationary (Stat) phases. Northern blot analyses were performed using α32P-labelled probes. Arrows on Northern blot picture indicate the sRNAs corresponding bands. The transcriptional start sites and terminators of sRNAs were determined by 5′ RACE and 3′ RACE or by in silico analysis using TransTerm software. The putative −10 and/or −35 promoter sequences are underlined, and the sRNA sequence is written in red letters. Putative 3′-ends of EF3314_EF335 sRNA (panel A) is indicated by stars (*). The 3′-end of the sequenceof EF0136_EF0137 (panel I) mentioned here corresponds to the 3′-end of the tiling array probe. Black arrows in the sequence indicate the predicted terminators. The fst gene is written in blue letters and direct repeats “a” and “b” (DRa and DRb) of par system are blue and green boxed, respectively (panels C, E, and J).
In order to determine the exact sequence of each sRNA candidate we identified the transcriptional start sites by 5′-RACE except for EFA0080_EFA0081 for which no result was gained. The 3′ ends of the transcripts were obtained either by 3′-RACE (Figure 1B, D, E, F, G, H, K, Table 1) or by combining transcript length data deduced from the Northern blots and computational prediction of transcriptional terminators [26] (Figure 1A, C, J, Table 1). Since neither putative terminator nor experimental data of the 3′ end of EF0136_EF0137 (Figure 1I) were obtained, the end of the sequence mentioned corresponds to the 3′ end of the tiling array probe. 5′-3′ RACE data of EF0820_EF0821 did not correlate to Northern blot results. From RACE-PCR, a 370 nt long sRNA was deduced that is larger than the predicted size (app. 100 nt) from Northern blot (using probe hybridizing on the 5′ region), suggesting that the large EF0820_EF0822 transcript was processed to short sRNA by modification of its 3′ end. Except for EF0820_EF0822, where the 99 last nucleotides correspond to the beginning sequence of EF0820, we could not identify obvious coding sequences (CdS), i.e. ORFs (open reading frames) with start codons connected to putative ribosome-binding sites in reasonable distances (around 8 nucleotides) inside the other sRNA candidates. Nevertheless, definitive exclusion of the presence of CdS in these regions needs experimental verification.
Altogether, the length range of the identified sRNAs was 87–628 nucleotides and the deduced sequences and promoter regions of the 11 sRNAs are shown in Figure 1. In comparison with sRNAs identified by Fouquier d'Hérouel et al. [23] using in silico prediction and “5′tag-RACE” strategy, only four overlap with our sRNA candidates (EF0605_EF0606, EF1097_EF1098, EF0869_EF0871, and EF2205_EF2206 corresponding to ref25C, ssrA, ref19C, and ffs, respectively). This shows that several techniques as well as different growth conditions (see below) are necessary for more exhaustive identification of sRNAs.
Features of sRNAs
As previously mentioned, an antisense RNA regulated addiction module named “par” system was described on the E. faecalis plasmid pAD1 [9]. The components of this toxin-antitoxin (TA) system are antisense RNA (RNA II) and its target, RNA I encoding the peptide toxin Fst. Such systems play a crucial role in plasmid stability by killing any daughter cells that fail to inherit a copy of the plasmid. Three putative sRNAs identified in our study (EFA0080_EFA0081 in pTEF1, EFB0062_EFB0063 in pTEF2, and EF0408_EF0409 in the chromosome) corresponded to the RNAI components of the TA systems already identified in E. faecalis V583 by Weaver and coworkers [11]. As shown in Figure 1 (C, E, J), RNA I (including fst toxin gene) and RNA II homologues had two direct repeat sequences and shared the same bidirectional terminator. One interesting question concerns the role of par addiction module located on the bacterial chromosome. Several studies revealed various roles such as in mobile element stability or stress response [12], [33], [34]. As pointed out, in the case of par EF0409 (including EF0408_EF0409 sRNA), its association with genes encoding phosphotransferase components homologous to a mannitol transport system suggests a potential function in nutritional uptake [11].
Northern blot and tiling microarray showed that EF1097_EF1098 was expressed in both growth and stationary phases and we were able to determine the exact sequence of this sRNA (Figure 1G). EF1097_EF1098 corresponds to E. faecalis tmRNA (ssrA) that is a unique bi-functional RNA acting as both a tRNA and an mRNA. It functions as the rescue system of ribosomes stalled on aberrant mRNAs and adds a peptide tag to nascent polypeptides for directed proteolysis (named trans-translation) [35], [36]. tmRNA is universally conserved and is one of the most abundant RNA in the cells [37]. It has not only an important role in mRNA turnover but also likely in monitoring protein folding (for review see [35]). Mutations that inactivate tmRNA are lethal for some species (ie, Neisseria gonnorhoeae, Haemophilus influenzae, Shigella flexneri) or, for others, affect bacterial physiology such as virulence (ie, Salmonella enterica, Yersinia pseudotuberculosis) or stress response (ie, E. coli, B. subtilis) [35], [37]. Determination of the impact of tmRNA deletion in E. faecalis is under investigation in our laboratory.
We used the Rfam database (a collection of non-coding RNA families) to determine the putative functions of characterized sRNAs [27]. We found that EF2205_EF2206 sRNA matched with the Signal Recognition Particle (SRP) functional category. SRP is a ribonucleoprotein complex that targets proteins for secretion through co-translational process and is composed of protein Ffh and 4.5S RNA in prokaryotes. Our analysis revealed that EF_1700 gene (ffh) product and EF2205_EF2206 correspond to the two components of the SRP in E. faecalis. Interestingly, a recent study demonstrated that mutation of the gene encoding 4.5S RNA in S. pyogenes (phylogenetically related to E. faecalis) results in reduction of virulence [38].
In order to predict target genes of the other sRNAs identified in this study, we performed in silico analysis (Table 2, Table S2). Two different softwares were used for a more precise identification. sRNATarget server is based on the Naive Bayes probabilistic method and take RNA secondary structure profile as the feature [28]. The second, IntaRNA, predicts interactions between two RNA molecules, and the scoring is based on hybridization free energy and accessibility of the interaction sites in both molecules [29]. Numerous putative target genes were obtained by combination of these two approaches (from 9 for EF3314_EF3315 to 81 for EF0136_EF0137) (Table 2, Table S2). In silico prediction (Table S2) as well as sequence analysis suggested antisense activity for EF1368_EF1369 and EF0136_EF0137. Indeed, EF1369 mRNA sequence, encoding a putative transcriptional regulator, was fully complementary to EF1368_EF1369 sRNA. Likewise, the first 136 nucleotides of EF0136_EF0137 were complementary with the beginning sequence of EF0137 mRNA. The combined in silico data constitute hypothetical regulons for the sRNA candidates that need to be experimentally verified.
Table 2. Number of putative target genes.
| Number of mRNA candidate | |||
| sRNA | sRNATarget (score>0.9)a | IntaRNAb | commonc |
| EF3314_EF3315 | 75 | 31d | 9 |
| EF0820_EF0822 | 176 | 213d | 44 |
| EF1368_EF1369 | 876 | 97e | 72 |
| EF0605_EF0606 | 210 | 85d | 24 |
| EF0869_EF0871 | 494 | 318d | 62 |
| EF0136_EF0137 | 1252 | 92e | 81 |
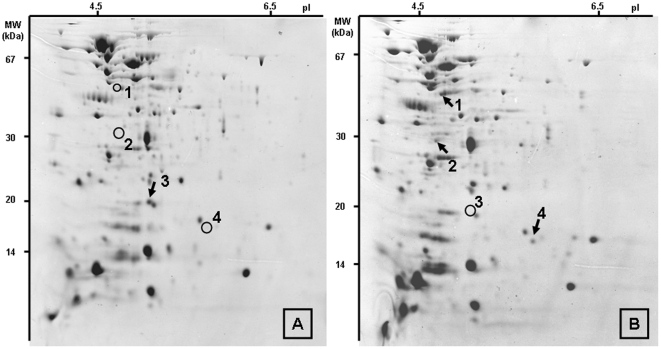
In general, sRNAs act at the post transcriptional level of regulation [1], [3]. Then, in order to observe a putative influence of one sRNA in E. faecalis, proteomic approach was undertaken comparing profiles of the ΔEF3314_EF3315 mutant and the parental strain. Two-dimensional gel electrophoresis of proteins from growing E. faecalis V19 and ΔEF3314_EF3315 mutant strains are shown in Figure 2. From two distinct experiments we observed that intensity of 4 spots were reproducibly different between the two strains. Numbers 1, 2, and 4 were only present in the mutant whereas number 3 was only seen in the wild type (Figure 2). By mass spectrometry, after extraction of proteins from the gel, we identified these polypeptides. Spots 1, 2, 3, and 4 correspond to DnaK (EF_1308, 63 kDa), ribosomal protein S1 (EF_1548, 43 kDa), ribosomal protein L6 (EF_0221, 19 kDa), and translation elongation factor Tu (EF_0221, 43 kDa), respectively. However, molecular weight (MW) deduced from the gels (around 45 kDa, 30 kDa, 15 kDa, for peptides 1, 2, and 4, respectively) did not correlated with the expected sizes. Therefore, peptides indentified from the mutant samples likely corresponded to protein degradation products. On the other hand, MW of spot number 3, which is absent in the mutant, was estimated at around 20 kDa in good accordance with the calculated size of the intact protein (19 kDa). These combined results suggested that EF3314_EF3315 might be involved in the turnover of some abundant proteins in E. faecalis, especially from the translational apparatus.
Figure 2. Two-dimensional gel electrophoresis of proteins from E. faecalis V19 (A) and ΔEF3314_EF335 mutant (B).
Arrows indicate polypeptides that are detected in one gel but not in the other. The position of the polypeptides absent in a given gel are indicated by circles.
Expression of sRNAs in different stress conditions
Generally, the expression of sRNAs are tightly regulated and induced by specific environmental condition [2]. We then performed tiling arrays with new RNA samples in order to analyze the transcription of sRNAs previously characterized under 11 different conditions of growth some of which may correspond to stresses encountered during intestinal colonization or during the infectious process (see Material and Methods). Expression patterns of the 11 sRNAs under H2O2, BS, and acid stress conditions, during growth in presence or absence of O2 and in serum and urine is presented in Table 3. EF0408_EF0409, EFA0080_EFA0081 and EFB0062_EFB0063, identified as members of TA systems were highly expressed at different stages of growth with oxygen (Table 3). Physiological significance of the induction of transcription of these three homologues especially in presence of oxygen remains unclear. However, the expressions of these paralogues appeared sequential during growth phases. EF0408_EF0409 was mainly transcribed during exponential phase, EFA0080_EFA0081 during early stationary phase, and EFB0062_EFB0063 after 24 h of stationary phase (Table 3). These observations suggest that the different TA systems may have different roles according to the growth phase of the bacteria.
Table 3. Expression patterns of sRNAs under different growth phases and stress conditions.
| sRNAs | Stress conditions | ||||||||||
| H2O2 | pH (acid) | BS | Expo | Early Stat | Stat | Expo | Early Stat | Stat | Urine | Serum | |
| with O2 | with O2 | with O2 | |||||||||
| EF3314_EF3315 | 127 | 2263 | 596 | 787 | 1756 | 64 | 3478 | 659 | 93 | 191 | 112 |
| EF0820_EF0822 | 114 | 150 | 135 | 370 | 253 | 47 | 473 | 605 | 51 | 59 | 122 |
| EFA0080_EFA0081 | 102 | 186 | 314 | 857 | 43619 | 2364 | 5034 | 1566 | 3886 | 1756 | 2178 |
| EF1368_EF1369 | 761 | 2817 | 874 | 1178 | 418 | 118 | 171 | 614 | 168 | 144 | 139 |
| EF0408_EF0409 | 1756 | 2916 | 649 | 20636 | 1916 | 283 | 1597 | 1909 | 136 | 954 | 257 |
| EF0605_EF0606 | 722 | 2056 | 246 | 1880 | 3246 | 214 | 261 | 113 | 326 | 802 | 129 |
| EF1097_EF1098 | 4535 | 32765 | 106115 | 1835 | 22301 | 2438 | 1518 | 1492 | 3977 | 41662 | 11483 |
| EF0869_EF0871 | 556 | 159 | 1236 | 196 | 7780 | 13465 | 374 | 119683 | 31710 | 5974 | 30293 |
| EF0136_EF0137 | 59 | 108 | 11 | 70 | 27 | 46 | 11 | 125 | 144 | 101 | |
| EFB0062_EFB0063 | 125 | 332 | 194 | 2817 | 802 | 20636 | 1756 | 1236 | 5503 | 211 | 179 |
| EF2205_EF2206 | 10724 | 21313 | 11296 | 9823 | 25155 | 10468 | 202452 | 11483 | 405266 | 29510 | 221227 |
E. faecalis tmRNA (EF1097_EF1098) and 4.5S RNA (EF2205_EF2206) showed a high intensity of hybridization under all conditions tested but BS and late stationary phase induced the highest level of tmRNA and 4.5S RNA expression, respectively. Furthermore, EF0869_EF0871 was highly expressed in urine and serum medium (Table 3). It has been shown that transcription of some genes encoding fitness and virulence factors are affected when E. faecalis is incubated in these biological media [39], [40]. It is then tempting to speculate that these sRNAs could play a crucial role in the cellular response triggered during the infectious process.
Surprisingly, for unexpected reason, signals corresponding to the two sRNAs EF0136_EF0137 and EF0820_EF0822 were very low in these tiling arrays experiments leading to unexploitable data. On the other hand, EF3314_EF3315, EF1368_EF1369 and EF0605_EF0606 sRNAs appeared moderately expressed but were obviously induced by acid stress (Table 3). However, exponential growth phase and early stationary phase in presence of oxygen were the most favorable conditions for EF3314_EF3315 and EF0605_EF0606 expressions, respectively (Table 3). This is in agreement with the induction of Ref25C (corresponding to EF0605_EF0606) in oxidative stress condition reported by Fouquier d'Hérouel et al. [23].
Our tiling arrays data using RNA samples obtained from cells incubated under 11 different growth conditions allowed us to identify 76 new IGRs with intensities of hybridization ten fold higher than signals from apparent untranslated regions. Probe sequences and tiling array data obtained with samples from stressed cells are shown in Table S3. A more detailed analysis of these new candidates is in progress in our laboratory. In addition, if the threshold was set to five-fold induction, 174 putative sRNAs were detected in our experiments. sRNAs are usually transcribed under specific growth conditions and it is likely that some could be expressed under stressing conditions not yet tested. Moreover, sRNAs may have been missed in our study due to experimental procedure since our chips only covered intergenic regions of the V583 genome and since fractionated RNAs have been used for the hybridizations. It has been generally predicted that genome sizes ranging from 3–4 Mbp may contain 80–300 sRNAs [14]. Taken together it is highly probable that the number of sRNA transcripts detected in E. faecalis will greatly increase in the near future.
Distribution of sRNAs among E. faecalis strains
To date, the whole genome sequence of 54 E. faecalis strains are available in the NCBI database. We performed standard BLAST analysis to detect the presence of the characterized sRNAs in these different E. faecalis strains (Table 4). Seven of them are highly conserved (90 to 100% identical) and present in all E. faecalis genomes (EF3314_EF3315, EF0820_EF0821, EF1368_EF1369, EF0408_EF0409, EF1097_EF1098, EF0869_EF0871 and EF2205_EF2206). The other four are not systematically observed because of their location on a mobile genetic element (EF0136_EF0137), in the pathogenicity island (PAI) (EF0605_EF0606) or on plasmids (EFA0080_EF0081 and EFB0062_EFB0063) [41]. sRNAs EF0605_EF0606, EF0136_EF0137, EFA0080_EFA0081 and EFB0062_EFB0063 homologues (at least 80% identical) are present in 9, 15, 35 and 23 strains of the 54 genomes analyzed, respectively (Table 4).
Table 4. Distribution of the 11 sRNAs among E. faecalis strains.
| sRNAs | |||||||||||
| E. faecalis | EF3314_ | EF0820_ | EFA0080_ | EF1368_ | EF0408_ | EF0605_ | EF1097_ | EF0869_ | EF0136_ | EFB0062_ | EF2205_ |
| strains | EF3315 | EF0822 | EFA0081 | EF1369 | EF0409 | EF0606 | EF1098 | EF0871 | EF0137 | EFB0063 | EF2206 |
| OGR1RF | 90 | 100 | 90 | 90 | 90 | 90 | 100 | ||||
| ARO1/DG | 90 | 100 | 90 | 90 | 100 | 90 | 80–90 P | 100 | |||
| ATCC 29200 | 100 | 100 | 80–90 G | 90 | 90 | 100 | 90 | 80–90 G | 100 | ||
| ATCC 4200 | 100 | 100 | 90 | 90 | 100 | 90 | 100 | ||||
| CH188 | 100 | 100 | 90 | 90 | 100 | 90 | 80–90 | 100 | |||
| D6 | 100 | 100 | 90 | 90 | 100 | 90 | 90 | 100 | |||
| DAPTP0512 | 90 | 100 | 80–90 G | 90 | 90 | 90 | 90 | 80–90 G | 90 | ||
| DAPTP0516 | 90 | 100 | 80–90 G | 90 | 90 | 90 | 90 | 80–90 G | 90 | ||
| DS5 | 100 | 100 | 100 P | 90 | 90 | 90 | 90 | 80–90 | 80–90 P | 100 | |
| E1Sol | 100 | 90 | 90 | 90 | 100 | 90 | 80–90 P | 100 | |||
| Fly1 | 90 | 90 | 90 | 90 | 90 | 90 | 100 | ||||
| HH22 | 100 | 100 | 100 G | 90 | 100 | 100 | 100 | 80–90 G | 100 | ||
| HIP11704 | 100 | 90 | 80–90 G | 90 | 90 | 90 | 90 | 80–90 G | 100 | ||
| JH1 | 100 | 100 | 80–90 P | 90 | 90 | 90 | 90 | 80–90 | 80–90 P | 100 | |
| Merz96 | 90 | 100 | 80–90 P | 90 | 90 | 90 | 90 | 80–90 P | 90 | ||
| PC1.1 | 100 | 100 | 90 | 90 | 90 | 90 | 80–90 | 100 | |||
| R712 | 90 | 100 | 80–90 G | 90 | 90 | 90 | 90 | 80–90 G | 90 | ||
| S613 | 90 | 100 | 80–90 G | 90 | 90 | 90 | 90 | 80–90 G | 90 | ||
| T1 | 100 | 100 | 90 | 90 | 90 | 90 | 90 P | 100 | |||
| T11 | 100 | 100 | 90 | 100 | 100 | 100 | 100 | ||||
| T2 | 100 | 100 | 80–90 P | 90 | 90 | 100 | 90 | 90 | 100 | 80–90 P | 100 |
| T3 | 100 | 100 | 90 | 90 | 90 | 90 | 80–90 P | 100 | |||
| T8 | 100 | 100 | 90 G | 90 | 90 | 90 | 90 | 80–90 G | 100 | ||
| TUSoD Ef11 | 100 | 90 | 80–90 | 90 | 90 | 90 | 90 | ||||
| TX0012 | 100 | 100 | 90 | 90 | 90 | 90 | 90 | ||||
| TX0017 | 100 | 100 | 80–90 G | 90 | 90 | 90 | 90 | 80–90 G | 100 | ||
| TX0027 | 100 | 100 | 80–90 G | 90 | 90 | 90 | 90 | 80–90 | 80–90 G | 100 | |
| TX0031 | 100 | 100 | 90 | 90 | 100 | 90 | 100 | ||||
| TX0043 | 100 | 100 | 90 | 90 | 100 | 90 | 90 | ||||
| TX0102 | 100 | 100 | 90 | 90 | 100 | 90 | 100 | ||||
| TX0104 | 100 | 100 | 80–90 G | 90 | 90 | 90 | 90 | 90 | 80–90 G | 100 | |
| TX0109 | 100 | 100 | 90 | 90 | 90 | 90 | 80–90 G | 90 | |||
| TX0309A | 100 | 100 | 90 | 100 | 100 | 100 | 100 | 80–90 | 90 G | 100 | |
| TX0309B | 100 | 100 | 90 | 100 | 100 | 100 | 100 | 80–90 | 90 G | 100 | |
| TX0312 | 100 | 100 | 90 | 90 | 100 | 90 | 90 | ||||
| TX0411 | 100 | 100 | 80–90 G | 90 | 90 | 100 | 90 | 90 G | 90 | ||
| TX0470 | 100 | 100 | 80–90 | 90 | 90 | 100 | 80–90 | 100 | |||
| TX0630 | 100 | 100 | 80–90 G | 90 | 90 | 100 | 90 | 100 | 90 G | 100 | |
| TX0635 | 100 | 100 | 90 G | 90 | 90 | 100 | 90 | 80–90 | 80–90 G | 100 | |
| TX0645 | 100 | 90 | 80–90 | 90 | 90 | 90 | 80–90 | 80–90 G | 100 | ||
| TX0855 | 100 | 90 | 80–90 G | 90 | 90 | 100 | 90 | 90 | 90 G | 100 | |
| TX0860 | 100 | 100 | 90 | 90 | 100 | 90 | 90 | 90 G | 100 | ||
| TX1302 | 100 | 100 | 90 | 90 | 90 | 90 | 100 | ||||
| TX1322 | 100 | 100 | 90 | 90 | 100 | 90 | 80–90 G | 100 | |||
| TX1341 | 100 | 100 | 90 | 90 | 100 | 90 | 100 | 80–90 | 80–90 G | 100 | |
| TX1342 | 100 | 100 | 90 | 90 | 90 | 90 | 100 | ||||
| TX1346 | 100 | 90 | 90 | 90 | 90 | 90 | 90 | ||||
| TX2134 | 100 | 100 | 80–90 G | 90 | 90 | 100 | 90 | 90 G | 100 | ||
| TX2137 | 100 | 100 | 80–90 G | 90 | 90 | 100 | 90 | 90 | 80–90 G | 100 | |
| TX2141 | 100 | 90 | 80–90 | 90 | 90 | 90 | 90 | ||||
| TX4000 | 100 | 100 | 90 | 90 | 90 | 90 | 100 | ||||
| TX4244 | 100 | 100 | 90 | 90 | 90 | 90 | 80–90 | 80–90 G | 100 | ||
| TX4248 | 100 | 100 | 80–90 G | 90 | 90 | 90 | 90 | 80–90 | 80–90 G | 100 | |
| X98 | 100 | 100 | 80–90 P | 90 | 90 | 100 | 90 | 80–90 P | 90 | ||
100 indicates 100% identity.
90 indicates more than >90% identity.
80–90 indicates between 80 and 90% identity.
White box indicates the absence of homology.
G: on genome.
P: on plasmid.
Homologues of EF0408_EF0409 (more than 90% identity) (member of TA system, see above) were systematically present in all E. faecalis genomes. Moreover, additional plasmidic EFA0080_EFA0081 and EFB0062_EFB0063 homologous were also observed in some chromosomes showing that most E. faecalis strains have several par systems arguing for a selective advantage for the bacterial cell.
Interestingly, EF0605_EF0606 is located in PAI between a gene encoding a Dps family protein (EF_0606) and an operon including a paralogue of gls24 (EF_0605-EF_0604). Dps is a protein involved in the protection of DNA against oxidative stress and Gls24 corresponds to a general stress protein that is a virulence factor in E. faecalis [42], [43], [44]. In S. pneumoniae, two sRNAs had demonstrated cis-acting effects on the transcription of adjacent genes [45]. From these observations and the fact that EF0605_EF0606 sRNA is induced under aerobic growth conditions, it may be hypothesized that it has a role in the control of expression of these enzymes and hence may be implicated in stress response and virulence of E. faecalis.
Perspectives
In this work we have determined the sequences, locations and expression patterns of 11 sRNAs in E. faecalis V583. These results provide a starting point towards understanding of the complex RNA regulatory network governing E. faecalis physiology and virulence. Recently, comparative genome-wide analysis of putative or characterized sRNAs of five major Gram-positive pathogens (L. monocytogenes EGD-e, Clostridium difficile 630, Staphylococcus aureus COL, S. pyrogenes M1 GAS, and E. faecalis V583) was reported [46]. This information will help to understand the molecular mechanisms of the pathogenic process which might be useful for the development of novel microbial diagnosis tools and anti-bacterial drugs such as antisense PNAs (peptide nucleic acids) [46].
Supporting Information
Primers and probes used in this study.
(DOC)
List of putative target genes of EF3314_EF3315, EF0820_EF0822, EF1368_EF1369, EF0605_EF0606, EF0869_EF0871, EF0136_EF0137 sRNA candidates.
(XLS)
Probe sequences and tiling array data obtained with samples from stressed cells of E. faecalis.
(XLS)
Acknowledgments
The expert technical assistance of Isabelle Rincé, Marie-Jeanne Pigny and Evelyne Marchand was greatly appreciated. We would like to thank all members of our labs for helpful discussions.
Footnotes
Competing Interests: Thomas Hartsch is an employee of Genedata AG (Basel, Switzerland), and internal legal approval was granted to publish this study. It was a collaborative effort between University of Caen, University of Giessen, and Genedata company groups and is not related to any products manufactured or marketed. There are no patents to declare. The authors hereby confirm that studies presented do not alter adherence to all the PLoS ONE policies on sharing data and materials, as detailed in the PLoS ONE guide for authors.
Funding: CM is funded by a thesis grant awarded by the Ministère de l'Enseignement Supérieur et de la Recherche. This study was supported by grants of the “Agence Nationale de la Recherche” in the frame of the transnational ERA-NET PathoGenoMics program (ANR-08-PATH-008-01), and by the Region of Lower Normandy (CRBN). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;20:615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Repoila F, Darfeuille F. Small regulatory non-coding RNAs in bacteria: physiology and mechanistic aspects. Biol Cell. 2009;101:117–131. doi: 10.1042/BC20070137. [DOI] [PubMed] [Google Scholar]
- 3.Storz G, Opdyke JA, Zhang A. Controlling mRNA stability and translation with small, noncoding RNAs. Curr Opin Microbiol. 2004;7:140–144. doi: 10.1016/j.mib.2004.02.015. [DOI] [PubMed] [Google Scholar]
- 4.Massé E, Escorcia FE, Gottesman S. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 2003;17:2374–2383. doi: 10.1101/gad.1127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nielsen JS, Lei LK, Ebersbach T, Olsen AS, Klitgaard JK, et al. Defining a role for Hfq in Gram-positive bacteria: evidence for Hfq-dependent antisense regulation in Listeria monocytogenes. Nucleic Acids Res. 2010;38:907–919. doi: 10.1093/nar/gkp1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun X, Zhulin I, Wartell RM. Predicted structure and phyletic distribution of the RNA-binding protein Hfq. Nucleic Acids Res. 2002;30:3662–3671. doi: 10.1093/nar/gkf508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Valentin-Hansen P, Eriksen M, Udesen C. The bacterial Sm-like protein Hfq: a key player in RNA transactions. Mol Microbiol. 2004;51:1525–1533. doi: 10.1111/j.1365-2958.2003.03935.x. [DOI] [PubMed] [Google Scholar]
- 8.Brantl S. Regulatory mechanisms employed by cis-encoded antisense RNAs. Curr Opin Microbiol. 2007;10:102–109. doi: 10.1016/j.mib.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 9.Weaver KE. Emerging plasmid-encoded antisense RNA regulated systems. Curr Opin Microbiol. 2007;10:110–116. doi: 10.1016/j.mib.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 10.Gerdes K, Thisted T, Martinussen J. Mechanism of post-segregational killing by the hok/sok system of plasmid R1: sok antisense RNA regulates formation of a hok mRNA species correlated with killing of plasmid-free cells. Mol Microbiol. 1990;4:1807–1818. doi: 10.1111/j.1365-2958.1990.tb02029.x. [DOI] [PubMed] [Google Scholar]
- 11.Weaver KE, Reddy SG, Brinkman CL, Patel S, Bayles KW, et al. Identification and characterization of a family of toxin-antitoxin systems related to the Enterococcus faecalis plasmid pAD1 par addiction module. Microbiology. 2009;155:2930–2940. doi: 10.1099/mic.0.030932-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerdes K, Wagner EG. RNA antitoxins. Curr Opin Microbiol. 2007;10:117–124. doi: 10.1016/j.mib.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 13.Livny J, Waldor MK. Identification of small RNAs in diverse bacterial species. Curr Opin Microbiol. 2007;10:96–101. doi: 10.1016/j.mib.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 14.Livny J, Teonadi H, Livny M, Waldor MK. High-throughput, kingdom-wide prediction and annotation of bacterial non-coding RNAs. PLoS One. 2008;3:e3197. doi: 10.1371/journal.pone.0003197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sharma CM, Vogel J. Experimental approaches for the discovery and characterization of regulatory small RNA. Curr Opin Microbiol. 2009;12:536–546. doi: 10.1016/j.mib.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 16.Sharma CM, Hoffmann S, Darfeuille F, Reignier J, Findeiss S, et al. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature. 2010;464:250–255. doi: 10.1038/nature08756. [DOI] [PubMed] [Google Scholar]
- 17.Landt SG, Abeliuk E, McGrath PT, Lesley JA, et al. Small non-coding RNAs in Caulobacter crescentus. Mol Microbiol. 2008;68:600–614. doi: 10.1111/j.1365-2958.2008.06172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perez N, Treviño J, Liu Z, Ho SC, Babitzke P, et al. A genome-wide analysis of small regulatory RNAs in the human pathogen group A Streptococcus. PLoS One. 2009;4:e7668. doi: 10.1371/journal.pone.0007668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar R, Shah P, Swiatlo E, Burgess SC, Lawrence ML, et al. Identification of novel non-coding small RNAs from Streptococcus pneumoniae TIGR4 using high-resolution genome tiling arrays. BMC Genomics. 2010;11:350. doi: 10.1186/1471-2164-11-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Toledo-Arana A, Dussurget O, Nikitas G, Sesto N, Guet-Revillet H, et al. The Listeria transcriptional landscape from saprophytism to virulence. Nature. 2009;459:950–956. doi: 10.1038/nature08080. [DOI] [PubMed] [Google Scholar]
- 21.Ogier JC, Serror P. Safety assessment of dairy microorganisms: the Enterococcus genus. Int J Food Microbiol. 2008;126:291–301. doi: 10.1016/j.ijfoodmicro.2007.08.017. [DOI] [PubMed] [Google Scholar]
- 22.Paulsen IT, Banerjei L, Myers GS, Nelson KE, Seshadri R, et al. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science. 2003;299:2071–2074. doi: 10.1126/science.1080613. [DOI] [PubMed] [Google Scholar]
- 23.Fouquier d'Hérouel A, Wessner F, Halpern D, Ly-Vu J, Kennedy SP, et al. A simple and efficient method to search for selected primary transcripts: non-coding and antisense RNAs in the human pathogen Enterococcus faecalis. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr012. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sahm DF, Kissinger J, Gilmore MS, Murray PR, Mulder R, et al. In vitro susceptibility studies of vancomycin-resistant Enterococcus faecalis. Antimicrob Agents Chemother. 1989;33:1588–1591. doi: 10.1128/aac.33.9.1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual, 2nd ed. 1989. Cold Spring Harbor Laboratory Press Cold Spring Harbor, N.Y.
- 26.Kingsford CL, Ayanbule K, Salzberg SL. Rapid, accurate, computational discovery of Rho-independent transcription terminators illuminates their relationship to DNA uptake. Genome Biol. 2007;8:R22. doi: 10.1186/gb-2007-8-2-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gardner PP, Daub J, Tate JG, Nawrocki EP, Kolbe DL, et al. Rfam: updates to the RNA families database. Nucleic Acids Res. 2009;37:D136–140. doi: 10.1093/nar/gkn766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cao Y, Zhao Y, Cha L, Ying X, Wang L, et al. sRNATarget: a web server for prediction of bacterial sRNA targets. Bioinformation. 2009;3:364–366. doi: 10.6026/97320630003364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith C, Heyne S, Richter AS, Will S, Backofen R. Freiburg RNA Tools: a web server integrating IntaRNA, ExpaRNA and LocARNA. Nucleic Acids Res. 2010;38(Suppl):373–377. doi: 10.1093/nar/gkq316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arnaud M, Chastanet A, Débarbouillé M. A new vector for efficient allelic replacement in naturally non transformable low GC% Gram-positive bacteria. Appl Environ Microbiol. 2004;70:6887–6891. doi: 10.1128/AEM.70.11.6887-6891.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Giard J-C, Laplace J-M, Rincé A, Pichereau V, Benachour A, et al. The Stress Proteome of Enterococcus faecalis. . Electrophoresis. 2001;14:2947–2954. doi: 10.1002/1522-2683(200108)22:14<2947::AID-ELPS2947>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 32.Budin-Verneuil A, Pichereau V, Auffray Y, Ehrlich DS, Maguin E. Proteomic characterization of the acid tolerance response in Lactococcus lactis MG1363. Proteomics. 2005;5:4794–4807. doi: 10.1002/pmic.200401327. [DOI] [PubMed] [Google Scholar]
- 33.Engelberg-Kulka H, Amitai S, Kolodkin-Gal I, Hazan R. Bacterial programmed cell death and multicellular behavior in bacteria. PLoS Genet. 2006;2:e135. doi: 10.1371/journal.pgen.0020135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fozo EM, Hemm MR, Storz G. Small toxic proteins and the antisense RNAs that repress them. Microbiol Mol Biol Rev. 2008;72:579–589. doi: 10.1128/MMBR.00025-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hayes CS, Keiler KC. Beyond ribosome rescue: tmRNA and co-translational processes. FEBS Lett. 2010;584:413–419. doi: 10.1016/j.febslet.2009.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dulebohn D, Choy J, Sundermeier T, Okan N, Karzai AW. Trans-translation: the tmRNA-mediated surveillance mechanism for ribosome rescue, directed protein degradation, and nonstop mRNA decay. Biochemistry. 2007;46:4681–93. doi: 10.1021/bi6026055. [DOI] [PubMed] [Google Scholar]
- 37.Keiler KC. Physiology of tmRNA: what gets tagged and why? Curr Opin Microbiol. 2007;10:169–175. doi: 10.1016/j.mib.2007.03.014. [DOI] [PubMed] [Google Scholar]
- 38.Treviño J, Perez N, Sumby P. The 4.5S RNA component of the signal recognition particle is required for group A Streptococcus virulence. Microbiology. 2010;156:1342–1350. doi: 10.1099/mic.0.036558-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shepard BD, Gilmore MS. Differential expression of virulence-related genes in Enterococcus faecalis in response to biological cues in serum and urine. Infect Immun. 2002;70:4344–4352. doi: 10.1128/IAI.70.8.4344-4352.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vebø HC, Solheim M, Snipen L, Nes IF, Brede DA. Comparative genomic analysis of pathogenic and probiotic Enterococcus faecalis isolates, and their transcriptional responses to growth in human urine. PLoS One. 2010;5:e12489. doi: 10.1371/journal.pone.0012489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McBride SM, Coburn PS, Baghdayan AS, Willems RJ, Grande MJ, et al. Genetic variation and evolution of the pathogenicity island of Enterococcus faecalis. J Bacteriol. 2009;191:3392–3402. doi: 10.1128/JB.00031-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Giard JC, Verneuil N, Auffray Y, Hartke A. Characterization of genes homologous to the general stress-inducible gene gls24 in Enterococcus faecalis and Lactococcus lactis. FEMS Microbiol Lett. 2002;206:235–239. doi: 10.1111/j.1574-6968.2002.tb11015.x. [DOI] [PubMed] [Google Scholar]
- 43.Giard JC, Rince A, Capiaux H, Auffray Y, Hartke A. Inactivation of the stress- and starvation-inducible gls24 operon has a pleiotrophic effect on cell morphology, stress sensitivity, and gene expression in Enterococcus faecalis. J Bacteriol. 2000;182:4512–4520. doi: 10.1128/jb.182.16.4512-4520.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Teng F, Nannini EC, Murray BE. Importance of gls24 in virulence and stress response of Enterococcus faecalis and use of the Gls24 protein as a possible immunotherapy target. J Infect Dis. 2005;191:472–480. doi: 10.1086/427191. [DOI] [PubMed] [Google Scholar]
- 45.Tsui HC, Mukherjee D, Ray VA, Sham LT, Feig AL, et al. Identification and characterization of noncoding small RNAs in Streptococcus pneumoniae serotype 2 strain D39. J Bacteriol. 2010;192:264–279. doi: 10.1128/JB.01204-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mraheil MA, Billion A, Kuenne C, Pischimarov J, Kreikemeyer B, et al. Comparative genome-wide analysis of small non-coding RNAs of major Gram-positive pathogens: from fundamentals to applications. Microbial Biotechnology. 2010;3:658–676. doi: 10.1111/j.1751-7915.2010.00171.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primers and probes used in this study.
(DOC)
List of putative target genes of EF3314_EF3315, EF0820_EF0822, EF1368_EF1369, EF0605_EF0606, EF0869_EF0871, EF0136_EF0137 sRNA candidates.
(XLS)
Probe sequences and tiling array data obtained with samples from stressed cells of E. faecalis.
(XLS)